Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Recent advances in microbial and enzymatic engineering for the biodegradation of micro- and nanoplastics
Jaewon Choi†
ab,
Hongbin Kim†ab,
Yu-Rim Ahnab,
Minse Kimab,
Seona Yuab,
Nanhyeon Kimab,
Su Yeon Limab,
Jeong-Ann Parkc,
Suk-Jin Haab,
Kwang Suk Lim*ab and
Hyun-Ouk Kim *ab
*ab
aDivision of Chemical Engineering and Bioengineering, College of Art, Culture and Engineering, Kangwon National University, Chuncheon, Korea. E-mail: kimhoman@kangwon.ac.kr; kslim@kangwon.ac.kr
bDepartment of Smart Health Science and Technology, Kangwon National University, Chuncheon, Korea
cDepartment of Environmental Engineering, Kangwon National University, Chuncheon 24341, Republic of Korea
First published on 25th March 2024
Abstract
This review examines the escalating issue of plastic pollution, specifically highlighting the detrimental effects on the environment and human health caused by microplastics and nanoplastics. The extensive use of synthetic polymers such as polyethylene (PE), polyethylene terephthalate (PET), and polystyrene (PS) has raised significant environmental concerns because of their long-lasting and non-degradable characteristics. This review delves into the role of enzymatic and microbial strategies in breaking down these polymers, showcasing recent advancements in the field. The intricacies of enzymatic degradation are thoroughly examined, including the effectiveness of enzymes such as PETase and MHETase, as well as the contribution of microbial pathways in breaking down resilient polymers into more benign substances. The paper also discusses the impact of chemical composition on plastic degradation kinetics and emphasizes the need for an approach to managing the environmental impact of synthetic polymers. The review highlights the significance of comprehending the physical characteristics and long-term impacts of micro- and nanoplastics in different ecosystems. Furthermore, it points out the environmental and health consequences of these contaminants, such as their ability to cause cancer and interfere with the endocrine system. The paper emphasizes the need for advanced analytical methods and effective strategies for enzymatic degradation, as well as continued research and development in this area. This review highlights the crucial role of enzymatic and microbial strategies in addressing plastic pollution and proposes methods to create effective and environmentally friendly solutions.
Introduction
Plastic, first introduced in the mid-20th century, has been essential to contemporary civilization due to its convenience and cost-effectiveness. Nevertheless, its extensive use has resulted in notable difficulties in waste management.1 Projections indicate that the worldwide output of plastic might exceed 8.3 billion tons, possibly leading to a concerning 12 billion tons of garbage by the year 2050.2 The buildup of waste in landfills and natural ecosystems poses significant environmental issues. The prevailing pattern highlights the need for sustainable solutions, given that conventional approaches such as recycling and burning have shown their insufficiency, resulting in a substantial buildup of plastic trash in landfills.3 The magnitude of plastic manufacturing, coupled with inadequate waste disposal methods, has resulted in the pervasive presence of plastic trash and subsequent environmental pollution caused by microplastics, which are formed as a consequence of the degradation of larger plastic objects.3,4 These present substantial hazards to ecosystems, human well-being, and safety.5,6 The increasing recognition of this ecological problem emphasizes the pressing need for effective degradation remedies.The widespread manufacturing and disposal of plastics have resulted in substantial contamination in land, water, and air environments, with microplastics being especially abundant. Inadequate waste management on land is a significant cause of marine plastic pollution, leading to an estimated 5.25 trillion microplastics and nanoplastics entering seas, soil, and air.7 The presence of these minuscule particles is worrisome because of their detrimental impact on soil fertility and the well-being of marine organisms. Microplastics and nanoplastics have a profound negative impact on ecosystems and the health of species. They disrupt natural processes, damage animals, and accumulate toxic compounds in their organs.8 The presence of microplastics has the potential to affect the capacity of soil to allow water to pass through, its density, and the movement of nutrients, which raises worries about the consequences of their presence.9,10 The entrance of micro- and nanoplastics into oceans is influenced by several processes, including UV photodegradation, mechanical forces, hydrolysis, and biological degradation.11,12 These minuscule particles disperse extensively across the ocean environment and have a notable influence on marine life and ecosystems. Due to their diminutive size, they can effortlessly infiltrate organisms and accumulate inside their organs, resulting in the accumulation of detrimental compounds that cause significant hazards to marine life.13
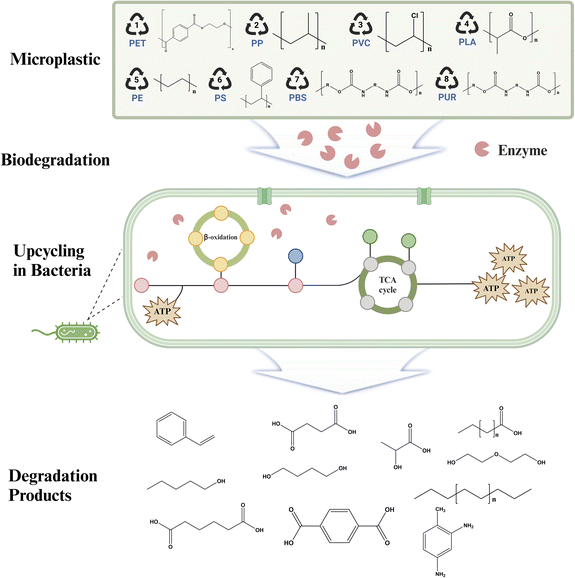
The widespread use of artificial polymers, which are crucial in contemporary society, has reached concerning levels and poses substantial ecological and physiological hazards in terrestrial, aquatic, and marine ecosystems. Due to their small size and large surface area, microplastics are very susceptible to absorbing harmful compounds including heavy metals, medicines, and flame retardants.14 Additionally, they are more easily consumed by living creatures. These activities result in a range of negative consequences for plants and animals, such as alterations in soil composition, increased growth of microorganisms, inflammation in fish, compromised immune systems, reproductive problems, and malfunction in organs.15–19 The heightened vulnerability of humans makes them particularly susceptible to the substantial risk posed by microplastic pollution. The pollutants have the ability to initiate inflammatory and neurotoxic effects by activating certain protein kinase pathways. The effects of microplastics among various creatures differ depending on their ability to withstand environmental stress and the ecological circumstances in which they live.20,21 The extensive use of these polymers and their inclination to amass contaminants emphasize the need for devising efficient degrading techniques. The aim of these techniques is to disassemble microplastics and reduce their presence in the environment and alleviate the harm they cause (Scheme 1).
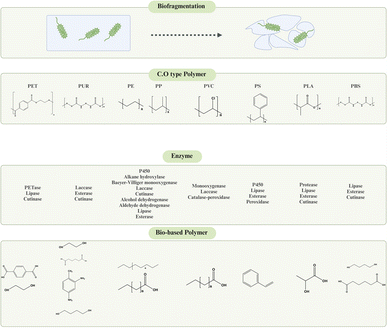
Plastics including polyethylene (PE), polyethylene terephthalate (PET), polyurethane (PU), polystyrene (PS), polypropylene (PP), and polyvinyl chloride (PVC) have notable environmental difficulties since they degrade slowly in nature.22 The degradation pathways of these polymers may be classified according to their chemical composition: those with a carbon–carbon backbone and those with heteroatoms in the main chain.23,24 The focus of this review is on the methodologies used by microorganisms to degrade synthetic polymers and the specific roles of various enzymes in this biological process. These technologies provide effective methods for transforming plastic trash into carbon atoms, carbon dioxide, and valuable chemicals. The use of enzyme technology is crucial in promoting environmental conservation by converting plastic waste into less detrimental compounds.25 This review will explore the most recent developments in enzymatic degradation techniques and assess their potential in reducing the environmental and health consequences of plastic pollution, with a specific emphasis on microplastics and nanoplastics.
The longevity and obstacles of synthetic polymers in the environment
Enzymatic degradation challenges and progress in common synthetic polymers
The extensive use of synthetic polymers such as PET, PE, PS, PP, and PVC has played a substantial role in the global surge of plastic production, which reached a staggering 335 million tons in 2016.26 These materials, which are used in numerous industries, present significant environmental obstacles because of their long-lasting and non-degradable characteristics.4 The increasing use of these polymers in our everyday lives calls for a careful evaluation of their environmental consequences, specifically in terms of energy usage and waste production.27 This situation has prompted a greater focus on the development of sustainable alternatives and recycling methods. The pressing concern lies in finding innovative solutions that can mitigate the adverse effects of plastic proliferation, while maintaining a balance between the utility of these polymers and their environmental footprint.The conversation surrounding the use of synthetic polymers revolves around their essential role in contemporary society and the resulting environmental hurdles. There are ongoing efforts to investigate environmentally friendly materials and improve recycling methods, but these endeavors encounter various obstacles, such as technological constraints and economic viability.28 The management of synthetic polymers' impact on the environment is a multifaceted challenge that necessitates a comprehensive approach.
Polyethylene (PE)
Dealing with polyethylene poses a serious challenge for enzymes when it comes to breaking down microplastics. This material is often used in packaging and containers because of its outstanding ability to resist microbial degradation. PE, especially HDPE, possesses a linear structure that enhances its resistance to enzymatic degradation. Exciting advancements in biotechnology have revealed the incredible capabilities of specific enzymes and microbial strains in effectively binding to and breaking down PE.29–31 Usually, these enzymes work by oxidizing the polymer chains, initiating a process of breaking them apart.32 Several environmental factors, such as exposure to UV light or physical abrasion, can contribute to this process.33 These factors enhance the surface area to promote enzymatic activity.Polyethylene terephthalate (PETE/PET)
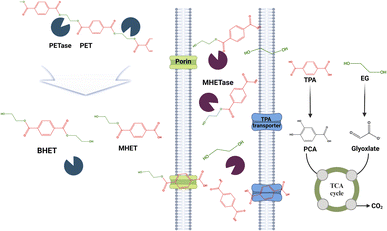
Extensive research has been conducted on the enzymatic degradation of PET, which is commonly found in beverage bottles and textiles. Researchers have made notable discoveries in the area of enzyme research and revealed the impressive capabilities of enzymes such as PETase and MHETase in efficiently breaking down PET into its fundamental components, ethylene glycol and terephthalic acid.34–36 These enzymes have been found in certain bacteria and fungi and their efficiency and longevity are being enhanced for industrial use.37–39 The potential of enzymatic degradation of PET for recycling operations is immense and offers a more environmentally friendly and efficient option compared to traditional mechanical and chemical recycling methods (Fig. 1).Polypropylene (PP)
PP is widely used in packaging and automotive components due to its impressive resistance to a range of chemical degradation processes.40,41 However, a recent study has uncovered the potential of enzymatic techniques in degrading PP. Certain microbial strains have been acknowledged for their remarkable capacity to adhere to and partially degrade PP.42,43 The challenge is to enhance the efficiency of these biological processes to make them more practical for widespread use. A study is being conducted to explore and improve enzymes that have the potential to enhance the efficiency of breaking down the chemical bonds in PP.44Polyvinyl chloride (PVC)
PVC, widely used in the construction and packaging industries, poses challenges for biodegradation because of its chlorine content.45 There is a lack of comprehensive scientific knowledge regarding the enzymatic degradation of PVC, especially when compared to other polymers. There has been a strong focus on identifying and managing microbial strains that can tolerate and break down the chlorine-based compounds commonly found in PVC.46,47 The study in this area is still in its initial phases with the goal of developing dependable and effective methods for the secure breakdown of PVC.Polystyrene (PS)
PS, a widely used material in packaging and insulation, is well-known for its remarkable durability due to its chemically stable aromatic structure.48,49 However, recent discoveries have revealed the presence of certain bacteria and fungi that can degrade PS using specialized enzymes.50–52 The enzymes effectively break down the styrene monomers, and the current pace and efficiency of the process meets expectations.53 The present study aims to enhance the efficiency and longevity of these enzymes to make the biodegradation of PS a more practical option.Polylactic acid (PLA)
PLA, a bioplastic derived from renewable sources, is inherently more prone to enzymatic degradation when compared to polymers made from petroleum.54 Research has shown that certain enzymes, such as proteinase K and lipase, have proven to be extremely efficient in the degradation of PLA.55–57 The degradation of PLA happens through the hydrolysis of ester bonds, leading to the formation of lactic acid, which can be subsequently metabolized by various microorganisms.58 PLA stands out for its positive environmental impact since it can be easily and completely broken down in composting facilities.Polybutylene succinate (PBS)
It has been observed that PBS, a type of biodegradable plastic, is more susceptible to enzymatic degradation compared to conventional polymers.2,59 There have been significant findings in the field of enzyme research, particularly in the area of PBS degradation.60,61 Notably, lipases have demonstrated remarkable efficiency in breaking down PBS. The degradation process involves the hydrolysis of ester bonds in the polymer, causing it to break down into smaller particles that can be easily broken down by biological processes.62 Because of its remarkable vulnerability to degradation, PBS is an extremely appealing material for endeavors that prioritize ecological sustainability.Polyurethane (PUR)
Enzymatic degradation of PUR can be quite complex due to its various chemical compositions.63,64 However, certain enzymes such as esterase and ureases show great potential in breaking down PUR.28,65,66 The enzymes have the ability to specifically target certain bonds in the PUR polymer, resulting in its degradation.28 Researchers in this field are focused on uncovering and enhancing the efficiency of these enzymes with the goal of developing effective techniques for breaking down PURs. The significance of this issue stems from the widespread use and persistent presence of PURs in the environment.Influence of chemical composition on plastic degradation dynamics
The dynamics of plastic degradation are closely connected to various internal and external factors, particularly in relation to enzymatic activity on microplastics.67–69 These factors encompass the intrinsic characteristics of the plastic, such as its molecular structure, composition, physical form, and the inclusion of additives.23,70,71 The rate and mechanism of biodegradation are greatly influenced by these characteristics. Note that the susceptibility of a plastic to enzymatic attack can be significantly affected by the presence of specific functional groups or the degree of polymer branching.Factors such as pH, temperature, oxygen levels, and light exposure are critical in influencing the effectiveness of enzymatic degradation. Certain enzymes exhibit improved performance or durability in particular environmental conditions, which can impact the rate of degradation.72–75 Environmental factors can also weaken the structure of plastics, making them more susceptible to enzymatic breakdown.
The fundamental chemical composition of plastics, specifically the types of bonds they possess, plays a crucial role in determining their degradability. Plastics with carbon–carbon (C–C) backbones, such as polyethylene and polypropylene, have a remarkable ability to resist microbial and enzymatic decomposition, which significantly slows down the degradation process.76 On the other hand, polymers that incorporate heteroatoms into their main chain, such as polyesters, are highly prone to enzymatic hydrolysis.77
The interaction between degrading enzymes and a plastic is greatly influenced by the chemical composition, which determines the surface hydrophobicity. Surfaces that repel water can also cause hydrophilic enzymes to be repelled, which presents initial obstacles in the biodegradation process. Effective enzymatic binding and action often require the formation of a biofilm.78,79
Given the origin of most modern plastics from petrochemical sources, the widespread existence of non-biodegradable plastics presents a notable environmental concern.13 To tackle this problem, it is important to keep working on improving our understanding and finding ways to enhance the degradation of microplastics by enzymes. Adapting enzymatic methods to effectively tackle the distinct obstacles posed by various plastic polymers is needed for advancing sustainable waste management and environmental conservation strategies.
Examining the enduring environmental effects of microplastics and nanoplastics
It is essential to have a comprehensive understanding of the physical characteristics and long-term effects of microplastics (MPs) and nanoplastics (NPs) in order to effectively tackle the ongoing issue of these pollutants in various ecosystems. The extensive variety of synthetic polymers used in the composition of MPs results in a wide range of physical forms, including foam, pellets, flakes, fibers, and films. It is important to note that a significant portion of these particles, approximately 60%, are smaller than 1 mm in size, primarily appearing as flakes and fibers.80–84 Nanoplastics possess distinct characteristics in comparison to MPs as a result of their reduced dimensions. The difference in size has a notable impact on their movement, behavior in the environment, interactions, availability to organisms, and potential harm.85–88Both industrial and residential activities contribute to the release of these pollutants, leading to the contamination of aquatic systems, air, and soil.89–91 The widespread pollution poses considerable threats to multiple aspects of the ecosystem, such as the food chain, plant life, marine organisms, and human populations.92,93 This pollution greatly affects both humans and the environment, as they both experience the consequences of it. The structural composition of polymers plays a key role in determining their environmental behavior and has a combination of ordered and disordered regions that have a significant impact.23 The current situation underscores the need for effective methods to address these persistent pollutants.
