Open Access Article
Open Access ArticleUnderstanding the decomposition process of the Pt1Ag24(SPhCl2)18 nanocluster at the atomic level†
Kefan Jianga,
Along Maab,
Yuansheng Lia,
Jiawei Wanga,
Zhengmao Yin*a and
Shuxin Wang *ab
*ab
aCollege of Materials Science and Engineering, Qingdao University of Science and Technology, Qingdao 266042, P. R. China. E-mail: shuxin_wang@qust.edu.cn; yzm198752@163.com
bKey Laboratory of Optic-Electric Sensing and Analytical Chemistry for Life Science, MOE, Shandong Key Laboratory of Biochemical Analysis, College of Chemistry and Molecular Engineering, Qingdao University of Science and Technology, Qingdao 266042, China
First published on 2nd April 2024
Abstract
We report the decomposition of the Pt1Ag24(SPhCl2)18 nanocluster into a crown-like Pt1Ag4(SR)8 (SR = 2,4-SPhCl2 and 4-SPhBr) complex. UV-vis spectra and single crystal X-ray diffraction were used to track the structure-conversion process. Based on the total structure, the differences in ligand exchange rates at different sites and the effects on the stability were mapped out. This work can not only help us understand the ligand exchange behavior of the clusters, but also provide experimental support for the design of stable metal clusters.
1 Introduction
Atomically precise metal nanoclusters can help elucidate the precise relationship between structure and properties,1–15 which is crucial to comprehend the nanocluster stability,16–19 transformation mechanism and growth mechanism in solution.20–25 The interaction between clusters not only can help us understand the interaction of nanoparticles at the atomic level,26,27 but also aids in designing and synthesizing clusters of specific sizes or characteristics.28–32 Indeed, based on the ligand-exchange-induced size/structure transformation (LEIST) method, a series of clusters with monodispersity have been synthesized and their structures determined by Single crystal X-ray diffraction (SC-XRD).33–40 It is worth noting that there are two major behaviours in LEIST: (i) conversion. For instance, the size transformation of the Au38(SC2H4Ph)24 nanocluster to the Au36(SPhtBu)24 nanocluster induced by HSPhtBu ligands.33 More importantly, due to the precise structure, the mechanism of such a process can be studied at the atomic level; (ii) decomposition. In the LESIT method, partial or complete decomposition usually occurs.22,41,42 For example, the synthesis of the monodisperse Pt1Au24(SC2H4Ph)18 nanocluster is accompanied by the decomposition of the Au25(SC2H4Ph)18 nanocluster.41 In past research, the process and mechanism of cluster size transformation in LESIT have been extensively studied.33,43–46 However, understanding the decomposition process of nanoclusters at the atomic level is crucial for us to understand the origin of stability, but there are few studies.Herein, we monitored the decomposition of Pt1Ag24(SPhCl2)18 nanoclusters with UV-vis spectra and SC-XRD.47 We obtained three crystals, including two intermediates (Pt1Ag24(SPhCl2)7.51(SPhtBr)12.49, denoted as NC-I; Pt1Ag24(SPhCl2)2.28(SPhtBr)17.72, denoted as NC-II) and one decomposed product (Pt1Ag4(SPhCl2)4.31(SPhtBr)3.69, denoted as Pt1Ag4).45 Through the structural analysis of these crystals and calculating the occupancy ratio of exchanged ligands, the ligand exchange rate at different positions on the clusters has been mapped out.48 It is worth noting that, although some ligands are located at symmetrical positions on the C3 axis, the ligand exchange rates show significant differences. These subtle differences lead to the decomposition of the cluster.
