Open Access Article
Open Access ArticleChemical composition and toxicity studies on Lantana camara L. flower essential oil and its in silico binding and pharmacokinetics to superoxide dismutase 1 for amyotrophic lateral sclerosis (ALS) therapy†
Abdullah Haikal *a and
Ahmed R. Ali
*a and
Ahmed R. Ali bc
bc
aDepartment of Pharmacognosy, Faculty of Pharmacy, Mansoura University, Mansoura 35516, Egypt. E-mail: abdullahhaikal@mans.edu.eg; abdullahhaikal9393@yahoo.com; Tel: +201129608369
bDepartment of Medicinal Chemistry, Faculty of Pharmacy, Mansoura University, Mansoura, 35516, Egypt
cDepartment of Pharmaceutical Chemistry, Faculty of Pharmacy, New Mansoura University, New Mansoura 7723730, Egypt. E-mail: ahmed_reda5588@mans.edu.eg; ahmed_reda551988@yahoo.com; Tel: +20-10-9838-4072
First published on 5th August 2024
Abstract
Using the gas chromatography mass spectrometry method, the chemical components of essential oil from flowers of Lantana camara growing in Egypt are analyzed. Through this investigation, 22 chemicals from floral oil were identified. Most of the oil is made up of sesquiterpene caryophyllene (15.51%) and monoterpene sabinene (14.90%). When the oil's composition was compared to oils extracted from the same plant on several continents, we observed that the essential components were largely the same with some difference in proportions and some compounds due to geographical differences. A molecular docking study of essential oil components was conducted with human superoxide dismutase 1, a target involved in the pathophysiology of amyotrophic lateral sclerosis (ALS). Isospathulenol showed a comparable docking score to the reference ligand bound to the dismutase enzyme. Isospathulenol showed a reasonable drug score with some safety concerns. In addition, isospathulenol is predicted to have high GI absorption, good permeability through the blood–brain barrier and reasonable bioavailability score with ease access to synthetic modifications. In addition, the same compound is devoid from any violation to Lipinski rules or any PAINS alerts. This may establish the promising characteristics of such a compound to be optimized into potential drug candidates for treatment of ALS.
1 Introduction
Amyotrophic lateral sclerosis (ALS) is a very debilitating, rapidly progressing, and lethal neurodegenerative illness characterized by the damage and death of lower motor neurons in the brain stem and spinal cord and upper motor neurons in the cerebral cortex.1 The hallmark of ALS is a gradual loss of voluntary motor function, which makes it difficult for the patient to work and carry out everyday tasks.2 Three to four years after the start of symptoms, the disease symptoms can exacerbate muscle weakness and the patient might experience respiratory paralysis leading to death.3 Total US national costs spanned ∼$212 million–∼$1.4 billion USD per year, and variably consisted of direct costs associated with healthcare resource use and indirect costs.4 From 0.26 per 100![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 person-years in Ecuador to 23.46 per 100
000 person-years in Ecuador to 23.46 per 100![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 person-years in Japan, there were differences in the incidence of ALS. The range of point prevalence was 1.57 per 100
000 person-years in Japan, there were differences in the incidence of ALS. The range of point prevalence was 1.57 per 100![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 in Iran and 11.80 per 100
000 in Iran and 11.80 per 100![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 in the US.5 The number of ALS patients is expected to increase by 69% in the next 20 years because of population ageing, especially in developing nations.6 ALS is still incurable, although there are some recognized medications. Physical therapy and symptomatic therapies are the only areas of focus for ALS patient care.7
000 in the US.5 The number of ALS patients is expected to increase by 69% in the next 20 years because of population ageing, especially in developing nations.6 ALS is still incurable, although there are some recognized medications. Physical therapy and symptomatic therapies are the only areas of focus for ALS patient care.7
Riluzole is an anti-glutamatergic medication that prevents glutamate from being released presynaptically.8 Edaravone received FDA approval in 2017 to treat ALS after it was approved in Japan in 2015 for marketing and manufacture.9 As a radical scavenger, antioxidant, and anti-inflammatory against oxidative stress and reactive oxygen species, edaravone provides its neuroprotective benefits. Furthermore, it halts the degeneration of motor neurons in the brain and spinal cord by lowering fas-associated death domain (FADD) through its anti-apoptotic action.8 Patients who receive riluzole and edaravone, the two medications licensed for the treatment of ALS, have a few-month survival advantage. Compared to patients who got a placebo, patients who received 100 mg per day of riluzole had a higher rate of survival. In one trial, the riluzole group's median survival time was 17.7 months, while the placebo group's was 14.9 months.10 Therefore, there is urgent need to develop more potent therapies for illnesses like ALS, but achieving so will necessitate developing a solid understanding of the fundamental mechanisms and reasons behind these disorders.11
The precise mechanism of ALS deterioration is still unknown because of its diverse and complicated character.12 Nevertheless, some of the primary etiological pathways involved in the pathophysiology of ALS are glutamate excitotoxicity, oxidative stress, inflammatory pathway, and protein aggregation.13 There are two types of ALS: familial (fALS) and sporadic (sALS). 95–90% of ALS cases are sporadic, whereas 5–10% are familial.14 SOD1 gene mutations account for around 7% of all sALS cases and 23% of fALS cases, making them common and significant causes of ALS.15 Superoxide dismutase 1 is a key cytoplasmic antioxidant enzyme that is encoded by the SOD1 gene. Superoxide radicals are converted to hydrogen peroxide and molecular oxygen by the Cu/Zn metalloenzyme SOD1.16 Through this process, cells are shielded from oxidative stress and the damaging effects of superoxide radicals.17 ALS has previously been linked to more than 185 SOD1 mutations.15 While these mutations impact distinct regions of the SOD1 structure,18 a significant number of them result in enhanced oxidative damage and protein aggregation, which are essential components of the pathogenicity of ALS.19,20
The entire drug development process, from discovery to clinical application, is fraught with difficulties and complications that stand in the way of finding compounds and developing a viable treatment for ALS.21 The pathophysiology of ALS is associated with a number of cellular pathways and a multitude of hereditary and environmental variables. It is still difficult to comprehend these systems and find trustworthy treatment targets. In addition, it might be challenging to demonstrate that modifying putative targets would have a beneficial therapeutic effect, even in cases when they are found. To evaluate the effectiveness of possible treatments in preclinical research, trustworthy biomarkers for disease progression and treatment response are required. In addition, SOD1 transgenic mice and other animal models of ALS have shed light on the disease, they fall short of accurately simulating human ALS. These models' outputs frequently don't transfer into successful therapeutic outcomes.22,23 In order to address these gaps, a multifaceted strategy including improvements in preclinical model creation, basic research advancements, creative trial designs, and successful collaborations is needed. That is why we see the urgent need to pursue a continuous virtual screening for novel sources for new hit compounds for ALS treatment. According to experimental research, targeting SOD1's Trp32 binding site enables it to function as an inhibitor by preventing the interaction between WT-SOD1 and mutant or misfolded SOD1. This is because a misfolded SOD1 and a natural SOD1 come into contact at the Trp32 residue of the Trp32 site. Furthermore, binding of the ligand at exposed hydrophobic residues such as Ile99 and Lys30 strengthens the resistance to misfolding and aggregation by decreasing unfavorable solvent interactions.24 This significantly promotes our searches for novel compounds targeting such binding site of SOD1.
