Open Access Article
Open Access ArticleA sequential liquid dispensing method in a centrifugal microfluidic device operating at a constant rotational speed for the multiplexed genetic detection of foodborne pathogens†
Daigo Natsuhara *a,
Yuka Kibab,
Ryogo Saitoc,
Shunya Okamoto
*a,
Yuka Kibab,
Ryogo Saitoc,
Shunya Okamoto c,
Moeto Nagai
c,
Moeto Nagai cd,
Yusuke Yamauchi
cd,
Yusuke Yamauchi *ae,
Masashi Kitamurab and
Takayuki Shibata
*ae,
Masashi Kitamurab and
Takayuki Shibata *c
*c
aDepartment of Materials Process Engineering, Graduate School of Engineering, Nagoya University, Nagoya, Aichi 464-8603, Japan. E-mail: d-natsuhara@mems.me.tut.ac.jp
bFaculty of Pharmacy and Pharmaceutical Sciences, Josai University, Sakado, Saitama 350-0295, Japan
cDepartment of Mechanical Engineering, Toyohashi University of Technology, Toyohashi, Aichi 441-8580, Japan. E-mail: shibata@me.tut.ac.jp
dInstitute for Research on Next-generation Semiconductor and Sensing Science (IRES2), Toyohashi University of Technology, Aichi 441-8580, Japan
eAustralian Institute for Bioengineering and Nanotechnology (AIBN), The University of Queensland, St. Lucia, QLD 4072, Australia. E-mail: y.yamauchi@uq.edu.au
First published on 17th July 2024
Abstract
This study proposes a sequential liquid dispensing method using a centrifugal microfluidic device operating at a constant rotational speed for the multiplexed genetic detection of nucleic acid targets across multiple samples in a single operation. A pair of passive valves integrated into each microchamber enabled the liquid to fill towards the center of rotation against the centrifugal force, facilitating the complete removal of air inside the microchamber. Liquid manipulation can be achievable without any surface coating of the device by exploiting the inherent hydrophobicity of the polymer. Furthermore, design guidelines for the optimization of microfluidic devices are clarified. Consequently, our proposed method allows direct liquid dispensing into the reaction chambers without cross-contamination while simultaneously metering the sample/reagent volume for the colorimetric loop-mediated isothermal amplification (LAMP) reaction. In addition, we demonstrated the simultaneous detection of four foodborne pathogens (Salmonella spp., Vibrio parahaemolyticus, Campylobacter spp., and norovirus genogroup II (GII)) across four samples in a centrifugal microfluidic device within 60 min. Furthermore, the device exhibited high quantitation (R2 > 0.98) of the DNA concentration in the sample. Our proposed method enables a more compact design by eliminating the need for metering chambers and offers a point-of-care testing platform with high simplicity as it operates at a constant rotational speed.
Introduction
Every year, unsafe foods cause millions of foodborne illnesses and deaths worldwide.1–4 The incidence of foodborne illnesses from consuming foods contaminated with harmful pathogens (viruses, bacteria, fungi, and parasites) has consistently increased in recent years.5 Rapidly identifying the sources of foodborne illnesses can aid in controlling risks and ensuring food safety. However, traditional pathogen detection methods rely on culture, followed by verification, which is time-consuming.6 Therefore, there is a demand for a rapid point-of-care testing tool to detect foodborne pathogens.7,8 The loop-mediated isothermal amplification (LAMP) method is promising for on-site detection due to its advantages, such as being performed at a constant temperature (60–65 °C) in a single step.9–11 The target nucleic acid can be amplified through the LAMP reaction by incubation with four to six primers and a thermostable DNA polymerase with strand displacement activity. Recently, many studies have demonstrated the feasibility of LAMP for detecting pathogens that cause infectious diseases in seafood,12–14 meat,15,16 and other foods.17–19 However, the LAMP assay in a conventional PCR tube requires the preparation and individual testing of multiple sample/reagent mixtures for each pathogen, which is a time-consuming process and consumes a substantial volume of reagents (typically 10–25 μL) for each test.Centrifugal microfluidic technology has been harnessed to address these concerns for the LAMP assays to detect multiple foodborne pathogens.20–22 A centrifugal microfluidic device utilizes centrifugal force to realize automated microfluidic operations such as pumping, sample mixing, timed valving, flow switching, volume metering, and dispensing into multiple chambers.23–27 As centrifugal microfluidic devices can introduce multiple samples/reagents simultaneously, they have the potential for on-site, high-throughput, and multiplexed LAMP assays in multiple samples. Sayad et al.28 detected nucleic acids derived from three pathogens (Salmonella spp., Vibrio cholerae, and Escherichia coli) using LAMP in a centrifugal microfluidic device. This device facilitated mixing LAMP reagents and primers, metering, and dispensing the mixture into six sets of five microchambers, followed by a sealing process to prevent liquid evaporation in the microchambers. After the LAMP reaction, the microchambers detecting the pathogens exhibited a color change, indicating a positive reaction without cross-contamination within 60 min. Furthermore, an endpoint detection system with an optical sensor was used to assess the color difference between the positive and negative reactions for amplicon analysis. Seo et al.29 developed a centrifugal microfluidic device for detecting colorimetric, high-throughput foodborne pathogens. The sample/reagent was divided into 24 aliquoted chambers and injected into each microchamber by passing it through individual capillary valves. This device demonstrated the multiplexed detection of Vibrio parahaemolyticus and E. coli within 60 min based on colorimetric LAMP. Furthermore, they recently developed a centrifugal microfluidic device aiming toward automated, high-throughput, sample-to-answer multiplexed nucleic acid detection in multiple samples.30–34 These advancements can significantly increase the throughput of on-site detection using large sample numbers. However, this device requires multiple operational steps to control the appropriate rotational speed and/or direction (clockwise and counterclockwise) for liquid manipulation. To ensure complete liquid loading, each step required a longer rotation and transition times associated with the changing steps, prolonging the entire process. At least two rotational speed steps are required to dispense the sample/reagents into the microchambers to prevent cross-contamination between the microchambers.35 Moreover, a hydrophobic coating on the capillary valves is necessary to withstand the centrifugal force for liquid metering before dispensing into the microchambers, whereas a hydrophilic coating on the siphon valve is necessary to ensure reliable capillary action.33,34 Consequently, the coating condition affects the stability and performance of the microfluidic device, making long-term storage challenging.
