Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Polysarcosine functionalised cationic polyesters efficiently deliver self-amplifying mRNA†
Hulya
Bayraktutan
 a,
Rafał J.
Kopiasz
a,
Rafał J.
Kopiasz
 ab,
Amr
Elsherbeny
ab,
Amr
Elsherbeny
 a,
Magda
Martinez Espuga
c,
Nurcan
Gumus
a,
Magda
Martinez Espuga
c,
Nurcan
Gumus
 a,
Umut Can
Oz
a,
Umut Can
Oz
 ad,
Krunal
Polra
e,
Paul F.
McKay
ad,
Krunal
Polra
e,
Paul F.
McKay
 e,
Robin J.
Shattock
e,
Paloma
Ordóñez-Morán
f,
Alvaro
Mata
e,
Robin J.
Shattock
e,
Paloma
Ordóñez-Morán
f,
Alvaro
Mata
 cg,
Cameron
Alexander
cg,
Cameron
Alexander
 a and
Pratik
Gurnani
a and
Pratik
Gurnani
 *h
*h
aDivision of Molecular Therapeutics and Formulation, Boots Science Building, School of Pharmacy, University of Nottingham, Nottingham, NG7 2RD, UK
bWarsaw University of Technology, Faculty of Chemistry, Noakowskiego 3 St., 00-664, Warsaw, Poland
cDivision of Regenerative Medicine and Cell Therapies, BioDiscovery Institute, School of Pharmacy, University of Nottingham, Nottingham, NG7 2RD, UK
dDepartment of Pharmaceutical Technology, Faculty of Pharmacy, Ankara University, Ankara, 06560, Türkiye
eDepartment of Infectious Diseases, Section of Immunology of Infection, Imperial College London, Norfolk Place, London, W21PG, UK
fCentre for Cancer Sciences, BioDiscovery Institute, Translational Medical Sciences Unit, School of Medicine, University of Nottingham, Nottingham, NG7 2RD, UK
gDepartment of Chemical and Environmental Engineering, University of Nottingham, Nottingham, NG7 2RD, UK
hUCL School of Pharmacy, University College London, 29-39 Brunswick Square, Bloomsbury, London, WC1N 1AX, UK. E-mail: p.gurnani@ucl.ac.uk
First published on 11th April 2024
Abstract
Messenger RNA (mRNA) technology is a potent platform for new vaccines and therapeutics. To deliver RNA medicines effectively, safe delivery vectors are crucial. Among these, poly(β-amino ester)s (PBAEs) complexes with nucleic acids have been shown as promising alternatives to conventional lipid nanoparticles, but toxicity and colloidal stability concerns still remain. Conjugation of poly(ethylene glycol) (PEG) has been used to mitigate these issues, but challenges associated with PEG-induced reactogenicity have emerged. Polysarcosine (pSar) has shown promise as a PEG alternative, demonstrating reduced reactogenicity when incorporated into lipid nanoparticles. We therefore developed a synthetic route to conjugate pSar on to PBAE end-groups, resulting in a pSar-PBAE-pSar triblock copolymer. Polyplexes containing various ratios (0, 20, 50, 80, 100% pSar) of the pSarylated and a non-pSarylated PBAE were prepared with self-amplifying messenger RNA, to examine the effect of surface pSar density. RNA encapsulation efficiencies, formulation properties and gene transfectability in vitro were then assessed indicating complete encapsulation of the cargo and sub-200 nm particle diameters in all cases. Despite high gene transfection in HEK293T cells, only the 50%-pSar displayed significantly higher expression of the reporter gene than the negative control in Caco-2. Further evaluation of 50%-pSar was conducted using 3D human colorectal cancer organoids, which displayed high transfection ability within the core of the organoid indicating high penetration of these nanoparticles. As a result, these findings indicate that pSarylated PBAEs have the potential to serve as a promising component material for enhancing delivery of nucleic acid therapeutics.
Introduction
Messenger RNA (mRNA) technology is now recognised as a versatile and potent medicinal platform that enables the transient expression of selective proteins either as vaccines against infectious disease, or therapeutics for a range of indications, including genetic disorders, and cancer.1,2 The successful demonstration of safety and efficacy for Moderna (SpikeVax) and BioNTech-Pfizer's (Comirnaty) mRNA-based vaccines against COVID-19, has significantly accelerated this technology, with many more mRNA vaccines and therapeutics now in clinical trials.3One of the most important components of an effective RNA medicine is a safe and efficient delivery system to protect the genetic sequence from endogenous nucleases and facilitate translocation to the cytosol of the target cells for expression.4–6 A variety of nonviral delivery strategies have therefore been investigated to deliver nucleic acids, including lipid nanoparticles (e.g. liposomes, cubosomes) and polyelectrolyte complexes (polyplexes).3,7–9 Polyplexes are self-assemblies of positively charged polymers which condense negatively charged nucleic acids into small (50–200 nm) nanoparticles driven by opposing electrostatic interactions. One of the promising classes of polycations broadly tested as RNA delivery vectors are poly(β-amino ester)s (PBAE)s.10,11 Due to ester bonds within the PBAEs’ main chain, these polymers are readily biodegradable via hydrolysis, decreasing their potential long-term accumulation and enhancing RNA release.12 These features, together with high flexibility of chemical functionality allowing for simple structural manipulation for a variety of nucleic acid types13,14 render PBAE-based polyplexes prospective candidates for RNA formulations. However, the polycationic and hydrophobic nature of some specific sub-classes of PBAEs can result in cytotoxicity via membrane disruption, which has prevented their clinical application. Previous reports have shown that PBAEs coupled with poly(ethylene glycol) (PEG) exhibit decreased surface cationic charges when complexed with nucleic acids, and this can mitigate toxicity as well as decrease interactions with serum proteins.15,16 Such an approach has been proven to prolong systemic circulation time of nanoparticles by shielding their surface from aggregation and phagocytosis and increase in-serum in vitro and in vivo transfection efficacy.17–20
Despite the popularity of PEG to confer “stealth” properties to nanoparticles in serum and the fact that PEG is classified as generally regarded as safe (GRAS)15 there is a growing number of reports on strong reactogenicity induced by PEG or PEGylated drugs.21 This issue has become more prevalent following the widespread use of PEGylated lipids within SpikeVax and Comirnaty COVID-19 vaccines, which have boosted pre-existing PEG antibody titers by up to 60-fold.22 This creates an urgent need to develop PEG alternatives, as future mRNA therapeutics which include PEG components may be highly immunogenic, thereby reducing potency and tolerability.23–25 One emerging PEG alternative is polysarcosine (pSar), a polypeptoid – a non-toxic, endogenous and non-proteinogenic amino acid which therefore endows it with key advantages over other potential “stealth” polymers such as poly(vinyl pyrrolidone)s,26 polyphosphoesters,27 polyglycerols,28 polypeptides,29 and poly(2-alkyl-oxazoline)s.30 Despite pSar's polyamide backbone, there is no evidence for degradability of pSar under physiological conditions to its monomeric form of sarcosine, unlike its counterpart polypeptides. However, as pSar is commonly synthesised at low molar masses this enables clearance via renal filtration potentially alleviating the need for biodegradability.31
Despite the similar physicochemical properties to PEG, recent examples of polysarcosinylated lipid nanoparticles have shown colloidal stability and, importantly, substantially reduced immunogenicity compared to PEGylated derivatives.32 However, to our knowledge pSar-PBAE conjugation has not been reported and may offer a versatile approach to a tuneable RNA delivery system combined with the advantages offered by pSar.
