Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Rediscovering the chemistry of the Cunninghamella species: potential fungi for metabolites and enzymes of biological, industrial, and environmental values
Hosam M. El-Seadawy†
 a,
Rehan M. El-Shabasy†b and
Ahmed Zayed
a,
Rehan M. El-Shabasy†b and
Ahmed Zayed *a
*a
aDepartment of Pharmacognosy, College of Pharmacy, Tanta University, El-Guish Street (Medical Campus), 31527, Tanta, Egypt. E-mail: ahmed.zayed1@pharm.tanta.edu.eg
bChemistry Department, Faculty of Science, Menofia University, 32512 Shebin El-Kom, Egypt
First published on 5th December 2024
Abstract
Endophytic fungi have a strong affinity for producing the same or comparable compounds to those produced by their hosts. Herein, genetic diversity and environmental adaptation of the Cunninghamella species were briefly investigated. The genetic flexibility in Cunninghamella represents an evolutionary mechanism that allows them to respond effectively to environmental changes. The current review paid much attention toward the phytochemical screening of Cunninghamella sp., revealing the presence of alkaloids, unsaturated sterols, fatty acids, polyphenols, and quinones. The intensive investigations clarified that Cunninghamella sp. are distinguished in producing several numbers of fatty acids, in particular polyunsaturated fatty acids (PUFA), in large quantities compared to other metabolites. The study demonstrated the effective role of Cunninghamella sp. in forming several bioactive metabolites owing to cytochrome P450 (CYP) that confirm significant value of such species for potential media biotransformation. The comparative investigations revealed that the isolation of flavonoids is yet to be reported, while the number of elucidated alkaloids and steroids is still limited. In contrast, successful results in the biotransformation of these metabolites were verified and showed a high affinity to convert simple substances to more valuable agents by Cunninghamella. The biomedical applications of naturally occurring compounds isolated from Cunninghamella were well documented; these included their antimicrobial, anti-cancer, anti-inflammatory, anti-Alzheimer, and antiaging properties. The antimicrobial activity was mostly attributed to the fatty acid contents in Cunninghamella sp. Moreover, tremendous attention was paid towards the agricultural and industrial usage of chitosan as it is one of the most crucial metabolites involved in wide applications. Chitosan is involved in food preservation for extending life storage period and utilized as biofertilizer, which enhances bacterial disease resistance. In addition, Cunninghamella is considered an important enzyme reservoir. Various Cunninghamella sp. produce several important enzymes, such as lignin peroxidase, catalase, cellulase, xylanase, laccase, and CYPs, that can be used for remediation, fertilization, preservation and medicinal purposes. Hence, further in-depth investigations are highly recommended to explore new insights into this potential reservoir of a wide spectrum of chemicals for industrial, medicinal, agricultural, and environmental applications.
1. Introduction
Endophytic fungi are microorganisms that reside within the tissues of the host plant without exhibiting any outward signs of illness.1 Several novel endophytic fungi have been recently isolated from different plant species and implemented for several medical and industrial purposes. Endophytes from plants are reported to be a source of several new lead compounds with potential uses in different fields, such as medicine, agriculture, environment, and industry.2 Endophytic fungi can produce metabolites like those of their host plants,2 such as flavonoids, alkaloids, terpenoids, coumarins, steroids, and lignans.3 Previously isolated fungal metabolites have verified that the diversity in biological activities encompasses antioxidant, anticancer, antimicrobial, anti-respiratory syncytial, antiproliferative and antibacterial functions.4,5. For instance, a myriad of biologically active components, including terpenoids, alkaloids and flavonoids, were isolated from the Phyllosticta sp. fungus, which showed potent antioxidant activity.6 In addition, a recent phytochemical study successfully utilized the endophytic fungi Coniolariella hispanica, Penicillium canescens, Paraphoma radicina and P. murcianum as precursors for producing cryptotanshinone as a terpene compound that considered the principal metabolite like the host Salvia abrotanoides plant.7 An additional distinctive bioactive agent was identified with 3-hydroxy-4-(hydroxy(4-hydroxyphenyl) methyl) dihydrofuran-2-on from Fusarium verticillioides, proving its antibacterial activity.8 Mellein and β-retinaldehyde were also isolated from the endophytic fungus Botryosphaeria fabicerciana, of which its potential efficacy as antioxidant and antimicrobial was revealed.9 Beside its effective role in biological activity, endophytic fungi also enhanced plant growth and development via the secretion of different enzymes involved in the biotechnological and industrial sectors.10Taking into consideration the potential attention paid towards endophytic fungi, Mucorales is the largest order.11 This order belongs to the phylum Zygomycota, subphylum Mucoromycotina11 that comprises 15 families, 57 genera and 334 species.12 Cunninghamella (family Cunninghamellaceae) was the most predominated genus belonging to Mucorales order and is one of the most potential fungi which has been investigated deeply.13 Cunninghamella was first established by Matruchot in 1903 upon collection of C. africana in the French Sudan, which would later be known as C. echinulata.14 Cunninghamella sp. has a strong ability to produce sporophores with uni-spored sporangia that are pedicellate on the vesicle surface and coated in spines. The sporophores have an uneven, pseudo-vertical, or verticillate branching shape.14 Furthermore, the majority of Cunninghamella