Open Access Article
Open Access ArticleHeterocyclic (pyrazine)carboxamide Ru(II) complexes: structural, experimental and theoretical studies of interactions with biomolecules and cytotoxicity†
Nokwanda Tsaulwayoa,
Reinner O. Omondi‡
 a,
Paranthaman Vijayan
a,
Paranthaman Vijayan a,
Nicole R. S. Sibuyi
a,
Nicole R. S. Sibuyi b,
Miché D. Meyerb,
Mervin Meyerb and
Stephen O. Ojwach
b,
Miché D. Meyerb,
Mervin Meyerb and
Stephen O. Ojwach *a
*a
aSchool of Chemistry and Physics, University of KwaZulu-Natal, Private Bag X01, Scottsville, Pietermaritzburg 3209, South Africa. E-mail: ojwach@ukzn.ac.za
bDepartment of Science and Innovation/Mintek Nanotechnology Innovation Centre, Biolabels Research Node, Department of Biotechnology, University of the Western Cape, Bag X17, Bellville, 7535, Cape Town, South Africa
First published on 11th March 2024
Abstract
Treatments of N-(1H-benzo[d]imidazol-2-yl)pyrazine-2-carboxamide (HL1) and N-(benzo[d]thiazol-2-yl)pyrazine-2-carboxamide carboxamide ligands (HL2) with [Ru(p-cymene)Cl2]2 and [Ru(PPh3)3Cl2] precursors afforded the respective Ru(II) complexes [Ru(L1)(p-cymene)Cl] (Ru1), [Ru(L2)(p-cymene)Cl] (Ru2), [Ru(L1)(PPh3)2Cl] (Ru3), and [Ru(L2)(PPh3)2Cl] (Ru4). These complexes were characterized by NMR, FT-IR spectroscopies, mass spectrometry, elemental analyses, and crystal X-ray crystallography for Ru2. The molecular structure of complex Ru2 contains one mono-anionic bidentate bound ligand and display pseudo-octahedral piano stool geometry around the Ru(II) atom. The interactions with calf thymus DNA (CT-DNA) and bovine serum albumin (BSA) were investigated by spectroscopic techniques. The experimental binding studies suggest that complexes Ru1–Ru4 interact with DNA, primarily through minor groove binding, as supported by molecular docking results. Additionally, these complexes exhibit strong quenching of the fluorescence of tryptophan residues in BSA, displaying static quenching. The in vitro cytotoxicity studies of compounds Ru1–Ru4 were assessed in cancer cell lines (A549, PC-3, HT-29, Caco-2, and HeLa), as well as a non-cancer line (KMST-6). Compounds Ru1 and Ru2 exhibited superior cytotoxicity compared to Ru3 and Ru4. The in vitro cytotoxicity and selectivity of compounds Ru1 and Ru2 against A549, PC-3, and Caco-2 cell lines surpassed that of cisplatin.
Introduction
Since the discovery of cisplatin in cancer therapy, research into metal-based chemotherapeutic agents has gained momentum.1–4 This endeavour is underpinned by the dual objectives of enhancing cytotoxicity against malignant cells, while concurrently striving to limit the side effects of these chemotherapeutic drugs.5–10 Among the numerous classes of metal complexes being investigated for their potential cancer therapy, ruthenium(II) complexes have emerged as a popular choice for non-platinum(II) based drugs. This is largely due to their desirable rate of ligand substitution kinetics, which are amenable to the dynamics observed in cellular processes, and thereby minimizing undesirable side reactions.11,12 Furthermore Ru(II) complexes have the ability to mimic iron in their interaction with serum transferrin and albumin proteins, which transport and solubilise iron within cytoplasm and thereby exploiting the body's inherent mechanism for the safe and non-toxic delivery of iron.13,14Within the “ruthenotherapy”,15–19 the sodium salt analogue NKP-1339 has progressed into clinical trials.20 Also, the ruthenium(II)-arene 1,3,5-triaza-7-phosphaadamantane (RAPTA) compound, exhibits substantial cytotoxicity against a number of cancer lines and induces cell death primarily via mitochondrial apoptosis.21,22 Moreover, both RM175 and ONCO4417 induce apoptosis through G2/M phase arrest. Noticeably, ONCO4417 also exhibits DNA-damaging properties at levels similar to cisplatin.23
Another class of ruthenium(II) based complexes which have gained significant interest in the development of anticancer agents are those supported on arene scaffolds.24–28 This is attributed to their distinct characteristics, including solubility, stability, lipophilicity, and accessibility via facile synthetic routes. One such example, is the investigations of the in vitro cytotoxicity of ruthenium(II)–arene complexes of N-heterocyclic carbene (NHC) ligands.29,30 These complexes exhibit good lipophilicity and show high accumulation within the A2780 cell line. It has also been observed that Ru(II)–arene complexes containing perfluorinated phosphine ligands, show improved cytotoxicity and selectivity.31,32 In another related study involving the assessment of antiproliferative activity and apoptosis mechanism of arene Ru(II) carbazole-based hydrazone complexes, it was found that the complexes display comparable cytotoxicity to cisplatin.33
To broaden the spectrum and chemical diversity of the currently available Ru(II) based anticancer drugs, we report the synthesis and structural studies of novel Ru(II) complexes functionalized on the (pyrazine)carboxamide scaffolds. The interactions of these complexes with DNA and protein were also explored using spectroscopic and molecular docking techniques. The cytotoxic effects of the complexes on a panel of six cell lines were also evaluated and are herein discussed.