In our previous studies,36–40 we developed a versatile microfluidic device for the multiplex detection of target nucleic acids based on the LAMP method. This technology has applications in a wide range of fields, including crop disease detection,36 rapid identification of toxic plants for emergency medical care,37 diagnosis of infectious disease,38 and food allergen detection.39 The microfluidic device allows the sequential dispensing of sample/reagent mixtures into an array of reaction microchambers in a single operation without surface coating by exploiting the inherent hydrophobicity of the polymer. In addition, we have developed a quantification system for nucleic acid targets by analyzing the color change in each reaction chamber from time-lapse images acquired during the LAMP reaction.40 This microfluidics-based system could be a platform for rapid and easy sample-to-answer diagnostics. However, using a syringe pump or electric pipette to introduce the sample/reagent mixture into the microfluidic device is time-consuming because multiple operations are required for each sample introduction. In this study, we developed a centrifugal microfluidic device capable of sequentially dispensing multiple sample/reagent mixtures into multiple microchambers not only at a constant rotational speed but also without any surface coating. First, we designed a microchamber integrated with a pair of passive valves capable of inwardly dispensing liquids into the microchambers. Secondly, we propose a microfluidic flow control theory for sequential liquid dispensing into multiple microchambers, which provides design guidelines for device optimization. Herein, we propose a sophisticated microchannel design to reduce the pressure applied to a passive valve for its rapid introduction into an array of microchambers. Moreover, we demonstrated the simultaneous detection of four foodborne pathogens (Salmonella spp., Vibrio parahaemolyticus, Campylobacter spp., and norovirus genogroup II (GII)) across four samples in a centrifugal microfluidic device within 60 min without any cross-contamination.
Experimental
Fabrication process of the centrifugal microfluidic device
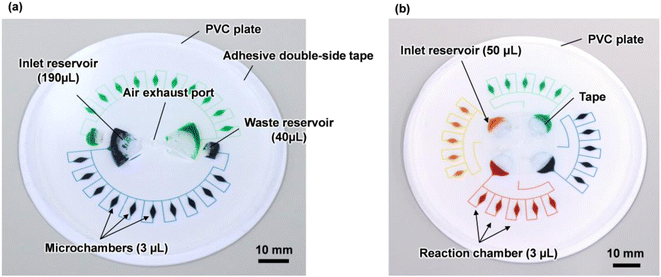
Fig. 1a shows a fabricated polydimethylsiloxane (PDMS)-based centrifugal microfluidic device for the multiplexed genetic diagnosis of foodborne pathogens, such as bacteria and viruses, to ensure food safety and sanitation. The diameter of the microfluidic device was approximately 80 mm. The device consisted of two sets of ten semi-elliptical microchambers (max. 1 mm in depth and 3 μL in volume) that were connected to individual one fan-shaped inlet reservoir (2 mm in depth and 190 μL in volume) and one fan-shaped waste reservoir (2 mm in depth and 40 μL in volume) via two independent rectangular microchannels (200 μm in width and 100 μm in height). The waste reservoir was connected to the air exhaust port to exhaust air from both the microchannel and microchambers during sequential liquid dispensing into an array of ten microchambers. This device allows simultaneous genetic testing of up to two samples and ten types of specific nucleic acid (DNA/RNA) targets in a single operation.The fabrication process is briefly illustrated as follows (Fig. S1†): initially, a thick negative photoresist (SU-8 3050; MicroChem, Newton, MA, USA) was patterned on a 4-inch single-crystal silicon wafer (e-Prize, Yokohama, Japan) as a mold using a single-step photolithography technique. To create deep microchamber structures (max. 1 mm in depth), pieces of wax of 2.7 mg (Ferris File-A-Wax; Freeman Manufacturing & Supply, Avon, OH, USA) were placed at the center of each SU-8 chamber pattern. A reflow process was then conducted by heating the mold on a hotplate at 120 °C for 15 min (EC1200-N; AS ONE, Osaka, Japan).39 Subsequently, 3D printed polylactic acid (PLA) parts (2 mm in depth) were fabricated using a fused filament fabrication 3D printer (Flashforge Adventurer 4; Apple Tree, Osaka, Japan) to create both the inlet and waste reservoirs (2 mm in depth). They were manually glued with an epoxy adhesive (Araldite, Huntsman Japan, Kobe, Japan) on the surface of the patterned SU-8 of inlet/waste reservoirs (Fig. S2a†). The SU-8 master mold with the semi-elliptical wax structures and 3D printed PLA parts was replicated in PDMS (Silpot 184; Dow Corning Toray, Tokyo, Japan) after curing on a hot plate at 80 °C for 40 min. After peeling off the PDMS from the SU-8 master mold, circular holes (with a diameter of 2.0 mm) for two inlet ports and one air exhaust port were punched into the PDMS device using a biopsy punch piercing tool (Kai Industries, Gifu, Japan). In the experiments, the sample/reagent mixture was introduced into each inlet reservoir by inserting a micropipette tip (ep Dualfilter T.I.P.S. 2–100 μL; Eppendorf, Wesseling-Berzdorf, Germany) through the punched inlet ports. Finally, both the microchambers and microchannels on the PDMS surface were sealed with a white polyvinyl chloride (PVC) plate (EB-235; Hikari, Osaka, Japan) using silicone-based adhesive double-sided tape (No. 5303 W; Nitto Denko, Osaka, Japan). This device was used to construct an optimal design theory for sequential liquid dispensing into an array of multiple microchambers using centrifugal force.
Furthermore, another microfluidic device consisting of four sets of five microchambers capable of simultaneous genetic testing of up to four samples and five types of specific nucleic acid targets was used for multiplexed colorimetric LAMP assays (Fig. 1b and S2b†). This device removes the waste reservoir and air exhaust port, and the air exhaust microchannel is connected to the inlet reservoir via a common central position. This configuration allowed air to be exhausted while the liquid was introduced into the microchambers to return to the empty space in the inlet reservoir. Thus, liquid dispensing can be achieved by centrifugal force even when the inlet port is sealed (without any air exhaust port), thereby mitigating the risk of contamination from leakage through the inlet ports (e.g. droplets and aerosols). The fabrication process of the inlet reservoirs was replaced by a reflow process using 40 mg of wax from the manual adhesive bonding process of the PLA parts, resulting in the creation of spherical inlet reservoirs (max. 1.8 mm in depth and 45 μL in volume), to improve the reproducibility of the process.