In this report, we demonstrate successful conjugation of pSar with a PBAE to yield a pSar-PBAE-pSar triblock copolymer (Fig. 1). To test application in an RNA delivery setting, the pSarylated PBAE was formulated with self-amplifying RNA, an alternative mRNA technology which includes replicase genes derived from the alphavirus genome within the construct. When saRNAs are translated, they not only express the encoded protein, but also cause replication of the RNA within the cytosol, enhancing protein expression levels at significantly lower doses, lengthening expression half-life and reducing the cost compared to mRNA.8,33–35 The saRNA constructs were formulated into polyplexes with varying content of pSarylated and non-pSarylated PBAEs to evaluate the effect of pSar density. The in vitro tolerability and transfection efficiency of these blended pSarylated-PBAE polyplexes was then examined in two immortalised cell lines, (HEK293T and Caco-2). Finally, the cellular uptake and transfection efficiency in 3D human colorectal cancer organoids were investigated.
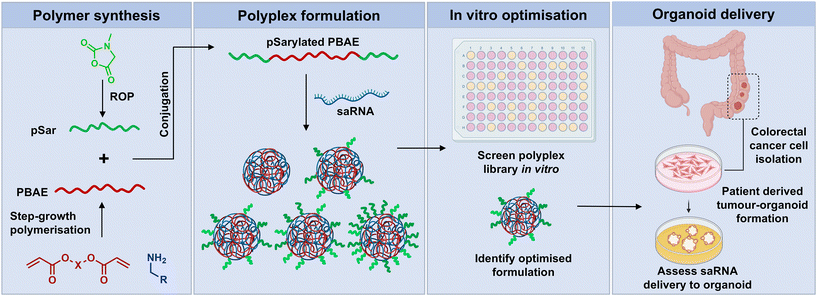 | ||
| Fig. 1 Schematic representation of PBAE synthesis, polyplex assembly, in vitro analysis and the cellular uptake and transfection efficiency in 3D colorectal cancer organoids. | ||
Materials and methods
Materials
1,4-Butanediol diacrylate (BDD), 5-aminopentan-1-ol (5-AP), triethylamine (TEA), N-Boc-sarcosine, fluorescein isothiocyanate (FITC), diethyl ether, tetrahydrofuran, sodium acetate (NaOAc) buffer (pH: 5.0), dye gel red and goat-anti-mouse IgG, Fc biotin and red CellMask™ plasma membrane stain were purchased from Sigma-Aldrich. Dichloromethane (DCM), N,N-dimethylformamide (DMF), acetonitrile (ACN), tetrahydrofuran (THF), hexane, diethyl ether, dimethyl sulfoxide (DMSO), loading buffer, Tris-acetate-EDTA (TAE) buffer, tris-(2-hydroxyethyl)phosphine (THP), Lipofectamine Messenger MAX™, Hoechst 33342, Lysotracker red and 96-well plates were obtained from ThermoFisher. Dulbecco's modified Eagle's medium (DMEM) cell culture medium, RPMI 1640 cell culture medium, fetal bovine serum (FBS) and Opti-MEM were bought from Gibco, Invitrogen, Carlsbad, CA, USA. PrestoBlue reagent and ONE-Glo D-luciferin substrate were purchased from Promega, UK. HEK293T and Caco-2 cell lines were purchased from the ATCC.Instrumentation and analysis
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 500–550 g mol−1. Using conventional calibration, Cirrus GPC software, dispersity (Đ) values, molecular weight (Mw) and experimental molar mass (Mn, SEC) were measured.
500–550 g mol−1. Using conventional calibration, Cirrus GPC software, dispersity (Đ) values, molecular weight (Mw) and experimental molar mass (Mn, SEC) were measured.
Polymer synthesis and characterisation
1H NMR (400 MHz, DMSO-d6) δ 2.81 (t, J = 6.6 Hz, 4H, –C![[H with combining low line]](https://www.rsc.org/images/entities/char_0048_0332.gif) 2-NH2), 2.73 (t, J = 6.6 Hz, 4H, S-CH2–), 2.20 (s, 4H, –NH2); 13C NMR (400 MHz, DMSO-d6) δ 42.09, 41.28.
2-NH2), 2.73 (t, J = 6.6 Hz, 4H, S-CH2–), 2.20 (s, 4H, –NH2); 13C NMR (400 MHz, DMSO-d6) δ 42.09, 41.28.
Calculation of the theoretical number average molar mass of
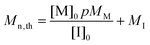 | (1) |
An experimental number average molar mass from 1H NMR spectroscopy Mn,NMR can also be calculated using by dividing integrals of methyl signals for the sarcosine repeating units at 2.80 ppm (3H) and cystamine signals 3.70 ppm (2H).
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 10 (moles of end amine
10 (moles of end amine![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) FITC) molar ratio and triethylamine was added to the mixture at a 0.1 molar ratio to polymer. The reaction mixture was allowed to stir for 24 hours in the dark at room temperature. The reaction mixture was then dialysed in the dark using molecular weight cut-off 3.5 kDa against 250 mL NaOAc buffer (pH: 5.0). Purification was continued for 4 days and the dialysis medium refreshed 2 times in a day. After freeze drying, FITC-labelled polymers were collected as a yellow solid and were stored at −20 °C as 100 mg mL−1 solution in DMSO.
FITC) molar ratio and triethylamine was added to the mixture at a 0.1 molar ratio to polymer. The reaction mixture was allowed to stir for 24 hours in the dark at room temperature. The reaction mixture was then dialysed in the dark using molecular weight cut-off 3.5 kDa against 250 mL NaOAc buffer (pH: 5.0). Purification was continued for 4 days and the dialysis medium refreshed 2 times in a day. After freeze drying, FITC-labelled polymers were collected as a yellow solid and were stored at −20 °C as 100 mg mL−1 solution in DMSO.
Calculation of buffering capacity of polymer between pH 7.4 to 5.0 normalised to mass of polymer
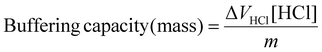 | (2) |
Calculation of buffering capacity of polymer between pH 7.4 to 5.0 normalised to moles of ionisable amines
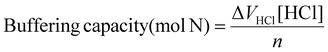 | (3) |
Synthesis of saRNA
For firefly luciferase saRNA synthesis, luciferase encoded pDNA was linearised with Mlul-HF (NEB, UK) for 3 hours at room temperature. Uncapped RNA was synthesised in a MEGAscript reaction (Ambion, UK) using 1 μg of linearised DNA as a template. The RNA was then purified by LiCl precipitation (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 volume ratio) for 24 hours at −20 °C. Then, transcripts were centrifuged at 14
1 volume ratio) for 24 hours at −20 °C. Then, transcripts were centrifuged at 14![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 rpm at 4 °C for 20 minutes, washed once with 70% ethanol, centrifuged for 5 minutes, the ethanol wash was removed, and the pellet was dried at room temperature for 5 minutes. The purified RNA was then capped using ScriptCap Cap 1 capping system kit (CellScript, Madison, WI, USA) and used according to the manufacturer's protocol. Capped transcripts were again purified by LiCl precipitation. The purified capped saRNA was resuspended in RNAase/DNAase free water and stored at −80 °C at 800 µg mL−1.
