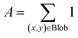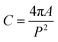Open Access Article
Open Access ArticleMorphological analysis of Pd/C nanoparticles using SEM imaging and advanced deep learning
Nguyen Duc Thuan *,
Hoang Manh Cuong,
Nguyen Hoang Nam,
Nguyen Thi Lan Huong and
Hoang Si Hong*
*,
Hoang Manh Cuong,
Nguyen Hoang Nam,
Nguyen Thi Lan Huong and
Hoang Si Hong*
School of Electrical and Electronic Engineering, Hanoi University of Science and Technology, Hanoi, Vietnam. E-mail: thuan.nguyenduc1@hust.edu.vn; hong.hoangsy@hust.edu.vn
First published on 5th November 2024
Abstract
In this study, we present a comprehensive approach for the morphological analysis of palladium on carbon (Pd/C) nanoparticles utilizing scanning electron microscopy (SEM) imaging and advanced deep learning techniques. A deep learning detection model based on an attention mechanism was implemented to accurately identify and delineate small nanoparticles within unlabeled SEM images. Following detection, a graph-based network was employed to analyze the structural characteristics of the nanoparticles, while density-based spatial clustering of applications with noise was utilized to cluster the detected nanoparticles, identifying meaningful patterns and distributions. Our results demonstrate the efficacy of the proposed model in detecting nanoparticles with high precision and reliability. Furthermore, the clustering analysis reveals significant insights into the morphological distribution and structural organization of Pd/C nanoparticles, contributing to the understanding of their properties and potential applications.
1. Introduction
The synthesis and characterization of nanoparticles are critical areas of research in nanotechnology, with applications spanning catalysis, energy storage, and biomedical fields.1,2 Among the various nanoparticles, palladium on carbon (Pd/C) nanoparticles have garnered significant attention due to their exceptional catalytic properties, making them indispensable in processes such as hydrogenation reactions, fuel cells, and environmental remediation.2–5 A comprehensive understanding of the morphology of these nanoparticles is essential for optimizing their performance and tailoring their properties for specific applications.6,7Traditional methods for characterizing nanoparticle morphology, such as transmission electron microscopy (TEM) and X-ray diffraction (XRD), provide valuable insights but often require extensive sample preparation and are time-consuming.8,9 Scanning electron microscopy (SEM) has emerged as a powerful alternative due to its relatively straightforward sample preparation, rapid imaging capabilities, and high spatial resolution.10,11 However, the manual analysis of SEM images is labor-intensive and subject to human error, necessitating the development of automated and accurate methods for morphological analysis.
In recent years, advances in deep learning have revolutionized various fields, including image analysis. Deep learning algorithms, particularly convolutional neural networks (CNNs), have demonstrated remarkable success in automatically identifying and classifying complex patterns in images.12–14 A significant challenge in applying deep learning to SEM image analysis is the requirement for large, labeled datasets.15,16 Labeling nanoparticles in SEM images is particularly difficult due to the high resolution and complexity of the images.17,18 Each nanoparticle must be accurately identified and annotated, a process that requires expert knowledge and is extremely time-consuming and prone to human error.
Furthermore, existing deep learning-based methods for nanoparticle analysis typically rely on SEM or TEM imaging, focusing primarily on size and shape analysis.19–23 While TEM provides higher resolution, it is difficult to capture the surface structures that are crucial for a comprehensive morphological analysis of nanoparticles like Pd/C. This limitation means that many current approaches fail to offer insights into both shape and structural characteristics, which are essential for understanding nanoparticle behavior and functionality. The importance of structure analysis cannot be overstated, particularly for Pd/C nanoparticles, where characteristics like surface defects, roughness, and atomic arrangements significantly impact catalytic activity, selectivity, and stability. Without proper structural analysis, key factors influencing the efficiency and durability of nanoparticles may be overlooked, which can lead to suboptimal performance.
To overcome the abovementioned limitations, this paper presents a novel approach for the morphological analysis of Pd/C nanoparticles by integrating SEM imaging with advanced deep learning techniques, specifically addressing the challenge of labeled data requirements. We propose a method that significantly reduces the need for extensive manual labeling while still achieving high accuracy in morphological characterization. Our approach leverages semi-supervised learning techniques, enabling the model to learn from partially labeled data, thus alleviating the dependency on labor-intensive and error-prone labeling processes. The proposed deep model is designed based on attention mechanism to improve the ability in detecting small nanoparticles. In addition to deep learning for detection, we employ advanced clustering algorithms to analyze the structure and distribution of the detected nanoparticles. These clustering methods allow us to uncover meaningful patterns and distributions that are crucial for understanding the morphological properties and behaviors of Pd/C nanoparticles. To highlight the distinctions between our method and existing approaches, Table 1 compares deep leaning-based methods for morphological analysis of nanoparticles.
| Reference | Nanoparticle type | Imaging modality | Analysis focus | |||
|---|---|---|---|---|---|---|
| Size | Shape | Distribution | Structure | |||
| Xu et al.19 | Al/SiO2/Si | TSOM | Yes | No | No | No |
| Wen et al.20 | Various materials | TEM | Yes | Yes | Yes | No |
| Sun et al.21 | NaGdF4: 49% Yb, 1% Tm | SEM/TEM | Yes | No | No | No |
| Lee et al.22 | HAuCl4·3H2O | TEM | Yes | Yes | No | No |
| Bals et al.23 | Various materials | SEM | No | Yes | Yes | No |
| Our work | Pd/C | SEM | Yes | Yes | Yes | Yes |
The structure of the paper is as follows: first, we describe the deep learning methodology, including data labeling, semi-supervised learning methods, training process, and clustering techniques. Next, we provide an overview of the synthesis and preparation of Pd/C nanoparticles and the acquisition of SEM images. We then present the results of our model's performance in detecting, analyzing the morphological features of Pd/C nanoparticles, and discuss the implications of these findings for future research and applications. Finally, we conclude with a summary of the contributions of this research.