Stroboscopic imaging equipment
The centrifugal microfluidic device was tested using a homemade stroboscopic imaging system (Fig. S3†). A spin coater (MS-B100; Mikasa, Tokyo, Japan) was used to precisely control the rotational speed of the microfluidic device. The still image per rotation of the device was acquired using a combination of a complementary metal-oxide semiconductor (CMOS) camera (MQ022CG-CM; Ximea, Münster, Germany) with a fixed focal lens (M112FM12; Tamron, Saitama, Japan) and a stroboscope (LH-15M/CF-15S60; Nisshin Electronic, Saitama, Japan). The shutter timings of the CMOS camera and the pulsed light of the stroboscope were synchronized using a microcomputer (Arduino Leonardo; Arduino, MA, USA) by detecting the slit formed at one location of the film mask using a photo interrupter (CNZ1023; Panasonic, Osaka, Japan). Using the imaging system, we observed the flow behavior during the sequential dispensing of the liquid to fill the microchambers under centrifugal force.Operating procedure for the colorimetric quantitative LAMP assay in the microfluidic device
The operating procedure for the multiplexed colorimetric LAMP assay in the centrifugal microfluidic device for the simultaneous genetic diagnosis of foodborne pathogens (three types of bacteria and one type of virus) is as follows: first, four specific primer sets (0.5 μL of Salmonella spp. and Vibrio parahaemolyticus, 0.3 μL of Campylobacter spp. and norovirus GII) designed to amplify the targeted nucleic acids were pre-spotted and dried in each reaction chamber (volume ≈ 3 μL) on a hot plate at 80 °C for 3 min. Subsequently, the microchambers and microchannels on the PDMS surface were sealed with a white PVC plate (diameter of 80 mm) using silicone-based adhesive double-sided tape. A mixture of a positive control template (DNA or RNA) and LAMP reagents (35 μL in volume) was pipetted through each inlet port into each inlet reservoir. Subsequently, the inlet ports were sealed with a silicone-based adhesive double-sided tape. Next, the device was rotated at a constant rotational speed of 1000 min−1 on the homemade stroboscopic imaging system, thus simultaneously dispensing the mixture to fill four sets of five microchambers within 50 s. Once the microchambers were filled, the device was secured using a homemade clamp (Fig. S4†). Finally, the device was immersed in a hot-water bath (TB-2N; AS ONE, Osaka, Japan) to amplify target nucleic acids via the LAMP reaction under isothermal conditions at 60 °C for 60 min. Time-lapse images were captured every 30 s during the LAMP reaction. After the LAMP assay, the time-dependent color change of each microchamber was automatically analyzed using a homebuilt hue-based analysis software from a series of time-lapse images of the device.40 The operating procedure is illustrated in Fig. S5.†In this study, the LAMP primer sets for detecting Salmonella spp. and Vibrio parahaemolyticus were obtained from the studies of Ou et al.41 and Shen et al.,42 previously. Each positive control template was designed based on V. parahaemolyticus OpaR gene (DDBJ/EMBL/GenBank accession no. AF035967) and S. typhimurium InvA (accession no. M90846). The LAMP primers and template sequences used to detect Salmonella and V. parahaemolyticus were listed in Tables S1 and S2,† respectively. The primers and templates were purchased from Eurofins Genomics (Tokyo, Japan). Commercially available diagnostic kits containing primer sets and positive control templates for detecting Campylobacter spp. and norovirus GII were purchased from Eiken Chemical Co. Ltd. (Tokyo, Japan). The Loopamp® RNA amplification kit (Eiken Chemical, Tokyo, Japan), including a 2× reaction mixture and enzyme mix (a mixture of Bst polymerase and AMV reverse transcriptase), was used to perform the Reverse Transcription LAMP reactions because the positive control for norovirus is provided in the form of an RNA template. Hydroxynaphthol blue (HNB; FUJIFILM Wako Pure Chemical, Osaka, Japan) was used as an indicator for colorimetric LAMP reactions, which showed a color change from violet to sky-blue, indicating a positive reaction. The final concentration of HNB in each microchamber was adjusted to 150 μM in the mixture of the template and LAMP reagents. A total of 37.5 μL of the mixture containing the nucleic acid sample and LAMP reagents was prepared for colorimetric LAMP assays in the microfluidic device (Table S3†). For comparison, conventional off-chip 25 μL LAMP assays (Table S4†) were conducted in 0.2 mL PCR tubes (SnapStrip® II PCR Tubes; Scientific Specialties, CA, USA) using a real-time turbidimeter (LoopampEXIA; Eiken Chemical, Tokyo, Japan).
Results and discussions
Outward sequential liquid dispensing into multiple microchambers
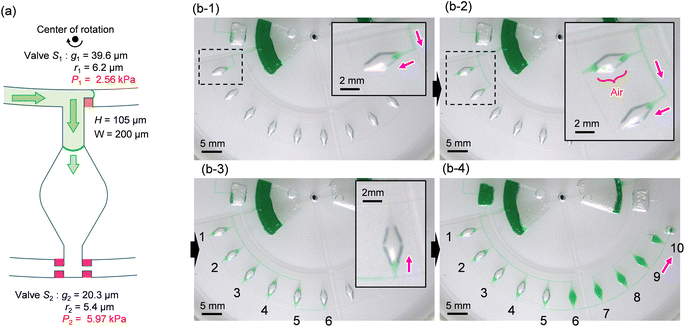
To ensure the sequential dispensing of a sample/reagent mixture into an array of reaction microchambers for the LAMP assay, it is necessary to completely eliminate air from the microchambers. After dispensing the mixture, the air in the microchamber can affect diagnostic accuracy because the residual air might act as nuclei for the growth of bubbles during the LAMP assay. Furthermore, it is essential to prevent liquid backflow between the microchambers. This prevents cross-contamination by leakage of LAMP primer sets fixed in the microchambers beforehand, thereby avoiding false diagnoses.First, we designed the microchambers integrated with a pair of passive valves (temporary stop valve S1 and permanent stop valve S2), as shown in Fig. 2a. In our previous studies,36–40 liquid was sequentially dispensed into multiple microchambers using an external pumping system (i.e., a syringe pump or electric pipette) by controlling the burst pressures of the passive valves integrated into each microchamber. In brief, the dispensing procedure was as follows. First, the liquid flow stopped after reaching the temporary stop valve S1 (burst pressure P1) because the liquid–air meniscus was pinned on the back edge of valve S1 and redirected toward the microchamber. After the microchamber was filled with liquid, the liquid flow was stopped at the permanent stop valve S2 (burst pressure P2), and the liquid flowed toward the next microchamber by passing through valve S1 because P1 < P2. Valve S2 also helps exhaust air in the microchamber. This process was repeated to sequentially fill all the microchambers with liquid. Here, the theoretical burst pressures of temporary stop valve S1 and permanent stop valve S2, which are designed as single-faced and double-faced stop valves, respectively, are described as follows:39
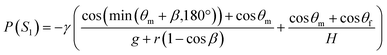 | (1) |
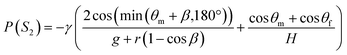 | (2) |
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) β) in the first term is the redefined gap distance between a specific position on the corner at an angle β and the vertical sidewall of the microchannel, where β is the angle between the direction perpendicular to the longitudinal direction of the microchannel and the position of the liquid–air meniscus corresponding to being pinned at the corner of the convex structure.
β) in the first term is the redefined gap distance between a specific position on the corner at an angle β and the vertical sidewall of the microchannel, where β is the angle between the direction perpendicular to the longitudinal direction of the microchannel and the position of the liquid–air meniscus corresponding to being pinned at the corner of the convex structure.
