Open Access Article
Open Access ArticleNew phenanthrenequinones from Cymbidium ensifolium roots and their anti-inflammatory activity on lipopolysaccharide-activated BV2 microglial cells†
May Thazin Thanta,
Hasriadi Hasriadib,
Preeyaporn Poldornc,
Siriporn Jungsuttiwong d,
Pornchai Rojsitthisak
d,
Pornchai Rojsitthisak ef,
Chotima Böttcherg,
Pasarapa Towiwat
ef,
Chotima Böttcherg,
Pasarapa Towiwat bh and
Boonchoo Sritularak
bh and
Boonchoo Sritularak *ae
*ae
aDepartment of Pharmacognosy and Pharmaceutical Botany, Faculty of Pharmaceutical Sciences, Chulalongkorn University, Bangkok 10330, Thailand. E-mail: boonchoo.sr@chula.ac.th
bDepartment of Pharmacology and Physiology, Faculty of Pharmaceutical Sciences, Chulalongkorn University, Bangkok 10330, Thailand
cHong Kong Quantum AI Laboratory, Ltd., Hong Kong 999077, China
dDepartment of Chemistry, Center of Excellence for Innovation in Chemistry, Faculty of Science, Ubon Ratchathani University, Ubon Ratchathani 34190, Thailand
eCenter of Excellence in Natural Products for Ageing and Chronic Diseases, Chulalongkorn University, Bangkok, 10330, Thailand
fDepartment of Food and Pharmaceutical Chemistry, Faculty of Pharmaceutical Sciences, Chulalongkorn University, Bangkok 10330, Thailand
gExperimental and Clinical Research Center, A Cooperation Between the Max Delbrück Center for Molecular Medicine in the Helmholtz Association and Charité – Universitätsmedizin Berlin, Berlin 13125, Germany
hAnimal Models of Chronic Inflammation-associated Diseases for Drug Discovery Research Unit, Chulalongkorn University, Bangkok, 10330, Thailand
First published on 5th September 2024
Abstract
The roots of Cymbidium ensifolium yielded a total of 17 compounds, comprising two new compounds (1–2), one new natural product (3), and 14 known compounds (4–17). The structures of new compounds were determined through the analysis of their spectroscopic data, including NMR, MS, UV, FT-IR, optical rotation, and CD. The anti-inflammatory activity of the isolated pure compounds was assessed using lipopolysaccharide-activated BV2 microglial cells. Compounds 1, 3, 6, 12, 14, and 16 showed the ability to reduce LPS induced NO release in BV2 microglial cells, with IC50 values of 9.95 ± 2.13, 8.77 ± 3.78, 2.39 ± 0.91, 6.69 ± 2.94, 2.96 ± 1.38, 8.42 ± 2.99 μM, respectively and reduced the secretion of proinflammatory mediators (TNF-α, IL-6, MCP-1) in a concentration-dependent manner. Furthermore, the mechanistic role of the compound 3 was determined, which demonstrated its ability to inhibit the nuclear factor-κB (NF-κB) pathway through decreasing phosphorylation of p65 subunits.
Introduction
Neuroinflammation has gained significant interest in recent years, particularly as it has been linked to chronic and progressive neurodegenerative conditions like Alzheimer's disease, Parkinson's disease, and multiple sclerosis.1 Microglia, the primary innate immune cells in the central nervous system, play crucial roles in supporting the health of neurons by contributing to cellular maintenance and innate immune responses.2 Microglia activation is evident in brain injuries and is triggered by various stimuli such as lipopolysaccharides (LPS), interferon-γ, or β-amyloid exposure.3 Activated microglia, considered a hallmark of neurodegeneration, can potentially exacerbate neurodegenerative processes by releasing pro-inflammatory and/or cytotoxic factors like IL-1β, TNF-α, NO, and reactive oxygen intermediates (ROS).4 These neurotoxic factors contribute to neuronal cell damage and the development of neurodegenerative diseases.5 Hence, the development of agents capable of inhibiting the production of inflammatory mediators holds promise as a therapeutic approach for treating neurodegenerative diseases.6Cymbidium, a genus belonging to the orchid family, is widely distributed throughout Southeast Asia, China, Japan, and Northern Australia.7 Certain species of Cymbidium, including Cymbidium finlaysonianum Lindl., Cymbidium aloifolium (L.) Sw., and Cymbidium ensifolium (L.) Sw., have been traditionally utilized as herbal remedies by Thai practitioners. Compounds isolated from C. finlaysonianum have been evaluated for cytotoxic activity against human small cell lung cancer (NCI-H187) cells.8 The ethanolic leaf extract of C. aloifolium has been documented for its analgesic and anti-inflammatory properties.9 The roots of Cymbidium ensifolium, referred to as “nang kham” or “chulan” in Thai, are utilized in traditional Thai medicine to alleviate liver dysfunction and nephropathy.10 In our previous studies, we investigated compounds isolated from the aerial parts of C. ensifolium for cytotoxic effects against lung cancer H460, breast cancer MCF7, and colon cancer CaCo2 cells.11 As part of our ongoing research on bioactive compounds from orchids,12–14 we investigated the chemical constituents of the roots of C. ensifolium and assessed their effectiveness in inhibiting neuroinflammation.
Results and discussion
Structural determination
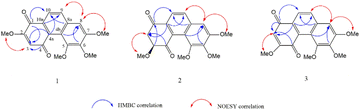
A phytochemical investigation of the methanolic extract of the roots of Cymbidium ensifolium resulted in the isolation of 17 compounds, including two new compounds (1–2), one new natural product (3), and 14 known compounds (4–17). The structures of known compounds were identified as 9,10-dihydro-2,5-dimethoxy-1,7-phenanthrenediol (4),15 p-hydroxybenzaldehyde (5),16 coelonin (6),17 hircinol (7),18 3,7-dihydroxy-2,4,6-trimethoxyphenanthrene (8),19 bulbophyllanthrin (9),20 p-hydroxybenzoic acid (10),21 vanillic acid (11),22 parviphenanthrine C (12),23 denthyrsinin (13),24 1-(4-hydroxylbenzyl)-4-methoxy 9,10-dihydrophenanthrene 2,7-diol (14),25 4-hydroxymethyl benzoic acid (15),26 blestriarene C (16),27 and N-trans-feruloyl tyramine (17)28 (Fig. 1).Compound 1 was obtained as an orange-colored amorphous solid. The molecular formula C18H16O6 was analyzed from its [M + H]+ at m/z 329.1019 (calcd for C18H17O6 at 329.1020) in the HR-ESI-MS. Furthermore, it had another major peak at m/z 679.1789 that coincided with the calculated 2M peak, [2M + Na]+ at m/z 679.1791. FT-IR revealed absorption bands at 3396 cm−1 (hydroxyl), 1726 cm−1 (carbonyl), 2922, 1461, and 1376 cm−1 (aromatic). The compound revealed UV absorptions at 210, 240, 305, and 395 nm indicated an aromatic system.29 The 13C NMR and DEPT spectra revealed eighteen carbon signals comprising four methoxy, four methines, and ten quaternary carbons. The presence of two carbonyl carbons can be supported by the chemical shifts δC 179.4 and 185.4. The 1H NMR spectrum of 1 also exhibited two singlet proton signals at δH 6.29 (1H, s, H-3) and 7.29 (1H, s, H-8), two ortho-coupled doublet protons at δH 8.01 (1H, d, J = 8.8 Hz, H-9) and 7.95 (1H, d, J = 8.8 Hz, H-10), four methoxy groups at δH 3.94 (3H, s, MeO-2), 4.05 (3H, s, MeO-5), 3.91 (3H, s, MeO-6) and 4.06 (3H, s, MeO-7). The HMBC correlations of C-1 (δC 179.4) with H-3 and H-10; C-4 (δC 185.4) with H-3 indicated the quinone structure at ring A of compound 1. The first methoxy group (δH 3.94) was positioned on ring A at C-2, as deduced from its HSQC correlation with MeO-2 carbon (δC 55.7), and NOESY interaction with H-3. It was supported by the HMBC correlation of C-2 (δC 158.2) with H-3 and MeO-2 protons. On ring B, one singlet proton at δH 7.29 was assigned as H-8 based on its HMBC correlation with C-9 (δC 131.2) and NOESY interaction with H-9 (Fig. 2). The position of second methoxy (δH 4.06) at C-7 was supported by its HSQC correlation with MeO-7 carbon (δC 60.8), HMBC correlation with C-7 (δC 156.2), and NOESY correlation with H-8. The third methoxy group (δH 3.91) was assigned to C-6 based on its HSQC correlation with MeO-6 carbon (δC 60.0); the HMBC correlations of C-6 (δC 143.8) with MeO-6 and H-8. The HSQC correlation between the methoxy group at δH 4.05 and MeO-5 carbon (δC 55.6); the HMBC correlation between MeO-5 protons and C-5 (δC 150.5) confirmed the fourth methoxy group at C-5. Based on the above spectra, compound 1 was characterized as 2,5,6,7-tetramethoxy-1,4-phenanthrenequinone and given the trivial name cymensifin D.