A growing number of natural compounds from plant origin like quercetin, ginkgolides, madecassoside, celastrol, curcumin, withaferin A, ferulic acid, huperzine A, berberine, and others have been investigated for their potential use in treating or controlling ALS via different mechanisms. For many years, these substances have also been utilized medically to treat wide range of illnesses.25 About 150 species of shrubs and herbaceous plants make up the genus Lantana, which is a member of the Verbenaceae family.26 Lantana camara is a fragrant, evergreen shrub of the genus Lantana that is one of the most important medicinal plants in the world.27,28 Although L. camara is native to tropical regions of America and Africa, it has been introduced as an ornamental plant to most countries of the world.29 In traditional medicine, L. camara has been widely used to treat a variety of conditions, including fevers, rheumatism, chicken pox, measles, ulcers, cancer, high blood pressure, tetanus, tumours, eczema, cuts, and catarrhal infections.26,30–32 It is a superior source for several groups of bioactive natural compounds, such as triterpenoids, flavonoids, steroids, iridoid glycosides, oligosaccharides, phenylpropanoid glycosides, and naphthoquinones.33–35 These phytomolecules exhibit a diverse range of biological activities, including hepatoprotective, leishmanicidal, anticancer, antibacterial, antioxidant, antimycobacterial, nematicidal, and antiulcer activities.30,32,35–39 Furthermore, it has been demonstrated that L. camara is among the most readily available and reasonably priced materials for the extraction of industrial essential oils, sometimes referred to as lantana oils.40,41 Previous studies on isolated essential oils from different parts of L. camara have shown them to possess a variety of biological activities, including anti-inflammatory,42 antibacterial,43 antioxidant,44 insecticidal,45 allelopathic,46 and larvicidal.47
One of the most commonly used approaches for developing new drugs, known as “structure-based drug design”, is concentrated on locating and characterizing biological targets, mostly proteins, and then matching those targets with small molecule compounds that possess the tight binding affinity to them.48–50 As a result, it's intriguing to examine how the components of the L. camara flowers essential oil may have potential for interaction with the protein superoxide dismutase 1 during their signaling cascade. In this study, we provide a thorough chemical examination of the volatile ingredients in the essential oils extracted from L. camara flowers grown in Egypt. Molecular docking simulations have also been employed to predict the possible binding of the natural components of L. camara flowers oil to proteins associated with the superoxide dismutase 1 receptor. On those identified compounds, we also carried out pharmacokinetics and ADME (absorption, distribution, metabolism, and excretion) investigations in addition to assessment of their toxicity.
2 Materials and methods
2.1. Plant material
The Lantana camara flowers were collected at the flowering stage in April from the medicinal plant station, Pharmacognosy Department, Faculty of Pharmacy, Mansoura University, Dakahlia, Egypt. The plant identity was confirmed by Prof. Ibrahim Mashaly, Professor of Ecology and Systematic Botany, Faculty of Science, Mansoura University. A voucher specimen (LCF221.2024) is deposited at the Pharmacognosy Department, Faculty of Pharmacy, Mansoura University.51,522.2. Hydro-distillation of the essential oil
Fresh Lantana camara flowers (250 g) were crushed into small pieces and subjected to hydro-distillation for 3 hours using Clevenger-type equipment according to European Pharmacopiea53 to get clear, yellow-colored oil. The obtained oil was collected and dehydrated using anhydrous sodium sulfate (El-Nasr Company for Pharmaceutical Chemicals, Egypt) and kept in a sealed vial at low temperatures in a refrigerator in the dark at 4 °C until use for GC/MS analysis and biological investigations.2.3. Gas chromatography-mass spectrometry (GC/MS) analysis
Thermo Scientific's Trace GC-ISQ mass spectrometer, which has an A3000 autosampler and a TG-5MS capillary column with a 30 m length, 0.25 mm i.d., and 0.25 m film thickness, was used for the GC/MS analysis. The temperature was scheduled in gradient mode (10 °C minutes−1) between 50 and 280 °C. Source temperature: 200 °C; interface temperature: 220 °C; injector temperature: 220 °C. Mass spectrometer adjusted in EI mode at 70 eV. One μL of diluted material was injected in splitless mode with a 50–600 amu mass scan. Helium was employed as the carrier gas (1 mL min−1). A combination of fatty acid methyl esters (C5–C20) was immediately fed into the GC injector under the aforesaid temperature program to compute the retention index of each molecule for the identification of the oil components based on their retention indices. All component retention indices were calculated using van Den Dool's technique.54 A comparison of the component's mass spectral fragmentation patterns and base peak with those found in the literature55,56 or in the mass spectral databases NIST and ChemStation data system further corroborated the identity of the components.2.4. Molecular docking and assessment of pharmacokinetic properties
AutoDock 4.2 (ref. 57) interfaced with LigandScout,58 which uses empirical scoring functions, the energy-minimized protein was employed for molecular docking. The grid box size was centered guided by the position of naphthalene–catechol linked reference compound. Genetic algorithm runs (default: 20) define the specified number of docking runs using the simulated annealing search engine. RMSD cluster tolerance (Å) (default: 2.0 Å) set the RMSD threshold (in Å) for joining together multiple docking results and show them as one entry in the list of docked poses. Number of individuals in population (default: 150) defines the size of the initial population for the genetic algorithm search. The maximum number of energy evaluations is the maximum number of energy evaluations performed during each genetic algorithm search run and was set to default value of 2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 500
500![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000. Maximum number of generations (default: 27
000. Maximum number of generations (default: 27![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000) set the maximum number of generations simulated during each genetic algorithm search run. SwissADME59 and the OSIRIS Property Explorer in DataWarrior60 were used to evaluate the absorption, distribution, metabolism, excretion, and toxicity (ADMET) properties. This was done in order to assess the chosen compounds' pharmacologic characteristics and drug-likeness.
000) set the maximum number of generations simulated during each genetic algorithm search run. SwissADME59 and the OSIRIS Property Explorer in DataWarrior60 were used to evaluate the absorption, distribution, metabolism, excretion, and toxicity (ADMET) properties. This was done in order to assess the chosen compounds' pharmacologic characteristics and drug-likeness.