sp. are saprobes, which are frequently founded in soil, stored grains, and other organic substrates.15 On the other hand, to identify additional species belonging to Cunninghamella, physical properties can be used, such as the colony color, texture, pattern of sporophore branching, vesicle form and size, sporangia shape and size, and the presence or absence, and length of spines in the sporangia.14 Based on the phylogenetic analyses and morphological characters, there are about 17 species of Cunninghamella that have been identified.16 In immunocompromised patients, such as those who have undergone hematopoietic stem cell transplants or hematological malignancies, some Cunninghamella sp. such as C. bertholletiae,17 C. blakesleeana,18 C. echinulata,19 C. elegans15 and C. arunalokei,20 can cause mucormycosis, an angioinvasive illness that primarily manifests as pulmonary and disseminated infections.21,22 Different Cunninghamella sp. can produce a variety of secondary metabolites with promising medical and industrial values.23 Intensive and recent investigations have triggered attention toward the successful utilization of the Cunninghamella species in drug fabrication, owing to the presence of CYP-450 monooxygenase systems that are analogous to those in mammals.24 In addition, Cunninghamella is regarded as a major source of a wide range of enzymes that can be employed in industry, bioremediation, and biotechnological features, including the biotransformation of various pharmaceutically significant substances like steroids and terpenoids,25 because it could convert the substrate into highly active compounds.26
The current review aims to highlight and extend the potential of the genus Cunninghamella in providing different classes of bioactive secondary metabolites. Interestingly, there is still limitation for direct isolation of lead compounds from Cunninghamella sp. However, most of the intensive investigations have paid more attention to the biotransformation process. Cunninghamella sp. has a strong capacity to use fewer effective compounds as the initial precursors, and convert them into structurally valuable molecules that are widely applied in drugs. This is attributed to the significant genes and enzymes included in Cunninghamella sp. that play an important role in biotransformation, and could be involved in industrial and biotechnological applications. In this review, a broad range of studies have been demonstrated for gaining comprehensive information about Cunninghamella.
2. Genetic diversity and functional capabilities of the Cunninghamella species
The genus Cunninghamella comprises several key species, including C. elegans, C. bertholletiae, and C. echinulata. These fungi exhibit significant genetic diversity, which influences their biochemical functions and potential applications in various fields.The CYP-450 genes are essential to the metabolic capabilities of Cunninghamella sp. For instance, the CYP5208A3 gene in C. elegans plays a crucial role in metabolizing a wide variety of chemical compounds.27 This gene contains coding regions that determine the protein's sequence, as well as regulatory elements such as promoters and enhancers, which control gene expression in response to environmental stimuli.28 The interaction between gene expression and other metabolic pathways enhances the enzyme's ability to chemical processing, demonstrating the organism's adaptability and potential applications, such as pollutant treatment and the development of new chemicals.29,30 Environmental adaptation is crucial for the survival and persistence of Cunninghamella sp. in diverse environments. These fungi possess a unique ability to adapt to various environmental conditions, such as temperature, humidity levels, and the presence of pollutants due to their genetic diversity, particularly CYP genes. This genetic diversity enables the fungi to process a wide range of chemical compounds, making them resilient and capable of adapting to environmental changes.31,32 For example, exposure to toxic compounds can trigger an increase in the expression of specific CYP genes, enhancing the fungi's ability to neutralize these compounds.33,34 The genetic adaptation can directly impact the survival of fungi in polluted or changing environments.35 Previous investigations suggested that these fungi possess advanced genetic modification capabilities, enhancing them to develop immediate and long-term responses against environmental changes.28 Studies revealed significant variation in the number of CYP genes amongst different Cunninghamella sp.; for example, C. bertholletiae possess 69 CYP genes, whereas C. elegans have only 32.36 The variation in gene numbers suggested a greater biochemical transformation capacity in Cunninghamella, influencing its ability to adapt to various chemical environments.26,37 Strains with extensive CYP gene families, like those found in C. bertholletiae, can efficiently degrade complex pollutants, thereby supporting their use in biotechnological and environmental contexts.38 This adaptability highlights the importance of Cunninghamella sp. in addressing pollution and chemical processing challenges.31,32
Cunninghamella sp. exhibit a remarkable ability to adapt to environmental conditions. This adaptability is attributed to the fungi's ability to modulate gene expression in response to environmental conditions. For instance, certain strains of Cunninghamella can modify their genetic makeup to develop resistance against chemical pollutants or environmental changes, giving them a competitive advantage in complex environments. Their ability to produce enzymes that can degrade environmental pollutants or transform industrial chemicals into less harmful substances is particularly notable. For example, C. elegans has been studied for its role in the biotransformation of pharmaceuticals, which highlights its potential in developing environmentally friendly waste treatment processes.