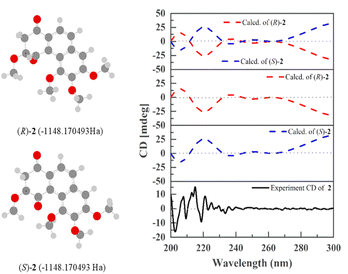
Compound 2 was obtained as a pale yellow amorphous solid. The molecular formula C18H18O6 was analyzed from its [M + H]+ at m/z 331.1176 (calcd for C18H19O6 331.1181) in the HR-ESI-MS. At m/z 683.2095, it likewise displayed a significant peak that matched the calculated 2M peak, [2M + Na]+ at m/z 683.2105. FT-IR revealed absorption bands at 3333 cm−1 (hydroxyl), 1717 and 1679 cm−1 (carbonyl), 2923, 1464, and 1316 cm−1 (aromatic).29 The UV absorption bands exhibited at 205, 230, 280 and 332 nm indicate an aromatic system.29 The 1H NMR spectrum revealed a pair of ortho-coupled protons at δH 7.84 (d, J = 8.8 Hz, H-9), and 7.94 (d, J = 8.5 Hz, H-10); an aromatic proton at δH 7.02 (s, H-8); one oxymethine at δH 4.67 (dd, J = 8.4, 6.0 Hz, H-3); four methoxy groups at δH 3.64 (s, MeO-3), 4.11 (s, MeO-5), 3.95 (s, MeO-6) and 4.05 (s, MeO-7); methylene protons at δH 3.42 (dd, J = 17.6, 6.0 Hz, H-2a) and 3.15 (dd, J = 17.6, 8.4 Hz, H-2b). The 13C NMR and DEPT spectra also exhibited eighteen carbon signals. Among them, two carbonyl carbons at δC 192.5 (C-1) and 201.3 (C-4); one methylene at δC 46.8 (C-2) and one oxymethine carbon at δC 82.2 (C-3) were positioned on ring A based on their HMBC correlations [C-1 with H2-2, H-3, and H-10; C-2 with H-3; C-3 with H2-2 and MeO-3; C-4 with H2-2] (Fig. 2). In the COSY spectrum, a correlation between H2-2 and H-3 was observed. The position of the methoxy group (δH 3.64) at C-3 was confirmed by its NOESY correlation with H2-2. The positions of the second (δH 4.05), third (δH 3.95) and fourth (δH 4.11) methoxy groups were placed on ring B at C-7, C-6, and C-5 based on their HMBC and NOESY interactions as similar as compound 1. The experimental electronic circular dichroism (ECD) spectrum of compound 2 exhibited a negative cotton effect at 202.7 nm, along with positive cotton effect at 215.0 nm. These spectral features were consistent with the (S)-2 curve in the calculated ECD (Fig. 3), suggesting that the absolute configuration at C-3 of compound 2 was proposed as S. On the basis of the spectral data, compound 2 was identified as 3,5,6,7-tetramethoxy-2,3-dihydro-1,4-phenanthrenequinone and named cymensifin E.
Compound 3 was obtained as a yellow amorphous solid. The molecular formula C18H16O6 was analyzed from its [M + H]+ at m/z 329.1020 (calcd for C18H17O6 at 329.1020) in the HR-ESI-MS. Moreover, it revealed a major peak at m/z 679.1783, which corresponded with the calculated 2M peak, [2M + Na]+ at m/z 679.1791. The UV absorptions and IR bands of 3 are similar to 1 and 2, suggesting of a phenanthrenequinone skeleton. The 1H, 13C NMR, and DEPT spectra of 3 also exhibited similarities with those of 1, except that the methoxy group is at C-3 in compound 3 instead of C-2 in compound 1 (Table 1). Compared with compound 1, the downfield chemical shift of C-3 in compound 3 (δC 163.5) was observed due to the presence of a methoxy group at that position. The position of the methoxy group (δH 4.00) at C-3 was confirmed by its HSQC correlation with MeO-3 carbon (δC 56.2), the HMBC correlations of C-3 with H-2 (δH 6.13) and MeO-3 and the NOESY correlation of H-2 and MeO-3. Similar to compound 1, the HMBC correlation of MeO-7 with C-7 (δC 155.7), NOESY correlation with H-8 (δH 7.32) and HSQC correlation with MeO-7 carbon (δC 60.5) confirmed the substitution of methoxy (δH 4.05) at C-7. The methoxy group (δH 3.92) was located at C-6 based on the HMBC correlations of C-6 (δC 143.9) with MeO-6 protons and H-8 (δH 7.32), as well as its HSQC correlation with the MeO-6 carbon (δC 60.1). The MeO-5 (δH 4.05) was verified at C-5 by its HMBC correlation with C-5 (δC 149.9) and HSQC correlation with MeO-5 carbon (δC 55.6). Based on the above spectral data, compound 3 was characterized as 3,5,6,7-tetramethoxy-1,4-phenanthrenequinone. Prior to this study, the natural occurrence of 3 was not known. So, the trivial name cymensifin F was given to this compound. However, the synthesis of compound 3 was previously reported by C. L. Lee et al., 2012.30
| Position | 1 (in acetone-d6) | 2 (in CDCl3) | 3 (in acetone-d6) | |||
|---|---|---|---|---|---|---|
| 1H (multiplicity, J in Hz) | 13C | 1H (multiplicity, J in Hz) | 13C | 1H (multiplicity, J in Hz) | 13C | |
| 1 | — | 179.4 | — | 192.5 | — | 183.7 |
| 2 | — | 158.2 | 3.42 (dd, 17.6, 6.0) | 46.8 | 6.13 (s) | 106.1 |
| 3.15 (dd, 17.6, 8.4) | ||||||
| 3 | 6.29 (s) | 110.1 | 4.67 (dd, 8.4, 6.0) | 82.2 | — | 163.5 |
| 4 | — | 185.4 | — | 201.3 | — | 181.6 |
| 4a | — | 134.3 | — | 137.3 | — | 132.5 |
| 4b | — | 119.8 | — | 120.1 | — | 119.8 |
| 5 | — | 150.5 | — | 149.6 | — | 149.9 |
| 6 | — | 143.8 | — | 142.9 | — | 143.9 |
| 7 | — | 156.2 | — | 156.2 | — | 155.7 |
| 8 | 7.29 (s) | 103.0 | 7.02 (s) | 102.8 | 7.32 (s) | 103.1 |
| 8a | — | 135.2 | — | 134.8 | — | 134.4 |
| 9 | 8.01 (d, 8.8) | 131.2 | 7.84 (d, 8.8) | 130.7 | 8.06 (d, 8.8) | 132.1 |
| 10 | 7.95 (d, 8.8) | 120.7 | 7.94 (d, 8.8) | 121.9 | 7.95 (d, 8.8) | 120.8 |
| 10a | — | 128.9 | — | 133.1 | — | 131.0 |
| MeO-2 | 3.94 (s) | 55.7 | — | — | — | — |
| MeO-3 | — | — | 3.64 (s) | 58.0 | 4.00 (s) | 56.2 |
| MeO-5 | 4.05 (s) | 55.6 | 4.11 (s) | 60.4 | 4.05 (s) | 55.6 |
| MeO-6 | 3.91 (s) | 60.0 | 3.95 (s) | 60.8 | 3.92 (s) | 60.1 |
| MeO-7 | 4.06 (s) | 60.8 | 4.05 (s) | 56.1 | 4.05 (s) | 60.5 |
Cytotoxicity profiles of the test compounds in BV2 microglial cells
