Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Design, synthesis, and in vitro and in silico study of 1-benzyl-indole hybrid thiosemicarbazones as competitive tyrosinase inhibitors†
Zahra Batoola,
Saeed Ullahb,
Ajmal Khanbf,
Farhan Siddiquec,
Sumaira Nadeemcd,
Abdulrahman Alshammarie,
Norah A. Albekairie,
Rimsha Taliba,
Ahmed Al-Harrasi*b and
Zahid Shafiq *a
*a
aInstitute of Chemical Sciences, Bahauddin Zakariya University, Multan-60800, Pakistan. E-mail: zahidshafiq@bzu.edu.pk
bNatural and Medical Sciences Research Centre, University of Nizwa, P.O. Box 33, PC 616, Birkat Al Mauz, Nizwa, Sultanate of Oman. E-mail: aharrasi@unizwa.edu.om
cDepartment of Pharmaceutical Chemistry, Faculty of Pharmacy, Bahauddin Zakariya University, Multan-60800, Pakistan
dDepartment of Pharmacy, The Women University, Multan, 60000, Pakistan
eDepartment of Pharmacology and Toxicology, College of Pharmacy, King Saud University, Post bezBox 2455, Riyadh, 11451, Saudi Arabia
fDepartment of Chemical and Biological Engineering, College of Engineering, Korea University, 145 Anam-ro, Seongbuk-gu, Seoul 02841, Republic of Korea
First published on 6th September 2024
Abstract
Developing new anti-tyrosinase drugs seems crucial for the medical and industrial fields since irregular melanin synthesis is linked to the resurgence of several skin conditions, including melanoma, and the browning of fruits and vegetables. A novel series of N-1 and C-3 substituted indole-based thiosemicarbazones 5(a–r) are synthesized and further analyzed for their inhibition potential against tyrosinase enzyme through in vitro assays. The synthesized compounds displayed very good to moderate inhibition with half maximal inhibitory concentration in the range of 12.40 ± 0.26 μM to 47.24 ± 1.27 μM. Among all the derivatives 5k displayed the highest inhibitory activity. According to SAR analysis, the derivatives with 4-substitution at the benzyl or phenyl ring of the thiosemicarbazones exhibited better inhibitory potential against tyrosinase. In silico analysis (including ADMET prediction and molecular docking) was conducted and compared with the standard drug (kojic acid). These findings may help the hunt for new melanogenesis inhibitors that the food and cosmetics industries may find valuable.
1. Introduction
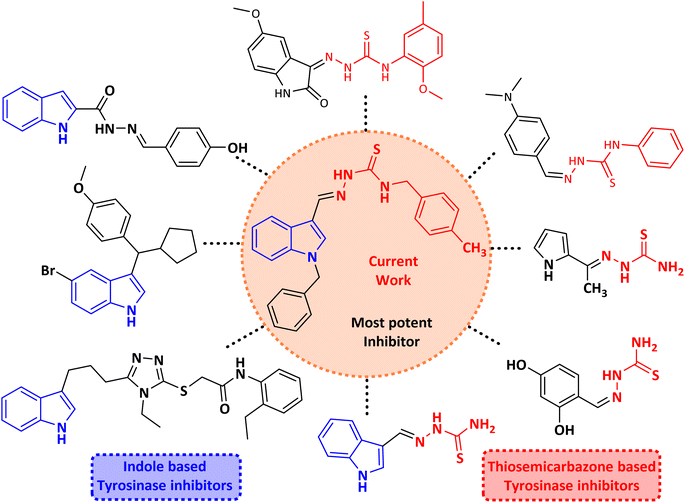
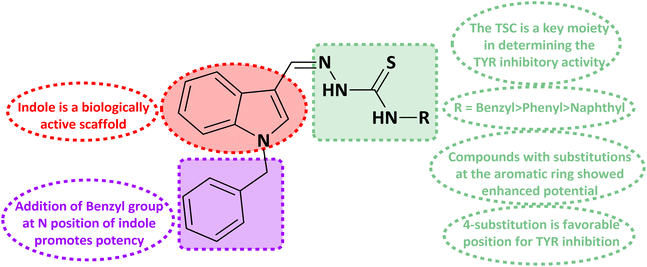
Melanocytes create melanin; tyrosine is a significant rate-limiting enzyme for the synthesis of melanin (EC 1.14.18.1) which has a vital function in shielding the skin from UV ray damage.1,2 Melanin is a vital biopolymer controlling skin, eye, and hair color.3–5 Tyrosinase is a metalloenzyme that has two bifunctional copper ions in its catalytic site.6,7 It controls the key steps in the melanin biosynthesis pathway.8,9 Excessive UV exposure results in aberrant melanin synthesis and deposition, often leading to skin pigmentation as in lentigo, freckles, melisma, and other skin conditions.10,11 It catalytically oxidizes L-tyrosine to dopaquinone, which regulates the first two steps of melanin biosynthesis:12 firstly, it converts L-tyrosine to 3,4-dihydroxyphenylalanine (L-DOPA); secondly, it further oxidizes L-DOPA to dopaquinone, which is a crucial intermediate in the biosynthesis of melanin and is subsequently converted to melanin pigments through related chemical reactions.13–15 Thus, tyrosinase inhibition is needed to reduce melanin production.16,17 Other than their use in pigmentation disorders, they are also crucial in the food sector. Enzymatic browning is also caused by tyrosinase's ability to catalyze the conversion of phenolic compounds found in fruits and vegetables into quinones. For this reason, regulation of tyrosinase activity is essential for maintaining the quality of fruits and vegetables.18–20 Numerous tyrosinase inhibitors, both natural and synthetic, with a broad range of structural variations have been found.21–26 However, only a few tyrosinase inhibitors, such as arbutin, glabridin, paeonol, and kojic acid, are employed as effective anti-tyrosinase agents in the food, cosmetics, and pharmaceutical sectors. However, they still have shortcomings, such as limited stability, skin irritation, and poor effectiveness. Thus, the development of safer and more potent tyrosinase inhibitors is necessary.The indole scaffold, which has a pyrrole structure parallel to benzene, is a principal structural nuclei in drug discovery because of its unique ability to mimic peptide structure and interact with enzymes.27–31 Additionally, it has been reported by previous researchers that the activity of indole analogs with N substitution, such as phenyl and benzyl substitution, has enhanced markedly.32,33 Recently, a series of novel 1-substituted indole-3-carboxaldehyde thiosemicarbazones were synthesized and investigated as potential anticancer and antimycobacterial agents.34 Another study reported two series of N-substituted indole derivatives i.e. N-substituted indole-based chalcones and N-substituted indole-based hydrazide–hydrazones which proved to be potential antimicrobial and antileishmanial agents.35 N-substituted indole derivatives showed increased anticancer activity against numerous cancer cell lines in comparison to the unsubstituted indole derivatives.36,37 Certain indole compounds have also been identified as prospective tyrosinase inhibitors in previous investigations.27,29–31 Furthermore, indole and its analogs are found in abundance in nature and exhibit high levels of safety. According to the aforementioned studies, indole can act as a fundamental framework for the development of novel tyrosinase inhibitors.
Thiosemicarbazones are a fascinating class of compounds. Numerous pharmacological and biological features have been discovered including their efficacy as antibacterial,38 antimalarial,39 and anticancer40 agents, etc. On the other hand, thiosemicarbazones also belong to one of the main families of tyrosinase inhibitors because of their typical structural component and capacity for the chelation of copper ions in the tyrosinase enzyme's active site. The structure–activity relationship studies demonstrated that thiosemicarbazide moiety was a crucial component for the determination of the inhibitory action of tyrosinase since the thiosemicarbazide scaffold has the potential to combine the copper ions at the tyrosinase active site successfully.41 A literature study has suggested that heterocyclic TSCs were more potent than their aromatic derivatives.42 Multiple researchers have recently reported that an extensive range of thiosemicarbazones are effective tyrosinase inhibitors (Fig. 1).43–51 In recent work, Masuri et al. evaluated the tyrosinase inhibition ability of two new bishydroxylated and two new monohydroxylated derivatives of (1E)-2-(1-(2-oxo-2H-chromen-3-yl)ethylidene)hydrazine-1-carbothioamide. Interestingly, compared to the reference substance, kojic acid, these compounds exhibit stronger tyrosinase–inhibitory activities.52 In another study, several thioquinolines conjugated to thiosemicarbazide and aryl-substituted were synthesized. The most potent derivative bearing 4-chlorophenyl ring demonstrated an IC50 value of 25.75 ± 0.19 μM compared to that of kojic acid as the positive control (IC50 = 34.93 ± 0.06 μM).53 In another work, methyl 4-pyridyl ketone thiosemicarbazone (4-PT), a potent and secure tyrosinase inhibitor and anti-browning agent, was synthesized.54
Molecular hybridization, the basis for drug development, is the process of fusing two or more active molecules to produce a new molecule that can inherit the beneficial structure of its parent molecule. This approach may boost the potency of the molecule. So, to keep striving towards the synthesis of potent tyrosinase inhibitors,55–57 we report herein the synthesis of novel N-benzyl indole-based thiosemicarbazones by the incorporation of N-benzyl indole with thiosemicarbazide derivatives. These compounds were also investigated for their potency against tyrosinase enzyme. Molecular docking, ADMET, kinetics, and structure–activity relationships were also studied. Tyrosinase is a multi-copper enzyme usually distributed in different organisms and plays a significant role in melanogenesis and enzymatic browning. Therefore, inhibitors of tyrosinase can be beneficial in cosmetics as well as medicinal industries due to its depigmentation nature, and also in food and agriculture industries as anti-browning compounds.58 Molecular docking is a versatile computational technique that has the potential to predict the binding affinity of a ligand to its corresponding receptor proteins. Glide docking studies (Table S1†) have been extensively employed to explore these ligand–receptor interactions.59 The use of molecular docking investigations will enable the identification of potential inhibitors60 and furnish an avenue to explore the binding mechanism of the inhibitors with the target protein. By comparing the efficacy of the newly discovered inhibitors with the standard drug, and the co-crystallized ligand, we can evaluate their probable role in tyrosinase enzyme inhibition.
2. Results and discussion
2.1. Chemistry
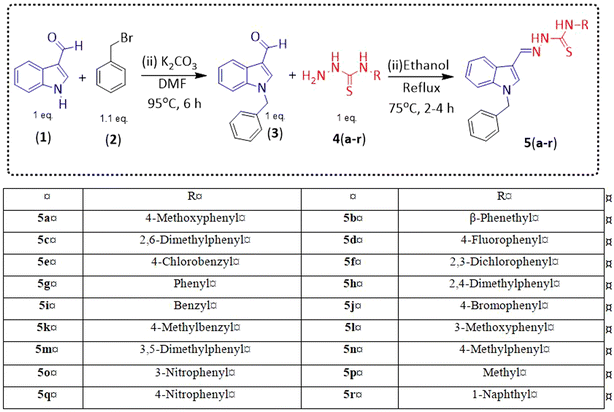
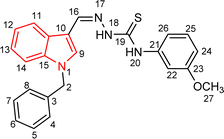
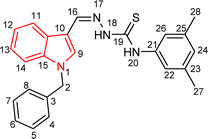
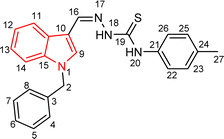
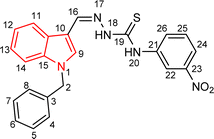
Indole and thiosemicarbazone skeletons are linked to various biological and pharmacological activities, therefore it's logical to incorporate the two moieties into a single molecular frame to synthesize more powerful biologically active molecules, like 5(a–r).61 The target molecules have been obtained by the modification at the N–H position of indole 3 carbaldehyde. The modification mainly includes the substitution by benzylation (benzyl bromide) in place of hydrogen at N–H of indole and the generation of thiosemicarbazones at C-3 with thiosemicarbazide to elevate its biological activity.The targeted compounds 5(a–s) were accomplished by the substitution at the N–H position which was done by the reaction of indole 3-carbaldehyde (1) with benzyl bromide (2) using anhydrous K2CO3 and DMF at 90 °C for 6 h, gives N-benzyl 3-formyl indole (3). The synthesis of thiosemicarbazones at C-3 was followed by the reaction of compound (3) with thiosemicarbazides 4(a–r). The synthetic route in the synthesis of 1-benzyl indole-based thiosemicarbazones 5(a–r) was displayed in Scheme 1. All the derivatives were characterized by various advanced spectroscopic techniques. A singlet is seen at 5.45–5.49 ppm which can be attributed to the CH2 group of the benzyl substitution at indole. A singlet is observed at 8.47–7.99 ppm range which can be attributed to the azomethine hydrogen, finalizing the condensation with aldehydes. NH hydrazine is also a significant signal found as a singlet in the 11.99–11.19 ppm range.
