Open Access Article
Open Access ArticleTracking trash to treasure: in situ monitoring of single microbial cell oil biosynthesis from waste cooking oil using Raman spectroscopy and imaging†
Jiro Karloa,
Victor Carrasco-Navarrob,
Arto Koistinenc and
Surya Pratap Singh *a
*a
aDepartment of Biosciences and Bioengineering, Indian Institute of Technology Dharwad, Dharwad 580011, Karnataka, India. E-mail: ssingh@iitdh.ac.in
bDepartment of Environmental and Biological Sciences, University of Eastern Finland, Kuopio Campus, Yliopistonranta 8, Kuopio 70210, Finland
cDepartment of Technical Physics, University of Eastern Finland, Kuopio 70210, Finland
First published on 21st October 2024
Abstract
Waste cooking oil is a major pollutant that contaminates terrestrial and aquatic bodies which is generated from household kitchens and eateries. The bioremediation of waste cooking oil (WCO) into microbial oil, also known as single microbial cell oil (SMCO), can be accomplished by oleaginous microbes. Conventional methods excel in SMCO analysis but lack efficacy for in situ or lysis-free monitoring of nascent SMCO synthesis and turnover. To bridge this knowledge gap, this study shows the applicability of Raman reverse stable isotope probing (RrSIP) in monitoring time-dependent nascent SMCO synthesis and assimilation in Yarrowia lipolytica, an oleaginous yeast grown in hydrophilic (glucose) as well as hydrophobic carbon sources (cooking oil and waste cooking oil). This study also combines the RrSIP approach with Raman imaging for temporal visualization of the distribution and turnover dynamics of the SMCO pool in a single cell. Our finding provides a unique perspective utilizing optical spectroscopy methods for lysis-free SMCO analysis and holds potential for prospective utility as an adjunct tool in bioprocess and biofuel industries.
1 Introduction
Waste cooking oil (WCO) is generated all over the world in domestic kitchens, food processing industries, restaurants, and others. It is considered one of the major environmental pollutants.1 Unprocessed WCO forms oily sludge and contaminates both aquatic as well as terrestrial habitats.2,3 Each year global WCO production is estimated to be around 15 million tons as per reported studies.4,5 Due to its relatively simple chemical composition, primarily made up of triglyceride esters of glycerol with long fatty acid chains, mainly oleic acid (C18![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) and linoleic acid (C18
1) and linoleic acid (C18![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2), WCO is considered a valuable feedstock from an industrial perspective.6 Multiple studies have reported its utility in producing value-added products such as biodiesel, bioplastics, biogas, biosurfactants, biopolymers, lubricants, and others.4,6–10 The bioremediation of WCO using oleaginous microorganisms is routinely performed, as it has a fast growth rate, the inherent ability of lipid storage, requires less labour and is highly scalable.11 This study utilizes Yarrowia lipolytica (YL), an oleaginous yeast categorised as safe by the Food and Drug Administration (FDA), USA which can synthesize important metabolites such as microbial oil containing high levels of unsaturated fatty acid (UFA), lipases, proteases and others.12–14 Due to its capability of synthesizing extracellular lipases, it stands out among yeasts to metabolize hydrophobic substrates such as WCO with the potential to accumulate lipids more than 30–45% of their dry weight.11 The accumulation of the lipids in oleaginous microbes is mostly in the form of triacylglycerides (TAGs) with YL strains reported to contain 88–89% of its total lipid content.15,16 In yeast, these neutral lipids are stored in lipid bodies (LBs) which also maintain a significant portion of free fatty acids and sterol esters.17,18
2), WCO is considered a valuable feedstock from an industrial perspective.6 Multiple studies have reported its utility in producing value-added products such as biodiesel, bioplastics, biogas, biosurfactants, biopolymers, lubricants, and others.4,6–10 The bioremediation of WCO using oleaginous microorganisms is routinely performed, as it has a fast growth rate, the inherent ability of lipid storage, requires less labour and is highly scalable.11 This study utilizes Yarrowia lipolytica (YL), an oleaginous yeast categorised as safe by the Food and Drug Administration (FDA), USA which can synthesize important metabolites such as microbial oil containing high levels of unsaturated fatty acid (UFA), lipases, proteases and others.12–14 Due to its capability of synthesizing extracellular lipases, it stands out among yeasts to metabolize hydrophobic substrates such as WCO with the potential to accumulate lipids more than 30–45% of their dry weight.11 The accumulation of the lipids in oleaginous microbes is mostly in the form of triacylglycerides (TAGs) with YL strains reported to contain 88–89% of its total lipid content.15,16 In yeast, these neutral lipids are stored in lipid bodies (LBs) which also maintain a significant portion of free fatty acids and sterol esters.17,18
In the previously reported studies, SMCO analysis has been performed using highly efficient methods such as gas chromatography-mass spectrometry (GC-MS) and gas chromatography flame ionization detection (GC-FID).16,19–21 These methods require the extraction of the lipids from cells. Fluorescence spectroscopy is another technique used for SMCO analysis.22–25 Intracellular SMCO estimation and localization can be performed using fluorescence microscopy and spectrofluorimetry.16,26 Lipid bodies of YL have been analysed in many studies using fluorescence imaging.16,23,25,27 SIMS (secondary ionisation mass spectrometry) is a highly sensitive technique used for analysing stable isotope-labelled metabolites but it is known to be inherently destructive.28,29 While the existing tools for SMCO analysis are efficient and sensitive, these methods cannot be applied for in situ monitoring of microbial oil synthesis and accumulation at the single-cell level due to the essential requirement of extraction from cells. To maximize the utilization of oleaginous microbes for bioremediation of WCO as efficient biomass, understanding the carbon flow, synthesis, and accumulation of SMCO at the single cell level a novel lysis-free approach is much needed. This study proposes the prospective utility of Raman spectroscopy and imaging in combination with the reverse stable isotope probe (RrSIP) method for the lysis-free monitoring of SMCO at the single-cell level.