Results and discussion
Synthesis and structural characterisation of the ruthenium(II) complexes
The carboxamide ligands N-(1H-benzo[d]imidazol-2-yl)pyrazine-2-carboxamide (HL1), and N-(benzo[d]thiazol-2-yl)pyrazine-2-carboxamide (HL2) were synthesized by following established literature procedures34,35 and were obtained in moderate to good yields of 52% (HL1) and 85% (HL2) as depicted in Scheme 1. Reactions of ligands HL1 and HL2 with [RuCl2(p-cymene)]2 in the presence of NaOMe in a 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2 mole ratio in absolute ethanol afforded complexes [Ru(L1)(p-cymene)Cl] (Ru1) and [Ru(L2)(p-cymene)Cl] (Ru2) in high yields of 72% and 84% respectively (Scheme 1). Similarly, reactions of equimolar amounts of ligands HL1 and HL2 with RuCl2(PPh3)3 in CH2Cl2 afforded the respective complexes [Ru(L1)(PPh3)2Cl] (Ru3) and [Ru(L2)(PPh3)2Cl] in moderate and low yields of 66% and 47% respectively (Scheme 1).
2 mole ratio in absolute ethanol afforded complexes [Ru(L1)(p-cymene)Cl] (Ru1) and [Ru(L2)(p-cymene)Cl] (Ru2) in high yields of 72% and 84% respectively (Scheme 1). Similarly, reactions of equimolar amounts of ligands HL1 and HL2 with RuCl2(PPh3)3 in CH2Cl2 afforded the respective complexes [Ru(L1)(PPh3)2Cl] (Ru3) and [Ru(L2)(PPh3)2Cl] in moderate and low yields of 66% and 47% respectively (Scheme 1).
Structural identities of ligands (HL1 and HL2) and their corresponding complexes Ru1–Ru4 were confirmed by a combination of 1H, 13C, 31P NMR, FT-IR spectroscopies, mass spectrometry, elemental analysis, and single crystal X-ray analysis (Ru2). For example, in the 1H NMR spectrum of HL1 (Fig. S1†), distinctive singlet at 12.20 ppm was observed, corresponding to two protons of the N–H from the benzimidazole and from the sec amide. Upon the formation of Ru1 (Fig. S3†) one signal disappeared, serving as clear evidence of the deprotonation of the N–H amide group during the metalation process. Similar observations were made in the 1H NMR spectra of complexes Ru2–Ru4 (Fig. S4–S6†). Furthermore, to confirm the identities of Ru3 and Ru4, we employed 31P NMR and sharp singlets were observed at 26.10 ppm for Ru3 and 26.47 ppm for Ru4 (Fig. S7 and S8†). These resonances are consistent with the presence of two magnetically equivalent PPh3 groups arranged in trans configuration, consistent with the proposed structures in Scheme 1.36,37 In addition, the formation and identity of complexes Ru1–Ru4, was also derived from comparisons of their 13C NMR spectral data to those of their respective ligands (Fig. S9–S13†). For instance, the carbonyl resonance peak at 161.6 ppm (HL1) is slightly shifted to 167.9 ppm in the corresponding complex Ru1 (Fig. S9 and S11†).
The chemical identities of compounds Ru1–Ru4 were additionally ascertained from their FT-IR spectra (Fig. S14–S19†). As an illustration, the C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O stretching band at 1691 cm−1 in HL1 (Fig. S14†) is shifted downfield to 1625 cm−1 in Ru1 (Fig. S16†). This observation is likely due to π-back donation, from the d-orbitals of the Ru(II) to the unoccupied π* orbitals of the ligands. Noteworthy, the C
O stretching band at 1691 cm−1 in HL1 (Fig. S14†) is shifted downfield to 1625 cm−1 in Ru1 (Fig. S16†). This observation is likely due to π-back donation, from the d-orbitals of the Ru(II) to the unoccupied π* orbitals of the ligands. Noteworthy, the C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O signals remained within the expected range for C
O signals remained within the expected range for C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O signal, indicating no keto–enol tautomerization, contradicting earlier reports by Gupta and their colleagues.38
O signal, indicating no keto–enol tautomerization, contradicting earlier reports by Gupta and their colleagues.38
Mass spectrometry was also utilized to elucidate the molecular composition of the isolated complexes (Fig. S20–S25†). The positive ion ESI-MS spectra show the base peaks at m/z = 510.05 (calcd 509.06, 100%) for Ru1, 527.03 (calcd 526.02, 100%) for Ru2, 899.13 (calcd 900.14) for Ru3, corresponding to the protonated molecular ion [M+H]+ ions. The positive HR-MS spectra of Ru4 showed a signal at m/z = 917.0967 (100%) and thus deviating from the calculated mass (917.0974) with an acceptable difference of 0.0007. Elemental analyses data of complexes Ru1–Ru4 were consistent with the empirical formulae of the proposed structures in Scheme 1 and proved their purity in bulk state.
