Open Access Article
Open Access ArticleIn vivo and in silico evaluation of the phytoalexin-eliciting activity in common bean (Phaseolus vulgaris L.) of jasmonoyl-L-isoleucine analogs having a pyrazolidin-3-one ring†
Samuel Vizcaíno Páez ab,
Diego Durango
ab,
Diego Durango b and
Wiston Quiñones
b and
Wiston Quiñones *a
*a
aQuímica Orgánica de Productos Naturales, Universidad de Antioquia, Medellín, 050010, Antioquia, Colombia. E-mail: wiston.quinones@udea.edu.co
bQuímica de los Productos Naturales y los Alimentos, Universidad Nacional de Colombia, Medellín, 050034, Antioquia, Colombia
First published on 12th December 2024
Abstract
Jasmonates are phytohormones derived from jasmonic acid that regulate metabolic processes involved in the chemical response of plants to biotic and abiotic stress. As part of this response, some species synthesize de novo compounds with biological activity against some pathogens. In this work, nine analogs of jasmonoyl-L-isoleucine containing a pyrazolidin-3-one core were tested in their activity to elicit the production of phytoalexins (daidzein, genistein, coumestrol, and phaseollin) in common bean (Phaseolus vulgaris L.) cultivars when added exogenously. Some variations in selected parts of the analogs, such as the side chain, the linker, or the conjugated amino acid, allowed the establishment of qualitative relations with the observed activity. The analogs were tested at two levels of concentration, and the observed activity was, in most cases, higher than the observed for methyl jasmonate at 0.5 mM, even at the lower level. Seedlings treated with most heterocyclic compounds exhibited significantly higher amounts of phaseollin than untreated seedlings. Jasmonoyl-L-isoleucine analogs having a pyrazolidin-3-one ring trigger the production of phytoalexins and can be used for crop protection. Additionally, the protein-complex receptor involved in the jasmonate signaling mechanism was modeled by homology for P. vulgaris, using that for Arabidopsis thaliana as a template. After being modeled, it was assessed and used to qualitatively correlate the observed activity values and the vina scores from the docking of the tested analogs.
1 Introduction
Jasmonates are a group of lipid-derived signalling ubiquitous phytohormones, including jasmonic acid (JA) and its derivatives (e.g. methyl jasmonate, MeJA; certain amino acid conjugates like jasmonoyl-L-isoleucine, JA-Ile; decarboxylated forms; glucose esters and hydroxylated forms). The JA-Ile has been reported to be the bioactive form of jasmonates in plants, as it acts as a ligand for the jasmonate receptor (the F-box protein CORONATINE INSENSITIVE 1 (COI1) and repressor protein JASMONATE ZIM-DOMAIN (JAZ) to form the COI1-JAZ coreceptor system).1 Jasmonates are known to be involved in different physiological mechanisms and metabolic pathways, and specially, the activation of a number of induced defensive responses in a wide array of different plant species.2,3 Compounds with this activity are called elicitors, and many have been commercially released in some countries as plant health promoters for use in crops (e.g. the chemical inducer benzo (1,2,3)-thiadiazole-7-carbothioic acid-S-methyl ester or ASM or Bion® or Actigard®). Since jasmonates are responsible for the activation of defensive responses against different biotic stresses including pests and diseases in plants, it was suggested that exogenous application of jasmonates could be an efficient alternative to chemical agents that act through biocidal mechanisms in food production systems.2 Several studies have shown that foliar spraying of plants and post-harvest treatment of harvested crops with jasmonates effectively improve the resistance mechanisms of the plant and the harvested crops.2 MeJA, the most widely studied and most commercially available jasmonate, has also been evaluated as a successful plant protection treatment in several pathogen- and pest insect systems.2 Unfortunately, exogenous application of MeJA in crop protection presents some challenges, due to its low water solubility, high volatility, strong and persistent odor, and because it can generate some phytotoxicity symptoms.4 Furthermore, since the cyclic structure (a disubstituted cyclopentanone) of jasmonates (MeJA, JA, and JA-Ile) has two chiral carbons, only certain configurations have been found to be active. Furthermore, since the cyclic structure (a disubstituted cyclopentanone) of jasmonates (MeJA, JA, and JA-Ile) has two chiral carbons, only certain configurations have been found to be active.5,6 This fact further limits the possible application of jasmonates in the field, since the synthesis and purification of the active stereoisomer can be complex and not commercially viable.On the other hand, common bean (Phaseolus vulgaris L., Fabaceae), is the most commonly consumed legume worldwide for its edible dry seeds or green, unripe pods. In fact, it is the most important legume produced for direct human consumption with a commercial value greater than that of all other leguminous crops combined. However, common bean is susceptible to many pests and diseases which significantly affect crop yield. Traditionally, the use of biocidal chemical agents (insecticides, fungicides, etc.) has been the primary approach to address these factors. New insect pest and disease control agents that act through more environmentally friendly mechanisms are needed. One such approach is the stimulation of the natural intrinsic mechanisms that plants possess to counteract the attack of insects and pathogenic microorganisms.