2 Experimental methods
2.1 Chemicals
All reagents were purchased from XiLong Scientific and used directly without further purification. Silver nitrate (AgNO3, 99%, metal basis), potassium tetrachloroplatinate (II) (K2PtCl4; 99%, metal basis), 2,4-dichlorobenzenethiol (HSPh2,4Cl2, 99%), 4-bromothiophenol (HSPhtBr, 99%), tetraphenylphosphonium bromide (PPh4Br, 98%), sodium borohydride (NaBH4, 98%), triethylamine (C6H15N, 99.5%), dichloromethane (CH2Cl2, HPLC), methanol (CH3OH, HPLC), N,N-dimethylformamide (DMF, HPLC), hexane (C6H14, HPLC), and diethyl ether ((CH3CH2)2O, HPLC) were used.2.2 Synthesis of Pt1Ag24(SPhCl2)18 nanocluster
The sample of Pt1Ag24(SPhCl2)18 was synthesized based on the previously reported process with minor modifications.47 Firstly, AgNO3 (10 mg, 0.0588 mmol) was dissolved in CH3OH. CH2Cl2 and K2PtCl4 (10 mg, 0.0240 mmol) were then added to the solution to form a pale-yellow mixture. In an ice bath, the mixture was cooled to 0 °C. Then, 2,4-dichlorobenzenethiol (10 μL, 0.0791 mmol) and tetraphenylphosphonium bromide (10 mg, 0.0238 mmol) were added in turn. After 30 min, 1 mL NaBH4 aqueous solution (40 mg mL−1, 1.057 mmol) and triethylamine (50 μL, 0.358 mmol) were added quickly to the mixture under vigorous stirring. This reaction was further aged for 12 h at 0 °C. Whereafter, the aqueous phase was removed. The mixture in the organic phase was washed three times with (CH3CH2)2O (2 × 10 mL). After a week, dark-red crystals were acquired in CH2Cl2/hexane at room temperature.2.3 Conversion from Pt1Ag24(SPhCl2)18 to Pt1Ag4(SR)8
Briefly, Pt1Ag24(SPhCl2)18 (10 mg, 0.00167 mmol) crystals were dissolved in 5 mL DMF, then HSPhtBr was added in different molar ratios. The progress of the reaction is less dependent on the time, and is highly dependent on the molar ratio of HSPhtBr mercaptan ligand added. After the addition of 200 equivalents of HSPhtBr, all Pt1Ag24(SPhCl2)18 nanoclusters were converted to Pt1Ag4(SR)8 complexes. The crude products of Pt1Ag4(SR)8 were washed two times with (CH3CH2)2O (2 × 10 mL). Single crystal growth of Pt1Ag24(SR)20 and Pt1Ag4(SR)8 was accomplished by vapor diffusion of (CH3CH2)2O into a concentrated solution of the Pt1Ag4(SR)8 in CH2Cl2 for 2 days.2.4 Synthesis of NC-I and NC-II
In the process of transforming Pt1Ag24(SPhCl2)18 to Pt1Ag4(SR)8, the reaction was halted at a distinctive juncture. For NC-I, after 60 equivalents of HSPhtBr was added and followed by stirring 3 minutes, the reaction was terminated. For NC-II, after 100 equivalents of HSPhtBr was added and followed by stirring for 3 minutes, the reaction was terminated. The crude products of NC-I and NC-II were washed two times with (CH3CH2)2O (2 × 10 mL). Single crystal growth of NC-I and NC-II was accomplished through vapor diffusion of (CH3CH2)2O into a concentrated solution of the NC-I and NC-II in CH2Cl2 for 2 days.2.5 Characterization
All UV-vis absorption spectra of the structure conversion process were acquired on UV-1810 at room temperature. The data collections for single crystal X-ray diffraction were carried out on a STOE stadivari diffractometer. Data reductions and absorption corrections were performed using the X-Area. The structure was solved by intrinsic phasing and refined with full-matrix least squares on F2 using the SHELXTL software package. All non-hydrogen atoms were refined anisotropically, and all the hydrogen atoms were set in geometrically calculated positions and refined isotopically using a riding model. In order to confirm the ligand type of the cluster, the Br position was refined as free variables.3 Results and discussion