Fig. 2b shows a typical experimental result, demonstrating outward sequential liquid dispensing into ten microchambers at a rotational speed of 1500 min−1, where green-colored water (100 μL) was introduced into the inlet reservoir. According to eqn (1) and (2), the resulting burst pressures of valves S1 and S2 were calculated to be 2.56 kPa and 5.97 kPa for the fabricated device, where for valves S1 and S2, the gap distances g1 = 39.6 μm and g2 = 20.3 μm, and the corner radius at the back edge of the convex structures was r1 = 6.2 μm (β = 62°) and r2 = 5.4 μm (β = 53°), respectively. The width and height of the microchannel were W = 200 μm and H = 105 μm, respectively. After the liquid reached valve S1 of the first chamber, the flow was redirected toward the chamber and reached the chamber entrance (b-1). However, the liquid droplets formed at the inlet side of the chamber were ejected, thereby blocking the outlet side of the chamber (b-2). As a result, air was trapped inside the chamber, preventing liquid filling. Immediately after air trapping, the flow toward the second chamber occurred by passing through S1 of the first chamber. The same unexpected behavior was observed up to the fifth chamber (b-3). In contrast, the sixth chamber was completely filled with liquid flowing from the outlet side of the fifth chamber. However, the flow direction of the liquid is inward, opposite to the direction of the centrifugal force (b-4). In addition, continuous liquid flow into the chamber occurred, completely exhausting the air inside the chamber. After the sixth chamber was filled, the flow direction was changed toward the seventh chamber by passing through valve S2 of the sixth chamber. Similarly, the liquid was sequentially dispensed inward up to the tenth chamber.
Inward sequential liquid dispensing into multiple microchambers
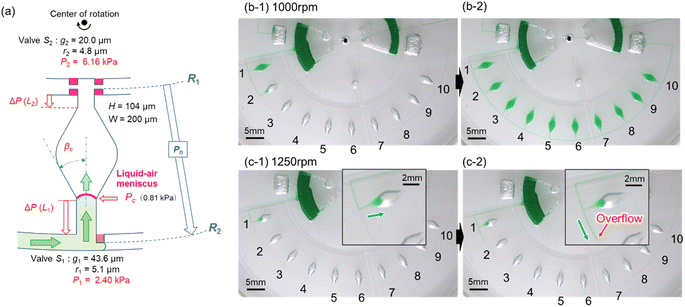
As described above, filling the liquid towards the center of rotation facilitated the complete air removal inside the microchamber. Therefore, we considered a newly designed valve configuration in which the arrangement of the two stop valves was reversed. As shown in Fig. 3a, temporary stop valve S1 and permanent stop valve S2 were located outward and inward, respectively, with respect to the center of rotation. The resulting burst pressure of valve S1 and S2 were calculated to be 2.41 kPa and 6.15 kPa for the fabricated device, where for valves S1 and S2, the gap distances were g1 = 43.6 μm and g2 = 20.0 μm, and the corner radius at the back edge of the convex structures was r1 = 5.1 μm (β = 65°) and r2 = 4.8 μm (β = 54°), respectively. The width and height of the microchannel were W = 200 μm and H = 104 μm, respectively.Fig. 3b shows a typical experimental result, demonstrating inward sequential liquid dispensing into ten microchambers at a rotational speed of 1000 min−1, where green-colored water (100 μL) was introduced into the inlet reservoir. As expected, the liquid flow stopped after reaching temporary stop valve S1 of the first chamber and was redirected toward the chamber. Subsequently, the first chamber was filled with liquid that continued to flow against the centrifugal force while simultaneously completely exhausting the air inside the chamber (b-1). Once the flow of the liquid was stopped at the permanent stop valve S2 of the first chamber, the liquid flowed toward the second chamber by passing through the temporary stop valve S1 of the first chamber. Similarly, all ten chambers were sequentially filled with green water within 164 s without air bubbles (b-2 and Video S1†).
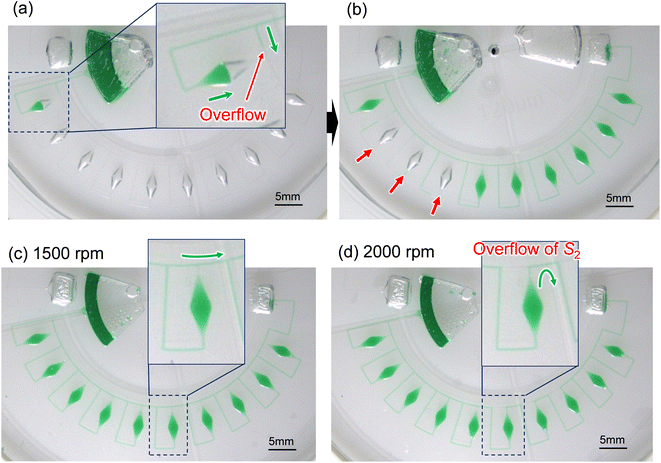
The experimental result of the liquid dispensing process, when the rotational speed was increased to 1250 min−1, is shown in Fig. 3c. The liquid flowed toward the first chamber after reaching the temporary stop valve S1 of the first chamber (c-1). However, before the first chamber was filled, the liquid overflowed toward the second chamber by passing through valve S1 in the first chamber (c-2). This is because when the liquid flows into the chamber in the radial direction opposite to the centrifugal force, the centrifugal force pushes back the leading edge of the liquid–air meniscus, resulting in this force acting on valve S1. The centrifugal pressure Ph is expressed as follows:25
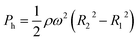 | (3) |
According to eqn (3), the maximum centrifugal pressure acting on valve S1 during liquid dispensing into the chamber is generated immediately after reaching valve S2 (R1 = 20.95 mm for the fabricated device). The estimated centrifugal pressure at a rotational speed of 1250 min−1 was Ph = 2.84 kPa, which was 1.2 times larger than the resulting value P1 = 2.40 kPa for the device. Therefore, contrary to our expectations, the liquid flow passed through valve S1 of the first chamber before filling it with liquid. In contrast, the estimated centrifugal pressure at a rotational speed of 1000 min−1 was Ph = 1.82 kPa, which can be within the range of the P1 value. Therefore, valve S1 did not leak until the chamber was filled, enabling sequential dispensing into all ten chambers. Based on the above considerations, the design constraints for valve S1 are as follows:
| P1 > Ph | (4) |
According to eqn (4), in this valve configuration, a significant constraint on the rotational speed makes it difficult to shorten the dispensing time.