3 Result and discussion
3.1. Essential oil composition of Lantana camara flowers
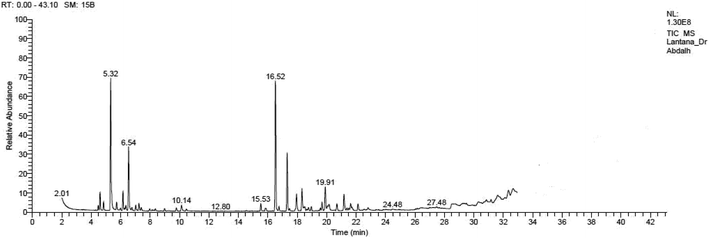
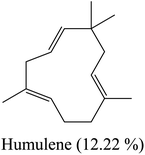
Yellow essential oil with a yield 0.1% v/w, lighter than water, was extracted from Lantana camara flowers. The oil components are shown in Table 1, together with their percentages and retention indices. A total of twenty-two compounds (97.87%) were found. The majority of the oil's ingredients, monoterpenes and sesquiterpenes, represent 48.91% and 48.96% of the mixture, respectively. Sabinene (14.90%) and eucalyptol (8.14%) are the principal components of monoterpenes, whereas caryophyllene (15.51%) and humulene (7.69%) are the principal components of sesquiterpenes. Out of all the sesquiterpenes that have been found, 15.26% are oxygenated sesquiterpenes, while about 10.57% are oxygenated monoterpenes. o-Cymene is the only aromatic volatile chemical that has been found among the ingredients (Table 1 and Fig. 1). L. camara flowers' essential oil is mostly composed of monoterpene sabinene (14.90%) and sesquiterpene caryophyllene (15.51%).| Peak no. | Retention time | Literature RI61 | Calculated RI | M+ peak | Base peak | Peak area % | Identified compounds | Structure |
|---|---|---|---|---|---|---|---|---|
| a Bold values point out the major components. | ||||||||
| 1 | 4.48 | 873 | 875 | 136 | 93 | 2.12 | 2-Thujene |  |
| 2 | 4.60 | 939 | 938 | 136 | 93 | 3.35 | α-Pinene | 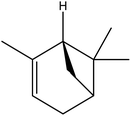 |
| 3 | 4.84 | 954 | 955 | 136 | 93 | 2.51 | Camphene | 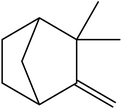 |
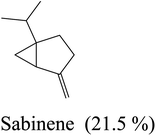
| 4 | 5.32 | 975 | 977 | 136 | 93 | 14.9 | Sabinene | 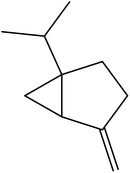 |
| 5 | 5.73 | 990 | 989 | 136 | 93 | 2.48 | β-Myrcene |  |
| 6 | 6.16 | 1011 | 1014 | 136 | 93 | 3.61 | 3-Carene | 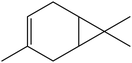 |
| 7 | 6.35 | 1026 | 1025 | 134 | 119 | 2.25 | o-Cymene | 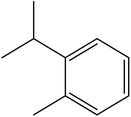 |
| 8 | 6.54 | 1031 | 1031 | 154 | 81 | 8.14 | Eucalyptol | 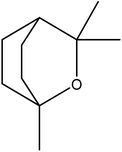 |
| 9 | 7.02 | 1050 | 1051 | 136 | 93 | 2.27 | β-cis-Ocimene | 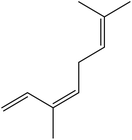 |
| 10 | 7.25 | 1059 | 1056 | 136 | 93 | 2.40 | γ-Terpinene | 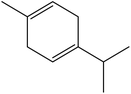 |
| 11 | 10.14 | 1164 | 1163 | 150 | 108 | 2.43 | Terpinen-4-ol | 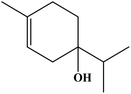 |
| 12 | 15.53 | 1376 | 1375 | 204 | 161 | 2.45 | α-Copaene | 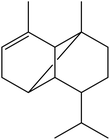 |
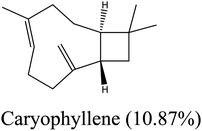
| 13 | 16.53 | 1419 | 1421 | 204 | 91 | 15.51 | Caryophyllene | 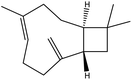 |
| 14 | 17.32 | 1438 | 1440 | 204 | 81 | 7.69 | Humulene | 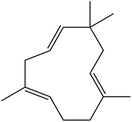 |
| 15 | 17.95 | 1441 | 1441 | 204 | 41 | 3.75 | Aromandendrene | 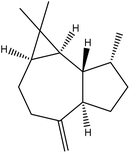 |
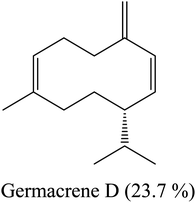
| 16 | 18.32 | 1485 | 1487 | 204 | 161 | 4.12 | Germacrene D | 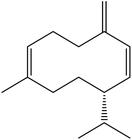 |
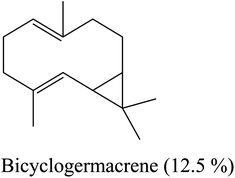
| 17 | 19.69 | 1500 | 1500 | 204 | 93 | 2.63 | Bicyclogermacrene | 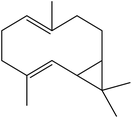 |
| 18 | 19.91 | 1563 | 1566 | 204 | 41 | 4.27 | trans-Nerolidol | 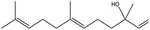 |
| 19 | 20.70 | 1585 | 1584 | 220 | 43 | 2.50 | Caryophyllene oxide | 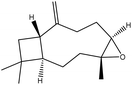 |
| 20 | 21.18 | 1623 | 1622 | 220 | 43 | 3.63 | Isospathulenol | 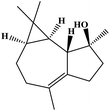 |
| 21 | 21.63 | 1631 | 1630 | 220 | 95 | 2.39 | Longifolenaldehyde | 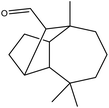 |
| 22 | 22.13 | 1672 | 1675 | 220 | 41 | 2.47 | Aromadendrene oxide I |  |
| 97.87 | ||||||||
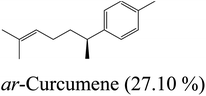
By comparing the percentages of compounds in the aforementioned plant oil with the percentages of compounds in the same oil from different geographical environments, we could come up with several observations. Sesquiterpenes caryophyllene and humulene represent the major constituents of the oil which are also collected in Egypt from the shrubs cultivated in the gardens of the National Gene Bank, National Institute of Horticulture, Faculty of Agriculture, Cairo University, in December 2004. Oil extracted from plant flowers in African lands like Cameroon is characterized by a high percentage of aromatic sesquiterpene (ar-Curcumene) and oxygenated sesquiterpene (Nerolidol), while sesquiterpenes caryophyllene and humulene represent the major constituents of the flowers oil from Côte d'Ivoire. The high percentage compounds of Nigerian flower oil are similar to the oil isolated in this study, but with different proportions.
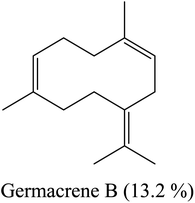
By heading to the continent of Asia, we find the major constituents of the oil isolated in the country of India: sesquiterpenes, caryophyllene and bicyclogermacrene. Indonesia is characterized by a high percentage of sesquiterpene caryophyllene and oxygenated sesquiterpene (davanone). In Saudi Arabia, oxygenated sesquiterpene caryophyllene oxide is the main constituent of L. camara flower essential oil. Finally, when we landed in South America, specifically in Brazil, we found sesquiterpenes germacrene D and germacrene B represent the major percentage of components in this oil. From this, it becomes clear to us the effect of different environments and climates on the composition of essential oils separated from plants. These results are summarized in Table 2.