3.1 Monitoring the transformation process from Pt1Ag24(SPhCl2)18 to Pt1Ag4(SR)8
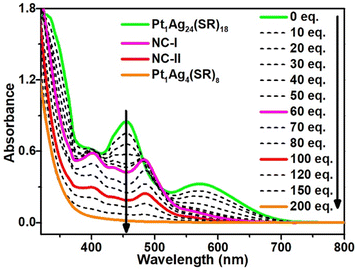
In this work, our focus centered on the decomposition of metal nanoclusters, and we selected Pt1Ag24(SPhCl2)18 (where, SPhCl2 = 2,4-dichlorothiolated) nanocluster as the model due to its metastability in the presence of aryl mercaptan.45,49–51 As shown in Fig. 1, the UV-vis absorption spectrum of Pt1Ag24(SPhCl2)18 nanocluster exhibits two characteristic absorption peaks at 455 nm and 570 nm, with a shoulder at 400 nm, consistent with the reported result.47 Pt1Ag24(SPhCl2)18 nanocluster was converted into Pt1Ag24(SR)20 (where SR is the mixture of 2,4-SPhCl2 and 4-SPhBr) nanocluster after adding less than 60 equivalents of 4-bromothiophenol ligand (HSPhtBr), which is similar to the reported results.45 Where characteristic absorptions of Pt1Ag24(SR)20 (at 400 nm, 435 nm, 480 nm, and 575 nm) became prominent, while those of Pt1Ag24(SPhCl2)18 (455 nm, 570 nm and 400 nm) diminished. In this series of spectra, four isosbestic points were discovered at 400, 420, 480, and 505 nm, signifying the process is quantitative. Simultaneously, the colour of the solution turned from atrovirens to reddish orange (Fig. S1†). The reason for the transformation from Pt1Ag24(SPhCl2)18 to Pt1Ag24(SR)20 is that three Ag2(SR)3 staple motifs of Pt1Ag24(SPhCl2)18 are aggregated to be one Ag6(SR)10 surface caps at nanocluster poles by the addition and induction of free thiol ligands. After adding 60 equivalents of 4-bromothiophenol ligand, the absorbance of the sample decreases as the amount of ligand increases. The constant peak position and the ratio of the different peaks indicate the clusters have been gradually decomposed into complexes. After adding 200 equivalents of HSPhtBr to the solution, there is no peak after 400 nm, indicating that the nanoclusters are completely decomposed into metal complexes. Concurrently, the colour of the solution turned from reddish orange to pale yellow (Fig. S1†).3.2 Crystal structure of Pt1Ag24(SR)20 and Pt1Ag4(SR)8
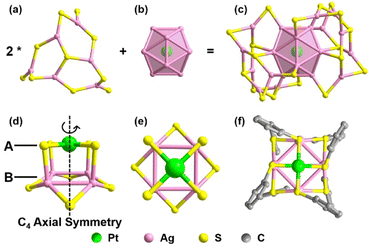
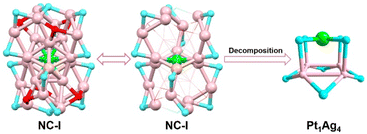
To elucidate the detailed decomposition process of this nanocluster, the reaction was stopped at the specific molar ratios.52 As shown in Fig. 1 (solid line), the crystal of Pt1Ag24(SPhCl2)7.51(SPhtBr)12.49 (magenta line, NC-I), Pt1Ag24(SPhCl2)2.28(SPhtBr)17.72 (red line, NC-II) and Pt1Ag4(SPhCl2)4.31(SPhtBr)3.69 (orange line, Pt1Ag4(SR)8) were obtained (Fig. S2†). SC-XRD was used to determine the overall structure of these crystals, respectively (Fig. S3†). Additionally, UV-vis spectra of NC-I, NC-II, and Pt1Ag4 crystals are shown in Fig. S4 and S5.† Results revealed that NC-I exhibits two strong absorption peaks at 400 nm and 482 nm, with two weaker peaks at 435 nm and 575 nm (Fig. S4†). Similarly, NC-II also showed two main absorption peaks (397 nm and 480 nm) and two weaker peaks (432 nm and 564 nm). Compared to NC-I, a blue shift was observed in the UV-vis spectrum of NC-II, attributed to different occupancy ratios of ligands. Pt1Ag4 composite only displayed a faint peak at 442 nm (Fig. S5†).The crystal structure analysis of Pt1Ag24(SR)20 and Pt1Ag4(SR)8 is presented in Fig. 2. For the Pt1Ag24(SR)20 nanocluster, the entire Pt1Ag24(SR)20 structure exhibits an icosahedral Pt1Ag12 metal kernel protected by two Ag6(SR)10 staple motifs (Fig. 2a–c).45 Consequently, the overall framework of Pt1Ag24(SR)20 can be regarded as “Pt1Ag12 + 2*Ag6(SR)10”. Each Ag6(SR)10 motif comprises three irregular hexagons (Fig. 2a), where three irregular