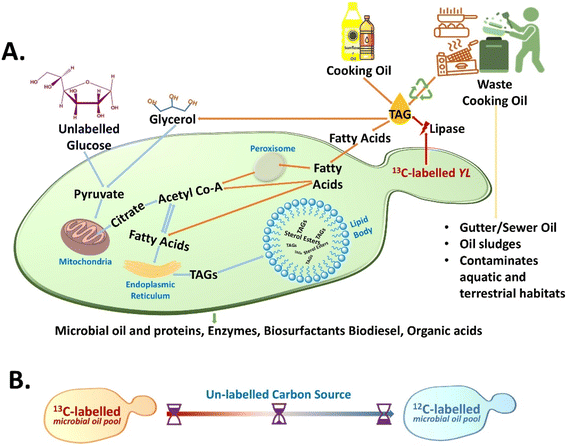
The unlabelled carbon sources are naturally higher in abundance and WCO generated is an unlabelled carbon source contributing to the effectiveness of the RrSIP methodology.30–34 In this approach, the 13C stable isotope-labelled cells are grown in the presence of 12C – unlabelled culture medium and with its metabolic incorporation into the newly synthesized metabolites leading to a blue shift in spectral bands is observed due to the isotopic effect.30–32 Monitoring this shifted band position with time serves as the spectral indicator for dynamic monitoring of nascent SMCO synthesis from de novo and ex novo pathways using hydrophilic and hydrophobic sources. The schematic diagram for the study is shown in Fig. 1(A) and (B). In previous studies, the SMCO composition of YL has been reported to be predominated by long-chain UFA making it a potential candidate for industrial use.14,35,36 Therefore, the UFA Raman band has been used as the representative spectral tracer for in situ SMCO analysis. The study validates the nascent UFA band as a lysis-free tracer for SMCO analysis and evaluated its applicability for monitoring the bioremediation or conversion of WCO.
2 Materials and methods
2.1 Yeast culture and growth conditions
Oleaginous yeast strain Yarrowia lipolytica NCIM 3472 was purchased from the National Collection of Industrial Microorganisms, Pune, India. YL was pre-cultured in a carbon-deficient synthetic broth medium (Sigma Aldrich) and supplied with 10 g L−1 glucose (Sigma Aldrich). Incubation parameters for the growth were set with the temperature at 28 °C and the rotational shaker set at 180 rpm. The growth evaluation of YL in a synthetic medium supplemented with unlabelled and uniformly labelled 13C glucose (99%, Cambridge Isotope Laboratories) was performed using Optical density at 600 nm (OD600) using UV-1900i UV-Vis Spectrophotometer (Shimadzu) at multiple incubation time intervals.2.2 Reverse stable isotope probing of cells
For reverse stable isotope probing, unlabelled cells were first grown in a synthetic broth medium containing uniformly labelled 13C glucose as the single carbon source. From the overnight culture, 13C labelled cells were washed and reinoculated in the same growth medium supplemented with the unlabelled carbon source and incubated in the optimized growth conditions mentioned previously. Unlabelled glucose, cooking oil and waste cooking oil were used as the carbon source. The cooking oil namely palmolein and sunflower oil was purchased from a local vendor and waste cooking oil was provided by a local eatery from IIT Dharwad. Oil samples (100 g L−1) were sterile syringe filtered before supplementing to the synthetic media as the sole carbon source before inoculation.2.3 Inhibition assay
For performing an inhibition assay, Cerulenin (Sigma Aldrich) a well-known fatty acid synthase inhibitor (50 μg ml−1) was used in the medium before inoculation. To monitor reverse the effect of the inhibition assay, exogenous mono- and poly-UFA supplements (oleic and linoleic acid) were provided in the 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 ratio. The standard fatty acids were obtained from Sisco Research Laboratories.
1 ratio. The standard fatty acids were obtained from Sisco Research Laboratories.
2.4 Lipid extraction
Yeast lipid extraction protocols were adapted from the previous report.37 The yeast cells were grown using the RrSIP methodology. After 24 h, aliquots from the broth culture were taken and centrifuged at 6000 rpm for 5 min at room temperature. The cell pellets were washed with PBS two times and resuspended in 1x-phosphate buffer saline (PBS, Sisco Research Laboratories). The 1 ml cell sample was first treated with chloroform/methanol (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2, 3.75 ml) and vortex for 15 min. The sample was incubated for 5 min on ice, followed by addition of chloroform (1.25 ml) and double distilled water (1.25 ml). The sample was vortexed for 5 minutes, centrifuged for 5 minutes at room temperature, and obtained a two-phase system. The organic bottom layer was taken by carefully removing the aqueous top and middle insoluble layer, which has to be performed very carefully, and it was dried using a speed vacuum (Eppendorf concentrator plus) and the lipid obtained was used for further Raman analysis.
2, 3.75 ml) and vortex for 15 min. The sample was incubated for 5 min on ice, followed by addition of chloroform (1.25 ml) and double distilled water (1.25 ml). The sample was vortexed for 5 minutes, centrifuged for 5 minutes at room temperature, and obtained a two-phase system. The organic bottom layer was taken by carefully removing the aqueous top and middle insoluble layer, which has to be performed very carefully, and it was dried using a speed vacuum (Eppendorf concentrator plus) and the lipid obtained was used for further Raman analysis.