2. Methodology
2.1. Automated labelling
Given the challenge of no labeled data for training, we employ an automated image-processing-based blob detection algorithm to label some of the nanoparticles. Blob detection is effective for labeling nanoparticles in unlabeled data because it automatically identifies localized features based on differences in intensity or color.24,25 Its scale-invariance detects nanoparticles of varying sizes, and its noise robustness filters out irrelevant details. This automation allows efficient processing of large datasets, making it a practical solution for generating labeled data and enabling further analysis and model training.26 Moreover, the blob detection algorithm is highly adaptable and can be generalized to analyze other types of nanoparticles by modifying key parameters such as blob size, and shape sensitivity (circularity, convexity, and inertia ratio).27 These adjustments allow the algorithm to accommodate a range of nanoparticle morphologies and material systems, making it versatile for detecting nanoparticles with varying sizes, shapes, and compositions. The blob detection process involves:24• Step 1. Thresholding: convert the grayscale image to a binary image by applying a threshold (50% of maximum intensity). Each pixel value is compared against a threshold value to classify it as either part of a blob or the background.
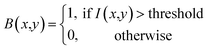 | (1) |
• Step 2. Connected components: identify connected regions (blobs) in the binary image. This involves finding all contiguous sets of pixels that form distinct blobs.
• Step 3. Blob filtering criteria: filter the identified blobs based on several criteria presented in Table 2. After filtering, detected blobs are labeled nanoparticles and served as training information for the deep learning model.
2.2. Model architecture
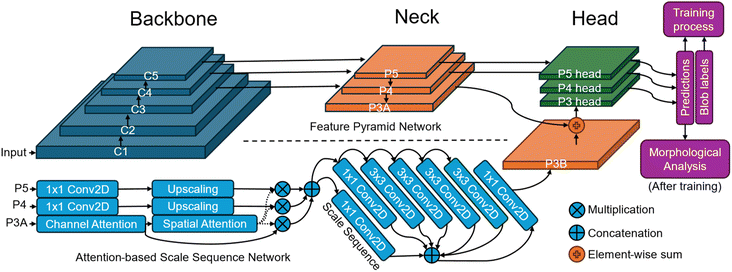
The proposed deep learning model is based on a feature pyramid network (FPN) architecture enhanced with an attention-based scale sequence network (ASSN) for better small object detection.28 The typical structure of a detector model comprises a backbone, neck, and head as illustrated in Fig. 1. This model begins with a backbone feature pyramid, where feature maps labeled C1 to C5 are extracted from CNNs. These feature maps progressively reduce in spatial resolution from C1 to C5, with C1 capturing low-level details like edges and textures, while C5 captures more abstract and semantic information. To ensure these features are manageable, 3 × 3 convolutional layers (Conv2D) are applied to reduce the depth (or number of channels) of each feature map. The middle section of the model is the feature neck, which includes feature maps labeled P3A, P4, and P5. These represent multi-scale features derived by merging earlier layers (C3 to C5) to create a scale sequence based on the scale space theory. The scale space theory generally suggests that objects should be detected across various scales to account for their size variations in images. This is particularly useful for small object detection, where capturing fine details at different scales can enhance recognition accuracy.The ASSN module, integrated into this structure, is designed to enhance small object detection by refining the feature pyramid.28 The ASSN module enhances the detection of small nanoparticles by applying attention mechanisms to the feature pyramid. Specifically, we denote the feature pyramid by different resolution feature maps from P3A to P5. Attention mechanisms are applied to the P3A feature map, which retains the most information about small objects. Channel attention in eqn (2) and spatial attention in eqn (3) are sequentially generated and applied to P3A based on CBAM:29
| ChannelAtt. = σ(MLP(AvgPool(P3A))) + σ(MLP(MaxPool(P3A))) | (2) |
| SpatialAtt. = σ(BN(f7 × 7(AvgPool(ChannelAtt.); MaxPool(ChannelAtt.)))) | (3) |
Finally, the right side of the model architecture focuses on detection heads for each scale (P3, P4, P5). These detection heads take the refined feature maps and make predictions, which include bounding box regressions and object classifications. The output of the model consists of vectors with a length of 5, corresponding to 4 values for the coordinates of the bounding box, and 1 value representing the confidence of the prediction. By having separate detection heads for each scale, the model can specialize in detecting objects that correspond to different sizes, improving accuracy for small objects. Details of the architecture is listed in Table 3 and principle of the proposed deep learning model can be found in previous works.28
| Layer | Type | Input layer | Output size | Kernel size | Stride | Activation |
|---|---|---|---|---|---|---|
| Input | — | — | 1280 × 886 × 1 | — | — | — |
| C1 | Conv2D | Input | 640 × 443 × 32 | 3 × 3 | 2 | ReLU |
| C2 | Conv2D | C1 | 320 × 221 × 64 | 3 × 3 | 2 | ReLU |
| C3 | Conv2D | C2 | 160 × 110 × 128 | 3 × 3 | 2 | ReLU |
| C4 | Conv2D | C3 | 80 × 55 × 256 | 3 × 3 | 2 | ReLU |
| C5 | Conv2D | C4 | 40 × 27 × 512 | 3 × 3 | 2 | ReLU |
| P3A | Down sampling | C3, C4, C5 | 160 × 110 × 896 | 2 for C4, 4 for C5 | — | Concat |
| P4 | Up/down sampling | C3, C4, C5 | 80 × 55 × 896 | 2 | — | Concat |
| P5 | Up sampling | C3, C4, C5 | 40 × 27 × 896 | 2 for C4, 4 for C3 | Concat | |
| P3B | Attention | P3A, P4, P5 | 160 × 110 × 896 | — | — | — |
| P3 head | Detection head | P3A, P3B | 160 × 110 × 5 | 3 × 3 | 1 | Sigmoid |
| P4 head | Detection head | P4 | 80 × 55 × 5 | 3 × 3 | 1 | Sigmoid |
| P5 head | Detection head | P5 | 40 × 27 × 5 | 3 × 3 | 1 | Sigmoid |
2.3. Training process