A modified method for inward sequential liquid dispensing into multiple microchambers
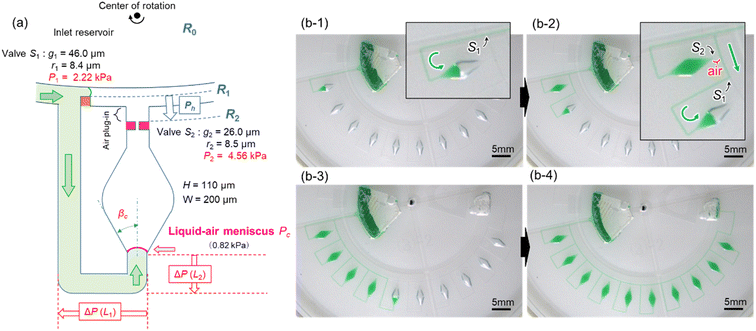
To reduce the centrifugal pressure exerted on stop valve S1 while filling the microchamber with liquid, we propose a modified valve configuration, as shown in Fig. 4a. Here, temporary stop valve S1 and permanent stop valve S2 were located inward and outward, respectively, concerning the center of rotation. This configuration allowed valves S1 and S2 to be positioned as close as possible to each other. The resulting burst pressure of valves S1 and S2 were calculated to be 2.22 kPa and 4.56 kPa for the fabricated device, where for valves S1 and S2, the gap distances were g1 = 46.0 μm and g2 = 26.0 μm, and the corner radius at the back edge of the convex structures was r1 = 8.4 μm (β = 61°) and r2 = 8.5 μm (β = 50°), respectively. The width and height of the microchannel were W = 200 μm and H = 110 μm, respectively.Fig. 4b demonstrates a modified method for inward sequential liquid dispensing into ten microchambers at a rotational speed of 1500 min−1, where green-colored water (100 μL) was introduced into the inlet reservoir. First, the liquid bypassed outward and entered inward into the channel (b-1). During this process, the liquid flow in the main microchannel was stopped at temporary stop valve S1 in the first chamber. After reaching the permanent stop valve S2 of the first chamber, the liquid flowed toward the second chamber by passing through valve S1 of the first chamber because the flow of the liquid was stopped at valve S1 of the second chamber (b-2). Furthermore, an air plug was trapped between valve S2 of the first chamber and the microchannel, thus significantly enhancing the pressure resistance performance (air plug-in valve reported in our previous study39). This valve configuration also effectively prevents cross-contamination between the reaction chambers during LAMP assays caused by the leakage of LAMP primers pre-fixed inside the chambers. Similarly, all ten chambers were sequentially filled with green water within 35 s without air bubbles (b-3, b-4, and Video S2†). Consequently, the dispensing time can be reduced to one-fifth of that achieved by this device (Fig. 3a).
The sequential liquid dispensing method proposed in this study offers the advantage of simultaneously measuring a mixture of the sample and LAMP reagents in each reaction chamber while dispensing, even at a constant rotational speed, without the need for a hydrophilic and/or hydrophobic coating on the surface of the device. This feature enables simple and rapid operation at a constant rotational speed and results in a low-cost and reliable device without requiring any surface coating. Furthermore, microfluidic devices can be designed to be more compact by eliminating the requirement for a metering chamber.
Fig. 5a shows an experimental result of sequential liquid dispensing into an array of 10 microchambers when the gap distance was increased to g = 120.1 μm (i.e., the burst pressure P1 = 1.14 kPa) for temporary stop valve S1. The device was rotated at a speed of 1500 min−1. Before the first chamber was filled, the liquid overflowed toward the second chamber by passing through valve S1, probably because of the insufficient pressure resistance of S1. However, sequential dispensing is achieved in the fifth and subsequent chambers (Fig. 5b). We derived a relationship to estimate the required burst pressure P1 of temporary stop valve S1 integrated into each chamber as follows:
| P1 > Pc + ΔP(L1) + ΔP(L2) | (5) |
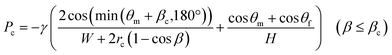 | (6) |
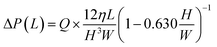 | (7) |
In this experiment, the estimated burst pressure Pc at the entrance of the microchamber was 0.81 kPa for rc = 6.3 μm βc = 19.8°. The estimated pressure drop ΔP(L1) and ΔP(L2) of the first chamber are 1.00 kPa and 0.28 kPa for L1 = 3.6 mm and L2 = 1.0 mm at a rotational speed of 1500 min−1, resulting in a measured volumetric flow rate Q = 204 μL min−1 for filling the first chamber. According to eqn (5), the pressure resistance performance of valve S1 should be designed to be at least 2.09 kPa or higher for dispensing into ten chambers at 1500 min−1. Therefore, liquid dispensing succeeded when using valve S1 with a burst pressure of P1 = 2.22 kPa (g1 = 46.0 μm), as shown in Fig. 4. In contrast, it failed when using valve S1 with a burst pressure of P1 = 1.14 kPa (g1 = 120.1 μm), as shown in Fig. 5a. Interestingly, we found that the centrifugal pressure did not act on temporary stop valve S1 until the chamber was filled, and the liquid flow stopped at permanent stop valve S2. Furthermore, the pressure drop ΔP(L) in the microchannel did not affect valve S1 while the liquid was flowing radially outward and, except in the circumferentially and radial inward direction. That is, the burst pressure of valve S1 should be designed to be higher than the pressure required for the liquid to flow into the chamber entrance. After reaching the fifth chamber, the pressure drops ΔP(L1) and ΔP(L2) were decreased to 0.24 kPa and 0.07 kPa, respectively, owing to a decrease in a measured volumetric flow rate Q = 50 μL min−1. As a result, the total pressure acting on valve S1 decreased to 1.12 kPa, according to eqn (5), which can be considered within the range of P1. Therefore, sequential dispensing could be achieved in the fifth and subsequent chambers.