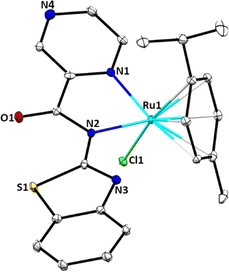
Molecular structure of complex Ru2
Single crystals suitable for X-ray analysis of complex Ru2 were grown by slow diffusion of diethyl ether into a concentrated solution of ethanol at room temperature. Fig. 1 depicts the molecular structure of Ru2, with thermal ellipsoids at 50%, while Table S1† contains crystallographic data and structure refinement parameters. The complex crystallizes in the monoclinic system with C2 space group. In the molecular structure of Ru2, the Ru(II) atom contains one mono-anionic bidentate bound ligand (L−2) and adopts a three-legged piano-stool arrangement. The coordination sphere around the Ru(II) atom in Ru2 is formed by the π-bonded p-cymene ring situated at the apex, while the N(1) and N(2) donor atoms of the bidentate ligand L2, along with the chlorido ligand, are conformationally located on the base of the piano stool-like structure. This arrangement forms a five-membered chelate ring with the Ru(II) ion. The dihedral angles for N(1)–Ru(1)–Cl(1) of 83.0(2)° and N(2)–Ru(1)–Cl(1), 82.31(19)° significantly depart from the expected 90°. Additionally, the bite angle for N(1)–Ru(1)–C(20), of 169.5(3)° deviates appreciably from linearity of 180°. Consequently, the geometry around the Ru(II) centre can best be described as distorted octahedral configuration. The N(1) and N(2) atoms are coordinated to the Ru(II) ion with almost equal bond distances (Ru(1)–N(1), 2.098(7); Ru(1)–N(2), 2.131(7)), signalling the absence of keto–enol tautomerization as observed in the spectral data. The bond length Ru(1)–N(1), 2.098(7) Å falls within the average value of 2.089 ± 0.014 Å reported for 18 related structures.39 The bond length Ru(1)–Cl(1), 2.404(2) Å correlate well with the mean distance of 2.411 ± 0.014 Å obtained for 33 similar structures.40Solution stability of complexes Ru1–Ru4
The solution stability of complexes Ru1–Ru4 was evaluated in both aqueous and DMSO media using UV-vis and 1H NMR spectroscopies, respectively. Fig. S26 and S28† show representative time-dependent UV-vis spectral scans of Ru4 and Ru2, respectively in PBS. The complexes showed characteristic peaks at 260 nm and their profiles remained largely invariant over the 72 h period. This implies that no structural changes occurred, hence stability of the complexes under the physiological conditions used in the DNA interactions and in vitro studies. Typical 1H NMR spectra of complexes Ru1 and Ru2 in DMSO-d6 are shown in Fig. S28 and S29,† respectively. Consistent with the UV-visible data, there were no noticeable changes in proton chemical shifts, suggesting the stability of the complexes in DMSO over the 48 h period.DNA binding studies
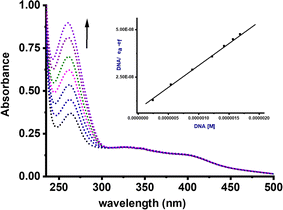
The Kb values for complexes Ru1–Ru4 fall within the range of 0.21–9.22 × 106 M−1 (Table 1). Strikingly, complexes Ru1 and Ru3, which contain the NH moiety, exhibited higher Kb values in comparison to the benzothiazole analogues Ru2 and Ru4. We have previously reported similar trends using related Pd(II) complexes.7 A plausible explanation for this phenomenon could be due to the hydrogen bonding interactions between the NH group of carboxamide and DNA nucleobases, thereby enhancing the strength and extent of these interactions. It is important to note that the Kb values for compounds Ru1 and Ru2, were higher than those of related Ru(II) cymene with (poly)cyclic aromatic diamine ligands (0.4–1.0 × 104).46 The negative ΔG values for compounds Ru1–Ru4 signify spontaneous interactions with CT-DNA.47
| Compound | UV-vis titration | EB fluorescence emission titration | |||||
|---|---|---|---|---|---|---|---|
| Kb 106 M−1 | ΔG‡25°C/kJ mol−1 | Ksv 104 M−1 | Kq 1011 M−1 | Kapp 106 M−1 | KF 102 M−1 | n | |
| Ru1 | 9.22 ± 0.75 | 39.73 | 1.01 ± 0.28 | 1.01 ± 0.29 | 2.96 ± 0.23 | 1.30 ± 0.10 | 0.99 |
| Ru2 | 0.32 ± 0.01 | 31.40 | 0.51 ± 0.07 | 0.51 ± 0.01 | 2.44 ± 0.12 | 0.51 ± 0.06 | 0.95 |
| Ru3 | 3.92 ± 0.46 | 37.61 | 1.84 ± 0.21 | 1.84 ± 0.11 | 3.41 ± 0.28 | 2.24 ± 0.16 | 0.99 |
| Ru4 | 0.21 ± 0.01 | 30.36 | 1.65 ± 0.22 | 1.65 ± 0.63 | 3.01 ± 0.28 | 1.13 ± 0.10 | 0.97 |
Ethidium competitive assay
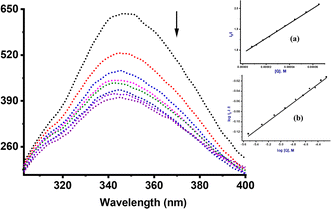
To confirm the interactions between compounds Ru1–Ru4 and CT-DNA, a fluorescent-quenching assay using EtBr dye was conducted at varying concentrations of metal complexes (Fig. S34–S37†). Interestingly, there was an approximate 18% reduction (hypochromic shift) observed at 600 nm, alluding to the limited ability of compounds Ru1–Ru4 to displace EtBr from the EtBr-CT-DNA adduct and effectively intercalate with the CT- DNA base pairs.48 The Stern–Volmer quenching constant (Ksv), bimolecular quenching rate constant (kq) and apparent binding affinity constants (Kapp) were derived from the Stern–Volmer equations eqn S3 and S4† (Table 1). Additionally, the Scatchard equation, eqn S5 was employed to calculate the association binding constant (KF) and the number of binding sites per nucleotide (n) as summarized in Table 1. The Ksv values for compounds Ru1–Ru4 ranged from 0.51 to 1.84 × 104 M−1. Noticeably, the values of Ksv are 103 to 104 fold lower than that of the classical intercalator EtBr (107 M−1),49 further hinting on the weaker interactions of compounds Ru1–Ru4 with CT-DNA in comparison EtBr. The obtained kq values, with a magnitude of 1011 M−1 are ten times higher than the upper limit (2.0 × 1010 L mol−1 s−1) observed for quenching rate constants in dynamic interactions.50 This substantial difference strongly suggests the existence of static quenching mechanism. Similarly, the magnitude of Kapp (106 M−1) is lower compared to the binding constants typically associated with the classical intercalators (107 M−1),51 further underscoring the existence of weak intercalative interaction. The KF values (101–102 M−1 magnitude) also indicate that the complexes have weak quenching efficiencies. The values of n ≈ 1, imply the presence of one binding site for every two base pairs.Hoechst 33258 displacement assay
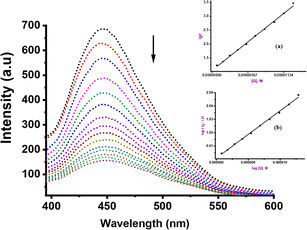
To further elucidate the precise mechanism of interaction between these complexes and CT-DNA, we performed competitive binding assay involving Hoechst 33258 dye. The characteristic emission curves of Hoechst-CT-DNA, in the presence of varying quantities of Ru(II) complexes are depicted in Fig. 3 (Ru2) and ESI Fig. S38–S41† for the other complexes. The intensity of the emissions at 450 nm decreases significantly by ca. 45% upon addition of increasing amounts of compounds Ru1–Ru4. This observation strongly confirm that the complexes participate in competitive binding with Hoechst 33258 and simultaneously interact with the minor groove of CT-DNA.52The values of the binding constants of Ksv, Kq, Kapp and KF with the Hoechst 33258 dye are presented in Table 2. These values are higher compared to those observed for EtBr displacement, signalling that compounds Ru1–Ru4 prefer a stronger and more favourable binding interaction through the minor groove of the CT-DNA rather than via an intercalative mode. The Ksv values ranged from 1.71 to 7.90 × 105 M−1 and are consistent with the affinity observed in other Ru(II) complexes known for groove binding mode.53 The magnitude of the kq value of 1012 M−1 considerably exceeds the collision quenching constant of biomolecules (2.0 × 1010 L mol−1 s−1), indicating the presence of static mode of quenching.54 We note that the Kapp values (magnitude 106–107 M−1) of compounds Ru1–Ru4 are greater than those of analogous compounds abound in literature, which fall within the range of 104–105 M−1.55 Both EtBr and Hoesch displacement studies reveal a dual mode of interactions between Ru1–Ru4 and CT-DNA, entailing partial intercalation and strong minor groove interactions.