Chemical defenses in plants involve induced secondary metabolites (called phytoalexins) and preformed ones (called phytoanticipins). Phytoalexins have been defined as antimicrobial compounds produced by plant tissues in response to microbial infection.7,8
In the case of leguminous plants such as P. vulgaris, these molecules belong to the isoflavonoid family, subgroups isoflavones, isoflavanones, isoflavans, coumestans, and pterocarpans, whose biosynthetic routes are described in Fig. 1. According to this, the known chalcone biosynthesis is the starting point, placing the narigenin chalcone (1) as common precursor. At this point, ring-closing and aryl migration lead to the corresponding isoflavone, but the hydroxylation in position 6′ or the lack of it, branches the route to the isoflavones daidzein (2) or genistein (3). From 2, another hydroxylation in the new 6′ position is a requirement to form the five-membered ring of the coumestan coumestrol (4) or the pterocarpan phaseollin (5). All these compounds have been isolated and tested for their biological activity. For instance, Van Etten et al.11 isolated phaseollin, kievitone, phaseollidin, and phaseollinisoflavan from hypocotyls of P. vulgaris infected with Rhizoctonia solani, and the ED50 for the inhibition of the radial growth of R. solani in solid media poisoned with these phytoalexins were 18, 36, 20 and 27 μg mL−1 respectively. These authors also examined the mode of action of phaseollin in R. solani, concluding that this should act on the plasmatic membrane or affect some processes needed for membrane functions.12 Coumestrol, genistein, and daidzein isolated from infected young twigs of Erythrina crista galli showed antimicrobial activity against Bacillus brevis with MIC values in the μM range.13 Genistein is also associated with the effective development of root nodules.14
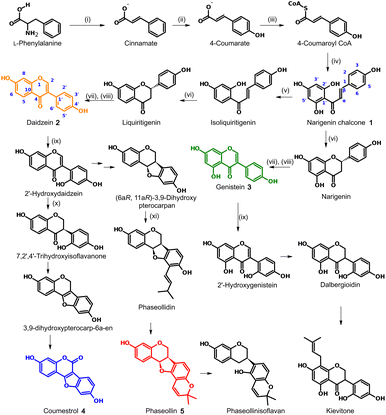 | ||
| Fig. 1 Biosynthetic pathway of the main phytoalexins in Phaseolus vulgaris.9,10 Enzymes: (i) phenylalanine ammonia-lyase (PAL); (ii) cinnamate 4-hydroxylase (C4H); (iii) 4-coumaroyl CoA ligase (4CL); (iv) chalcone synthase (CHS); (v) chalcone reductase (CHR); (vi) chalcone isomerase (CHI); (vii) isoflavone synthase (IFS); (viii) 2-hydroxyflavanone dehydratase (HID); (ix) isoflavone 2′-hydroxylase (IFH); (x) isoflavone reductase (IFR); (xi) prenyl transferase (PT). | ||
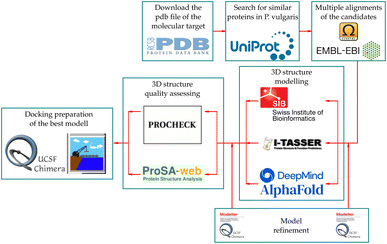
The induction of phytoalexin production in P. vulgaris has been tested with amino-sugars,15 analogs of salicylic acid,9 derivatives of indanone with alkyl and amino acidic side chains,16–19 most of them compared to the activity of methyl jasmonate (6). However, no jasmonate analogs have been tested in this species. Structures containing pyrazolidin-3-one core, similar to the synthesized analogs 7a–i (Fig. 2), have been reported as a potent and selective agonist of Prostaglandin E2 (PGE2), a group of mammal hormone-like compounds with similar biosynthetic origin as jasmonates in plants. The molecular target of jasmonates has been devised since 2009 by homology with auxin mechanism, and taking advantage of the potent activity of the phytotoxin coronatine (COR, 8) (a super strong jasmonate mimetic produced by Pseudomonas syringae) as an agonist of jasmonates, the (+)-7-iso-jasmonoyl-L-isoleucine (9) could be addressed as the main endogenous bioactive jasmonate.1 Finally, in 2010, the protein complex of COronatine-Insensitive 1 (COI1) and the degron of JAsmonate Zim-domain (JAZ) was co-crystallized with both 8 and 9 by Sheard et al.,20 enabling the study of molecular interactions through computational approaches such as docking or molecular dynamics.21 To understand the molecular interactions between jasmonates and structurally related compounds and the COI1-JAZ co-receptor in bean, a 3D model of the protein complex is demanded. In addition, a molecular modelling could be used to identify potential elicitors in common bean and may be helpful to explain the experimental results. In this work, we present the test of jasmonate analogs containing a pyrazolidin-3-one ring as potential elicitors of phytoalexin production in a specific variety of P. vulgaris, along with the construction and evaluation of the corresponding protein-complex target in the common bean by homology to that reported for Arabidopsis thaliana, and its use as jasmonate receptor for docking screening.
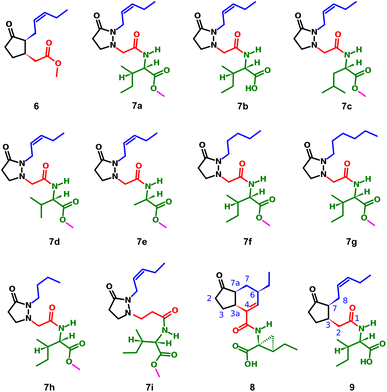 | ||
| Fig. 2 Structure of methyl jasmonate (6), the tested compounds (7a–i), coronatine (8) and (+)-7-iso-jasmonoyl-L-isoleucine (9). | ||
2 Results and discussion
2.1 Phytoalexin-eliciting activity of the pyrazolidin-3-one analogs in P. vulgaris
Fig. 3 top displays the quantification of the elicited phytoalexins as the mean of the two replicas and the standard deviations as the vertical error bars.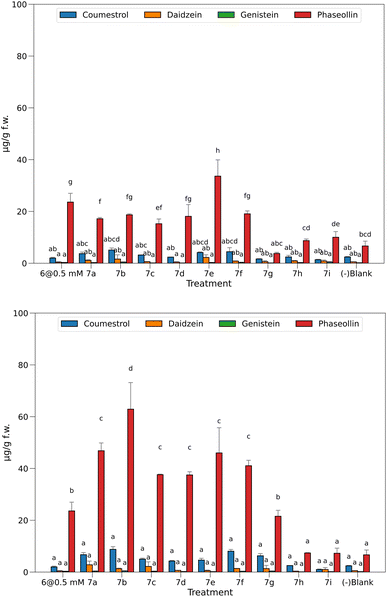 | ||
| Fig. 3 Phytoalexins from P. vulgaris produced by the treatments at 0.05 mM (top) and 0.5 mM (bottom). Bars sharing the same letter has no significant differences. | ||
With the solutions of the analogs set at 0.05 mM, the levels of isoflavones 2 and 3 were relatively low, 7e being the one with the highest production. However, these amounts do not exceed 2.2 μg g−1 f.w. for 2 or 0.4 μg g−1 f.w. for 3. According to our previous observations in time course experiments, the amount of the precursors tends to decrease after the first 24 hours as the plant produces pterocarpans, coumestans, or their prenyl derivatives in the corresponding biosynthetic route.16 The amounts of pterocarpan and coumestan were comparatively higher than that of isoflavones at this time of post-induction, reaching values of 5.1 μg g−1 f.w. for 4 when induced with 7b and 33.6 μg g−1 f.w. for 5 when 7e was the inductor.