000 rpm at 4 °C for 20 minutes, washed once with 70% ethanol, centrifuged for 5 minutes, the ethanol wash was removed, and the pellet was dried at room temperature for 5 minutes. The purified RNA was then capped using ScriptCap Cap 1 capping system kit (CellScript, Madison, WI, USA) and used according to the manufacturer's protocol. Capped transcripts were again purified by LiCl precipitation. The purified capped saRNA was resuspended in RNAase/DNAase free water and stored at −80 °C at 800 µg mL−1.
Preparation of polyplexes
Polyplexes were prepared by electrostatic interactions between positively charged polymers and negatively charged saRNA by mixing various w/w ratios (32, 64 and 128). Firstly, the working dilutions of polymers and saRNA were prepared in 25 mM NaOAc buffer (pH: 5.0). Depending on the desired w/w ratio, differing amounts of polymer stock solutions (100 mg mL−1 in DMSO) were mixed with nucleic acids in the buffer, gently mixed using a pipette and incubated at room temperature for 30 minutes. Whilst preparing the PBAE/saRNA complexes, taking a w![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) w 64 ratio as an example, 10 µg mL−1 of the RNA stock solution was mixed with 2.560 mg mL−1 PBAE by pipette using a 25 mM NaOAc buffer.
w 64 ratio as an example, 10 µg mL−1 of the RNA stock solution was mixed with 2.560 mg mL−1 PBAE by pipette using a 25 mM NaOAc buffer.
Encapsulation efficiency determination using ribogreen assay
A fluorescence-based Quant-iT RiboGreen assay (ThermoFisher, UK) was utilised to be able to detect free RNA in solution after polymer encapsulation. Samples were diluted in 1× TE buffer (v/v 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) and the assay was performed according to the manufacturer's protocol. Samples were then loaded on a black, 96-well plate, and analysed for fluorescence on a microplate reader at an excitation of 485 nm and emission at 528 nm.
1) and the assay was performed according to the manufacturer's protocol. Samples were then loaded on a black, 96-well plate, and analysed for fluorescence on a microplate reader at an excitation of 485 nm and emission at 528 nm.
In vitro experiments
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 500) dye for 30 minutes at room temperature in order to help with gating on live cells. Then, cells were fixed with 100 μL of 4% paraformaldehyde in PBS for 20 minutes. Fixed cells were centrifuged, and pellets resuspended in 50 μL of PBS. Data were acquired using an Image Stream 100 (Amnis, Seattle, US) and on single cell in focus events was used for analysis. Ideas Software (Amnis, Seattle, WA, USA) was used for data analysis. Total cell fluorescence was calculated by default total cell masks for the measurement of the nanoparticle internalisation and cell interiors were identified using the area erode tool (brightfield channel). Internalisation index was shown as percentage of maximum internalisation (percentage of interior cell fluorescence to total cell fluorescence).
500) dye for 30 minutes at room temperature in order to help with gating on live cells. Then, cells were fixed with 100 μL of 4% paraformaldehyde in PBS for 20 minutes. Fixed cells were centrifuged, and pellets resuspended in 50 μL of PBS. Data were acquired using an Image Stream 100 (Amnis, Seattle, US) and on single cell in focus events was used for analysis. Ideas Software (Amnis, Seattle, WA, USA) was used for data analysis. Total cell fluorescence was calculated by default total cell masks for the measurement of the nanoparticle internalisation and cell interiors were identified using the area erode tool (brightfield channel). Internalisation index was shown as percentage of maximum internalisation (percentage of interior cell fluorescence to total cell fluorescence).
Statistical analysis
Graphs and statistics were prepared in GraphPad Prism 9.5.1 version or higher. Statistical differences were analysed using either one-way or two-way ANOVA adjusted for multiple comparison (either Tukey's or Dunnet's), p value lower than 0.05 was considered as statically significant and the levels of statistical significance were set p* < 0.05, p** < 0.01, p*** < 0.001 and p**** < 0.0001.Results and discussion
Synthesis and characterisation of PBAEs
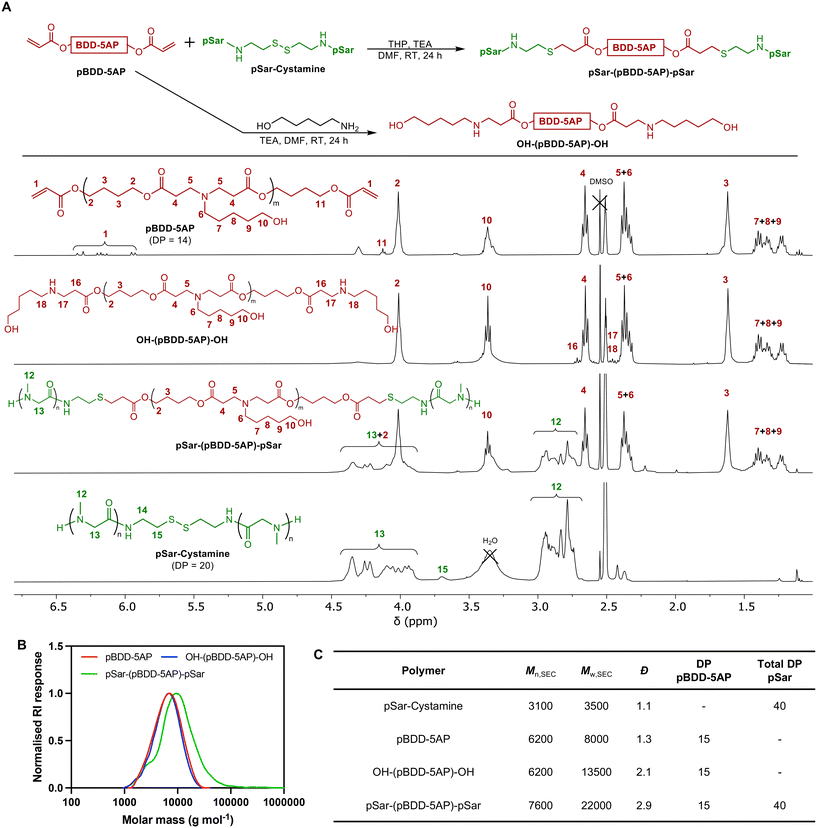
In the first step a poly(β-amino ester) was prepared from 1,4-butanediol diacrylate (BDD) and 5-aminopentanol (5AP), via a Michael-addition based step-growth polymerization with 10 mol% excess of BDD in relation to 5AP, to ensure termination of the polymer with acrylate groups (pBDD-5AP). After purification by precipitation in diethyl ether, acrylate end groups (6.1 ppm) were confirmed using 1H NMR spectroscopy which when integrated against signals from the methylene BDD-5AP repeating unit (1.62 ppm), revealed an average DP of 15 for the pBDD-5AP PBAE (Fig. 2A and Scheme S1† and spectrum S1). SEC analysis revealed an Mn,SEC = 6200 g mol−1 compared to an Mn,NMR = 4500 g mol−1 and Đ = 1.3. Although Đ = 2 is the expected theoretical value for step-growth polymerisations at high conversion, the lower Đ observed is likely due to the precipitation procedure removing low molar mass oligomers thus narrowing the molar mass distribution and increasing the average molar masses, which was indeed observed. The selection of this PBAE composition was based on prior pre-clinical investigations, which demonstrated that pBDD-5AP PBAEs can display good formulation characteristics (such as small particle size, high colloidal stability, and high zeta potentials) and potent transfection performance, both in vitro and in vivo, with various types of nucleic acid cargoes.36Before producing our pSar conjugated PBAE, we first generated a sarcosine N-carboxyanhydride (SarNCA) from N-Boc-sarcosine (Boc-Sar) using a moisture-tolerant protocol developed by Lu and co-workers (Scheme S1† and Fig. 2A).37 SarNCA was then converted into pSar-cystamine, a pSar derivative containing a single disulfide bond in the middle of the chain, using ring opening polymerisation from a bifunctional cystamine initiator targeting a DP of 18 on each side (DP = 36 in total). This relatively low DP was chosen to mimic the low molar masses used for the DMG-PEG lipid (Mn = 2000 g mol−1) in the lipid nanoparticles for the commercially available SARS-CoV-2 mRNA vaccines.38 After purification and isolation through precipitation in diethyl either, pSar-cystamine was characterised by 1H NMR spectroscopy revealing an experimental DP of 20 on each side of the disulfide. This was calculated using integrals of cystamine signals at 3.70 ppm and methyl signals for the sarcosine repeating units at 2.80 ppm (Fig. 2A and spectrum S2). The higher-than-expected DP may be due to a slight initiator inefficiency or impurities within cystamine for NCA polymerisation. DMF-SEC analysis of pSar-cystamine exhibited a monomodal chromatogram with low dispersity (Đ = 1.1) which as expected for NCA polymerisation. The Mn,SEC = 3100 g mol−1 was close to theoretical values for DP40 pSar + cystamine (Mn,th = 3700 g mol−1) however the deviation observed is likely due to the use of PMMA standards in the SEC calibration which do not accurately represent pSar (Fig. S1†).