The training process begins with initializing the model weights using pre-trained YOLOv7/YOLOv8 weights to accelerate convergence. A composite loss function, including localization loss and confidence loss, is defined to measure the accuracy of the model's predictions with blob labels presented in section 3.1. The formulas for the localization loss and confidence loss are presented in the corresponding eqn (4) and (5) as follows:
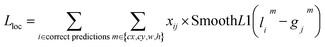 | (4) |
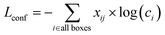 | (5) |
The dataset used for experiments consists of 1000 images, split into an 80![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 20 ratio for training and validation. The input image size is 1280 × 886 pixels, selected to maintain sufficient resolution for detecting nanoparticles, and the output consists of bounding boxes for nanoparticle localization corresponding confidence scores. The Adam optimizer is employed with an initial learning rate of 0.001, which decays over time for gradual fine-tuning. A batch size of 16 was chosen, and training was conducted over 300 epochs. Other hyperparameters were configured to match the default settings of YOLOv7 and YOLOv8, with momentum set to 0.937 and weight decay set to 0.0005. Data augmentation techniques including rotation (±20°) and flipping (vertically and horizontally) were applied to ensure better model generalization.
20 ratio for training and validation. The input image size is 1280 × 886 pixels, selected to maintain sufficient resolution for detecting nanoparticles, and the output consists of bounding boxes for nanoparticle localization corresponding confidence scores. The Adam optimizer is employed with an initial learning rate of 0.001, which decays over time for gradual fine-tuning. A batch size of 16 was chosen, and training was conducted over 300 epochs. Other hyperparameters were configured to match the default settings of YOLOv7 and YOLOv8, with momentum set to 0.937 and weight decay set to 0.0005. Data augmentation techniques including rotation (±20°) and flipping (vertically and horizontally) were applied to ensure better model generalization.
Model performance is evaluated using precision and recall for detection accuracy and mean absolute error (MAE) for assessing centroid location predictions. A prediction is considered correct if the predicted bounding box shares at least one pixel with the ground truth bounding box. Non-maximum suppression (NMS) is applied as a post-processing step to filter out overlapping bounding boxes. Details of the training process of the proposed deep learning model can be found in previous works.28
2.4. Morphological analysis
For visualizing structured or ordered nanoparticles, a graph-based (GB) network construction approach is employed. In this method, detected nanoparticles are treated as nodes in a graph, with edges representing the spatial relationships between them. The process begins by identifying the positions of nanoparticles in the SEM images using the deep learning model. These positions are then used to construct a graph where the connectivity between nodes reflects the physical arrangement of nanoparticles. The graph construction involves defining a distance threshold to determine which nanoparticles are considered neighbors. Then, the adjacency matrix is constructed based on the threshold and the distances between nodes:
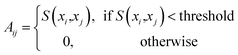 | (6) |
For non-structured or disordered nanoparticle distributions, the density-based spatial clustering of applications with noise (DBSCAN) algorithm is used for clustering and visualization.30 DBSCAN is a robust clustering method that groups points (in this case, nanoparticles regardless of their shape and size) based on their density in the spatial domain, making it well-suited for identifying clusters in irregularly distributed data. The process begins by applying the deep learning model to detect nanoparticles in the SEM images, obtaining their coordinates. DBSCAN then processes these coordinates, identifying clusters based on two parameters: epsilon (ε), the maximum distance between two points to be considered neighbors (in this research, ε = 30 nm), and minPts, the minimum number of points required to form a dense region (in this research, minPts = 5). A point p is considered core point if it has at least minPts neighbors. Each point r is called p-reachable if there exist n points from p1 to pn satisfying:30
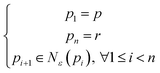 | (7) |
3. Results and discussion
3.1. Synthesis and SEM imaging of Pd/C nanoparticles
Commercially obtained carbon materials were used in this study to evaluate the effect of structural differences on reactivity. The palladium complex, Pd2dba3, was synthesized following a previously reported procedure,31 with its purity confirmed via NMR spectroscopy and elemental analysis. The Pd2dba3 complex was chosen for its ability to form small nanoparticles under mild conditions.32 The reaction of palladium deposition is shown as follows:| Pd2dba3 + C → Pd/C + dba | (8) |
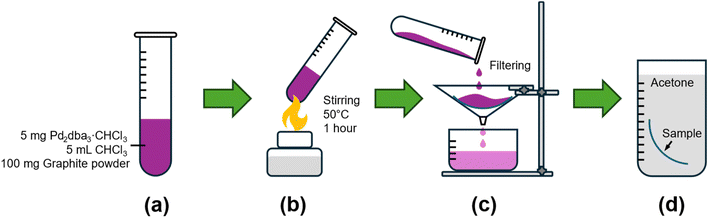
To deposit palladium on carbon surfaces, a direct process was employed to avoid introducing impurities or requiring additional reagents and high temperatures, which could affect the sample's morphology. For samples with an ordered distribution of Pd nanoparticles, a screw cap tube was charged with Pd2dba3·CHCl3 (5 mg), graphite powder (100 mg), and CHCl3 (5 mL). This mixture was stirred at 50 °C for 1 hour, followed by filtration to separate the transparent solution from the carbon material. The material was then dried and, if necessary, washed with acetone to remove any residual dba ligand. The process of palladium deposition on carbon surface is illustrated in Fig. 2.