| Compound | Hoechst 33258 displacement | |||
|---|---|---|---|---|
| Ksv 105 M−1 | Kq 1012 M−1 | Kapp 107 M−1 | KF 104 M−1 | |
| Ru1 | 7.90 ± 0.29 | 7.90 ± 0.66 | 2.27 ± 0.15 | 0.64 ± 0.07 |
| Ru2 | 1.71 ± 0.01 | 1.71 ± 0.14 | 0.44 ± 0.02 | 0.12 ± 0.01 |
| Ru3 | 9.74 ± 0.25 | 9.74 ± 0.76 | 1.44 ± 0.17 | 6.1 ± 0.55 |
| Ru4 | 1.82 ± 0.01 | 1.82 ± 0.14 | 0.51 ± 0.06 | 4.33 ± 0.39 |
Protein interactions
Serum albumin primarily functions as a carrier for a wide range of molecules, including drugs, within the bloodstream and thus ensuring the delivery of the pharmaceuticals to their intended targets within the body.56 Our choice of the bovine serum albumin (BSA) as a model protein was driven by its structural similarity, sharing approximately 76% homology to the human serum albumin (HSA).56,57 The emission as a result of the presence of tryptophan (Trp) amino acid residues, specifically Trp-212 and Trp-134 located within the subdomains IIA and 1B, respectively was observed and recorded at 350 nm.58,59 The spectra were recorded over a range spanning from 300 to 420 nm, while varying the concentration of the metal complexes (Fig. S42–S45†). A considerable reduction in the fluorescence intensity was observed (Fig. 4) and attributed to binding with Trp-134 and Trp-213 residues situated within the hydrophobic cavity.60,61The values for Ksv, kq, KF and n are presented in Table 3. The Ksv values, ranging from 0.72 to 4.01 × 104 M−1, are approximately three to four orders of magnitude lower than those observed for classical intercalators, falling in the range of 107 M−1.62 The lower Ksv values show that the interaction between the complexes and BSA is not fully controlled by diffusion. This is augmented by the higher kq values, with magnitude of 1011 M−1 s−1, which surpasses those of known associative biopolymers (2.0 × 1010 M−1 s−1), and points to the involvement of static quenching mechanism.63,64 The KF values, with a magnitude of 101–102 M are significantly lower than the association constant of 1015 M−1 for irreversible interactions. This disparity suggests that the complexes bind reversibly to BSA.65,66 The n values are close to 1, indicating that each complex has only one binding site in the BSA.
| Compound | Fluorescence titration | |||
|---|---|---|---|---|
| Ksv 104 M−1 | Kq 1011 M−1 | KF 102 M−1 | n | |
| Ru1 | 4.01 ± 0.24 | 4.01 ± 0.28 | 2.61 ± 0.19 | 0.83 |
| Ru2 | 0.72 ± 0.05 | 0.72 ± 0.15 | 0.13 ± 0.01 | 0.71 |
| Ru3 | 1.96 ± 0.13 | 1.96 ± 0.38 | 1.01 ± 0.10 | 0.93 |
| Ru4 | 1.43 ± 0.13 | 1.43 ± 0.37 | 1.42 ± 0.11 | 0.85 |
Molecular docking
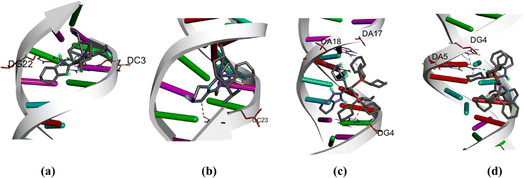
Molecular docking techniques was used to predict the preferred orientation of the molecules when bound to the receptor to form stable complexes. The approach is used to further substantiate the observed experimental binding constants, mode of interactions and location of binding in the biomolecules. Specifically, we conducted simulations of compounds Ru1–Ru4 within the DNA dodecamer and BSA protein. Distinctively, the docked poses of Ru1–Ru4 demonstrate binding to the nucleotide residues within the minor groove of DNA double helix (Fig. 5), in tandem with the experimental results. Conventional hydrogen bond interactions participate with DG22 (Ru1, 2.85 Å), and DA5 (Ru4, 3.08 Å). The stability of compound Ru1 is also contributed by the favourable-acceptor–acceptor interaction formed with DC3 (2.94 Å). Similarly, carbon hydrogen bond interactions (with DC23, 3.46 Å) stabilize Ru2 in the groove. Nucleotides DG4 (for Ru3, 5.18 Å and Ru4, 5.11 Å) and DA17 (for Ru3, 5.45 Å) are involved in pi–alkyl interactions. Additionally, Ru3 is surrounded by DA18 (1.80 Å) via unfavourable donor–donor interaction. The binding scores of −8.2 kcal mol−1 (Ru1), −8.0 kcal mol−1 (Ru2), −7.7 kcal mol−1 (Ru3), and −7.4 kcal mol−1 (Ru4), show a rough positive correlation with the spectroscopic binding constants as provided in Tables 2 and 3The interactions of compounds Ru1–Ru4 with BSA receptor are depicted in Fig. 6. The poses of the complexes are predominantly surrounded by the hydrophobic amino acid residues, characterized by hydrophobicity values ranging from −3.00 to 3.00 (Fig. S46 and S47† for Ru1 and Ru2, respectively). Alkyl interactions dominate compound Ru1 (with ILE522, 5.05 Å; VAL423, 4.98 Å; PRO420, 5.09 Å), Ru2 (ILE522, 5.02 Å; VAL423, 5.06 Å), Ru3 (CYS447, 4.90 Å; TYR451, 5.34 Å; LYS294, 4.44 Å) and Ru4 (ILE522, 4.75 Å; PRO420, 4.96 Å; LYS114, 4.73 Å active residues). Conventional hydrogen bonding interactions are observed in Ru1 (ARG458, 2.47 Å), Ru2 (GLU424, 2.13 Å; HIS145, 3.44 Å; ARG144, 2.71 Å) and Ru4 (LEU112, 2.51 Å). Furthermore, Ru1 interacts associatively with ASP108 (3.78 Å) through carbon hydrogen interaction. Moreover, Ru4 is confined in the neighbourhood of LEU112 (3.10 Å) via sulfur interactions. The docking scores of −8.0, −7.6, −6.6–6.3 kcal mol−1 for Ru1, Ru2, Ru3 and Ru4 (Fig. 5), nearly match the trend for experimental values, shown in Table 3.