To conjugate pSar to the acrylate functional PBAE pBDD-5AP, first the disulphide bridge was reduced in situ into reactive thiols using tris(hydroxypropyl)phosphine (THP) to initiate a base catalysed thia-Michael addition reaction between the pSar chains and acrylate functional PBAE. The conjugation reaction was conducted with an excess of pSar per acrylate end-group. Following the reaction, pSar-(pBDD-5AP)-pSar was isolated and excess pSar was removed through dialysis against DMF, followed by precipitation in a diethyl ether. 1H NMR spectroscopy confirmed the successful conjugation of pSar to the PBAE, generating pSar-(pBDD-5AP)-pSar (Fig. 2 and spectrum S3), revealing quantitative conversion of acryloyl groups (no visible signals at 5.9–6.4 ppm) and the expected ratio between pBDD-5AP and pSar repeating units, calculated from the pBDD-5AP methylene signals at 1.62 ppm and 2.80 ppm from methyl groups from pSar. This also confirms the removal of excess pSar that was available in the conjugation reaction. SEC analysis revealed a clear shift towards higher molar mass (Mn,SEC = 7600 g mol−1) following the reaction, indicating successful pSar conjugation.
As a control polymer without pSar, we also synthesised a hydroxyl terminated PBAE, OH-(pBDD-5AP)-OH, by endcapping the acrylate functional pBDD-5AP with further 5-AP, with 1H NMR spectroscopy confirming full disappearance of acrylate end groups (Fig. 2A and spectrum S4). Size exclusion chromatographs of the synthesised polymers revealed the pre-functionalised PBAE, pBDD-5AP, exhibited Mn,SEC of 6200 g mol−1 and Đ = 1.3, similar to other reported PBAEs (Fig. 2B and C). Upon functionalisation with 5AP to produce OH-(pBDD-5AP)-OH, there was no significant increase in molar mass (Mn,SEC = 6200 g mol−1, and Đ = 2.1). Overall, our characterisation data confirmed the successful synthesis of our targeted pSar conjugated PBAE and control polymers.
The buffering capacities were assessed for OH-(pBDD-5AP)-OH and pSar-(pBDD-5AP)-pSar as the pKa of polycations has been recognised as crucial for enhancing endosomal buffering and endo/lysosomal escape of polymer-based nucleic acid delivery systems. Acid–base titration in the pH range of 10.0–3.0 for PBAEs with and without pSarylation displayed similar titration curves, implying that pSarylation did not impact their buffering capacity (Fig. S2 and Table S1†). Specifically looking at the range related to endosomal acidification, considered between pH 7.4 and pH 5.0,39 PEI displayed a buffering capacity 12 mol of H+ per mol of ionisable nitrogen in this pH range, whilst OH-(pBDD-5AP)-OH and pSar-(pBDD-5AP)-pSar displayed buffering capacities of 4.0 and 3.5 respectively. The weaker buffering capacities observed for the PBAEs, compared to a branched PEI control, was likely due to PEI's structure containing a high density of primary, secondary and tertiary amines compared to our PBAEs only exhibiting tertiary amines at a reduced density across the polymer chain. This therefore broadens the buffering capacity of PEI and the high density also imparts a neighbouring group effect commonly seen with ionisable polycations.
Effect of pSar density on polyplex physicochemical properties
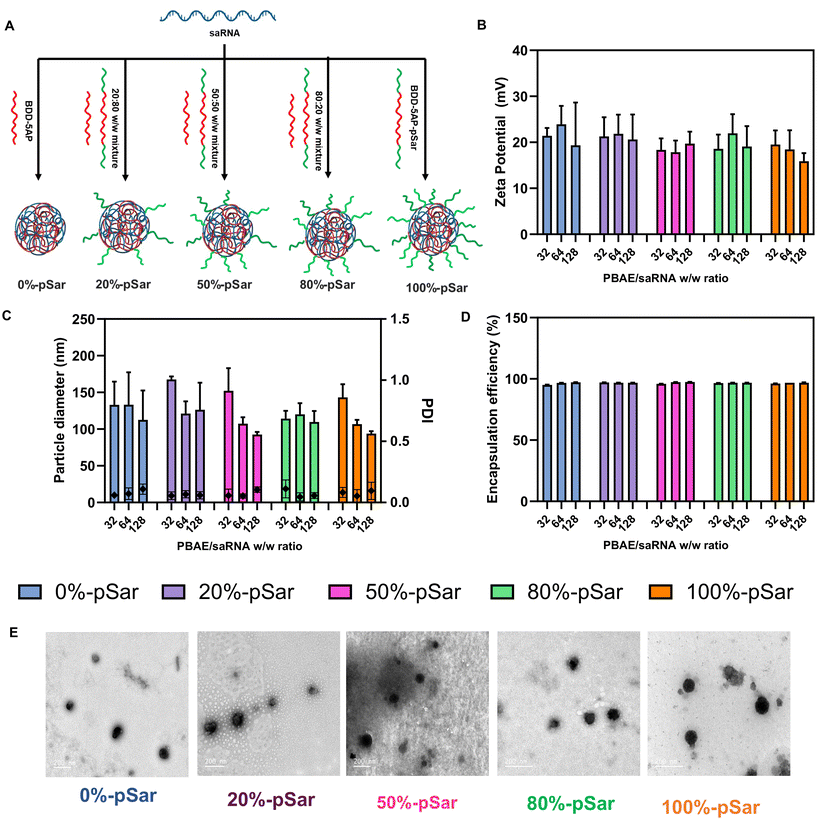
Polyplex preparation was performed at three different polymer/fluc saRNA weight-to-weight ratios (w/w ratios of 32, 64 and 128) for OH-(pBDD-5AP)-OH and pSar-(pBDD-5AP)-pSar PBAEs to optimise the physicochemical properties of the polyplex. These include particle size, surface charge, encapsulation efficiency, biocompatibility, and RNA expression. To assess the effect of pSar density on the nanoparticle surface, we also prepared polyplexes prepared with 20%-pSar, 50%-pSar and 80%-pSar which are mixtures of pSar-(pBDD-5AP)-pSar and OH-(pBDD-5AP)-OH as a weight% (Fig. 3A). Briefly polyplexes were prepared by mixing the desired polymers and saRNA in pH 5.0 sodium acetate buffer. Varying quantities of polymer stock solutions, depending on the desired