The completeness of the reaction was confirmed by two methods. Visually, the post-deposition solution appeared completely transparent, indicating no remaining Pd2dba3, which is deep red. Additionally, using a Bruker DRX500 spectrometer, 1H NMR spectrum of the supernatant confirmed the complete consumption of the palladium complex and the release of the dba ligand in its free form. ICP-AES (inductively coupled plasma atomic emission spectrometer) analysis using a JY 38 (Jobin Yvon) spectrometer verified the deposition of palladium on the carbon surface, showing a palladium content of 0.98 wt% in the Pd/C sample, indicating that 99% of the palladium was successfully deposited.
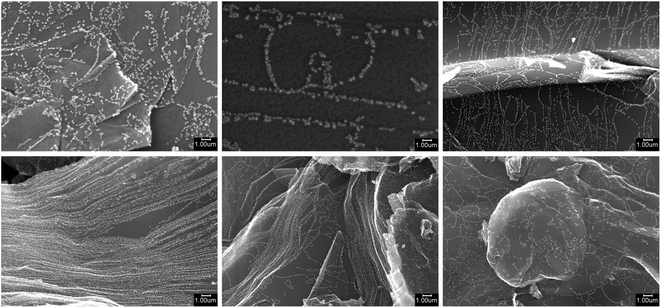
To verify the presence of palladium nanoparticles and study their morphology, SEM imaging and EDX spectroscopy were employed. Samples were mounted on an aluminum stub and fixed with conductive graphite adhesive tape, then observed under native conditions using a Hitachi SU8000 field-emission scanning electron microscope (FE-SEM). Images were acquired in secondary electron mode with an accelerating voltage of 10–30 kV and a working distance of 6–12 mm. The presence of palladium was confirmed using an X-max EDX system (Oxford Instruments, UK). The SEM images revealed the surface topography and morphology of the carbon materials with deposited Pd nanoparticles. EDX analysis confirmed the presence of palladium across all samples. Comparison of images before and after the deposition process demonstrated the formation of new Pd nanoparticles on the carbon surface. Fig. 3 shows the examples of SEM images acquired from the microscope.
The SEM imaging process involves a total of 1000 images of carbon materials with deposited palladium nanoparticles. Each image is 1280 × 1024 pixels in size and presented in TIFF format (.tif). The images are named to facilitate identification with the provided data. A 134-pixel wide digital caption at the bottom of each image indicates key parameters such as the accelerating voltage, working distance, magnification, mode of operation, type of detector, and scale. These indicators, along with additional acquisition parameters and sample names, are available in a separate CSV file, which includes the following details: sample number, acceleration voltage (V), magnification, working distance (μm), emission current (nA), lens mode, and area code. The dataset is available at: https://doi.org/10.6084/m9.figshare.11783661.
3.2. Detection performance
This section presents the detection performance of nanoparticles using the proposed deep learning method. A separate validation set which comprises 20% of the entire dataset and was not used during training, is employed to evaluate the model. Fig. 4(a–c) illustrates the effectiveness of the proposed deep learning model in detecting nanoparticles in SEM images. Fig. 4(a) presents the original SEM image of the nanoparticles, which serves as the baseline for comparison. The image contains numerous nanoparticles distributed across the surface, which are challenging to identify manually due to their small size and overlapping nature. Fig. 4(b) shows the results of a traditional blob detection algorithm applied to the same SEM image. This algorithm uses image-processing techniques to identify potential nanoparticles by detecting regions that differ in intensity compared to the background. The identified nanoparticles are marked with red circles. While the blob detection algorithm successfully identifies many nanoparticles, it misses several smaller particles and misidentifies some areas as particles.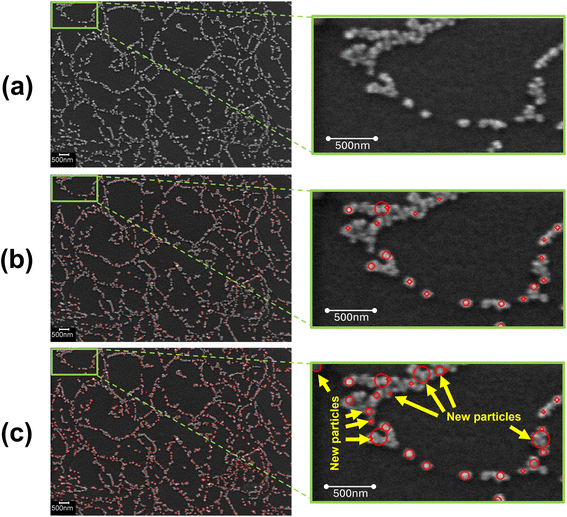 | ||
| Fig. 4 (a) Original SEM image. (b) Nanoparticles detected by blob detection algorithm. (c) Nanoparticles detected by proposed deep learning model. | ||
In Fig. 4(c), the deep learning model's detection results are displayed. This model was trained using data labeled by the blob detection algorithm, providing a visual representation of nanoparticles for the model to learn. The detected nanoparticles are again highlighted with red circles. The rightmost image zooms in on a specific region, showing the new particles identified by the deep learning model that were missed by the blob detection algorithm. These new particles are marked with yellow arrows, indicating the model's superior performance in detecting smaller and less distinct nanoparticles. The deep learning model's enhanced detection capability is evident in the increased number of detected particles, particularly the smaller ones that the blob detection algorithm failed to recognize. This improvement demonstrates the model's ability to learn from the labeled data and generalize its knowledge to detect nanoparticles more accurately on the validation set.