Treatments without elicitors (-blank) were employed as witnesses to compare the base levels of phytoalexins in the seedlings. These were set to the same conditions but using 0.5% EtOH solution instead of the potential elicitor. This way, a base amount of coumestan and pterocarpan was observed, yielding 2.4 μg g−1 f.w. of 4 and 6.7 μg g−1 f.w. of 5. These values constitute a baseline for these compounds, demonstrating that the chosen variety of P. vulgaris (ICA Quimbaya) was sensitive enough to the unavoidable stress conditions of the experiment.
In general, the amount of phytoalexins in common bean seedlings treated with methyl jasmonate and the heterocyclic analogs of JA-Ile was dependent on the concentration of the potential elicitor. With the solutions of the heterocyclic analogs set at 0.05 mM, the levels of the isoflavones 2 and 3 were relatively low, and did not exceed 2.2 μg g−1 f.w. for 2 or 0.4 μg g−1 f.w. for 3. The amounts of pterocarpan and coumestan were comparatively higher than that of isoflavones at this time of post induction, reaching values of 5.1 μg g−1 f.w. for 4 when induced with 7b and 33.6 μg g−1 f.w. for 5 when 7e was the inductor. These findings are consistent with previous research on elicitation in common bean in time course experiments, when the amount of the precursor isoflavone tends to decrease after the first 24 hours as the plant produces pterocarpans, coumestan, or their prenyl derivatives in the corresponding biosynthetic route. It was reported that the difference in chemical response to diseases between susceptible and resistant common bean varieties does not lie in the concentration of precursors, but is more marked in the later stages of the biosynthetic pathway. Thus, it was proposed that phaseollin may be a chemical indicator of resistance in common bean. It has also been suggested that susceptibility in common bean to pathogenic microorganisms may be associated with failures and delays in the prenylation process, which leads to the formation of the phytoalexins phaseollin and kievitone. Methyl jasmonate (6) is a well-known elicitor of this kind of activity, a fact that was also seen in this bioassay because treatment with solutions at 0.5 mM does elicit biosynthesis of 5 up to 23.6 μg g−1 f.w., more than in untreated plants (-blank). However, it was not higher than that in 7e. Considering the significant differences between the means, a group of three treatments did not surpass the baseline (7g–i), while the other six induced the biosynthesis of 5 in a ratio of 2.3 to 5 times more than in the untreated ones.
Interestingly, concentration of 5 was significantly higher in common bean seedlings treated with 7e at 0.05 mM than with methyl jasmonate.
Fig. 3 bottom shows the results for the quantified amounts of phytoalexins after inducing with elicitors at 0.5 mM.
The higher levels of 4 and 5 compared with precursor 2 indicate a high efficiency in conversion to more advanced stages in the biosynthesis as a result of the treatments. Regarding 4 and 5, most of the tested elicitors showed a dose-dependent behavior, excepting 7h and 7i, which were also inactive at 0.05 mM. In a second group, 7g was inactive at 0.05 mM, but the increment in concentration induced the production of 5 at a similar amount (21.5 μg g−1 f.w.) than for the well-known elicitor 6 (23.6 μg g−1 f.w.). Surprisingly, the rest of the heterocyclic jasmonoyl analogs are in a third group, yielding 5 in ratios of 5.6 to 7 times the value of the untreated seedlings (-blank), and from 1.6 to 2.7 times than with the elicitor 6. Interestingly, the free acid 7b produced the highest amount of 5 at this concentration, 62.9 μg g−1 f.w., representing almost ten times more than in the negative blank.
A direct comparison of 7b with 7a reveals a 34% increment when the carboxylic group is unprotected. However, a more comprehensive analysis of the relation between the structural features and the observed activity is addressed in Fig. 4.
A fact to be considered is that the routes toward the coumestan 4 and the pterocarpan 5 come from the same precursor 2, but the tested elicitors selectively stimulated the phaseollin biosynthetic pathway.
This effect contrasts with that reported for the elicitation of common bean with salicylic acid and derivatives, and isonicotinic acid and derivatives, in which the levels of coumestrol are similar or slightly higher than phaseollin.9 Another noteworthy fact is that no necrosis of the root tissues was observed at the highest concentration tested, contrasting with previous biotests performed with synthetic analogs in our laboratory.18,19
In common bean seedlings, methyl jasmonate at 1.0 mM and above has shown symptoms of phytotoxicity.
In general, all the heterocyclic analogs of JA-Ile exhibited higher water solubility, compared to methyl jasmonate. The presence of the amino acid or its methyl ester, and the two nitrogen atoms in the cyclic system increase the polarity of the compounds and favoured the formation of hydrogen bonds with water. In addition, the aqueous solutions of pyrazolidin-3-ones did not emit strong or persistent odors. The fact that jasmonate-mediated responses are dependent on the configuration of the chiral carbons highlights the rarity of heterocyclic analogues displaying such strong phytoalexin-eliciting activity. This can be rationalized by considering that nitrogen atoms in cyclic systems can undergo pyramidal inversion, where the three groups connected to a nitrogen atom with a lone pair of electrons switch positions. This inversion affects the chirality of the molecule, causing it to become a mixture of stereoisomers. Thus, the use of pyrazolidine-3-ones as JA-Ile analogues could be an alternative to overcome the difficulties of jasmonates for field application, related to their low water solubility, high volatility, and strong odor. Additionally, the synthesis of pyrazolidine-3-ones is simpler and their amino acid conjugates are more readily available, given the complexities associated with the synthesis and purification of the active configuration of jasmonates.
2.2 Structure activity relationship (SAR)
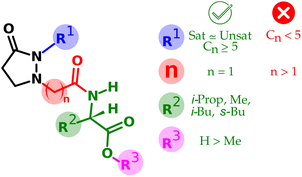
The structural variations in the tested compounds and the results of the elicitor effect of phytoalexins allowed the establishment of some relationships, also summarized in Fig. 4.• From R1: replacement of the pentenyl side chain by a saturated one only works while preserving the chain length.