3.2. Molecular docking study
Superoxide dismutase 1 (SOD1) in humans converts the superoxide anion radical to H2O2, shielding cells from reactive oxygen species.70 The neurodegenerative condition known as amyotrophic lateral sclerosis (ALS) was first associated with the gene SOD1. More than 20 years ago, mutations in SOD1 were also shown to be the cause of ALS. The loss of motor neurons in ALS is a deadly neurological disease that eventually results in paralysis and death.71 In order to determine the mechanism of interaction between two interacting chemical entities (inhibitors and receptor sites), we conducted a molecular modeling study. In this work, we used the LigandScout to predict 3D binding modes by computational docking simulation.58 In this work, we estimated the binding affinity in kcal mol−1 of our isolated compounds against superoxide dismutase 1 in (PDB code: 5YTO). This can aid in the interpretation of the biological activity for such isolated natural compounds and will help with future structural optimization efforts to produce more potent inhibitors. The X-ray crystal structure of superoxide dismutase 1 in complex with naphthalene–catechol linked compound (PDB code: 5YTO) was chosen for molecular docking investigations of our isolated phytochemicals. The X-ray crystal structure of superoxide dismutase 1 (PDB code: 5YTO) was downloaded from the Protein Data Bank (PDB) database (http://www.rcsb.org/). This specific isoform was selected due to its good parameters for experimental resolution (1.9 Å), R-value work of 0.179, and R-value free of 0.215.72 The reference naphthalene compound formed hydrophobic interactions primarily between Trp32 and aromatic rings, a positive ionizable interaction between Glu21 and the secondary amine, and hydrogen bond interactions between Glu100 and the phenolic hydroxyl group, according to the interactions derived from the X-ray derived structure (Fig. 2). In order to confirm that the docking program could reproduce the bioactive configuration of the naphthalene–catechol linked compound, we first verified our docking method by removing the reference compound from the superoxide dismutase 1 crystallographic structure and docking it into the binding site before running the molecular docking simulation. In comparison to the initial X-ray determined conformation, the best-docked ligand conformation displayed in Fig. 2 exhibited a root mean square deviation (RMSD) of 1.2 Å.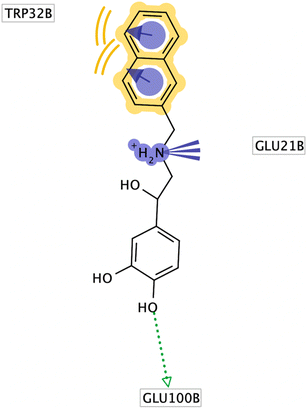 | ||
| Fig. 2 2D binding pose for reference naphthalene compound in human superoxide dismutase 1 active site (PDB Id: 5YTO). | ||
Targeting superoxide dismutase 1, molecular docking has been investigated in the identification of potential lead compounds between the screened phytochemicals. The protein–ligand complex binding energies were taken into consideration when shortlisting the hits in elucidated natural compounds. Twenty-two compounds were docked into the catalytic domain of superoxide dismutase 1, guided by the site of the co-crystallized naphthalene ligand, and their binding energies were found to be either lower or like those of naphthalene–catechol linked compound (Fig. 2). The binding energies of entries 1–11 (Table 3) were ranging from −4.31 to −5.26 kcal mol−1 lower than the binding energy of the reference compound. Those entries showed a common hydrophobic interaction with Trp32. Some additional interactions like hydrophobic interaction with Val31 and Ile99 with α-pinene and π-cation interaction with o-cymene. However, those additional interactions with such entries did not improve the binding affinity. For example, α-pinene and terpinen-4-ol, showing interaction with Val29, still has a weak binding affinity (−5.03 and −5.26 kcal mol−1). They did not show a significant increase (less than 10% increase) when compared with 2-thujene which had a binding affinity of −4.92 kcal mol−1. Entries 12–21 showed a promising docking score between −6.18 to −7.45 kcal mol−1 which is lower or comparable to docking score of the reference ligand.
| Entry | Comp. id | Estimated ΔG (kcal mol−1) | Entry | Comp. id | Estimated ΔG (kcal mol−1) |
|---|---|---|---|---|---|
| 1 | 2-Thujene | −4.92 | 13 | Caryophyllene | −6.78 |
| 2 | α-Pinene | −5.03 | 14 | Humulene | −6.46 |
| 3 | Camphene | −5.15 | 15 | Germacrene D | −6.90 |
| 4 | Sabinene | −4.92 | 16 | Bicyclogermacrene | −6.90 |
| 5 | β-Myrcene | −4.31 | 17 | Aromandendrene | −6.72 |
| 6 | 3-Carene | −5.17 | 18 | trans-Nerolidol | −6.18 |
| 7 | o-Cymene | −4.45 | 19 | Caryophyllene oxide | −6.58 |
| 8 | Eucalyptol | −5.03 | 20 | Longifolenaldehyde | −6.86 |
| 9 | β-cis-Ocimene | −4.43 | 21 | Isospathulenol | −7.45 |
| 10 | γ-Terpinene | −4.43 | 22 | Aromadendrene oxide I | −6.74 |
| 11 | Terpinen-4-ol | −5.26 | 23 | Reference ligand | −7.78 |
| 12 | α-Copaene | −7.02 |
The hydrophobic interaction with Trp32 still prevailed in that specific series of compounds and most of them did not form a hydrogen bond with any of the amino acid residues at the binding site. This was demonstrated by visual inspection of the docking modes shown in Fig. 3. Among the 23 poses shown for the essential oil components and the reference compound, we could find that only two compounds, terpinen-4-ol and isospathulenol, did not show this interaction. In addition, previous studies have shown that targeting this Trp32-containing binding site will inhibit SOD1 misfolding/aggregation by stabilizing solvent-accessible hydrophobic residues.24 Isospathulenol is the only exception of the previous description and showed the highest binding energy with a value of −7.45 kcal mol−1. Such compound was able to form hydrophobic interaction with the key amino acid residue (Trp32) and other non-polar side chain of Val 29 and Val31. In addition, isospathulenol was able to interact with Glu100 through hydrogen bond in a similar pattern to the reference ligand with the naphthalene–catechol core. This unique well-adjusted interaction pattern of isospathulenol mimics the reference ligand with similar binding energy. As a result, a hydrophobic framework and hydroxy substituent properly positioned were essential structural features that could be extracted from the screened phytochemicals to attain binding affinity to our target enzyme. Thus, by substituting the hydrophobic core and adding extended substitutions appropriate locations, we can improve the hydrophobicity surrounding the central scaffold and obtain more potent analogs with tight binding affinity to superoxide dismutase 1. Fig. 3 shows the binding poses for the screened phytochemical compounds against human superoxide dismutase 1 (PDB Id: 5YTO).
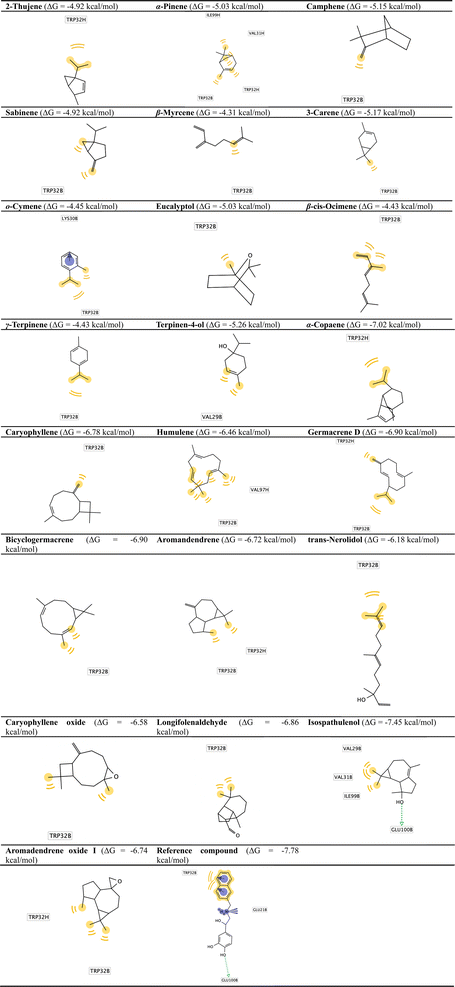 | ||
| Fig. 3 Docked poses of isolated phytochemicals in human superoxide dismutase I active site (PDB Id: 5YTO). | ||
3.3. Assessment of toxicity risks, druglikeness, and drug score for isolated phytochemicals
Using the open-source OSIRIS Property Explorer (https://www.organic-chemistry.org/prog/), we assessed the toxicity risks, drug-relevant properties, druglikeness, and drug score for isolated compounds in this work. Table 4 displayed the computed properties for the isolated phytochemicals. For this chemical series, the log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) P value (log(Coctanol/Cwater)) offers a trustworthy estimation of hydrophilicity. Low hydrophilicities brought on by a high log
P value (log(Coctanol/Cwater)) offers a trustworthy estimation of hydrophilicity. Low hydrophilicities brought on by a high log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) P value may indicate ineffective penetration or absorption of drug candidate. Compounds with a log
P value may indicate ineffective penetration or absorption of drug candidate. Compounds with a log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) P value of less than 5.0 are probably going to be absorbed effectively. Table 4 demonstrated low lipophilicity of our identified natural compounds (less than 5) except for entries 13–16 (caryophyllene humulene, germacrene D and bicyclogermacrene). Another factor provided by OSIRIS Property Explorer that affects drug absorption and distribution is aqueous solubility, denoted by log
P value of less than 5.0 are probably going to be absorbed effectively. Table 4 demonstrated low lipophilicity of our identified natural compounds (less than 5) except for entries 13–16 (caryophyllene humulene, germacrene D and bicyclogermacrene). Another factor provided by OSIRIS Property Explorer that affects drug absorption and distribution is aqueous solubility, denoted by log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) S. Inadequate water solubility of a chemical may lead to inadequate absorption. Log
S. Inadequate water solubility of a chemical may lead to inadequate absorption. Log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) S values are represented in mol liter−1 by the OSIRIS program. Eighty percent of the marketed drugs have log
S values are represented in mol liter−1 by the OSIRIS program. Eighty percent of the marketed drugs have log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) S values greater than −4. Based on the results on Table 4, log
S values greater than −4. Based on the results on Table 4, log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) S larger than −4 is present in all the 22 entries dictating the need for further structural modifications to obtain good drug candidate. The ideal option for molecular weights is to keep them as low as possible (less than 500) in order to enable optimal delivery and absorption to the site of action. All of the identified phytochemicals have low molecular weights not exceeding 500 securing a chemical space for molecular modifications. The total polar surface area (TPSA) of a molecule is computed by adding up its polar moieties of the molecular surface. It has a strong correlation with many bioavailability characteristics, including blood–brain barrier penetration and intestinal absorption. The molecules are likely to pass through membranes with ease if the total of all the polar atoms exposed to the molecule's surface is approximately 80 or 100 Å2. It is more likely that the other isolated chemicals won't have any problems with bioavailability after oral delivery since the TPSA value for all of them is less than 140 Å2.