Fig. 5c and d show the influence of the burst pressure P2 of permanent stop valve S1 on the performance of sequential liquid dispensing at rotational speeds of 1500 min−1 and 2000 min−1, respectively. Here, a single-faced stop valve (g2 = 29.9 μm) was used as the permanent stop valve S2 to decrease the bust pressure to 3.23 kPa compared to that of the double-faced stop valve S2 (P2 = 4.56 kPa), as shown in Fig. 4. On the other hand, the burst pressure of the temporary stop valve S1 (P1 = 2.64 kPa for g1 = 38.6 μm) was designed to be approximately the same compared to that of the single-faced stop valve S1 (P1 = 2.22 kPa), as shown in Fig. 4. The liquid was successfully dispensed into all ten chambers sequentially at a rotational speed of 1500 min−1 (Fig. 5c), whereas the liquid was flowed over the valve S2 of the sixth chamber at a rotational speed of 2000 min−1 (Fig. 5d). This unfavorable flow may cause the pre-fixed LAMP primer inside the chambers to leak, leading to cross-contamination and potentially resulting in a false diagnosis. Therefore, we estimated the required burst pressure P2 for permanent stop valve S2 by deriving the following equation:
| P2 > P1 + Ph | (8) |
Multiplexed detection of foodborne pathogens by colorimetric LAMP
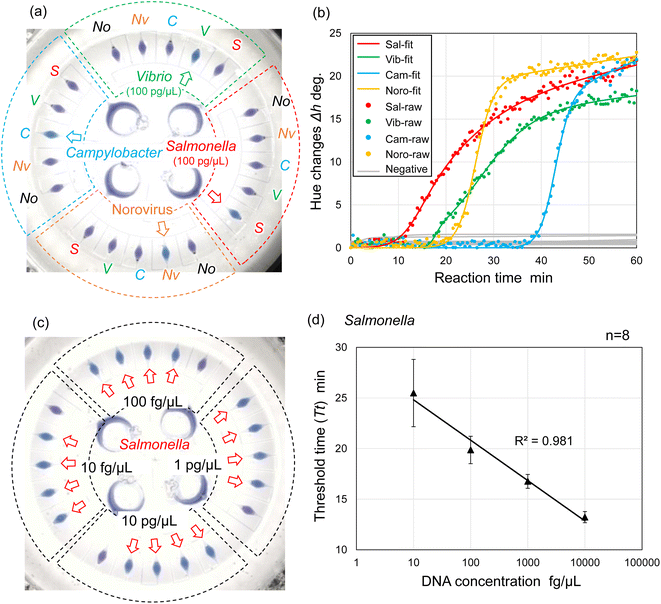
We explored the possibility of rapid, simultaneous detection of multiple foodborne pathogens, that is, specific DNA targets from Salmonella spp., V. parahaemolyticus, Campylobacter spp., and norovirus GII by the colorimetric LAMP assay. To enable the multiplexed detection of multiple samples, a centrifugal microfluidic device consisting of four sets of five microchambers was fabricated, as shown in Fig. 1b. As mentioned in the previous section, the device does not have a waste reservoir or air exhaust port, and each air exhaust microchannel is connected to its respective inlet reservoir via a common central position. The colored water was dispensed sequentially into multiple chambers at a rotational speed of 1500 min−1, even though the inlet ports had been sealed with double-sided adhesive tape (Fig. S7 and Video S3†). In this device, the gap distances of valves S1 and S2 were g1 = 40.8 μm (r1 = 3.9 μm) and g2 = 20.9 μm (r2 = 4.2 μm), respectively. The width and height of the microchannel were W = 200 μm and H = 108 μm, respectively. The microscopy images of the detailed fabricated PDMS device are shown in Fig. S8.†Fig. 6a shows the experimental results of colorimetric LAMP detection of Salmonella, V. parahaemolyticus, Campylobacter, and norovirus GII in the centrifugal microfluidic device. S, V, C, and Nv indicate that Salmonella-specific, V. parahaemolyticus-specific, Campylobacter-specific, and norovirus-specific primer sets were pre-spotted and dried in each microchamber, respectively. The symbol No was used if no primer was pre-spotted in the chamber. Salmonella DNA and Vibrio DNA templates with each concentration of 100 pg μL−1 were introduced into the five microchambers surrounded by red and green dashed lines, respectively. In addition, positive controls of Campylobacter DNA template and norovirus RNA template from each Eiken detection kit, diluted to 1/10 the concentration of the standard recipe, were introduced into the five microchambers surrounded by the blue and yellow dashed lines, respectively. Note that to successfully dispense a mixture of the sample and LAMP reagents into five microchambers, it was required to reduce the rotational speed from 1500 min−1 for water to 1000 min−1 because the contact angles of the LAMP reagents containing a surfactant (0.1% w/v Tween 20) were reduced to θm = 79.0° and θf = 82.0°. In addition, the surface tension might be reduced to approximately 0.04 N m−1, according to a previous study.44 As predicted by eqn (1) and (2), the burst pressures of valves S1 and S2 are estimated to be decreased to 0.59 kPa and 2.57 kPa, respectively. Therefore, to satisfy the constraint of eqn (5) and (8), the required pressure resistance performance of valves S1 and S2 should be decreased to 0.50 kPa and 0.67 kPa, respectively, by reducing the rotational speed to 1000 min−1. Here, the estimated pressure drop ΔP(L1) and ΔP(L2) of the first chamber were 0.39 kPa and 0.11 kPa for L1 = 3.6 mm and L2 = 1.0 mm, respectively, and the centrifugal pressure Ph was 0.08 kPa between valves S1 (R1 = 20.19 mm) and S2 (R2 = 20.55 mm). Because the Ps value (=−0.06 kPa) in eqn (5) is negative, it does not need to be considered as required pressure resistance. The above result indicates that the proposed dispensing theory provides comprehensive design guidelines for universally applicable microfluidic devices, considering both the properties of the liquid and the material characteristics of the microchannel. After running the LAMP assay at 60 °C for 60 min, the color of four microchambers with the LAMP primers matched to the introduced DNA or RNA templates was changed from violet to sky blue, indicating positive reactions. The DNA amplification curves representing hue angle changes in the CIE L*a*b* color space were calculated using a home-built analysis software from the time-lapse images acquired during the LAMP reaction. The curves were fitted to the following theoretical function:40
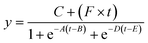 | (9) |
Furthermore, we investigated the linear dynamic range of quantitation and limit of detection (LOD) of the LAMP assays in a centrifugal microfluidic device. Fig. 6c shows an example of experimental results showing the detection of Salmonella DNA templates with varying DNA concentrations ranging from 10 fg μL−1 to 10 pg μL−1. In this experiment, the Salmonella-specific LAMP primer set was pre-spotted in all the chambers (indicated by red arrows) except for one chamber with no pre-spotted primers in each independent compartment. Two experiments were performed under the same conditions, and then the threshold time (Tt value) was determined from each DNA amplification curve of eight chambers at each concentration level ranging from 1 fg μL−1 to 100 pg μL−1. Tt values were automatically calculated as the peak value of the second derivative of each DNA amplification curve using a home-built analysis software.40 As a result, the LOD of the Salmonella DNA template was estimated to be 1 fg μL−1 in the centrifugal microfluidic device because positive reactions were detected in all eight chambers (Fig. S10a†). Subsequently, to assess the linear dynamic range of quantitation in the device, the correlation coefficient (R2) of the standard curve was evaluated from the resulting Tt values; a range of R2 > 0.98 was within the quantitative range.45 At DNA concentrations ranging from 1 fg μL−1 to 100 pg μL−1, the resulting standard curve exhibited poor quantification (R2 = 0.897) because the Tt values obtained at a high concentration of 100 pg μL−1 deviated from the standard curve. There was significant variation in the Tt values obtained at a low concentration of 1 fg μL−1 (Fig. S10b†). Similarly, the correlation coefficients of the standard curves were estimated to be R2 = 0.864 and R2 = 0.974 at DNA concentrations ranging from 10 fg μL−1 to 100 pg μL−1 (Fig. S10c†) and from 1 fg μL−1 to 10 pg μL−1 (Fig. S10d†), respectively. As a result, the linear dynamic range of the Salmonella DNA template was determined to be between the concentration of 10 fg μL−1 and 10 pg μL−1 with a strong negative correlation (R2 = 0.981), as shown in Fig. 6d.