• From R1: in the case of saturated side chains, the shortening drops the activity more than the lengthening.
• From R2: the change of the amino acid moiety did not alter the observed activity, at least for those tested with lipophilic side chain.
• From n: lengthening the linker between the ring and the amino acid makes the activity drop thoroughly.
• From R3: protection of the carboxylic group of the amino acid portion is tolerated; however, such activity decreases ca. one-third of the corresponding free acid version.
2.3 Protein-complex receptor in P. vulgaris
The UniProt search based on the COI1 protein from A. thaliana (henceforth COI1_ARATH) yielded 240 hits, two of them from the species of interest with entry names V7CZF7_PHAVU and V7BBE9_PHAVU. The logo constructed with these protein sequences (see ESI†) shows high conservation between them, since the LRR motif, which allows the characteristic horseshoe conformation, is present in all of them.The pdb file of COI1_ARATH has a small section without coordinates (R549-E563), which does not directly interfere with the ligand–protein or protein–protein interactions of the complex but matters to serve as a modeling template. To correct this, this random coil was modeled using the MODELLER tool of UCSF Chimera, choosing the one with the lowest zDOPE value (−1.27) that does not clash with the JAZ degron. Afterward, these two candidates were modeled using the three online tools, giving six models with the following code names: V7CZF7_PHAVU_SM, V7BBE9_PHAVU_SM, V7CZF7_PHAVU_IT, V7BBE9_PHAVU_IT, V7CZF7_PHAVU_AF and V7BBE9_PHAVU_AF. The values from the model quality assessment of the six proteins (Table 1) show that less than 2% of the residues are in disallowed regions according to the phi (ϕ) and psi (ψ) dihedral angles. In addition, G-factors from IT models are more negative than the others, by the higher percentage (0.80% and 1.50%) of residues in disallowed regions. Meanwhile, the models from AF had the highest G-factors, even more than the template protein used as a reference. However, the mean square shows how distant a model is to the reference, helping to choose the model that resembles the most COI_ARATH, in this case, V7CZF7_PHAVU_SM.
| Protein | Seq. identity | Ramachandran plot | G-Factors | Z-Score | Mean square | |||||
|---|---|---|---|---|---|---|---|---|---|---|
| Core% | Allowed% | Generally allowed% | Disallowed% | Dihedrals | Covalent | Overall | ||||
| COI1_ARATH | 100.0 | 80.0 | 19.2 | 0.80 | 0.00 | −0.41 | 0.47 | −0.07 | −8.54 | 0.000 |
| V7CZF7_PHAVU_SM | 68.9 | 86.2 | 13.3 | 0.40 | 0.20 | −0.35 | −0.01 | −0.20 | −8.69 | 0.042 |
| V7CZF7_PHAVU_IT | 68.9 | 73.7 | 23.6 | 1.90 | 0.80 | −0.75 | −0.05 | −0.45 | −8.50 | 0.071 |
| V7CZF7_PHAVU_AF | 68.9 | 89.4 | 10.4 | 0.20 | 0.00 | −0.11 | 0.30 | 0.06 | −9.26 | 0.085 |
| V7BBE9_PHAVU_SM | 70.7 | 86.2 | 13.4 | 0.40 | 0.00 | −0.36 | −0.01 | −0.21 | −8.90 | 0.053 |
| V7BBE9_PHAVU_IT | 70.7 | 75.7 | 20.8 | 1.90 | 1.50 | −0.66 | −0.04 | −0.39 | −8.10 | 0.079 |
| V7BBE9_PHAVU_AF | 70.7 | 88.9 | 10.7 | 0.40 | 0.00 | −0.12 | 0.30 | 0.06 | −9.25 | 0.082 |
In addition to the quality values, the characterization of the amino acid residues within the pocket gave an impression of how similar this critical part is, compared to COI1_ARATH. In Table 2 there is a list of the residues within a sphere of 4.0 Å radius from the centroid of JA-Ile in the crystal. Here, the absence of one or more residues makes a difference that can be crucial for the binding energy measurements and, thereby can be a criterion for discarding a model. Thus, among the surviving models V7CZF7_PHAVU_SM was chosen, considering the similitude with COI1_ARATH observed for the quality assessment values.
| COI1 ARAT crystal | V7CZF7 PHAVU SM | V7CZF7 PHAVU AF | V7CZF7 PHAVU IT | V7BBE9 PHAVU SM | V7BBE9 PHAVU AF | V7BBE9 PHAVU IT |
|---|---|---|---|---|---|---|
| R85 | R81 | R81 | R81 | R76 | R76 | R76 |
| A86 | A82 | A82 | A82 | A77 | A77 | A77 |
| F89 | F85 | F85 | F85 | F80 | F80 | F80 |
| L91 | L87 | L87 | L87 | L82 | L82 | L82 |
| R348 | R341 | R341 | — | R336 | R336 | — |
| E350 | E343 | E343 | — | E338 | E338 | E338 |
| A384 | A377 | A377 | A377 | A372 | A372 | A372 |
| V385 | V378 | V378 | V378 | V373 | V373 | V373 |
| Y386 | Y379 | Y379 | Y379 | Y374 | Y374 | — |
| R409 | R402 | R402 | R402 | R397 | R397 | R397 |
| V411 | V404 | V404 | V404 | V399 | V399 | V399 |
| Y444 | Y437 | Y437 | Y437 | Y432 | — | Y432 |
| L469 | L462 | L462 | L462 | L457 | L457 | L457 |
| R496 | R489 | R489 | R489 | R484 | R484 | R484 |
| W519 | W513 | W513 | W513 | W508 | W508 | W508 |
For JAZ protein, the BLASTP found many orthologs of TI10A_ARATH, two for P. vulgaris with entry names V7AYI4_PHAVU and V7CCD0_PHAVU. The logo made for 20 of them (see ESI†) also depicts a highly conserved sequence, particularly in the region labeled as Jas motif. The template from A. thaliana only contains the degron, thus the orthologs in P. vulgaris were shortened as well, ensuring the inclusion of the Jas motif in all of them. Each model built for the two proteins was labeled with a suffix according to the tool used, namely _SM, _AF, or _IT. The N-terminus of the JAZ degron breaks the helical conformation between E200 and R206 to become stretched. Sheard et al. pointed out the importance of this portion for the ligand–protein interaction, consequently, refinements of the JAZ degrons of the PHAVU models were performed with MODELLER tool to fit such a conformation.20 Fig. 5 shows an overlapping of the best outcome from MODELLER for each protein model, and Table 3 summarizes the Root Mean Square Deviation (RMSD [Å] eqn (1)) calculated between the α-C of each model compared with the template.