A series of concentrations of isolated compounds from the roots of Cymbidium ensifolium underwent testing in BV2 microglial cells. Treatment with compounds over a range of concentrations (0, 5, 10, 20, 40, 80 μM). The concentration that did not cause toxicity to the cells was considered a safety concentration for the cells. In the present study, concentrations below 20 μM were observed as the safety thresholds for all tested compounds and were subsequently selected for examination in activated microglial cells. Compounds 2 and 13 were not assessed due to insufficient amounts (Table 2).| Compound | Percentage viability (%) | |||||
|---|---|---|---|---|---|---|
| 0 μM | 5 μM | 10 μM | 20 μM | 40 μM | 80 μM | |
| a Data are expressed as mean ± SD (n = 3). The differences in treatment with the vehicle group were statistically assessed using one-way ANOVA followed by Dunnett post hoc test. Statistically significant reductions in cell viability are denoted as *, **, *** representing significance levels of p < 0.05, p < 0.01, and p < 0.001, respectively. | ||||||
| 1 | 100 ± 0 | 100.67 ± 3.33 | 101.51 ± 0.67 | 104.57 ± 7.09 | 101.75 ± 8.6 | 73.72 ± 18.53* |
| 3 | 100 ± 0 | 99.88 ± 1.96 | 95.62 ± 7.44 | 94.93 ± 3.76 | 88.84 ± 6.76 | 55.15 ± 26.41** |
| 4 | 100 ± 0 | 102.2 ± 3.53 | 99.34 ± 5.77 | 101.48 ± 2.3 | 95.34 ± 3.88 | 49.15 ± 24.65*** |
| 5 | 100 ± 0 | 105.43 ± 1.81 | 104.16 ± 7.68 | 100.52 ± 7.37 | 101.3 ± 14.9 | 94.45 ± 21.56 |
| 6 | 100 ± 0 | 106.33 ± 4.65 | 108.08 ± 5.54 | 105.55 ± 7.76 | 76.69 ± 2.86** | 44.78 ± 13.23*** |
| 7 | 100 ± 0 | 105.04 ± 4.37 | 108.16 ± 3.16 | 112.38 ± 2.51 | 111.09 ± 1.88 | 103.99 ± 9.12 |
| 8 | 100 ± 0 | 106.26 ± 0.36 | 100.28 ± 6.16 | 108.21 ± 0.53 | 113.19 ± 1.57 | 94.43 ± 21.2 |
| 9 | 100 ± 0 | 101.76 ± 2.99 | 101.31 ± 4.09 | 104.51 ± 3.47 | 104.84 ± 5.59 | 111.8 ± 5.8 |
| 10 | 100 ± 0 | 101.21 ± 2.85 | 96.63 ± 1.58 | 103.54 ± 7.79 | 95.79 ± 4.1 | 85.14 ± 9.87* |
| 11 | 100 ± 0 | 102.94 ± 0.86 | 99.73 ± 1.55 | 98.77 ± 0.86 | 94.8 ± 2.82* | 86.43 ± 4.09*** |
| 12 | 100 ± 0 | 103.63 ± 3.27 | 101.21 ± 5.91 | 98.78 ± 4.41 | 94.28 ± 4.79 | 87.5 ± 8.17* |
| 14 | 100 ± 0 | 107.82 ± 3.2 | 97.91 ± 8.91 | 91.92 ± 1.47 | 65.89 ± 4.08*** | 26.73 ± 14.68*** |
| 15 | 100 ± 0 | 103.32 ± 1.97 | 105 ± 3.91 | 111.46 ± 2.26 | 107.65 ± 2.12 | 104.46 ± 3.05 |
| 16 | 100 ± 0 | 104.25 ± 2.01 | 98.35 ± 3.48 | 96.41 ± 0.97 | 44.5 ± 11.83*** | 10.42 ± 5.37*** |
| 17 | 100 ± 0 | 106.54 ± 6.44 | 97.31 ± 7.01 | 100.77 ± 8.73 | 101.42 ± 5.33 | 84.92 ± 5.53* |
The effects of the test compounds on nitric oxide release in activated microglial cells
Nitric oxide is one of the inflammatory mediators that plays an essential role in CNS diseases.31 Physiologically, nitric oxide acts to regulate neurotransmitter release and communication between neurons.32 However, an excessive amount of nitric oxide facilitates the progression of neurodegenerative diseases by promoting excitotoxic cell death and inducing mitochondrial dysfunction.33 As such, alleviating nitric oxide is an available option to improve and delay the progression of CNS diseases. In the present study, a series of active compounds from the roots of Cymbidium ensifolium were assessed in LPS-induced microglial cells. As shown in Table 3, cymensifin D (1), cymensifin F (3), coelonin (6), parviphenanthrine C (12), (1-(4-hydroxylbenzyl)-4-methoxy 9,10-dihydrophenanthrene 2,7-diol) (14) and blestriarene C (16) demonstrated promising IC50 (50% inhibition of nitric oxide releases) values with the percentage nitric oxide inhibition is 9.95 ± 2.13, 8.77 ± 3.78, 2.39 ± 0.91, 6.69 ± 2.94, 2.96 ± 1.38, and 8.42 ± 2.99 μM, respectively.| Compound | IC50 ± SD (μM) |
|---|---|
| a The data are expressed as mean ± SD (n = 3). | |
| 1 | 9.95 ± 2.13 |
| 3 | 8.77 ± 3.78 |
| 4 | >20 |
| 5 | >20 |
| 6 | 2.39 ± 0.91 |
| 7 | >20 |
| 8 | >20 |
| 9 | >20 |
| 10 | >20 |
| 11 | >20 |
| 12 | 6.69 ± 2.94 |
| 14 | 2.96 ± 1.38 |
| 15 | >20 |
| 16 | 8.42 ± 2.99 |
| 17 | >20 |
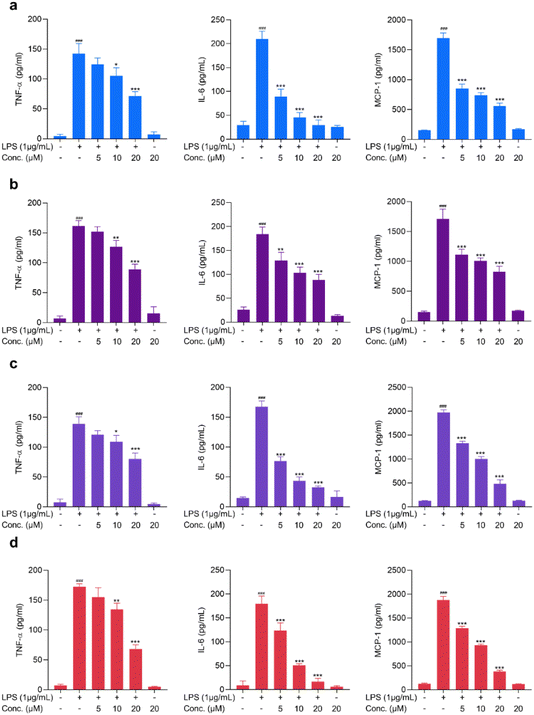
The effects of the test compounds on the expression of proinflammatory mediators (TNF-α, IL-6 and MCP-1) in activated microglial cells
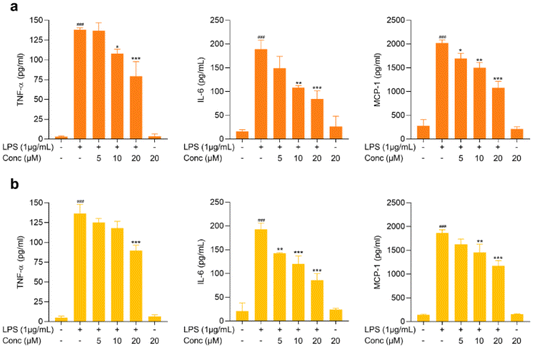
Proinflammatory cytokines, including TNF-α and IL-6, are the major contributing factors in neuroinflammation and have been reported to contribute to the pathogenesis of CNS diseases, including Alzheimer's disease, Parkinson's disease, and other neuropsychiatric disorders.34,35 Microglia, CNS-resident immune cells, are the cells that contribute to the significant release of proinflammatory cytokines and are pathophysiologically involved in the progression of diseases.36 In addition to cytokines, chemokines, including MCP-1, have been reported to cause neuronal loss and have also been reported to be involved in CNS-associated diseases.37 Hence, exploring active compounds with the characteristic of suppressing proinflammatory mediators is essential for discovering neuroinflammation-modulating agents. These proinflammatory mediators are significantly elevated upon LPS stimulation.38 In the present study, the potential activity of the test compounds on modulating TNF-α, IL-6 and MCP-1 was assessed in LPS-stimulated BV2 cells. As shown in Fig. 4 and 5, LPS treatment increased the release of proinflammatory mediators, yet pretreatment with cymensifin D (1), cymensifin F (3), coelonin (6), parviphenanthrine C (12), (1-(4-hydroxylbenzyl)-4-methoxy 9,10-dihydrophenanthrene 2,7-diol) (14) and blestriarene C (16) significantly reduced proinflammatory mediator secretions in activated microglia in a concentration-dependent manner. Coelonin (6), a dihydrophenanthrene compound isolated from Bletilla striata, has been reported for its anti-inflammatory activity on LPS-induced RAW264.7 cells.39 Numerous studies have illustrated the remarkable anti-inflammatory activity of phenanthrene, dihydrophenanthrene, and phenanthrenequinone derivatives.39,40 Interestingly, compounds 1, 3, 6, 12, 14, and 16 belong to those categories. Taken together, these compounds possess the ability to modulate proinflammatory mediators in activated microglia and could potentially be further used as neuroinflammation-modulating agents.Cymensifin F (3) modulates NF-κB activation in LPS-treated microglia cells