3. Previously isolated metabolites from Cunninghamella species
Cunninghamella sp. are a valuable source for a plethora of naturally occurring compounds with promising efficiency in biological, industrial, agricultural, and environmental applications. Relevant and recent studies have described the isolation of different secondary metabolites like fatty acids, sterols, phenolic acids, quinones, polysaccharides and nitrogenous compounds from Cunninghamella sp. It is worth mentioning that few compounds have been isolated from Cunninghamella sp., compared to the high numbers of biotransformed metabolites, as presented in Table 1. Cunninghamella has a strong ability to metabolize numerous chemical agents in regio- and stereo-selective strategies, according to the phase I (oxidative) and phase II (conjugative) biotransformation mechanisms. Concurrently, Cunninghamella is considered as one of the significant and distinguished biotechnological techniques regarding biotransformation.39| Cunninghamella species | Precursors | Producing metabolites | Spectroscopic analysis | Application | Ref. |
|---|---|---|---|---|---|
| a NA: not available. | |||||
| C. elegans | Coumarins | 3, 4-Dihydrocoumarin | NAa | Cytotoxic | 46 |
| Umbelliferone | |||||
| Dicoumarol | 4-Hydroxycoumarin | ||||
| C. blakesleeana | Icariin, epimedin C, epimedoside A, epimedin A, epimedin B | Icariside II | NMR | Anti-osteoporosis | 43 |
| 2-O-Rhamnosylikarisoside II | |||||
| Epimedoside b | |||||
| Baohuoside VII | |||||
| Sagittatoside B | |||||
| C. blakesleeana | Norkurarinone | Kurarinone | NAa | Cytotoxic | 44 |
| 4′′,5′′-Dihydroxykurarinone | |||||
| 6′′-Hydroxyl-2′-methoxyl-norkurarinone 7-O-β-D-glucoside | |||||
| 6′′-Hydroxylnorkurarinone 4′-O-β-D-glucoside | |||||
| 7-Methoxyl-4′′,5′′-dihydroxynorkurarinone | |||||
| C. echinulata | Kurarinone | 6′′-Hydroxykurarinone | NAa | Cytotoxicity | 45 |
| 4′′,5′′,8′′-Trihydroxynorkurarinone | |||||
| Norkurarinone | |||||
| Kurarinone 7-O-β-D-glucoside | |||||
| C. blakesleeana | Silybin | Silybin-7-sulfate | NMR | Antioxidant | 47 |
| 2,3-Dehydrosilybin-7-sulfate | |||||
3.1. Flavonoids
The bioactive flavonoids represent the most crucial phytochemicals isolated from plants, fungi and other natural sources, revealing a wide range of biological benefits for the human body. The complex structures of flavonoids make their extraction from plants difficult, and there are additional problems associated with the chemical synthesis due to the utilization of several toxic solvents. Microbial production is considered a preferable method for the isolation of these metabolites for industrial applications, as it is a more economical, sustainable and eco-friendly approach.40 However, with the advances in isolation techniques, microbial production is still limited on a laboratory scale. Furthermore, its enhancement and large-scale fabrication remain a great challenge. Interestingly, despite their potential activity and low toxicity, none of these compounds have yet been isolated directly from Cunninghamella sp., as introduced in Fig. 1. Clearly, based on the literature review, most reported studies have discussed the vital role of Cunninghamella in the successful biotransformation of less effective compounds used as the substrate and converted them into more valuable flavonoids and flavonoid derivatives to maximize their impact in drug discovery (Table 1).41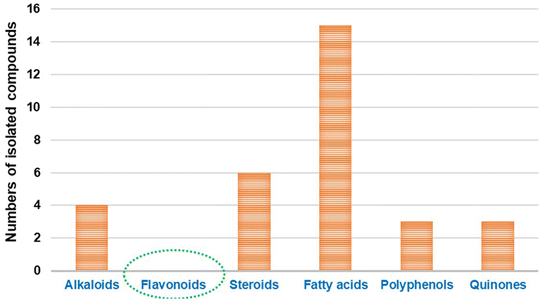 | ||
| Fig. 1 Comparison between the previously identified metabolites isolated from the Cunninghamella species. | ||
Compared to the isolated flavonoids, several numbers of bio-transformed flavonoids have been facilely produced by Cunninghamella. Noticeably, the basic reactions associated with microbial biotransformation included glycosylation/deglycosylation, carbonyl reduction, hydroxylation/dehydroxylation, O-methylation/O-demethylation, cyclization, hydrogenation/dehydrogenation, sulfation and C ring cleavage of the benzo-γ-pyrone system.42 Aspergillus, Penicillium and Cunninghamella sp. are the most prevalent genera in flavonoid biotransformation, and they are distinguished in their ability to perform nearly the entire reactions with significant yields.42 For instance, C. blakesleeana has been incorporated in the biotransformation of the principal flavonoid glycoside shown in Fig. 2, and is isolated from the herb epimedii for producing a number of rare flavonoids with excellent yield (<95%).43 Cunninghamella boosts the potential selectivity of C-7 hydrolysis to form a number of unexpected flavonoid glycosides, including icariside II (95.1%), 2-O-rhamnosylikarisoside II (97.7%), epimedoside b (93.7%), baohuoside VII (95.8%) and sagittatoside B (96.4%), as presented in Fig. 2.43 C. blakesleeana was also utilized in the biotransformation of norkurarinone to kurarinone, 4′′,5′′ dihydroxynorkurarinone, 7-methoxyl-norkurarinone, 6′′-hydroxyl-2′-methoxyl-norkurarinone-7-O-β-D-glucoside, 6′′-hydroxylnorkurarinone-4′-O-β-D-glucoside, 4′′,5′′-dihydroxykurarinone and 7-methoxyl-4′′,5′′-dihydroxynorkurarinone, as shown in Fig. 3.44 Furthermore, kurarinone was biotransferred to afford flavonoid derivatives using C. echinulata via hydroxylation, dihydroxylation on the C4′′ = C5′′, O-methylation and glycosylation reactions45 (Fig. 3) (Table 1).