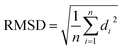 | (1) |
| Degron model | RMSD [Å] |
|---|---|
| V7AYI4_PHAVU_AF | 4.089 |
| V7AYI4_PHAVU_IT | 2.814 |
| V7AYI4_PHAVU_SM | 0.330 |
| V7CCD0_PHAVU_AF | 6.189 |
| V7CCD0_PHAVU_IT | 4.548 |
| V7CCD0_PHAVU_SM | 3.820 |
From this information, V7AYI4_PHAVU_SM stands out as the best model for the JAZ degron, and along with V7CZF7_PHAVU_SM allowed the construction of the homologous protein-complex receptor model in P. vulgaris (Fig. 6). Both proteins were combined in a single pdb file, and the coordinate system was taken from the template, for simplicity of further calculations.
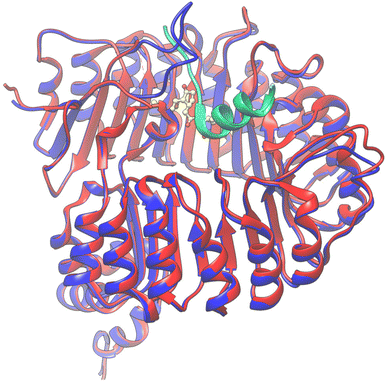 | ||
| Fig. 6 Overlapping of the 3D structures of both models interacting with 9 (yellow) in the pocket. COI1_ARATH (red), JAZ_degron_ARATH (light blue), V7CZF7_PHAVU_SM (blue) and V7AYI4_PHAVU_SM (green). | ||
2.4 Docking validation
Some redocking experiments were performed to test the ability of the built model to reproduce the pose of the natural ligands 8 and 9 in the co-crystallized protein–ligand complex. The center of the box was set as the centroid of the corresponding natural ligand (x = 0.199783, y = 4.377261 and z = 47.645130). The results were analyzed regarding RMSD, which measures the deviation of the superimposed atomic coordinates from each pose and those from the corresponding ligand in the crystal. The results were compared regarding the binding energies [kcal mol−1] and data dispersion to have an impression of expected values from a good hit. The conditions for the redocking are listed in Table 4.| Entry | Ligand | Conformation | Rounds | Box size [Å3] | Exhaustiveness |
|---|---|---|---|---|---|
| 1 | 9 | From crystal | 20 | 30 × 30 × 30 | 32 |
| 2 | 9 | From crystal | 20 | Auto | 32 |
| 3 | 9 | PM6 opt. | 20 | 30 × 30 × 30 | 32 |
| 4 | 9 | PM6 opt. | 20 | Auto | 32 |
| 5 | 8 | From crystal | 20 | 30 × 30 × 30 | 32 |
| 6 | 8 | From crystal | 20 | Auto | 32 |
| 7 | 8 | PM6 opt. | 20 | 30 × 30 × 30 | 32 |
| 8 | 8 | PM6 opt. | 20 | Auto | 32 |
The results after redock 8 and 9 in the COI1-JAZ_degron complex are shown in Fig. 7.
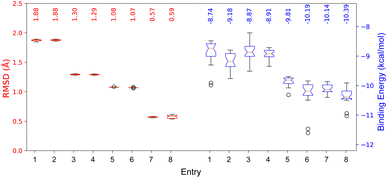 | ||
| Fig. 7 RMSD and binding energies from redocking the native ligands 8 and 9 in the COI1-JAZ_degron protein complex. | ||
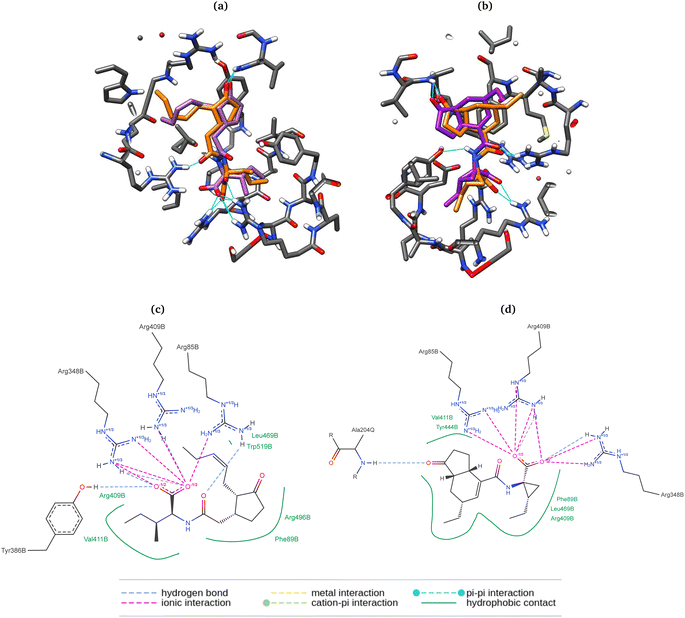
Here, the differences between the binding energy of both ligands reflect the widely observed fact that the phytotoxin 8 is more active than the native ligand 9. Regardless of taking the same conformation as in the crystallized ligands (entries 1, 2, 5, and 6), RMSD values were ca. 0.5 Å higher than those for the previously optimized geometries with the PM6 basis set. Furthermore, experiments with box size automation (even entries) showed more negative binding energies and some with less data dispersion. Another interesting observation is that the RMSD is directly affected by the number of rotating bonds of the structure since the partial rigidity of the rings of 8 allows to get poses in the pocket close to the one in the crystal. All RMSD values were below the accepted threshold of 2.0 Å, and the best results (entries 4 and 8) were obtained when the geometry was previously optimized, and the box size automated. 2D depictions of the best poses for 8 and 9 (Fig. 8) have a planar representation of the hydrogen bonds between the carboxylic groups and the arginines 85, 348, 409, as well as the hydrophobic contact of the amino acid residues, the side chain and the ring with F89, R409, V411, Y444, L469, R496, W519. The key interaction between the carbonyl group of 8 and A204 from JAZ degron is also present. The mentioned conditions were implemented for the screening with both A. thaliana and P. vulgaris receptor models. 3D overlapped representations of redocking (Fig. 8) show how close the coordinates are from the crystallized natural ligands (orange) to those of the poses achieved from redocking (purple). H-bond interactions (cyan solid lines) with amino acid residues of the pocked are also depicted.