Exploring the ecological and health implications of microplastic and nanoplastic pollution
The presence of MPs and NPs in different ecosystems, whether due to natural processes such as erosion or human activities such as industrial emissions, results in a wide range of harmful consequences.94–96 These particles have been associated with serious health risks, such as the potential to cause cancer and interfere with endocrine systems.97,98 Their environmental impact is exacerbated by their capacity to attract and transport other detrimental substances, including persistent organic pollutants and heavy metals.99,100 In addition, MPs and NPs can carry harmful bacteria, including antibiotic-resistant strains, which poses a considerable threat to both wildlife and human communities.The management and regulation of MPs and NPs require advanced analytical and quantitative methods, particularly when addressing complex environmental matrices. The challenge is made more difficult by the complexity of recycling these particles.101 A deep understanding of the physical and chemical properties of MPs and NPs is needed to develop effective strategies for their enzymatic degradation.
In order to effectively tackle the negative impacts of MPs and NPs, it is important to acknowledge their wide-ranging implications and establish dependable approaches for their identification, assessment, and breakdown. This field of study not only provides potential solutions for addressing the environmental and health risks linked to microplastic and nanoplastic pollution, but also emphasizes the importance of ongoing efforts to comprehend and combat this widespread problem.
The role of microbes in plastic degradation
The crucial role of microorganisms in plastic degradation
As we strive to find sustainable and environmentally friendly solutions to the problem of plastic pollution, the importance of microorganisms in breaking down microplastics (MPs) and nanoplastics (NPs) has become a significant focus.102–106 This area of study investigates the capacity of different microorganisms and insects to transform plastic into eco-friendly substances.92,93 Several microorganisms, such as bacteria, fungi, and actinomycetes, have been recognized for their enzymatic capability to degrade the intricate chemical bonds of microplastics into simpler compounds.107–109These microorganisms, found in various habitats such as soil, aquatic environments, and even air, have an important role in the natural breakdown of organic matter. An excellent illustration is the breakdown of PE, a commonly used plastic, by a range of microorganisms.29,31 One of the most fascinating findings in this field involves the gut bacteria found in the larvae of the large waxworm, Galleria mellonella.110,111 These bacteria have demonstrated impressive effectiveness in breaking down PE through a process known as hydrolysis.112
This research on the interaction between microbes and plastic has uncovered exciting possibilities for addressing plastic waste. It has the potential to transform discarded petroleum-based polymers into reusable materials or feedstock for biomass production, and offers new ways to manage plastic waste effectively.24,113 The success of these groundbreaking solutions depends on a comprehensive grasp of the distinct microbial enzymes implicated in the degradation process. A deep understanding of this subject is crucial to create more efficient and sustainable approaches to tackle the growing issue of plastic pollution and advance the field of enzyme-based microplastic degradation.
Dynamics of the microbial degradation of plastics
The breakdown of microplastics (MPs) by microorganisms is a sophisticated process driven by enzymes. Many factors can influence the process, including the molecular weight and chemical composition of the microplastic, environmental conditions, the specific microbial species, and the physical properties of the plastic, such as crystallinity and the presence of functional groups or additives.8,9 The degradation process usually occurs in a sequence of stages beginning with the development of a biofilm on the surface of the plastic. This is then followed by biodegradation, biotransformation, and finally mineralization.35,114 Irrespective of the natural degradability of the polymer, the first step involves hydrolysis, which breaks down microplastics into smaller molecular fragments. This step is crucial and relies on the microorganisms at hand (Fig. 2).Microorganisms play a key role in the intricate biochemical reactions that occur during the aerobic biodegradation of plastics. Microorganisms use oxidizing enzymes to generate carbonyl groups, which are subsequently oxidized into carboxylic acids.115 This results in the hydrolysis of the polymer chain, which facilitates degradation. The microorganisms metabolize the small hydrocarbon fragments produced in a highly efficient manner.116 The last step involves converting the hydrolysis products into microbial biomass, resulting in the release of water and carbon dioxide.93,117 The enzymes involved in this degradation process can be classified into two main categories: those that alter the surface of microplastics to enhance their solubility in water, and those that break down the plastic into smaller components for microbial metabolism.118
An understanding of the intricate processes involved in the breakdown of microplastics by microorganisms is needed to devise successful approaches to minimize the negative effects of microplastic pollution on the environment. It is also crucial for making progress in enzyme-based techniques for microplastic degradation and gaining valuable insights into potential solutions for this urgent environmental issue (Table 1).
| Microorganism | Enzyme | Temperature range (°C) | Biological effects (results) | Ref. |
|---|---|---|---|---|
| Polyethylene terephthalate | ||||
| Ideonella sakaiensis 201-F6 | PETase | 20∼45 | Almost completely degraded after 6 weeks | 34 |
| Thermobifida fusca DSM43793 | TfH | 30–60 | 50% degraded after 3 weeks | 119 |
| Fusarium solani pisi | FsC | 30–60 | 97% weight loss after 96 hours | 120 |
| Thermobifida cellulosilytica DSM44535 | Thc_Cut1 | 50 | Increase of reactive hydroxyl or carboxyl groups | 121 |
| Saccharomonospora viridis AHK190 | Cut190 S226P/R228S | 60–65 | 27% weight loss after 3 days | 122 |
| Bacillus subtilis 4P3-11 | BsEstB | 40–45 | Introduction of novel carboxyl and hydroxyl groups | 123 |
| Thermomonospora curvata DSM 43183 | Tcur0390 | 50 | Stronger substrate affinity and increase of the H–S distance | 124 |
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) |
||||
| Polypropylene | ||||
| Pseudomonas aeruginosa WGH-6 | AH alkane hydroxylase | 30 | 17.2% weight loss after 40 days | 44 |
| Aneurinibacillus spp. | Lipase | 50 | 44.2% weight loss after 140 days | 125 |
| Brevibacillus spp. | ||||
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) |
||||
| Polyethylene | ||||
| Microbacterium paraoxydans | Lac | Room temperature | 61% weight loss after 2 months | 126 |
| Alternaria alternata FB1 | 153 potential enzymes | 30 | 62.79% decreased after 28 days | 127 |
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) |
||||
| Polyvinyl chloride | ||||
| Klebsiella sp. EMBL-1 | Catalase-peroxidase | 30 | 19.57% weight loss after 3 months | 46 |
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) |
||||
| Polyurethane | ||||
| Rhodococcus equi TB-60 | Urethane hydrolase | 30 | 70% degradation after 10 days | 128 |
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) |
||||
| Polystyrene | ||||
| Pseudomonas aeruginosa DSM 50071 | SGT/SH | 25 | WCA decreased from 91.56° to 79.8° after 2 months | 129 |
| Bacillus paralicheniformis G1 | Alkane monooxygenase/cytochrome P450 | 30 | 34% weight loss after 2 months | 53 |
Determinants of microbial efficacy in plastic degradation
The degradation process of microplastics is influenced by a variety of factors including microbial growth kinetics, the properties of microplastics themselves, and the environmental conditions at hand. The structural properties of microplastics, their material composition, shape, and the presence of additives play a main role in determining their vulnerability to biodegradation.External environmental factors, including pH levels, temperature, oxygen availability, exposure to light, and the presence of other substances, are crucial in influencing the process. The interaction between temperature and pH is key since pH levels have a significant impact on the electrostatic interactions between the microplastic surface and microorganisms and chemicals in soil or water.130 Degradation can be slowed down under certain conditions, while enzyme activity may be affected in different environments. For the most effective biological degradation of microplastics, it is important to maintain optimal conditions such as low pH levels and low temperatures. The rate of degradation can be influenced by a range of chemical and physical properties of microplastics, including density, molecular weight, degree of crystallinity, and the presence of specific functional groups or substituents.131 Microplastics with carbon–carbon (C–C) bonds provide enhanced durability against microbial attack, whereas microplastics with ester bonds are more prone to the effects of hydrolytic enzymes.116
The inclusion of plasticizers or other additives can greatly affect the biodegradability of microplastics. These additives have the potential to either enhance or impede microbial colonization depending on their unique characteristics and the composition of the microbial community they are targeting.116 The presence of external substances on the surface of microplastics can slow microbial degradation. On the other hand, nutritional supplements that are high in carbon and nitrogen can enhance the growth of microorganisms on microplastic surfaces, thereby speeding up the degradation process.131 These different factors are important in the development of enzyme-based approaches for efficient microplastic degradation (Fig. 3).
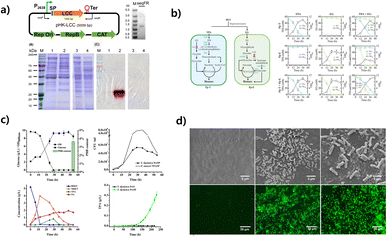 | ||
| Fig. 3 Microplastic decomposition using microbial engineering. (a) Construction of LCC-expressing plasmid pHK-LCC and extracellular expression of active LCC in C. thermocellum. Reproduced with permission from ref. 36. Copyright (2020) John Wiley & Sons, Inc. (b) Schematic of the designer T–E consortium composed of two strains Pp-T and Pp-E. Pp-T specializes in TPA degradation, which was developed by deleting the ped operon and constitutively expressing the genes tpaAa, tpaAb, tpaB, tpaC, and tpaK. Reproduced with permission from ref. 132. Copyright (2023) Springer Nature. (c) Co-cultivation of Y. lipolytica Po1fP and P. stutzeri TPA3P. (A) OD, glucose consumption, and PHB content. (B) BHET hydrolysis curve. Reproduced with permission from ref. 133. Copyright (2021) Elsevier. (d) Biofilm formation and viability. Morphotypes of the cells in the mature biofilm on the PE sheet. Fluorescent microscopic images of biofilms, which show cell viability after the 28 day incubation. Reproduced with permission from ref. 110. Copyright (2014) American Chemical Society. | ||
Evaluating the effectiveness and challenges of biological plastic degradation
Progress in the degradation of plastics by microorganisms, particularly with enzyme-based methods, has been impressive. Biocatalysts have demonstrated significant potential in breaking down microplastics (MP) into smaller particles such as nanoplastics (NP). There is increasing interest in these biocatalysts due to their ability to greatly decrease the size of plastic particles. Nevertheless, the efficiency of microorganisms in degrading plastics is influenced by the various characteristics of the materials and is contingent upon the size of the plastic particles, ranging from larger flakes to minuscule dimensions.134Several biodegradation methods are currently under investigation, such as pure bacterial cultures, fungal cultures, bacterial consortia, and the use of specific enzymes.135 Microorganisms such as bacteria, fungi, and algae have shown the ability to break down complex plastic polymer chains into smaller units, or monomers.136,137 This ability is derived from the production of certain enzymes or metabolites that aid in the degradation process. One of the difficulties of microbial degradation is the considerable amount of time needed to break down contaminants. Even with the demonstrated efficacy of bacteria, the process of breaking down microplastics can be time-consuming in certain instances and often takes several months. Efforts are currently being made to enhance environmental conditions and optimize microbial strains to accelerate the degradation process and improve the efficiency of bacterial action on MPs and NPs. In light of the difficulties posed by degradation rates, there are continuous endeavors to investigate the thorough biodegradation of microplastics and nanoplastics.
Approaches for plastic degradation using enzymes
The crucial role of microorganisms in plastic degradation
The issue of plastic pollution has become more pressing, and the development of creative and environmentally friendly solutions, particularly in the area of plastic degradation, is needed. Enzymatic methods for plastic degradation have become increasingly recognized as very effective and eco-friendly alternatives to traditional plastic treatment methods. This strategy, grounded in a commitment to environmental sustainability, offers a way to transform plastics into reusable monomers or create valuable bioproducts by converting them into carbon dioxide, water, and new biomass.138,139Microorganisms are crucial in this process since they produce specialized enzymes that can effectively break down polymers and support the metabolism of the resulting hydrolyzates. The enzymatic biodegradation of polymers usually involves important reactions such as hydrolysis and oxidation, which break down polymer chains into smaller oligomers and monomers.140,141 These reactions are essential for the breakdown of various polymer bonds, such as ester, carbonate, amide, and glycoside bonds, resulting in the creation of monomers. In terms of their susceptibility to enzymatic degradation, petroleum-based polymers can be categorized into two types: hydrolyzable and non-hydrolyzable. Examples of hydrolyzable polymers include polyethylene terephthalate (PET) and polyurethane (PUR), while non-hydrolyzable polymers include polyethylene (PE), polystyrene (PS), polypropylene (PP), and polyvinyl chloride (PVC).141,142 Enzymes from the hydrolytic enzyme group, including esterases, lipases, depolymerizers, and fetases, have a high proficiency in breaking down the carbon structure of plastics. They focus on the ester bonds and carbonyl carbon atoms that are created when the oxidation process occurs, transforming the polymer into individual monomers.143
Nevertheless, the degradation of non-hydrolyzable polymers such as PE, PS, PP, and PVC poses a considerable obstacle. The chemical degradation of the carbon backbone is a complex area that still requires further research. Ongoing progress in enzymatic degradation is needed to preserve the environment and improve recycling techniques.
Enzyme specificity in plastic polymer degradation
In the area of enzyme-mediated microplastic degradation, past research has revealed the involvement of numerous microorganisms and enzymes in the breakdown of polyethylene (PE). These organisms, such as actinomycetes, bacteria, and fungi, play distinct roles in the degradation of PE.144 Prominent enzymes have been identified as having a crucial role in the process, including manganese peroxidase from fungi such as Phanerochaete chrysosporium and Trametes versicolor, and alkane hydrolase from Pseudomonas species (Fig. 4).145–147The degradation mechanisms for various forms of PE, including LDPE and HDPE, are currently being studied. Enzymes like laccase and alkane hydrolases have demonstrated potential in initiating PE degradation. Laccase, in particular, aids in oxidizing the PE surface and creating carbonyl regions that are more prone to subsequent enzymatic activity. Alkane hydrolases have shown great efficacy in breaking down heat-treated PE.135,148–151 Enzymes involved in PE degradation have been isolated from a wide range of microbial sources, including Proteobacteria, Firmicutes, and Actinobacteria. These microorganisms have shown promise in aiding the breakdown of microplastics. However, there is still much to learn about the exact process by which these enzymes degrade microplastics, and further research is needed.152 Furthermore, there have been notable advancements in the field of microbial enzymes that possess the ability to break down lignin polymers containing oxidative C–C bonds. Notable examples include manganese peroxidase, lignin peroxidase, and laccase. The understanding of these enzymes in the area of polyethylene biodegradation is continuously developing, and additional research is necessary to gain a complete grasp of their functions and mechanisms. It is important that future studies focus on gaining a deeper understanding and characterization of the different enzymes involved in polyethylene degradation.153,154
Focused enzymatic activity targeting polyethylene terephthalate
The field of enzymatic microplastic degradation has made significant progress, especially in targeting polyethylene terephthalate (PET). This is based on extensive research on Thermobifida fusca hydrolase, which has led to the discovery of various enzymes that can break down PET. Out of all the enzymes, the PETase enzyme from Ideonella sakaiensis 201-F6 is particularly142 noteworthy for its impressive capacity to effectively degrade PET into various intermediates such as BHET, MHET, and TPA. MHETase, an additional enzyme, carries out further processing of MHET to produce terephthalic acid and ethylene glycol.34,155Recent advancements have resulted in the creation of stronger versions of PETase, which have improved stability and efficiency in breaking down materials. In addition, a 25 kDa suberinase from Streptomyces scabies has demonstrated considerable potential in the degradation of PET.156,157 Thorough analyses of the structure and mechanisms have uncovered distinct interactions between PETase, cutinase, and PET, which occur through an induced fit mechanism. PETase is capable of breaking down a wide range of polycyclic aromatic microplastics due to its impressive versatility.158,159
Efforts to enhance the efficiency of PETase have involved various techniques such as mutagenesis, overexpression, and the use of microalgae for transformation. These advancements are crucial in tackling the environmental issues caused by PET pollution. This research is a significant advancement in developing enzymatic methods to effectively break down PET microplastics and makes a valuable contribution to environmental remediation endeavors.160,161
Enzymatic strategies for polystyrene degradation
Important advancements have been made in the field of enzymatic microplastic degradation, with a particular focus on polystyrene (PS). Progress in this field is evident from the fact that a wide range of microorganisms, such as bacteria and fungi, have the ability to break down PS. Nevertheless, the precise enzymes responsible for initiating this degradation process remain to be fully understood.135,162Prior research has emphasized the significance of extracellular esterases from Lentinus tigrinus in the breakdown of PS. It has also mentioned that polymerases from Bacillus and Pseudomonas species play a role in the breakdown of PS.163,164 In addition, there have been reports on enzymes that are involved in breaking down styrene, such as styrene monooxygenase, styrene oxide isomerase, phenylacetaldehydrogenase, and phenylacetyl coenzyme A ligase.165–169 These enzymes are essential for the conversion of PS polymers through a series of reactions. It starts with the transformation of monomers into styrene, followed by oxidation to phenylacetate, and ultimately the integration of phenylacetate into the Krebs cycle.170
An in-depth knowledge and thorough analysis of these enzymes is essential to enhance the enzymatic breakdown of PS microplastics. This research explores new possibilities for tackling the environmental issues caused by PS waste and offers creative and eco-friendly solutions for managing PS waste. Efforts to identify and optimize these enzymes are needed to develop effective strategies to address the impacts of PS pollution.