2.5 Protein extraction
The yeast protein extraction protocol was adapted from the previous study.38 The yeast cells were grown using the RrSIP methodology. After 24 h, aliquots from the broth culture were taken and centrifuged at 6000 rpm for 5 minutes at room temperature. The supernatant was discarded, and the cells were washed with PBS (2x). The cell pellet obtained was resuspended in 200 μl lysis buffer (0.1 M NaOH, 0.05 M EDTA, 2% SDS and 2% β-mercaptoethanol) and kept in dry bath for 10 minutes at 90 °C. 4 M acetic acids (5 μl) was added and vortex for 0.5 minutes and again kept in dry bath for 10 minutes at 90 °C. Clear lysate was obtained using centrifugation. Methanol (4 vols), chloroform (1 vols) and water (3 vols) were added to lysate in the following order followed by vortex after each addition. Centrifugation was done for 5 minutes, and the aqueous phase was removed. Methanol (3 vols) was added followed by vortex and centrifuge. The pellet was dried using a speed vacuum (Eppendorf concentrator plus) and the pellet obtained was used for further Raman analysis.2.6 Sample preparation, single-cell Raman spectroscopy and data analysis
For the Single-cell RrSIP experiments, the aliquots were washed with 1x-PBS. To obtain individual single cells on the ethanol-washed calcium fluoride slide appropriate dilutions were performed at different time intervals. A total of 25 to 30 single cells were used for Raman spectral acquisition from two biological replicates (∼15 single cells × 2x) at different time points. WiTec confocal micro-Raman spectrometer, equipped with a 532 nm laser source and 100× objective lens was used for acquiring a single spectrum from each cell. Each spectrum acquired was averaged over 5 accumulations with a 20 seconds exposure time. The MATLAB software version R2021B was used for processing the spectral data. Denoising of the spectral data was done using the Savitzky Golay filter, baseline corrected with 5th order polynomial and unit normalized. The mean spectra along with standard deviation and all the other plots were generated using Origin Pro 2023b Software.2.7 Single-cell Raman imaging and analysis
The single cell was obtained and mounted on a calcium fluoride substrate as per the previously mentioned procedure.28 For the Raman imaging, the step size of the scan was ∼0.3 microns with a 5 seconds laser exposure time. Each single-cell Raman image took around 3 to 5 hours to acquire depending on the cell size and scan area. The Raman hyperspectral imaging spectral data was pre-processed via routine pre-processing events including filtering, baseline correction and unit normalization using MATLAB 2021B. Raman images based on SMCO tracer bands were generated using in-house codes and k means cluster analysis using MATLAB 2021B software-based GUI.39 Other plots related to hyperspectral data were generated using Origin Pro 2023b Software.3 Results and discussion
3.1 In situ monitoring de novo synthesis of single microbial cell oil and its turnover
UFA is an integral part of YL microbial cell oil composition. In a previous study, the SMCO content of YL 3472 was reported to be consisting of the highest amount of mono-UFA (for cells grown in glucose) and the highest poly-UFA in cells grown in waste cooking oil.16 Band position at 1655 cm−1 and 1605 cm−1 (cis C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C stretching vibration) in the Raman spectra of cells have been reported as unlabelled and 13C labelled UFAs. 1445 cm−1 has been used to analyse the total lipid pool which should be the ideal candidate for SMCO synthesis analysis, but this peak has not been reported to show any isotopic substitution shift to the best of our knowledge.28,30,40–44
C stretching vibration) in the Raman spectra of cells have been reported as unlabelled and 13C labelled UFAs. 1445 cm−1 has been used to analyse the total lipid pool which should be the ideal candidate for SMCO synthesis analysis, but this peak has not been reported to show any isotopic substitution shift to the best of our knowledge.28,30,40–44
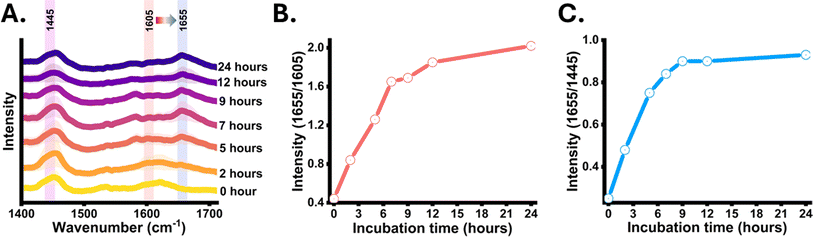
Firstly, the growth pattern of YL was evaluated grown in carbon-free synthetic media supplemented with unlabelled and 13C glucose through optical density measurements at different time points 0, 2, 5, 7, 9, 12, 24, 48 h as shown in Fig. S1(A).† The growth pattern of the cells was similar to the cells growing in carbon-source-rich media, confirming unaltered metabolic activity due to 13C substitution. Average Raman spectra from yeast cells along with standard deviation at different incubation time intervals (0, 2, 5, 7, 9, 12 and 24 h) are shown in Fig. 2(A). At 0 h, the inoculated cells are 13C-labelled, an intensified 13C-labelled UFA band signal at 1605 cm−1 was observed and a minor signal at 1655 cm−1 indicating that almost the entire SMCO UFA pool is 13C-labelled. At 2 h, increased band intensity at the 1655 cm−1 region indicated the 13C carbon flow from culture media to cell cytoplasm and active de novo UFA biosynthesis. At 5 h a similar intensity of 1605 cm−1 and 1655 cm−1 were observed and with the increase in the incubation time, the intensity at 1655 cm−1 was enhanced while that of 1605 cm−1 diminished. Further at 24 h, a highly intense signal at 1655 cm−1 was observed whereas at 1605 cm−1 diminished, but the residual intensity remains as the position is slightly influenced by ergosterol at 1602 cm−1 and nearby biomolecular signals.45 This shows that the 1655 cm−1 band can be a possible tracer of nascent UFA, but further validation is required. Additionally, the ratiometric integrated intensity at 1655/1605 bands at different incubation time intervals provides a quantitative insight into metabolic carbon incorporation and turnover dynamics of nascent UFA. As shown in Fig. 2(B), an increase in 1655/1605 is seen in earlier time points indicating the newly synthesized unlabelled UFA gradually replacing the old pool of UFA. The curve reaches a plateau in the late incubation hours indicating almost the entire old pool of UFA is substituted by the nascent UFA. Fig. 2(C) shows ratiometric intensity changes in 1655/1445 showing the turnover of nascent UFA in the total lipid pool of the single YL cells. However, this time-dependent blueshift of the UFA band may change from strain to strain, in other species and growth parameters which needs to be optimized accordingly. As UFA are an essential component of SMCO composition and lipid body (also known as lipid droplet, oil body), therefore time-dependent intensity dynamics of the nascent UFA band at 1655 cm−1 can be the evident indicative tracer for de novo SMCO synthesis and nascent SMCO turnover similar to the phenylalanine (1003 cm−1)31–33,42,46 and amide I (1667 cm−1)31–33,42,46 acting as Raman representative protein bands.