2.2. Pharmacology
| Compounds | Structure | Yield (%) | Percent inhibition (0.5 mM) | IC50 ± μM (SEM) |
|---|---|---|---|---|
| a Reaction conditions (i) K2CO3, DMF, 95 °C, 6 h (ii) ethanol, 75 °C reflux, 2–4 h. | ||||
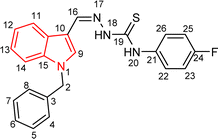
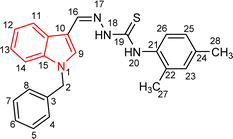
| 5a | 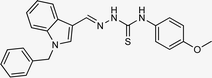 |
81 | 87.15 | 24.16 ± 0.38 |
| 5b | 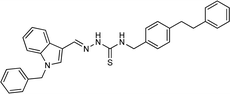 |
87 | 78.35 | 45.38 ± 0.75 |
| 5c | 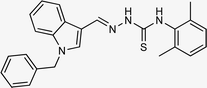 |
92 | 85.66 | 27.40 ± 0.60 |
| 5d | 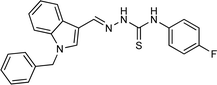 |
83 | 90.47 | 16.76 ± 0.38 |
| 5e | 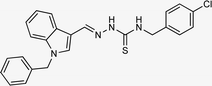 |
77 | 84.73 | 33.64 ± 1.10 |
| 5f | 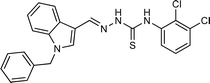 |
84 | 90.83 | 15.28 ± 0.37 |
| 5g | 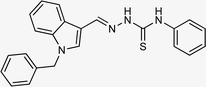 |
87 | 81.47 | 38.26 ± 1.34 |
| 5h | 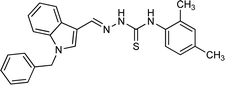 |
86.5 | 83.76 | 29.71 ± 0.72 |
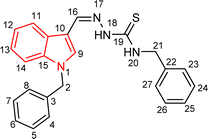
| 5i | 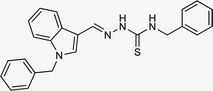 |
66 | 82.71 | 36.72 ± 1.48 |
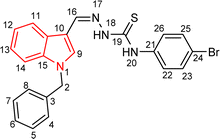
| 5j | 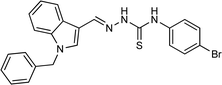 |
100 | 84.19 | 26.83 ± 0.29 |
| 5k | 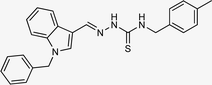 |
87 | 90.84 | 12.40 ± 0.26 |
| 5l | 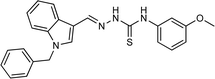 |
75 | 84.39 | 31.64 ± 1.26 |
| 5m | 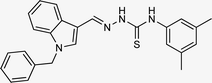 |
59 | 76.35 | 47.24 ± 1.27 |
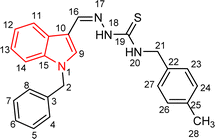
| 5n | 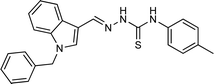 |
72 | 86.54 | 26.11 ± 0.47 |
| 5o | 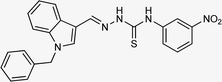 |
89 | 91.34 | 17.10 ± 0.28 |
| 5p | 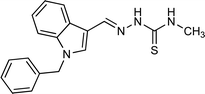 |
72 | 46.20 | N/A |
| 5q | 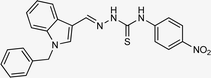 |
67 | 91.53 | 15.26 ± 0.30 |
| 5r |  |
66 | 77.86 | 34.58 ± 0.81 |
| Standard |  |
Kojic acid | 18.30 ± 0.41 | |
5e IC50 = 33.64 ± 1.10 μM
5r IC50 = 34.58 ± 0.81 μM
5i IC50 = 36.72 ± 1.48 μM
5g IC50 = 38.26 ± 1.34 μM
5b IC50 = 45.38 ± 0.75 μM
Compound 5m was the least potent member of the series with an IC50 value of 47.24 ± 1.27 μM.
In a study by Thanigaimalai et al. designed a series of naphthaldehyde-based thiosemicarbazones, the finding suggested that the hydrophobicity of the substituent on hydrazine had a significant role in the inhibitory effect against melanogenesis. Moreover, the activity was greatly enhanced when either of these hydrogens was replaced at N1 or N3.62 Based on this study, we will investigate the effect of substitutions of varying hydrophobicity on the inhibitory potential against tyrosinase.
The R group is varied with aromatic, non-aromatic, and aliphatic groups. The compounds 5i and 5g with benzyl and phenyl rings respectively showed almost similar inhibitory potential. The inhibitory potential is further collapsed in compound 5r when 1-naphthyl is used. Interestingly in compound 5p when methyl group is used, no inhibitory activity is seen. On increasing the hydrophobicity from methyl to phenyl and benzyl, a significant increase can be seen in the potency. While increasing it further to naphthyl substitution, the potency is collapsed which can be attributed to the steric hindrance with the active site of the tyrosinase enzyme.
The compounds 5q and 5o with a nitro group substitution on the phenyl ring displayed excellent inhibitory potency better than the reference standard kojic acid (IC50 = 18.30 ± 0.41 μM). However, a nitro group at the para position 5q (IC50 = 15.26 ± 0.30 μM) slightly outshined 5o (IC50 = 17.10 ± 0.28 μM) with the nitro group at the meta position. Compounds 5a and 5l containing methoxy substitution at the para and meta positions of the phenyl ring respectively also followed the same pattern. 5a showed better activity with IC50 value 24.16 ± 0.38 μM than 5l with IC50 value of 31.64 ± 1.26 μM. In the case of methyl substitution on the phenyl ring, the compound 5n with methyl substitution at the para position (IC50 = 26.11 ± 0.47 μM) put on similar inhibitory potency against tyrosinase as the kojic acid. Interestingly with the increase in no. of substitutions, potency is somewhat decreased. Compound 5c with 2,6 dimethyl substitution (IC50 = 27.40 ± 0.60 μM) and 5h with 2,4 dimethyl substitution (IC50 = 29.71 ± 0.72 μM) showed moderately lower activity than 5n with 4 methyl substitution. While in compound 5m with 3,5 dimethyl substitutions, activity showed a steep decline with an IC50 value of 47.24 ± 1.27 μM. In the case of our study, a certain relationship can be seen between the inhibition and the structure as shown in Fig. 2. In all of the above-mentioned cases, compounds with p-substitution showed superior potency.
In the case of halogen-substituted derivatives, compound 5d, which is the third most potent derivative of the series with p-fluoro substitution on the phenyl ring displayed higher potency than 5j containing p-bromo substitution. The result is consistent with the findings of Liu et al. who synthesized novel thiourea derivatives with sulfur-containing heterocyclic scaffolds. The study also reported that the compounds containing fluorine atoms at the phenyl ring exhibited higher tyrosinase inhibitory activities.63
Compound 5k with 4-methyl benzyl substitution displayed the highest potency in the entire series. While compound 5e with 4-chloro benzyl substitution displayed moderately lower activity. It can be seen that with the increase in electronegativity of the substituents, inhibitory potential is decreased. In the case of unsubstituted benzyl in compound 5i, activity is lower than both the substituted derivatives 5k and 5e. It may be suggested that the substitutions favor inhibitory potency.
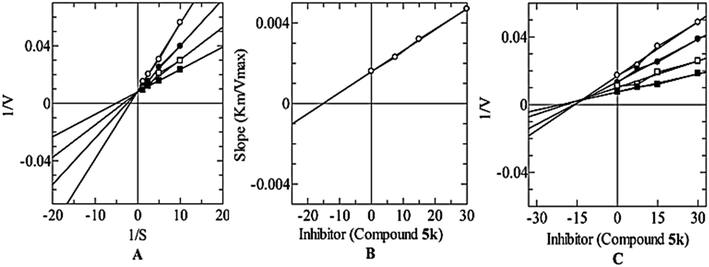
2.3. Kinetics
A kinetic study of the most potent compound 5k was performed to explore its mechanism of action. Derivative 5k was identified as a competitive type of inhibitor with a Ki value of 10.20 ± 0.006 μM. Such type of inhibitor binds with the active site residues of the tyrosinase hence, usually, this type of inhibition increases the Km value of the enzyme while there is no effect on the Vmax value of the enzyme and remains constant (Fig. 3).2.4. Molecular docking
Molecular docking is a worth mentioning method to evaluate the binding affinity, as well as the interaction parameters of investigated ligands in this study. This tool presents significant insights into the intended binding capabilities of the ligands under consideration with the specified target protein.64 The glide module was incorporated to execute a molecular docking study, and the outcomes were then assessed based on corresponding glide score values as shown in Table 2 for the hit compounds 5k, the standard drug kojic acid, and the co-crystallized ligand of the chosen tyrosinase target protein, the glide score data of all the studied ligands is presented in (Table S2†).| Ligands | gScore (kcal mol−1) | Emodel (kcal mol−1) | HBI residue (distance Å) | Polar interacting amino acids | Hydrophobic interacting amino acid residues | RMSD (Å) |
|---|---|---|---|---|---|---|
| a Abbreviation: HBI, hydrogen bonding interactions. | ||||||
| 5k | −5.157 | −63.013 | Asp357 (2.02) | Gln307, Thr308, Thr360, Ser364 | Ile17, Tyr311, Trp358, Phe368, Val371 | 2.71 |
| Kojic acid | −4.549 | −34.051 | Thr308 (1.96), Asp312 (2.00), Glu356 (2.08) | Gln307, Thr308 | Ala304, Tyr311, Tyr314, Trp358 | 3.08 |
| Co-crystallized ligand | −0.129 | −19.187 | Asp357 (1.94), Glu359 (1.98), Lys379 (1.85) | Ser364 | Trp358, Phe368, Val371 | |
The poses retrieved after docking were examined visually, and the associated binding interactions of the synthesized compounds with the binding pocket residues were analyzed with the help of ligand–interaction diagrams (3D & 2D). The docking scores are tabulated as negative values, the lower the docking score value and glide energy value, the higher would be the binding affinity and vice versa. It was thus established that most of the reported compounds displayed high glide docking scores against the chosen target protein under study and all the synthesized compounds are superior in binding affinity as compared to the co-crystallized ligand, whereas the compounds 5k, 5c, 5b, and 5a are even better than the standard drug (kojic acid) under investigation as displayed in Table S1.†
The hit compound 5k showed an appreciable binding interactive profile with tyrosinase target protein having a gScore of −5.157 kcal mol−1. Asp357 (2.02 Å) amino acid residue is engaged in hydrogen bonding and Gln307, Thr308, Thr360, and Ser364 are the polar interacting amino acid residues. Ile17, Tyr311, Trp358, Phe368, and Val371 are the hydrophobic interacting residues. The hydrophobic interactions are of vital significance for drug-target binding as they are involved in stabilization of the ligand at the binding interface. The visual display of standard drug-kojic acid for the said tyrosinase inhibition activity shows a gScore of −4.549 kcal mol−1. It establishes hydrogen bonding with Thr308 (1.96 Å), Asp312 (2.00 Å), and Glu356 (2.08 Å). In addition, Ala304, Tyr311, Tyr314, Trp358 are hydrophobically engaged amino acids whereas Gln307, and Thr308 are polar amino acids as shown in Fig. 4.These interactions highlight the consistent binding pattern between the hit compound (5k), the standard drug (kojic acid) and the co-crystallized ligand. These findings thus establishes that the 5k exhibits the superior tyrosinase inhibition activity as compared to the standard drug and the co-crystallized ligand among all the investigated ligands. The Asp357 is participant in hydrogen bonding in the co-crystallized ligand and the hit compound, 5k whereas the Thr308 is commonly observed polar contacts in both 5k and the standard drug whereas the Ser364 is found in both the 5k and co-crystallized. Furthermore, as far as the hydrophobic profile is concerned, Tyr311 is commonly visualized in both the 5k and the standard drug, whereas Phe368, and Val371 are the common residues in the 5k and the co-crystallized. Trp358 is commonly viewed hydrophobic interactions in all these investigated compounds. Fig. S2† further highlights various interactions in the other hit compounds.