Advancing enzyme engineering for plastic degradation
Another important development involves enhancing the heat tolerance of enzymes that target plastics. This is particularly important for plastics with high glass transition temperatures (Tg), as their crystallinity decreases as the temperature rises.173 This can make it easier for enzymes to access the plastics and speed up degradation. Plastics are easier to process by enzymes when they become more flexible and mobile at temperatures near or above their Tg.173 Nevertheless, a serious challenge arises when it comes to naturally occurring enzymes such as PETase since they tend to lose their efficiency when exposed to high temperatures, which restricts their ability to maintain thermal stability.174 To tackle this problem, previous studies have primarily concentrated on developing different versions of PETase and similar enzymes that possess the ability to endure elevated temperatures, thus aligning with the Tg of various types of plastics. This requires a thorough examination of the distinctive characteristics of thermophilic proteins and using this knowledge to enhance the ability of plastic-degrading enzymes to withstand high temperatures. The ultimate objective is to maximize the efficiency of these enzymes, particularly in environments that closely align with the Tg of the specific plastic being targeted.13,122
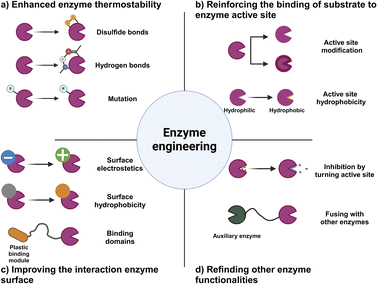
With the development of enhanced enzymes, we can create enzymes with greater power and efficiency that enables them to break down a wider variety of plastics. Enhancement of the thermal stability and pH tolerance of these enzymes has the potential to significantly improve their practicality in environmental cleanup and recycling processes. Continual endeavors in enzyme engineering focus on surpassing the constraints of natural enzymes, while also customizing enzymes to fulfil precise needs for plastic degradation.175 Our work involves enhancing enzymes to target specific types of plastics, enhancing their selectivity, and maximizing their catalytic efficiency. The improvement of the stability and activity of enzymes is crucial for the progress of enzyme-based microplastic degradation.
Enhancing enzymatic thermal stability through the use of disulfide bonds for microplastic degradation
The enhancement of enzyme thermal stability has become a key method in enzymatic microplastic degradation, particularly in the area of PET hydrolysis. One effective approach involves incorporating disulfide bonds and salt bridges. This process requires meticulous adjustment of the protein structure at specific locations or in its overall arrangement. As an illustration, the replacement of amino acid residues in metal-binding regions with disulfide bonds has been demonstrated to greatly improve the ability of enzymes to withstand high temperatures.176The significance of disulfide bridges is especially notable in PET hydrolases, which possess numerous sites for binding divalent metals, as evidenced by the crystal structure of the Cut190 enzyme. It has been noted that the inclusion of divalent ions such as calcium (Ca2+) or magnesium (Mg2+) has the dual effect of enhancing the thermal stability of the enzyme and optimizing its temperature range for operation. Methods such as circular dichroism (CD) have shown that the melting temperature of the enzyme is significantly raised with the addition of calcium ions. Further insights from molecular dynamics (MD) simulations and X-ray structural analysis have uncovered the significance of Ca2+ binding in triggering essential conformational alterations in the enzyme, which results in enhanced catalytic efficiency.122 The presence of intramolecular disulfide bridges, specifically DS1 and DS2, plays an important role in maintaining the functional integrity of the catalytic triple bond in enzymes such as PETase. One aspect that enhances the flexibility of the loop is DS1, which leads to a boost in enzyme activity. On the other hand, DS2 is crucial in preserving structural stability. Focused enhancement of these sites has been demonstrated to greatly improve the capacity of the enzyme to break down PET.177,178 Investigation of the crystal structures of different enzymes, such as LCC, Tf cutinase, and IsPETase, has opened up possibilities for developing innovative approaches to enhance their resistance to heat. As an illustration, enzyme activity has been enhanced by replacing the divalent metal binding site of LCC with a disulfide bridge and introducing targeted mutations.179 Currently, there are ongoing efforts to enhance the thermal stability of enzymes that play a role in breaking down plastics, specifically PET. This task requires the generation of enzyme mutants using directed evolution techniques, as well as the identification of optimal sites for calcium binding and disulfide bridge formation.
The strategic use of disulfide bonds is demonstrating the enhancement of the thermal stability of enzymes employed in the degradation of microplastics. These advancements are crucial for the development of stronger and more effective enzymatic solutions to tackle the environmental issues caused by plastic pollution.
Advancements in enzymatic stability through hydrogen bonding and electrostatic interactions
Significant progress has been achieved in enhancing the stability of protein structures with a dedicated emphasis on hydrogen bonding and hydrophobic interactions. The advancements in the strategic use of proline and its surrounding residues are very noteworthy. Prolines, known for their unique cyclic side chains, have played an important role in enhancing the structural rigidity and thermal stability of PET hydrolases.The substitution of serine with proline in bacterial PET hydrolases such as Est119 from Thermobifida alba AHK119 and Cut190 from Saccharomonospora viridis AHK190 results in a notable enhancement in heat resistance and PET degradation efficiency. In the same way, the incorporation of proline into enzymes such as LCCICCG tetramer and Thermobifida alba cutinase has a notable effect on their melting temperature and boosts their hydrolytic activity toward PET. Enhancement of the hydrogen bonding network in enzymes has been a key area of study, and PETase is a prominent illustration of this. Modifications made to the flexible regions of these enzymes have led to the development of variants that exhibit enhanced rigidity and thermal stability.176,180 Notable examples include the IsPETase S121E/D186H double variant and the ThermoPETase triple variant. These variants exhibit higher melting temperatures and stronger binding to PET, which enhances their suitability for industrial applications.181 FAST-PETase mutants, such as S132E, D186H, R224Q, N233K, and R280A, have been found to exhibit exceptional kinetics and performance at elevated temperatures, surpassing the capabilities of the original enzyme in terms of hydrolysis efficiency. Crystal structure analysis of these mutants has confirmed their noteworthy contribution to enhancing the thermal stability of the enzyme.182
Nevertheless, challenges persist in breaking down highly crystalline plastics, which hinders the extensive use of these enzymes. Research has indicated that FAST-PETase can fully break down PET after thermal pretreatment, suggesting that these enzymes are successful in the recycling process.182 However, their use in the environmental degradation of crystalline polymers is still somewhat restricted.
Enhancing enzymatic stability with glycosylation in microplastic degradation
Enhancement of the thermal stability of enzymes used in microplastic degradation has gained significant attention, and glycosylation has emerged as a promising technique in this regard. This process has demonstrated encouraging results in enhancing thermal stability, particularly when used with enzymes expressed in eukaryotic microbial cells. Understanding glycosylation, a crucial post-translational modification, is essential to maintain protein stability and prevent thermal aggregation.An excellent illustration is the PET hydrolase LCC, renowned for its exceptional thermal stability. The expression of LCC in Pichia pastoris led to glycosylation, which resulted in an enzyme form that exhibits increased resistance to high-temperature aggregation. This variant of LCC has enhanced efficiency in breaking down PET at elevated temperatures.183 Nevertheless, achieving precise control over the glycosylation sites on the enzyme's surface poses a serious challenge, given its potential impact on the interaction of the enzyme with the PET substrate. Precise positioning of these glycosylation sites is needed to prevent negative impacts on the active site and overall functionality of the enzyme. Cutting-edge computational methods, such as GRAPE, have been employed to enhance enzymes such as IsPETase, leading to the development of variants like DuraPETase. These variants exhibit a remarkable increase in melting temperature, thereby enhancing their stability and effectiveness in degrading PET.184
In a previously reported study, neural networks were used to conduct extensive in silico mutagenesis and experimental validation. The aim was to develop enzymes with enhanced thermal stability. During this study, a number of mutations were discovered in PETase that greatly enhanced its performance in high temperature conditions. The enzyme FAST-PETase demonstrated a notable enhancement in the rate of hydrolysis when compared to its original form. While it is important to consider the potential impact on the catalytic efficiency of the enzyme,182 it is also beneficial to increase thermal stability. The function of the enzyme can be influenced by structural modifications in the active site. As an illustration, replacing Ala with Arg280 in PETase enhances the speed of PET degradation.
Glycosylation is a crucial element in enhancing enzyme thermal stability, and it is essential to carefully select a glycosylation site that does not disrupt the catalytic activity of the enzyme. A meticulous approach to enzyme modification is needed for the advancement of enzyme-based strategies for microplastic degradation (Fig. 5).
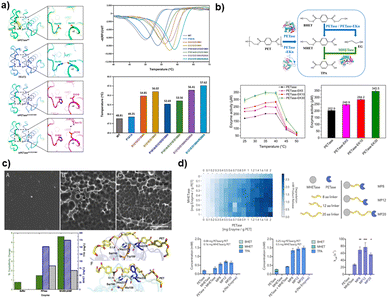 | ||
| Fig. 5 Strategies for enhancing thermal stability and catalytic activity of plastic degrading enzymes using protein engineering. (a) The IsPETase mutant, characterized by increased thermal stability and a higher Tm value compared to the wild-type IsPETase, achieves this improvement through the stabilization of the β6–β7 linked loop. Reproduced with permission from ref. 181. Copyright (2019) American Chemical Society. (b) Enhanced catalytic activity of a PETase-EKn variant with a more open substrate binding pocket resulting from C-terminal fusion of PETase with a zwitterionic polypeptide consisting of glutamic acid (E) and lysine (K) residues. Reproduced with permission from ref. 185. Copyright (2021) American Chemical Society. (c) PETase with a narrow active site due to the double mutation S238F/W159H shows a higher loss of crystallinity in PET and improved aromatic interaction with the substrate compared to wild-type PETase. Reproduced with permission from ref. 186. Copyright (2018) PNAS. (d) Synergistic depolymerization efficacy of MHETase-PETase, a chimeric enzyme linking the C terminus of MHETase to the N terminus of PETase, on PET films. Reproduced with permission from ref. 186. Copyright (2020) PNAS. | ||
Enhancing microplastic degradation through improved enzyme–substrate dynamics
The effectiveness of enzyme-based microplastic degradation relies heavily on the intricate interactions between enzymes and their specific substrates, particularly when it comes to PET hydrolases. This aspect is of the utmost importance in heterogeneous catalysis because it involves the solubility of the enzyme in an aqueous system, which is in stark contrast to the insolubility of the PET chains.187,188 This imbalance frequently results in substantial adsorption on the PET surface, which can have a negative impact on the catalytic efficiency of the enzyme.189 It is worth noting that the structure of PETase does not inherently facilitate substrate binding. This discovery has sparked a surge of scientific curiosity in hydrophobic polymers that resemble naturally occurring substances such as carbohydrate-active enzymes. The interaction between enzymes and PET substrates is mainly influenced by the electrostatic and hydrophobic interactions among the amino acid residues.190,191The improvement of the interaction between enzymes and substrates is a main area of emphasis. One efficient approach is to alter the hydrophobic surface and/or electrostatic properties of the enzyme. Through an analysis of the charged and solvent-exposed amino acid residues of proteins, studies are striving to minimize the electrostatic repulsion between the PET substrate and the enzyme surface.121,192,193 These adjustments can enhance the binding affinity and boost the degradation efficiency of PET. These advancements are crucial in the development of enzyme-based methods for microplastic degradation and have a significant role in reducing the environmental impact of plastic pollution.
The enhancement of enzyme–substrate interactions is not just a scientific endeavor, but an important aspect of promoting environmental sustainability. Through careful optimization of these interactions, enzymes can be customized to better target and break down particular forms of microplastics, resulting in the higher overall efficiency of the degradation process. This approach shows immense potential in addressing the growing concern of microplastic pollution and offers a more efficient and eco-conscious solution to this worldwide problem. With the ongoing advancements in research, the potential for developing highly effective and specialized enzymes for microplastic degradation is growing.
Enhancing the active site of PET hydrolase to enhance microplastic degradation
The enzymatic degradation of polyethylene terephthalate (PET) microplastics is intricately linked to the interaction between the active site of PET hydrolases and the PET substrate.194 In order to optimize the catalytic activity of these hydrolases, it is often necessary to make precise modifications in the active site region. These modifications may involve reconfiguring the substrate, adjusting the cofactor specificity, or introducing mutations in the active site that have been proven to greatly affect the overall reactivity.195,196 One important objective in PET-degrading enzyme engineering is to enhance the accessibility of the active site to the plastic surface.197 This is usually achieved by broadening the range of substrates that can bind to the enzyme. The approach used involved manipulating Fusarium solani cutinase to create the L182A mutant. This mutant was designed to have enhanced hydrolytic activity against PET by making specific amino acid modifications to expand the active site niche.198Enzymes such as PETase, Cut190, MHETase, and Pseudomonas aestusnigri hydrolase have employed comparable techniques to selectively target residues in the substrate binding site by means of structural mutagenesis.199 Recent modifications have significantly enhanced the binding of PET substrate and minimized the interference caused by degradation byproducts. As a result, the efficiency of PET depolymerization has been greatly improved.200 Efforts are currently being made to enhance the hydrophobic characteristics of the binding site or optimize the active site. These efforts strive to enhance or diminish the bond of substances, ultimately resulting in more efficient PET degradation.185,198,201–203
In addition, modifying the structure of the active site of the enzyme can assist in reducing the inhibition caused by PET degradation intermediates or products.204 These different strategies work together to improve the interaction between PET hydrolase and its substrate for more efficient degradation of PET. This is a significant contribution to tackling the environmental issues associated with microplastics made from PET. The improvement of the active site of PET hydrolases is a crucial step in the development of more efficient enzymatic solutions for microplastic degradation (Table 2).