3.2 Validation of the nascent unsaturated fatty acid band from the cells
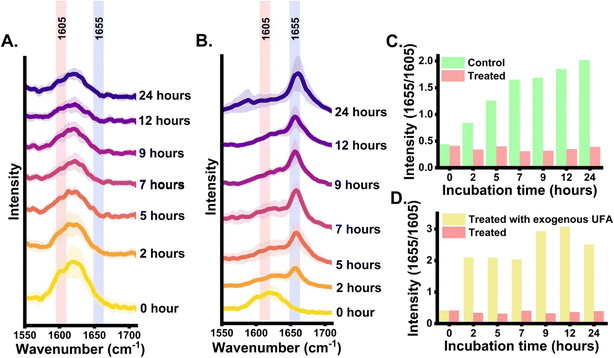
Further to validate that the detected nascent Raman band signal at 1655 cm−1 position is coming from unlabelled fatty acid an inhibition assay was performed (loss of function) using cerulenin. It is a well-known inhibitor of fatty acid synthase (FAS), an important enzyme in the de novo fatty acid synthesis pathway.47 Firstly to confirm that the yeast strain is not resistant to cerulenin (50 μg ml−1), the growth monitoring and the treatment were done at 7 h post-inoculation when cells are in the exponential phase of division, Fig. S1(B).† A clear inhibitory effect of cerulenin on the growth of yeast was observed. In the next step, treatment was performed at 0 h and the average Raman spectra of cerulenin-treated cells at multiple incubation time points were acquired. It shows a drastic reduction in signal at 1655 cm−1 attributed to nascent unsaturated band position as shown in Fig. 3(A). The peak intensity at 1655 cm−1 was negligible in the cerulenin-treated cells. This supports the possible correlation between the peak at 1655 cm−1 and the nascent UFA band in Raman spectra as cerulenin inhibits fatty acid synthesis. The mechanism of action of cerulenin involves inhibition of the FAS enzyme leading to halted nascent fatty acid generation in cells which makes the cells survive by depleting the stored cellular SMCO and ultimately leading to cell death. The origin of 1655 cm−1 was also validated through a “gain of function” assay. Here, the inhibitory effect of cerulenin on the cells was reversed by supplementing them with exogenous UFA.47,48 This rescue of the treated cells by exogenous UFA supplementation occurs due to its preferential uptake for fulfilling the fatty acid scarcity due to inhibition of fatty acid synthesis.47 The study hypothesized that when treated cells incorporate external UFA for its rescue, a sudden spike in peak intensity at 1655 cm−1 will be observed from cells at early time points which is 2 h, Fig. 3(B).Before supplementing the exogenous fatty acid to cells, the validation study to confirm that 1655 cm−1 is unique to UFA only was done. To address this concern, Raman spectra of some well-known standard saturated fatty acids (SFA) namely myristic acid (C14![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0), pentadecylic acid (C15
0), pentadecylic acid (C15![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0), palmitic acid (C16
0), palmitic acid (C16![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0), stearic acid (C18
0), stearic acid (C18![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0) and UFA namely oleic acid (C18
0) and UFA namely oleic acid (C18![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1), linoleic acid (C18
1), linoleic acid (C18![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2) were acquired and ratiometric intensity quantification was performed to unravel the degree of unsaturation Fig. S2.† The revived growth of cerulenin-treated cells by supplemented exogenous UFA can be seen in Fig. S1(B)† which can be further enhanced when provided with SFA. Raman spectra of rescued cells at 2 h post-treatment, show a highly intensified band at 1655 cm−1 which does not originate from de novo synthesis but from the uptake of exogenous UFA Fig. 3(B). This provides further evidence in support of the fact that the Raman band position at 1655 cm−1 of cells indeed arises from unlabelled UFA. Additionally, quantitative insight into the ratiometric UFA band intensity (1655/1605) dynamics between the untreated cells and cerulenin-treated cells demonstrating the relative difference in nascent UFA content over time is shown in Fig. 3(C). Similarly, a relative quantitative ratio-metric analysis was performed for cerulenin-treated and rescued cells as shown in Fig. 3(D). Furthermore, the band at 1655 cm−1 region is broad and this is well-known that the spectral region is associated with adjacent protein band at around 1667 cm−1 (amide I).32,49 To address this aspect and validate that the band position 1655 cm−1 is predominately from UFA, the proteins and lipids were extracted from the reversely unlabelled cells after 24 h and recorded Raman spectra. The spectral range interpolated to the region of interest covering the UFA and amide I band is shown in Fig. S3.† The highlighted peak position from the extracted unlabelled lipid is observed around 1655 cm−1 and extracted unlabelled protein at around 1667 cm−1. Further, 1003 cm−1 peak of phenylalanine was only observed in Raman spectra of extracted unlabelled protein which acts as a control.32
2) were acquired and ratiometric intensity quantification was performed to unravel the degree of unsaturation Fig. S2.† The revived growth of cerulenin-treated cells by supplemented exogenous UFA can be seen in Fig. S1(B)† which can be further enhanced when provided with SFA. Raman spectra of rescued cells at 2 h post-treatment, show a highly intensified band at 1655 cm−1 which does not originate from de novo synthesis but from the uptake of exogenous UFA Fig. 3(B). This provides further evidence in support of the fact that the Raman band position at 1655 cm−1 of cells indeed arises from unlabelled UFA. Additionally, quantitative insight into the ratiometric UFA band intensity (1655/1605) dynamics between the untreated cells and cerulenin-treated cells demonstrating the relative difference in nascent UFA content over time is shown in Fig. 3(C). Similarly, a relative quantitative ratio-metric analysis was performed for cerulenin-treated and rescued cells as shown in Fig. 3(D). Furthermore, the band at 1655 cm−1 region is broad and this is well-known that the spectral region is associated with adjacent protein band at around 1667 cm−1 (amide I).32,49 To address this aspect and validate that the band position 1655 cm−1 is predominately from UFA, the proteins and lipids were extracted from the reversely unlabelled cells after 24 h and recorded Raman spectra. The spectral range interpolated to the region of interest covering the UFA and amide I band is shown in Fig. S3.† The highlighted peak position from the extracted unlabelled lipid is observed around 1655 cm−1 and extracted unlabelled protein at around 1667 cm−1. Further, 1003 cm−1 peak of phenylalanine was only observed in Raman spectra of extracted unlabelled protein which acts as a control.32