Fig. 5 presents a detailed analysis of detected nanoparticles, combining visual examples with statistical summaries to illustrate the model's detection capabilities. Fig. 5(a) displays an original SEM image alongside the detected nanoparticles, demonstrating the model's proficiency in accurately identifying nanoparticles against a complex background. Fig. 5(b) provides histograms of various shape statistics for the detected nanoparticles in Fig. 5(a). The particle size histogram reveals that most nanoparticles range between 2 and 10 nm, indicating the model's ability to detect a broad size spectrum. The circularity histogram shows that most particles have circularity values between 0.7 and 0.9, suggesting that most nanoparticles are nearly round. The convexity histogram illustrates that detected nanoparticles generally have convex shapes, with values clustered around 0.8 to 1.0. The inertia ratio histogram indicates a diverse distribution of mass within the nanoparticles, with a peak around 0.7 to 0.8, reflecting the model's effectiveness in identifying particles with varying mass distributions. Fig. 5(c) showcases enlarged images of 50 detected nanoparticles in Fig. 5(a), emphasizing the diversity in shape and size. These examples underline the model's robustness in identifying nanoparticles with different characteristics, further validating its reliability in real-world applications.
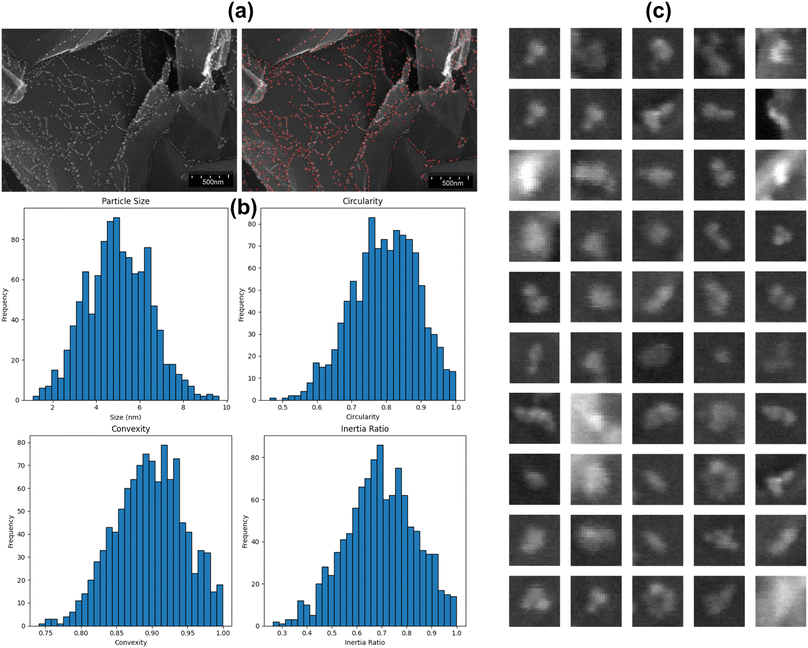 | ||
| Fig. 5 (a) Original SEM image and detected nanoparticles. (b) Shape statistics of detected nanoparticles. (c) Enlarged examples of detected nanoparticles. | ||
Table 4 provides a comprehensive summary of key statistical metrics for the detected nanoparticles in the validation dataset. The particle size was determined from the dimensions of the bounding box, while circularity, convexity, and inertia ratio were inferred by analyzing the geometric properties of the detected particles within the bounding boxes. The particle size statistics show a mean size of 5.54 nm, with a median of 5.18 nm and a standard deviation of 3.22 nm, spanning a range from 0.52 to 10.48 nm. Circularity metrics reveal a mean value of 0.82 and a median of 0.86, with most values falling between 0.54 and 0.99. The convexity of nanoparticles shows a mean of 0.89 and a median of 0.91, with a standard deviation of 0.06, indicating that most detected particles are generally convex. The inertia ratio has a mean of 0.75 and a median of 0.70, with values ranging from 0.36 to 0.99, showcasing the diversity in mass distribution within the detected nanoparticles. The combination of visual examples and detailed statistical summaries confirms the model's ability to detect a wide range of nanoparticles accurately. This comprehensive analysis demonstrates the model's effectiveness and reliability, making it a valuable tool for nanoparticle characterization in SEM images.
| Statistics | Particle size | Circularity | Convexity | Inertia ratio |
|---|---|---|---|---|
| Mean | 5.54 nm | 0.82 | 0.89 | 0.75 |
| Median | 5.18 nm | 0.86 | 0.91 | 0.70 |
| Standard deviation | 3.22 nm | 0.12 | 0.06 | 0.15 |
| Range | 0.52–10.48 nm | 0.54–0.99 | 0.74–0.99 | 0.36–0.99 |
To validate the reliability of our results, including the detected nanoparticle images and the associated morphological statistics, we manually labeled 10 random images for evaluation purposes. This manual labeling was carried out by experts in the field to ensure accuracy. Using these manually labeled images as ground truth, we compared the proposed method with thresholding and blob detection techniques in nanoparticle detection tasks. The comparison results, detailed in Table 5, reveal that the proposed method significantly outperforms both thresholding and blob detection.
| Metric | Thresholding | Blob detection | Proposed method |
|---|---|---|---|
| Precision | 0.78 | 0.86 | 0.92 |
| Recall | 0.54 | 0.79 | 0.95 |
| Mean absolute error for centroid location | 1.22 nm | 0.42 nm | 0.35 nm |
In terms of precision, the proposed method achieved a score of 0.92, which is substantially higher than the 0.86 obtained by blob detection and the 0.78 from thresholding. This indicates that the proposed method is more effective in accurately identifying true nanoparticles and reducing false positives. For recall, the proposed method scored 0.95, surpassing the 0.79 of blob detection and the 0.54 of thresholding, demonstrating its superior capability in detecting all actual nanoparticles and minimizing false negatives. Additionally, the proposed method exhibited the lowest mean absolute error for centroid location at 0.35 nm, compared to 0.42 nm for blob detection and 1.22 nm for thresholding. This highlights its enhanced spatial accuracy in locating nanoparticles.