hexagons are fused together by sharing the Ag–SR–Ag edges and terminated with three SR ligands. In the Pt1Ag4(SR)8 complex (Fig. 2d–f), the Pt atom exists independently of the Ag4 plane, with an average distance of 3.4 Å. Among the eight bridging thiolates, four bridging thiolates are bonded to Pt and Ag atoms, while the others link between two Ag atoms. Alternatively, the structure of Pt1Ag4(SR)8 can be viewed as two layers: the upper layer (A layer) resembles “a cross” with Pt1(SR)4, and the lower layer (B layer) is Ag4(SR)4, akin to “a plate”. Notably, the entire structure of Pt1Ag4(SR)8 exhibits C4 Axial Symmetry (Fig. 2d). The Ag–Ag bonds in the Pt1Ag4 kernel range from 3.067 to 3.165 Å (average: 3.115 Å), surpassing the Ag–Ag bond distance (2.889 Å) in bulk silver. Additionally, the Pt–S bonds range from 2.327 to 2.334 Å (average: 2.331 Å). There are two kinds of bridging thiolates in Pt1Ag4(SR)8 complex,53 with average Pt–S–Ag angle of 87.64 to 89.03° (average: 88.33°) and Ag–S–Ag angle of 77.22 to 79.63° (average: 78.26°). The clockwise and anticlockwise of the thiol ligands reveal the Pt1Ag4(SR)8 complex is racemic (Fig. 2f and S6†).25
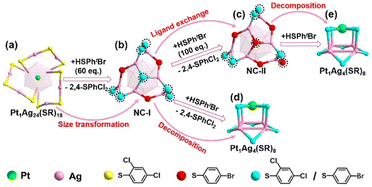
Building upon the aforementioned results, the transformation from Pt1Ag24(SPhCl2)18 to Pt1Ag4(SR)8 can be classified into two stages. A comprehensive diagram of the entire transformation process is depicted in Fig. 3. According to the symmetry of the crystal, only half of the Ag2(SR)3 motif for Pt1Ag24(SR)18 and the Ag6(SR)10 motif for Pt1Ag24(SR)20 are shown in Fig. 3 for clarity. To better illustrate the variation in ligand occupancy ratio and species during the conversion process, S atoms were used as substitutes for the entire mercaptan ligands. And all ligands were divided into three categories: ligands completely occupied by 2,4-SPhCl2 were highlighted in yellow, ligands absolutely occupied by 4-SPhBr were represented by red atoms and ligands occupied by both 2,4-SPhCl2 and 4-SPhBr were highlighted in turquois.
Stage 1: In the first stage (from 0 eq. to 60 eq.), Pt1Ag24(SPhCl2)18 was converted to Pt1Ag24(SPhCl2)7.51(SPhtBr)12.49 (NC-I), matching the UV-vis spectra from 0 eq. to 60 eq. (Fig. 1) and the transformation process from Pt1Ag24(SPhCl2)18 (Fig. 3a) to NC-I (Fig. 3b). The average number of 2,4-SPhCl2 reached ca. 7.51 in NC-I. Stage 2: In the second stage (from 60 eq. to 200 eq.), as shown in Fig. 3b–e, Pt1Ag24(SPhCl2)7.51(SPhtBr)12.49 (NC-I) was decomposed into Pt1Ag4(SR)8 or exchanged by the HSPhtBr ligand into Pt1Ag24(SPhCl2)2.28(SPhtBr)17.72 (NC-II) with the addition of HSPhtBr mercaptan. The average number of 2,4-SPhCl2 in NC-II reached a maximum of approximately 2.28. Further, NC-II was also decomposed into Pt1Ag4(SR)8. Detailed crystal parameters were provided in Tables S1–S3.† Additionally, the disparate occupancy ratios of ligands within NC-I and NC-II nanoclusters result in their respective crystallization in distinct crystal systems—triclinic and monoclinic systems (Fig. S7†). Furthermore, this further impacts the arrangement of cluster molecules, as although the cluster molecules of NC-I and NC-II are arranged in an “ABAB” pattern, the orientation of the molecules' arrangement is entirely different (Fig. S8†).15,54
3.3 The conversion mechanism from Pt1Ag24(SR)18 to Pt1Ag4(SR)8
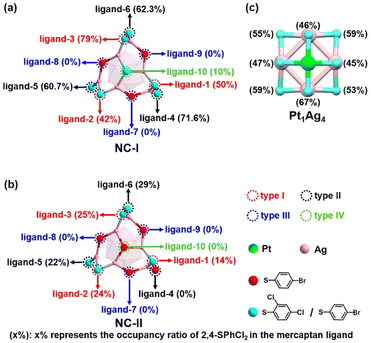