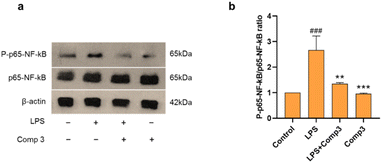
To study the anti-inflammatory mechanism of the novel phenanthrenequinones, cymensifin F (3) was selected based on its IC50 value (8.77 ± 3.78 μM). The potential anti-inflammatory mechanism of cymensifin F (3) was determined using immunoblotting. Activated microglia in CNS diseases, manifested with increased releases of proinflammatory mediators, are associated with the stimulation of the NF-κB pathway.41 NF-κB activation controls the production of TNF-α, IL-6, and MCP-1.6 Moreover, pro-inflammatory cytokines can be downregulated by inhibiting NF-κB transcriptional activity in the microglial nucleus.42 Consequently, we investigated the potential anti-inflammatory effect of cymensifin F (3) on NF-κB activity. In our study, NF-κB (p65) was phosphorylated in response to LPS. Furthermore, pretreatment with cymensifin F (3) significantly suppressed LPS-induced NF-κB phosphorylation (Fig. 6). These findings suggest that the anti-neuroinflammatory properties of cymensifin F (3) may be attributed to its ability to inhibit NF-κB phosphorylation.Experimental
General experimental procedures
Optical rotation was measured by a Jasco P-2000 digital polarimeter (Easton, MD, USA). UV spectra were captured using a Milton Roy Spectronic 3000 Array spectrophotometer (Rochester, Monroe, NY, USA). CD spectra were obtained from a Jasco J-815 CD spectrophotometer (Hachioji, Tokyo, Japan). FT-IR spectra were acquired using a PerkinElmer FT-IR 1760X spectrophotometer (Boston, MA, USA). Mass spectra were recorded by a Bruker MicroTOF mass spectrometer (ESI-MS) (Billerica, MA, USA). NMR spectra were obtained by a Bruker Avance Neo 400 MHz NMR spectrometer (Billerica, MA, USA). Vacuum-liquid chromatography (VLC) and column chromatography (CC) were performed by silica gel 60 (no. 1.07734.2500), size 0.063–0.200 mm, and (no. 1.09385.2500), size 0.040–0.063 mm (Merck, NJ, USA). Gel filtration chromatography was conducted by Sephadex LH-20 (Merck, NJ, USA). The initial assessment of the purity of isolated compounds was conducted by thin-layer chromatography (TLC), silica gel 60 F254 plates (Merck, NJ, USA) under UV light. 3-(4,5-Dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT), lipopolysaccharide (LPS), and other chemicals were acquired from Sigma-Aldrich (St. Louis, MO, USA). NF-κB p65 (cat. no. 8252S), P-NF-κB p65 (cat. no. 3033S), β-actin (cat. no. 4970S), and anti-rabbit antibody (cat. no. 7074S) were purchased from Cell Signaling Technology (Massachusetts, USA). Nitrocellulose membrane (0.45 μ) was obtained from HiMedia Laboratories Pvt. Ltd. (Mumbai, India). Clarity Western ECL Substrate (cat. no. 1705061) was acquired from Bio-Rad Laboratories, United States.Plant material
The roots of Cymbidium ensifolium were purchased from Chatuchak Market in September 2020. The authentication was performed by Dr Boonchoo Sritularak. A voucher specimen BS-CE-092563 has been deposited at the Department of Pharmacognosy and Pharmaceutical Botany, Faculty of Pharmaceutical Sciences, Chulalongkorn University.Extraction and isolation
Dried roots of Cymbidium ensifolium (1 kg) were macerated with methanol (MeOH) (5 × 3 L), and the methanolic extract (92.2 g) was obtained. The methanolic extract was subsequently subjected to vacuum liquid chromatography (silica gel, EtOAc–hexane, gradient) to give seven fractions (A–G). Fraction B (1.5 g) was separated by Sephadex LH-20 (acetone) to yield 5 fractions (BA–BE). Fraction BC (92 mg) was separated on silica gel eluting with dichloromethane (CH2Cl2) to yield compounds 4 (3 mg) and 5 (5.4 mg). Fraction BE (73.1 mg) was separated on a silica gel column (acetone–CH2Cl2, 0.2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 9.8) to yield compound 6 (3.2 mg). Fraction BD (76.4 mg) was fractionated on a silica gel column (acetone–CH2Cl2, 0.2
9.8) to yield compound 6 (3.2 mg). Fraction BD (76.4 mg) was fractionated on a silica gel column (acetone–CH2Cl2, 0.2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 9.8) to give 2 fractions (BDA–BDB). BDB (30 mg) was purified on Sephadex LH-20 (acetone) to yield compound 7 (5.2 mg). Fraction C was separated on a silica gel column (acetone–CH2Cl2, 0.5
9.8) to give 2 fractions (BDA–BDB). BDB (30 mg) was purified on Sephadex LH-20 (acetone) to yield compound 7 (5.2 mg). Fraction C was separated on a silica gel column (acetone–CH2Cl2, 0.5![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 9.5), and six fractions (CA–CF) were obtained. CA fraction (261.9 mg) was separated on Sephadex LH-20 (acetone) to yield compound 8 (5.9 mg) and CAA fraction which was separated again on a silica gel column (acetone–hexane, 2.5
9.5), and six fractions (CA–CF) were obtained. CA fraction (261.9 mg) was separated on Sephadex LH-20 (acetone) to yield compound 8 (5.9 mg) and CAA fraction which was separated again on a silica gel column (acetone–hexane, 2.5![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 7.5) to produce compound 9 (1.3 mg). The CC fraction (103.7 mg) was separated on Sephadex LH-20 (acetone) and then silica gel (acetone–CH2Cl2, 0.2