| Enzyme | Source | Substrate (ligand) | Engineering strategies | Mutation(s)/modification(s) | Biological effects (results) | Ref. |
|---|---|---|---|---|---|---|
| Enhancing enzymatic thermal stability | ||||||
| Cut190 | Saccharomonospora viridis AHK190 | PET | Substitutions of amino acid through site-directed mutagenesis | S226P/R228S | Increase of Tm value by 3.7![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) °C with higher compared with the wild-type Cut190 °C with higher compared with the wild-type Cut190 |
122 |
| IsPETaseTM | Ideonella sakaiensis | PET | Substitutions of amino acid through site-directed mutagenesis | S121E/D186H/R280A | Increase of Tm value by 8.81 °C at 40 °C and 14-fold improved PET degradation activity compared with the IsPETaseWT | 181 |
| Est1 | Thermobifida alba AHK119 | PET | Introducing proline and valine residues | A68V/T253P | The introduction of proline enhances heat resistance and activity in Est1 and Est119 compared to their respective enzyme types without proline | 180 |
| Est119 | ||||||
| LCC-G | Leaf and branch compost genome | PET | Introducing glycan | Glycosylation at N197 and N266 | Resistance to thermal aggregation and improved PET hydrolysis activity at 10 °C higher than LCC enzymes | 183 |
| LCC | Leaf and branch compost genome | PET | Introducing disulfide bridge | D238C/S283C | Increase of Tm value by 9.8![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) °C higher than that of w/t LCC °C higher than that of w/t LCC |
179 |
| Cut190* | Saccharomonospora viridis AHK190 | PET | Introducing disulfide bridge | Q138A/D250C-E296C/Q123H/N202H | Improvement in PET film degradation by approximately 3 times and more than 30% at 70 °C, compared to the degradation of Cut190* at 63 °C | 205 |
| DuraPETase | Ideonella sakaiensis | PET | Accumulation of a substantial number of mutations using the GRAPE strategy | S214H-I168RW159H-S188Q-R280A-A180I-G165A-Q119Y-L117F-T140D | Maintains thermal stability at high temperatures of 60 °C for 3 days | 184 |
| ICCG variants | Leaf and branch compost genome | PET | Triple residues mutation | A59K/V63I/N248P | Higher Tm values (98, 98.9, 98.6 °C vs. 95.2 °C) of ICCG variants (RIP, KIP, KRP) compared with ICCG | 176 |
| A59P/V63I/N248P | ||||||
| A59K/V63R/N248P | ||||||
| HotPETase | Ideonella sakaiensis | PET | Induction of mutations by evolutionary pressure | 21 mutations | Highest Tm value (82.5 °C) recorded to date among IsPETase derivatives | 206 |
| PES-H1R204C/S250C | Leaf and branch compost genome | PET | Introducing disulfide bridge | R204C/S250C | Increase of Tm value by 6.4![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) °C higher than that of w/t LCC °C higher than that of w/t LCC |
207 |
| PETaseiS136E | Ideonella sakaiensis | PET | Using the PROSS (Protein Repair One Stop Shop) approach, which recommends amino acid substitutions related to enzyme stabilization | S136E | Increase of Tm value by 1.1 °C and activity half-life t1/2 by 2.4 times higher than that of w/t IsPETase | 181 |
| Cut190*SS | Saccharomonospora viridis AHK190 | PET | Multiple residues mutation | Q138A/D250C-E296C/Q123H/N202H/S226P/R228S | Increase of Tm value by 30 °C higher than that of Cut190* (Cut190S226P/R228S mutant) | 208 |
| TfCut2D204C/E253C | Thermobifida fusca KW3 | PET | Replacement of calcium binding site with disulfide bridge | D204C/E253C- | Increase of Tm value up to 94.7 °C (w/t TfCut2: 69.8 °C) and half-inactivation temperature up to 84.6 °C (w/t TfCut2: 67.3 °C) | 209 |
| IsPETaseTMK95N/F201I | Ideonella sakaiensis | PET | Amino acid substitutions through site-directed mutagenesis | K95N/F201I | Increase of Tm value by 5.0 °C higher than that of IsPETaseTM (IsPETase triple mutant) | 210 |
| TtC-G | Thielavia terrestris (TtC) | PET | Introducing glycosylation | Glycosylation at N127 and N195 | Increase of Tm value by 3.0 °C higher than that of TtC-nonglycosylated forms | 211 |
| PET2 7M | Ideonella sakaiensis 201-F6 | PET | Introducing disulfide bridge | R47C/G89C/F105R/E110K/S156P/G180A/T297P | Increase of Tm value by 6.7 °C higher than w/t PET2 | 212 |
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) |
||||||
| Improving interaction with substrate through modification of enzyme active site and surface | ||||||
| Cutinase variants | Fusarium solani pisi | PET | Expanding the active site using site-directed mutagenesis | L81A/N84A/L182A/V184A/L189A | Increase of hydrolytic activity by 4- and 5-fold on PET fibers compared with w/t cutinase | 198 |
| PE-H(Y250S) | Pseudomonas aestusnigri VGXO14T genome | PET | Expansion of the active flanking niche site using site directed mutagenesis | Y250S | Increase of enzymatic activity by 3-fold on pNPB compared with PE-H | 201 |
| PETaseS238F/W159H | Ideonella sakaiensis 201-F6 | PET | Narrowing the substrate binding gap through site-directed mutation of active site residues | S238F/W159H | Increase of absolute crystallinity loss by 4.13% in PET films higher than that of w/t PETase | 186 |
| PETaseI179F | Ideonella sakaiensis | PET | Improvement of enzyme hydrophobicity and steric hindrance through site-directed mutagenesis around the active site | R61A, L88F, I179F | Increase of Kcat/KM by 15.6-fold compared with w/t PETase | 202 |
| Tfu_0883Q132A/T101A | Thermobifida fusca | PET | Expansion of the active flanking niche site using site directed mutagenesis | Q132A/T101A | Increase of the amount of degradation product (TPA) by 10-fold and hydrolysis rate k2 by 10-fold compared with the w/t Tfu_0883 | 203 |
| PETase-EK30 | Ideonella sakaiensis | PET | Changing the substrate binding pocket of the PETase molecule to a more open structure using EKylation | EKylation | Increase of total released compounds by 11.5 times and enhance catalyst performance by 7.7 times compare with w/t PETase | 185 |
| Thc_Cut2 | Thermobifida cellulosilytica DSM44535 | PET | Increasing protein surface hydrophobic interactions through amino acid mutations located outside the active site | Arg29Asn or Ala30Val | Increase of Kcat/KM by 30-fold in PNPB substrate compared with w/t Thc_Cut2 | 192 |
| PhaZRpiT1 | Ralstonia pickettii T1 | PHB | Amino acid residue substitution to increase hydrophobic interactions between substrates | Y443F | Increase of maximum degradation rate up to 43 ± 3.7 mg min−1 compared with w/t 35 ± 0.5 mg min−1 | 213 |
| del71Cbotu_EstA | Clostridium botulinum (Cbotu_EstA) | PET | Improving adsorption of the enzyme to the substrate by removing 71 residues at the N-terminus of the enzyme | Remove 71 residues from the N-terminus | Increase of total released compounds by more than 8 times, KM by 4 times and Kcat by 2 times compared with the w/t enzyme | 214 |
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) |
||||||
| Enzyme accessory module fusion and chimeric protein construction | ||||||
| PA_PBM | Nocardia farcinica | PU | Attaching the hydrophobic binding module of polyhydroxyalkanoate depolymerase (PBM) to polyamidase (PA) | Fuse PBM to PA C terminus | 7.5 times more total released compounds and increase of enzymatic activity by 4-fold compared with w/t PA | 215 |
| Thc_Cut1_HFB | Thermobifida cellulosilytica | PET | Linking a partial region (P263 to P287) hydrophobin of cellobiohydrolase I from T. reesei to Thc_Cut1 | Connecting hydrophobin to the N terminus of Thc_Cut1 | Increase of hydrolysis product release rate by 16-fold compared with the w/t Thc_Cut1 | 72 |
| Cutinase-CBM | Thermobifida fusca | PET | Fusing the carbohydrate binding module (CBM) of T. fusca cellulase Cel6A (CBM(Cel6A)) and Cellulomonas fimi cellulase CenA (CBM(CenA)) to the C terminus of T. fusca cutinase | Fusing the CBM module to the cutinase C terminus | Increase of the amount of released fatty acids up to 3-fold compared with the w/t Thc_Cutinase | 216 |
| Thc_Cut1-PBM | Thermomyces cellullosylitica | PET | Fusing the polyhydroxyalkanoate depolymerase of Alcaligenes faecalis to the C terminus of Thc_Cut1 | Fusing the PBM to the C terminus of Thc_Cut1 | Increase of the amount of released hydrolysis products by 3.75-fold compared with the w/t Thc_Cut1 | 217 |
| IsPETaseEHA_CBM | Ideonella sakaiensis | PET | Fusing the Cellulose binding domain (CBM) to the IsPETaseS121E/D186H/R280A (IsPETaseEHA) variants C terminus of Thc_Cut1 | Fusing the CBM to the C terminus of IsPETaseEHA | Increase of hydrolytic activity up to 71.5% higher than that of IsPETaseEHA | 218 |
| LCCICCG-ChBD | Leaf and branch compost genome | PET | Fusing the chitin binding domain (ChBD) of Chitinolyticbacter meiyuanensis SYBC-H1 to the C terminus of the LCCICCG | Fusing the ChBD to the C terminus of the LCCICCG variant | Increase of depolymerization performance by 11.6-fold compared with the LCCICCG without module | 219 |
| DSI-Tfuc2 | Thermobifida fusca | PET | Fusing the Dermaseptin SI (DSI) to the N-terminus of Thermobifida fusca cutinase D204C/E253C (Tfuc2) | Fusing the DSI to the N terminus of the Tfuc2 variant | Increase of PET decomposition rate by 22.7 fold compared with the Tfuc2 variant | 220 |
| Lip–Cut | Thermomyces lanuginosus | PCL | End-to-end fusion of bifunctional lipase–cutinase (Lip–Cut) | Fusing the lipase and the cutinase | Increase of the weight loss of PCL film by 13.3 times, 11.8 times, and 5.7 times higher than that caused by Lip, Cut, and Lip/Cut blends, respectively | 221 |
| MHETase:PETase | Ideonella sakaiensis | PET | Covalently links the C terminus of MHETase to the N terminus of PETase | Fusing the MHETase and the PETase | Increase of hydrolysis product release rate by 3-fold compared with the single enzyme (MHETase) | 222 |
Enhancing enzyme surface properties to enhance efficiency in degrading microplastics
In the area of enzyme-based microplastic degradation, numerous endeavors have been undertaken to enhance the efficiency of plastic degradation. This involves modifying the surface properties and charge characteristics of the active site of PET hydrolases. The enhancement of the hydrophobicity of the active site has been shown to be a successful approach to improving binding to plastic substrates and consequently boosting degradation efficiency. As an illustration, when the cutinase Cbotu_EstA is adjusted,214 it reveals a greater portion of its hydrophobic surface. This leads to improved adsorption to PET substrates and an increase in hydrolytic activity. Nevertheless, an overabundance of hydrophobic residues may result in undesirable consequences like enzyme aggregation or structural instability.In a previously reported study, certain mutations that greatly improve the activity of PETase were discovered. This was achieved with the use of molecular docking and crystallographic analysis. Several mutations, including R61A, L88F, and I179F, have been found to greatly enhance enzyme efficiency. In a similar vein, modification of the thermostable LCC and Tf Cut2 PET hydrolases by substituting His/Phe with Ser/Ile enhances their ability to break down PET at lower temperatures, resulting in more efficient depolymerization.202 The improvement of hydrophobicity can also enhance enzyme–substrate interactions. This was observed in the PHB-degrading enzyme from R. pickettii T1, where the substitution of serine and tyrosine with hydrophobic residues increased adhesion to the PHB surface. As a result, the efficiency of plastic hydrolysis was significantly improved.223
Recent developments in enzyme design have led to notable improvements in catalytic activity and a reduction in byproduct inhibition. For instance, the Tfu_0883 cutinase underwent a double mutation (Q132A/T101A), resulting in notable enhancements. The replacement of amino acid residues in the active site of the TfCut2 cutinase with residues from the LCC cutinase led to enhanced PET degradation at higher temperatures.203 Furthermore, the192 substitution of a mutation, Ile179, in the PETase enzyme with the more hydrophobic Phe resulted in an enhanced catalytic efficiency toward PET substrates at 30 °C.202 The modifications made enhanced the alignment of the binding sites and resulted in more robust interactions between the enzyme and substrate. These recent advancements underscore the significance of thoughtfully planned enzyme modifications in enhancing the breakdown of PET and other plastic materials. These strategic modifications in enzyme surface properties demonstrate promise for developing more efficient solutions to the urgent issue of microplastic pollution.185,192,213
Improving enzyme functionality with the use of accessory binding domains
In order to enhance the efficiency of enzyme-based microplastic degradation, especially for PET, previous studies have investigated the integration of accessory binding domains that draw inspiration from the intricate structure of cellulases. These auxiliary modules, referred to as carbohydrate-binding modules (CBMs), are segments present in carbohydrate-active enzymes that facilitate the breakdown of natural biopolymers. The integration of CBMs into PET hydrolysing enzymes seeks to optimize the interaction of the enzyme with PET, leading to improved degradation efficiency. CBMs are highly regarded for their excellent compatibility with a wide range of natural polymers and synthetic plastics. Nevertheless, predicting the protein sequences that determine the function of PET-binding modules poses a serious challenge due to their inherent complexity.224,225Notable advancements have been achieved in this field, such as the successful combination of a cutinase from Thermobifida fusca with CBMCenA from Cellulomonas fimi to enhance the breakdown of PET fibers.216 An alteration was made to CBMCenA by introducing a single tryptophan mutation. This modification aimed to enhance its adherence to PET fibers and enable more efficient enzymatic degradation.226 Moreover, the combination of Thc_Cut1 cutinase from Thermobifida cellulosiliqua and CBM trCBH from Hypocrea jecorina notably boosts the binding of the enzyme to the PET surface, thereby enhancing its capacity to break down the material. The combination of PET hydrolases and CBM has demonstrated encouraging outcomes in enhancing the efficiency of PET degradation in various PET feedstocks.227
Additional cutting-edge techniques being investigated involve the use of polyhydroxyalkanoate binding modules (PBMs), hydroponics, and amphiphilic anchor peptides for chimeric fusion. These methods focus on enhancing the attachment of enzymes to PET to improve the degradation of polyester–PU nanoparticles. As an illustration, the combination of hydrophobic, a protein with hydrophobic properties, and PETase has proven to be effective in improving the binding and degradation of PET films. These strategies, which aim to improve the binding ability of PET-degrading enzymes, show significant potential in optimizing PET binding and enhancing hydrolysis efficiency. This offers a fresh approach to tackling the issue of microplastic pollution.72,215,217,219,220
Factors influencing the efficiency of enzymatic plastic degradation
In order to gain insights into the enzymatic degradation efficiency of microplastics, an extensive analysis was carried out to examine the distinctive properties of plastic polymers. The chemical structure, molecular weight, and crystallinity of these polymers play a crucial role in determining their degradation rate. Polymers containing ester bonds, such as polyester polyurethane, typically demonstrate greater biodegradability in comparison to polymers lacking these bonds.228 Biodegradation of high molecular weight plastics can be more challenging, but there are additives available that can assist in the process. The intricate composition of microplastics, with their symmetrical shapes, strong hydrogen bonds, and regular units, can often impede enzymatic degradation.229Environmental factors are also influential in the degradation process. Temperature is important since elevated temperatures can speed up degradation and affect oxidation mechanisms. Accurate pH levels are also important in assessing the activity and growth of microorganisms that contribute to degradation. Additionally, different pH levels can affect the structure of plastic and its vulnerability to degradation. UV exposure and biodegradation are key factors contributing to the breakdown of plastic. Other factors, such as mechanical shredding, temperature, pH, and catalysts, also play a role in this process. Humidity plays a significant role in the biodegradation process. Higher levels of humidity generally support biodegradation, while excessive moisture can actually impede the process.140,230–232
The degradation kinetics of plastics are influenced by a variety of factors, resulting in a complex and multifaceted degradation process. This review highlights the significance of creating efficient enzyme-based strategies that are customized to the distinct characteristics of various microplastics to enhance their degradation efficiency.