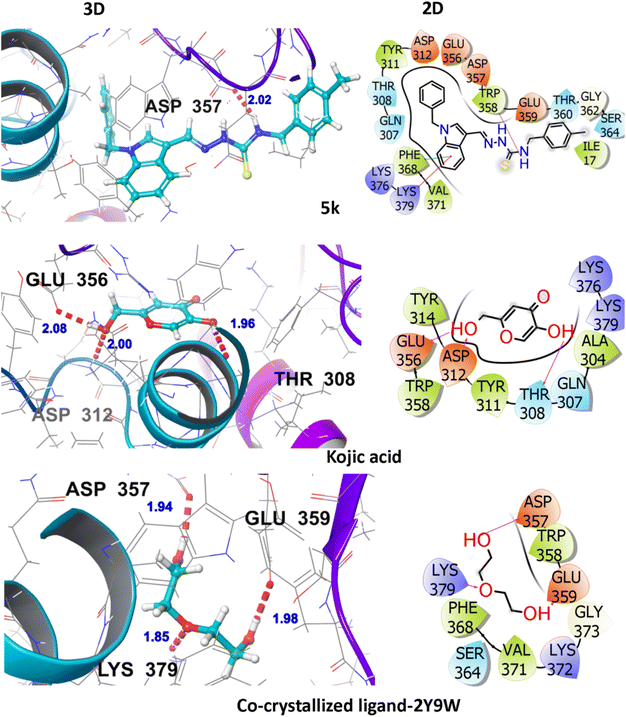 | ||
| Fig. 4 3D and 2D visualization of compound-5k, kojic acid and co-crystalized ligand with target protein (PDB ID: 2Y9W). | ||
2.5. Validation of the docking protocol
For each ligand–reference pair, whenever a pose is classified as a good solution, it predicts that the scoring function regenerates the crystallographic binding orientation.The RMSD values for the hit compound are within the acceptable limit of less than 3 Å, with 5k at 2.71 Å. The reference compound kojic acid touches the standard limit, with an RMSD of 3.08 Å as seen in Table 2 and Fig. 5.
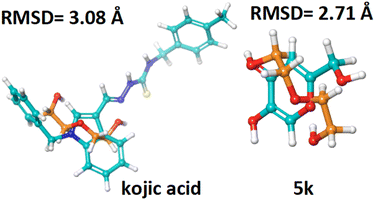 | ||
| Fig. 5 RMSD values of the hit compound (5k), and the reference compound kojic acid (cyan color) interacting with the co-crystallized ligand (orange color). | ||
2.6. ADMET analysis
Our aims and objectives emphasized to assess the ADMET properties of the investigated compounds, the hit compound 5k is showing good binding interactions with the target receptor in glide molecular docking assay along with the ADMET profile of standard drug, kojic acid as presented in Table 3 while the detailed ADMET properties of all investigated ligands are shown in Table S3†. The compound (5e) demonstrated considerable lipid solubility followed by compounds 5i, and 5a (Table S3†) with XLOGP3 as high as 6.09. It was then evaluated that the investigated compounds exhibit poor absorption when their TPSA is greater than 140 Å2, and all the investigated compounds showed compliance with this standard. In addition, all these compounds have no more than 5 HBDs, no more than 10 HBAs, and molecular weight is less than 500 dalton. The favorable “drug-likeness” is adhered to Lipinski, Ghose, Veber, and Egan rules with a zero-violation trend, shown by most of these compounds. The synthesized compounds under study possess a high likelihood of passive absorption through the gastrointestinal tract with 0.55 bioavailability, and they do not cross the blood–brain barrier (except compound 5p). In addition, these compounds are not the substrates for p-glycoprotein, which is a key factor in drug efflux except for the compounds (5b, 5c, 5e, 5i, and 5k). Most of these compounds showed inhibition of CYP enzymes, thus exhibiting a significant interactive profile with drug-metabolizing enzymes, and associated toxicity (Table 3). In addition, all these compounds have no more than 5 HBDs, no more than 10 HBAs, and a corresponding molecular weight is less than 500 dalton presenting them the suitable drug candidates. In addition, the boiled-egg plot was executed to validate the GI absorption, as well as the BBB permeation characteristics of the hit compound 5e, experimentally proved hit compound 5k, and standard kojic acid. In addition, it is established from the plot that none of the compounds has passed through the BBB but they lie within the GI absorption premises (Fig. 6). Fig. S3† shows that compound 5p is the only compound from the investigated list of compounds that can cross the BBB.| Absorption | Metabolism | |||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| a Abbreviations: TPSA, topological polar surface area; BBB, blood–brain barrier; BA, bioavailability; SA, synthetic accessibility; ADMET, absorption, distribution, metabolism, excretion, and toxicity; GI, gastrointestinal tract. | ||||||||||||||
| Ligand | TPSA | ESOL LogS | XLOGP3 | GI absorption | BBB | P-gp substrate | BA | CYP1A2 inhibitor | CYP2C19 inhibitor | CYP2C9 inhibitor | CYP2D6 inhibitor | CYP3A4 inhibitor | Lipinski # violations | SA |
| Kojic acid | 33.13 | −0.7 | −0.64 | High | No | No | 0.55 | No | No | No | No | No | 0 | 2.53 |
| 5k | 73.44 | −5.62 | 5.13 | High | No | Yes | 0.55 | Yes | Yes | Yes | Yes | Yes | 0 | 3.3 |
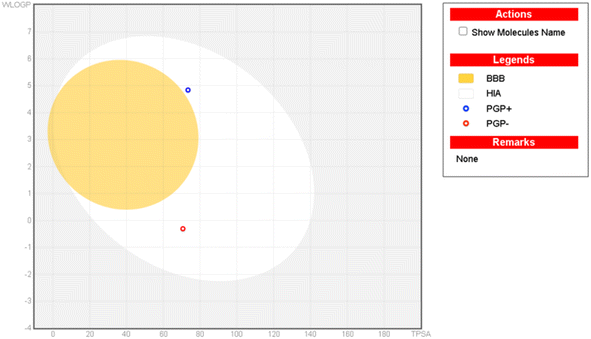 | ||
| Fig. 6 Boiled-egg plot (plot of WLOGP against TPSA) of the hit compound-5k and standard-kojic acid from SwissADME. | ||
3. Conclusion
In the current study, a series of N-benzyl indole-based thiosemicarbazones were designed, synthesized, and characterized by spectroscopic techniques (1H NMR and 13C NMR). The derivates were screened for their inhibition potential against tyrosinase enzyme. All synthesized compounds present an outstanding to moderate inhibition potency compared to kojic acid as a positive control. These encouraging findings may lead to preclinical testing of compounds 5k in the hopes of creating a novel class of tyrosinase inhibitors. Because they address the need for effective and safe therapies for abnormal pigmentation and associated health risks, these results have implications for the pharmaceutical and cosmetics sectors.4. Experimental
4.1. Chemistry
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 petroleum ether and ethyl acetate as eluent). The solid formed was filtered, washed with ethanol, and dried to yield targeted thiosemicarbazones 5(a–r). Characterization data of each compound synthesized is given below:
1 petroleum ether and ethyl acetate as eluent). The solid formed was filtered, washed with ethanol, and dried to yield targeted thiosemicarbazones 5(a–r). Characterization data of each compound synthesized is given below:
4.1.3.1 (E)-2-[(1-benzyl-1H-indol-3-yl)methylene]-N-(4-methoxyphenyl)hydrazinecarbothioamide (5a).
Color: off-white, yield: 81%, melting point: 217–218 °C. δH (600 MHz, DMSO-d6) 11.51 (1H, s, H-18), 9.50 (1H, s, H-20), 8.41 (1H, s, H-9), 8.28 (1H, d, J = 7.8 Hz, Ar-H), 8.08 (1H, s, H-16), 7.53 (1H, d, J = 8.1 Hz, Ar-H), 7.49–7.43 (2H, m, Ar-H), 7.33 (2H, t, J = 7.5 Hz, Ar-H), 7.30–7.25 (3H, m, Ar-H), 7.22 (1H, t, J = 7.6, Ar-H), 7.17 (1H, t, J = 7.5 Hz, Ar-H), 6.98–6.91 (2H, m, H-23, 25), 5.47 (2H, s, N-CH2), 3.78 (3H, s, –OCH3); 13C NMR (151 MHz, DMSO) δ 175.55 (C-19), 157.30 (C-16), 140.91 (CH), 137.92 (C), 137.36 (C), 134.56 (CH), 132.70 (C), 129.12 (C-5, 7), 128.07 (CH), 127.71 (C-4, 8), 127.65 (CH), 125.22 (C), 123.36 (CH), 122.72 (CH), 121.50 (CH), 113.79 (CH), 111.18 (C), 111.15 (C), 55.74 (C-2), 49.89 (C-27). HPLC: CH3CN
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) H2O = 80
H2O = 80![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 20; tR: 2.051 min, purity: 99.3%. TOF MS ES+ (m/z): [M + H]+, calcd: 415.1592, found: 415.1598, anal. calcd for C24H22N4OS: C, 69.54; H, 5.35; N, 13.52; found: C, 69.59; H, 5.42; N, 13.58.
20; tR: 2.051 min, purity: 99.3%. TOF MS ES+ (m/z): [M + H]+, calcd: 415.1592, found: 415.1598, anal. calcd for C24H22N4OS: C, 69.54; H, 5.35; N, 13.52; found: C, 69.59; H, 5.42; N, 13.58.
4.1.3.2 (E)-2-[(1-benzyl-1H-indol-3-yl)methylene]-N-phenethylhydrazinecarbothioamide (5b).
Color: white, yield: 87%, melting point: 198–201 °C. δH (600 MHz, DMSO-d6) 11.27 (1H, s, H-18), 8.31 (1H, s, H-9), 8.05 (1H, d, J = 7.9 Hz, Ar-H), 7.99 (1H, s, H-16), 7.87 (1H, t, J = 5.8 Hz, H-20), 7.52 (1H, d, J = 8.2 Hz, Ar-H), 7.33 (6H, dd, J = 10.0, 5.7 Hz, Ar-H), 7.29–7.21 (5H, m, Ar-H), 7.17 (1H, t, J = 7.5 Hz, Ar-H), 5.45 (2H, s, H-2), 3.88 (2H, q, J = 6.7 Hz, H-21), 2.96 (2H, t, J = 7.3 Hz, H-22); 13C NMR (151 MHz, DMSO) δ 176.40 (C-19), 140.62 (C-16), 139.76 (CH), 137.91 (C), 137.38 (C), 134.56 (CH), 129.11 (C-5, 7), 129.09 (CH), 128.97 (CH), 128.06 (CH), 127.66 (C-4, 8), 126.70 (CH), 125.01 (C), 123.37 (CH), 122.67 (CH), 121.49 (CH), 111.23 (C), 111.14 (C), 49.85 (C-2), 45.25 (C-21), 35.38 (C-22). HPLC: CH3CN
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) H2O = 80
H2O = 80![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 20; tR: 1.814 min, purity: 99.3%. TOF MS ES+ (m/z): [M + H]+, calcd: 413.1799, found 413.1798, anal. calcd for C25H24N4S: C 72.78; H, 5.86; N, 13.58; found: C, 72.83; H, 5.92; N, 13.61.
20; tR: 1.814 min, purity: 99.3%. TOF MS ES+ (m/z): [M + H]+, calcd: 413.1799, found 413.1798, anal. calcd for C25H24N4S: C 72.78; H, 5.86; N, 13.58; found: C, 72.83; H, 5.92; N, 13.61.
4.1.3.3 (E)-2-[(1-benzyl-1H-indol-3-yl)methylene]-N-(2,6-dimethylphenyl)hydrazinecarbo thioamide (5c).
Color: white, yield: 92%, melting point: 225–227 °C. δH (600 MHz, DMSO-d6) 11.45 (1H, s, H-18), 9.28 (1H, s, H-20), 8.42 (1H, s, Ar-H), 8.40 (1H, s, H-9), 8.06 (1H, s, H-16), 7.50 (1H, d, J = 8.2 Hz, Ar-H), 7.33 (2H, t, J = 7.4 Hz, Ar-H), 7.26 (3H, d, J = 7.7 Hz, Ar-H), 7.21 (1H, t, J = 7.6 Hz, Ar-H), 7.15–7.10 (4H, Ar-H), 5.47 (2H, s, H-2), 2.23 (6H, s, H-27, 28); 13C NMR (151 MHz, DMSO) δ 176.12 (C-19), 140.80 (C-16), 138.11 (C), 137.99 (C), 137.35 (C), 137.16 (CH), 134.62 (CH), 129.10 (CH), 128.04 (CH), 128.00 (CH), 127.58 (CH), 127.24 (CH), 125.17 (C), 123.32 (CH), 121.38 (CH), 111.35 (C), 110.97 (C), 49.85 (C-2), 18.66 (C-27, 28). HPLC: CH3CN
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) H2O = 80
H2O = 80![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 20; tR: 2.444 min, purity: 100.0%. TOF MS ES+ (m/z): [M + H]+, calcd: 413.1799, found: 413.1799, anal. calcd for C25H24N4S: C, 72.78; H, 5.86; N, 13.58; found: C, 72.83; H, 5.92; N, 13.60.