7.5) to produce compound 9 (1.3 mg). The CC fraction (103.7 mg) was separated on Sephadex LH-20 (acetone) and then silica gel (acetone–CH2Cl2, 0.2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 9.8) to produce three fractions (CCA–CCC) fractions. CCA fraction (10 mg) underwent purification five times on preparative thin layer chromatography (PTLC) using dichloromethane as the mobile phase and compound 1 (3 mg) was produced. CCB fraction (3 mg) was purified on PTLC (acetone–hexane, 2
9.8) to produce three fractions (CCA–CCC) fractions. CCA fraction (10 mg) underwent purification five times on preparative thin layer chromatography (PTLC) using dichloromethane as the mobile phase and compound 1 (3 mg) was produced. CCB fraction (3 mg) was purified on PTLC (acetone–hexane, 2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 8, 2 times) to furnish compound 2 (0.8 mg). Fraction CCC (103.7 mg) was separated on Sephadex LH-20 (acetone) and then purified on purified on PTLC (EtOAc–CH2Cl2, 0.2
8, 2 times) to furnish compound 2 (0.8 mg). Fraction CCC (103.7 mg) was separated on Sephadex LH-20 (acetone) and then purified on purified on PTLC (EtOAc–CH2Cl2, 0.2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 9.8) to produce compound 3 (2.3 mg). Fraction CD (389.3 mg) was purified on Sephadex LH20 (methanol), and then Sephadex LH-20 (acetone) to yield CDA (22.6 mg) which was further separated on a silica gel column (acetone–CH2Cl2, 1
9.8) to produce compound 3 (2.3 mg). Fraction CD (389.3 mg) was purified on Sephadex LH20 (methanol), and then Sephadex LH-20 (acetone) to yield CDA (22.6 mg) which was further separated on a silica gel column (acetone–CH2Cl2, 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 9) to produce compound 10 (1.5 mg). Fraction CF (131.7 mg) was separated on Sephadex LH-20 (acetone) and 2 fractions (CFA and CFB) were obtained. CFA (40 mg) was separated on a silica gel column (EtOAc–CH2Cl2, 2
9) to produce compound 10 (1.5 mg). Fraction CF (131.7 mg) was separated on Sephadex LH-20 (acetone) and 2 fractions (CFA and CFB) were obtained. CFA (40 mg) was separated on a silica gel column (EtOAc–CH2Cl2, 2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 8) and then purified again on Sephadex LH-20 (methanol) to give compound 11 (3.4 mg). CFB (29 mg) was separated on a silica gel column (MeOH–CH2Cl2, 0.1
8) and then purified again on Sephadex LH-20 (methanol) to give compound 11 (3.4 mg). CFB (29 mg) was separated on a silica gel column (MeOH–CH2Cl2, 0.1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 9.9) and then purified again on PTLC by developing four times with dichloromethane to yield compound 12 (1.7 mg). Fraction D (1.3 g) was separated on a silica gel column (acetone–CH2Cl2) with gradient elution, and 10 fractions (DA–DJ) were obtained. Fraction DA (17 mg) was separated on a silica gel column by eluting with dichloromethane to produce compound 13 (0.8 mg). Compound 14 (1.5 mg) was obtained by purifying DI fraction (87 mg) with Sephadex LH-20 (acetone) and then Sephadex LH-20 (methanol). Compound 15 (3 mg) was produced by purifying DJ (157.3 mg) with Sephadex LH-20 (methanol). Fraction E (1.5 g) was separated on a silica gel column (acetone–CH2Cl2, 0.5
9.9) and then purified again on PTLC by developing four times with dichloromethane to yield compound 12 (1.7 mg). Fraction D (1.3 g) was separated on a silica gel column (acetone–CH2Cl2) with gradient elution, and 10 fractions (DA–DJ) were obtained. Fraction DA (17 mg) was separated on a silica gel column by eluting with dichloromethane to produce compound 13 (0.8 mg). Compound 14 (1.5 mg) was obtained by purifying DI fraction (87 mg) with Sephadex LH-20 (acetone) and then Sephadex LH-20 (methanol). Compound 15 (3 mg) was produced by purifying DJ (157.3 mg) with Sephadex LH-20 (methanol). Fraction E (1.5 g) was separated on a silica gel column (acetone–CH2Cl2, 0.5![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 9.5), and 5 fractions (EA–EE) were obtained. Fraction EE (137.9 mg) was purified on Sephadex LH-20 (acetone) to produce EEA and EEB. Fraction EEB (15.3 mg) was separated again on Sephadex LH-20 (methanol) to furnish compound 16 (1.7 mg). Fraction F (2.8 g) was separated on a silica gel column (EtOAc–hexane) with gradient elution to give three fractions (FA–FC). Compound 17 (2.7 mg) was obtained from fraction FC (10 mg) by purifying with Sephadex LH-20 (acetone) and then Sephadex LH-20 (methanol).
9.5), and 5 fractions (EA–EE) were obtained. Fraction EE (137.9 mg) was purified on Sephadex LH-20 (acetone) to produce EEA and EEB. Fraction EEB (15.3 mg) was separated again on Sephadex LH-20 (methanol) to furnish compound 16 (1.7 mg). Fraction F (2.8 g) was separated on a silica gel column (EtOAc–hexane) with gradient elution to give three fractions (FA–FC). Compound 17 (2.7 mg) was obtained from fraction FC (10 mg) by purifying with Sephadex LH-20 (acetone) and then Sephadex LH-20 (methanol).
Cymensifin D (1): orange-coloured amorphous solid; UV (MeOH) λmax (log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) ε) 210 (3.96), 240 (4.10), 305 (3.85), 395 (3.04); HR-ESI-MS: [M + H]+ at m/z 329.1019 (calcd for C18H17O6 329.1020); IR: νmax at 3396, 2922, 1726, 1461, 1376 cm−1.
ε) 210 (3.96), 240 (4.10), 305 (3.85), 395 (3.04); HR-ESI-MS: [M + H]+ at m/z 329.1019 (calcd for C18H17O6 329.1020); IR: νmax at 3396, 2922, 1726, 1461, 1376 cm−1.