Tackling PET pollution with enzymatic solutions
Degradation process of PET by hydrolytic enzymes
Polyethylene terephthalate (PET), a widely used synthetic plastic found in disposable beverage containers, has experienced a notable surge in global production. In 2013, a staggering 56 million tons of PET were manufactured.233 The durability of PET, stemming from its aromatic ring structure and ester bonds, plays a significant role in the issue of plastic pollution. Improper disposal of single-use plastics further worsens this problem. In contrast to other biodegradable polyesters such as polyhydroxyalkanoate, PCL, polybutylene succinate, and poly(butylene adipate-coterephthalate) (PBAT), PET is recognized for its resistance to natural degradation processes. Recent scientific advances have revealed fascinating insights into certain microorganisms capable of breaking down PET polymers.34,234–236 One such microorganism, Ichneumonella sakaiensis 201-F6, has demonstrated the ability to utilize the terephthalate component in its metabolic activity. This discovery offers a fresh perspective on the degradation of PET (Fig. 6).34A study involving Pseudomonas putida GO16 highlights the significant progress made in biotechnology, specifically in converting PET into more environmentally friendly materials such as polyhydroxyalkanoates. This process, which uses pyrolysis, demonstrates potential for the recycling of PET waste.237 The degradation rate of PET films is influenced by various factors including crystallinity, purity, and the orientation of the polymer chains. These factors greatly affect the efficiency of the degradation process. PET microplastics present a serious environmental concern due to their potential effects on human health, specifically in relation to the endocrine system and estrogen regulation.238,239 This situation highlights the urgent demand for innovative and efficient degradation and recycling methods for PET and other synthetic polymers. Enzyme-based strategies for microplastic degradation have gained recognition as a viable solution, providing a sustainable approach to address the environmental consequences of PET pollution.
Enhancing enzyme technology for efficient PET degradation
Remarkable advancements have been achieved in enhancing the performance of PETase, the key enzyme in the breakdown of polyethylene terephthalate (PET), in the area of enzyme-based microplastic degradation. Prior research has primarily concentrated on enhancing the interaction of PETase with PET substrates.240,241 As an illustration, PETase was modified with double mutations to enhance the efficiency of PET degradation. We conducted extensive research on double mutations in Thermobifida fusca to gain a deeper understanding of the enzyme's degradation capabilities.203The activity of Cut190, a cutinase variant from S. viridis, was found to be influenced by the presence of Ca2+ in the binding site. Prior research has discovered three calcium binding sites in Cut190, each exerting distinct effects on the active site of the enzyme. By making modifications to these sites, the thermal stability of the enzyme was enhanced, and the degradation of PET was greatly increased.242 The main function of PETase is to transform PET into intermediate compounds such as mono-(2-hydroxyethyl) terephthalate (MHET) and bis-(2-hydroxyethyl) terephthalate (BHET). These compounds are subsequently broken down by MHETase into ethylene glycol (EG) and terephthalic acid (TPA). These products are subsequently involved in the tricarboxylic acid (TCA) cycle. PETase functions best in a pH range of 7–9 and maintains its stability in a pH range of 6–10.243 An optimal pH of 9.0 and a temperature of 30 °C were determined to be the most effective conditions for the variant of PETase. Efforts to enhance the stability of Ideonella sakaiensis PETase involved targeted genetic modifications to enhance its resistance to heat and prolong its effectiveness (Fig. 7).244
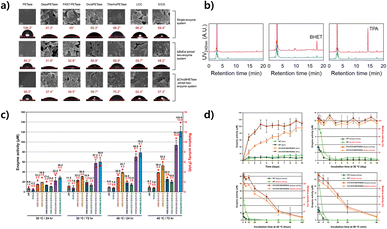 | ||
| Fig. 7 Increased PET decomposition efficiency by PETase deformation. (a) The SEM images (up panel) and water contact angle analysis (down panel) of the PET film in a single-enzyme degradation system, two-enzyme degradation system with ΔBsEst, and a two-enzyme degradation system with ΔChryBHETase after 48 h at 60 °C. Reproduced with permission from ref. 245. Copyright (2023) Springer Nature. (b) HPLC profiles of PETase powder incubation experiments: 2 weeks, 3 weeks and 4 weeks after incubation. Green and red lines indicate CC-124 wild type and CC-124_PETase #11 lysates. Reproduced with permission from ref. 246. Copyright (2020) Springer Nature. (c) PETase activity of the variants, PET degradation activity of IsPETaseWT and variants, the enzyme activity is the sum of MHET and TPA. Reproduced with permission from ref. 181. Copyright (2019) American Chemical Society. (d) Enzyme activity for 10 days and heat-inactivation experiment of IsPETaseWT and IsPETaseS121E/D186H/R280A. Reproduced with permission from ref. 181. Copyright (2019) American Chemical Society. | ||
In a previous study, it was discovered that a specific mutation, R280A, demonstrated exceptional efficacy. This mutation showed enhanced activity and improved hydrolysis efficiency when BHET was used as a substrate. Additional examination of PETase resulted in the discovery of two mutations that enhance stability through the formation of hydrogen bonds: the substitution of serine at247 position 121 with glutamic or aspartic acid, and the substitution of aspartate at position 186 with histidine. The double mutation (serine 121 and aspartate 186) led to the development of a PETase R280A variant that exhibited a remarkable enhancement in degradation capacity, resulting in a 13.9-fold increase in efficiency.182 Recent research has also investigated the alteration of the protein structure to enhance plastic degradation. By integrating a poly-3-hydroxybutyrate (PBM) binding domain into the enzyme, its capacity to break down PET was greatly enhanced. With professional experimentation, it was discovered that incorporating a CBM domain into the mutant greatly enhanced the speed at which PET degradation occurred. In fact, the rate increased by 2.28 times when compared to the original variant. These advancements highlight the potential of engineered enzymes in effectively tackling the issue of PET microplastic pollution.219,225
Approaches to address microplastic and nanoplastic pollution
Exploring the environmental dynamics of micro- and nanoplastics
With the rise in plastic production worldwide, it is concerning to see how plastics are breaking down into microplastics due to various environmental factors such as ultraviolet light, temperature fluctuations, soil biological activity, and human involvement. Microplastics can undergo a process where they break down into even smaller particles called nanoplastics, which worsens the issue of plastic pollution. These microplastics can take the form of fibers, pellets, spheres, or flakes. The presence of micro- and nanoplastics is widespread in different environments, including landfills, wastewater systems, industrial and agricultural wastes, and polymer coatings.248,249 An in-depth understanding of the sources, properties, and distribution of these microplastic particles is crucial in the context of enzyme-based microplastic degradation strategies.250 This knowledge is vital for developing efficient measures to minimize their environmental impact. The extensive dispersion of these pollutants and their challenging degradation and recycling underscore the pressing need to tackle this worldwide environmental issue. An all-encompassing approach is required to take on the extensive dispersion of micro- and nanoplastics and minimize their effects on ecosystems.Biological effects of micro- and nanoplastics
Recent studies have uncovered the profound effects of microplastics and nanoplastics on the biological functions and health of a wide range of organisms. These studies have highlighted the negative impacts on ecosystem health and physiological processes of living organisms. They have emphasized the detrimental impact of large quantities of microplastics on the regular functioning of aquatic organisms,251–253 particularly fish. These minuscule particles, consumed through food and inhalation, result in harm to the tissues of fish. Giacomo Limonta's research on zebrafish revealed noteworthy alterations in the expression of immune response genes following exposure to low concentrations of microplastics. In Mehdi Banaei's research on Cyprinus carpio, significant findings were observed regarding gene expression and enzyme activity associated with oxidative stress and detoxification. These findings highlight the biochemical disruptions caused by microplastics in aquatic organisms.254–256Microplastics present a significant danger to various forms of marine life, extending beyond just fish. Microplastics pose a serious ecological concern due to their capacity to adsorb heavy metals, harbor bacterial pathogens, carry multidrug-resistant E. coli, and act as a vector for persistent pollutants.257–260 The risks are further intensified by the combined effects of manufacturing chemicals and organic contaminants that are attached to microplastics. Microplastics also support the development of various microbial communities and create biofilms made up of algae, bacteria, and fungi. This occurrence has the potential to amplify the transmission of microbial pathogens and antimicrobial resistance.261,262
The potential health effects of microplastics on humans are currently receiving significant attention. It has been estimated that a considerable quantity of microplastics is consumed by humans annually, potentially resulting in various health issues including intestinal blockages, inflammatory reactions, and alterations in the gut microbiome. This growing research highlights the importance of further investigating the environmental and health effects of microplastics and nanoplastics.263,264 In particular, there is an urgent demand for the advancement and improvement of enzyme-based techniques to effectively break down these prevalent pollutants.
Using microbial capabilities for plastic degradation
The search for microorganisms and enzymes capable of degrading plastics is gaining importance in the battle against plastic pollution. This requires the application of molecular cloning and culture techniques to improve the enzymatic and metabolic abilities of these microorganisms, thereby enhancing their potential for plastic degradation.107 The use of molecular biology tools, specifically polymerase-based rapid cloning, has been key in the discovery of polymer-degrading enzymes and the mapping of the genes responsible for these functions. These tools enable the manipulation of gene expression through genetic engineering to improve enzyme production, resulting in more efficient degradation in both the natural environment and composting facilities.265Extensive microbial libraries have been developed to identify and validate microbes that excel at degrading plastics. Studies of the 16S rRNA gene in these libraries have yielded valuable insights into the microbial ecosystems linked to plastics. They have shed light on the interactions between these communities and factors such as substrate type, geographic location, and seasonal variations. The degradation of plastics by microbes is influenced by various factors, including the chemical composition of the polymer, environmental conditions, and the inherent characteristics of the microbes themselves. The chemical composition of the polymer is important in determining its biodegradability, with environmental conditions playing a supporting role in promoting degradation. Microbial enzymes are crucial in this process since they selectively target substrates for biodegradation. It is necessary to understand the metabolic pathways of microorganisms that efficiently degrade polymers to develop targeted strategies to combat microplastic pollution. This involves studying bacteria and fungi that play a key role in this process. This occurrence has the potential to amplify the transmission of microbial pathogens and antimicrobial resistance.266,267
The potential health effects of microplastics on humans are currently being closely examined. It is estimated that people consume substantial quantities of microplastics annually, resulting in potential health issues such as intestinal blockages, inflammatory reactions, and alterations in the gut microbiome. This emerging research highlights the importance of ongoing investigation into the environmental and health effects of microplastics and nanoplastics. There is an urgent demand for the advancement and improvement of enzyme-based techniques to effectively break down these prevalent pollutants.
Conclusions
It is imperative that we address the pressing issue of plastic pollution caused by the widespread presence of microplastics and nanoplastics with immediate and efficient measures. This review has emphasized the significant role of enzymatic and microbial strategies in tackling these global environmental and health challenges. The use of enzymes such as PETase and MHETase, along with microbial degradation pathways, presents exciting possibilities for breaking down tough polymers such as PE, PET, and PS into more environmentally friendly substances. Despite the notable progress made in understanding and enhancing the capabilities of specific enzymes and microbes, there are still challenges that need to be addressed. Factors to consider are the efficiency of the degradation process, the scalability of these solutions, and the varying properties of plastic polymers. The significance of microplastics on the environment and health, specifically on marine life and human health, underscores the importance of implementing efficient degradation and recycling technologies. A combination of disciplines is needed to address plastic pollution and find effective solutions. Further studies and developments are necessary to enhance the effectiveness and real-world implementation of enzymatic and microbial degradation methods. Enzymatic and microbial strategies show great potential in addressing plastic pollution.Author contributions
J. C., H. K.; writing—review and editing, M. K., Y.-R. A.; conceptualization and methodology, S. Y., N. K. and S. Y. L.; visualization and investigation, J.-A. P. and S.-J. H; validation, K. S. L. and H.-O. K.; project administration. All authors have read and agreed to the published version of the manuscript.Conflicts of interest
There are no conflicts to declare.Acknowledgements
This work was supported by a National Research Foundation of Korea grant funded by the Korean government through the Korea Institute of Planning (NRF-2021M3E5E3083400, 2022R1A4A1019201 and NRF-2022R1A2C1007354).Notes and references
- V. C. Sekhar, K. M. Nampoothiri, A. J. Mohan, N. R. Nair, T. Bhaskar and A. Pandey, J. Hazard. Mater., 2016, 318, 347–354 CrossRef CAS PubMed.
- R. Geyer, J. R. Jambeck and K. L. Law, Sci. Adv., 2017, 3, e1700782 CrossRef PubMed.
- J. Kaushal, M. Khatri and S. K. Arya, Clean. Eng. Technol., 2021, 2, 100083 CrossRef.
- M. Shimao, Curr. Opin. Biotechnol., 2001, 12, 242–247 CrossRef CAS PubMed.
- A. I. Osman, C. Farrell, A. H. Al-Muhtaseb, A. S. Al-Fatesh, J. Harrison and D. W. Rooney, Environ. Sci. Eur., 2020, 32, 1–12 CrossRef.
- U. Qasim, A. I. Osman, A. H. Al-Muhtaseb, C. Farrell, M. Al-Abri, M. Ali, D.-V. N. Vo, F. Jamil and D. W. Rooney, Environ. Chem. Lett., 2021, 19, 613–641 CrossRef CAS.
- M. Eriksen, L. C. Lebreton, H. S. Carson, M. Thiel, C. J. Moore, J. C. Borerro, F. Galgani, P. G. Ryan and J. Reisser, PLoS One, 2014, 9, e111913 CrossRef PubMed.
- J. Fabres, P. Villarrubia-Gómez and S. E. Cornella, Marine Policy, 2018, 96, 213–220 CrossRef.
- A. A. de Souza Machado, C. W. Lau, J. Till, W. Kloas, A. Lehmann, R. Becker and M. C. Rillig, Environ. Sci. Technol., 2018, 52, 9656–9665 CrossRef CAS PubMed.
- S. W. Kim and Y.-J. An, Environ. Int., 2019, 126, 699–706 CrossRef PubMed.
- Y. K. Song, S. H. Hong, M. Jang, G. M. Han, S. W. Jung and W. J. Shim, Environ. Sci. Technol., 2017, 51, 4368–4376 CrossRef CAS PubMed.
- G. Vered, A. Kaplan, D. Avisar and N. Shenkar, Mar. Pollut. Bull., 2019, 138, 618–625 CrossRef CAS PubMed.
- D. Danso, J. Chow and W. R. Streit, Appl. Environ. Microbiol., 2019, 85, e01095 CrossRef CAS PubMed.
- M. Kedzierski, J. Villain, M. Falcou-Préfol, M. E. Kerros, M. Henry, M. L. Pedrotti and S. Bruzaud, PLoS One, 2019, 14, e0212088 CrossRef CAS PubMed.
- J. S. Choi, Y.-J. Jung, N.-H. Hong, S. H. Hong and J.-W. Park, Mar. Pollut. Bull., 2018, 129, 231–240 CrossRef CAS PubMed.
- O. Güven, K. Gökdağ, B. Jovanović and A. E. Kıdeyş, Environ. Pollut., 2017, 223, 286–294 CrossRef PubMed.
- W. Huang, X. Wang, D. Chen, E. G. Xu, X. Luo, J. Zeng, T. Huan, L. Li and Y. Wang, J. Hazard. Mater., 2021, 417, 126003 CrossRef CAS PubMed.
- A. Bour, A. Haarr, S. Keiter and K. Hylland, Environ. Pollut., 2018, 236, 652–660 CrossRef CAS PubMed.
- J. Heo, J. Choi, J. Y. Kim, H. Jeong, D. Choi, U. Han, J. H. Park, H. H. Park and J. Hong, J. Hazard. Mater., 2021, 414, 125472 CrossRef CAS PubMed.
- C.-B. Jeong, H.-M. Kang, M.-C. Lee, D.-H. Kim, J. Han, D.-S. Hwang, S. Souissi, S.-J. Lee, K.-H. Shin and H. G. Park, Sci. Rep., 2017, 7, 41323 CrossRef CAS PubMed.
- M. Hu and D. Palić, Redox Biol., 2020, 37, 101620 CrossRef CAS PubMed.
- K. Ragaert, L. Delva and K. Van Geem, Waste Manage., 2017, 69, 24–58 CrossRef CAS PubMed.