S larger than −4 is present in all the 22 entries dictating the need for further structural modifications to obtain good drug candidate. The ideal option for molecular weights is to keep them as low as possible (less than 500) in order to enable optimal delivery and absorption to the site of action. All of the identified phytochemicals have low molecular weights not exceeding 500 securing a chemical space for molecular modifications. The total polar surface area (TPSA) of a molecule is computed by adding up its polar moieties of the molecular surface. It has a strong correlation with many bioavailability characteristics, including blood–brain barrier penetration and intestinal absorption. The molecules are likely to pass through membranes with ease if the total of all the polar atoms exposed to the molecule's surface is approximately 80 or 100 Å2. It is more likely that the other isolated chemicals won't have any problems with bioavailability after oral delivery since the TPSA value for all of them is less than 140 Å2.
| Entry | Comp. id | c![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) Log Log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) P P |
Log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) S S |
Mol. weight | TPSA (Å2) | Druglike-nesss | Drug-score | Toxicity risks (mutagenicity, tumorigenicity, irritancy, reproductive effects) |
|---|---|---|---|---|---|---|---|---|
| 1 | 2-Thujene | 2.61 | −2.55 | 136.0 | 0 | −1.61 | 0.53 | None |
| 2 | α-Pinene | 2.72 | −2.52 | 136.0 | 0 | −1.8 | 0.31 | Irritant |
| 3 | Camphene | 2.8 | −2.69 | 136.0 | 0 | −5.86 | 0.27 | Mutagenic |
| 4 | Sabinene | 2.86 | −2.69 | 136.0 | 0 | −6.78 | 0.45 | None |
| 5 | β-Myrcene | 4.29 | −2.5 | 136.0 | 0 | −7.82 | 0.09 | Tumorigenic, irritant, and reproductive effects |
| 6 | 3-Carene | 2.72 | −2.52 | 136.0 | 0 | −2.9 | 0.17 | Tumorigenic and irritant |
| 7 | o-Cymene | 3.19 | −2.83 | 134.0 | 0 | −2.42 | 0.47 | None |
| 8 | Eucalyptol | 2.11 | −2.48 | 154.0 | 9.23 | −3.21 | 0.17 | Mutagenic and reproductive effects |
| 9 | β-cis-Ocimene | 4.23 | −2.33 | 136.0 | 0 | −5.46 | 0.41 | None |
| 10 | γ-Terpinene | 3.05 | −2.37 | 136.0 | 0 | −2.82 | 0.29 | Irritant |
| 11 | Terpinen-4-ol | 2.34 | −2.19 | 154.0 | 20.23 | −7.41 | 0.28 | Irritant |
| 12 | α-Copaene | 3.98 | −3.62 | 204.0 | 0 | −6.25 | 0.23 | Irritant |
| 13 | Caryophyllene | 5.49 | −3.66 | 204.0 | 0 | −6.49 | 0.18 | Irritant |
| 14 | Humulene | 6.24 | −3.4 | 204.0 | 0 | −4.72 | 0.28 | None |
| 15 | Germacrene D | 5.96 | −3.55 | 204.0 | 0 | −10.29 | 0.28 | None |
| 16 | Bicyclogermacrene | 5.53 | −3.49 | 204.0 | 0 | −4.88 | 0.07 | Mutagenic, tumorigenic, and irritant effects |
| 17 | Aromandendrene | 4.0 | −3.79 | 204.0 | 0 | −7.14 | 0.14 | Tumorigenic and irritant |
| 18 | trans-Nerolidol | 5.4 | −3.12 | 222.0 | 20.23 | −6.38 | 0.19 | Irritant |
| 19 | Caryophyllene oxide | 4.06 | −3.56 | 220.0 | 12.53 | −4.77 | 0.25 | None |
| 20 | Longifolenaldehyde | 2.95 | −3.61 | 220.0 | 17.07 | −7.3 | 0.25 | Irritant |
| 21 | Isospathulenol | 3.27 | −3.14 | 220.0 | 20.23 | −1.87 | 0.17 | Tumorigenic and irritant |
| 22 | Aromadendrene oxide I | 2.92 | −3.41 | 220.0 | 12.53 | −4.97 | 0.09 | Mutagenic, tumorigenic, and irritant |
Utilizing fragment-based druglikeness, OSIRIS Explorer utilizes druglikeness scores for 5300 distinct substructure moieties. Subunits from 15K commercial chemical compounds (Fluka) and 3300 marketed pharmaceuticals were gathered to create the OSIRIS fragment library. Based on statistical analysis of druglikeness scores, nearly 80% of drug molecules are found in the positive range, whereas the majority of Fluka compounds are found in the negative range. Therefore, keeping the druglikeness value of potential drug candidate in the positive range is a smart choice. As we can see, screened phytochemical compounds have drug likeness values in the negative range. Isospathulenol, showing the highest binding affinity with superoxide dismutase 1, has a promising druglikeness value of −1.87 which is the upper negative range. The OSIRIS explorer tool generates toxicity risk alerts, which indicate that the provided structure may have hazardous effects related to a certain risk category. The submission of compounds with a substructure that generates toxicity alerts is detected by the OSIRIS explorer. The majority of the derived results were predicated on the notion that most marketed drugs are toxicity-free. Risky fragments are those that indicate a dangerous compound's substructure and are infrequently or never present in medications that are on the market. It is important to remember that the same component may or may not have harmful effects depending on whether risk alerts are present. The entries 1, 4, 7, 9, 13, 14, 15, 16, and 19 did not exhibit any discernible tendency to cause toxic consequences, however the remaining isolated compounds were observed to have potential toxicity hazards. Typically, c![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) log
log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) P, log
P, log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) S, molecular weight, druglikeness, and toxicity risks are combined into one value to generate the drug-score in OSIRIS explorer. A medicinal chemist might use this useful parameter to find out if the compound qualifies as a drug candidate. As we can see, the isolated phytochemicals have drug scores ranging from 0.09 to 0.53. Isospathulenol, with tight binding affinity with superoxide dismutase 1, has a moderate dug score value of 0.17. This could be promising, which needs further improvement to get rid of toxicity risks for that particular compound.