Cymensifin E (2): pale yellow amorphous solid; [α]20D −21.5 (c 0.04, MeOH); UV (MeOH) λmax (log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) ε) 205 (3.92), 230 (3.83), 280 (3.92), 332 (3.22); HR-ESI-MS; [M + H]+ at m/z 331.1176 (calcd for C18H19O6 331.1181); ECD (MeOH): λmax (Δε) 202.7 (−1.61), 215.0 (+1.52) nm; IR: νmax at 3333, 2919, 2923, 1717, 1679, 1464, 1316 cm−1.
ε) 205 (3.92), 230 (3.83), 280 (3.92), 332 (3.22); HR-ESI-MS; [M + H]+ at m/z 331.1176 (calcd for C18H19O6 331.1181); ECD (MeOH): λmax (Δε) 202.7 (−1.61), 215.0 (+1.52) nm; IR: νmax at 3333, 2919, 2923, 1717, 1679, 1464, 1316 cm−1.
Cymensifin F (3): yellow amorphous solid; UV (MeOH) λmax (log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) ε) 220 (3.91), 245 (4.09), 300 (3.77), 390 (3.01); HR-ESI-MS: [M + H]+ at m/z 329.1020 (calcd for C18H17O6 329.1020); IR: νmax at 3333, 2921, 1732, 1470, 1378 cm−1.
ε) 220 (3.91), 245 (4.09), 300 (3.77), 390 (3.01); HR-ESI-MS: [M + H]+ at m/z 329.1020 (calcd for C18H17O6 329.1020); IR: νmax at 3333, 2921, 1732, 1470, 1378 cm−1.
Computational detail of ECD calculation
In this study, computational approaches were employed to optimize various configurations of compound 2. Density Functional Theory (DFT) calculations were conducted at the B3LYP/6-31g(d,p) level. Subsequently, Electron Circular Dichroism (ECD) spectra were computed using time-dependent DFT (TD-DFT) at the B3LYP/6-31+G(d,p) level, incorporating solvation effects modeled with the Continuum Model (PCM) employing methanol. All computational analyses were performed using Gaussian16 software.43 Additionally, the ECD spectra were simulated using overlapping Gaussian functions parameterized with a fitting parameter (σ = 0.25 eV) via the SpecDis 1.64 program,44 utilizing the length gauge representation to enhance reliability.Cell culture
BV2 microglial cells were obtained from AcceGen Biotechnology (Fairfield, NJ, USA). The cells were grown in DMEM media (PAN Biotech, Aidenbach, Germany) and supplemented with 10% fetal bovine serum (PAN Biotech, Aidenbach, Germany), 1% antibiotics (penicillin–streptomycin) (GIBCO, Carlsbad, CA, USA). The cells were kept at 37 °C, 5% CO2, and 95% humidity. The cells were passaged at 70–80% confluence.Cell treatment
For cytotoxicity, the cells were seeded at a concentration of 2 × 104 cells per well in 96-well plates for 24 hours. The cells were then treated with various concentrations of the test compounds (0, 5, 10, 20, 40 and 80 μM) for another 24 hours. The selected safety concentrations were further used to evaluate the efficacy of the test compounds on proinflammatory mediator release in LPS-stimulated cells. Briefly, cells were pretreated with the safety concentrations of the test compounds for 1 hour followed by the addition of 1 μg per mL LPS for 24 hours. The media was then collected for further assessment of the proinflammatory mediators (NO, TNF-α, IL-6, and MCP-1). In the western blot, the cells were pretreated with the highest safety dose of the test compound for 1 hour and exposed to LPS for another 1 hour. The cells were collected and extracted for further use in the western blot.Cell viability assay
The cytotoxicity of the test compound was determined by assessing the viability of the cells using the MTT assay. After treatment, the cells were added with MTT solution at a concentration of 500 μg mL−1 for 3 hours. The crystal formazan was then dissolved with DMSO, followed by the measurement of the absorbance in the microplate reader. The absorbance was determined at a wavelength of 570 nm.Nitrite assay
The levels of nitric oxide were measured using a nitrite assay. Griess reagent (Sigma-Aldrich®, St. Louis, MO, USA) was used to measure nitrite levels in the media as an indicator of nitric oxide releases. Briefly, after treatment, 100 μL of media was mixed with 100 μL Griess reagent and incubated for 10 min. The absorbance was then measured on a microplate reader at a wavelength of 520 nm.ELISA assay
Released inflammatory mediators, including TNF-α, IL-6, and MCP-1, after treatment with the test compounds in activated microglia were determined using an ELISA assay (Biolegend, San Diego, USA). The assays were performed according to the manufacturer's instructions.Western blot analysis
The cells were seeded at a density of 2 × 106 cells in 6 cm cell culture dishes. Following the treatments, the cell culture medium was removed, and the cells were washed with PBS. The cells were then added with 60 μL of lysis buffer (supplemented with 1% protease/phosphatase inhibitor) to the cells to cause lysing. The cell lysates were collected using scrapers, centrifuged (4 °C, 12![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 rpm, 10 min), and the supernatants were collected. A Bradford assay was used to calculate each lysate's protein content. An equal amount of proteins (40 μg) were loaded and electrophoresed on 10% sodium dodecyl sulfate–polyacrylamide gels (SDS–PAGE). A semi-dry transfer machine was used to transfer proteins onto nitrocellulose membranes, which were then treated with 5% skim milk in TBST for 1 hour at room temperature with gentle agitation in order to prevent non-specific binding sites. Following an overnight incubation at 4 °C with primary antibodies; P-p65-NF-κB (1
000 rpm, 10 min), and the supernatants were collected. A Bradford assay was used to calculate each lysate's protein content. An equal amount of proteins (40 μg) were loaded and electrophoresed on 10% sodium dodecyl sulfate–polyacrylamide gels (SDS–PAGE). A semi-dry transfer machine was used to transfer proteins onto nitrocellulose membranes, which were then treated with 5% skim milk in TBST for 1 hour at room temperature with gentle agitation in order to prevent non-specific binding sites. Following an overnight incubation at 4 °C with primary antibodies; P-p65-NF-κB (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1000), p65-NF-κB (1
1000), p65-NF-κB (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1000) and β-actin (1
1000) and β-actin (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2000), the membranes were treated for 2 hours at room temperature with horseradish peroxidase-labeled anti-rabbit secondary antibodies (1
2000), the membranes were treated for 2 hours at room temperature with horseradish peroxidase-labeled anti-rabbit secondary antibodies (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1000). After washing with TBST, the chemiluminescence (ECL) solution was allowed to incubate on the membranes. Subsequently, a medical X-ray cassette, Kodak Green 400 Screen (Rochester, NY, USA), was used to observe the protein bands. ImageJ software was used to quantify the intensity of the protein bands.
1000). After washing with TBST, the chemiluminescence (ECL) solution was allowed to incubate on the membranes. Subsequently, a medical X-ray cassette, Kodak Green 400 Screen (Rochester, NY, USA), was used to observe the protein bands. ImageJ software was used to quantify the intensity of the protein bands.