3.3 Visualisation and distribution of the de novo synthesized single microbial cell oil and its turnover over time
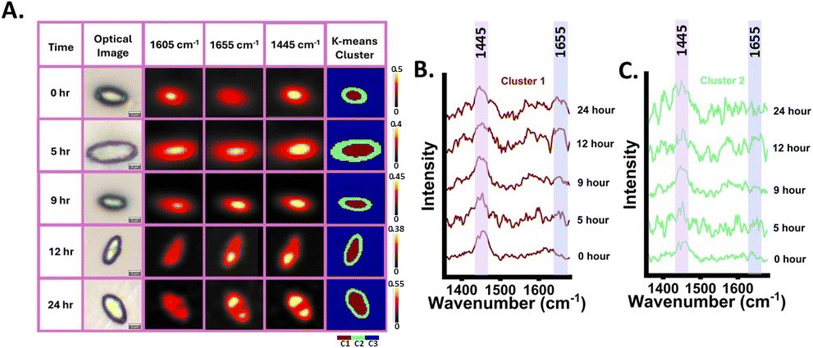
After validating that the band at 1655 cm−1 is from nascent UFA and a potential tracer for SMCO synthesis, this band was utilized to map the distribution and dynamic of SMCO using RrSIP imaging, Fig. 4(A). Raman images of YL were generated from SMCO representative bands normalised intensity using false colour at different time intervals (0, 5, 9, 12 and 24 h). Corresponding optical image of the cells is also presented for a better understanding of the cellular morphology. The Raman images of the total lipid signal at 1445 cm−1 and UFA signals (1605 and 1655 cm−1) are giving coinciding signals in co-localized distribution and have high signal intensity at specific regions of cells, which can be interpreted as lipid body as per previous Raman images lipid body studies.42,43,50,51 Low-intensity SMCO band signal can be seen all over cellular regions indicating the presence of fatty acid as its synthesis occurs in the cytoplasm which is later stored as TAG in lipid bodies and other forms of lipids as shown in Fig. 4(A). At 0 h the image shows high signal intensity distribution at 1605 cm−1. At 5 h equivalent intensity distribution from both 1605 and 1655 cm−1 was seen. However, over time the distribution of 1655 cm−1 in the Raman image becomes significantly more and 1605 cm−1 reduces. This further demonstrates that by using the tracer band at 1655 cm−1, the nascent SMCO distribution and turnover dynamics can be monitored over time. Further Raman image of the cells was generated by K-means cluster analysis (k = 3). This unsupervised analysis of the hyperspectral image data set was done at each time interval without specifying any biomolecular bands. The hyperspectral image data set is decomposed into three different clusters with each cluster having statistically similar spectral information. Cluster 1 was seen to be originating from the intracellular region and the corresponding spectral component of cluster 1 is shown in Fig. 4(A) and (B). The cluster 1 spectral component of each time point indicates the dynamic changes in the intensity of UFA at 1655 cm−1 from 0 h to later incubation time points. As discussed earlier, at residual intensity remains at 1605 cm−1 as it is influenced by the presence of ergosterol at 1602 cm−1 and other biomolecular bands. The spectral component of cluster 2 also shows biomolecular information shown in Fig. 4(C) and cluster 3 shows signals from the background as shown in Fig. S4.†3.4 In situ monitoring of ex novo synthesis of single microbial cell oil synthesis from cooking oil and waste cooking oil source
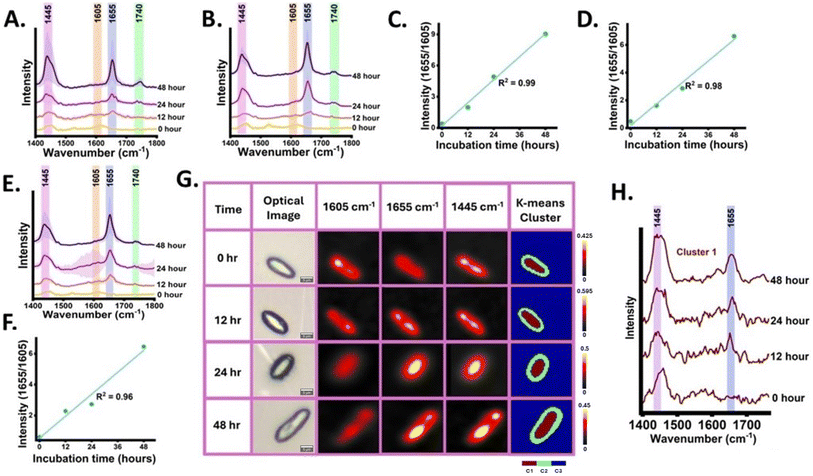
Ex novo single-cell microbial oil synthesis is the capability of microbial cells to metabolize and incorporate the hydrophobic carbon sources such as oils, and alkanes and utilize them for cellular structure, metabolism, and energy storage.52 Firstly, 13C labelled YL was grown in the carbon-free synthetic medium with cooking oil namely palmolein oil and sunflower oil as the single supplemented carbon source and incubated for 48 h. Cooking oils are majorly composed of TAGs containing UFA chains. Raman spectra of palmolein oil and sunflower oil used in this study as the hydrophobic carbon sources are shown in Fig. S5,† YL can secrete extracellular lipases which break down TAGs into glycerol and fatty acids. The Raman spectra at 0, 12, 24 and 48 h were acquired and monitored the lysis-free carbon flow from cooking oil to the SMCO pool validating ex novo SMCO synthesis and accumulation by tracking the dynamics of nascent 1655 cm−1 band. At 0 h, residual band intensity at 1655 cm−1 was observed indicating the cell's SMCO pool is 13C labelled. Post inoculation at 12 h intensified band signal at 1655 cm−1 was detected as shown in Fig. 5(A) and (B). Over time, at 24 and 48 h post-inoculation the intensified signal at SMCO bands at 1655 cm−1 and 1445 cm−1 was observed and a rise in band intensity at 1740 cm−1 which has been assigned to the ester bond. This indicative increase in band intensity corroborates with the previous findings which have shown that the ex novo synthesis induces higher assimilation of TAGs in YL.52,53 This result aligns with the prior study, which indicates feeding YL with oil as a carbon source has higher biomass yield when compared to YL grown in glucose with minimal medium.54 To provide relative quantitative insight into the accumulation of the UFA from the ex novo SMCO synthesis in the already existing UFA pool, the ratiometric integrated intensity at 1655/1605 was plotted with incubation time showing a linear increment with time with R square value of 0.99 and 0.98 as shown in Fig. 5(C) and (D).After monitoring the de novo and ex novo SMCO synthesis in microbial cells targeting the tracer UFA band, the next goal was to analyse SMCO synthesis grown in synthetic media with WCO as the only carbon source. It is significant as SMCO produced from WCO helps in bioremediation as well as serves as sustainable biomass in biofuel and bioprocess industries. The WCO carbon source notably increases SMCO accumulation by folds when compared to cells grown in glucose which has been reported in a previous study.11 Free fatty acids in WCO can be directly used for biodiesel production using conventional well-known methodologies such as enzymatic catalysis and noncatalytic transesterification routes. However, it is not a very feasible approach as WCO also contains, water, glycerides, aldehydes, organic acids and other organic compounds which hinders desirable fatty acid methyl esters (FAME) yield and affects the quality of biodiesel. Therefore, microbial-based bioremediation has been utilized for a cost-effective and improved FAME yield.11 The Raman spectra of waste cooking oil are shown in Fig. S5.