S, molecular weight, druglikeness, and toxicity risks are combined into one value to generate the drug-score in OSIRIS explorer. A medicinal chemist might use this useful parameter to find out if the compound qualifies as a drug candidate. As we can see, the isolated phytochemicals have drug scores ranging from 0.09 to 0.53. Isospathulenol, with tight binding affinity with superoxide dismutase 1, has a moderate dug score value of 0.17. This could be promising, which needs further improvement to get rid of toxicity risks for that particular compound.
3.4. Assessment of cellular permeability, pharmacokinetics, and medicinal chemistry friendliness of isolated phytochemical compounds
We employed SwissADME, a free web server, to compute the necessary factors for our separated phytochemicals to be used as a viable drug candidate. The calculated properties are shown in Table 5. A drug molecule needs to reach its action site at a high enough concentration and remain there long enough to cause the intended biological reaction. Because of this, medicinal chemists frequently use various computer models to estimate various physicochemical characteristics for drug compounds. The extracted phytochemicals were submitted to be evaluated for ADME, pharmacokinetics, and medicinal chemistry friendliness using the online SwissADME web browser (http://www.swissadme.ch/).59 Both passive permeability in gastrointestinal absorption and penetration through the blood–brain barrier (BBB) were determined using the BOILED-Egg model.73 With the exception of eucalyptol, terpinen-4-ol, trans-nerolidol, caryophyllene oxide, longifolenaldehyde, isospathulenol, and aromadendrene oxide I, the identified phytochemical compounds exhibit low GIT permeability. Moreover, the identified natural compounds showed strong BBB permeability with the exception of caryophyllene, humulene, germacrene D, and bicyclogermacrene (Table 5). A drug molecule's potential to develop into an oral drug candidate with adequate bioavailability may be indicated by its drug-likeness value. The identified twenty-two compounds was assessed based on Lipinski's rule of five (Ro5). The Ro5 states that a candidate meets the following requirements to be considered drug-like and orally active: molecular weight <500 Da, octanol–water partition coefficient (log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) P ≤ 5), hydrogen-bond donor (HBD) ≤ 5, and hydrogen-bond acceptor (HBA) ≤ 10.74 All of the identified phytochemical compounds displayed 0–1 violations of Lipinski's rule of five.
P ≤ 5), hydrogen-bond donor (HBD) ≤ 5, and hydrogen-bond acceptor (HBA) ≤ 10.74 All of the identified phytochemical compounds displayed 0–1 violations of Lipinski's rule of five.
| Entry | Comp. id | GI absorption | BBB permeant | Lipinski N. violations | PAINS N. alert | Bioavailability score | Synthetic accessibility |
|---|---|---|---|---|---|---|---|
| 1 | 2-Thujene | Low | Yes | 1 | 0 | 0.55 | 4.19 |
| 2 | α-Pinene | Low | Yes | 1 | 0 | 0.55 | 4.44 |
| 3 | Camphene | Low | Yes | 1 | 0 | 0.55 | 3.50 |
| 4 | Sabinene | Low | Yes | 1 | 0 | 0.55 | 2.87 |
| 5 | β-Myrcene | Low | Yes | 0 | 0 | 0.55 | 2.85 |
| 6 | 3-Carene | Low | Yes | 1 | 0 | 0.55 | 3.84 |
| 7 | o-Cymene | Low | Yes | 1 | 0 | 0.55 | 1.00 |
| 8 | Eucalyptol | High | Yes | 0 | 0 | 0.55 | 3.65 |
| 9 | β-cis-Ocimene | Low | Yes | 0 | 0 | 0.55 | 3.63 |
| 10 | γ-Terpinene | Low | Yes | 0 | 0 | 0.55 | 3.11 |
| 11 | Terpinen-4-ol | High | Yes | 0 | 0 | 0.55 | 3.28 |
| 12 | α-Copaene | Low | Yes | 1 | 0 | 0.55 | 4.62 |
| 13 | Caryophyllene | Low | No | 1 | 0 | 0.55 | 4.51 |
| 14 | Humulene | Low | No | 1 | 0 | 0.55 | 3.66 |
| 15 | Germacrene D | Low | No | 1 | 0 | 0.55 | 4.55 |
| 16 | Bicyclogermacrene | Low | No | 1 | 0 | 0.55 | 4.34 |
| 17 | Aromandendrene | Low | Yes | 1 | 0 | 0.55 | 3.70 |
| 18 | trans-Nerolidol | High | Yes | 0 | 0 | 0.55 | 3.53 |
| 19 | Caryophyllene oxide | High | Yes | 0 | 0 | 0.55 | 4.35 |
| 20 | Longifolenaldehyde | High | Yes | 0 | 0 | 0.55 | 3.45 |
| 21 | Isospathulenol | High | Yes | 0 | 0 | 0.55 | 4.35 |
| 22 | Aromadendrene oxide I | High | Yes | 0 | 0 | 0.55 | 4.03 |
Another computational item used by medicinal chemists to direct their efforts in creating novel chemical entities is “medicinal chemistry friendliness”. The capacity to identify potentially troublesome substructure or patterns and decide about which compounds to pursue in order of priority is a fundamental and crucial stage in every drug discovery endeavor. PAINS (also known as frequent hitters, promiscuous compounds, or pan assay interference compounds) are chemicals with substructure moieties that, regardless of the target protein, exhibit a potent and measurable response in several assays. The fragments would be considered a possible threat to furnish frequent hitters if they resulted in false positives in six orthogonal assays.75 In the event that the submitted molecule has such substructures, the SwissADME server would raise alarms. All isolated phytochemical compounds displayed zero warnings for the PAINS filter. The computation of a compound's likelihood of having at least 10% oral bioavailability in rats or the Caco-2 permeability model is the basis of the Abbot Bioavailability Score. A violation of the Lipinski rule of five, total charge, and TPSA all affect this bioavailability score. Additionally, this score might be useful in grouping submitted compounds into four probability groups: 11%, 17%, 56%, or 85%. All of our extracted phytochemical components displayed a score of 0.55. The SwissADME service also offers a synthetic accessibility (SA) factor that aids medicinal chemists in determining whether the submitted molecule may be readily synthesized and tested in bio-assays. It ranges from 1 (very simple) to 10 (extremely difficult). 1024 Building blocks contributed to the calculation of synthetic accessibility range relying in a database of building blocks with different size and complexity.76 Table 5 displays the different SA scores of the extracted phytochemical compounds ranging from 1.00 to 4.62.
4 Conclusion
GC/MS analysis showed that the essential oil of farmed Lantana camara is mostly composed of a combination of 22 components, the two main ones being the sesquiterpene caryophyllene (15.51%) and monoterpene sabinene (14.90%). Molecular docking against human superoxide dismutase 1 was performed revealing isospathulenol with promising binding affinity. The same compounds showed high potential with good PK profile and ease of synthetic diversification. This establishes a future position for such compound for development into drug candidates for treatment or control ALS.Consent to participate
All authors worked on the final manuscript.Consent for publication
All authors agreed to publish the final manuscript.Data availability
The data that support the findings of this study are available from the corresponding author upon reasonable request.Author contributions
Abdullah Haikal: conceptualization, collected the plant, prepared the essential oil, wrote the first draft and revised the final draft of the manuscript; Ahmed R. Ali: ran computational studies, wrote the first draft and revised the final draft of the manuscript.Conflicts of interest
The Authors declare that there is no conflict of interest.Acknowledgements
This research did not receive any specific grant from funding agencies in the public, commercial, or not-for-profit sectors.References
- Q. Q. Tao and Z. Y. Wu, China Med. J., 2017, 130, 2269–2272 Search PubMed.