- Y. Tokiwa, B. P. Calabia, C. U. Ugwu and S. Aiba, Int. J. Mol. Sci., 2009, 10, 3722–3742 CrossRef CAS PubMed.
- Z. Montazer, M. B. Habibi Najafi and D. B. Levin, Can. J. Microbiol., 2019, 65, 224–234 CrossRef CAS PubMed.
- S. Grima, V. Bellon-Maurel, P. Feuilloley and F. Silvestre, J. Polym. Environ., 2000, 8, 183–195 CrossRef CAS.
- S. A. Acharjee, P. Bharali, B. Gogoi, V. Sorhie, B. Walling and B. W. Alemtoshi, Water, Air, Soil Pollut., 2023, 234, 21 CrossRef CAS PubMed.
- A. A. Shah, F. Hasan, A. Hameed and S. Ahmed, Biotechnol. Adv., 2008, 26, 246–265 CrossRef CAS PubMed.
- A. Loredo-Treviño, G. Gutiérrez-Sánchez, R. Rodríguez-Herrera and C. N. Aguilar, J. Polym. Environ., 2012, 20, 258–265 CrossRef.
- B. Nowak, J. Pająk, M. Drozd-Bratkowicz and G. Rymarz, Int. Biodeterior. Biodegrad., 2011, 65, 757–767 CrossRef CAS.
- A. Gajendiran, S. Krishnamoorthy and J. Abraham, 3 Biotech, 2016, 6, 1–6 CrossRef PubMed.
- S. K. Sen and S. Raut, J. Environ. Chem. Eng., 2015, 3, 462–473 CrossRef.
- A.-C. Albertsson, S. O. Andersson and S. Karlsson, Polym. Degrad. Stab., 1987, 18, 73–87 CrossRef CAS.
- A. Ammala, S. Bateman, K. Dean, E. Petinakis, P. Sangwan, S. Wong, Q. Yuan, L. Yu, C. Patrick and K. Leong, Prog. Polym. Sci., 2011, 36, 1015–1049 CrossRef CAS.
- S. Yoshida, K. Hiraga, T. Takehana, I. Taniguchi, H. Yamaji, Y. Maeda, K. Toyohara, K. Miyamoto, Y. Kimura and K. Oda, Science, 2016, 351, 1196–1199 CrossRef CAS PubMed.
- W. J. Li, L. N. Jayakody, M. A. Franden, M. Wehrmann, T. Daun, B. Hauer, L. M. Blank, G. T. Beckham, J. Klebensberger and N. Wierckx, Environ. Microbiol., 2019, 21, 3669–3682 CrossRef CAS PubMed.
- F. Yan, R. Wei, Q. Cui, U. T. Bornscheuer and Y. J. Liu, Microb. Biotechnol., 2021, 14, 374–385 CrossRef CAS PubMed.
- W. Maity, S. Maity, S. Bera and A. Roy, Appl. Biochem. Biotechnol., 2021, 193, 2699–2716 CrossRef CAS PubMed.
- B. Zhu, D. Wang and N. Wei, Trends Biotechnol., 2022, 40, 22–37 CrossRef CAS PubMed.
- V. Pirillo, M. Orlando, D. Tessaro, L. Pollegioni and G. Molla, Int. J. Mol. Sci., 2021, 23, 264 CrossRef PubMed.
- D. F. Williams, Eng. Med., 1981, 10, 5–7 CrossRef.
- A. Nakkabi, M. Sadiki, M. Fahim, N. Ittobane, S. IbnsoudaKoraichi, H. Barkai and S. El Abed, Int. J. Environ. Res., 2015, 9, 157–162 CAS.
- I. Cacciari, P. Quatrini, G. Zirletta, E. Mincione, V. Vinciguerra, P. Lupattelli and G. Giovannozzi Sermanni, Appl. Environ. Microbiol., 1993, 59, 3695–3700 CrossRef CAS PubMed.
- A. Arkatkar, A. A. Juwarkar, S. Bhaduri, P. V. Uppara and M. Doble, Int. Biodeterior. Biodegrad., 2010, 64, 530–536 CrossRef CAS.
- P. Wang, J. Zhao, Y. Ruan, X. Cai, J. Li, L. Zhang and H. Huang, J. Polym. Environ., 2022, 30, 3949–3958 CrossRef CAS.
- D. Braun, J. Polym. Sci., Part A: Polym. Chem., 2004, 42, 578–586 CrossRef CAS.
- Z. Zhang, H. Peng, D. Yang, G. Zhang, J. Zhang and F. Ju, Nat. Commun., 2022, 13, 5360 CrossRef CAS PubMed.
- L. Giacomucci, N. Raddadi, M. Soccio, N. Lotti and F. Fava, New Biotechnol., 2019, 52, 35–41 CrossRef CAS PubMed.
- P. Europe, An Analysis of European Latest Plastics Production, Demand and Waste Data, 2016 Search PubMed.
- Y. Yang, J. Yang, W.-M. Wu, J. Zhao, Y. Song, L. Gao, R. Yang and L. Jiang, Environ. Sci. Technol., 2015, 49, 12080–12086 CrossRef CAS PubMed.
- N. Shimpi, M. Borane, S. Mishra and M. Kadam, Macromol. Res., 2012, 20, 181–187 CrossRef CAS.
- O. Motta, A. Proto, F. De Carlo, F. De Caro, E. Santoro, L. Brunetti and M. Capunzo, Int. J. Hyg. Environ. Health, 2009, 212, 61–66 CrossRef CAS PubMed.
- L. Yang, J. Gao, Y. Liu, G. Zhuang, X. Peng, W.-M. Wu and X. Zhuang, Chemosphere, 2021, 262, 127818 CrossRef CAS PubMed.
- A. G. Kumar, M. Hinduja, K. Sujitha, N. N. Rajan and G. Dharani, Sci. Total Environ., 2021, 774, 145002 CrossRef PubMed.
- R. A. Gross and B. Kalra, Science, 2002, 297, 803–807 CrossRef CAS PubMed.
- M. Stepczyńska and P. Rytlewski, Int. Biodeterior. Biodegrad., 2018, 126, 160–166 CrossRef.
- N. Hegyesi, Y. Zhang, A. Kohári, P. Polyák, X. Sui and B. Pukánszky, Ind. Crops Prod., 2019, 141, 111799 CrossRef CAS.
- X. Hu, T. Su, P. Li and Z. Wang, Polym. Bull., 2018, 75, 533–546 CrossRef CAS.
- Z. Saadi, A. Rasmont, G. Cesar, H. Bewa and L. Benguigui, J. Polym. Environ., 2012, 20, 273–282 CrossRef CAS.
- S. A. Rafiqah, A. Khalina, A. S. Harmaen, I. A. Tawakkal, K. Zaman, M. Asim, M. Nurrazi and C. H. Lee, Polymers, 2021, 13, 1436 CrossRef CAS PubMed.
- L. Zheng, Z. Wang, S. Wu, C. Li, D. Zhang and Y. Xiao, Ind. Eng. Chem. Res., 2013, 52, 6147–6155 CrossRef CAS.
- K. Shi, T. Su and Z. Wang, Polym. Degrad. Stab., 2019, 164, 55–60 CrossRef CAS.
- F. I. Khan, D. Lan, R. Durrani, W. Huan, Z. Zhao and Y. Wang, Front. Bioeng. Biotechnol., 2017, 5, 16 Search PubMed.
- I. A. Ignatyev, W. Thielemans and B. Vander Beke, ChemSusChem, 2014, 7, 1579–1593 CrossRef CAS PubMed.
- R. N. C. Utomo, W.-J. Li, T. Tiso, C. Eberlein, M. Doeker, H. J. Heipieper, A. Jupke, N. Wierckx and L. M. Blank, ACS Sustain. Chem. Eng., 2020, 8, 17466–17474 CrossRef.
- T. Nakajima-Kambe, Y. Shigeno-Akutsu, N. Nomura, F. Onuma and T. Nakahara, Appl. Microbiol. Biotechnol., 1999, 51, 134–140 CrossRef CAS PubMed.
- G. T. Howard, Int. Biodeterior. Biodegrad., 2002, 49, 245–252 CrossRef CAS.
- J.-D. Gu, Environ. Sci. Pollut. Res., 2021, 28, 1278–1282 CrossRef PubMed.
- G. Kale, T. Kijchavengkul, R. Auras, M. Rubino, S. E. Selke and S. P. Singh, Macromol. Biosci., 2007, 7, 255–277 CrossRef CAS PubMed.
- T. Kijchavengkul and R. Auras, Polym. Int., 2008, 57, 793–804 CrossRef CAS.
- V. Siracusa, Polymers, 2019, 11, 1066 CrossRef CAS PubMed.
- K. N. Fotopoulou and H. K. Karapanagioti, Hazardous Chemicals Associated with Plastics in the Marine Environment, 2019, pp. 71–92 Search PubMed.
- D. Ribitsch, E. Herrero Acero, A. Przylucka, S. Zitzenbacher, A. Marold, C. Gamerith, R. Tscheließnig, A. Jungbauer, H. Rennhofer and H. Lichtenegger, Appl. Environ. Microbiol., 2015, 81, 3586–3592 CrossRef CAS PubMed.
- Y.-S. Ho and G. McKay, Process Biochem., 1999, 34, 451–465 CrossRef CAS.
- J. Pospíšil, J. Pilař, N. Billingham, A. Marek, Z. Horak and S. Nešpůrek, Polym. Degrad. Stab., 2006, 91, 417–422 CrossRef.
- A. Booth, S. Kubowicz, C. Beegle-Krause, J. Skancke, T. Nordam, E. Landsem, M. Throne-Holst and S. Jahren, Miljødirektoratet M-918, 2017, pp. 1–147 Search PubMed.
- B. Zhang, L. Chen, J. Chao, X. Yang and Q. Wang, Environ. Sci. Pollut. Res., 2020, 27, 31046–31060 CrossRef CAS PubMed.
- L. Liu, M. Xu, Y. Ye and B. Zhang, Sci. Total Environ., 2022, 806, 151312 CrossRef CAS PubMed.
- S. S. Ali, T. Elsamahy, E. Koutra, M. Kornaros, M. El-Sheekh, E. A. Abdelkarim, D. Zhu and J. Sun, Sci. Total Environ., 2021, 771, 144719 CrossRef CAS PubMed.
- J.-d. Gu and R. Mitchell, The Prokaryotes: an Evolving Electronic Resource for the Microbiological Community, 2001 Search PubMed.
- G. Chen, Z. Fu, H. Yang and J. Wang, TrAC, Trends Anal. Chem., 2020, 130, 115981 CrossRef CAS.
- S. Selvam, K. Jesuraja, S. Venkatramanan, P. D. Roy and V. J. Kumari, J. Hazard. Mater., 2021, 402, 123786 CrossRef CAS PubMed.
- M. Tiwari, T. Rathod, P. Ajmal, R. Bhangare and S. Sahu, Mar. Pollut. Bull., 2019, 140, 262–273 CrossRef CAS PubMed.
- R. Karthik, R. Robin, R. Purvaja, D. Ganguly, I. Anandavelu, R. Raghuraman, G. Hariharan, A. Ramakrishna and R. Ramesh, Sci. Total Environ., 2018, 645, 1388–1399 CrossRef CAS PubMed.
- S. Lambert, C. Scherer and M. Wagner, Integr. Environ. Assess. Manage., 2017, 13, 470–475 CrossRef CAS PubMed.
- J. Yuan, J. Ma, Y. Sun, T. Zhou, Y. Zhao and F. Yu, Sci. Total Environ., 2020, 715, 136968 CrossRef CAS PubMed.
- D. Barcelo and Y. Picó, Case Stud. Chem. Environ. Eng., 2020, 2, 100019 CrossRef.
- K. Boyle and B. Örmeci, Water, 2020, 12, 2633 CrossRef.
- K. D. Cox, G. A. Covernton, H. L. Davies, J. F. Dower, F. Juanes and S. E. Dudas, Environ. Sci. Technol., 2019, 53, 7068–7074 CrossRef CAS PubMed.
- A. L. Andrady, Mar. Pollut. Bull., 2011, 62, 1596–1605 CrossRef CAS PubMed.
- D. M. Mitrano, P. Wick and B. Nowack, Nat. Nanotechnol., 2021, 16, 491–500 CrossRef CAS PubMed.
- W. J. Shim, S. H. Hong and S. E. Eo, Anal. Methods, 2017, 9, 1384–1391 RSC.
- S. Dey, A. K. Rout, B. K. Behera and K. Ghosh, Environ. Microbiome, 2022, 17, 1–21 CrossRef PubMed.
- H. S. Zurier and J. M. Goddard, Curr. Opin. Food Sci., 2021, 37, 37–44 CrossRef CAS.
- Y. Matsuguma, H. Takada, H. Kumata, H. Kanke, S. Sakurai, T. Suzuki, M. Itoh, Y. Okazaki, R. Boonyatumanond and M. P. Zakaria, Arch. Environ. Contam. Toxicol., 2017, 73, 230–239 CrossRef CAS PubMed.
- J. M. Hipfner, M. Galbraith, S. Tucker, K. R. Studholme, A. D. Domalik, S. F. Pearson, T. P. Good, P. S. Ross and P. Hodum, Environ. Pollut., 2018, 239, 215–222 CrossRef CAS PubMed.
- A. G. Caron, C. R. Thomas, K. L. Berry, C. A. Motti, E. Ariel and J. E. Brodie, Mar. Pollut. Bull., 2018, 127, 743–751 CrossRef CAS PubMed.
- S. L. Wright, R. C. Thompson and T. S. Galloway, Environ. Pollut., 2013, 178, 483–492 CrossRef CAS PubMed.
- Y. Chae, D. Kim and Y.-J. An, Aquat. Toxicol., 2019, 216, 105296 CrossRef CAS PubMed.
- M. B. Sathicq, R. Sabatino, G. Corno and A. Di Cesare, Environ. Pollut., 2021, 279, 116896 CrossRef CAS PubMed.
- M. Cole, P. Lindeque, C. Halsband and T. S. Galloway, Mar. Pollut. Bull., 2011, 62, 2588–2597 CrossRef CAS PubMed.
- L. Hou, D. Kumar, C. G. Yoo, I. Gitsov and E. L.-W. Majumder, Chem. Eng. J., 2021, 406, 126715 CrossRef CAS.
- Z. Chen, W. Zhao, R. Xing, S. Xie, X. Yang, P. Cui, J. Lü, H. Liao, Z. Yu and S. Wang, J. Hazard. Mater., 2020, 384, 121271 CrossRef CAS PubMed.
- A. Paço, K. Duarte, J. P. da Costa, P. S. Santos, R. Pereira, M. Pereira, A. C. Freitas, A. C. Duarte and T. A. Rocha-Santos, Sci. Total Environ., 2017, 586, 10–15 CrossRef PubMed.
- S. Y. Park and C. G. Kim, Chemosphere, 2019, 222, 527–533 CrossRef CAS PubMed.
- C. Sánchez, Biotechnol. Adv., 2020, 40, 107501 CrossRef PubMed.
- K. Vogel, R. Wei, L. Pfaff, D. Breite, H. Al-Fathi, C. Ortmann, I. Estrela-Lopis, T. Venus, A. Schulze and H. Harms, Sci. Total Environ., 2021, 773, 145111 CrossRef CAS PubMed.
- A. Amobonye, P. Bhagwat, S. Singh and S. Pillai, Sci. Total Environ., 2021, 759, 143536 CrossRef CAS PubMed.
- P. Chandra and D. P. Singh, in Microorganisms for Sustainable Environment and Health, Elsevier, 2020, pp. 431–467 Search PubMed.
- H. S. Auta, C. U. Emenike, B. Jayanthi and S. H. Fauziah, Mar. Pollut. Bull., 2018, 127, 15–21 CrossRef CAS PubMed.
- J. Yang, Y. Yang, W.-M. Wu, J. Zhao and L. Jiang, Environ. Sci. Technol., 2014, 48, 13776–13784 CrossRef CAS PubMed.