w/w ratio, were then combined with saRNA manually using a micropipette and allowed to incubate at ambient temperature for 30 minutes. It should also be noted that in the literature there are different approaches to modulate the ratio of nucleic acid cargo to the polymer delivery vector. One common approach is a simple weight ratio between the nucleic acid and polymer, which is more common in the PBAE literature for pharmaceutical development. However, many researchers also consider the nitrogen to phosphate ratio (N/P) as a way to identify a charge ratio and normalise for different materials. In this study we opted for weight to weight ratios to be consistent with previous literature on PBAEs and polymer-conjugated PBAEs,40 however we have also reported the N/P ratios for comparison with other literature (Table S2†).Hydrodynamic diameters of all nanoparticles were observed to be between 100–150 nm with low polydispersity index (PDI) values (<0.2), regardless of pSar content, determined by dynamic light scattering (DLS) (Fig. 3C). Some particles e.g. 50%-pSar and 100%-pSar possessed a significantly decreasing size with w/w ratio. In addition, zeta potential of the particles was measured to be between 20–25 mV and no significant difference was observed between the groups (Fig. 3B). The encapsulation efficiency of nanoparticles was analysed utilizing Ribogreen assay and revealed full RNA encapsulation for nanoparticles produced at all pSar-(pBDD-5AP)-pSar/OH-(pBDD-5AP)-OH ratios (Fig. 3D). Finally, TEM micrographs of all polyplexes prepared at a w/w ratio of 128 displayed spherical complexes between 35 to 220 nm in diameter consistent with DLS analysis and taking into account some particle shrinking in the dry state (Fig. 3E). Furthermore, as DLS analysis can over represent larger particles present in heterogeneous particle distributions, it is likely that any smaller species visible by TEM may not have been included in the particle size averages.41 Overall, these results indicate that all five formulations were capable of encapsulating saRNA and producing particle sizes below the typical 200 nm diameter threshold for efficient gene delivery.
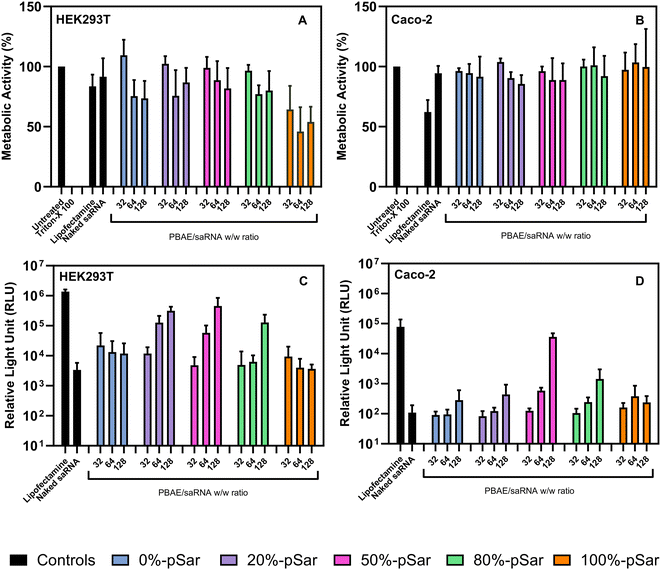
In vitro evaluation of saRNA delivery
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 50) displayed the most effective gene delivery vector for glioblastoma cells.40
50) displayed the most effective gene delivery vector for glioblastoma cells.40
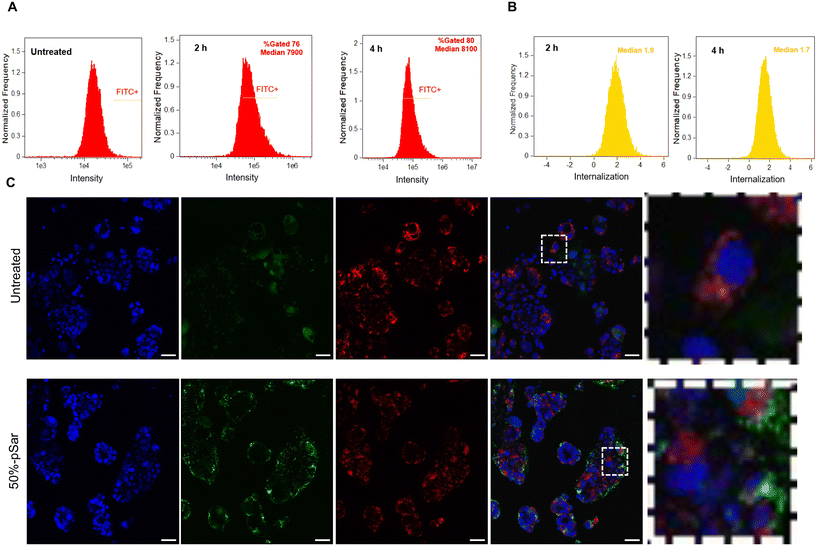
To assess further the intracellular internalisation mechanism of the pSar-(pBDD-5AP)-pSar in Caco-2 cells we employed confocal microscopy to evaluate colocalization with a lysosomal (Lysotracker™ red) and nuclear (Hoecht 33342) stain (Fig. 5C). After 4 h post administration, confocal micrographs showed that polyplexes successfully internalised within Caco-2 and merged images demonstrated with minimal co-localisation with the acidic lysosomes with image analysis indicating Pearson's correlation coefficient between FITC labelled polyplexes and Lysotracker™ stained lysosomes were below 0.5. Given the ∼100 nm particle diameters it is likely that these formulations were internalised by an endocytosis mechanism, however the apparent low colocalization with the acidic lysosomes may indicate that the particles escaped before endosomal maturation to lysosomal compartments. Although further endosomal escape studies would be needed to investigate this phenomenon in more detail, it is possible that enhanced endosomal escape was a factor in the high expression efficiencies observed for these complexes. However, it should also be noted that fluorescein is a pH responsive dye hence may exhibit reduced fluorescence within the acidic lysosomal compartments, adding an uncertainty to measures of dye co-localisation, nonetheless the high transfection efficiencies observed for this formulation indicates clear propensity of endosomal escape which may have all escaped within the first 2 hours.