- S. Zarei, K. Carr, L. Reiley, K. Diaz, O. Guerra, P. F. Altamirano, W. Pagani, D. Lodin, G. Orozco and A. Chinea, Surg. Neurol. Int., 2015, 6, 171 CrossRef PubMed.
- M. A. van Es, O. Hardiman, A. Chio, A. Al-Chalabi, R. J. Pasterkamp, J. H. Veldink and L. H. van den Berg, Lancet, 2017, 390, 2084–2098 CrossRef PubMed.
- J. D. Berry, M. Blanchard, K. Bonar, E. Drane, M. Murton, U. Ploug, K. Ricchetti-Masterson, N. Savic, E. Worthington and T. Heiman-Patterson, Amyotrophic Lateral Scler. Frontotemporal Degener., 2023, 24, 436–448 CrossRef PubMed.
- C. Wolfson, D. E. Gauvin, F. Ishola and M. Oskoui, Neurology, 2023, 101, e613–e623 CrossRef PubMed.
- K. C. Arthur, A. Calvo, T. R. Price, J. T. Geiger, A. Chio and B. J. Traynor, Nat. Commun., 2016, 7, 12408 CrossRef CAS PubMed.
- V. D. Bello-Haas, Degener. Neurol. Neuromuscular Dis., 2018, 8, 45–54 Search PubMed.
- H. Cho and S. Shukla, Pharmaceuticals, 2021, 14(1), 29 CrossRef CAS PubMed.
- G. L. Pattee, A. Genge, P. Couratier, C. Lunetta, G. Sobue, M. Aoki, H. Yoshino, C. E. Jackson, J. Wymer, A. Salah and S. Nelson, Expert Rev. Neurother., 2023, 23, 859–866 CrossRef CAS PubMed.
- M. K. Jaiswal, Med. Res. Rev., 2019, 39, 733–748 CrossRef PubMed.
- A. D. Gitler, P. Dhillon and J. Shorter, Dis. Models Mech., 2017, 10, 499–502 CrossRef CAS PubMed.
- R. Mancuso and X. Navarro, Prog. Neurobiol., 2015, 133, 1–26 CrossRef PubMed.
- J. Lee, S. J. Hyeon, H. Im, H. Ryu, Y. Kim and H. Ryu, Exp. Neurobiol., 2016, 25, 233–240 CrossRef PubMed.
- A. M. Dekker, M. Seelen, P. T. van Doormaal, W. van Rheenen, R. J. Bothof, T. van Riessen, W. J. Brands, A. J. van der Kooi, M. de Visser, N. C. Voermans, R. J. Pasterkamp, J. H. Veldink, L. H. van den Berg and M. A. van Es, Neurobiol. Aging, 2016, 39, 220 CrossRef PubMed.
- L. Tang, Y. Ma, X. L. Liu, L. Chen and D. S. Fan, Transl. Neurodegener., 2019, 8, 2 CrossRef PubMed.
- V. Rodriguez-Sureda, A. Vilches, O. Sanchez, L. Audi and C. Dominguez, Oxid. Med. Cell. Longev., 2015, 2015, 509241 Search PubMed.
- S. J. Kaur, S. R. McKeown and S. Rashid, Gene, 2016, 577, 109–118 CrossRef CAS PubMed.
- G. R. C. Pereira, A. N. R. Da Silva, S. S. Do Nascimento and J. F. De Mesquita, J. Cell. Biochem., 2019, 120, 3583–3598 CrossRef CAS PubMed.
- O. Pansarasa, M. Bordoni, L. Diamanti, D. Sproviero, S. Gagliardi and C. Cereda, Int. J. Mol. Sci., 2018, 19(5), 1345 CrossRef PubMed.
- A. A. Brasil, M. D. C. de Carvalho, E. Gerhardt, D. D. Queiroz, M. D. Pereira, T. F. Outeiro and E. C. A. Eleutherio, Proc. Natl. Acad. Sci. U. S. A., 2019, 116, 25991–26000 CrossRef CAS PubMed.
- U. Shareef, A. Altaf, M. Ahmed, N. Akhtar, M. S. Almuhayawi, S. K. Al Jaouni, S. Selim, M. A. Abdelgawad and M. K. Nagshabandi, Saudi Pharmaceut. J., 2024, 32, 101913 CrossRef CAS PubMed.
- P. Soares, C. Silva, D. Chavarria, F. S. G. Silva, P. J. Oliveira and F. Borges, Ageing Res. Rev., 2023, 83, 101790 CrossRef CAS PubMed.
- A. DeLoach, M. Cozart, A. Kiaei and M. Kiaei, Expert Opin. Drug Discovery, 2015, 10, 1099–1118 CrossRef CAS PubMed.
- A. Rahman, B. Saikia and A. Baruah, Phys. Chem. Chem. Phys., 2023, 25, 26833–26846 RSC.
- V. Kumar, P. Gupta and M. I. Hassan, J. Proteins Proteomics, 2019, 10, 131–147 Search PubMed.
- E. L. Ghisalberti, Fitoterapia, 2000, 71, 467–486 CrossRef CAS PubMed.
- O. P. Sharma, A. Singh and S. Sharma, Fitoterapia, 2000, 71, 487–491 CrossRef CAS PubMed.
- S. K. Srivastava, M. Khan and S. P. S. Khanuja, US Pat., 6,893,668, 2005 Search PubMed.
- O. P. Sharma, H. P. Makkar, R. K. Dawra and S. S. Negi, Clin. Toxicol., 1981, 18, 1077–1094 CrossRef CAS PubMed.
- R. Sathish, B. Vyawahare and K. Natarajan, J. Ethnopharmacol., 2011, 134, 195–197 CrossRef CAS PubMed.
- L. Sagar, R. Sehgal and S. Ojha, BMC Complementary Altern. Med., 2005, 5, 18 CrossRef PubMed.
- M. D. Day, C. J. Wiley, J. Playford and M. P. Zalucki, Aust. Cent. Int. Agric. Res., 2003, 102 Search PubMed.
- E. O. Sousa, T. S. Almeida, I. R. Menezes, F. F. Rodrigues, A. R. Campos, S. G. Lima and J. G. da Costa, Rec. Nat. Prod., 2012, 6, 144–150 CAS.
- O. P. Sharma, S. Sharma, V. Pattabhi, S. B. Mahato and P. D. Sharma, Crit. Rev. Toxicol., 2007, 37, 313–352 CrossRef CAS PubMed.
- S. Begum, A. Ayub, S. Qamar Zehra, B. Shaheen Siddiqui and M. Iqbal, Choudhary and Samreen, Chem. Biodiversity, 2014, 11, 709–718 CrossRef CAS PubMed.
- F. Qamar, S. Begum, S. M. Raza, A. Wahab and B. S. Siddiqui, Nat. Prod. Res., 2005, 19, 609–613 CrossRef CAS PubMed.
- J. M. Herbert, J. P. Maffrand, K. Taoubi, J. M. Augereau, I. Fouraste and J. Gleye, J. Nat. Prod., 1991, 54, 1595–1600 CrossRef CAS PubMed.
- S. Begum, A. Wahab and B. S. Siddiqui, Nat. Prod. Res., 2008, 22, 467–470 CrossRef CAS PubMed.
- S. Begum, S. M. Raza, B. S. Siddiqui and S. Siddiqui, J. Nat. Prod., 2004, 58, 1570–1574 CrossRef.