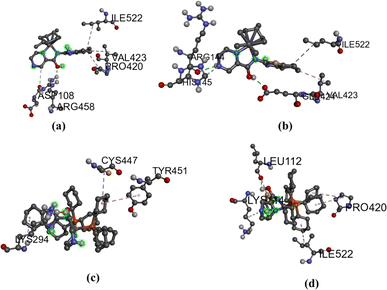 | ||
| Fig. 6 BSA interactions diagram (scaled ball and stick), (a) Ru1 (b) Ru2 (c) Ru3 and Ru4 (d), with binding scores of −8.0 (Ru1), −7.6 kcal mol−1 (Ru2), 6.6 kcal mol−1 (Ru3) and −6.3 kcal mol−1 (Ru4). | ||
In vitro cytotoxicity
The cytotoxic effects of Ru1–Ru4 complexes were assessed against six human cell lines; lung cancer (A549), prostate cancer (PC-3), colon cancer (HT-29 and Caco-2), Caco-2 (colon cancer), cervical cancer (HeLa) and normal skin fibroblast (KMST-6) cells. Cell viability was quantified by MTT assay following 24 h treatment. Since DMSO was used as the solvent for the complexes at 0.1%, it was also incorporated as a vehicle control. Percentage cell survival studies clearly indicated that Ru1–Ru4 complexes exhibited a dose-dependent cytotoxic activity (Fig. S48†). Cytotoxicity of these complexes increased proportionally as their concentration increased from 6.25 to 100 μg mL−1. The resultant IC50 values were calculated using dose–response curve fitting analysis, and the values were subsequently compared with those of cisplatin (Table 4). Significantly, Ru1 and Ru2 complexes displayed remarkable potency, surpassing the efficacy of cisplatin in certain cases, especially against A549, PC-3, HT-29, and Caco-2 cells (for Ru1) cell (Table 4). Moreover, both Ru1 and Ru2 exhibited SI > greater than 2, indicating their selectivity towards the cancer cells over the normal cells.| Compound | IC50 (μM) | Selective index (SI) | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| KMST-6 | A549 | PC-3 | HT-29 | Caco-2 | HeLa | A549 | PC-3 | HT-29 | Caco-2 | HeLa | |
| a >100 denotes that IC50 was undetermined at the test concentrations (0–100 μM). | |||||||||||
| Ru1 | 20.31 ± 2.68 | 3.29 ± 1.01 | 0.08 ± 0.03 | 7.54 ± 2.87 | 5.38 ± 0.02 | 5.47 ± 0.58 | >2 | >2 | >2 | >2 | >2 |
| Ru2 | >100 | 2.38 ± 0.20 | 6.26 ± 1.41 | 21.84 ± 3.47 | 17.16 ± 1.53 | 31.09 ± 4.81 | >2 | >2 | >2 | >2 | >2 |
| Ru3 | 6.78 ± 0.19 | >100 | >100 | >100 | >100 | >100 | 0.06 | 0.06 | 0.06 | 0.06 | 0.06 |
| Ru4 | >100 | 22.88 ± 0.15 | >100 | >100 | >100 | >100 | >2 | 1 | 1 | 1 | 1 |
| Cisplatin | 38.8 ± 13.29 | 91.8 ± 5.89 | 9.2 ± 1.73 | 17.4 ± 4.54 | 18.16 ± 0.86 | 3.76 ± 0.93 | 0.51 | >2 | >2 | >2 | >2 |
Evidently, the introduction of p-cymene appears to enhance the biological activity of the complexes. In HeLa cells for example, the p-cymene Ru1 complex showed lower percentage cell viability and IC50 of 5.47 μg mL−1 compared to the value of >100 μg mL−1 displayed by the corresponding PPh3 Ru3 complex (Table 4). These results mirrored the previous findings and has been associated with the improved stability of the arene complexes, in addition to improved lipophilic properties.66,67 In the case of Ru3 and Ru4 complexes, bearing the PPh3 co-ligands, their poor cytotoxicity could also be attributed to the steric crowding effect of the bulky PPh3 group, which ultimately limited the interaction of the complexes with the DNA and BSA protein. However, this explanation contradicts their derived spectroscopic constants, which depict higher values compared to their counterparts Ru1 and Ru2. In addition to the poor cytotoxicity of Ru3 and Ru4, their poor selectivity (SI ≤ 1) imply that these PPh3 complexes are not good candidates for cancer treatment.