- B. J. Cassone, H. C. Grove, O. Elebute, S. M. Villanueva and C. M. LeMoine, Proc. R. Soc. B, 2020, 287, 20200112 CrossRef CAS PubMed.
- B. Eyheraguibel, M. Traikia, S. Fontanella, M. Sancelme, S. Bonhomme, D. Fromageot, J. Lemaire, G. Lauranson, J. Lacoste and A. Delort, Chemosphere, 2017, 184, 366–374 CrossRef CAS PubMed.
- Z. Montazer, M. B. Habibi Najafi and D. B. Levin, Polymers, 2020, 12, 123 CrossRef CAS PubMed.
- A.-C. Albertsson and S. Karlsson, Prog. Polym. Sci., 1990, 15, 177–192 CrossRef CAS.
- X.-F. Wei, M. Bohlén, C. Lindblad, M. Hedenqvist and A. Hakonen, Water Res., 2021, 198, 117123 CrossRef CAS PubMed.
- A. Chamas, H. Moon, J. Zheng, Y. Qiu, T. Tabassum, J. H. Jang, M. Abu-Omar, S. L. Scott and S. Suh, ACS Sustain. Chem. Eng., 2020, 8, 3494–3511 CrossRef CAS.
- S. Miri, R. Saini, S. M. Davoodi, R. Pulicharla, S. K. Brar and S. Magdouli, Chemosphere, 2022, 286, 131670 CrossRef CAS PubMed.
- M. Nag, D. Lahiri, B. Dutta, G. Jadav and R. R. Ray, Environ. Sci. Pollut. Res., 2021, 28, 41365–41379 CrossRef CAS PubMed.
- R. J. Müller, H. Schrader, J. Profe, K. Dresler and W. D. Deckwer, Macromol. Rapid Commun., 2005, 26, 1400–1405 CrossRef.
- Å. M. Ronkvist, W. Xie, W. Lu and R. A. Gross, Macromolecules, 2009, 42, 5128–5138 CrossRef.
- E. Herrero Acero, D. Ribitsch, G. Steinkellner, K. Gruber, K. Greimel, I. Eiteljoerg, E. Trotscha, R. Wei, W. Zimmermann and M. Zinn, Macromolecules, 2011, 44, 4632–4640 CrossRef CAS.
- F. Kawai, M. Oda, T. Tamashiro, T. Waku, N. Tanaka, M. Yamamoto, H. Mizushima, T. Miyakawa and M. Tanokura, Appl. Microbiol. Biotechnol., 2014, 98, 10053–10064 CrossRef CAS PubMed.
- D. Ribitsch, S. Heumann, E. Trotscha, E. Herrero Acero, K. Greimel, R. Leber, R. Birner-Gruenberger, S. Deller, I. Eiteljoerg and P. Remler, Biotechnol. Prog., 2011, 27, 951–960 CrossRef CAS PubMed.
- R. Wei, T. Oeser, J. Then, N. Kühn, M. Barth, J. Schmidt and W. Zimmermann, AMB Express, 2014, 4, 1–10 CrossRef CAS PubMed.
- S. Skariyachan, A. A. Patil, A. Shankar, M. Manjunath, N. Bachappanavar and S. Kiran, Polym. Degrad. Stab., 2018, 149, 52–68 CrossRef CAS.
- H. Rajandas, S. Parimannan, K. Sathasivam, M. Ravichandran and L. S. Yin, Polym. Test., 2012, 31, 1094–1099 CrossRef CAS.
- R. Gao, R. Liu and C. Sun, J. Hazard. Mater., 2022, 431, 128617 CrossRef CAS PubMed.
- Y. Akutsu-Shigeno, Y. Adachi, C. Yamada, K. Toyoshima, N. Nomura, H. Uchiyama and T. Nakajima-Kambe, Appl. Microbiol. Biotechnol., 2006, 70, 422–429 CrossRef CAS PubMed.
- H. R. Kim, H. M. Lee, H. C. Yu, E. Jeon, S. Lee, J. Li and D.-H. Kim, Environ. Sci. Technol., 2020, 54, 6987–6996 CrossRef CAS PubMed.
- E. B. M. Tamnou, A. T. Arfao, M. E. Nougang, C. S. Metsopkeng, O. V. N. Ewoti, L. M. Moungang, P. A. Nana, L.-R. A. Takang-Etta, F. Perrière and T. Sime-Ngando, Environ. Challenges, 2021, 3, 100056 CrossRef.
- S. Shabbir, M. Faheem, N. Ali, P. G. Kerr, L.-F. Wang, S. Kuppusamy and Y. Li, Sci. Total Environ., 2020, 717, 137064 CrossRef CAS PubMed.
- P. Liu, T. Zhang, Y. Zheng, Q. Li, T. Su and Q. Qi, Eng. Microbiol., 2021, 1, 100003 CrossRef CAS.
- T. Bao, Y. Qian, Y. Xin, J. J. Collins and T. Lu, Nat. Commun., 2023, 14, 5712 CrossRef CAS PubMed.
- J. Gigault, H. El Hadri, B. Nguyen, B. Grassl, L. Rowenczyk, N. Tufenkji, S. Feng and M. Wiesner, Nat. Nanotechnol., 2021, 16, 501–507 CrossRef CAS PubMed.
- Y. Zhang, J. N. Pedersen, B. E. Eser and Z. Guo, Biotechnol. Adv., 2022, 60, 107991 CrossRef CAS PubMed.
- A. R. Othman, H. A. Hasan, M. H. Muhamad, N. I. Ismail and S. R. S. Abdullah, Environ. Chem. Lett., 2021, 19, 3057–3073 CrossRef CAS.
- A. A. Koelmans, P. E. Redondo-Hasselerharm, N. H. M. Nor, V. N. de Ruijter, S. M. Mintenig and M. Kooi, Nat. Rev. Mater., 2022, 7, 138–152 CrossRef.
- Z. Wu, W. Shi, T. G. Valencak, Y. Zhang, G. Liu and D. Ren, Sci. Total Environ., 2023, 163908 CrossRef CAS PubMed.
- J.-P. Lange, ACS Sustain. Chem. Eng., 2021, 9, 15722–15738 CrossRef CAS.
- N. Mohanan, Z. Montazer, P. K. Sharma and D. B. Levin, Front. Microbiol., 2020, 11, 580709 CrossRef PubMed.
- E. M. Blair, K. L. Dickson and M. A. O'Malley, Curr. Opin. Microbiol., 2021, 64, 100–108 CrossRef CAS PubMed.
- I. Vroman and L. Tighzert, Materials, 2009, 2, 307–344 CrossRef CAS.
- J. Lai, H. Huang, M. Lin, Y. Xu, X. Li and B. Sun, Front. Microbiol., 2023, 13, 1113705 CrossRef PubMed.
- S. Ghatge, Y. Yang, J.-H. Ahn and H.-G. Hur, Appl. Biol. Chem., 2020, 63, 1–14 Search PubMed.
- Y. Iiyoshi, Y. Tsutsumi and T. Nishida, J. Wood Sci., 1998, 44, 222–229 CrossRef CAS.
- J. B. van Beilen and B. Witholt, in Pseudomonas: Volume 3 Biosynthesis of Macromolecules and Molecular Metabolism, Springer, 2004, pp. 397–423 Search PubMed.
- I. Kotova, Y. V. Taktarova, E. Tsavkelova, M. Egorova, I. Bubnov, D. Malakhova, L. Shirinkina, T. Sokolova and E. Bonch-Osmolovskaya, Microbiology, 2021, 90, 671–701 CAS.
- M. Fujisawa, H. Hirai and T. Nishida, J. Polym. Environ., 2001, 9, 103–108 CrossRef CAS.
- M. G. Yoon, H. J. Jeon and M. N. Kim, J. Biorem. Biodegrad., 2012, 3, 1–8 Search PubMed.
- H. J. Jeon and M. N. Kim, Int. Biodeterior. Biodegrad., 2015, 103, 141–146 CrossRef CAS.
- J. Zhao, Z. Guo, X. Ma, G. Liang and J. Wang, J. Appl. Polym. Sci., 2004, 91, 3673–3678 CrossRef CAS.
- Z. Cai, M. Li, Z. Zhu, X. Wang, Y. Huang, T. Li, H. Gong and M. Yan, Microorganisms, 2023, 11, 1661 CrossRef CAS PubMed.
- X. Qin, X. Sun, H. Huang, Y. Bai, Y. Wang, H. Luo, B. Yao, X. Zhang and X. Su, Biotechnol. Biofuels, 2017, 10, 1–13 CrossRef PubMed.
- P. Chowdhary, G. Shukla, G. Raj, L. F. R. Ferreira and R. N. Bharagava, SN Appl. Sci., 2019, 1, 1–12 CAS.
- C. Roth, R. Wei, T. Oeser, J. Then, C. Föllner, W. Zimmermann and N. Sträter, Appl. Microbiol. Biotechnol., 2014, 98, 7815–7823 CrossRef CAS PubMed.
- R. Jabloune, M. Khalil, I. E. B. Moussa, A.-M. Simao-Beaunoir, S. Lerat, R. Brzezinski and C. Beaulieu, Microb. Environ., 2020, 35, ME19086 CrossRef PubMed.
- D. Komeil, A.-M. Simao-Beaunoir and C. Beaulieu, Can. J. Microbiol., 2013, 59, 294–303 CrossRef CAS PubMed.
- P. J. Baker, C. Poultney, Z. Liu, R. Gross and J. K. Montclare, Appl. Microbiol. Biotechnol., 2012, 93, 229–240 CrossRef PubMed.
- X. Han, W. Liu, J.-W. Huang, J. Ma, Y. Zheng, T.-P. Ko, L. Xu, Y.-S. Cheng, C.-C. Chen and R.-T. Guo, Nat. Commun., 2017, 8, 2106 CrossRef PubMed.
- A. Duplan and B. L. Hall, FASEB J., 2018, 32, 796.712 Search PubMed.
- A. Rupert, T. Khong, A. Duplan, R. Fenton and B. Hall, FASEB J., 2021, 35 Search PubMed.
- M. Sielicki, D. Focht and J. Martin, Can. J. Microbiol., 1978, 24, 798–803 CrossRef CAS PubMed.
- L. Tahir, M. I. Ali, M. Zia, N. Atiq, F. Hasan and S. Ahmed, Pol. J. Microbiol., 2013, 62, 101–108 CAS.
- A. J. Mohan, V. C. Sekhar, T. Bhaskar and K. M. Nampoothiri, Bioresour. Technol., 2016, 213, 204–207 CrossRef CAS PubMed.
- D. Tischler, D. Eulberg, S. Lakner, S. R. Kaschabek, W. J. van Berkel and M. Schlömann, J. Bacteriol., 2009, 191, 4996–5009 CrossRef CAS PubMed.
- K. O'Connor, C. M. Buckley, S. Hartmans and A. Dobson, Appl. Environ. Microbiol., 1995, 61, 544–548 CrossRef PubMed.
- F. Beltrametti, A. M. Marconi, G. Bestetti, C. Colombo, E. Galli, M. Ruzzi and E. Zennaro, Appl. Environ. Microbiol., 1997, 63, 2232–2239 CrossRef CAS PubMed.
- N. Itch, K. Hayashi, K. Okada, T. Ito and N. Mizuguchi, Biosci., Biotechnol., Biochem., 1997, 61, 2058–2062 CrossRef CAS PubMed.
- G. Bestetti, P. Di Gennaro, A. Colmegna, I. Ronco, E. Galli and G. Sello, Int. Biodeterior. Biodegrad., 2004, 54, 183–187 CrossRef CAS.
- L. Hou and E. L.-W. Majumder, Materials, 2021, 14, 503 CrossRef CAS PubMed.
- R. Wei and W. Zimmermann, Microb. Biotechnol., 2017, 10, 1308–1322 CrossRef CAS PubMed.
- A. Barclay and K. R. Acharya, Biomolecules, 2023, 13, 1407 CrossRef CAS PubMed.
- N. Alves, J. F. Mano, E. Balaguer, J. M. Dueñas and J. G. Ribelles, Polymer, 2002, 43, 4111–4122 CrossRef CAS.
- F. Kawai, T. Kawabata and M. Oda, Appl. Microbiol. Biotechnol., 2019, 103, 4253–4268 CrossRef CAS PubMed.
- M. Goldsmith and D. S. Tawfik, Curr. Opin. Struct. Biol., 2017, 47, 140–150 CrossRef CAS PubMed.
- W. Zeng, X. Li, Y. Yang, J. Min, J.-W. Huang, W. Liu, D. Niu, X. Yang, X. Han and L. Zhang, ACS Catal., 2022, 12, 3033–3040 CrossRef CAS.
- S. Joo, I. J. Cho, H. Seo, H. F. Son, H.-Y. Sagong, T. J. Shin, S. Y. Choi, S. Y. Lee and K.-J. Kim, Nat. Commun., 2018, 9, 382 CrossRef PubMed.
- B. Liu, L. He, L. Wang, T. Li, C. Li, H. Liu, Y. Luo and R. Bao, ChemBioChem, 2018, 19, 1471–1475 CrossRef CAS PubMed.
- V. Tournier, C. Topham, A. Gilles, B. David, C. Folgoas, E. Moya-Leclair, E. Kamionka, M.-L. Desrousseaux, H. Texier and S. Gavalda, Nature, 2020, 580, 216–219 CrossRef CAS PubMed.
- U. Thumarat, T. Kawabata, M. Nakajima, H. Nakajima, A. Sugiyama, K. Yazaki, T. Tada, T. Waku, N. Tanaka and F. Kawai, J. Biosci. Bioeng., 2015, 120, 491–497 CrossRef CAS PubMed.
- H. F. Son, I. J. Cho, S. Joo, H. Seo, H.-Y. Sagong, S. Y. Choi, S. Y. Lee and K.-J. Kim, ACS Catal., 2019, 9, 3519–3526 CrossRef CAS.
- H. Lu, D. J. Diaz, N. J. Czarnecki, C. Zhu, W. Kim, R. Shroff, D. J. Acosta, B. R. Alexander, H. O. Cole and Y. Zhang, Nature, 2022, 604, 662–667 CrossRef CAS PubMed.
- A. N. Shirke, C. White, J. A. Englaender, A. Zwarycz, G. L. Butterfoss, R. J. Linhardt and R. A. Gross, Biochemistry, 2018, 57, 1190–1200 CrossRef CAS PubMed.
- Y. Cui, Y. Chen, X. Liu, S. Dong, Y. e. Tian, Y. Qiao, R. Mitra, J. Han, C. Li and X. Han, bioRxiv, 2019, preprint, 787069, DOI:10.1101/787069.
- K. Chen, Y. Hu, X. Dong and Y. Sun, ACS Catal., 2021, 11, 7358–7370 CrossRef CAS.
- H. P. Austin, M. D. Allen, B. S. Donohoe, N. A. Rorrer, F. L. Kearns, R. L. Silveira, B. C. Pollard, G. Dominick, R. Duman and K. El Omari, Proc. Natl. Acad. Sci. U. S. A., 2018, 115, E4350–E4357 CrossRef CAS PubMed.
- R. Wei, T. Oeser and W. Zimmermann, Adv. Appl. Microbiol., 2014, 89, 267–305 Search PubMed.
- L. D. Ellis, N. A. Rorrer, K. P. Sullivan, M. Otto, J. E. McGeehan, Y. Román-Leshkov, N. Wierckx and G. T. Beckham, Nat. Catal., 2021, 4, 539–556 CrossRef CAS.
- J. Kari, J. P. Olsen, K. Jensen, S. F. Badino, K. B. Krogh, K. Borch and P. Westh, ACS Catal., 2018, 8, 11966–11972 CrossRef CAS.
- A. Berselli, M. J. Ramos and M. C. Menziani, ChemBioChem, 2021, 22, 2032–2050 CrossRef CAS PubMed.
- F. Kawai, Catalysts, 2021, 11, 206 CrossRef CAS.
- E. Herrero Acero, D. Ribitsch, A. Dellacher, S. Zitzenbacher, A. Marold, G. Steinkellner, K. Gruber, H. Schwab and G. M. Guebitz, Biotechnol. Bioeng., 2013, 110, 2581–2590 CrossRef CAS PubMed.