3.3. Morphological analysis
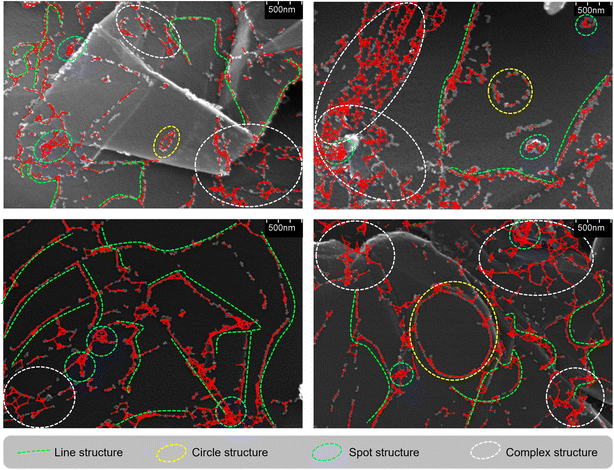
This section illustrates detailed morphological analyses of nanoparticles observed through SEM images obtained from the proposed model. Fig. 6 visualizes the structured nanoparticles by highlighting different types of formations within the GB network. The red markings in the images highlight the edges identified by the GB network, forming the basis of the network analysis. Line structures, represented by green dashed lines, depict elongated, linear formations. Circle structures, shown with yellow dashed circles, indicate circular or ring-like features. Spot structures, marked with cyan dashed circles, represent small, isolated spots likely corresponding to tiny clusters. Complex structures, enclosed within white dashed ellipses, demonstrate intricate formations that combine elements of lines, circles, and spots, indicating highly interconnected networks. These visual representations help in distinguishing and categorizing various structural formations within the nanoparticle ensemble, facilitating a deeper understanding of their spatial distribution and connectivity.Table 6 presents statistical data derived from the GB network constructed using the validation dataset. The degree metric shows a mean of 3.1, suggesting that each node (nanoparticle) has, on average, about three connections, with a range from isolated nodes (degree 0) to highly connected ones (degree 11). The clustering coefficient, with a mean of 0.58, indicates a moderate level of clustering, suggesting that features tend to form localized clusters within the network. The path length metric reveals a mean of 14.6 nm, providing insight into the typical separation between connected features, with a range of 1 to 30 nm, indicating variability from very short to relatively long connections. This analysis offers a comprehensive understanding of the structural characteristics and connectivity patterns within the nanoparticle network, shedding light on their morphological complexity.
| Statistics | Degree | Clustering coefficient | Path length |
|---|---|---|---|
| Mean | 3.1 | 0.58 | 14.6 nm |
| Median | 2.9 | 0.56 | 16.8 nm |
| Standard deviation | 1.8 | 0.22 | 8.3 nm |
| Range | 0–11 | 0–1 | 1–30 nm |
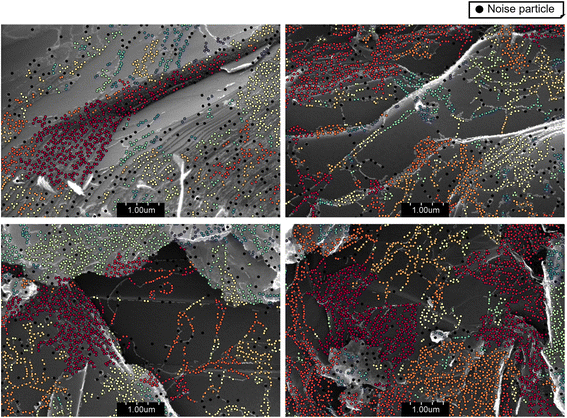
Fig. 7 illustrates the clustering of nanoparticles based on the DBSCAN algorithm. The visualization, on SEM images derived from the proposed detection model, shows the clusters formed by the DBSCAN algorithm. Different clusters are highlighted using various colors, with noise particles indicated by black dots. This method allows for the identification of distinct groups of nanoparticles and provides insight into their distribution and density. The application of DBSCAN for morphological analysis of unstructured nanoparticles has proven effective in clustering and identifying noise in SEM images.
Table 7 presents a detailed statistical analysis of the clustering results from the validation dataset. The data indicates that the mean number of clusters identified is approximately 24.8, with an average cluster size of 367 nanoparticles. The percentage of noise, or particles not belonging to any cluster, is relatively low, averaging around 3.5%. These results suggest that the DBSCAN algorithm is effective in distinguishing meaningful clusters from background noise, providing a clear and organized view of nanoparticle distribution. The median values offer further insights, with a median cluster count of 26.9 and a median cluster size of 314 nanoparticles. This slight variation from the mean indicates some degree of skewness in the data, possibly due to the presence of a few very large or very small clusters. The standard deviation values highlight the variability in cluster size and noise percentage, with standard deviations of 12.2 and 102, respectively, for the number of clusters and cluster size, and 1.0 for the percentage of noise. The range of values observed 2 to 42 clusters, cluster sizes from 2 to 785 nanoparticles, and noise percentages from 0.7% to 6.5% demonstrates the diversity in the dataset and the ability of the DBSCAN algorithm to adapt to different clustering scenarios. The statistical data supports the robustness of the clustering method, with low noise percentages and consistent identification of clusters.