2.5 Docking
The binding energies form the docking of the synthesized molecules and the natural ligands in both receptors are plotted in Fig. 9.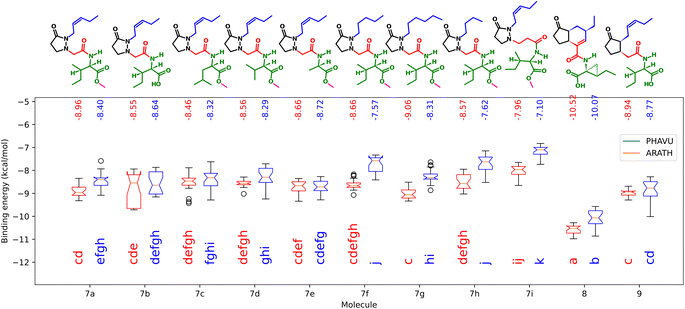 | ||
| Fig. 9 Binding energies from the synthesized molecules docked in the protein models from A. thaliana (blue) and P. vulgaris (red). Boxes sharing the same letters have no significant differences. | ||
Here, as expected, the more negative values correspond to 8, having significant differences with 9. As observed in the biological test, performance of the analogs did not change significantly when the amino acid residue was changed. For the free acid 7b, lower energy values were achieved in the case of ARATH, but the data were more dispersed for both receptors, with no significant differences with the corresponding ester 7a. On the other hand, results for 7i also correspond to the observed in the biotest, suggesting that the lengthening of the linker disfavors the protein–ligand interactions in the pocket of the P. vulgaris receptor, releasing only −7.1 kcal mol−1. In addition, the replacement of the side chain by a saturated one did not imply major changes in binding energies, according to the results in both receptors. The lower activity of compound 7h was better represented by the results from PHAVU receptor, with a behavior similar to that in the bio-assay, the shorter the chain, the worse the activity.
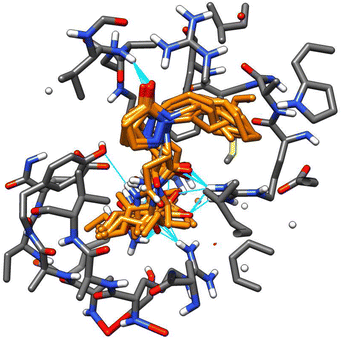
A remarkable fact is that all tested molecules showed similar performances in both receptors, with a Pearson's coefficient of 0.722 (see ESI†). An overlapped 3D representation of the best poses of all molecules (Fig. 10) allows us to appreciate the key H-bond between the pyrazolidin-3-one carbonyl and the amide of the alanine residue in JAZ degron.
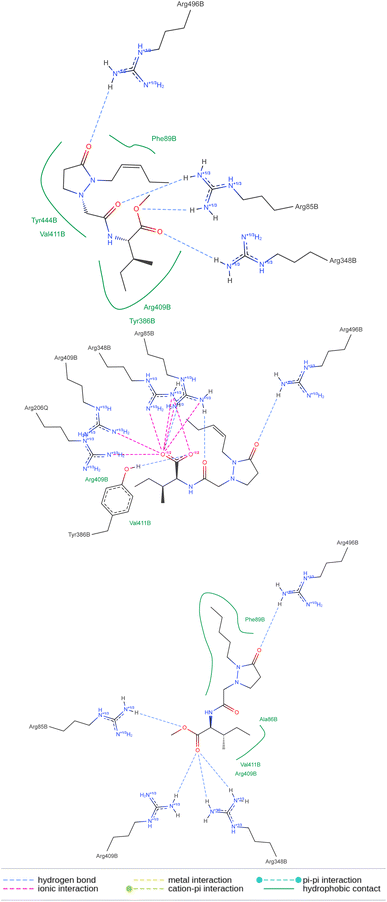
The above means that the protein receptor modeled for P. vulgaris is suitable for estimating the possible interactions if potential ligands reach the pocket of this target. The 2D representation of the docked molecules (Fig. 11) shows both sets of interactions in the active site of the protein complex, the hydrogen bonds between the carbonyls and the group of arginines, and the lipophilic contact between the bunch of methylenes in each molecule and the less polar amino acid residues. Despite the number of polar interactions being heigher for the free acid, they are not absent in the ester groups, which may justify the tolerance of this protecting group in the active site. To the best of our knowledge, the 3D structure of the protein-complex receptor in P. vulgaris is not known. It was obtained by homology modeling of the best hits from the search of orthologs of COI1 in P. vulgaris, using as a template the two co-crystalized structures available in the Protein Data Bank (PDB codes: 3ogk and 3ogl) for A. thaliana. The 3D models built were then used to have insight of the molecular interactions between these proteins and the tested ligands (JA-Ile, COR, and the pyrazolidin-3-one analogs) by the docking approach. Homology modeling using COI1-JAZ co-receptor has been used for different plant species.22–24 Our results may be helpful for further experimental studies on promising scaffolds for the development of more specific elicitors, capable of triggering the chemical defense machinery of plants without detrimental consequences for their growth.