20; tR: 2.444 min, purity: 100.0%. TOF MS ES+ (m/z): [M + H]+, calcd: 413.1799, found: 413.1799, anal. calcd for C25H24N4S: C, 72.78; H, 5.86; N, 13.58; found: C, 72.83; H, 5.92; N, 13.60.
4.1.3.4 (E)-2-[(1-benzyl-1H-indol-3-yl)methylene]-N-(4-fluorophenyl)hydrazinecarbothioamide (5d).
Color: white, yield: 83%, melting point: 221–223 °C. δH (600 MHz, DMSO-d6) 11.61 (1H, s, H-18), 9.62 (1H, s, H-20), 8.42 (1H, s, H-9), 8.29 (1H, d, J = 7.9 Hz, Ar-H), 8.10 (1H, s, H-16), 7.60 (2H, dt, J = 8.8, 3.7 Hz, Ar-H), 7.53 (1H, d, J = 8.3 Hz, Ar-H), 7.36–7.31 (2H, m, Ar-H), 7.28–7.27 (3H, m, Ar-H), 7.24–7.16 (4H, m, Hz, Ar-H), 5.48 (2H, s, H-2); 13C NMR (151 MHz, DMSO) δ 175.05 (C-19), 160.36 (C-16), 158.76 (d, JC–F = 241.6 Hz, C-24), 140.85 (CH), 137.47 (C), 136.93 (C), 135.74(CH), 134.28 (CH), 128.68 (C-5, 7), 127.79 (CH), 127.64 (CH), 127.21 (CH), 124.78 (C), 122.94 (CH), 122.35 (CH), 121.08 (CH), 114.80 (CH), 114.65 (C), 110.70 (C), 49.46 (C-2). HPLC: CH3CN
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) H2O = 80
H2O = 80![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 20; tR: 2.204 min, purity: 99.7%. TOF MS ES+ (m/z): [M + H]+, calcd: 403.1392, found: 403.11395, anal. calcd for C23H19FN4S: C, 68.63; H, 4.76; N, 13.92; found: C, 68.69; H, 4.83; N, 13.99.
20; tR: 2.204 min, purity: 99.7%. TOF MS ES+ (m/z): [M + H]+, calcd: 403.1392, found: 403.11395, anal. calcd for C23H19FN4S: C, 68.63; H, 4.76; N, 13.92; found: C, 68.69; H, 4.83; N, 13.99.
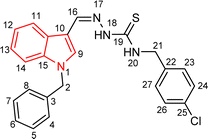
4.1.3.5 (E)-2-[(1-benzyl-1H-indol-3-yl)methylene]-N-(4-chlorobenzyl)hydrazinecarbothioamide (5e).
Color: white, yield: 77%, melting point: 225–227 °C. δH (600 MHz, DMSO-d6) 11.37 (1H, s, H-18), 8.50 (1H, t, J = 6.4 Hz, H-20), 8.36 (1H, s, H-9), 8.28 (1H, d, J = 7.9 Hz, Ar-H), 8.02 (1H, s, H-16), 7.51 (1H, d, J = 8.2 Hz, Ar-H), 7.41–7.38 (4H, m, Ar-H), 7.32 (2H, t, J = 7.6 Hz, Ar-H), 7.26 (3H, d, J = 7.5 Hz, Ar-H), 7.22 (1H, t, J = 7.7 Hz, Ar-H), 7.16 (1H, t, J = 7.5 Hz, Ar-H), 5.46 (2H, s, H-2), 4.89 (2H, d, J = 6.2 Hz, H-21); 13C NMR (151 MHz, DMSO) δ 177.05 (C-19), 140.95 (C-16), 139.35 (CH), 137.93 (C), 137.37 (C), 134.55 (CH), 131.62 (C), 129.44 (C-5, 7), 129.11 (CH), 128.59 (CH), 128.05 (CH), 127.64 (C-4, 8), 125.10 (C), 123.36 (CH), 122.91 (CH), 121.43 (CH), 111.24 (C), 111.10 (C), 49.85 (C-2), 46.48 (C-21); HPLC: CH3CN
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) H2O = 80
H2O = 80![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 20; tR: 2.690 min, purity: 99.3%. TOF MS ES+ (m/z): [M + H]+, calcd: 433.1253, found 433.1253, anal. calcd for C24H21ClN4S; C, 60.93; H, 4.00; N, 12.36; found; C, 60.98; H, 4.12; N, 12.46.
20; tR: 2.690 min, purity: 99.3%. TOF MS ES+ (m/z): [M + H]+, calcd: 433.1253, found 433.1253, anal. calcd for C24H21ClN4S; C, 60.93; H, 4.00; N, 12.36; found; C, 60.98; H, 4.12; N, 12.46.
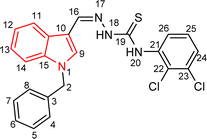
4.1.3.6 (E)-2-[(1-benzyl-1H-indol-3-yl)methylene]-N-(2,3-dichlorophenyl)hydrazinecarbo thioamide (5f).
Color: greenish white, yield: 84%, melting point: 213–215 °C. δH (600 MHz, DMSO-d6) 11.92 (1H, s, H-18), 9.75 (1H, s, H-20), 8.45 (1H, s,H-9), 8.35 (1H, d, J = 7.9 Hz, Ar-H), 8.12–8.11 (1H, m, Ar-H), 8.01 (1H, s, H-16), 7.54 (2H, dd, J = 8.1, 1.6 Hz, Ar-H), 7.42 (1H, t, J = 8.1 Hz, Ar-H), 7.33 (2H, dd, J = 8.3, 6.8 Hz, Ar-H), 7.30–7.26 (3H, m, Ar-H), 7.25–7.24 (1H, m, Ar-H), 7.20–7.15 (1H, m, Ar-H), 5.48 (2H, s, H-2); 13C NMR (151 MHz, DMSO) δ 174.82 (C-19), 142.05 (C-16), 138.91 (CH), 137.84 (C), 137.45 (C), 135.53 (CH), 131.98 (C), 129.13 (C-5, 7), 128.09 (CH), 127.94 (C-4, 8), 127.69 (C), 127.67 (CH), 127.15 (C), 125.03 (C), 123.53 (CH), 122.81 (CH), 121.57 (CH), 111.31 (C), 110.96 (CH), 49.92 (C-2). HPLC: CH3CN
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) H2O = 80
H2O = 80![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 20; tR: 4.467 min, purity: 97.7%. TOF MS ES+ (m/z): [M + H]+, calcd: 453.0707, found: 453.0704, anal. calcd for C23H18Cl2N4S: C, 60.93; H, 4.00; N, 12.36; found: C, 60.99; H, 4.12; N, 12.42.
20; tR: 4.467 min, purity: 97.7%. TOF MS ES+ (m/z): [M + H]+, calcd: 453.0707, found: 453.0704, anal. calcd for C23H18Cl2N4S: C, 60.93; H, 4.00; N, 12.36; found: C, 60.99; H, 4.12; N, 12.42.
4.1.3.7 (E)-2-[(1-benzyl-1H-indol-3-yl)methylene]-N-phenylhydrazinecarbothioamide (5g).
Color: white, yield: 87%, melting point: 203–205 °C. δH (600 MHz, DMSO-d6) 11.62 (1H, s, H-18), 9.64 (1H, s, H-20), 8.43 (1H, s, H-9), 8.27 (1H, d, J = 7.8 Hz, Ar-H), 8.11 (1H, s, H-16), 7.66 (2H, d, J = 7.9 Hz, Ar-H), 7.54 (1H, d, J = 8.1 Hz, Ar-H), 7.38 (2H, t, J = 7.7 Hz, Ar-H), 7.33 (2H, t, J = 7.5 Hz, Ar-H), 7.28–7.26 (3H, m, Ar-H), 7.24–7.21 (3H, m, Ar-H), 5.48 (2H, s, H-2); 13C NMR (151 MHz, DMSO) δ 175.02 (C-19), 141.15 (C-16), 139.77 (CH), 137.90 (C), 137.39 (C), 134.73 (CH), 129.13 (C-5, 7), 128.62 (CH), 128.08 (C), 127.65 (C-4, 8), 125.63 (CH), 125.45 (C), 125.23 (CH), 123.40 (CH), 122.66 (CH), 121.58 (CH), 111.21 (C), 111.14 (CH), 49.91 (C-2). HPLC: CH3CN
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) H2O = 80
H2O = 80![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 20; tR: 2.284 min, purity: 99.9%. TOF MS ES+ (m/z): [M + H]+, calcd: 385.1486, found: 385.1887, anal. calcd for C23H20N4S: C, 71.85; H, 5.24; N, 14.57; found: C, 71.90; H, 5.29; N, 14.62.
20; tR: 2.284 min, purity: 99.9%. TOF MS ES+ (m/z): [M + H]+, calcd: 385.1486, found: 385.1887, anal. calcd for C23H20N4S: C, 71.85; H, 5.24; N, 14.57; found: C, 71.90; H, 5.29; N, 14.62.
4.1.3.8 (E)-2-[(1-benzyl-1H-indol-3-yl)methylene]-N-(2,4-dimethylphenyl)hydrazinecarbo thioamide (5h).
Color: white, yield: 86.5%, melting point: 180–182 °C. δH (600 MHz, DMSO-d6) 11.52 (1H, s, H-18), 9.34 (1H, s, H-20), 8.42 (1H, s, H-9), 8.37–8.29 (1H, m, Ar-H), 8.07 (1H, s, H-16), 7.57–7.48 (1H, m), 7.36–7.21 (3H, m), 7.27–7.20 (4H, m), 7.16–7.02 (3H, m), 5.47 (2H, s, H-2), 2.31 (3H, d, J = 3.6 Hz), 2.25 (3H, d, J = 3.6 Hz); 13C NMR (151 MHz, DMSO) δ 175.92 (C-19), 140.89 (CH), 137.94 (C-16), 137.38 (C), 136.11 (C), 135.82 (CH), 134.82 (C), 134.71 (CH), 131.04 (C), 129.11 (C-5, 7), 128.50 (CH), 128.06 (C), 127.64 (C-4, 8), 126.80 (CH), 125.16 (C), 123.37 (CH), 122.93 (CH), 121.43 (CH), 111.25 (CH), 111.12 (C), 49.88 (C-2), 21.08 (C-27), 18.27 (C-28). HPLC: CH3CN
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) H2O = 80
H2O = 80![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 20; tR: 2.806 min, purity: 100.0%. TOF MS ES+ (m/z): [M + H]+, calcd: 413.1799, found: 413.1799, anal. calcd for C25H24N4S: C, 72.78; H, 5.86; N, 13.58; found: C, 72.82; H, 5.91; N, 13.63.
20; tR: 2.806 min, purity: 100.0%. TOF MS ES+ (m/z): [M + H]+, calcd: 413.1799, found: 413.1799, anal. calcd for C25H24N4S: C, 72.78; H, 5.86; N, 13.58; found: C, 72.82; H, 5.91; N, 13.63.
4.1.3.9 (E)-N-benzyl-2-[(1-benzyl-1H-indol-3-yl)methylene]hydrazinecarbothioamide (5i).
Color: white, yield: 66%, melting point: 193–195 °C. δH (600 MHz, DMSO-d6) 11.34 (1H, s, H-18), 8.43 (1H, t, J = 6.2 Hz, H-20), 8.35 (1H, s, H-9), 8.24 (1H, d, J = 7.9 Hz, Ar-H), 8.02 (1H, s, H-16), 7.51 (1H, d, J = 8.2 Hz, Ar-H), 7.39–7.30 (6H, m, Ar-H), 7.28–7.20 (4H, m, Ar-H), 7.21 (1H, t, J = 7.7 Hz, Ar-H), 7.13 (1H, t, J = 7.5 Hz, Ar-H), 5.46 (2H, s, H-2), 4.91 (2H, d, J = 6.1 Hz, H-21); 13C NMR (151 MHz, DMSO) δ 177.00 (C-19), 140.76 (C-16), 140.17 (CH), 137.94 (C), 137.36 (C), 134.51 (CH), 129.11 (C-5, 7), 128.68 (CH), 128.05 (CH), 127.64 (C-4, 8), 127.53 (CH), 127.16 (C), 125.09 (C), 123.34 (CH), 122.81 (CH), 121.40 (CH), 111.25 (C), 111.12 (CH), 49.84 (C-2), 47.11 (C-21). HPLC: CH3CN
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) H2O = 80
H2O = 80![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 20; tR: 2.237 min, purity: 100.0%.TOF MS ES+ (m/z): [M + H]+, calcd: 399.1643, found: 399.1643, anal. calcd for C24H22N4S: C, 72.33; H, 5.56; N, 14.06; found: C, 72.39; H, 5.62; N, 14.10.