As illustrated in Fig. 3, both NC-I and NC-II nanoclusters show similar structures. The variance in the occupancy ratio of 2,4-SPhCl2 and 4-SPhBr ligands reveals the ligand exchange process of nanoclusters (Table S4†). Notably, NC-I and NC-II, along with Pt1Ag4, were examined for the occupancy ratios (x%) of 2,4-SPhCl2 in mercaptan ligands, as depicted in Fig. 4.45,55,56 To enhance clarity, we categorized ten mercaptan ligands into four types in NC-I and NC-II: type I (ligand-1, ligand-2, and ligand-3), type II (ligand-4, ligand-5, and ligand-6), type III (ligand-7, ligand-8, and ligand-9), and type IV (ligand-10).The occupancy ratios of 2,4-SPhCl2 and 4-SPhBr units in NC-I, NC-II, and Pt1Ag4 were summarized in Table S4,† and detailed information about related bonds is shown in Table S5.† Obviously, the exchange sites of type III (ligand-7, ligand-8 and ligand-9) were completely exchanged by 4-SPhBr ligands in NC-I (Fig. 4a), indicating that thiol ligands in type III are more susceptible to be exchanged. Similarly, the occupancy ratio of 2,4-SPhCl2 to type IV (ligand-10) is only 10% in NC-I (Fig. 4a), and is fully exchanged by 4-SPhBr in NC-II (Fig. 4b). The occupancy ratios of 2,4-SPhCl2 at these four ligand sites in NC-I and NC-II suggest that they are not critical for the decomposition of the Pt1Ag24(SR)20 nanocluster. For the ligands of type I, the occupancy ratio of 2,4-SPhCl2 to ligand-1 in NC-I was 50%, decreasing to 14% in NC-II. Concurrently, the ratio of ligand-2 and ligand-3 also decreased. Fig. 4 and Table S4† showed the following ratio of ligand-2: NC-I/NC-II = 42%/24%, and ligand-3: NC-I/NC-II = 79%/25%. Regarding the ligands of type II, at these positions, the occupancy ratios of 2,4-SPhCl2 in these three ligands significantly decrease from NC-I to NC-II (Fig. 4a, b and Table S4†). Specifically, in NC-I and NC-II, the occupancy ratios of 2,4-SPhCl2 in ligand-4 are 71.6% and 0% (Fig. 4a, b and Table S4†), respectively, indicating that 2,4-SPhCl2 units were fully exchanged by 4-SPhBr in NC-II. Fig. 4 and Table S4† showed the following occupancy ratios of 2,4-SPhCl2 in the ligands of type II, ligand-5: NC-I/NC-II = 60.7%/22% and ligand-6: NC-I/NC-II = 62.3%/29%, respectively. However, the relevant S–Ag bond lengths in NC-I and NC-II did not change significantly (Table S5†).
According to the analysis of the occupancy ratios of 2,4-SPhCl2 in the ten ligands, we found that four ligands of type III and type IV were independent of the decomposition of Pt1Ag24(SR)20. Additionally, due to the significantly higher occupancy ratios of 2,4-SPhCl2 in Pt1Ag4 compared to NC-II (Fig. 4b, c and Table S4†), and considering the abundant presence of HSPhtBr thiol ligands in the solution, it is impossible that the 2,4-SPhCl2 and 4-SPhBr units in Pt1Ag4 to originate from NC-II. Furthermore, for the six mercaptan ligands of type I and type II, the average occupancy ratio of the 2,4-SPhCl2 unit in NC-I (average: 0.609) is similar to the average occupancy ratio of 2,4-SPhCl2 in Pt1Ag4(SR)8 (average: 0.539) in Fig. 4a, c and Table S4.† Based on existing technology, it is difficult to directly observe the transfer process of a single ligand/metal in the nanocluster transformation process. It is reasonable to assume that the most likely reason for the similar ratio is that the ligands on the Pt1Ag4(SR)8 complex are inherited from the ligands of type I and type II in NC-I. We proposed the possible decomposition mechanism of the Pt1Ag24(SR)18 nanocluster (Fig. 5). One Pt atom, four Ag atoms and eight mercaptan ligands containing 2,4-SPhCl2 and 4-SPhBr units formed Pt1Ag4(SR)8. And one central Pt atom, twelve thiol ligands of type I and type II, and sixteen Ag atoms connected to twelve thiol ligands in NC-I are potential sources of one Pt atom, eight thiol ligands and four silver atoms in Pt1Ag4.