3 Conclusions
Nine synthetic jasmonoyl-L-isoleucine hetero-analogs were tested as elicitors of phytoalexin production in P. vulgaris. Most of the tested molecules elicited selective production of the bioactive metabolite phaseollin when added exogenously, even at a low concentration of 0.05 mM. The active molecules showed a dose-dependent behavior, and the structural differences between them allowed to establish tolerable changes such as the saturation or the elongation of the side chain, or exchange of the amino acid moiety as long as it remains lipophilic; While other changes dropped the activity, such as side chain shortening or linker elongation. The molecular receptor of jasmonate-like molecules was modeled by homology for P. vulgaris using the one of A. thaliana as a template. After assessing the built model, a docking screening of the tested molecules showed similar behavior for vina scores regarding the observed activity in the biological model. The 3D model of the protein-complex receptor in P. vulgaris provides novel insight into the molecular interactions among JA-Ile receptor, COR, and a series of pyrazolidin-3-ones that displayed phytoalexin-eliciting activity. Further studies should focus on determining the phytoalexin-eliciting potential of JA-Ile pyrazolidin-like analogs in other agriculturally important species, evaluating the effects of elicitor concentration, application time, phytotoxicity, improved resistance, among others. Furthermore, the findings reported in this article open the possibility for other heterocyclic nuclei to be considered in developing new synthetic elicitors for agriculture.4 Experimental
4.1 General
Previous work describes the synthesis of pyrazolidine-like analogs of jasmonoyl-L-isoleucine.25 Reagent-grade methyl jasmonate (6) and the standard-grade isoflavones daidzein (2) and genistein (3) were purchased in Sigma-Aldrich. The purification and characterization of coumestrol (4) and phaseollin (5) were described in a previous work.15 HPLC-grade solvents were used for chromatography runs and sample preparation. The solutions of the potential elicitors were prepared with purified type II water and bidistilled commercial ethanol. Qualitative filter paper and PTFE 0.22 μm microfilters were used to filtrate the extracts and the HPLC samples, respectively. Seeds of Phaseolus vulgaris L. ICA Quimbaya variety were provided by Semillas el bosque S.A.S. High-Performance Liquid Chromatograph (HPLC) Hitachi Chromaster with a Diode Array Detector (DAD) was used for liquid chromatography. 3D protein and ligand visualizations were done in UCSF Chimera v 1.15 (https://www.cgl.ucsf.edu/chimera/).26 The search for the orthologs of each protein was performed in the UniProt (https://www.uniprot.org/) database, while Clustal Omega (https://www.ebi.ac.uk/jdispatcher/) was the tool for multiple alignments. For protein modeling, SWISS-MODEL (https://swissmodel.expasy.org/) (SM), I-TASSER (https://zhanggroup.org/I-TASSER/) (IT), and AlphaFold (https://alphafold.ebi.ac.uk/) (AF) servers were chosen. The protein refinement was performed with the MODELLER (https://salilab.org/modeller/) tool inside UCSF Chimera software. The numerical values for the assessment of the models were obtained from the ProSA-web27 (https://prosa.services.came.sbg.ac.at/prosa.php) and the SAVES v6.0 (https://saves.mbi.ucla.edu/) servers. All docking calculations were performed in Autodock Vina v1.2.3 and the 2D diagram of the ligand–protein interactions was modeled using the PoseView (https://www.zbh.uni-hamburg.de/en/forschung/amd/server/poseview.html) online free tool.28 A list with the names, descriptions, and references is available online (https://www.rdkit.org/docs/GettingStartedInPython.html). Logos from the protein sequence alignments were performed with the WebLogo 3 (https://weblogo.threeplusone.com/create.cgi) online tool. For the ligand preparation, including input of the molecules and conversion to Mol2 and pdbqt formats, the RDKit (https://www.rdkit.org/) cheminformatics tool was used, along with Open Babel (https://openbabel.org/) the API toolkit.4.2 Biological activity on bean cultivars
A specific variety of beans was chosen to evaluate the capacity of the synthesized analogs to trigger a chemical response through phytoalexin production. The ICA Quimbaya (aka red quartz) is recognized as an anthracnose-resistant variety, a property linked to the quality of having the biochemical machinery to produce isoflavonoid phytoalexins with proven antimicrobial and antifungal activities.![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 dilution of each filtrated with MeOH.
1 dilution of each filtrated with MeOH.| Time [min] | 0.5% HOAc aq. [%] | MeOH [%] | Flow [mL min−1] |
|---|---|---|---|
| Init. | 90.0 | 10.0 | 0.7 |
| 20.0 | 40.0 | 60.0 | 0.7 |
| 25.0 | 00.0 | 100.0 | 0.7 |
| 30.0 | 00.0 | 100.0 | 0.7 |
| 33.0 | 90.0 | 10.0 | 0.7 |
| 35.0 | 90.0 | 10.0 | 0.7 |
4.3 Construction of a docking receptor
Examining the potential protein–ligand interactions between the synthesized compounds and the active site of the target in P. vulgaris requires a suitable receptor. However, there are no reports of the three-dimensional structure of this protein complex for this species. Notwithstanding, two co-crystallized structures are available in the Protein Data Bank (PDB) for A. thaliana. These are the COI1-ASK1-COR-JAZ1_degron (PDB code: 3ogk) and the COI1-ASK1-JA-Ile-JAZ1_degron (PDB code: 3ogl) complexes, with X-ray diffraction resolutions of 3.18 Å and 2.80 Å respectively. This protein complex served as a template for the construction of the homologous target in the species under study, following the workflow described in Fig. 12.4.4 Docking experiments
The ligand preparation also included the addition of hydrogens and charges and conversion to the pdbqt inputs via the Mol2 file. However, Open Babel software was used in this case.
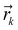 corresponds to the cartesian coordinates of each heavy atom, and
corresponds to the cartesian coordinates of each heavy atom, and 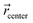 are the coordinates of the centroid of the ligand.
are the coordinates of the centroid of the ligand.
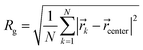 | (2) |
Data availability
The data supporting this article have been included as part of the ESI.† Source code of the software for docking automation and data processing is part of an ongoing personal, non-profit project, and it can be found here (https://github.com/samuelvp360/SVP360CalculationController/tree/new_version) with DOI: https://doi.org/10.5281/zenodo.13698825. The version of the code employed for this study is version 1.0. The data analysis script to plot the results with the Tukey test for the inductions is available in the interactive notebook Google Collab here (https://colab.research.google.com/drive/1DwaLIqt8J-pQjs8zquOKyTtSBsD5VSol?usp=sharing). The data analysis script to plot the results from docking is available here (https://colab.research.google.com/drive/1FFWQhORWVdZHdYb07raLdisMhgvSXq7x?usp=sharing). To reproduce the plots in both cases, first open the Collab session, upload the corresponding raw csv data provided in the ESI,† then run all cells.Author contributions
Concept: W. Q. F., and D. D.; experiments: S. V. P.; supervision and resources: D. D. and W. Q. F.; writing: S. V. P., D. D. and W. Q. F.Conflicts of interest
There are no conflicts to declare.Acknowledgements
The authors thanks Minciencias (Ministerio de Ciencia Tecnología e Innovación de Colombia) for financial support (Grant No. 111874558342; FP44842-057-2017).Notes and references
- S. Fonseca, A. Chini, M. Hamberg, B. Adie, A. Porzel, R. Kramell, O. Miersch, C. Wasternack and R. Solano, Nat. Chem. Biol., 2009, 5, 344–350 CrossRef CAS PubMed.