Conclusions
In summary, the present stud describes in detail the synthesis and structural characterization of heterocyclic (pyrazine)carboxamide Ru(II) complexes. The spectroscopic methods indicate that the complexes are sufficiently stable in aqueous and physiological conditions. The interactions between the Ru(II) complexes (Ru1–Ru4) and CT-DNA reveal two distinct binding modes, namely minor groove binding and partial intercalation. When interacting with BSA protein, both experimental and molecular docking results show that the complexes exhibit favourable non-covalent interactions with the Trp residues within the hydrophobic cavity. The p-cymene complexes Ru1 and Ru2 demonstrated higher cytotoxic efficacy than the PPh3 counterparts Ru3 and Ru4. More significantly, the cytotoxicity and selectivity of complexes Ru1 and Ru2 against A549, PC-3, and Caco-2 cell lines surpassed that of cisplatin.Experimental section
Synthesis of Ru(II) complexes
![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O), 1542 (C
O), 1542 (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N). TOF MS-ES, m/z (%) 509.0454. Found 510.0543 (M++H). Anal calcd (%) for C22H22N5ClRuO: C, 51.39; N, 9.85; H, 3.95% found: C, 51.62; N, 9.76; H, 3.60%.
N). TOF MS-ES, m/z (%) 509.0454. Found 510.0543 (M++H). Anal calcd (%) for C22H22N5ClRuO: C, 51.39; N, 9.85; H, 3.95% found: C, 51.62; N, 9.76; H, 3.60%.![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O), 1586 (C
O), 1586 (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N). TOF MS-ES, m/z (%) 526.0245; found 527.0325 (M++H). Anal calcd (%) for C22H21N4ClRuOS: C, 50.04; N, 10.17; H, 3.86% found: C, 50.23; N, 10.65; H, 4.02%.
N). TOF MS-ES, m/z (%) 526.0245; found 527.0325 (M++H). Anal calcd (%) for C22H21N4ClRuOS: C, 50.04; N, 10.17; H, 3.86% found: C, 50.23; N, 10.65; H, 4.02%.![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O), 1566 (C
O), 1566 (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N). TOF MS-ES, m/z (%) 899.1335; found 900.1415 (M++H) anal calcd (%) for C48H38N5ClOP2Ru: C, 58.87; N, 7.01; H, 3.86; found: C, 58.71; N, 7.39; H, 4.06%.
N). TOF MS-ES, m/z (%) 899.1335; found 900.1415 (M++H) anal calcd (%) for C48H38N5ClOP2Ru: C, 58.87; N, 7.01; H, 3.86; found: C, 58.71; N, 7.39; H, 4.06%.![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O), 1591 (C
O), 1591 (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N). MS spectrum, m/z: calcd: 916.1012 found 917.0967 (M++H). Anal calcd (%) for C46H37N4ClOP2RuS: C, 57.86; N, 9.69; H, 3.98; found: C, 57.91; N, 9.41; H, 3.93%.
N). MS spectrum, m/z: calcd: 916.1012 found 917.0967 (M++H). Anal calcd (%) for C46H37N4ClOP2RuS: C, 57.86; N, 9.69; H, 3.98; found: C, 57.91; N, 9.41; H, 3.93%.Conflicts of interest
The authors have no known conflicts of interests to declare.Acknowledgements
The authors wish to acknowledge the National Research Foundation (NRF-South Africa, DAAD), for the PhD bursary to NT, DST-National Research Foundation (SA) for the CPRR research grant to SO and DSI/Mintek Nanotechnology Innovation Centre for the research grant to MM.References
- P. Chinmay, Results Chem., 2023, 6, 101149 CrossRef.
- S. Shirvalilou, Z. T. Parsaei, M. H. Parsaei, S. Sargazi, R. Sheervalilou, M. Shirvaliloo, H. Ghaznavi and S. Khoei, Wiley Interdiscip. Rev.: Nanomed., 2023, e1922 CAS.
- V. Voicu, F. M. Brehar, C. Toader, R.-A. Covache-Busuioc, A. D. Corlatescu, A. Bordeianu, H. P. Costin, B.-G. Bratu, L.-A. Glavan and A. V. Cuirea, Biomolecules, 2023, 13, 1388 CrossRef CAS PubMed.
- L. De Michieli, M. D. Gaspari, G. Sinigiani, A. Lupi, L. Vedovelli, A. Salvalaggio, M. D. Barbera, S. Rizzo, K. Pilichou and D. Cecchin, Int. J. Cardiol., 2023, 389, 131204 CrossRef PubMed.
- Y. Wang, Y. Hou, S. Wang, T. Zheng and W. Du, ChemBioChem, 2023, 24, e202300395 CrossRef CAS PubMed.
- R. O. Omondi, D. Jaganyi and S. O. Ojwach, BioMetals, 2023, 1–15 Search PubMed.
- R. O. Omondi, A. O. Fadaka, A. A. Fatokun, D. Jaganyi and S. O. Ojwach, J. Biol. Inorg. Chem., 2022, 27, 653–664 CrossRef CAS PubMed.
- S. Kumar, M. K. Shukla, A. K. Sharma, G. K. Jayaprakash, R. K. Tonk, D. K. Chellappan, S. K. Singh, K. Dua, F. Ahmed and S. Bhattacharyya, Med. Commun., 2023, 4, e253 CAS.
- J. J. Wilson and T. C Johnstone, Curr. Opin. Chem. Biol., 2023, 76, 102363 CrossRef CAS PubMed.
- Y. Zhang, B. T. Doan and G. Gasser, Chem. Rev., 2023, 123, 10135–10155 CrossRef CAS PubMed.
- R. O. Omondi, D. Jaganyi, S. O. Ojwach and A. A. Fatokun, Inorg. Chim. Acta, 2018, 482, 213–220 CrossRef CAS.