For comparison, conventional off-chip LAMP assays were performed using real-time turbidity measurements in PCR tubes (25 μL reaction volume), with three experimental replicates for each DNA concentration. The LOD was 0.1 fg μL−1, one order of magnitude lower than that obtained in the microfluidic device (Fig. S11a†). The linear dynamic range of quantitation was between 1 and 10 pg (R2 = 0.986), which was one order of magnitude wider than that obtained using the microfluidic device (Fig. S11b†). To potentially enhance the LOD of microfluidic devices, future studies should investigate methods to prevent the adsorption of target DNA and/or enzymes on the PDMS surface. In addition, preheating the DNA sample with the LAMP primer for denaturation and enhancing primer annealing could improve the LOD.46,47
Conclusions
We developed a centrifugal microfluidic device capable of sequential liquid dispensing into multiple chambers integrated with a pair of passive valves for the multiplexed LAMP detection of foodborne pathogens. The proposed microchannel design, which enables the liquid to fill towards the center of rotation against the centrifugal force, facilitates the complete removal of air from inside the microchamber without surface coating by exploiting the inherent hydrophobicity of the polymer. The design constraints for each passive valve were clarified, and the experimental results were verified using theoretical models. This enabled us to predict the rotational speed at which the liquid could be dispensed into the microchambers from the properties of the liquid and its contact angle with the device surface. Furthermore, using the colorimetric LAMP assay, we successfully demonstrated multiplexed detection of four foodborne pathogens (Salmonella spp., V. parahaemolyticus, Campylobacter spp., and norovirus GII) across four samples. The LOD for the detection of Salmonella DNA template was estimated to be 1 fg μL−1. High quantitation was observed in DNA concentration ranging from 10 fg μL−1 to 10 pg μL−1 with a strong negative correlation (R2 > 0.98). Consequently, the sequential liquid dispensing method proposed here allows direct liquid dispensing into the reaction chambers without cross-contamination while simultaneously metering the LAMP reaction volume. This method provides a simple and rapid operation at a constant rotational speed and a low-cost and reliable device without any surface coating. Furthermore, microfluidic devices can be designed to be more compact by eliminating the requirement for a metering chamber.In future studies, to develop a sample-to-answer platform, we will focus on integrating the necessary steps for genetic diagnosis, including nucleic acid extraction, purification, and mixing of the sample and LAMP reagents directly onto the microfluidic device. The autonomous operation of all fluid manipulation processes in the device, which operates at a constant rotational speed, can make the diagnostic system rapid, easy to use, low-cost, and highly robust. This system offers flexibility for customizing the types of nucleic acid targets (DNA/RNA) of interest by varying the primer types and combinations pre-spotted in the reaction chambers. Thus, our versatile platform is powerful for the on-site testing of a wide range of infectious agents (viruses, bacteria, fungi, and parasites) in humans, animals, and plants, as well as allergens and illegal substances, without laborious and multiple manual procedures.
Data availability
The authors confirm that the data supporting the findings of this study are available within the main article and ESI.†Author contributions
Daigo Natsuhara; methodology, investigation, writing – original draft preparation, Yuka Kiba; methodology, investigation, Ryogo Saito; methodology, investigation, Shunya Okamoto; methodology, writing – review and editing, supervision, Moeto Nagai; writing – review and editing, supervision, Yusuke Yamauchi; writing – review and editing, supervision, Masashi Kitamura; methodology, writing – review and editing, supervision, Takayuki Shibata; conceptualization, writing – review and editing, supervision, project administration funding acquisition. All authors have read and agreed to the published version of the manuscript.Conflicts of interest
The authors declare no conflicts of interests.Acknowledgements
This research was partially supported by “Knowledge Hub Aichi”, Priority Research Project from Aichi Prefectural Government, the Toyo Suisan Foundation, and JSPS KAKENHI Grant Numbers JP22KJ1627, JP24K00776. We would like to thank Editage (https://www.editage.com) for English language editing.References
- R. V. Tauxe, Int. J. Food Microbiol., 2002, 78, 31–41 CrossRef CAS PubMed.
- E. Scallan, R. M. Hoekstra, F. J. Angulo, R. V. Tauxe, M.-A. Widdowson, S. L. Roy, J. L. Jones and P. M. Griffin, Emerging Infect. Dis., 2011, 17, 7–15 CrossRef.
- M. K. Thomas, R. Murray, L. Flockhart, K. Pinter, A. Fazil, A. Nesbitt, B. Marshall, J. Tataryn and F. Pollari, Foodborne Pathog. Dis., 2015, 12, 820–827 CrossRef.
- T. Martinovic, U. Andjelković, M. Š. Gajdošik, D. Rešeter, D. Josić and D. Josic, J. Proteomics, 2016, 146, 226–235 CrossRef PubMed.
- W. Su, D. Liang and M. Tan, Trends Food Sci. Technol., 2021, 113, 97–109 CrossRef CAS.
- X. Zhao, C.-W. Lin, J. Wang and D. H. Oh, J. Microbiol. Biotechnol., 2014, 24, 297–312 CrossRef CAS PubMed.
- S. Petrucci, C. Costa, D. Broyles, E. Dikici, S. Daunert and S. Deo, Trends Food Sci. Technol., 2021, 115, 409–421 CrossRef CAS PubMed.
- A. Bosch, E. Gkogka, F. S. L. Guyader, F. L. Hamon, A. Lee, L. van Lieshout, B. Marthi, M. Myrmel, A. Sansom, A. C. Schultz and A. Winker, Int. J. Food Microbiol., 2018, 285, 110–128 CrossRef PubMed.
- T. Notomi, H. Okayama, H. Masubuchi, T. Yonekawa, K. Watanabe, N. Amino and T. Hase, Nucleic Acids Res., 2000, 28, e63 CrossRef CAS.
- T. Notomi, Y. Mori, N. Tomita and H. Kanda, J. Microbiol., 2015, 53, 1–5 CrossRef CAS PubMed.
- Y. Mori and T. Notomi, J. Infect. Chemother., 2009, 15, 62–69 CrossRef CAS.
- F. Han, F. Wang and B. Ge, Appl. Environ. Microbiol., 2011, 77, 2589–2595 CrossRef CAS.
- X. Sun, Q. Xu, Y. Pan, W. Lan, Y. Zhao and V. C. H. Wu, Ann. Microbiol., 2012, 62, 263–271 CrossRef CAS.