| Statistics | Number of clusters | Cluster size | Percentage of noise |
|---|---|---|---|
| Mean | 24.8 | 367 | 3.5 |
| Median | 26.9 | 314 | 3.6 |
| Standard deviation | 12.2 | 102 | 1.0 |
| Range | 2–42 | 2–785 | 0.7–6.5 |
3.4. Implications for future research
The findings of this research have several important implications for future studies in the field of nanotechnology and materials science. Firstly, the successful application of deep learning for nanoparticle detection opens new avenues for automating and enhancing the accuracy of nanoparticle analysis in SEM images. Future research could focus on refining and expanding the deep learning model to detect a wider variety of nanoparticles across different substrates and imaging conditions. Additionally, integrating this model with other imaging techniques, such as transmission electron microscopy (TEM) or atomic force microscopy (AFM), could provide a more comprehensive understanding of nanoparticle morphology and behavior.The use of the GB network for structural analysis and DBSCAN for clustering offers a robust framework for understanding nanoparticle organization. Further research could explore the application of these techniques to different types of nanoparticles and composite materials, potentially leading to the discovery of new material properties and behaviors. Moreover, advancing the clustering algorithms to include dynamic and temporal analysis could provide insights into the formation and evolution of nanoparticle clusters over time, which is crucial for applications in catalysis, drug delivery, and sensor technologies.33
Another important direction for future research is the investigation of the relationship between nanoparticle morphology and their functional properties. By correlating the morphological data obtained from this study with experimental measurements of catalytic activity, electrical conductivity, or other relevant properties, researchers can develop a deeper understanding of how nanoparticle structure influences their performance. This knowledge could guide the design and synthesis of nanoparticles with tailored properties for specific applications.
4. Conclusion
In this study, we have successfully demonstrated a novel approach for the morphological analysis of Pd/C nanoparticles using SEM imaging combined with advanced deep learning and clustering techniques. Our deep learning detection model effectively identified and delineated small nanoparticles within unlabeled SEM images, showcasing high precision and reliability in nanoparticle detection. The subsequent application of a GB network facilitated the structural analysis of these nanoparticles, while the use DBSCAN provided valuable insights into the clustering patterns and distributions of the detected nanoparticles. Our findings underscore the potential of integrating deep learning models with clustering algorithms to automate and improve the accuracy of nanoparticle analysis. This approach can significantly advance research in nanotechnology and materials science by providing a more detailed and quantitative understanding of nanoparticle structures and distributions. In the future, we will consider the shape of nanoparticles to enhance our understanding of their distribution and properties.Data availability
Data for this article, including Scanning Electron Microscopy (SEM) images are available at Figshare at: https://doi.org/10.6084/m9.figshare.11783661.Conflicts of interest
There are no conflicts of interest to declare.Acknowledgements
This research is funded by Hanoi University of Science and Technology (HUST) under project number T2023-TĐ-003.References
- V. Bommakanti, M. Banerjee, D. Shah, K. Manisha, K. Sri and S. Banerjee, An overview of synthesis, characterization, applications and associated adverse effects of bioactive nanoparticles, Environ. Res., 2022, 214, 113919 CrossRef.
- D. MubarakAli, H. Kim, P. S. Venkatesh, J. W. Kim and S. Y. Lee, A Systemic Review on the Synthesis, Characterization, and Applications of Palladium Nanoparticles in Biomedicine, Appl. Biochem. Biotechnol., 2023, 195, 3699–3718 CrossRef.
- S. Dey and G. C. Dhal, Highly Active Palladium Nanocatalysts for Low-Temperature Carbon Monoxide Oxidation, Polytechnica, 2019, 3, 1–25 CrossRef.
- C. K. Y. Law, L. Bonin, B. De Gusseme, N. Boon and K. Kundu, Biogenic synthesis of palladium nanoparticles: new production methods and applications, Nanotechnol. Rev., 2022, 11, 3104–3124 CrossRef.
- M. Alaqarbeh, S. F. Adil, T. Ghrear, M. Khan, M. Bouachrine and A. Al-Warthan, Recent Progress in the Application of Palladium Nanoparticles: A Review, Catalysts, 2023, 13, 1343 CrossRef.
- R. Abbasi, G. Shineh, M. Mobaraki, S. Doughty and L. Tayebi, Structural parameters of nanoparticles affecting their toxicity for biomedical applications: a review, J. Nanopart. Res., 2023, 25, 1–35 CrossRef.
- V. Harish, M. M. Ansari, D. Tewari, A. B. Yadav, N. Sharma, S. Bawarig, M. L. García-Betancourt, A. Karatutlu, M. Bechelany and A. Barhoum, Cutting-edge advances in tailoring size, shape, and functionality of nanoparticles and nanostructures: a review, J. Taiwan Inst. Chem. Eng., 2023, 149, 105010 CrossRef.
- A. Haider, M. Ikram and A. Rafiq, Characterization of Nanomaterials, in Green Nanomaterials as Potential Antimicrobials, 2023, pp. 61–86 Search PubMed.
- R. B. Patil and A. D. Chougale, Analytical methods for the identification and characterization of silver nanoparticles: a brief review, Mater. Today Proc., 2021, 47, 5520–5532 CrossRef.
- R. M. Patil, P. P. Deshpande, M. Aalhate, S. Gananadhamu and P. K. Singh, An Update on Sophisticated and Advanced Analytical Tools for Surface Characterization of Nanoparticles, Surf. Interfaces, 2022, 33, 102165 CrossRef CAS.
- M. Botifoll, I. Pinto-Huguet and J. Arbiol, Machine learning in electron microscopy for advanced nanocharacterization: current developments, available tools and future outlook, Nanoscale Horiz., 2022, 7, 1427–1477 RSC.