Data and statistical analysis
The data were analyzed and visualized using GraphPad Prism (San Diego, CA, USA). The data shown in the graphs are expressed as means ± SD. The statistical differences between groups were analyzed using a one-way ANOVA followed by Dunnett, Bonferroni post hoc test. A p-value below 0.05 was considered statistically significant.Conclusions
In the present study, a total of 17 compounds were extracted from the roots of Cymbidium ensifolium, including two new phenanthrenequinones (1–2), one new natural product phenanthrenequinone (3), and 14 known compounds (4–17). The isolated pure compounds were assessed for their anti-neuroinflammatory properties using lipopolysaccharide-activated BV2 microglial cells. Compounds 1, 3, 6, 12, 14, and 16 demonstrated the capability to decrease lipopolysaccharide-induced nitric oxide (NO) release in BV2 microglial cells. Additionally, they reduced the secretion of proinflammatory mediators such as TNF-α, IL-6, and MCP-1 in activated microglia in a concentration-dependent manner. Cymensifin F (3) was further investigated for its possible anti-neuroinflammatory mechanism. In LPS-stimulated BV2 microglia, the anti-inflammatory effect of cymensifin F (3) is mediated through the downregulation of phosphorylated NF-κB. All of these findings suggest that C. ensifolium roots contain bioactive compounds that may alleviate neuroinflammation and aid in the treatment of neurodegenerative diseases.Data availability
The authors confirm that the data supporting the findings of this study are available within the article and its ESI.†Author contributions
B. S. conceived, designed, and supervised the research project, as well as revised the manuscript; P. T. supervised the anti-neuroinflammatory activity and revised the manuscript; H. H. supervised and assisted on anti-neuroinflammatory activity as well as prepared the manuscript; M. T. T. performed the experiments and prepared the manuscript; P. P. and S. J. analyzed the ECD experimental data; P. R. and C. B. provided comments and suggestions on the preparation of the manuscript and performing the experiments.Conflicts of interest
The authors declare no conflict of interest.Acknowledgements
M. T. T. is grateful to Chulalongkorn University for a C2F (Second Century Fund) postdoctoral fellowship under the supervision of B. S. P. P. expresses gratitude to Hong Kong Quantum AI Laboratory, Ltd for providing a postdoctoral researcher scholarship.References
- Q. Zheng, W. Sun and M. Qu, Anti-neuro-inflammatory effects of the bioactive compound capsaicin through the NF-κB signaling pathway in LPS-stimulated BV2 microglial cells, Pharmacogn. Mag., 2018, 14, 489–494 CrossRef CAS.
- A. Salemme, A. R. Togna, A. Mastrofrancesco, V. Cammisotto, M. Ottaviani, A. Bianco and A. Venditti, Anti-inflammatory effects and antioxidant activity of dihydroasparagusic acid in lipopolysaccharide-activated microglial cells, Brain Res. Bull., 2016, 120, 151–158 CrossRef CAS PubMed.
- C. Y. Jin, J. D. Lee, C. Park, Y. H. Choi and G. Y. Kim, Curcumin attenuates the release of pro-inflammatory cytokines in lipopolysaccharide-stimulated BV2 microglia, Acta Pharmacol. Sin., 2007, 28, 1645–1651 CrossRef CAS PubMed.
- M. Candiracci, E. Piatti, M. Dominguez-Barragan, D. Garcia-Antras, B. Morgado, D. Ruano, J. F. Gutierrez, J. Parrado and A. Castano, Anti-inflammatory activity of a honey flavonoid extract on lipopolysaccharide-activated N13 microglial cells, J. Agric. Food Chem., 2012, 60, 12304–12311 CrossRef CAS PubMed.
- C. I. Yu, C. I. Cheng, Y. F. Kang, P. C. Chang, I. P. Lin, Y. H. Kuo, A. J. Jhou, M. Y. Lin, C. Y. Chen and C. H. Lee, Hispidulin inhibits neuroinflammation in lipopolysaccharide-activated BV2 microglia and attenuates the activation of Akt, NF-κB, and STAT3 pathway, Neurotoxic. Res., 2020, 38, 163–174 CrossRef CAS PubMed.
- C. Wang-Sheng, A. Jie, L. Jian-Jun, H. Lan, X. Zeng-Bao and L. Chang-Qing, Piperine attenuates lipopolysaccharide (LPS)-induced inflammatory responses in BV2 microglia, Int. Immunopharmacol., 2017, 42, 44–48 CrossRef CAS PubMed.
- W. J. Kim, H. S. Cha, M. H. Lee, S. Y. Kim, S. H. Kim and T. J. Kim, Effects of Cymbidium root ethanol extract on atopic dermatitis, J. Evidence-Based Complementary Altern. Med., 2016, 1, 5362475 CrossRef PubMed.
- N. Lertnitikul, C. Pattamadilok, C. Chansriniyom and R. Suttisri, A new dihydrophenanthrene from Cymbidium finlaysonianum and structure revision of cymbinodin-A, J. Asian Nat. Prod. Res., 2018, 22, 83–90 CrossRef PubMed.
- M. A. Howlader, M. Alam, A. KhT, F. Khatun and A. S. Apu, Antinociceptive and anti-inflammatory activity of the ethanolic extract of Cymbidium aloifolium (L.), Pak. J. Biol. Sci., 2011, 14, 909–911 CrossRef PubMed.
- J. A. Takahashi, Isolation, structure elucidation and biological activity of natural products, Molecules, 2023, 28, 5392 CrossRef CAS PubMed.
- T. O. Jimoh, B. C. Costa, C. Chansriniyom, C. Chaotham, P. Chanvorachote, P. Rojsitthisak, K. Likhitwitayawuid and B. Sritularak, Three new dihydrophenanthrene derivatives from Cymbidium ensifolium and their cytotoxicity against cancer cells, Molecules, 2022, 27, 2222 CrossRef CAS PubMed.
- B. Rivai, H. Hasriadi, P. W. D. Wasana, C. Chansriniyom, P. Towiwat, Y. Punpreuk, K. Likhitwitayawuid, P. Rojsitthisak and B. Sritularak, Potential role of a novel biphenanthrene derivative isolated from Aerides falcata in central nervous system diseases, RSC Adv., 2023, 13, 10757–10767 RSC.
- V. Kongkatitham, A. Dehlinger, C. Chaotham, K. Likhitwitayawuid, C. Böttcher and B. Sritularak, Diverse modulatory effects of bibenzyls from Dendrobium species on human immune cell responses under inflammatory conditions, PLoS One, 2024, 19, e0292366 CrossRef CAS PubMed.
- M. T. Thant, N. Bhummaphan, J. Wuttiin, C. Puttipanyalears, W. Chaichompoo, P. Rojsitthisak, Y. Punpreuk, C. Böttcher, K. Likhitwitayawuid and B. Sritularak, New phenolic glycosides from Coelogyne fuscescens Lindl. var. brunnea and their cytotoxicity against human breast cancer cells, ACS Omega, 2024, 9, 7679–7691 CAS.
- P. Tuchinda, J. Udchachon, K. Khumtaveeporn, W. C. Taylor, L. M. Engelhardt and A. H. White, Phenanthrenes of Eulophia nuda, Phytochemistry, 1988, 27, 3267–3271 CrossRef CAS.
- T. L. Palama, A. Khatib, Y. H. Choi, B. Payet, I. Fock, R. Verpoorte and H. Kodja, Metabolic changes in different developmental stages of Vanilla planifolia pods, J. Agric. Food Chem., 2009, 57, 7651–7658 CrossRef CAS PubMed.
- D. C. Rueda, A. Schöffmann, M. De Mieri, M. Raith, E. A. Jähne, S. Hering and M. Hamburger, Identification of dihydrostilbenes in Pholidota chinensis as a new scaffold for GABAA receptor modulators, Bioorg. Med. Chem., 2014, 22, 1276–1284 CrossRef CAS PubMed.
- M. H. Fisch, B. H. Flick and J. Arditti, Structure and antifungal activity of hircinol, loroglossol and orchinol, Phytochemistry, 1973, 12, 437–441 CrossRef CAS.