20; tR: 2.237 min, purity: 100.0%.TOF MS ES+ (m/z): [M + H]+, calcd: 399.1643, found: 399.1643, anal. calcd for C24H22N4S: C, 72.33; H, 5.56; N, 14.06; found: C, 72.39; H, 5.62; N, 14.10.
4.1.3.10 (E)-2-[(1-benzyl-1H-indol-3-yl)methylene]-N-(4-bromophenyl)hydrazinecarbothioamide (5j).
Color: greenish white, yield: 100%, melting point: 217–218 °C. δH (600 MHz, DMSO-d6) 11.69 (1H, s, H-18), 9.68 (1H, s, H-20), 8.43 (1H, s, H-9), 8.30–8.24 (1H, m), 8.10 (1H, s, H-16), 7.66–7.61 (2H, m), 7.58–7.51 (3H, m), 7.37–7.30 (2H, m), 7.27–7.25 (3H, m), 7.22 (1H, ddd, J = 8.3, 7.0, 1.3 Hz), 7.18 (1H, ddd, J = 8.1, 7.1, 1.1 Hz), 5.48 (2H, s, H-2); 13C NMR (151 MHz, DMSO) δ 174.92 (C-19), 141.49 (C-16), 139.26 (CH), 137.89 (C), 137.38 (C), 134.83 (CH), 131.36 (C), 129.13 (C-5, 7), 128.08 (CH), 127.69 (C-4, 8), 127.65 (CH), 125.22 (C), 123.41 (CH), 122.74 (CH), 121.56 (CH), 117.58 (CH), 111.19 (C), 111.08 (CH), 49.90 (C-2). HPLC: CH3CN
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) H2O = 80
H2O = 80![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 20; tR: 3.159 min, purity: 97.3%. TOF MS ES+ (m/z): [M + H]+, calcd: 463.0592, found: 463.0561, anal. calcd for C23H19BrN4S: C, 59.61; H, 4.13; N, 12.09; found: C, 59.74; H, 4.25; N, 12.14.
20; tR: 3.159 min, purity: 97.3%. TOF MS ES+ (m/z): [M + H]+, calcd: 463.0592, found: 463.0561, anal. calcd for C23H19BrN4S: C, 59.61; H, 4.13; N, 12.09; found: C, 59.74; H, 4.25; N, 12.14.
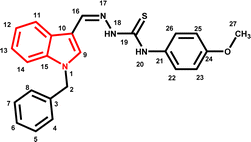
4.1.3.11 (E)-2-[(1-benzyl-1H-indol-3-yl)methylene]-N-(4-methylbenzyl)hydrazinecarbothioamide (5k).
Color: white, yield: 87%, melting point: 223–225 °C. δH (600 MHz, DMSO-d6) 11.34 (1H, s, H-18), 8.38 (1H, t, J = 6.2 Hz, H-20), 8.36 (1H, s, H-9), 8.23 (1H, d, J = 7.9 Hz, Ar-H), 8.01 (1H, s, H-16), 7.51 (1H, d, J = 8.2 Hz, Ar-H), 7.34–7.30 (2H, m, Ar-H), 7.30–7.24 (5H, m, Ar-H), 7.22 (1H, t, J = 7.6 Hz, Ar-H), 7.18–7.11 (3H, m, Ar-H), 5.45 (2H, s, H-2), 4.87 (2H, d, J = 6.1 Hz, H-21), 2.28 (3H, s); 13C NMR (151 MHz, DMSO) δ 176.91 (C-19), 140.68 (C-16), 137.93 (CH), 137.36 (C), 137.07 (C), 136.22, 134.46 (CH), 129.24 (C-5, 7), 129.10, 128.05 (CH), 127.64 (C-4, 8), 127.57 (CH), 125.10 (C), 123.35 (CH), 122.78 (CH), 121.41 (CH), 111.27 (CH), 111.13 (C), 49.85 (C-2), 46.89 (C-21), 21.17 (C-28). HPLC: CH3CN
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) H2O = 80
H2O = 80![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 20; tR: 2.615 min, purity: 100%. TOF MS ES+ (m/z): [M + H]+, calcd: 413.1799, found: 413.1798, anal. calcd for C25H24N4S: C, 72.78; H, 5.86; N, 13.58; found C, 72.82; H, 5.92; N, 13.62.
20; tR: 2.615 min, purity: 100%. TOF MS ES+ (m/z): [M + H]+, calcd: 413.1799, found: 413.1798, anal. calcd for C25H24N4S: C, 72.78; H, 5.86; N, 13.58; found C, 72.82; H, 5.92; N, 13.62.
4.1.3.12 (E)-2-[(1-benzyl-1H-indol-3-yl)methylene]-N-(3-methoxyphenyl)hydrazinecarbothioamide (5l).
Color: off-white, yield: 75%, melting point: 187–189 °C. δH (600 MHz, DMSO-d6) 11.63 (1H, s, H-18), 9.60 (1H, s, H-20), 8.42 (1H, s, H-9), 8.23 (1H, dd, J = 7.6, 1.3 Hz, Ar-H), 8.11 (1H, s, H-16), 7.54 (1H, dt, J = 8.3, 1.0 Hz, Ar-H), 7.42 (1H, t, J = 2.2 Hz, Ar-H), 7.36–7.31 (2H, m, Ar-H), 7.30–7.25 (3H, m, Ar-H), 7.25–7.17 (3H, m, Ar-H), 6.78–6.76 (1H, m, Ar-H), 5.48 (2H, s, H-2), 3.78 (3H, s, OCH3); 13C NMR (151 MHz, DMSO) δ 174.71, 159.55, 141.14, 140.84, 137.90, 137.38, 134.74, 129.36, 129.13, 128.08, 127.64, 125.23, 123.40, 122.55, 121.59, 117.44, 111.24, 111.09, 110.93, 110.82, 55.61, 49.90. HPLC: CH3CN
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) H2O = 80
H2O = 80![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 20; tR: 2.293 min, purity: 99.8%. TOF MS ES+ (m/z): [M + H]+, calcd: 415.159, found: 415.1590, anal. calcd for C24H22N4OS: C, 69.54; H, 5.35; N, 13.52; found: C, 69.59; H, 5.42; N, 13.58.
20; tR: 2.293 min, purity: 99.8%. TOF MS ES+ (m/z): [M + H]+, calcd: 415.159, found: 415.1590, anal. calcd for C24H22N4OS: C, 69.54; H, 5.35; N, 13.52; found: C, 69.59; H, 5.42; N, 13.58.
4.1.3.13 (E)-2-[(1-benzyl-1H-indol-3-yl)methylene]-N-(3,5-dimethylphenyl)hydrazine carbo thioamide (5m).
Color: white, yield: 59%, melting point: 221–223 °C. δH (600 MHz, DMSO-d6) 11.46 (1H, s, H-18), 9.29 (1H, s, H-20), 8.43 (1H, s, Ar-H), 8.41 (1H, s, Ar-H), 8.06 (1H, s, H-16), 7.50 (1H, d, J = 8.2 Hz, Ar-H), 7.34–7.31 (2H, m, Ar-H), 7.30–7.24 (3H, m, Ar-H), 7.24–7.19 (1H, m, Ar-H), 7.17–7.10 (4H, m, Ar-H), 5.47 (2H, s, H-2, Ar-H), 2.23 (6H, s, H-27, 28, Ar-H); 13C NMR (151 MHz, DMSO) δ 176.14 (C-19), 140.80 (C-16), 138.12 (CH), 137.99 (C), 137.36 (C), 137.17 (CH), 134.62 (C), 129.10 (C-5, 7), 128.04 (CH), 128.01 (C), 127.58 (C-4, 8), 127.24 (CH), 125.18 (C), 123.33 (CH), 121.39 (CH), 111.37 (CH), 110.97 (C), 49.86 (C-2), 18.67 (–CH3). HPLC: CH3CN
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) H2O = 80
H2O = 80![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 20; tR: 2.441 min, purity: 99.8%. TOF MS ES+ (m/z): [M + H]+, calcd: 413.1799, found: 413.1799, anal. calcd for C25H24N4S: C, 72.78; H, 5.86; N, 13.58; found: C, 72.83; H, 5.91; N, 13.63.
20; tR: 2.441 min, purity: 99.8%. TOF MS ES+ (m/z): [M + H]+, calcd: 413.1799, found: 413.1799, anal. calcd for C25H24N4S: C, 72.78; H, 5.86; N, 13.58; found: C, 72.83; H, 5.91; N, 13.63.
4.1.3.14 (E)-2-[(1-benzyl-1H-indol-3-yl)methylene]-N-(p-tolyl)hydrazinecarbothioamide (5n).
Color: white, yield: 72%, melting point: 221–223 °C. δH (600 MHz, DMSO-d6) 11.57 (1H, s, H-18), 9.55 (1H, s, H-20), 8.43 (1H, s, H-9), 8.26 (1H, dd, J = 7.7, 1.3 Hz, Ar-H), 8.10 (1H, s, H-16), 7.53 (1H, dt, J = 8.2, 0.9 Hz, Ar-H), 7.52–7.48 (2H, m, Ar-H), 7.35–7.31 (2H, m, Ar-H), 7.29–7.25 (3H, m, Ar-H), 7.23 (1H, ddd, J = 8.3, 7.0, 1.3 Hz, Ar-H), 7.21–7.16 (3H, m, Ar-H), 5.47 (2H, s, H-2), 2.32 (3H, s, CH3); 13C NMR (151 MHz, DMSO) δ 175.13 (C-19), 140.98 (C-16), 137.91 (CH), 137.38 (C), 137.20 (C), 134.64 (CH), 129.12 (C), 129.07 (C-5, 7), 128.07 (CH), 127.65 (C-4, 8), 125.70 (CH), 125.24 (C), 123.37 (CH), 122.62 (CH), 121.55 (CH), 111.20 (CH), 111.16 (C), 49.90 (C-2), 21.06 (C-27). HPLC: CH3CN
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) H2O = 80
H2O = 80![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 20; tR: 2.652 min, purity: 99.1%. TOF MS ES+ (m/z): [M + H]+, calcd: 399.1643, found: 399.1643, anal. calcd for C24H22N4S: C, 72.33; H, 5.56; N, 14.06; found: C, 72.39; H, 5.62; N, 14.12.
20; tR: 2.652 min, purity: 99.1%. TOF MS ES+ (m/z): [M + H]+, calcd: 399.1643, found: 399.1643, anal. calcd for C24H22N4S: C, 72.33; H, 5.56; N, 14.06; found: C, 72.39; H, 5.62; N, 14.12.
4.1.3.15 (E)-2-[(1-benzyl-1H-indol-3-yl)methylene]-N-(3-nitrophenyl)hydrazinecarbothioamide (5o).
Color: dark yellow, yield: 89%, melting point: 208–210 °C. δH (600 MHz, DMSO-d6) 11.87 (1H, s, H-18), 9.99 (1H, s, H-20), 8.75 (1H, t, J = 2.2 Hz, Ar-H), 8.47 (1H, s, H-9), 8.32 (1H, d, J = 7.8 Hz, Ar-H), 8.12 (1H, s, H-16), 8.04 (1H, ddd, J = 8.2, 2.3, 1.0 Hz, Ar-H), 7.66 (1H, t, J = 8.1 Hz, Ar-H), 7.57–7.51 (1H, m, Ar-H), 7.36–7.31 (2H, m, Ar-H), 7.28–7.26 (3H, m, Ar-H), 7.24 (1H, ddd, J = 8.2, 7.0, 1.3 Hz, Ar-H), 7.20 (1H, td, J = 7.5, 7.1, 1.1 Hz, Ar-H), 5.49 (2H, s, H-2); 13C NMR (151 MHz, DMSO) δ 174.95 (C-19), 147.77 (C-16), 142.09 (C), 141.17 (CH), 137.87 (C), 137.41 (C), 135.03 (CH), 132.03 (C), 129.69 (C-5, 7), 129.13 (CH), 128.09 (CH), 127.65 (C-4, 8), 125.24 (C), 123.46 (CH), 122.85 (CH), 121.59 (CH), 119.77 (CH), 111.20 (CH), 111.04 (C), 49.93 (C-2). HPLC: CH3CN
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) H2O = 80
H2O = 80![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 20; tR: 2.205 min, purity: 99.8%. TOF MS ES+ (m/z): [M + H]+, calcd: 430.1337, found: 430.1337, anal. calcd for C23H19N5O2S: C, 64.32; H, 4.46; N, 16.31; O, 7.45; S, 7.47; found: C, 64.38; H, 4.52; N, 16.37.