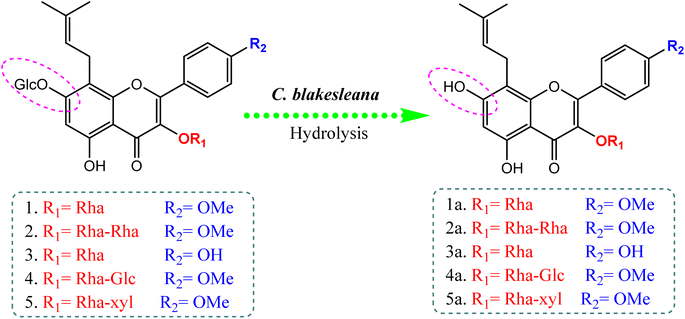 | ||
| Fig. 2 Bio-transformation of flavonoids to rare flavonoid glycosides by Cunninghamella blakesleeana via hydrolysis reaction.43 | ||
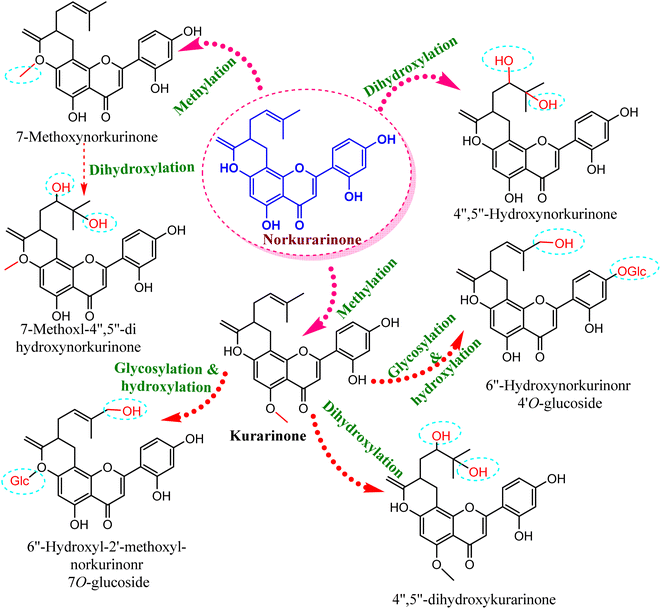 | ||
| Fig. 3 Biotransformation reaction of norkurarinone by Cunninghamella blakesleeana and C. echinulata. | ||
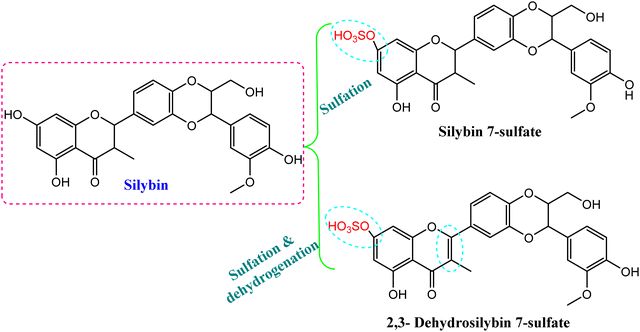
Another study has introduced the efficient metabolized coumarin into 3,4-dihydrocoumarin, umbelliferone and trans-cinnamic acid using C. elegans NRRL 1392 and dicoumarol, which was transformed into 4-hydroxycoumarin. The produced compounds were characterized by different spectroscopic techniques (e.g., NMR, mass spectrometry) and showed cytotoxic activity46 (Table 1). In addition, microbial biotransformation of the major flavolignan founded in milk thistle, silybin, by C. blakesleeana resulted in isolation of 2,3-dehydrosilybin 7-sulfate and silybin 7-sulfate, as shown in Fig. 4.47 A sulfation reaction was induced on C-7 of silybin, which potentially reduced the DPPH free radical scavenging activity. Meanwhile, dehydrogenation at C2 = C3 produced 2,3-dehydrosilybin 7-sulfate, which significantly enhanced the antioxidant activity.47
As a result, it was clearly observed that Cunninghamella sp. is distinguished for producing diverse and rare flavonoids via the biotransformation process, in comparison to the isolated metabolites.