† The band intensity dynamics at 1655 cm−1 as a reliable efficient tracer to monitor the SMCO synthesis and accumulation was validated and described in the previous section. The Raman spectra from the cells grown in WCO were acquired at 0, 12, 24 and 48 h as shown in Fig. 5(E). When compared to 0 h, the increased peak intensity of 1655 cm−1 band at 12 h and highly intensified peaks at 24 and 48 h were observed. At 24 and 48 h intensified peaks at 1445 and 1740 cm−1 were also detected. This shows that the SMCO synthesis and assimilation from waste cooking oil. To provide relative quantitative validation into the assimilation of UFA and its turnover, the ratiometric integrated intensity at 1655/1605 was plotted with incubation time showing a linear increment with time with an R square value of 0.96 as shown in Fig. 5(F). Further Raman image of the cells from the hyperspectral image data was generated at different time intervals from 0, 12, 24 and 48 h as shown in Fig. 5(G). The distribution of the intensity corresponding to SMCO which is 1605 cm−1, 1655 cm−1 and 1445 cm−1 was monitored investigating the distribution of UFA and nascent UFA assimilation dynamics. The Raman image gives us insight into SMCO distribution and localization of lipid bodies. The SMCO tracer band at 1655 cm−1 shows residual intensity distribution in Raman image at 0 h which gets intensified with the incubation time, aids in visualizing the nascent SMCO assimilation and turnover from WCO. Further hyperspectral image data was subjected to k means (k = 3) unsupervised algorithm forming 3 clusters of different colours, with each cluster having statistically similar information. The k means cluster image was generated with 3 clusters and the corresponding spectral components as shown in Fig. 5(G). The spectral pattern of components of different time intervals of cluster 1 is shown in Fig. 5(H) which can be seen to be the group of pixels inside the cells corroborating with the dynamics of 1655 cm−1 with time. The spectral component of cluster 2 also shows biomolecular information and cluster 3 shows signals from the background Fig. S6.† These observations support the fact that RrSIP-based in situ monitoring of SMCO synthesized from waste cooking oil can provide qualitative as well as semi-quantitative insight in a lysis-free and non-destructive manner.
Previously, Raman spectroscopy and imaging have been used to study lipids in different microbes. In vivo lipid profiling of oil-producing microalgae N. oleoabundans, B. braunii, B. sudeticus, C. reinhardtii, and T. minutus was demonstrated by Seema et al. using single-cell laser trapping Raman spectroscopy and quantitative assessment of the degree of unsaturation was performed using intensity ratio (I1650/I1440).44 Cheng et al. have observed lipid droplets (lipid body) have a higher contrast at 1654 cm−1 position when compared to cytoplasm because of the lower contribution from the amide I band.43 Hemanth et al. have used Raman stable isotope probing-based imaging of single yeast cell Schizosaccharomyces pombe (S. Pombe) cells showing protein localization on lipid bodies targeting 13C labelled nascent protein band at 967 cm−1 and lipid droplets at 1602 cm−1 and also reported band at 1654 cm−1 as unlabelled unsaturated lipid band.42 Raman image of the cell can provide information about spatial biomolecular distribution.55 Using Raman imaging of single cell S. Pombe, Shinshuke et al. have shown the influence of high phospholipid contribution in 1655 cm−1 bands correlating it with the image from lipid band at 1440 cm−1.41 Shao et al. studied the intracellular distribution and accumulation of lipids in nitrogen deficient condition of freshwater algae Scenedesmus obliquus using Raman spectroscopy and imaging targeting band at 1445 cm−1.40 Tsuyoshi et al. have shown the generated Raman images of the lipid body of oleaginous diatom Fistularia Solaris Raman band at 1445 cm−1 was observed using Raman imaging and quantitative analysis of lipid unsaturation was studied using ratiometric intensity I1656/I1445. Similarly, waste cooking oil has been analysed using Raman spectroscopy without using microbial mediator studying vibrational mode at 1441, 1657 and 1747 cm−1.56
Even though the importance of lysis-free in situ monitoring of fatty acid biosynthesis is well known, there have not been many efforts. Point Raman spectroscopy allows a rapid detection of nascent SMCO turnover dynamics over time. In contrast, Raman imaging even though time-consuming can provide crucial spatial information that can help in the visualization of nascent subcellular biomolecule distribution and localization. Further studies are required to advance this approach to qualitative and quantitatively monitor the presence of different mono- and poly-unsaturated fatty acids, their synthesis and turnover dynamics in SMCO inside the single cell in a lysis-free manner. Raman-based approach has its advantages and limitations. The authors believe that point Raman spectroscopy can be an adjunct rapid detection tool for SMCO bio-process industrial applications in future as it requires less sample preparation and can be used for in-line monitoring. However, improved preprocessing algorithms such as efficient removal of the signal interference from the growth medium and its integration with data acquisition are essential.
The study though a preliminary investigation presents a unique approach of applying reverse stable isotope probing with Raman spectroscopy and imaging, for in situ monitoring and visualization of single microbial cell oil synthesis. The RrSIP-based approach can be an intriguing adjunct for monitoring the WCO metabolic incorporation and nascent microbial oil dynamics in an extraction-free approach.