- P. Weyerstahl, H. Marschall, A. Eckhardt and C. Christiansen, Flavour Fragrance J., 1999, 14, 15–28 CrossRef CAS.
- J.-A. Randrianalijaona, P. A. R. Ramanoelina, J. R. E. Rasoarahona and E. M. Gaydou, Anal. Chim. Acta, 2005, 545, 46–52 CrossRef CAS.
- J. Benites, C. Moiteiro, G. Miguel, L. Rojo, J. LÓPez, F. VenÂNcio, L. Ramalho, S. Feio, S. Dandlen, H. Casanova and I. Torres, J. Chil. Chem. Soc., 2009, 54, 379–384 CAS.
- N. R. Tesch, F. Mora, L. Rojas, T. Díaz, J. Velasco, C. Yánez, N. Rios, J. Carmona and S. Pasquale, Nat. Prod. Commun., 2011, 6, 1031–1034 CrossRef CAS PubMed.
- E. O. Sousa, J. B. T. Rocha, L. M. Barros, A. R. C. Barros and J. G. M. Costa, Ind. Crops Prod., 2013, 43, 517–522 CrossRef CAS.
- S. Zoubiri and A. Baaliouamer, J. Essent. Oil Res., 2012, 24, 377–383 CrossRef CAS.
- M. Verdeguer, M. A. Blázquez and H. Boira, Biochem. Syst. Ecol., 2009, 37, 362–369 CrossRef CAS.
- V. K. Dua, A. C. Pandey and A. P. Dash, Indian J. Med. Res., 2010, 131, 434–439 CAS.
- A. Haikal, N. Thangavel, M. Albratty, A. Najmi, H. A. Al Hazmi, D. Sivadasan, G. Khuwaja and I. M. Shamkh, Lett. Drug Des. Discov., 2024, 21, 1569–1581 CrossRef CAS.
- M. Elkazzaz, A. Ahmed, Y. E. E. Abo Amer, T. Haydara, W. A. Eltayb, M. S. Khan, K. Bhattacharya, S. Alkhammash, H. Mattar, S. Beigh, M. F. Abo El Magd, A. Haikal, I. Abid, A. S. Abouzied and I. M. Shamkh, Indian J. Pharm. Educ. Res., 2023, 57, 1044–1052 CrossRef CAS.
- I. M. Shamkh, M. Elkazzaz, E. S. Radwan, J. Najeeb, M. T. Rehman, M. F. AlAjmi, M. Shahwan, M. Sufyan, N. K. Alaqeel, I. A. Ibrahim, B. Jabbar, M. S. Khan, T. M. Karpinski, A. Haikal, R. M. Aljowaie, S. M. Almutairi and A. Ahmed, Curr. Pharm. Des., 2023, 29, 2752–2762 CrossRef CAS PubMed.
- World Checklist of Selected Plant Families (WCSP)., Lanatana camara L. in the plant list, https://www.gbif.org/species/2925303, accessed on 6/5/2024 3:00 pm.
- African Plant Database., Lantana camara L., https://africanplantdatabase.ch/en/nomen/specie/155431/lantana-camara-l, accessed on 6/5/2024 3:00 pm.
- European Directorate for the Quality of Medicines, European pharmacopopoeia, European Council, Maisonneuve, France, 1975, vol. 3, pp. 68–71 Search PubMed.
- H. van Den Dool and P. D. Kratz, J. Chromatogr., 1963, 11, 463–471 CrossRef CAS PubMed.
- A. Haikal, A. A. Galala, M. Elshal, Y. Amen and A. A. Gohar, J. Ethnopharmacol., 2024, 318, 117000 CrossRef CAS PubMed.
- A. Haikal, M. El-Neketi, W. F. Awadin, M. A. Hassan and A. A. Gohar, J. King Saud Univ., Sci., 2022, 34(8), 102356 CrossRef.
- G. M. Morris, R. Huey, W. Lindstrom, M. F. Sanner, R. K. Belew, D. S. Goodsell and A. J. Olson, J. Comput. Chem., 2009, 30, 2785–2791 CrossRef CAS PubMed.
- G. Wolber and T. Langer, J. Chem. Inf. Model., 2005, 45, 160–169 CrossRef CAS PubMed.
- A. Daina, O. Michielin and V. Zoete, Sci. Rep., 2017, 7, 42717 CrossRef PubMed.
- T. Sander, J. Freyss, M. von Korff and C. Rufener, J. Chem. Inf. Model., 2015, 55, 460–473 CrossRef CAS PubMed.
- R. P. Adams, Identification of essential oil components by gas chromatography/mass spectrometry, Texensis Publishing, Gruver, TX USA, 2017 Search PubMed.
- N. M. Abdel-Hady, A. S. Abdei-Halim and A. M. Al-Ghadban, J. Egypt. Soc. Parasitol., 2005, 35, 687–698 Search PubMed.
- C. d. S. Ricardo, A. d. M. F. Antonio, A. C. Edvan, A. T. Jacqueline, P. F. Vany, M. F. a. n. Ismael, R. E. R. Pedro, C. G. c. Ana and C. H. Luciana, Res. J. Med. Plant, 2015, 9, 922–928 CrossRef.
- M. B. Ngassoum, S. Yonkeu, L. Jirovetz, G. Buchbauer, G. Schmaus and F.-J. Hammerschmidt, Flavour Fragrance J., 1999, 14, 245–250 CrossRef CAS.
- F. Nea, D. A. Kambire, M. Genva, E. A. Tanoh, E. L. Wognin, H. Martin, Y. Brostaux, F. Tomi, G. C. Lognay, Z. F. Tonzibo and M. L. Fauconnier, Molecules, 2020, 25(10), 2400 CrossRef CAS PubMed.
- L. Misra and A. K. Saikia, J. Essent. Oil Res., 2011, 23, 1–5 CrossRef CAS.
- C. F. Zuhra and Y. S. Sembiring, Biomedika, 2021, 14, 19–28 CrossRef.
- A. A. Kasali, O. Ekundayo, C. Paul, W. A. Koenig, A. O. Eshilokun and P. Yadua, J. Essent. Oil Res., 2004, 16, 582–584 CrossRef.
- M. Khan, A. Mahmood and H. Z. Alkhathlan, Arab. J. Chem., 2016, 9, 764–774 CrossRef CAS.
- S. G. Rhee, K. S. Yang, S. W. Kang, H. A. Woo and T. S. Chang, Antioxid. Redox Signaling, 2005, 7, 619–626 CrossRef CAS PubMed.
- D. R. Rosen, T. Siddique, D. Patterson, D. A. Figlewicz, P. Sapp, A. Hentati, D. Donaldson, J. Goto, J. P. O'Regan and H. X. Deng, et al., Nature, 1993, 362, 59–62 CrossRef CAS PubMed.
- R. Manjula, G. S. A. Wright, R. W. Strange and B. Padmanabhan, FEBS Lett., 2018, 592, 1725–1737 CrossRef CAS PubMed.
- A. Daina and V. Zoete, ChemMedChem, 2016, 11, 1117–1121 CrossRef CAS PubMed.
- C. A. Lipinski, F. Lombardo, B. W. Dominy and P. J. Feeney, Adv. Drug Deliv. Rev., 2001, 46, 3–26 CrossRef CAS PubMed.
- J. B. Baell and G. A. Holloway, J. Med. Chem., 2010, 53, 2719–2740 CrossRef CAS PubMed.
- Y. Fukunishi, T. Kurosawa, Y. Mikami and H. Nakamura, J. Chem. Inf. Model., 2014, 54, 3259–3267 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d4ra04281f |
| This journal is © The Royal Society of Chemistry 2024 |