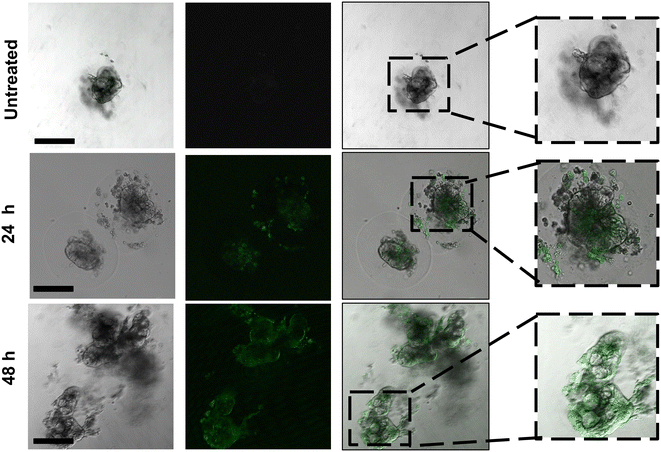
We therefore evaluated our chosen formulation 50%-pSar (w/w 128) encapsulating a GFP-saRNA in human colon colorectal organoids to monitor saRNA expression via confocal microscopy. The 50%-pSar saRNA formulation was incubated with organoids once they had reached the desired compaction level. After 4 h, nanoparticles were removed, and fresh media was replaced onto the organoids. GFP expression was determined after 24 and 48 h. Confocal microscopy revealed GFP expression within the central regions of the organoids at 24 h, with increasing fluorescence intensity observable after 48 h (Fig. 6). Previous studies have shown expression of exogenously delivered nucleic acids at the surface of cancer organoids, however the 50%-pSar PBAE formulations were substantially improved compared to the prior systems in terms of generating high expression levels deep within the core of the organoid, indicating high penetration of these nanoparticles within the dense matrix of cells.46,47 These results indicate that pSarylated PBAEs may represent promising new materials for tumour directed delivery of nucleic acid therapeutics. Additionally, the full preservation of the organoid structure over the 48 h transfection experiment may suggest that these materials could be used to aid delivery of mRNA and saRNA to further organoid models to examine how expression of exogenously administered RNA varies within the surface and core of these tumour models.
Conclusions
In this work we have developed a new methodology to produce polysarcosinylated poly(beta amino esters) as an alternative steric stabilising group to PEG. We used this new pSar-PBAE conjugate to produce a library of polyplexes with different pSar densities on the nanoparticle surface by mixing pSarylated PBAEs with analogous unfunctionalized PBAEs in different ratios. We observed that pSarylation had minimal effect on polyplex physicochemical properties, while all pSar densities yielded higher transfection efficiencies than the positive control lipofectamine in HEK293T cells. In contrast, only the 50%-pSar polyplexes produced saRNA expression comparable to lipofectamine in colorectal cancer cells (Caco-2), with uptake studies indicating fast cellular uptake and limited colocalization in lysosomes indicating potential for endosomal escape. Finally, the 50%-pSar polyplexes showed potential translation for tumour delivery, exhibiting high expression of saRNA within the core colorectal cancer organoids within 24 h, likely due to the enhanced permeability of these polyplexes endowed with the pSar coating. The efficient diffusion to the organoid core may make these formulations amenable to intratumoural delivery of genetic material encoding for pro-inflammatory cytokines which are required for stimulation of the immune system around the local tumour environment in immuno-oncology applications. Overall, these results indicate that pSarylation is a suitable alternative for PEGylation of PBAEs, yielding potent gene delivery vehicles. As pSar is an established PEG alternative, future research should focus on studying the differences in immunogenicity between PEG-PBAE and pSar-PBAE conjugates, which is especially important for vaccine and immunotherapy nucleic acid formulations.Data access statement
All relevant data can be obtained upon request from the corresponding author at p.gurnani@ucl.ac.uk.Author contributions
Conceptualization, H. B. and P. G.; data curation and formal analysis, H. B., R. K. J., P. G.; writing original draft preparation, H. B.; writing review and editing, H. B., R. K. J., A. E., M. M., N. G., U. C. O., K. P., P. F. M., R. J. S., P. O. M., A. M., C. A., P. G.; supervision, A. M., C. A., P. G.; project administration, C. A., P. G.; funding acquisition, C. A. All authors have read and agreed to the published version of the manuscript.Conflicts of interest
All authors declare that they have no conflicts of interest.Acknowledgements
We thank the Royal Society [Wolfson research merit award WM150086 to CA]. Work on this project is supported by Wellcome Leap as part of the R3 Program [Biofoundry-in-a-Box], the Turkish Ministry of National Education, and an Engineering and Physical Sciences Research Council ICASE 2021 Studentship. Umut Can Oz is thankful for a fellowship (2219) from the TUBITAK. We also thank Tom Hyde, Esme Ireson and Paul Cooling for expert technical support. The Nanoscale & Macroscale Research Centre (NMRC) is acknowledged for providing the facilities for TEM and related analysis particularly to Denise McLean. We thank the School of Life Sciences imaging facility (SLIM) and their staff, particularly Dr Tim Self, for use of their facilities and expert guidance. We thank the MINIGUTS team (consultants, pathologists, scientists and nurses) for recruiting patients, and helping with the collection of colon cancer patients’ biopsies at Queen's Medical Centre (QMC), Nottingham.References
- Y. Zhong, S. Du and Y. Dong, mRNA delivery in cancer immunotherapy, Acta Pharm. Sin. B, 2023, 13(4), 1348–1357 CrossRef CAS PubMed.
- N. Pardi, M. J. Hogan, F. W. Porter and D. Weissman, mRNA vaccines - a new era in vaccinology, Nat. Rev. Drug Discovery, 2018, 17(4), 261–279 CrossRef CAS PubMed.
- D. Li, C. Liu, Y. Li, R. Tenchov, J. M. Sasso, D. Zhang, D. Li, L. Zou, X. Wang and Q. Zhou, Messenger RNA-Based Therapeutics and Vaccines: What's beyond COVID-19?, ACS Pharmacol. Transl. Sci., 2023, 6(7), 943–969 CrossRef CAS PubMed.
- B. Fayed, J. Jagal, R. Cagliani, R. A. Kedia, A. Elsherbeny, H. Bayraktutan, G. Khoder and M. Haider, Co-administration of amoxicillin-loaded chitosan nanoparticles and inulin: A novel strategy for mitigating antibiotic resistance and preserving microbiota balance in Helicobacter pylori treatment, Int. J. Biol. Macromol., 2023, 253, 126706 CrossRef CAS PubMed.
- Z. A. Azrak, M. S. Taha, J. Jagal, A. Elsherbeny, H. Bayraktutan, M. H. AbouGhaly, A. H. Elshafeey, K. Greish and M. Haider, Optimized mucoadhesive niosomal carriers for intranasal delivery of carvedilol: A quality by design approach, Int. J. Pharm., 2024, 123935 CrossRef CAS PubMed.
- A. K. Blakney, Y. Zhu, P. F. McKay, C. R. Bouton, J. Yeow, J. Tang, K. Hu, K. Samnuan, C. L. Grigsby, R. J. Shattock and M. M. Stevens, Big Is Beautiful: Enhanced saRNA Delivery and Immunogenicity by a Higher Molecular Weight, Bioreducible, Cationic Polymer, ACS Nano, 2020, 14(5), 5711–5727 CrossRef CAS PubMed.
- S. Taranejoo, J. Liu, P. Verma and K. Hourigan, A review of the developments of characteristics of PEI derivatives for gene delivery applications, J. Appl. Polym. Sci., 2015, 132(25), 42096 CrossRef.