4 Conclusions
Bioremediation of waste cooking oil is one of the best possible ways to produce microbial-derived value-added products. A lysis-free approach that can be used for monitoring this bioremediation is crucial for evaluating the efficiency of microbial biomass for industrial applications. This study has shown a non-destructive Raman spectroscopy and imaging approach for monitoring the synthesis and assimilation of SMCO using carbon sources such as glucose and waste cooking oil. Further, minimal usage of chemical reagents aids in minimal sample preparation. The overall findings suggest that the RrSIP coupled with Raman imaging holds immense potential as an adjunct lysis-free assay to visualize and monitor SMCO distribution and dynamics. This approach can serve as a versatile cost-effective tool in lipidomics for biofuel and bioprocess industrial applications. In future, this proposed analytical methodology can have a wide range of applications including sensing of commercially relevant sustainable microbial metabolite build-up from the trash.Data availability
The Raman spectral data and MATLAB codes used in this study can be accessed via the link: Tracking-Trash-to-Treasure-In-situ-monitoring-of-SMCO-biosynthesis-from-WCO-using-RS-and-RI (https://github.com/jirokarlo/Tracking-Trash-to-Treasure-In-situ-monitoring-of-SMCO-biosynthesis-from-WCO-using-RS-and-RI).Conflicts of interest
The author has no conflicts of interest to declare.Acknowledgements
This work was carried out under research grant project no (37/1739/23/EMR-II) supported by the Council of Scientific and Industrial Research (CSIR), Government of India and project no. IIRP-2023-1734 from the Indian Council of Medical Research (ICMR), Government of India. The authors are grateful for the seed funding to conduct the CESMI project at the University of Eastern Finland, provided by the Finnish Ministry of Education and Culture through the Finnish Indian Consortia for Research and Education (FICORE) Global pilot network. We also thank the Finnish National Agency for Education's Team Knowledge Finland (TFK) programme for their financial support to the project ‘DEDUCE’ (grant number 78/116/2022).References
- A. Mannu, M. Ferro, M. E. Di Pietro and A. Mele, Sci. Prog., 2019, 102, 153–160 CrossRef PubMed.
- O. Awogbemi, D. V. Von Kallon, V. S. Aigbodion and S. Panda, Case Stud. Chem. Environ. Eng., 2021, 4, 100158 CrossRef CAS.
- X. Cui, N. Sun, P. Cao, J. Guo and P. Ming, J. Manuf. Process., 2022, 77, 508–524 CrossRef.
- G. De Feo, C. Ferrara, L. Giordano and L. S. Ossèo, Recycling, 2023, 8, 64 CrossRef.
- A. H. Hashem, A. M. Khattab and M. Abdelraof, Biomass Convers. Biorefin., 2023, 13, 16711–16721 CrossRef CAS.
- Q.-Q. Zhang, B.-X. Cai, W.-J. Xu, H.-Z. Gang, J.-F. Liu, S.-Z. Yang and B.-Z. Mu, Sci. Rep., 2015, 5, 9971 CrossRef CAS.
- A. Mannu, S. Garroni, J. Ibanez Porras and A. Mele, Processes, 2020, 8, 366 CrossRef CAS.
- Y. Li, Z. Cheng, C. Zhao, C. Gao, W. Song, L. Liu and X. Chen, ACS Synth. Biol., 2021, 10, 1966–1979 CrossRef CAS PubMed.
- R. Marchetti, C. Vasmara, L. Bertin and F. Fiume, Appl. Microbiol. Biotechnol., 2020, 104, 2833–2856 CrossRef CAS PubMed.
- J. Karlo, R. Prasad and S. P. Singh, J. Agric. Food Res., 2023, 11, 100482 CAS.
- G. Raut, S. Jagtap, V. R. Kumar and A. RaviKumar, Biomass Convers. Biorefin., 2022, 1–18 Search PubMed.
- B. Zieniuk and A. Fabiszewska, World J. Microbiol. Biotechnol., 2018, 35, 10 CrossRef PubMed.
- A. Rywińska, P. Juszczyk, M. Wojtatowicz, M. Robak, Z. Lazar, L. Tomaszewska and W. Rymowicz, Biomass Bioenergy, 2013, 48, 148–166 CrossRef.
- Y.-L. Jia, L.-R. Wang, Z.-X. Zhang, Y. Gu and X.-M. Sun, Crit. Rev. Food Sci. Nutr., 2022, 62, 8920–8934 CrossRef CAS PubMed.
- C. Ratledge and J. P. Wynn, Adv. Appl. Microbiol., 2002, 1–52 CAS.
- G. Katre, C. Joshi, M. Khot, S. Zinjarde and A. RaviKumar, AMB Express, 2012, 2, 36 CrossRef.
- P. Gajdoš, R. Ledesma-Amaro, J.-M. Nicaud, M. Čertík and T. Rossignol, FEMS Yeast Res., 2016, 16, fow062 CrossRef PubMed.
- A. Beopoulos, Z. Mrozova, F. Thevenieau, M.-T. Le Dall, I. Hapala, S. Papanikolaou, T. Chardot and J.-M. Nicaud, Appl. Environ. Microbiol., 2008, 74, 7779–7789 CrossRef CAS PubMed.
- Y. Pang, Y. Zhao, S. Li, Y. Zhao, J. Li, Z. Hu, C. Zhang, D. Xiao and A. Yu, Biotechnol. Biofuels, 2019, 12, 241 CrossRef PubMed.
- L. Mitrea, L.-F. Călinoiu, B.-E. Teleky, K. Szabo, A.-G. Martău, B.-E. Ştefănescu, F.-V. Dulf and D.-C. Vodnar, Environ. Technol. Innovation, 2022, 28, 102943 CrossRef CAS.
- M. Groenewald, T. Boekhout, C. Neuvéglise, C. Gaillardin, P. W. M. van Dijck and M. Wyss, Crit. Rev. Microbiol., 2014, 40, 187–206 CrossRef CAS PubMed.
- F. Thevenieau and J.-M. Nicaud, OCL, 2013, 20, D603 CrossRef.
- T. M. N. Ta, C. Romero-Guido, T. H. Phan, H. D. Tran, H. T. Dinh and Y. Waché, AIMS Biophys., 2022, 9, 257–270 CAS.