- S. B. Jeon, D. J. Seo, H. Oh, D. H. Kingsley and C. Choi, Food Control, 2017, 73, 1002–1009 CrossRef CAS.
- F. Wang, L. Jiang, W. Yang, W. Prinyawiwatkul and B. Ge, Appl. Environ. Microbiol., 2012, 78, 2727–2736 CrossRef CAS.
- S. Wachiralurpan, T. Sriyapai, S. Areekit, T. Kaewphinit, P. Sriyapai, S. Santiwatanakul and K. Chansiri, Food Anal. Methods, 2017, 10, 3763–3772 CrossRef.
- L. Niessen, J. Luo, C. Denschlag and R. F. Vogel, Food Microbiol., 2013, 36, 191–206 CrossRef CAS PubMed.
- Y. Li, P. Fan, S. Zhou and L. Zhang, Microb. Pathog., 2017, 107, 54–61 CrossRef CAS.
- Q. Yang, K. J. Domesle and B. Ge, Foodborne Pathog. Dis., 2018, 15, 309–331 CrossRef.
- A. A. Sayad, F. Ibrahim, S. M. Uddin, K. X. Pei, M. S. Mohktar, M. Madou and K. L. Thong, Sens. Actuators, B, 2016, 227, 600–609 CrossRef CAS.
- L. Zhang, F. Tian, C. Liu, Q. Feng, T. Ma, Z. Zhao, T. Li, X. Jiang and J. Sun, Lab Chip, 2018, 18, 610–619 RSC.
- S. J. Oh, B. H. Park, G. Choi, J. H. Seo, J. H. Jung, J. S. Choi, D. H. Kim and T. S. Seo, Lab Chip, 2016, 16, 1917–1926 RSC.
- M. Madou, J. Zoval, G. Jia, H. Kido, J. Kim and N. Kim, Annu. Rev. Biomed. Eng., 2006, 8, 601–628 CrossRef CAS PubMed.
- R. Gorkin, J. Park, J. Siegrist, M. Amasia, B. S. Lee, J.-M. Park, J. Kim, H. Kim, M. Madou and Y.-K. Cho, Lab Chip, 2010, 14, 1758–1773 RSC.
- O. Strohmeier, M. Keller, F. Schwemmer, S. Zehnle, D. Mark, F. von Stetten, R. Zengerle and N. Paust, Chem. Soc. Rev., 2015, 44, 6187–6229 RSC.
- J. Ducrée, S. Haeberle, S. Lutz, S. Pausch, F. von Stetten and R. Zengerle, J. Manuf. Syst., 2007, 17, S103–S115 Search PubMed.
- M. Tang, G. Wang, S.-K. Kong and H.-P. Ho, Micromachines, 2016, 7, 26 CrossRef PubMed.
- A. Sayad, F. Ibrahim, S. M. Uddin, J. Cho, M. Madou and K. L. Thong, Biosens. Bioelectron., 2018, 100, 96–104 Search PubMed.
- J. H. Seo, B. H. Park, S. J. Oh, G. Choi, D. H. Kim, E. Y. Lee and T. S. Seo, Sens. Actuators, B, 2017, 246, 146–153 CrossRef CAS.
- S. J. Oh, B. H. Park, J. H. Jung, G. Choi, D. C. Lee, D. H. Kim and T. S. Seo, Biosens. Bioelectron., 2016, 75, 293–300 CrossRef CAS.
- H. V. Nguyen, V. D. Nguyen, E. Y. Lee and T. S. Seo, Biosens. Bioelectron., 2019, 136, 132–139 Search PubMed.
- V. M. Phan, S. W. Kang, Y. H. Kim, M. Y. Lee, H. V. Nguyen, Y. L. Jeon, W. I. Lee and T. S. Seo, Sens. Actuators, B, 2023, 390, 133962 Search PubMed.
- V. M. Phan, H. W. Nguyen, K. H. Bui, H. V. Nguyen and T. S. Seo, Sens. Actuators, B, 2023, 394, 134362 CrossRef CAS.
- H. V. Nguyen, V. M. Phan and T. S. Seo, Sens. Actuators, B, 2024, 339, 1334771 Search PubMed.
- D. Mark, P. Weber, S. Lutz, M. Focke, R. Zengerle and F. von Stetten, Microfluid. Nanofluid., 2011, 10, 1279–1288 CrossRef CAS.
- D. Natsuhara, K. Takishita, K. Tanaka, A. Kage, R. Suzuki, Y. Mizukami, N. Saka, M. Nagai and T. Shibata, Micromachines, 2020, 11, 540 CrossRef PubMed.
- S. Misawa, D. Natsuhara, Y. Kiba, T. Yamamuro, R. Suzuki, T. Shibata and M. Kitamura, Forensic Toxicol., 2021, 39, 259–265 CrossRef CAS.
- D. Natsuhara, R. Saito, H. Aonuma, T. Sakurai, S. Okamoto, M. Nagai, H. Kanuka and T. Shibata, Lab Chip, 2021, 21, 4779–4790 RSC.
- D. Natsuhara, S. Misawa, R. Saito, K. Shirai, S. Okamoto, M. Nagai, M. Kitamura and T. Shibata, Sci. Rep., 2022, 12, 12852 CrossRef CAS.
- D. Natsuhara, A. Miyajima, T. Bussho, S. Okamoto, M. Nagai, M. Ihira and T. Shibata, Analyst, 2024, 149, 3335–3345 RSC.
- H. Ou, Y. Wang, J. Gao, J. Bai, Q. Zhang, L. Shi, X. Wang and C. Wang, Ann. Palliat. Med., 2021, 10, 6580–6858 Search PubMed.
- Z. Shen, Y. Liu and L. Chen, Pathogens, 2022, 11, 10 CrossRef CAS.
- E. Yildrim, S. J. Trietsch, J. Joore, A. van den Berg, T. Hankemeier and P. Vulto, Lab Chip, 2014, 14, 3334–3340 RSC.
- A. Bąk and W. Podgórska, Colloids Surf., A, 2016, 504, 414–425 Search PubMed.
- T. S. Kang and T. Tanaka, Food Chem., 2018, 269, 549–558 Search PubMed.
- J. Wang, J. P. Steheli, A. Wu, J. E. Kreutz, Q. Hu, J. Wang, T. Schneider, B. S. Fujimoto, Y. Qin, G. S. Yen, B. Weng, K. Shibley, H. Haynes, R. L. Winer, Q. Feng and D. T. Chiu, Anal. Chem., 2021, 93, 3266–3272 CrossRef CAS PubMed.
- F. Schuler, C. Siber, S. Hin, S. Wadle, N. Paust, R. Zengerle and F. von Stetten, Anal. Methods, 2016, 8, 2750–2755 RSC.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d4ra04055d |
| This journal is © The Royal Society of Chemistry 2024 |