20; tR: 2.205 min, purity: 99.8%. TOF MS ES+ (m/z): [M + H]+, calcd: 430.1337, found: 430.1337, anal. calcd for C23H19N5O2S: C, 64.32; H, 4.46; N, 16.31; O, 7.45; S, 7.47; found: C, 64.38; H, 4.52; N, 16.37.
4.1.3.16 (E)-2-[(1-benzyl-1H-indol-3-yl)methylene]-N-methylhydrazinecarbothioamide (5p).
Color: white, yield: 72%, melting point: 220–221 °C. δH (600 MHz, DMSO-d6) 11.19 (1H, s, H-18), 8.32 (1H, S, H-9), 7.99 (1H, s, H-16), 7.94 (1H, q, J = 4.5 Hz, H-20), 7.51 (1H, d, J = 8.1 Hz, Ar-H), 7.33–7.31 (2H, m, Ar-H), 7.27–7.26 (3H, m, Ar-H), 7.25–7.21 (1H, m, Ar-H), 7.18 (1H, t, J = 7.5 Hz, Ar-H), 5.45 (2H, s, H-2), 3.09 (3H, d, J = 4.5 Hz, H-21); 13C NMR (151 MHz, DMSO) δ 177.27 (C-19), 140.35 (C-16), 137.94 (C), 137.35 (C), 134.31 (CH), 129.11 (C-5, 7), 128.05 (CH), 127.68 (C-4, 8), 125.08 (C), 123.33 (CH), 123.04 (CH), 121.38 (CH), 111.35 (CH), 111.04 (C), 49.84 (C-2), 31.52 (C-21). HPLC: CH3CN
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) H2O = 80
H2O = 80![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 20; tR: 1.592 min, purity: 100.0%. TOF MS ES+ (m/z): [M + H]+, calcd: 323.1330, found: 323.1330, anal. calcd for C18H18N4S: C, 67.05; H, 5.63; N, 17.38; S, 9.94; found: C, 67.12; H, 5.69; N, 17.45.
20; tR: 1.592 min, purity: 100.0%. TOF MS ES+ (m/z): [M + H]+, calcd: 323.1330, found: 323.1330, anal. calcd for C18H18N4S: C, 67.05; H, 5.63; N, 17.38; S, 9.94; found: C, 67.12; H, 5.69; N, 17.45.
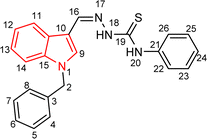
4.1.3.17 (E)-2-[(1-benzyl-1H-indol-3-yl)methylene]-N-(4-nitrophenyl)hydrazinecarbothioamide (5q).
Color: dark yellow, yield: 67%, melting point: 223–225 °C. δH (600 MHz, DMSO-d6) 11.99 (1H, s, H-18), 10.10 (1H, s, H-20), 8.47 (1H, s, H-9), 8.29–8.21 (3H, m), 8.14 (1H, s, H-16), 8.11 (2H, d, J = 9.0 Hz), 7.54 (1H, d, J = 8.1 Hz), 7.33 (2H, t, J = 7.5 Hz), 7.29–7.26 (3H, m), 7.26–7.22 (1H, m), 7.22–7.18 (1H, m), 5.49 (2H, s, H-2); 13C NMR (151 MHz, DMSO) δ 174.19 (C-19), 146.25 (C-16), 143.49 (C), 142.27 (CH), 137.84 (C), 137.43 (C), 135.23 (CH), 129.13 (C-5, 7), 128.10 (CH), 127.65 (C-4, 8), 125.23 (C), 124.39 (C), 123.86 (CH), 123.49 (CH), 122.68 (CH), 121.67 (CH), 111.28 (CH), 110.96 (C), 49.94 (C-2). HPLC: CH3CN
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) H2O = 80
H2O = 80![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 20; tR: 2.363 min, purity: 88.5%. TOF MS ES+ (m/z): [M + H]+, calcd: 430.1337, found: 430.1338, anal. calcd for C23H19N5O2S: C, 64.32; H, 4.46; N, 16.31; found: C, 64.39; H, 4.53; N, 16.36.
20; tR: 2.363 min, purity: 88.5%. TOF MS ES+ (m/z): [M + H]+, calcd: 430.1337, found: 430.1338, anal. calcd for C23H19N5O2S: C, 64.32; H, 4.46; N, 16.31; found: C, 64.39; H, 4.53; N, 16.36.
4.1.3.18 (E)-2-[(1-benzyl-1H-indol-3-yl)methylene]-N-(naphthalen-1-yl)hydrazinecarbothioamide (5r).
Color: white, yield: 66%, melting point: 199–201 °C. δH (600 MHz, DMSO-d6) 11.69 (1H, s, H-18), 9.88 (1H, s, H-20), 8.49 (1H, s, H-9), 8.39 (1H, d, J = 8.0 Hz), 8.12 (1H, s, H-16), 8.02–7.97 (1H, m), 7.96–7.93 (1H, m), 7.90 (1H, d, J = 8.2 Hz), 7.66 (1H, dt, J = 7.3, 1.0 Hz), 7.60–7.49 (4H, m), 7.35–7.32 (2H, m), 7.28–7.26 (3H, m), 7.21 (1H, ddd, J = 8.2, 7.0, 1.2 Hz), 7.11 (1H, ddd, J = 8.0, 7.0, 1.0 Hz), 5.48 (2H, s, H-2); 13C NMR (151 MHz, DMSO) δ 176.81 (C-19), 141.24 (C-16), 137.95 (CH), 137.40 (C), 136.23 (C), 134.86 (CH), 134.20 (C), 130.94 (CH), 129.12 (C-5, 7), 128.56 (CH), 128.07 (CH), 127.65 (C-4, 8), 127.07 (CH), 126.58 (CH), 126.52 (CH), 126.47 (CH), 125.91 (C), 125.19 (C), 123.49 (C), 123.37 (CH), 123.04 (CH), 121.42 (CH), 111.26 (CH), 111.11 (C), 49.89 (C-2). HPLC: CH3CN
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) H2O = 80
H2O = 80![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 20; tR: 2.554 min, purity: 100.0%. TOF MS ES+ (m/z): [M + H]+, calcd: 435.1643, found: 435.1638, anal. calcd for C27H22N4S: C, 74.63; H, 5.10; N, 12.89; found: C, 74.69; H, 5.18; N, 12.95.
20; tR: 2.554 min, purity: 100.0%. TOF MS ES+ (m/z): [M + H]+, calcd: 435.1643, found: 435.1638, anal. calcd for C27H22N4S: C, 74.63; H, 5.10; N, 12.89; found: C, 74.69; H, 5.18; N, 12.95.
4.2. Tyrosinase inhibition assay
Using a spectrophotometer microplate reader, the 96-well plate tyrosinase inhibition experiment was performed by Masamoto et al.'s protocol.65,66 First, 10 μL of the test compounds and the positive control, “kojic acid” were added to the 96-well microplate. Then, add 60 μL of phosphate buffer and 10 μL of mushroom tyrosinase (30 U mL−1 in phosphate buffer) to the reaction mixture incubated for 10 min at 25 °C. Following incubation, 20 μL of substrate L-DOPA (0.5 mM) in phosphate buffer was added. The end product dopachrome formation was measured at 480 nm for continues 10 min with one min interval time using 96-well ELISA plate reader (Bio-Rad xMARK microplate spectrophotometer). All of the experiments were carried out in triplicate to account for potential mistakes, and the mean findings were reported in SEM. All of the in vitro experiment results, including % inhibition and IC50 values, were examined using EZ-Fit, an enzyme kinetics tool that fits curves (Perrella Scientific Inc., Amherst, USA).The in vitro mechanistic analysis was carried out under comparable experimental settings to get insight into the mechanism of action of the inhibitors that were found. Four distinct doses of the substrate L-Dopa 0.1, 0.2, 0.4, and 0.8 mM were used in the kinetic analysis.
The % inhibition was computed using the formula provided below.
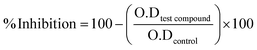 | (1) |
EZ-FIT (Perrella Scientific, Inc., USA) was used to calculate each tested compound's IC50 value. All the experiments were performed in triplicate to reduce predicted errors, and differences in the outcomes are expressed as Standard Error of Mean values (SEM).
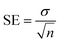 | (2) |
4.3. Kinetic analysis
The in vitro mechanistic study was carried out by following a similar inhibitory procedure with a similar experimental temperature and time period. The addition of four different concentrations of the substrate L-Dopa including 0.1, 0.2, 0.4, and 0.8 mM was used in the kinetic analysis.4.4. Molecular docking
To explore the in silico tyrosinase inhibition activity, the X-ray crystal structure of tyrosinase inhibitor (PDB ID: 2Y9W) which is the crystal structure of PPO3, a tyrosinase from Agaricus bisporus, in deoxy-form that contains additional unknown lectin-like subunit with a resolution of 2.30 Å, was retrieved from RCSB protein data bank67 in PDB format. The protein was processed for molecular docking with the aid of a protein preparation wizard from the Schrodinger suite (http://www.schrodinger.com). The processing includes the stepwise rectification of missing side chains or loops, and then allocation of bond orders. The protonation states for each of the hetero-atoms were detected based on the number of hydrogen bonds, as well as the score Epik penalty. Consequently, the protein structure was refined. In addition, the partial atomic charges were computed precisely by using the OPLS4 force field. For a cluster of hydrogen-bonded species, an interactive optimization was executed, and finally, the optimized structure was saved as.mae (Maestro) file format within the specified already chosen working directory.68 The two-dimensional (2D) structures of the investigated compounds (ligands) were sketched by ChemDraw 20.1.1, followed by energy minimization via Chem3D 20.1.1, and then saved in.sdf file format. Furthermore, the LigPrep module of Schrödinger Release 2016-4 was incorporated to generate, and optimize their three-dimensional (3D) structures, employing the OPLS4 force field.69 The receptor grid generation tool within the Schrödinger suite was incorporated to make a grid box, the dimensions of which were accustomed to cover the co-crystallized ligand within the binding pocket of the chosen target protein. The docking process was conducted by the Glide module in standard precision (SP) mode.70 The glide and Emodel scores were tabulated for each ligand's optimal conformational pose and were compared with the scores of the corresponding co-crystallized ligand and the standard drug.4.5. Validation of docking protocol
In our experiment, each ligand from Table 2 was docked via the Maestro glide software with the co-crystallized ligand of the target protein (reference). The hit compound 5k (according to the scoring function energies) was chosen for further structural analysis by calculating the root mean square deviation (RMSD) of each pose against its conformation in the crystal structure.71 Here, three different RMSD classifications for docking solutions: (i) good solutions when RMSD ≤ 2.0 Å, (ii) acceptable solutions when RMSD is between 2.0 and 3.0 Å, and (iii) bad solutions when RMSD ≥ 3.0 Å.4.6. ADMET analysis
ADMET study is designed to predict ADMET properties (absorption, distribution, metabolism, excretion, and toxicity) of investigated compounds via the SwissADME web server.72 The compounds were evaluated by ADMET analysis based on established rules established by Lipinski et al.,73 Muegge et al.,74 Ghose et al.,75 Egan et al.,76 and Veber et al.77 In addition, a range of a variety of other characteristics was also analyzed, such as the number of hydrogen bond donors, hydrogen bond acceptors, rotatable bonds, total polar surface area, and synthetic accessibility to determine the drug likeliness of synthesized compounds.Data availability
The data supporting this article have been included as part of the ESI.†Conflicts of interest
The authors have declared no conflict of interest.Acknowledgements
Authors are thankful to the Researchers Supporting Project number (RSPD2024R1035), King Saud University, Riyadh, Saudi Arabia.References
- N. H. Nasab, H. Raza, Y. S. Eom, M. Hassan, A. Kloczkowski and S. J. Kim, Synthesis and Discovery of Potential Tyrosinase Inhibitor of New Coumarin-Based Thiophenyl-Pyrazolylthiazole Nuclei: In Vitro Evaluation, Cytotoxicity, Kinetic and Computational Studies, Chemical Biology & Drug Design 2023 Search PubMed.
- Z. Najafi, F. Rafiei, A. Ghafouri-Khosrowshahi, M. Mahdavi, M. Dianatpour and A. Iraji, Design, Synthesis, and Molecular Dynamics Simulation Studies of New Chalcone-Based 2-Arylidene-1, 3-indandiones as Tyrosinase Inhibitors, Chemistryselect, 2023, 8(33), e202302192 CrossRef CAS.