- G. Sahu, S. A. Patra, S. Lima, S. Das, H. Görls, W. Plass and R. Dinda, Chem.–Eur. J., 2023, 29, e202202694 CrossRef CAS PubMed.
- R. K. Gupta, R. Pandey, G. Sharma, R. Prasad, B. Koch, S. Srikrishna, P.-Z. Li, Q. Xu and D. S. Pandey, Inorg. Chem, 2013, 52, 3687–3698 CrossRef CAS PubMed.
- T. E. Wood and A. Thompson, Chem. Rev., 2007, 107, 1831–1861 CrossRef CAS PubMed.
- G. H. Ribeiro, A. P. Guedes, T. D. de-Oliveira, C. R. de Correia, L. Colina-Vegas, M. A. Lima, J. A. Nobrega, M. R. Cominetti, F. V. Rocha and A. G. Ferreira, Inorg. Chem., 2020, 59, 15004–15018 CrossRef CAS PubMed.
- N. Alatrash, F. H. Issa, N. S. Bawazir, S. J. West, K. E. Van Manen-Brush, C. P. Shelor, A. S. Dayoub, K. A. Myers, C. Janetopoulos and E. A. Lewis, Chem. Sci., 2020, 11, 264–275 RSC.
- S. Seyed Alinaghi, A. Karimi, H. Mojdeganlou, S. Alilou, S. P. Mirghaderi, T. Noori, A. Shamsabadi, O. Dadras, F. Vahedi and P. Mohammadi, Sci. Rep., 2022, 5, 13004–13018 Search PubMed.
- J. M. Rademaker-Lakhai, D. V.-D. Bongard, D. Pluim, J. H. Beijnen and J. H. Schellens, Clin. Cancer Res., 2004, 10, 3717–3727 CrossRef CAS PubMed.
- C. G. Hartinger, M. A. Jakupec, S. Zorbas-Seifried, M. Groessl, A. Egger, W. Berger, H. Zorbas, P. J. Dyson and B. K. Keppler, Chem. Biodiversity, 2008, 5, 2140–2155 CrossRef CAS PubMed.
- A. K. Bytzek, G. Koellensperger, B. K. Keppler and C. G. Hartinger, J. Inorg. Biochem., 2016, 160, 250–255 CrossRef CAS PubMed.
- C. Scolaro, A. Bergamo, L. Brescacin, R. Delfino, M. Cocchietto, G. Laurenczy, T. J. Geldbach, G. Sava and P. J. Dyson, J. Med. Chem., 2005, 48, 4161–4171 CrossRef CAS PubMed.
- C. A. Vock, C. Scolaro, A. D. Phillips, R. Scopelliti, G. Sava and P. J. Dyson, J. Med. Chem., 2006, 49, 5552–5561 CrossRef CAS PubMed.
- R. E. Aird, J. Cancer, 2002, 86, 1652–1657 CrossRef CAS PubMed.
- N. Alahmadi, H. S. Alhasan, H. Gomaa, A. A. Abdelwahab and M. Y. Emran, RSC Med. Chem., 2022, 183, 107909 CAS.
- M. V. Babak, D. Plażuk, S. M. Meier, H. J. Arabshahi, J. Reynisson, B. Rychlik, A. Błauż, K. Szulc, M. Hanif and S. Strobl, Chem.–Eur. J., 2015, 21, 5110–5117 CrossRef CAS PubMed.
- P. Hikisz, E. Namiecińska, P. Paneth and E. Budzisz, Molecules, 2023, 28, 3969 CrossRef CAS PubMed.
- N. K. Singh, Y. Kumar, R. P. Paitandi, R. K. Tiwari, A. Kumar and D. S. Pandey, Inorg. Chim. Acta., 2023, 545, 121241 CrossRef CAS.
- A. Alguacil, F. Scalambra, A. M. Romerosa-Nievas, A. Bento-Oliveira and F. Marques, Bioinorg. Chem. Appl., 2023, 2023, 15 Search PubMed.
- C. Chen, C. Xu, T. Li, S. Lu, F. Luo and H. Wang, Eur. J. Med. Chem., 2020, 203, 112605 CrossRef CAS PubMed.
- C. A. Annunziata, M. E. Cucciolito, M. D. Ronza, G. Ferraro, M. Hadiji, A. Merlino, D. Ortiz, R. Scopelliti, F. F. Tirani, P. J. Dyson and F. Ruffo, Organometallics, 2023, 42, 952–964 CrossRef.
- S. Nikolic, J. Arakelyan, V. Kushnarev, S. M. Alfadul, D. Stankovic and Y. I. Kraynik, Inorg. Chem., 2023, 62, 8188–8199 CrossRef CAS PubMed.
- T. S. Kamatchi, M. K. M. Subarkhan, R. Ramesh, H. Wang and J. G. Małecki, Dalton Trans., 2020, 49, 11385–11395 RSC.
- A. Arunachalam, R. Rengan, D. Umapathy and J. V. Arockiam, Organometallics, 2022, 41, 2474–2486 CrossRef CAS.
- C. Panda, A. Sarkar and S. S. Gupta, Coord. Chem. Rev., 2020, 417, 213314 CrossRef CAS.
- D. Bansal and R. Gupta, Dalton Trans., 2016, 45, 502–507 RSC.
- P. Vijayan, S. Yadav, S. Yadav and R. Gupta, Inorg. Chim. Acta, 2020, 502, 119285 CrossRef CAS.
- P. Vijayan, P. Viswanathamurthi, P. Sugumar, M. N. Ponnuswamy, M. D. Balakumaran, P. T. Kalaichelvan, K. Velmurugan, R. Nandhakumar and R. J. Butcher, Inorg. Chem. Front., 2015, 2, 620–639 RSC.
- S. Biswas, S. Das, T. Gupta, S.K. Singh, M. Pissas and G. Rajaraman, Chem.–Eur. J., 2016, 22, 18532–18550 CrossRef CAS PubMed.
- S. R. I. Cooper, CrystEngComm, 2020, 22, 7186 RSC.