- D. Ribitsch, A. Hromic, S. Zitzenbacher, B. Zartl, C. Gamerith, A. Pellis, A. Jungbauer, A. Łyskowski, G. Steinkellner and K. Gruber, Biotechnol. Bioeng., 2017, 114, 2481–2488 CrossRef CAS PubMed.
- T. Fecker, P. Galaz-Davison, F. Engelberger, Y. Narui, M. Sotomayor, L. P. Parra and C. A. Ramírez-Sarmiento, Biophys. J., 2018, 114, 1302–1312 CrossRef CAS PubMed.
- D. Quaglia, M. C. Ebert, P. F. Mugford and J. N. Pelletier, PLoS One, 2017, 12, e0171741 CrossRef PubMed.
- Q. Liu, G. Xun and Y. Feng, Biotechnol. Adv., 2019, 37, 530–537 CrossRef CAS PubMed.
- J. Wang, H. Zhang, D. Yin, X. Xu, T. Tan and Y. Lv, Synth. Syst. Biotechnol., 2021, 6, 163–172 CrossRef CAS PubMed.
- R. Araújo, C. Silva, A. O'Neill, N. Micaelo, G. Guebitz, C. M. Soares, M. Casal and A. Cavaco-Paulo, J. Biotechnol., 2007, 128, 849–857 CrossRef PubMed.
- F. Liu, T. Wang, W. Yang, Y. Zhang, Y. Gong, X. Fan, G. Wang, Z. Lu and J. Wang, Front. Bioeng. Biotechnol., 2023, 11, 1263996 CrossRef PubMed.
- Z. Chen, R. Duan, Y. Xiao, Y. Wei, H. Zhang, X. Sun, S. Wang, Y. Cheng, X. Wang and S. Tong, Nat. Commun., 2022, 13, 7138 CrossRef CAS PubMed.
- A. Bollinger, S. Thies, E. Knieps-Grünhagen, C. Gertzen, S. Kobus, A. Höppner, M. Ferrer, H. Gohlke, S. H. Smits and K.-E. Jaeger, Front. Microbiol., 2020, 11, 114 CrossRef PubMed.
- Y. Ma, M. Yao, B. Li, M. Ding, B. He, S. Chen, X. Zhou and Y. Yuan, Engineering, 2018, 4, 888–893 CrossRef CAS.
- C. Silva, S. Da, N. Silva, T. Matamá, R. Araújo, M. Martins, S. Chen, J. Chen, J. Wu and M. Casal, Biotechnol. J., 2011, 6, 1230–1239 CrossRef CAS PubMed.
- R. Wei, T. Oeser, J. Schmidt, R. Meier, M. Barth, J. Then and W. Zimmermann, Biotechnol. Bioeng., 2016, 113, 1658–1665 CrossRef CAS PubMed.
- M. Oda, Y. Yamagami, S. Inaba, T. Oida, M. Yamamoto, S. Kitajima and F. Kawai, Appl. Microbiol. Biotechnol., 2018, 102, 10067–10077 CrossRef CAS PubMed.
- E. L. Bell, R. Smithson, S. Kilbride, J. Foster, F. J. Hardy, S. Ramachandran, A. A. Tedstone, S. J. Haigh, A. A. Garforth and P. J. Day, Nat. Catal., 2022, 5, 673–681 CrossRef CAS.
- L. Pfaff, J. Gao, Z. Li, A. Jäckering, G. Weber, J. Mican, Y. Chen, W. Dong, X. Han and C. G. Feiler, ACS Catal., 2022, 12, 9790–9800 CrossRef CAS PubMed.
- M. Oda, Biophys. Physicobiol., 2021, 18, 168–176 CrossRef CAS PubMed.
- J. Then, R. Wei, T. Oeser, A. Gerdts, J. Schmidt, M. Barth and W. Zimmermann, FEBS Open Bio, 2016, 6, 425–432 CrossRef CAS PubMed.
- S. Brott, L. Pfaff, J. Schuricht, J. N. Schwarz, D. Böttcher, C. P. Badenhorst, R. Wei and U. T. Bornscheuer, Eng. Life Sci., 2022, 22, 192–203 CrossRef CAS PubMed.
- A. N. Shirke, D. Basore, S. Holton, A. Su, E. Baugh, G. L. Butterfoss, G. Makhatadze, C. Bystroff and R. A. Gross, Appl. Microbiol. Biotechnol., 2016, 100, 4435–4446 CrossRef CAS PubMed.
- A. Nakamura, N. Kobayashi, N. Koga and R. Iino, ACS Catal., 2021, 11, 8550–8564 CrossRef CAS.
- T. Hiraishi, N. Komiya and M. Maeda, Polym. Degrad. Stab., 2010, 95, 1370–1374 CrossRef CAS.
- A. Biundo, D. Ribitsch, G. Steinkellner, K. Gruber and G. M. Guebitz, Biotechnol. J., 2017, 12 CAS.
- C. Gamerith, E. H. Acero, A. Pellis, A. Ortner, R. Vielnascher, D. Luschnig, B. Zartl, K. Haernvall, S. Zitzenbacher and G. Strohmeier, Polym. Degrad. Stab., 2016, 132, 69–77 CrossRef CAS.
- Y. Zhang, S. Chen, M. Xu, A. Cavoco-Paulo, J. Wu and J. Chen, Appl. Environ. Microbiol., 2010, 76, 6870–6876 CrossRef CAS PubMed.
- D. Ribitsch, A. O. Yebra, S. Zitzenbacher, J. Wu, S. Nowitsch, G. Steinkellner, K. Greimel, A. Doliska, G. Oberdorfer and C. C. Gruber, Biomacromolecules, 2013, 14, 1769–1776 CrossRef CAS PubMed.
- L. Dai, Y. Qu, J.-W. Huang, Y. Hu, H. Hu, S. Li, C.-C. Chen and R.-T. Guo, J. Biotechnol., 2021, 334, 47–50 CrossRef CAS PubMed.
- R. Xue, Y. Chen, H. Rong, R. Wei, Z. Cui, J. Zhou, W. Dong and M. Jiang, Front. Bioeng. Biotechnol., 2021, 9, 762854 CrossRef PubMed.
- Z. Liu, Y. Zhang and J. Wu, Enzyme Microb. Technol., 2022, 156, 110004 CrossRef CAS PubMed.
- M. Liu, T. Zhang, L. Long, R. Zhang and S. Ding, Polym. Degrad. Stab., 2019, 160, 120–125 CrossRef CAS.
- B. C. Knott, E. Erickson, M. D. Allen, J. E. Gado, R. Graham, F. L. Kearns, I. Pardo, E. Topuzlu, J. J. Anderson and H. P. Austin, Proc. Natl. Acad. Sci. U. S. A., 2020, 117, 25476–25485 CrossRef CAS PubMed.
- T. Hiraishi, Y. Hirahara, Y. Doi, M. Maeda and S. Taguchi, Appl. Environ. Microbiol., 2006, 72, 7331–7338 CrossRef CAS PubMed.
- A. Sidar, E. D. Albuquerque, G. P. Voshol, A. F. Ram, E. Vijgenboom and P. J. Punt, Front. Bioeng. Biotechnol., 2020, 8, 871 CrossRef PubMed.
- J. Weber, D. Petrović, B. Strodel, S. H. Smits, S. Kolkenbrock, C. Leggewie and K.-E. Jaeger, Appl. Microbiol. Biotechnol., 2019, 103, 4801–4812 CrossRef CAS PubMed.
- Y. Zhang, L. Wang, J. Chen and J. Wu, Carbohydr. Polym., 2013, 97, 124–129 CrossRef CAS PubMed.
- C.-D. Tang, J.-F. Li, X.-H. Wei, R. Min, S.-J. Gao, J.-Q. Wang, X. Yin and M.-C. Wu, PLoS One, 2013, 8, e64766 CrossRef CAS PubMed.
- Y. Zhong, P. Godwin, Y. Jin and H. Xiao, Adv. Ind. Eng. Polym. Res., 2020, 3, 27–35 Search PubMed.
- G. E. Luckachan and C. Pillai, J. Polym. Environ., 2011, 19, 637–676 CrossRef CAS.
- M. E. Peterson, R. M. Daniel, M. J. Danson and R. Eisenthal, Biochem. J., 2007, 402, 331–337 CrossRef CAS PubMed.
- P. R. Sutkar, R. D. Gadewar and V. P. Dhulap, J. Hazard. Mater. Adv., 2023, 100343 CrossRef CAS.
- S. N. Dimassi, J. N. Hahladakis, M. N. D. Yahia, M. I. Ahmad, S. Sayadi and M. A. Al-Ghouti, Arabian J. Chem., 2022, 104262 CrossRef CAS.
- L. N. Ji, Appl. Mech. Mater., 2013, 312, 406–410 Search PubMed.
- D. Damayanti and H.-S. Wu, Polymers, 2021, 13, 1475 CrossRef PubMed.
- H. K. Webb, J. Arnott, R. J. Crawford and E. P. Ivanova, Polymers, 2012, 5, 1–18 CrossRef.
- D. Myalenko and O. Fedotova, Polymers, 2023, 15, 1619 CrossRef CAS PubMed.
- T. Narancic, M. Salvador, G. M. Hughes, N. Beagan, U. Abdulmutalib, S. T. Kenny, H. Wu, M. Saccomanno, J. Um and K. E. O'Connor, Microb. Biotechnol., 2021, 14, 2463–2480 CrossRef CAS PubMed.
- J. Oliveira, A. Belchior, V. D. da Silva, A. Rotter, Ž. Petrovski, P. L. Almeida, N. D. Lourenço and S. P. Gaudêncio, Front. Mar. Sci., 2020, 7, 567126 CrossRef.
- A. I. Osman, M. Hosny, A. S. Eltaweil, S. Omar, A. M. Elgarahy, M. Farghali, P.-S. Yap, Y.-S. Wu, S. Nagandran and K. Batumalaie, Environ. Chem. Lett., 2023, 21, 2129–2169 CrossRef CAS PubMed.
- M. E. Sevilla, M. D. Garcia, Y. Perez-Castillo, V. Armijos-Jaramillo, S. Casado, K. Vizuete, A. Debut and L. Cerda-Mejía, Polymers, 2023, 15, 1779 CrossRef CAS PubMed.
- X. Qi, Y. Ma, H. Chang, B. Li, M. Ding and Y. Yuan, Front. Microbiol., 2021, 12, 778828 CrossRef PubMed.
- A. Senga, Y. Hantani, G.-J. Bekker, N. Kamiya, Y. Kimura, F. Kawai and M. Oda, J. Biochem., 2019, 166, 149–156 CAS.
- N. F. S. Khairul Anuar, F. Huyop, G. Ur-Rehman, F. Abdullah, Y. M. Normi, M. K. Sabullah and R. Abdul Wahab, Int. J. Mol. Sci., 2022, 23, 12644 CrossRef CAS PubMed.
- N. A. Samak, Y. Jia, M. M. Sharshar, T. Mu, M. Yang, S. Peh and J. Xing, Environ. Int., 2020, 145, 106144 CrossRef CAS PubMed.
- A. Li, Y. Sheng, H. Cui, M. Wang, L. Wu, Y. Song, R. Yang, X. Li and H. Huang, Nat. Commun., 2023, 14, 4169 CrossRef CAS PubMed.
- J. W. Kim, S.-B. Park, Q.-G. Tran, D.-H. Cho, D.-Y. Choi, Y. J. Lee and H.-S. Kim, Microb. Cell Factories, 2020, 19, 1–9 CrossRef PubMed.
- W. Yang, Y. Li and D. Boraschi, Int. J. Mol. Sci., 2023, 24, 4065 CrossRef CAS PubMed.
- A. Amobonye, P. Bhagwat, S. Raveendran, S. Singh and S. Pillai, Front. Microbiol., 2021, 12, 3728 Search PubMed.
- A. Rafiq and J.-L. Xu, Waste Manage., 2023, 171, 54–70 CrossRef CAS PubMed.
- C. N. Reddy, P. Kallem, K. Mounika, A. Muqeet, J. C. J. Raj, C. Aishwarya, R. K. Gupta, V. Polisetti, B. Mishra and Y. Rajasri, Polym. Test., 2023, 108223 CrossRef CAS.
- M. S.-L. Yee, L.-W. Hii, C. K. Looi, W.-M. Lim, S.-F. Wong, Y.-Y. Kok, B.-K. Tan, C.-Y. Wong and C.-O. Leong, Nanomaterials, 2021, 11, 496 CrossRef CAS PubMed.
- L. C. De Sá, M. Oliveira, F. Ribeiro, T. L. Rocha and M. N. Futter, Sci. Total Environ., 2018, 645, 1029–1039 CrossRef PubMed.
- K. V. Malafeev, A. Apicella, L. Incarnato and P. Scarfato, Polymers, 2023, 15, 3680 CrossRef CAS PubMed.
- W. Liu, H. Liao, M. Wei, M. Junaid, G. Chen and J. Wang, TrAC, Trends Anal. Chem., 2023, 117477 Search PubMed.
- G. Limonta, A. Mancia, A. Benkhalqui, C. Bertolucci, L. Abelli, M. C. Fossi and C. Panti, Sci. Rep., 2019, 9, 15775 CrossRef PubMed.
- M. C. Guerrera, M. Aragona, C. Porcino, F. Fazio, R. Laurà, M. Levanti, G. Montalbano, G. Germanà, F. Abbate and A. Germanà, Appl. Sci., 2021, 11, 5768 CrossRef CAS.
- S. Liu, J. Shi, J. Wang, Y. Dai, H. Li, J. Li, X. Liu, X. Chen, Z. Wang and P. Zhang, Front. Microbiol., 2021, 12, 652520 CrossRef PubMed.
- Q. Chen, H. Zhao, Y. Liu, L. Jin and R. Peng, Toxics, 2023, 11, 490 CrossRef CAS PubMed.
- M. K. Viršek, M. N. Lovšin, Š. Koren, A. Kržan and M. Peterlin, Mar. Pollut. Bull., 2017, 125, 301–309 CrossRef PubMed.
- V. Zadjelovic, R. J. Wright, C. Borsetto, J. Quartey, T. N. Cairns, M. G. Langille, E. M. Wellington and J. A. Christie-Oleza, Microbiome, 2023, 11, 225 CrossRef CAS PubMed.
- P. Cholewińska, H. Moniuszko, K. Wojnarowski, P. Pokorny, N. Szeligowska, W. Dobicki, R. Polechoński and W. Górniak, Int. J. Environ. Res. Public Health, 2022, 19, 8137 CrossRef PubMed.
- A. Khan, Z. Jie, J. Wang, J. Nepal, N. Ullah, Z.-Y. Zhao, P.-Y. Wang, W. Ahmad, A. Khan and W. Wang, Sci. Total Environ., 2023, 165688 CrossRef CAS PubMed.
- S. Alqahtani, S. Alqahtani, Q. Saquib and F. Mohiddin, Front. Toxicol., 2023, 5, 1193386 CrossRef PubMed.
- W. Yang, N. Jannatun, Y. Zeng, T. Liu, G. Zhang, C. Chen and Y. Li, Front. Toxicol., 2022, 4, 956885 CrossRef PubMed.
- H. Rafeeq, N. Afsheen, S. Rafique, A. Arshad, M. Intisar, A. Hussain, M. Bilal and H. M. Iqbal, Chemosphere, 2023, 310, 136751 CrossRef CAS PubMed.
- J. S. Johnson, D. J. Spakowicz, B.-Y. Hong, L. M. Petersen, P. Demkowicz, L. Chen, S. R. Leopold, B. M. Hanson, H. O. Agresta and M. Gerstein, Nat. Commun., 2019, 10, 5029 CrossRef PubMed.
- T. Tinta, T. Kogovšek, K. Klun, A. Malej, G. J. Herndl and V. Turk, Mar. Drugs, 2019, 17, 94 CrossRef CAS PubMed.
Footnote |
| † These authors contributed equally. |
| This journal is © The Royal Society of Chemistry 2024 |