4 Conclusions
In summary, we have presented a study on the decomposition of Pt1Ag24(SPhCl2)18 nanocluster leading to the formation of the Pt1Ag4(SR)8 complex. This process was monitored using UV-vis spectra and SC-XRD. The pathway of transformation can be roughly divided into two stages: (I) HSPhtBr-triggered size transformation of Pt1Ag24(SPhCl2)18 forms NC-I; (II) structural decomposition of NC-I to Pt1Ag4(SR)8 and ligand exchange of NC-I to NC-II. And Pt1Ag4(SR)8 complex is inherited from the Pt atom, partial Ag atoms, and the ligand containing 2,4-SPhCl2 and 4-SPhBr in NC-I.Conflicts of interest
There are no conflicts to declare.Acknowledgements
We acknowledge the financial support provided by the National Natural Science Foundation of China (21803001 and 21771156) and Start-up Funding from Qingdao University of Science and Technology.Notes and references
- B. Bhattarai, Y. Zaker, A. Atnagulov, B. Yoon, U. Landman and T. P. Bigioni, Acc. Chem. Res., 2018, 51, 3104–3113 CrossRef CAS PubMed.
- A. W. Cook and T. W. Hayton, Acc. Chem. Res., 2018, 51, 2456–2464 CrossRef CAS PubMed.
- K. Konishi, M. Iwasaki and Y. Shichibu, Acc. Chem. Res., 2018, 51, 3125–3133 CrossRef CAS PubMed.
- Z. Lei, X. K. Wan, S. F. Yuan, Z. J. Guan and Q. M. Wang, Acc. Chem. Res., 2018, 51, 2465–2474 CrossRef CAS PubMed.
- R. L. Whetten, H. C. Weissker, J. J. Pelayo, S. M. Mullins, X. Lopez-Lozano and I. L. Garzon, Acc. Chem. Res., 2019, 52, 34–43 CrossRef CAS PubMed.
- Q. F. Yao, T. K. Chen, X. Yuan and J. P. Xie, Acc. Chem. Res., 2018, 51, 1338–1348 CrossRef CAS PubMed.
- Q.-F. Zhang, X. Chen and L.-S. Wang, Acc. Chem. Res., 2018, 51, 2159–2168 CrossRef CAS PubMed.
- Y. Z. Lu and W. Chen, Chem. Soc. Rev., 2012, 41, 3594–3623 RSC.
- S. Wang, S. Jin, S. Yang, S. Chen, Y. Song, J. Zhang and M. Zhu, Sci. Adv., 2015, 1, e1500441 CrossRef PubMed.
- L. Tang, Y. Luo, X. Ma, B. Wang, M. Ding, R. Wang, P. Wang, Y. Pei and S. Wang, Angew. Chem., Int. Ed., 2023, 62, e202300553 CrossRef CAS PubMed.
- L. Tang, A. Ma, C. Zhang, X. Liu, R. Jin and S. Wang, Angew. Chem., Int. Ed., 2021, 60, 17969–17973 CrossRef CAS PubMed.
- Y. Yun, H. Sheng, K. Bao, L. Xu, Y. Zhang, D. Astruc and M. Zhu, J. Am. Chem. Soc., 2020, 142, 4126–4130 CrossRef CAS PubMed.
- S. Wang, X. Meng, A. Das, T. Li, Y. Song, T. Cao, X. Zhu, M. Zhu and R. Jin, Angew. Chem., Int. Ed., 2014, 53, 2376–2380 CrossRef CAS PubMed.
- X. Wang, B. Yin, L. Jiang, C. Yang, Y. Liu, G. Zou, S. Chen and M. Zhu, Science, 2023, 381, 784–790 CrossRef CAS PubMed.
- A. Ma, J. Wang, Y. Wang, Y. Zuo, Y. Ren, X. Ma and S. Wang, Polyoxometalates, 2024, 3, 9140054 CrossRef.
- H. Shen, L. Wang, O. López-Estrada, C. Hu, Q. Wu, D. Cao, S. Malola, B. K. Teo, H. Häkkinen and N. Zheng, Nano Res., 2021, 14, 3303–3308 CrossRef CAS.