- N. Morin, Q. Czerwiec, J. Nicaud, C. Neuvéglise and T. Rossignol, Yeast, 2020, 37, 348–355 CrossRef CAS PubMed.
- A. Daskalaki, N. Perdikouli, D. Aggeli and G. Aggelis, Appl. Microbiol. Biotechnol., 2019, 103, 8585–8596 CrossRef CAS PubMed.
- P. Radha, S. Narayanan, A. Chaudhuri, S. Anjum, D. L. Thomas, R. Pandey and K. Ramani, Biomass Convers. Biorefin., 2023, 13, 1–12 CrossRef.
- X. Niehus, A.-M. Crutz-Le Coq, G. Sandoval, J.-M. Nicaud and R. Ledesma-Amaro, Biotechnol. Biofuels, 2018, 11, 11 CrossRef.
- Y. Wang, W. E. Huang, L. Cui and M. Wagner, Curr. Opin. Biotechnol., 2016, 41, 34–42 CrossRef CAS PubMed.
- S. A. Eichorst, F. Strasser, T. Woyke, A. Schintlmeister, M. Wagner and D. Woebken, FEMS Microbiol. Ecol., 2015, 91, fiv106 CrossRef PubMed.
- K. Jiro, A. Gupta and S. S. Pratap, Analyst, 2024, 149, 2833–2841 RSC.
- Y. Wang, Y. Song, Y. Tao, H. Muhamadali, R. Goodacre, N.-Y. Zhou, G. M. Preston, J. Xu and W. E. Huang, Anal. Chem., 2016, 88, 9443–9450 CrossRef CAS PubMed.
- J. Karlo, A. K. Dhillon, S. Siddhanta and S. P. Singh, J. Biophotonics, 2023, 17(2), e202300341 CrossRef PubMed.
- M. Chisanga, H. Muhamadali, D. McDougall, Y. Xu, N. Lockyer and R. Goodacre, Analyst, 2021, 146, 1734–1746 RSC.
- J. Karlo, A. Vijay, M. S. Phaneeswar and S. P. Singh, ACS Omega, 2024, 9, 23753–23760 CrossRef CAS PubMed.
- V. Tsakraklides, A. Kamineni, A. L. Consiglio, K. MacEwen, J. Friedlander, H. G. Blitzblau, M. A. Hamilton, D. V. Crabtree, A. Su, J. Afshar, J. E. Sullivan, W. G. LaTouf, C. R. South, E. H. Greenhagen, A. J. Shaw and E. E. Brevnova, Biotechnol. Biofuels, 2018, 11, 131 CrossRef.
- E. Carsanba, S. Papanikolaou, P. Fickers and H. Erten, Microorganisms, 2020, 8(7), 1054 CrossRef CAS PubMed.
- R. M. Olayide Israel, Extraction of Lipids from Yeast Search PubMed.
- T. von der Haar, PLoS One, 2007, 2, e1078 CrossRef PubMed.
- N. Mobaraki and J. M. Amigo, Chemom. Intell. Lab. Syst., 2018, 172, 174–187 CrossRef CAS.
- Y. Shao, H. Fang, H. Zhou, Q. Wang, Y. Zhu and Y. He, Biotechnol. Biofuels, 2017, 10, 300 CrossRef PubMed.
- C.-K. Huang, H. Hamaguchi and S. Shigeto, Chem. Commun., 2011, 47, 9423 RSC.
- H. N. Noothalapati Venkata and S. Shigeto, Chem. Biol., 2012, 19, 1373–1380 CrossRef CAS PubMed.
- M. N. Slipchenko, T. T. Le, H. Chen and J.-X. Cheng, J. Phys. Chem. B, 2009, 113, 7681–7686 CrossRef CAS PubMed.
- H. Wu, J. V. Volponi, A. E. Oliver, A. N. Parikh, B. A. Simmons and S. Singh, Proc. Natl. Acad. Sci., 2011, 108(9), 3809–3814 CrossRef CAS.
- L. Chiu, F. Hullin-Matsuda, T. Kobayashi, H. Torii and H. Hamaguchi, J. Biophotonics, 2012, 5, 724–728 CrossRef CAS PubMed.
- M. Chisanga, H. Muhamadali, D. McDougall, Y. Xu, N. Lockyer and R. Goodacre, Analyst, 2021, 146, 1734–1746 RSC.
- L. N. Nguyen and J. D. Nosanchuk, Commun. Integr. Biol., 2011, 4, 631–632 CrossRef PubMed.
- J. Awaya, T. Ohno, H. Ohno and S. Ōmura, Biochim. Biophys. Acta, Lipids Lipid Metab., 1975, 409, 267–273 CrossRef CAS PubMed.
- J. Karlo, A. K. Dhillon, S. Siddhanta and S. P. Singh, J. Biophotonics, 2022, 16(4), e202200341 CrossRef PubMed.
- M. Uematsu and T. Shimizu, Commun. Biol., 2021, 4, 1176 CrossRef CAS PubMed.
- K. Kochan, H. Peng, B. R. Wood and V. S. Haritos, Biotechnol. Biofuels, 2018, 11, 106 CrossRef PubMed.
- A. Fabiszewska, M. Stępień, P. Goryca, B. Zieniuk and B. Florjańczyk, Biomolecules, 2019, 9, 685 CrossRef CAS PubMed.
- A. Beopoulos, T. Chardot and J.-M. Nicaud, Biochimie, 2009, 91, 692–696 CrossRef CAS PubMed.
- A. M. Worland, J. J. Czajka, Y. Xing, W. F. Harper, A. Moore, Z. Xiao, Z. Han, Y. Wang, W. W. Su and Y. J. Tang, Metab. Eng. Commun., 2020, 11, e00130 CrossRef PubMed.
- J. Karlo, A. K. Dhillon, S. S. Razi, S. Siddhanta and S. P. Singh, Raman Spectroscopy: Advances and Applications, Springer Nature Singapore, Singapore, 2024, pp. 349–375 Search PubMed.
- H. Jin, H. Li, Z. Yin, Y. Zhu, A. Lu, D. Zhao and C. Li, Food Chem., 2021, 362, 130191 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d4ra05187d |
| This journal is © The Royal Society of Chemistry 2024 |