- A. K. Blakney, P. F. McKay, K. Hu, K. Samnuan, N. Jain, A. Brown, A. Thomas, P. Rogers, K. Polra, H. Sallah, J. Yeow, Y. Zhu, M. M. Stevens, A. Geall and R. J. Shattock, Polymeric and lipid nanoparticles for delivery of self-amplifying RNA vaccines, J. Controlled Release, 2021, 338, 201–210 CrossRef CAS PubMed.
- P. Gurnani, A. K. Blakney, J. Yeow, C. R. Bouton, R. J. Shattock, M. M. Stevens and C. Alexander, An improved synthesis of poly(amidoamine)s for complexation with self-amplifying RNA and effective transfection, Polym. Chem., 2020, 11(36), 5861–5869 RSC.
- H. Bayraktutan, P. Symonds, V. Brentville, A. Mata, L. Durrant, C. Alexander and P. Gurnani, Sparsely Pegylated Poly (Beta-Amino Esters) Polyplexes Enhance Antigen Specific T-Cell Response of a Bivalent Sars-Cov-2 DNA Vaccine. Available at SSRN 4695805.
- X. Wang, Z. Zhang and N. Hadjichristidis, Poly(amino ester)s as an emerging synthetic biodegradable polymer platform: Recent developments and future trends, Prog. Polym. Sci., 2023, 136, 101634 CrossRef CAS.
- N. K. Dastgerdi, N. Gumus, H. Bayraktutan, D. Jackson, K. Polra, P. F. McKay, F. Atyabi, R. Dinarvand, R. J. Shattock and L. Martinez-Pomares, Charge neutralized poly (β-amino ester) polyplex nanoparticles for delivery of self-amplifying RNA, Nanoscale Adv., 2024, 6, 1409–1422 RSC.
- Y. Rui, D. R. Wilson, S. Y. Tzeng, H. M. Yamagata, D. Sudhakar, M. Conge, C. A. Berlinicke, D. J. Zack, A. Tuesca and J. J. Green, High-throughput and high-content bioassay enables tuning of polyester nanoparticles for cellular uptake, endosomal escape, and systemic in vivo delivery of mRNA, Sci. Adv., 2022, 8(1), eabk2855 CrossRef CAS PubMed.
- A. Akinc, D. M. Lynn, D. G. Anderson and R. Langer, Parallel synthesis and biophysical characterization of a degradable polymer library for gene delivery, J. Am. Chem. Soc., 2003, 125(18), 5316–5323 CrossRef CAS PubMed.
- J. S. Suk, Q. Xu, N. Kim, J. Hanes and L. M. Ensign, PEGylation as a strategy for improving nanoparticle-based drug and gene delivery, Adv. Drug Delivery Rev., 2016, 99(Pt A), 28–51 CrossRef CAS PubMed.
- Y. Liu, Y. Li, D. Keskin and L. Shi, Poly(beta-Amino Esters): Synthesis, Formulations, and Their Biomedical Applications, Adv. Healthcare Mater., 2019, 8(2), e1801359 CrossRef PubMed.
- J. Kim, S. K. Mondal, S. Y. Tzeng, Y. Rui, R. Al-Kharboosh, K. K. Kozielski, A. G. Bhargav, C. A. Garcia, A. Quinones-Hinojosa and J. J. Green, Poly(ethylene glycol)-Poly(beta-amino ester)-Based Nanoparticles for Suicide Gene Therapy Enhance Brain Penetration and Extend Survival in a Preclinical Human Glioblastoma Orthotopic Xenograft Model, ACS Biomater. Sci. Eng., 2020, 6(5), 2943–2955 CrossRef CAS PubMed.
- J. H. Brumbach, C. Lin, J. Yockman, W. J. Kim, K. S. Blevins, J. F. Engbersen, J. Feijen and S. W. Kim, Mixtures of poly(triethylenetetramine/cystamine bisacrylamide) and poly(triethylenetetramine/cystamine bisacrylamide)-g-poly(ethylene glycol) for improved gene delivery, Bioconjugate Chem., 2010, 21(10), 1753–1761 CrossRef CAS PubMed.
- C. Zhu, M. Zheng, F. Meng, F. M. Mickler, N. Ruthardt, X. Zhu and Z. Zhong, Reversibly shielded DNA polyplexes based on bioreducible PDMAEMA-SS-PEG-SS-PDMAEMA triblock copolymers mediate markedly enhanced nonviral gene transfection, Biomacromolecules, 2012, 13(3), 769–778 CrossRef CAS PubMed.
- C. Wang, X. Wang, L. Du, Y. Dong, B. Hu, J. Zhou, Y. Shi, S. Bai, Y. Huang, H. Cao, Z. Liang and A. Dong, Harnessing pH-Sensitive Polycation Vehicles for the Efficient siRNA Delivery, ACS Appl. Mater. Interfaces, 2021, 13(2), 2218–2229 CrossRef CAS PubMed.
- G. T. Kozma, T. Shimizu, T. Ishida and J. Szebeni, Anti-PEG antibodies: Properties, formation, testing and role in adverse immune reactions to PEGylated nano-biopharmaceuticals, Adv. Drug Delivery Rev., 2020, 154–155, 163–175 CrossRef CAS PubMed.
- Y. Ju, W. S. Lee, E. H. Pilkington, H. G. Kelly, S. Li, K. J. Selva, K. M. Wragg, K. Subbarao, T. H. O. Nguyen, L. C. Rowntree, L. F. Allen, K. Bond, D. A. Williamson, N. P. Truong, M. Plebanski, K. Kedzierska, S. Mahanty, A. W. Chung, F. Caruso, A. K. Wheatley, J. A. Juno and S. J. Kent, Anti-PEG Antibodies Boosted in Humans by SARS-CoV-2 Lipid Nanoparticle mRNA Vaccine, ACS Nano, 2022, 16(8), 11769–11780 CrossRef CAS PubMed.
- M. Barz, R. Luxenhofer, R. Zentel and M. J. Vicent, Overcoming the PEG-addiction: well-defined alternatives to PEG, from structure–property relationships to better defined therapeutics, Polym. Chem., 2011, 2(9), 1900–1918 RSC.
- Y. W. Kong and E. C. Dreaden, PEG: Will It Come Back to You? Polyethelyne Glycol Immunogenicity, COVID Vaccines, and the Case for New PEG Derivatives and Alternatives, Front. Bioeng. Biotechnol., 2022, 10, 879988 CrossRef PubMed.
- A. Elsherbeny, H. Bayraktutan, U. C. Oz, C. Moloney, J. C. Ashworth, A. M. Grabowska and C. Alexander, Responsive Nanomaterial Delivery Systems for Pancreatic Cancer Management, Adv. Ther., 2023, 2300330 Search PubMed.
- M. Berger, F. Toussaint, S. B. Djemaa, J. Laloy, H. Pendeville, B. Evrard, C. Jerome, A. Lechanteur, D. Mottet, A. Debuigne and G. Piel, Poly(vinyl pyrrolidone) derivatives as PEG alternatives for stealth, non-toxic and less immunogenic siRNA-containing lipoplex delivery, J. Controlled Release, 2023, 361, 87–101 CrossRef CAS PubMed.