- Y. Li, H. K. Kim, R. D. McGillicuddy, S.-L. Zheng, K. J. Anderton, G. J. Stec, J. Lee, D. Cui and J. A. Mason, J. Am. Chem. Soc., 2023, 145, 9304–9312 CrossRef CAS PubMed.
- S. K. Barik, T.-H. Chiu, Y.-C. Liu, M.-H. Chiang, F. Gam, I. Chantrenne, S. Kahlal, J.-Y. Saillard and C. W. Liu, Nanoscale, 2019, 11, 14581–14586 RSC.
- H. Chen, L. Peng, Y. Bian, X. Shen, J. Li, H.-C. Yao, S.-Q. Zang and Z. Li, Appl. Catal., B, 2021, 284, 119704 CrossRef CAS.
- L. Tang, X. Kang, X. Wang, X. Zhang, X. Yuan and S. Wang, Inorg. Chem., 2021, 60, 3037–3045 CrossRef CAS PubMed.
- R. Itteboina, U. D. Madhuri, P. Ghosal, M. Kannan, T. K. Sau, T. Tsukuda and S. Bhardwaj, J. Phys. Chem. A, 2018, 122, 1228–1234 CrossRef CAS PubMed.
- Y. Negishi, H. Horihata, A. Ebina, S. Miyajima, M. Nakamoto, A. Ikeda, T. Kawawaki and S. Hossain, Chem. Sci., 2022, 13, 5546–5556 RSC.
- Y. Tan, Y. Lv, L. Xu, Q. Li, J. Chai, S. Yang, H. Yu and M. Zhu, J. Am. Chem. Soc., 2023, 145, 4238–4245 CrossRef CAS PubMed.
- S. Chen, S. Wang, J. Zhong, Y. Song, J. Zhang, H. Sheng, Y. Pei and M. Zhu, Angew. Chem., Int. Ed., 2015, 54, 3145–3149 CrossRef CAS PubMed.
- A. Ma, W. Du, J. Wang, K. Jiang, C. Zhang, W. Sheng, H. Zheng, R. Jin and S. Wang, J. Phys. Chem. Lett., 2023, 14, 5095–5101 CrossRef CAS PubMed.
- K. R. Krishnadas, A. Ghosh, A. Baksi, I. Chakraborty, G. Natarajan and T. Pradeep, J. Am. Chem. Soc., 2016, 138, 140–148 CrossRef CAS PubMed.
- K. R. Krishnadas, A. Baksi, A. Ghosh, G. Natarajan and T. Pradeep, Nat. Commun., 2016, 7, 13447 CrossRef CAS PubMed.
- L. Xu, Q. Li, T. Li, J. Chai, S. Yang and M. Zhu, Inorg. Chem. Front., 2021, 8, 4820–4827 RSC.
- Y.-D. Cao, D. Yin, S. Li, X.-Y. Dong, Y. Feng, H. Liu, L.-L. Fan, G.-G. Gao and S.-Q. Zang, Angew. Chem., Int. Ed., 2023, 62, e202307678 CrossRef CAS PubMed.
- H. Shan, J. Shi, T. Chen, Y. Cao, Q. Yao, H. An, Z. Yang, Z. Wu, Z. Jiang and J. Xie, ACS Nano, 2023, 17, 2368–2377 CrossRef CAS PubMed.
- X. Wei, X. Kang, Z. Zuo, F. Song, S. Wang and M. Zhu, Natl. Sci. Rev., 2020, 8, nwaa077 CrossRef PubMed.
- S. Chen, W. Du, C. Qin, D. Liu, L. Tang, Y. Liu, S. Wang and M. Zhu, Angew. Chem., Int. Ed., 2020, 59, 7542–7547 CrossRef CAS PubMed.
- C. J. Zeng, C. Y. Liu, Y. Pei and R. C. Jin, ACS Nano, 2013, 7, 6138–6145 CrossRef CAS PubMed.
- M. S. Bootharaju, C. P. Joshi, M. J. Alhilaly and O. M. Bakr, Chem. Mater., 2016, 28, 3292–3297 CrossRef CAS.
- H. W. Dong, L. W. Liao and Z. K. Wu, J. Phys. Chem. C, 2017, 8, 5338–5343 CAS.