- Y. G. Chen, J. J. Xu, H. Yu, C. Qing, Y. L. Zhang, Y. Liu and J. H. Wang, 3,7-Dihydroxy-2,4,6-trimethoxyphenanthrene, a new phenanthrene from Bulbophyllum odoratissimum, J. Korean Chem. Soc., 2007, 51, 352–355 CrossRef CAS.
- P. L. Majumder, A. Kar and J. N. Shoolery, Bulbophyllanthrin, a phenanthrene of the orchid Bulbophyllum leopardium, Phytochemistry, 1985, 24, 2083–2087 CrossRef CAS.
- J. M. Lee, D. G. Lee, K. H. Lee, S. H. Cho, K. W. Nam and S. Lee, Isolation and identification of phytochemical constituents from the fruits of Acanthopanax senticosus, Afr. J. Pharm. Pharmacol., 2013, 7, 294–301 CrossRef CAS.
- A. C. González-Baró, B. S. Parajón-Costa, C. A. Franca and R. Pis-Diez, Theoretical and spectroscopic study of vanillic acid, J. Mol. Struct., 2008, 889, 204–210 CrossRef.
- S. Z. Huang, F. D. Kong, G. Chen, X. H. Cai, L. M. Zhou, Q. Y. Ma, Q. Wang, W. L. Mei, H. F. Dai and Y. X. Zhao, A phytochemical investigation of Stemona parviflora roots reveals several compounds with nematocidal activity, Phytochemistry, 2019, 159, 208–215 CrossRef CAS PubMed.
- G. N. Zhang, L. Y. Zhong, S. A. Bligh, Y. L. Guo, C. F. Zhang, M. Zhang, Z. T. Wang and L. S. Xu, Bi-bicyclic and bi-tricyclic compounds from Dendrobium thyrsiflorum, Phytochemistry, 2005, 66, 1113–1120 CrossRef CAS PubMed.
- L. Bai, T. Kato, K. Inoue, M. Yamaki and S. Takagi, Stilbenoids from Bletilla striata, Phytochemistry, 1993, 33, 1481–1483 CrossRef CAS.
- S. P. Tan, K. H. Cho and M. A. Nafiah, A leaf wax and small metabolites from the leaves of Alphonsea cylindrica, Chem. Nat. Compd., 2023, 59, 167–169 CrossRef CAS.
- M. Yamaki, L. Bai, K. Inoue and S. Takagi, Biphenanthrenes from Bletilla striata, Phytochemistry, 1989, 28, 3503–3505 CrossRef CAS.
- M. Forino, L. Tartaglione, C. Dell'Aversano and P. Ciminiello, NMR-based identification of the phenolic profile of fruits of Lycium barbarum (goji berries). Isolation and structural determination of a novel N-feruloyl tyramine dimer as the most abundant antioxidant polyphenol of goji berries, Food Chem., 2016, 194, 1254–1259 CrossRef CAS PubMed.
- C. L. Lee, K. Nakagawa-Goto, D. Yu, Y. N. Liu, K. F. Bastow, S. L. Morris-Natschke, F. R. Chang, Y. C. Wu and K. H. Lee, Cytotoxic calanquinone A from Calanthe arisanensis and its first total synthesis, Bioorg. Med. Chem. Lett., 2008, 18, 4275–4277 CrossRef CAS PubMed.
- C. L. Lee, Y. T. Lin, F. R. Chang, G. Y. Chen, A. Backlund, J. C. Yang, S. L. Chen and Y. C. Wu, Synthesis and biological evaluation of phenanthrenes as cytotoxic agents with pharmacophore modeling and ChemGPS-NP prediction as topo II inhibitors, PLoS One, 2012, 7, e37897 CrossRef CAS PubMed.
- S. Murphy, Production of nitric oxide by glial cells: regulation and potential roles in the CNS, Glia, 2000, 29, 1–13 CrossRef CAS PubMed.
- V. Calabrese, C. Mancuso, M. Calvani, E. Rizzarelli, D. A. Butterfield and A. M. Giuffrida Stella, Nitric oxide in the central nervous system: neuroprotection versus neurotoxicity, Nat. Rev. Neurosci., 2007, 8, 766–775 CrossRef CAS PubMed.
- A. B. Knott and E. Bossy-Wetzel, Nitric oxide in health and disease of the nervous system, Antioxid. Redox Signaling, 2009, 11, 541–553 CrossRef CAS PubMed.
- H. Hong, B. S. Kim and H. I. Im, Pathophysiological role of neuroinflammation in neurodegenerative diseases and psychiatric disorders, Int. Neurourol. J., 2016, 20, S2–S7 CrossRef PubMed.
- H. S. Kwon and S. H. Koh, Neuroinflammation in neurodegenerative disorders: the roles of microglia and astrocytes, Transl. Neurodegener., 2020, 9, 42 CrossRef PubMed.
- W. J. Streit, R. E. Mrak and W. S. T. Griffin, Microglia and neuroinflammation: a pathological perspective, J. Neuroinflammation, 2004, 1, 14 CrossRef PubMed.
- A. J. Sawyer, W. Tian, J. K. Saucier-Sawyer, P. J. Rizk, W. M. Saltzman, R. V. Bellamkonda and T. R. Kyriakides, The effect of inflammatory cell-derived MCP-1 loss on neuronal survival during chronic neuroinflammation, Biomaterials, 2014, 35, 6698–6706 CrossRef CAS PubMed.
- H. Hasriadi, P. W. D. Wasana, W. Thongphichai, Y. Samun, S. Sukrong and P. Towiwat, Curcuma latifolia Roscoe extract reverses inflammatory pain in mice and offers a favorable CNS safety profile, J. Ethnopharmacol., 2024, 318, 116877 CrossRef PubMed.
- F. Jiang, M. Li, H. Wang, B. Ding, C. Zhang, Z. Ding, X. Yu and G. Lv, Coelonin, an anti-inflammation active component of Bletilla striata and its potential mechanism, Int. J. Mol. Sci., 2019, 20, 4422 CrossRef CAS PubMed.
- P. J. Chen, I. L. Ko, C. L. Lee, H. C. Hu, F. R. Chang, Y. C. Wu, Y. L. Leu, C. C. Wu, C. Y. Lin, C. Y. Pan and Y. F. Tsai, Targeting allosteric site of AKT by 5, 7-dimethoxy-1, 4-phenanthrenequinone suppresses neutrophilic inflammation, EBioMedicine, 2019, 40, 528–540 CrossRef PubMed.
- Q. Xie, G. Z. Wu, N. Yang, Y. H. Shen, J. Tang and W. D. Zhang, Delavatine A, an unusual isoquinoline alkaloid exerts anti-inflammation on LPS-induced proinflammatory cytokines production by suppressing NF-κB activation in BV2 microglia, Biochem. Biophys. Res. Commun., 2018, 502, 202–208 CrossRef CAS PubMed.
- H. Hasriadi, P. W. Dasuni Wasana, O. Vajragupta, P. Rojsitthisak and P. Towiwat, Mechanistic insight into the effects of curcumin on neuroinflammation-driven chronic pain, Pharmaceuticals, 2021, 14, 777 CrossRef PubMed.
- M. J. Frisch, G. W. Trucks, H. B. Schlegel, G. E. Scuseria, M. A. Robb, J. R. Cheeseman, G. Scalmani, V. Barone, G. A. Petersson, H. Nakatsuji and X. Li, Gaussian Inc. 16, Revision A. 03, Gaussian Inc., Wallingford, CT, 2016 Search PubMed.
- T. Bruhn, A. N. U. Schaumlöffel, Y. Hemberger and G. Bringmann, SpecDis: quantifying the comparison of calculated and experimental electronic circular dichroism spectra, Chirality, 2013, 25, 243–249 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d4ra04761c |
| This journal is © The Royal Society of Chemistry 2024 |