- A. M. Reilly, R. I. Cooper, C. S. Adjiman, S. Bhattacharya, A. D. Boese, J. G. Brandenburg, P. J. Bygrave, R. Bylsma, J. E. Campbell and R. Car, Acta Crystallogr., Sect. B: Struct. Sci., Cryst. Eng. Mater., 2016, 72, 439–459 CrossRef CAS PubMed.
- N. Muhammad, M. Hanif and P. Yang, Coord. Chem. Rev., 2024, 499, 215507 CrossRef CAS.
- T. S. Prathima, B. Choudhry, M. G. Ahmad, K. Chanda and M. M. Balamurali, Coord. Chem. Rev., 2023, 490, 215231 CrossRef CAS.
- R. O. Omondi and S. O. Ojwach, Inorg. Chim. Acta, 2020, 512, 119883 CrossRef CAS.
- K. M. Oliveira, J. Honorato, G. R. Goncalves, M. R. Cominetti, A. A. Batista and R. S. Correa, Dalton Trans., 2020, 49, 12643–12652 RSC.
- A. Zahirović, S. Roca, E. Kahrović and A. Višnjevac, J. Mol. Struct., 2021, 1236, 130326 CrossRef.
- M. S. Alsaeedi, B. A. Babgi, M. H. Abdellattif, A. Jedidi, M. G. Humphrey and M. A. Hussien, Molecules, 2020, 26, 76 CrossRef PubMed.
- C.-V. Legna, W. Villarreal, M. Navarro, C. R. de Oliveira, A. E. Graminha, P. I. da, S. Maia, V. M. Deflon, A. G. Ferreira, M. R. Cominetti and A. A. Batista, J. Inorg. Biochem., 2015, 153, 150–161 CrossRef PubMed.
- P. O. Vardevanyan, A. P. Antonyan, M. A. Parsadanyan, H. G. Davtyan and A. T. Karapetyan, Exp. Mol. Med., 2003, 35, 527–533 CrossRef CAS PubMed.
- D. İnci, R. Aydin and Y. Zorlu, Eur. Biophys. J., 2021, 50, 771–785 CrossRef PubMed.
- J. R Lakowicz and G. Weber, Biochemistry, 1973, 12, 4161–4170 CrossRef PubMed.
- M. Cory, D. D. Mckee, J. Kagan, D. W. Henry and J. A. Allen Miller, J. Am. Chem. Soc., 1985, 107, 2528–2536 CrossRef CAS.
- S. Kumar and M. S. Nair, RSC Adv., 2021, 11, 29354–29371 RSC.
- M. Medjedović, A. R. Simovic, D. Ćoćić, M. Milutinović, L. Senft, S. Blagojević, N. Milivojević and B. Petrović, Polyhedron, 2020, 178, 114334 CrossRef.
- A. A. Adeleke, M. S. Islam, K. Olofinsan, V. F. Salau, C. Mocktar and B. Omondi, New J. Chem., 2021, 45, 17827–17846 RSC.
- S. Balou, A. Zarkadoulas, M. Koukouvitaki, L. Marchiò, E. K. Efthimiadou and C. A. Mitsopoulou, Bioinorg. Chem. Appl., 2021, 2021 Search PubMed.
- S. A. Elsayed, I. M. Elnabky, A. di Biase and A. M. El-Hendawy, Appl. Organomet. Chem., 2022, 36, e6481 CrossRef CAS.
- H. Edelhoch, Biochem, 1967, 6, 1948–1954 CrossRef CAS PubMed.
- C. Kakoulidou, C. T. Chasapis, A. G. Hatzidimitriou, K. C. Fylaktakidou and G. Psomas, Dalton Trans., 2022, 16688–16705 RSC.
- S. Gurusamy, K. Krishnaveni, M. Sankarganesh, R. N. Asha and A. Mathavan, J. Mol. Liq., 2022, 345, 117045 CrossRef CAS.
- P. Y. Wang, C. T. Yang and L. K. Chu, Chem. Phys. Lett., 2021, 781, 138998 CrossRef CAS.
- K. L. Bell and H. C. Brenner, Biochemistry, 1982, 21, 799–804 CrossRef CAS PubMed.
- M. M. Milutinović, A. Rilak, I. Bratsos, O. Klisurić, M. Vraneš, N. Gligorijević, S. Radulović and Ž. D. Bugarčić, J. Inorg. Biochem., 2017, 169, 1–12 CrossRef PubMed.
- M. Ganeshpandian, R. Loganathan, E. Suresh, A. Riyasdeen, M. A. Akbarsha and M. Palaniandavar, Dalton Trans., 2014, 43, 1203–1219 RSC.
- R. S. Correa, K. M. de Oliveira, F. G. Delolo, A. Alvarez, R. Mocelo, A. M. Plutin, M. R. Cominetti, E. E. Castellano and A. A. Batista, J. Inorg. Biochem., 2015, 150, 63–71 CrossRef CAS PubMed.
- T. Topală, A. Bodoki, L. Oprean and R. O. Radu, Clujul Med., 2014, 87, 215 Search PubMed.
- L. Fetzer, M. Ali, M. Xiangjun, J. P. Collin, C. Sirlin, C. Gaiddon and M. Pfeffere, Dalton Trans., 2011, 40, 8869–8878 RSC.
- P. Hikisz, E. Namiecińska, P. Paneth and E. Budzisz, Molecules, 2023, 28, 3969 CrossRef CAS PubMed.
Footnotes |
| † Electronic supplementary information (ESI) available: Experimental methodologies and analytical findings, which encompass NMR and FT-IR spectroscopic spectra, mass spectrometry data, X-ray crystallography data, and associated files. Additionally, it includes electronic and emission curves for DNA and protein interactions, in silico molecular docking diagrams, and various biological graphs. The crystallographic data entry for Ru2 is made available through the deposition number. CCDC 2304942. For ESI and crystallographic data in CIF or other electronic format see DOI: https://doi.org/10.1039/d4ra00525b |
| ‡ Current address: Department of Chemistry, University of Cape Town, Rondebosch, 7701, South Africa. |
| This journal is © The Royal Society of Chemistry 2024 |