- A. Baghdasaryan, E. Brun, Y. Wang, G. Salassa, J. Lacour and T. Buergi, Chem. Sci., 2021, 12, 7419–7427 RSC.
- C. L. Heinecke, T. W. Ni, S. Malola, V. Makinen, O. A. Wong, H. Hakkinen and C. J. Ackerson, J. Am. Chem. Soc., 2012, 134, 13316–13322 CrossRef CAS PubMed.
- Z. H. Tang, T. Ahuja, S. M. Wang and G. L. Wang, Nanoscale, 2012, 4, 4119–4124 RSC.
- W. Du, S. Jin, L. Xiong, M. Chen, J. Zhang, X. Zou, Y. Pei, S. Wang and M. Zhu, J. Am. Chem. Soc., 2017, 139, 1618–1624 CrossRef CAS PubMed.
- X. Zou, X. Kang and M. Zhu, Chem. Soc. Rev., 2023, 52, 5892–5967 RSC.
- H. F. Qian, D. E. Jiang, G. Li, C. Gayathri, A. Das, R. R. Gil and R. C. Jin, J. Am. Chem. Soc., 2012, 134, 16159–16162 CrossRef CAS PubMed.
- Y. Niihori, W. Kurashige, M. Matsuzaki and Y. Negishi, Nanoscale, 2013, 5, 508–512 RSC.
- T. Dainese, S. Antonello, S. Bogialli, W. W. Fei, A. Venzo and F. Maran, ACS Nano, 2018, 12, 7057–7066 CrossRef CAS PubMed.
- Y. A. Wang, B. Nieto-Ortega and T. Burgi, Chem. Commun., 2019, 55, 14914–14917 RSC.
- X. W. X. Kang, S. Jin, S. Wang and M. Zhu, CCS Chem., 2021, 3, 1929–1939 CrossRef.
- J.-H. Yu, Z.-R. Yuan, J. Xu, J.-G. Wang, M. Azam, T.-D. Li, Y.-Z. Li and D. Sun, Chem. Sci., 2023, 14, 6564–6571 RSC.
- J. Z. Yan, H. F. Su, H. Y. Yang, S. Malola, S. C. Lin, H. Hakkinen and N. F. Zheng, J. Am. Chem. Soc., 2015, 137, 11880–11883 CrossRef CAS PubMed.
- K. H. Wijesinghe, N. A. Sakthivel, T. Jones and A. Dass, J. Phys. Chem. C, 2020, 11, 6312–6319 CAS.
- H. Shen, E. Selenius, P. Ruan, X. Li, P. Yuan, O. Lopez-Estrada, S. Malola, S. Lin, B. K. Teo, H. Häkkinen and N. Zheng, Chem.–Eur. J., 2020, 26, 8465–8470 CrossRef CAS PubMed.
- Q. Z. Li, S. Yang, T. Chen, S. Jin, J. S. Chai, H. Zhang and M. Z. Zhu, Nanoscale, 2020, 12, 23694–23699 RSC.
- C. Zhu, T. Duan, H. Li, X. Wei, X. Kang, Y. Pei and M. Zhu, Inorg. Chem. Front., 2021, 8, 4407–4414 RSC.
- S. Wang, Y. Tan, T. Li, Q. Zhou, P. Li, S. Yang, H. Yu and M. Zhu, Inorg. Chem., 2022, 61, 18450–18457 CrossRef CAS PubMed.
- C. Zeng, T. Li, A. Das, N. L. Rosi and R. Jin, J. Am. Chem. Soc., 2013, 135, 10011–10013 CrossRef CAS PubMed.
- A. Ma, J. Wang, J. Kong, Y. Ren, Y. Wang, X. Ma, M. Zhou and S. Wang, Phys. Chem. Chem. Phys., 2023, 25, 9772–9778 RSC.
- N. A. Sakthivel, M. Stener, L. Sementa, M. Medves, G. Ramakrishna, A. Fortunelli, A. G. Oliver and A. Dass, J. Phys. Chem. C, 2019, 123, 29484–29494 CrossRef CAS.
- C. Kumara, C. M. Aikens and A. Dass, J. Phys. Chem. Lett., 2014, 5, 461–466 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available: Details of the experiment, characterization and supporting figures. CCDC 2327471–2327473. For ESI and crystallographic data in CIF or other electronic format see DOI: https://doi.org/10.1039/d4ra01274g |
| This journal is © The Royal Society of Chemistry 2024 |