- M. Reyes-Díaz, T. Lobos, L. Cardemil, A. Nunes-Nesi, J. Retamales, M. Alberdi and A. Ribera-Fonseca, Molecules, 2016, 21(6), 567 CrossRef PubMed.
- G. Haider, T. von Schrader, M. Füßlein, S. Blechert and T. M. Kutchan, Biol. Chem., 2000, 381, 741 CAS.
- U. Hartmond, R. Yuan, J. K. Burns, A. Grant and W. J. Kender, J. Am. Soc. Hortic. Sci., 2000, 125(5), 547–552 CAS.
- L. Holbrook, P. Tung, K. Ward, D. M. Reid, S. Abrams, N. Lamb, J. W. Quail and M. M. Moloney, Plant Physiol., 1997, 114, 419–428 CrossRef CAS PubMed.
- G. Flores, G. P. Blanch and M. L. Ruiz del Castillo, Food Chem., 2013, 141, 2982–2987 CrossRef CAS.
- K. Müller and H. Börger, Experimentelle Untersuchungen über die Phytophthora – Resistenz der Kartoffel, 1939 Search PubMed.
- H. D. VanEtten, J. W. Mansfield, J. A. Bailey and E. E. Farmer, Plant Cell, 1994, 6, 1191–1192 CrossRef CAS.
- D. Durango, N. Pulgarin, F. Echeverri, G. Escobar and W. Quiñones, Molecules, 2013, 18, 10609–10628 CrossRef CAS PubMed.
- L. D. Cox, S. Munholland, L. Mats, H. Zhu, W. L. Crosby, L. Lukens, K. P. Pauls and G. G. Bozzo, Metabolites, 2021, 11, 1–25 CrossRef.
- D. A. Smith, H. D. Van Etten and D. F. Bateman, Physiol. Plant Pathol., 1975, 5, 51–64 CrossRef CAS.
- H. D. VanEtten and D. F. Bateman, Phytopathology, 1971, 61, 1363–1372 CrossRef CAS.
- F. Redko, M. L. Clavin, D. Weber, F. Ranea, T. Anke and V. Martino, Z. Naturforsch. C, 2007, 62, 164–168 CrossRef CAS PubMed.
- F. Zhang and D. L. Smith, J. Exp. Bot., 1996, 47, 785–792 CrossRef CAS.
- D. Durango, W. Quiñones, F. Torres, Y. Rosero, J. Gil and F. Echeverri, Molecules, 2002, 7, 817 CrossRef CAS.
- L. Botero, S. Vizcaíno, W. Quiñones, F. Echeverri, J. Gil and D. Durango, Biotechnol. Rep., 2021, 29, e00601 CrossRef CAS.
- D. Aristizábal, J. Gil, W. Quiñones and D. Durango, Molecules, 2022, 27, 3500 CrossRef.
- F. E. Quenguan, Derivados del acido 1-oxo-4-indanoil carboxílico como potenciales elicitores de defensas química en leguminosas cultivadas en Colombia., 2020, available at https://repositorio.unal.edu.co/handle/unal/80087.
- K. Gómez, F. Quenguan, D. Aristizabal, G. Escobar, W. Quiñones, O. García-Beltrán and D. Durango, Heliyon, 2022, 8, e08979 CrossRef.
- L. B. Sheard, X. Tan, H. Mao, J. Withers, G. Ben-Nissan, T. R. Hinds, Y. Kobayashi, F.-F. Hsu, M. Sharon, J. Browse, S. Y. He, J. Rizo, G. A. Howe and N. Zheng, Nature, 2010, 468, 400 CrossRef CAS PubMed.
- J. Yan, C. Zhang, M. Gu, Z. Bai, W. Zhang, T. Qi, Z. Cheng, W. Peng, H. Luo, F. Nan, Z. Wang and D. Xie, The Plant Cell, 2009, 21, 2220–2236 CrossRef CAS.
- F. Valenzuela-Riffo, A. Garrido-Bigotes, P. M. Figueroa, L. Morales-Quintana and C. R. Figueroa, J. Mol. Graphics Modell., 2018, 85, 250–261 CrossRef CAS PubMed.
- I. Monte, J. Caballero, A. M. Zamarreño, G. Fernández-Barbero, J. M. García-Mina and R. Solano, Proc. Natl. Acad. Sci. U. S. A., 2022, 119, e2212155119 CrossRef CAS.
- Y. Nakamura, C. Paetz, W. Brandt, A. David, M. Rendón-Anaya, A. Herrera-Estrella, A. Mithöfer and W. Boland, J. Chem. Ecol., 2014, 40, 687–699 CrossRef CAS.
- S. Vizcaíno Páez, D. Durango, C. J. Müller, M. Breuning and W. Quiñones Fletcher, RSC Adv., 2024, 4, 3790–3797 RSC.
- E. F. Pettersen, T. D. Goddard, C. C. Huang, G. S. Couch, D. M. Greenblatt, E. C. Meng and T. E. Ferrin, J. Comput. Chem., 2004, 25, 1605–1612 CrossRef CAS PubMed.
- M. J. Sippl, Proteins: Struct., Funct., Bioinf., 1993, 17, 355–362 CrossRef CAS.
- K. Stierand and M. Rarey, ACS Med. Chem. Lett., 2010, 1, 540–545 CrossRef CAS PubMed.
- W. P. Feinstein and M. Brylinski, J. Cheminf., 2015, 7, 18 Search PubMed.
Footnotes |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d4ra06461e |
| ‡ This value was selected according to previous studies in our group. |
| This journal is © The Royal Society of Chemistry 2024 |