- R. Aversa, P. Coronica, C. De Nobili and S. Cozzini, Deep Learning, Feature Learning, and Clustering Analysis for SEM Image Classification, Data Intelligence, 2020, 2, 513–528 CrossRef.
- G. Piazza, C. Valsecchi and G. Sottocornola, Deep Learning Applied to SEM Images for Supporting Marine Coralline Algae Classification, Diversity, 2021, 13, 640 CrossRef.
- M. Ge, F. Su, Z. Zhao and D. Su, Deep learning analysis on microscopic imaging in materials science, Mater. Today Nano, 2020, 11, 100087 CrossRef.
- F. López de la Rosa, R. Sánchez-Reolid, J. L. Gómez-Sirvent, R. Morales and A. Fernández-Caballero, A Review on Machine and Deep Learning for Semiconductor Defect Classification in Scanning Electron Microscope Images, Appl. Sci., 2021, 11, 9508 CrossRef.
- Z. Liu, L. Jin, J. Chen, Q. Fang, S. Ablameyko, Z. Yin and Y. Xu, A survey on applications of deep learning in microscopy image analysis, Comput. Biol. Med., 2021, 134, 104523 CrossRef PubMed.
- A. Cid-Mejías, R. Alonso-Calvo, H. Gavilán, J. Crespo and V. Maojo, A deep learning approach using synthetic images for segmenting and estimating 3D orientation of nanoparticles in EM images, Comput. Methods Progr. Biomed., 2021, 202, 105958 CrossRef PubMed.
- B. Lee, S. Yoon, J. W. Lee, Y. Kim, J. Chang, J. Yun, J. C. Ro, J. S. Lee and J. H. Lee, Statistical Characterization of the Morphologies of Nanoparticles through Machine Learning Based Electron Microscopy Image Analysis, ACS Nano, 2020, 14, 17125–17133 CrossRef CAS PubMed.
- Y. Xu, D. Xu, N. Yu, B. Liang, Z. Yang, M. S. Asif, R. Yan and M. Liu, Machine Learning Enhanced Optical Microscopy for the Rapid Morphology Characterization of Silver Nanoparticles, ACS Appl. Mater. Interfaces, 2023, 15, 18244–18251 CrossRef CAS PubMed.
- H. Wen, J. M. Luna-Romera, J. C. Riquelme, C. Dwyer and S. L. Y. Chang, Statistically Representative Metrology of Nanoparticles via Unsupervised Machine Learning of TEM Images, Nanomaterials, 2021, 11, 2706 CrossRef CAS.
- Z. Sun, J. Shi, J. Wang, M. Jiang, Z. Wang, X. Bai and X. Wang, A deep learning-based framework for automatic analysis of the nanoparticle morphology in SEM/TEM images, Nanoscale, 2022, 14, 10761–10772 RSC.
- B. Lee, S. Yoon, J. W. Lee, Y. Kim, J. Chang, J. Yun, J. C. Ro, J. S. Lee and J. H. Lee, Statistical Characterization of the Morphologies of Nanoparticles through Machine Learning Based Electron Microscopy Image Analysis, ACS Nano, 2020, 14, 17125–17133 CrossRef CAS PubMed.
- J. Bals and M. Epple, Deep learning for automated size and shape analysis of nanoparticles in scanning electron microscopy, RSC Adv., 2023, 13, 2795–2802 RSC.
- T. Lindeberg, Feature Detection with Automatic Scale Selection, Int. J. Comput. Vis., 1998, 30, 79–116 CrossRef.
- L. Vincent, L. Vincent and P. Soille, Watersheds in Digital Spaces: An Efficient Algorithm Based on Immersion Simulations, IEEE Trans. Pattern Anal. Mach. Intell., 1991, 13, 583–598 CrossRef.
- P. F. Felzenszwalb and D. P. Huttenlocher, Efficient graph-based image segmentation, Int. J. Comput. Vis., 2004, 59, 167–181 CrossRef.
- B. Jähne, Digital Image Processing, 1995, DOI:10.1007/978-3-662-03174-2.
- Y.-W. Lee and B.-G. Kim, Attention-based scale sequence network for small object detection, Heliyon, 2024, 10, e32931 CrossRef PubMed.
- S. Woo, J. Park, J. Y. Lee and I. S. Kweon, in CBAM: Convolutional Block Attention Module, Lecture Notes in Computer Science (Including Subseries Lecture Notes in Artificial Intelligence and Lecture Notes in Bioinformatics), LNCS, 2018, vol. 11211, pp. 3–19 Search PubMed.
- M. Ester, H.-P. Kriegel, J. Sander and X. Xu, in Proceedings of the Second International Conference on Knowledge Discovery and Data Mining, AAAI Press, 1996, pp. 226–231 Search PubMed.
- S. S. Zalesskiy and V. P. Ananikov, Pd2(dba)3 as a precursor of soluble metal complexes and nanoparticles: determination of palladium active species for catalysis and synthesis, Organometallics, 2012, 31, 2302–2309 CrossRef CAS.
- A. S. Galushko, V. V. Ilyushenkova, J. V. Burykina, R. R. Shaydullin, E. O. Pentsak and V. P. Ananikov, The Fast Formation of a Highly Active Homogeneous Catalytic System upon the Soft Leaching of Pd Species from a Heterogeneous Pd/C Precursor, Inorganics, 2023, 11, 260 CrossRef CAS.
- D. Astruc, Introduction: Nanoparticles in Catalysis, Chem. Rev., 2020, 120, 461–463 CrossRef PubMed.
| This journal is © The Royal Society of Chemistry 2024 |