3.2. Alkaloids
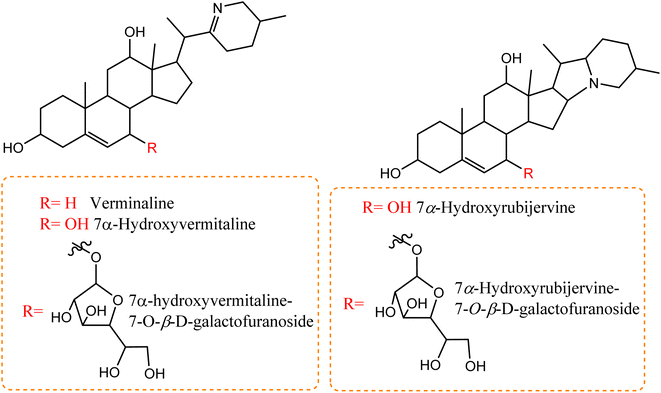
A number of recent studies have focused on the isolation of newly discovered alkaloids from plants. However, very few of these molecules have been isolated from Cunninghamella sp. For instance, Inoue et al. discussed the isolation of a new tricyclic alkaloid named ICM020 from Cunninghamella sp. F-1490.48 The structure of this compound was determined to be (3S, 10aR)-3,4a-dihydroxy-2,3,4,4a-tetrahydro-2H-pyrano[3,2-b]benzo[e]morpholine-9-carboxylic acid using various spectroscopic techniques.49 This compound was found to have osteoclastogenic inhibitory activity. Using the parathyroid hormone related peptide (PTHrP) test method and tartrate-restricted acid phosphatase (TRAP)-positive multinucleated cells formation assay method, a significant inhibitory impact was demonstrated against the development of osteoclasts in mouse bone marrow cells with an IC50 value of 0.78 μg mL−1. Furthermore, this compound showed weak cytotoxicity against bone marrow cell lines using the trypan blue exclusion assay, which suggested that its cytotoxicity was not a factor in the prevention of osteoclastogenesis.48,50 Some nucleosides and nucleotides were also reported from Cunninghamella sp., such as adenosine, uridine and uracil, which were isolated and fully characterized from C. elegans.51 Adenosine is known as an antiarrhythmic drug, and is found commercially as Adenocard®. The identified metabolites were investigated for their wound healing efficiency against pigs.51 To the best of our knowledge, this is the comprehensive series of isolated alkaloids derived from Cunninghamella sp., and future research may reveal new bioactive molecules.Compared to the isolated alkaloids from Cunninghamella, biotransformation has become the leading strategy for producing several compounds that have significant biodiversity. For example, Chalom et al. reported on the oxidative transformation of stemofoline during fermentation with C. elegans TISTR. The study resulted in three bioactive alkaloid derivatives (shown in Fig. 5) that displayed potential inhibition against acetylcholinesterase with (IC50 = 11.01 ± 1.49 mM) compared to the precursor (IC50 = 45.1 ± 5.46 mM).52 Lü et al. have also investigated the biotransformation of vermitaline via C. echinulata (ACCC 30369), producing new alkaloid derivatives that were characterized by NMR and mass spectra.53 The biotransformed compounds were identified as 7α-hydroxyvermitaline-7-O-β-D-galactofuranoside, 7α-hydroxyrubijervine-7-O-β-D-galactofuranoside, and 7α-hydroxyrubijervine, 7α-hydroxyvermitaline, as shown in Fig. 6.53
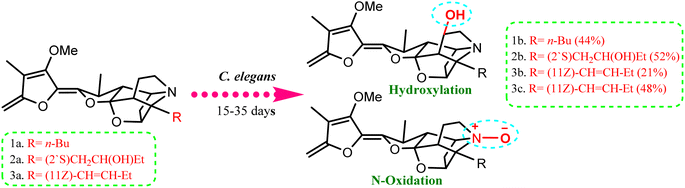 | ||
| Fig. 5 Oxidation and hydroxylation biotransformation of stemofoline alkaloids through fermentation by Cunninghamella elegans TIST 3370. | ||
Based on the literature, it was clearly noted that until now, few reports have discussed the isolation and biotransformation of alkaloids by Cunninghamella sp., despite their potential efficacy in drugs (Table 2). A huge gap is still being observed between Cunninghamella-derived alkaloids compared to the estimated number of plant-derived alkaloids represented by <60% of all potential drugs.54 Hence, further investigations regarding Cunninghamella-derived alkaloids are highly recommended, which could be a fruitful source for new bioactive agents for efficient drugs.
| Cunninghamella species | Precursors | Producing metabolites | Spectroscopic analysis | Application | Ref. |
|---|---|---|---|---|---|
| a ND: not determined. | |||||
| C. elegans TISTR 3370 | Stemofoline | (6R)-Hydroxystemofoline | NMR | Inhibitor against acetylcholinesterase | 52 |
| (2′S)-Hydroxystemofoline | |||||
| (11Z)-1′,2′-Didehydrostemofoline & 1′,2′-didehydrostemofoline-N-oxide | |||||
| C. echinulata (ACCC 30369) | Vermitaline | 7a-Hydroxyrubijervine | MS, NMR | NDa | 53 |
| 7α-Hydroxyrubijervine-7-O-β-D-galactofuranoside | |||||
| 7α-Hydroxyvermitaline | |||||
| 7α-Hydroxyrubijervine-7-O-β-D-galactofuranoside | |||||
| 6β,17β-Dihydroxy-7α,17α-dimethylestr-4-en-3-one | |||||
| 6β,10β,17β-Trihydroxy-7α,17α-dimethylestr-4-en-3-one | |||||
3.3. Fatty acids