- R. Romagnoli, P. Oliva, F. Prencipe, S. Manfredini, M. P. Germanò, L. De Luca, F. Ricci, D. Corallo, S. Aveic and E. Mariotto, Cinnamic acid derivatives linked to arylpiperazines as novel potent inhibitors of tyrosinase activity and melanin synthesis, Eur. J. Med. Chem., 2022, 231, 114147 CrossRef CAS PubMed.
- H. Moreiras, F. J. Pereira, M. V. Neto, L. Bento-Lopes, T. C. Festas, M. C. Seabra and D. C. Barral, The exocyst is required for melanin exocytosis from melanocytes and transfer to keratinocytes, Pigm. Cell Melanoma Res., 2020, 33(2), 366–371 CrossRef CAS PubMed.
- J. Su, D. Li, Y. Hu, X. You, X. Guo, X. Li and B. Chen, A novel C6-sulfonated celastrol analog as a tyrosinase and melanin inhibitor: Design, synthesis, biological evaluation and molecular simulation, J. Mol. Struct., 2023, 1283, 135288 CrossRef CAS.
- G. Wang, M. He, Y. Huang and Z. Peng, Synthesis and biological evaluation of new kojic acid-1, 3, 4-oxadiazole hybrids as tyrosinase inhibitors and their application in the anti-browning of fresh-cut mushrooms, Food Chem., 2023, 409, 135275 CrossRef CAS PubMed.
- M. He, M. Fan, W. Yang, Z. Peng and G. Wang, Novel kojic acid-1, 2, 4-triazine hybrids as anti-tyrosinase agents: Synthesis, biological evaluation, mode of action, and anti-browning studies, Food Chem., 2023, 419, 136047 CrossRef CAS PubMed.
- M. Hassan, S. Shahzadi and A. Kloczkowski, Tyrosinase inhibitors naturally present in plants and synthetic modifications of these natural products as anti-melanogenic agents: a review, Molecules, 2023, 28(1), 378 CrossRef CAS PubMed.
- J. Li, L. Feng, L. Liu, F. Wang, L. Ouyang, L. Zhang, X. Hu and G. Wang, Recent advances in the design and discovery of synthetic tyrosinase inhibitors, Eur. J. Med. Chem., 2021, 224, 113744 CrossRef CAS PubMed.
- S. Chortani, A. Hajlaoui, S. Jlizi, A. H. Harrath, H. B. Jannet and A. Romdhane, Access to new phosphonate-and imidazolidine-benzopyrimidinone derivatives as antityrosinase and anti-acetylcholinesterase agents: Design, synthesis and molecular docking, J. Mol. Struct., 2022, 1268, 133693 CrossRef CAS.
- L. Ielo, B. Deri, M. P. Germano, S. Vittorio, S. Mirabile, R. Gitto, A. Rapisarda, S. Ronsisvalle, S. Floris and Y. Pazy, Exploiting the 1-(4-fluorobenzyl) piperazine fragment for the development of novel tyrosinase inhibitors as anti-melanogenic agents: Design, synthesis, structural insights and biological profile, Eur. J. Med. Chem., 2019, 178, 380–389 CrossRef CAS PubMed.
- S. Xue, Z. Li, X. Ze, X. Wu, C. He, W. Shuai, M. Marlow, J. Chen, D. Scurr and Z. Zhu, Design, Synthesis, and Biological Evaluation of Novel Hybrids Containing Dihydrochalcone as Tyrosinase Inhibitors to Treat Skin Hyperpigmentation, J. Med. Chem., 2023, 66(7), 5099–5117 CrossRef CAS PubMed.
- M. R. de Barros, T. M. Menezes, Y. S. Garcia and J. L. Neves, Inhibitory effects of iron-based carbonaceous nanocomposites on mushroom tyrosinase activity: molecular aspects and mechanistic insights, New J. Chem., 2023, 47(19), 9134–9142 RSC.
- Z. Najafi, A. Ebadi, G. Chehardoli, M. Ziaei, T. Akbarzadeh, M. Saeedi, P. Gholamhoseini and M. Mahdavi, Design, synthesis, in vitro, and in silico studies of novel benzylidene 6-methoxy-1-tetralone linked to benzyloxy and benzyl-1, 2, 3-triazole rings as potential tyrosinase inhibitors, J. Mol. Struct., 2023, 1271, 134018 CrossRef CAS.
- H. Dong, X. Wang, G. Li, L. Zhao and C. Zhao, Synthesis, characterization, antioxidant and tyrosinase inhibitory activities of hesperetin derivatives, Synth. Commun., 2023, 53(19), 1579–1587 CrossRef CAS.
- B. Roulier, I. Rush, L. M. Lazinski, B. Pérès, H. Olleik, G. Royal, A. Fishman, M. Maresca and R. Haudecoeur, Resorcinol-based hemiindigoid derivatives as human tyrosinase inhibitors and melanogenesis suppressors in human melanoma cells, Eur. J. Med. Chem., 2023, 246, 114972 CrossRef CAS PubMed.
- A. Korkmaz and E. Bursal, Benzothiazole sulfonate derivatives bearing azomethine: Synthesis, characterization, enzyme inhibition, and molecular docking study, J. Mol. Struct., 2022, 1257, 132641 CrossRef CAS.
- X. Zhou, A. Iqbal, J. Li, C. Liu, A. Murtaza, X. Xu, S. Pan and W. Hu, Changes in browning degree and reducibility of polyphenols during autoxidation and enzymatic oxidation, Antioxidants, 2021, 10(11), 1809 CrossRef CAS PubMed.
- Y. T. Wen, Y. Q. Liang, W. M. Chai, Q. M. Wei, Z. Y. Yu and L. J. Wang, Effect of ascorbic acid on tyrosinase and its anti-browning activity in fresh-cut Fuji apple, J. Food Biochem., 2021, 45(12), e13995 CrossRef CAS PubMed.
- I. Cabezudo, I. A. Ramallo, V. L. Alonso and R. L. Furlan, Effect directed synthesis of a new tyrosinase inhibitor with anti-browning activity, Food Chem., 2021, 341, 128232 CrossRef CAS PubMed.
- D. Yoon, H. J. Jung, J. Lee, H. J. Kim, H. S. Park, Y. J. Park, M. K. Kang, G. Y. Kim, D. Kang and Y. Park, In vitro and in vivo anti-pigmentation effects of 2-mercaptobenzimidazoles as nanomolar tyrosinase inhibitors on mammalian cells and zebrafish embryos: Preparation of pigment-free zebrafish embryos, Eur. J. Med. Chem., 2024, 116136 CrossRef CAS PubMed.
- S. Vittorio, C. Dank and L. Ielo, Heterocyclic Compounds as Synthetic Tyrosinase Inhibitors: Recent Advances, Int. J. Mol. Sci., 2023, 24(10), 9097 CrossRef CAS PubMed.
- A. Korkmaz and E. Bursal, Synthesis, biological activity and molecular docking studies of novel sulfonate derivatives bearing salicylaldehyde, Chem. Biodiversity, 2022, 19(6), e202200140 CrossRef CAS PubMed.
- A. Korkmaz and E. Bursal, An in vitro and in silico study on the synthesis and characterization of novel bis (sulfonate) derivatives as tyrosinase and pancreatic lipase inhibitors, J. Mol. Struct., 2022, 1259, 132734 CrossRef CAS.
- A. Korkmaz, G. Kurtay, E. Kaya and E. Bursal, Design, synthesis, spectroscopic characterizations, in vitro pancreatic lipase as well as tyrosinase inhibition evaluations and in silico analysis of novel aryl sulfonate-naphthalene hybrids, J. Biomol. Struct. Dyn., 2023, 41(15), 7128–7143 CrossRef CAS PubMed.
- A. Korkmaz, E. Bursal and G. Kurtay, In Vitro and In Silico Evaluation of Amylase, Tyrosinase, and Pancreatic Lipase Inhibitions of Novel Benzothiazole-Sulfonate Derivatives, ChemistrySelect, 2023, 8(42), e202302936 CrossRef CAS.
- L. Lu, C. Hu, X. Min, Z. Liu, X. Xu and L. Gan, In Vitro and In Vivo Biological Evaluation of Indole-thiazolidine-2, 4-dione Derivatives as Tyrosinase Inhibitors, Molecules, 2023, 28(22), 7470 CrossRef CAS PubMed.
- T. Ma, X.-P. Wei, X.-C. Wang, X.-X. Qiao, G. Li, Y. He and X.-J. Zhao, Highly Enantioselective Synthesis of 3 a-Fluorofuro [3, 2-b] indolines via Organocatalytic Aza-Friedel–Crafts Reaction/Selective C–F Bond Activation, Org. Lett., 2023, 25(48), 8666–8671 CrossRef CAS PubMed.
- Z. Kudličková, R. Michalková, A. Salayová, M. Ksiažek, M. Vilková, S. Bekešová and J. Mojžiš, Design, Synthesis, and Evaluation of Novel Indole Hybrid Chalcones and Their Antiproliferative and Antioxidant Activity, Molecules, 2023, 28(18), 6583 CrossRef PubMed.
- A. Citarella, D. Moi, M. Pedrini, H. Pérez-Peña, S. Pieraccini, A. Dimasi, C. Stagno, N. Micale, T. Schirmeister and G. Sibille, Synthesis of SARS-CoV-2 M pro inhibitors bearing a cinnamic ester warhead with in vitro activity against human coronaviruses, Org. Biomol. Chem., 2023, 21(18), 3811–3824 RSC.
- S. Garrepalli, R. Gudipati, R. Kapavarapu, K. Ravindhranath and M. Pal, Synthesis and characterization of two known and one new impurities of dolutegravir: In silico evaluation of certain intermediates against SARS CoV-2 O-ribose methyltransferase (OMTase), J. Mol. Struct., 2023, 1271, 133992 CrossRef CAS PubMed.
- J. Badiger, K. Manjulatha, M. Girish, A. Sharif and M. Purohit, Synthesis and biological evaluation of some N-substituted indoles, ARKIVOC, 2009, 2009(xii), 217–231 Search PubMed.
- S. Ölgen, Z. Kiliç, A. O. Ada and T. Çoban, Synthesis and evaluation of novel N–H and N-substituted indole-2-and 3-carboxamide derivatives as antioxidants agents, J. Enzyme Inhib. Med. Chem., 2007, 22(4), 457–462 Search PubMed.
- V. Mashayekhi, K. Haj Mohammad Ebrahim Tehrani, P. Azerang, S. Sardari and F. Kobarfard, Synthesis, antimycobacterial and anticancer activity of novel indole-based thiosemicarbazones, Arch. Pharmacal Res., 2021, 44, 1–13 Search PubMed.
- M. Arshad, Synthesis, characterization, antimicrobial and computational studies of some sulfonamide derivatives possessing thiadiazole and indole nucleus, Eur. J. Pharm. Med. Res., 2017, 4(12), 511–517 Search PubMed.
- S. Umamatheswari, B. Balaji, M. Ramanathan and S. Kabilan, Synthesis, stereochemistry, antimicrobial evaluation and QSAR studies of 2, 6-diaryltetrahydropyran-4-one thiosemicarbazones, Eur. J. Med. Chem., 2011, 46(4), 1415–1424 CrossRef CAS PubMed.
- N. Balakrishnan, J. Haribabu, R. E. Malekshah, S. Swaminathan, C. Balachandran, N. Bhuvanesh, S. Aoki and R. Karvembu, Effect of N-benzyl group in indole scaffold of thiosemicarbazones on the biological activity of their Pd (II) complexes: DFT, biomolecular interactions, in silico docking, ADME and cytotoxicity studies, Inorg. Chim. Acta, 2022, 534, 120805 CrossRef CAS.
- M. Yusuf and P. Jain, Synthesis and biological significances of 1, 3, 4-thiadiazolines and related heterocyclic compounds, Arabian J. Chem., 2014, 7(5), 525–552 CrossRef CAS.
- P. Chellan, S. Nasser, L. Vivas, K. Chibale and G. S. Smith, Cyclopalladated complexes containing tridentate thiosemicarbazone ligands of biological significance: Synthesis, structure and antimalarial activity, J. Organomet. Chem., 2010, 695(19–20), 2225–2232 CrossRef CAS.
- M. Jarestan, K. Khalatbari, A. Pouraei, S. A. Sadat Shandiz, S. Beigi, M. Hedayati, A. Majlesi, F. Akbari and A. Salehzadeh, Preparation, characterization, and anticancer efficacy of novel cobalt oxide nanoparticles conjugated with thiosemicarbazide, 3 Biotech, 2020, 10, 1–9 CrossRef PubMed.
- A. You, J. Zhou, S. Song, G. Zhu, H. Song and W. Yi, Structure-based modification of 3-/4-aminoacetophenones giving a profound change of activity on tyrosinase: from potent activators to highly efficient inhibitors, Eur. J. Med. Chem., 2015, 93, 255–262, DOI:10.1016/j.ejmech.2015.02.013.