- C. Pelosi, M. R. Tinè and F. R. Wurm, Main-chain water-soluble polyphosphoesters: Multi-functional polymers as degradable PEG-alternatives for biomedical applications, Eur. Polym. J., 2020, 141, 110079 CrossRef CAS.
- M. Tully, M. Dimde, C. Weise, P. Pouyan, K. Licha, M. Schirner and R. Haag, Polyglycerol for Half-Life Extension of Proteins-Alternative to PEGylation?, Biomacromolecules, 2021, 22(4), 1406–1416 CrossRef CAS PubMed.
- P. Zhang, P. Jain, C. Tsao, Z. Yuan, W. Li, B. Li, K. Wu, H. C. Hung, X. Lin and S. Jiang, Polypeptides with High Zwitterion Density for Safe and Effective Therapeutics, Angew. Chem., Int. Ed., 2018, 57(26), 7743–7747 CrossRef CAS PubMed.
- A. Mero, G. Pasut, L. Dalla Via, M. W. Fijten, U. S. Schubert, R. Hoogenboom and F. M. Veronese, Synthesis and characterization of poly(2-ethyl 2-oxazoline)-conjugates with proteins and drugs: suitable alternatives to PEG-conjugates?, J. Controlled Release, 2008, 125(2), 87–95 CrossRef CAS PubMed.
- A. Birke, J. Ling and M. Barz, Polysarcosine-containing copolymers: Synthesis, characterization, self-assembly, and applications, Prog. Polym. Sci., 2018, 81, 163–208 CrossRef CAS.
- S. S. Nogueira, A. Schlegel, K. Maxeiner, B. Weber, M. Barz, M. A. Schroer, C. E. Blanchet, D. I. Svergun, S. Ramishetti, D. Peer, P. Langguth, U. Sahin and H. Haas, Polysarcosine-Functionalized Lipid Nanoparticles for Therapeutic mRNA Delivery, ACS Appl. Nano Mater., 2020, 3(11), 10634–10645 CrossRef CAS.
- A. J. Geall, A. Verma, G. R. Otten, C. A. Shaw, A. Hekele, K. Banerjee, Y. Cu, C. W. Beard, L. A. Brito, T. Krucker, D. T. O'Hagan, M. Singh, P. W. Mason, N. M. Valiante, P. R. Dormitzer, S. W. Barnett, R. Rappuoli, J. B. Ulmer and C. W. Mandl, Nonviral delivery of self-amplifying RNA vaccines, Proc. Natl. Acad. Sci. U. S. A., 2012, 109(36), 14604–14609 CrossRef CAS PubMed.
- P. Gurnani, A. K. Blakney, R. Terracciano, J. E. Petch, A. J. Blok, C. R. Bouton, P. F. McKay, R. J. Shattock and C. Alexander, The In Vitro, Ex Vivo, and In Vivo Effect of Polymer Hydrophobicity on Charge-Reversible Vectors for Self-Amplifying RNA, Biomacromolecules, 2020, 21(8), 3242–3253 CrossRef CAS PubMed.
- F. Saviano, T. Lovato, A. Russo, G. Russo, C. R. Bouton, R. J. Shattock, C. Alexander, F. Quaglia, A. K. Blakney, P. Gurnani and C. Conte, Ornithine-derived oligomers and dendrimers for in vitro delivery of DNA and ex vivo transfection of skin cells via saRNA, J. Mater. Chem. B, 2020, 8(22), 4940–4949 RSC.
- R. A. Cordeiro, A. Serra, J. F. Coelho and H. Faneca, Poly (β-amino ester)-based gene delivery systems: From discovery to therapeutic applications, J. Controlled Release, 2019, 310, 155–187 CrossRef CAS PubMed.
- Z. Y. Tian, Z. Zhang, S. Wang and H. Lu, A moisture-tolerant route to unprotected alpha/beta-amino acid N-carboxyanhydrides and facile synthesis of hyperbranched polypeptides, Nat. Commun., 2021, 12(1), 5810 CrossRef CAS PubMed.
- C. Hald Albertsen, J. A. Kulkarni, D. Witzigmann, M. Lind, K. Petersson and J. B. Simonsen, The role of lipid components in lipid nanoparticles for vaccines and gene therapy, Adv. Drug Delivery Rev., 2022, 188, 114416 CrossRef CAS PubMed.
- A. Alazzo, N. Gumus, P. Gurnani, S. Stolnik, R. Rahman, K. Spriggs and C. Alexander, Investigating histidinylated highly branched poly(lysine) for siRNA delivery, J. Mater. Chem. B, 2022, 10(2), 236–246 RSC.
- J. Kim, S. K. Mondal, S. Y. Tzeng, Y. Rui, R. Al-kharboosh, K. K. Kozielski, A. G. Bhargav, C. A. Garcia, A. Quiñones-Hinojosa and J. J. Green, Poly(ethylene glycol)–Poly(beta-amino ester)-Based Nanoparticles for Suicide Gene Therapy Enhance Brain Penetration and Extend Survival in a Preclinical Human Glioblastoma Orthotopic Xenograft Model, ACS Biomater. Sci. Eng., 2020, 6(5), 2943–2955 CrossRef CAS PubMed.
- S. K. Filippov, R. Khusnutdinov, A. Murmiliuk, W. Inam, L. Y. Zakharova, H. Zhang and V. V. Khutoryanskiy, Dynamic light scattering and transmission electron microscopy in drug delivery: a roadmap for correct characterization of nanoparticles and interpretation of results, Mater. Horiz., 2023, 10(12), 5354–5370 RSC.
- C. H. Jones, M. Chen, A. Ravikrishnan, R. Reddinger, G. Zhang, A. P. Hakansson and B. A. Pfeifer, Mannosylated poly (beta-amino esters) for targeted antigen presenting cell immune modulation, Biomaterials, 2015, 37, 333–344 CrossRef CAS PubMed.
- V. Hiebl, D. Schachner, A. Ladurner, E. H. Heiss, H. Stangl and V. M. Dirsch, Caco-2 cells for measuring intestinal cholesterol transport-possibilities and limitations, Biol. Proced. Online, 2020, 22(1), 1–18 CrossRef PubMed.
- Y. Phanse, A. E. Ramer-Tait, S. L. Friend, B. Carrillo-Conde, P. Lueth, C. J. Oster, G. J. Phillips, B. Narasimhan, M. J. Wannemuehler and B. H. Bellaire, Analyzing cellular internalization of nanoparticles and bacteria by multi-spectral imaging flow cytometry, J. Visualized Exp., 2012,(64), e3884 Search PubMed.
- M. J. Munro, S. T. Tan and C. Gray, Applications for Colon Organoid Models in Cancer Research, Organoids, 2023, 2(1), 37–49 CrossRef.
- B. Laperrousaz, S. Porte, S. Gerbaud, V. Härmä, F. Kermarrec, V. Hourtane, F. Bottausci, X. Gidrol and N. Picollet-D′hahan, Direct transfection of clonal organoids in Matrigel microbeads: a promising approach toward organoid-based genetic screens, Nucleic Acids Res., 2018, 46(12), e70–e70 CrossRef PubMed.
- L. Wasungu, J. M. Escoffre, A. Valette, J. Teissie and M. P. Rols, A 3D in vitro spheroid model as a way to study the mechanisms of electroporation, Int. J. Pharm., 2009, 379(2), 278–284 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d4py00064a |
| This journal is © The Royal Society of Chemistry 2024 |