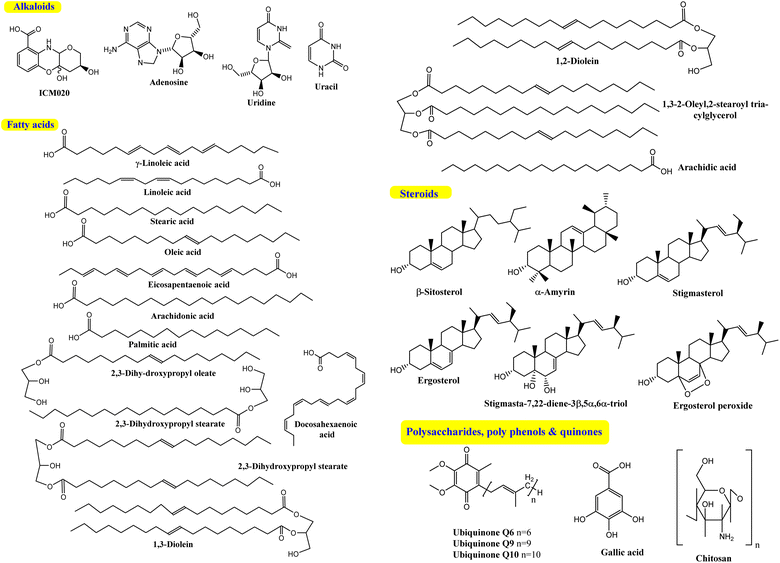
Several Cunninghamella sp., like C. elegans, C. echinulata and C. bainieri, have shown a potential affinity toward the fabrication of a plethora of lipid compounds.55 Polyunsaturated fatty acids (PUFA) can be accumulated in significant quantities by Cunninghamella, such as γ-linoleic acid (GLA).56 Different chromatographic and spectroscopic methods were used to determine the PUFA profile of various Cunninghamella sp. One of the main PUFA in Cunninghamella is GLA, which represents 21.7% and 21.1% of the total lipid content in C. echinulata and C. blakesleeana, respectively.57–59 GLA plays a crucial role in brain function and normal growth with developments.60 An additional investigation recently analyzed the fatty acid (FA) contents in C. echinulata using GC-MS. Experimentally, the chemical profile analysis demonstrated the variation percentages of several FAs; saturated FAs comprised 44.49%, monounsaturated FAs comprised 19.91%, and PUFAs comprised 35.61%. Linoleic acid recorded the highest value at 13.79%, followed by stearic (12.78%) and then oleic acid (12.50%), while GLA yielded a suitable percent (8.02%). Additional types of PUFAs appeared, including eicosapentaenoic acid, arachidonic acid, and docosahexaenoic acid.61 The production yield of FAs could be increased through optimization of the cultivation conditions. A comparative study after several trials revealed that lipids (1.43 g L−1) could be accumulated in Cunninghamella sp. under the conditions of 5 g peptone and 20 g sucrose for 9 days.62 Under the optimum conditions after 6 days, the majority of the produced FA was saturated. Meanwhile, the PUFAs were the minor component, including palmitic acid, followed by oleic acid and GLA. In contrast, the FA profile was wholly changed under different conditions (9 days, followed by 3 days at 15 °C) such that oleic acid was predominant with increasing percent of GLA from 1.14 to 3.22%.62In the continued search for isolated FAs from Cunninghamella sp., Salicorn 5 was also employed in the isolation of a number of PUFAs, like linoleic acid, GLA, oleic acid and other lipids.63 Although the amount of lipids produced by Salicorn 5 (Cunninghamella sp.) was relatively small compared to those formed in vegetables (e.g., rapeseed oil at 35–40%), the short generation time and high growth rate of fungi make their continued investigation worthwhile.63 Additional reports have investigated the isolation and identification of stearic, palmitic and oleic acids from C. blakesleeana and C. elegans extracts using column chromatography.51,64,65 Gas chromatography (GC) was also successfully employed for the characterization of several FAs in various quantities from C. blakesleeana biomass. The major FA was stearic acid (74.61%), followed by palmitic acid (10.35%), whereas the lowest percent was characterized by arachidic acid.64 Consequently, based on the relevant results, Cunninghamella sp. could be used as a commercial source for these types of secondary metabolites.
An in-depth investigation showed that Cunninghamella is superior in the isolation of several FAs, while the biotransformed compounds associated with this class have yet to be reported. Finally, it is worth mentioning that essential FAs cannot be synthesized by humans, and it must be obtained from diets. Analysis of the FA composition showed that PUFAs represented 87.03% of the total FAs, comprising oleic acid (35.57%), linoleic acid (21.58%), palmitoleic acid (16.31%), and linolenic acid (13.28%), while eicosenoic acid, stearic acid, myristic acid, and arachidic acid were found in much lower amounts.63 A relevant investigation revealed that the lipid compositions were comparable to those of the edible oils and fats; hence, Cunninghamella sp. could be successfully applied as an important source of edible oils.63
3.4. Steroids
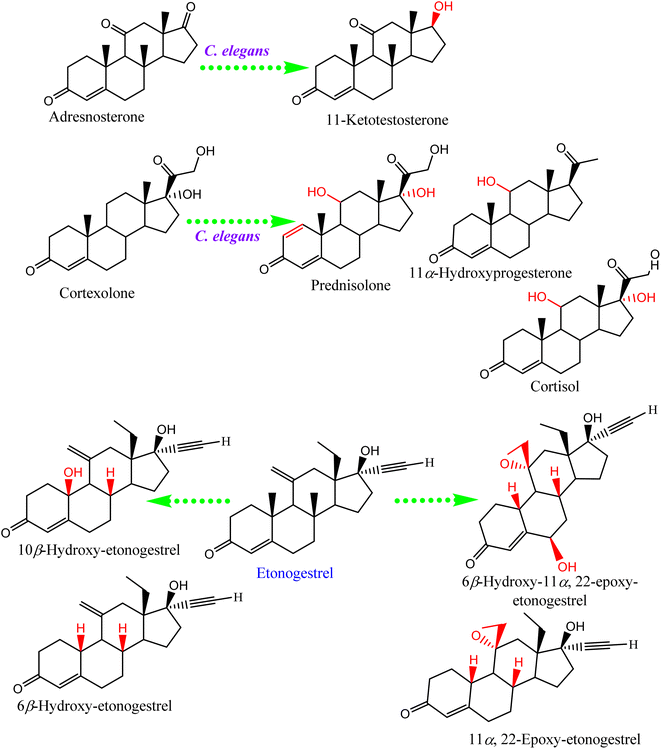