- A. C. Sartorelli, E. C. Moore, M. S. Zedeck and K. C. Agrawal, Inhibition of ribonucleoside diphosphate reductase by 1-formylisoquinoline thiosemicarbazone and related compounds, Biochemistry, 1970, 9(23), 4492–4498 CrossRef CAS PubMed.
- Y.-J. Zhu, K.-K. Song, Z.-C. Li, Z.-Z. Pan, Y.-J. Guo, J.-J. Zhou, Q. Wang, B. Liu and Q.-X. Chen, Antityrosinase and antimicrobial activities of trans-cinnamaldehyde thiosemicarbazone, J. Agric. Food Chem., 2009, 57(12), 5518–5523 CrossRef CAS PubMed.
- Z.-C. Li, L.-H. Chen, X.-J. Yu, Y.-H. Hu, K.-K. Song, X.-W. Zhou and Q.-X. Chen, Inhibition kinetics of chlorobenzaldehyde thiosemicarbazones on mushroom tyrosinase, J. Agric. Food Chem., 2010, 58(23), 12537–12540 CrossRef CAS PubMed.
- L.-H. Chen, Y.-H. Hu, W. Song, K.-K. Song, X. Liu, Y.-L. Jia, J.-X. Zhuang and Q.-X. Chen, Synthesis and antityrosinase mechanism of benzaldehyde thiosemicarbazones: novel tyrosinase inhibitors, J. Agric. Food Chem., 2012, 60(6), 1542–1547 CrossRef CAS PubMed.
- J. Liu, W. Yi, Y. Wan, L. Ma and H. Song, 1-(1-Arylethylidene) thiosemicarbazide derivatives: A new class of tyrosinase inhibitors, Bioorg. Med. Chem., 2008, 16(3), 1096–1102 CrossRef CAS PubMed.
- W. Yi, R.-H. Cao, Z.-Y. Chen, L. Yu, L. Ma and H.-C. Song, Design, synthesis and biological evaluation of hydroxy-or methoxy-substituted phenylmethylenethiosemicarbazones as tyrosinase inhibitors, Chem. Pharm. Bull., 2009, 57(11), 1273–1277 CrossRef CAS PubMed.
- W. Yi, R. Cao, Z. Chen, L. Yu, H. Wen, Q. Yan, L. Ma and H. Song, Rational design and synthesis of 4-o-substituted phenylmethylenethiosemicarbazones as novel tyrosinase inhibitors, Chem. Pharm. Bull., 2010, 58(5), 752–754 CrossRef CAS PubMed.
- P. Thanigaimalai, K.-C. Lee, V. K. Sharma, E. Roh, Y. Kim and S.-H. Jung, Ketonethiosemicarbazones: Structure–activity relationships for their melanogenesis inhibition, Bioorg. Med. Chem. Lett., 2011, 21(12), 3527–3530 CrossRef CAS PubMed.
- K.-C. Lee, P. Thanigaimalai, V. K. Sharma, M.-S. Kim, E. Roh, B.-Y. Hwang, Y. Kim and S.-H. Jung, Structural characteristics of thiosemicarbazones as inhibitors of melanogenesis, Bioorg. Med. Chem. Lett., 2010, 20(22), 6794–6796 CrossRef CAS PubMed.
- H. Dong, J. Liu, X. Liu, Y. Yu and S. Cao, Combining molecular docking and QSAR studies for modeling the anti-tyrosinase activity of aromatic heterocycle thiosemicarbazone analogues, J. Mol. Struct., 2018, 1151, 353–365 CrossRef CAS.
- S. Masuri, B. Era, F. Pintus, E. Cadoni, M. G. Cabiddu, A. Fais and T. Pivetta, Hydroxylated coumarin-based thiosemicarbazones as dual antityrosinase and antioxidant agents, Int. J. Mol. Sci., 2023, 24(2), 1678 CrossRef CAS PubMed.
- M. Noori, R. Sabourian, A. Tasharoie, M. Safavi, A. Iraji, M. Khalili Ghomi, N. Dastyafteh, C. Irajie, E. Zarenezhad and S. M. Mostafavi Pour, Thioquinoline derivatives conjugated to thiosemicarbazide as potent tyrosinase inhibitors with anti-melanogenesis properties, Sci. Rep., 2023, 13(1), 2578 CrossRef CAS PubMed.
- N. A. Hassanuddin, E. Normaya, H. Ismail, A. Iqbal, M. B. M. Piah, S. Abd Hamid and M. N. Ahmad, Methyl 4-pyridyl ketone thiosemicarbazone (4-PT) as an effective and safe inhibitor of mushroom tyrosinase and antibrowning agent, Int. J. Biol. Macromol., 2024, 255, 128229 CrossRef CAS PubMed.
- K. Tilekar, O. Shelke, N. Upadhyay, A. Lavecchia and C. Ramaa, Current status and future prospects of molecular hybrids with thiazolidinedione (TZD) scaffold in anticancer drug discovery, J. Mol. Struct., 2022, 1250, 131767 CrossRef CAS.
- J. Ko, J. Lee, H. J. Jung, S. Ullah, Y. Jeong, S. Hong, M. K. Kang, Y. J. Park, Y. Hwang and D. Kang, Design and synthesis of (Z)-5-(substituted benzylidene)-3-cyclohexyl-2-thioxothiazolidin-4-one analogues as anti-tyrosinase and antioxidant compounds: In vitro and in silico insights, Antioxidants, 2022, 11(10), 1918 CrossRef CAS PubMed.
- S. H. Kim, Y. M. Ha, K. M. Moon, Y. J. Choi, Y. J. Park, H. O. Jeong, K. W. Chung, H. J. Lee, P. Chun and H. R. Moon, Anti-melanogenic effect of (Z)-5-(2, 4-dihydroxybenzylidene) thiazolidine-2, 4-dione, a novel tyrosinase inhibitor, Arch. Pharmacal Res., 2013, 36, 1189–1197 CrossRef CAS PubMed.
- S. Zolghadri, A. Bahrami, M. T. Hassan Khan, J. Munoz-Munoz, F. Garcia-Molina, F. Garcia-Canovas and A. A. Saboury, A comprehensive review on tyrosinase inhibitors, J. Enzyme Inhib. Med. Chem., 2019, 34(1), 279–309 CrossRef CAS PubMed.
- M. Wajid, M. Uzair, G. Muhammad, F. Siddique, M. Bashir, S. Nadeem, A. Ashraf, N. Assad, A. Mushtaq, M. Z. Rafay and A. Aqdas, Sulfamethoxazole-derived Schiff bases: synthesis, characterization, biological activities, molecular docking, DFT, and ADME studies, J. Mol. Struct., 2024, 1312, 138640, DOI:10.1016/j.molstruc.2024.138640.
- F. Siddique, O. Daoui, M. Ayoub, S. Elkhattabi, S. Chtita, S. Afzal, A. Mohyuddin, I. Kaukab, S. A. Ejaz and A. M. Salamatullah, Identification and in silico screening of natural phloroglucinols as potential PI3Kα inhibitors: A computational approach for drug discovery, Open Chem., 2024, 22(1), 20240064 CrossRef.
- C. Viegas-Junior, A. Danuello, V. da Silva Bolzani, E. J. Barreiro and C. A. M. Fraga, Molecular hybridization: a useful tool in the design of new drug prototypes, Curr. Med. Chem., 2007, 14(17), 1829–1852 CrossRef CAS PubMed.
- P. Thanigaimalai, E. V. Rao, K.-C. Lee, V. K. Sharma, E. Roh, Y. Kim and S.-H. Jung, Structure–activity relationship of naphthaldehydethiosemicarbazones in melanogenesis inhibition, Bioorg. Med. Chem. Lett., 2012, 22(2), 886–889 CrossRef CAS PubMed.
- P. Liu, C. Shu, L. Liu, Q. Huang and Y. Peng, Design and synthesis of thiourea derivatives with sulfur-containing heterocyclic scaffolds as potential tyrosinase inhibitors, Bioorg. Med. Chem., 2016, 24(8), 1866–1871 CrossRef CAS PubMed.
- R. El-Mernissi, N. El Menyiy, R. Moubachir, A. Zouhri, Y. El-Mernissi, F. Siddique, S. Nadeem, H. Ibork, A. El Barnossi and G. F. Wondmie, Cannabis sativa L. essential oil: Chemical composition, anti-oxidant, anti-microbial properties, and acute toxicity: In vitro, in vivo, and in silico study, Open Chem., 2024, 22(1), 20230214 CrossRef CAS.
- Y. Masamoto, H. Ando, Y. Murata, Y. Shimoishi, M. Tada and K. Takahata, Mushroom tyrosinase inhibitory activity of esculetin isolated from seeds of Euphorbia lathyris L., Biosci., Biotechnol., Biochem., 2003, 67(3), 631–634, DOI:10.1271/bbb.67.631.
- A. R. Pasha, M. Khan, A. Khan, J. Hussain, M. Al-Rashida, T. Islam, Z. Batool, H. Kashtoh, M. H. Abdellattif, A. Al-Harrasi and Z. Shafiq, Synthesis, in vitro, and in silico study of novel pyridine based 1, 3-diphenylurea derivatives as tyrosinase inhibitors, Bioorg. Chem., 2024, 107724, DOI:10.1016/j.bioorg.2024.107724.
- S. K. Burley, H. M. Berman, C. Bhikadiya, C. Bi, L. Chen, L. Di Costanzo, C. Christie, K. Dalenberg, J. M. Duarte and S. Dutta, RCSB Protein Data Bank: biological macromolecular structures enabling research and education in fundamental biology, biomedicine, biotechnology and energy, Nucleic Acids Res., 2019, 47(D1), D464–D474 CrossRef CAS PubMed.
- J. J. Sahayarayan, K. S. Rajan, R. Vidhyavathi, M. Nachiappan, D. Prabhu, S. Alfarraj, S. Arokiyaraj and A. N. Daniel, In-silico protein-ligand docking studies against the estrogen protein of breast cancer using pharmacophore based virtual screening approaches, Saudi J. Biol. Sci., 2021, 28(1), 400–407 CrossRef CAS PubMed.
- D. Vassetti, M. Pagliai and P. Procacci, Assessment of GAFF2 and OPLS-AA general force fields in combination with the water models TIP3P, SPCE, and OPC3 for the solvation free energy of druglike organic molecules, J. Chem. Theory Comput., 2019, 15(3), 1983–1995 CrossRef CAS PubMed.
- M. P. Repasky, R. B. Murphy, J. L. Banks, J. R. Greenwood, I. Tubert-Brohman, S. Bhat and R. A. Friesner, Docking performance of the glide program as evaluated on the Astex and DUD datasets: a complete set of glide SP results and selected results for a new scoring function integrating WaterMap and glide, J. Comput.-Aided Mol. Des., 2012, 26, 787–799 CrossRef CAS PubMed.
- H. Gohlke, M. Hendlich and G. Klebe, Knowledge-based scoring function to predict protein-ligand interactions, J. Mol. Biol., 2000, 295(2), 337–356, DOI:10.1006/jmbi.1999.3371.
- A. Daina, O. Michielin and V. Zoete, SwissADME: a free web tool to evaluate pharmacokinetics, drug-likeness and medicinal chemistry friendliness of small molecules, Sci. Rep., 2017, 7(1), 42717 CrossRef PubMed.
- C. A. Lipinski, F. Lombardo, B. W. Dominy and P. J. Feeney, Experimental and computational approaches to estimate solubility and permeability in drug discovery and development settings, Adv. Drug Delivery Rev., 2012, 64, 4–17 CrossRef.
- I. Muegge, S. L. Heald and D. Brittelli, Simple selection criteria for drug-like chemical matter, J. Med. Chem., 2001, 44(12), 1841–1846 CrossRef CAS PubMed.
- A. K. Ghose, V. N. Viswanadhan and J. J. Wendoloski, A knowledge-based approach in designing combinatorial or medicinal chemistry libraries for drug discovery. 1. A qualitative and quantitative characterization of known drug databases, J. Comb. Chem., 1999, 1(1), 55–68 CrossRef CAS PubMed.
- W. J. Egan, K. M. Merz and J. J. Baldwin, Prediction of drug absorption using multivariate statistics, J. Med. Chem., 2000, 43(21), 3867–3877 CrossRef CAS PubMed.
- D. F. Veber, S. R. Johnson, H.-Y. Cheng, B. R. Smith, K. W. Ward and K. D. Kopple, Molecular properties that influence the oral bioavailability of drug candidates, J. Med. Chem., 2002, 45(12), 2615–2623 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d4ra05015k |
| This journal is © The Royal Society of Chemistry 2024 |