Steroids have emerged in living organisms with diverse and potential activity. Phytosterol compounds were reported from various Cunninghamella sp., including β-sitosterol and α-amyrin from C. elegans and C. blakesleeana, in addition to stigmasterol from Cunninghamella sp. In general, these compounds are commonly known in the plant kingdom as having a wide range of medical purposes.55 For instance, the leaves of Odontonema strictum are rich in β-sitosterol and stigmasterol valued at 60% and 40%, respectively.66 α-Amyrin has emerged in several plant species with potential activities (e.g., anti-inflammatory, antitumor, hepatoprotective and anxiolytic). However, the accurate determination of this compound in plant sources is still limited due to problems associated with efficient isolation, identification and quantification. Besides that, the isolation of such compounds for examination against diseases has not been specifically established.67 As a result, Cunninghamella could be an alternative source for such compounds and a suitable candidate for discovering new bioactive agents. In addition, a previous study has reported that ergosterol, stigmasta-7,22-diene-3β,5α,6α-triol and stigmasterol have been isolated from Salicorn 5 (Cunninghamella sp.).63 These compounds were also produced from halophyte Salicornia bigelovii,68 explaining that endophytic fungi have a potential affinity to create the same chemical compounds or comparable ones produced by their hosts.Conversely, biotransformation action plays a vital role in revealing the biodiversity in steroid production by Cunninghamella (Table 3). For instance, C. echinulata was involved in the isolation of ergosterol, besides two novel adipate esters from fusidic acid.69 The species was also used in a formylation reaction to obtain a unique fusidic acid derivative identified as 3-O-formyl-27-hydroxyfusidic acid.70 The chemical structures were elucidated by intensive spectroscopic methods like 1D, 2D-NMR and HRESIMS. In silico studies induced a significant agonist/antagonist effect through binding to the μ opioid receptor and antidiabetic activity via aldose reductase inhibitory action.69 It was observed that fusidic acid in mammals can be metabolized via C-3 or C-27 oxidation and glucuronide conjugation. Compared to mammals, microbes used C-3 and C-6 oxidation, C-6 and C-7 hydroxylation, and deacetylation of C-16, and then spontaneous lactone formation.71,72 Interestingly, modification on the side chain of fusidic acid rarely occurred. However, among several organisms, C. echinulata was the most efficient fungi in biotransformation, causing oxidation successfully at C-26 and C-27.73 Furthermore, a number of bioactive stereoselective derivatives were created such as fermentation of mesterolone by C. blakesleeana, producing a number of stereoselective steroids (as shown in Fig. 7a), which were investigated against different activities like anticancer, phosphodiesterase-5 enzymes, and oxidative burst.74 Moreover, three additional new steroids were obtained from the biotransformed androgenic steroid mibolerone with C. blakesleeana and C. echinulata.75 Cunninghamella sp. showed high capacity to catalyze the hydroxylation at the allylic positions of C-1, C-6, C-10, C-11, and C-20. C-6, C-10, and C-11 were the sites for β-hydroxylation, whereas α-hydroxylation occurred at C-1, as shown in Fig. 7b. The produced compounds were investigated against different activities, including β-glucuronidase inhibitory, anticancer and leishmanicidal activity.75 By the same way, mestanolone was also hydroxylated and transformed by C. blakesleeana to afford new steroidal derivatives, as shown in Fig. 7b. A number of steroidal derivatives were fabricated via microbial transformation of etonogestrel utilizing C. echinulata and C. blakesleeana, which showed cytotoxic activity.76 The transformed molecules were biosynthesized through epoxidation and hydroxylation at C-6, C-10, and C-15, whereas the epoxy ring was formed between the C-11 and C-22 positions (Fig. 8).76 Moreover, the biotransformation of adrenosterone and cortexolone resulted in the production of new derivatives with C. elegans via hydroxylation, as presented in Fig. 8.77,78 Notably, from the relevant and aforementioned studies, it was observed that steroidal biotransformation basically occurred via hydroxylation at different positions of the steroid skeleton.
| Cunninghamella species | Precursors | Producing metabolites | Spectroscopic analysis | Application | Ref. |
|---|---|---|---|---|---|
| a ND: not determined.b NA: not available. | |||||
| C. blakesleeana | Mesterolone | 1α-Methyl-1β,11β,17β-trihydroxy-5α-androstan-3-one | 1D, 2D-NMR, HRESI-MS | Anti-cancer, phosphodiesterase-5 enzymes, oxidative burst | 74 |
| 1α-Methyl-7α,11β,17β-trihydroxy-5α-androstan-3-one | |||||
| 1α-Methyl-1β,6α,17β-trihydroxy-5α-androstan-3-one | |||||
| 1α-Methyl-1β,11α,17β-trihydroxy-5α-androstan-3-one | |||||
| 1α-Methyl-11α,17β-dihydroxy-5α-androstan-3-one | |||||
| 1α-Methyl-6α,17β-dihydroxy-5α-androstan-3-one | |||||
| 1α-Methyl-7α,17β-dihydroxy-5α-androstan-3-one | |||||
| C. blakesleeana and C. echinulata | Mibolerone | 10β,17β-Dihydroxy-7α,17α-dimethylestr-4-en-3-one | 1D & 2D-NMR | β-Glucuronidase inhibitory, anticancer and leishmanicidal activity | 75 |
| 6β,17β-Dihydroxy-7α,17α-dimethylestr-4-en-3-one | |||||
| 6β,10β,17β-Trihydroxy-7α,17α-dimethylestr-4-en-3-one | |||||
| C. blakesleeana | Mestanolone | 9α,11β,17β-Trihydroxy-17α-methyl-5α-androstan-3-one | MS, 1H-, 13C and 2D-NMR, X-ray diffraction | Anticancer, immunomodulatory | 79 |
| 1β,11α,17β-Trihydroxy-17α-methyl-5α-androstan-3-one | |||||
| C. blakesleeana & C. echinulata | Etonogestrel | 6β-Hydroxy-11,22-Epoxy-etonogestrel | HREI-MS, UV, IR | Inhibition of β-glucuronidase enzyme & cytotoxic | 76 |
| 14α-Hydroxy-etonogestrel | |||||
| 10β-Hydroxy-etonogestrel | |||||
| 11,22-Epoxy-etonogestrel | |||||
| 6β-Hydroxy-etonogestrel | |||||
| Cunninghamella sp. (Salicorn 5) | — | Ergosterol, stigmasta-7,22-diene-3β,5α,6α-triol and stigmasterol | Electrospray ionization mass spectrometry (ESI-MS) | NDa | 63 |
| C. echinulata | 3-O-Formyl-27-hydroxyfusidic acid | 1D, 2D-NMR, HRESIMS | NDa | 70 | |
| C. echinulata NRRL 1382 | Fusidic acid | Ergosterol | 1D, 2D-NMR | NDa | 69 |
| C. elegans | Adrenosterone | 11-Ketotestosterone | Single-crystal X-ray diffraction | NDa | 77 |
| C. elegans | Cortexolone | Prednisolone | NAb | NDa | 78 and 79 |
| 11α-Hydroxyprogesterone | |||||
| Cortisol | |||||
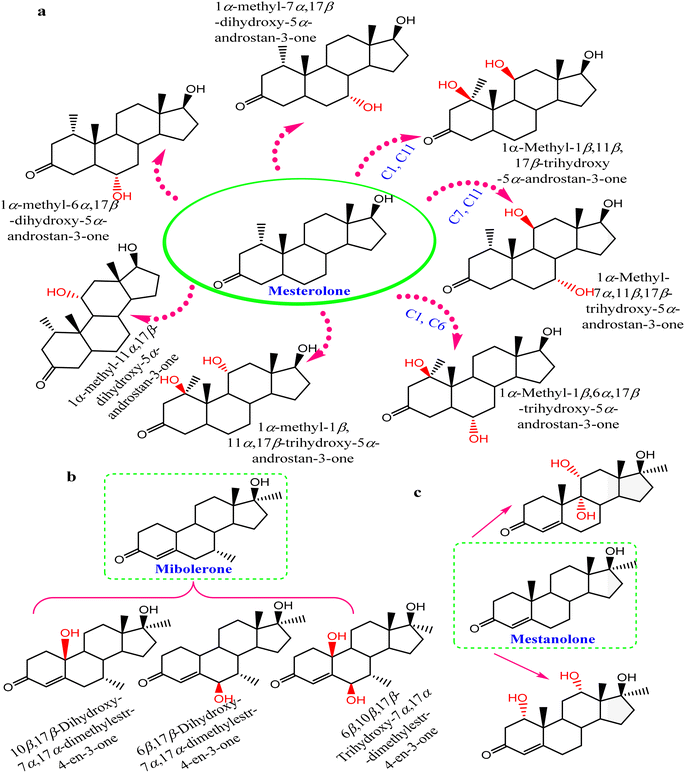 | ||
| Fig. 7 Steroidal hydroxylated biotransformation of (a) mesterolone, (b) mibolerone, and (c) mestanolone with C. blakesleeana and C. echinulata. | ||
3.5. Polysaccharides, polyphenols and quinones
Cunninghamella is thought to be a possible source for polysaccharides (Fig. 9) like chitosan, which can be recovered in significant amounts from fungal cell walls.80 Chitosan is also produced from other fungal sources, like Rhizopus arrhizus, Mucor rouxii, Aspergillus niger, Penicillium notatum and Absidia orchidis.81 Fungal chitosan is preferred over animal chitosan because the latter builds up in the renal tissue and increases the excretion of calcium in urine, which might result in kidney stone formation.82,83 Using three in vitro tests, fungal oligochitosan isolated from C. elegans was shown to exhibit more potent antioxidant activity than animal chitosan (reducing power, hydroxyl radical chelation and iron chelation) with a total antioxidant capacity of 17%, 40% and 13%, respectively. However, animal chitosan showed no activity in these tests.84 Gallic acid (GA) was also isolated from C. elegans, which induced a variety of biological activity to be described later.51Another Cunninghamella metabolite, ubiquinone, was used as a biochemical marker for the classification and identification of Cunninghamella sp.85 This methodology helps with morphological taxonomy to tackle a number of issues pertaining to classification and phylogenetics. Cunninghamella has three types of ubiquinones based on the carbon numbers in their side chain (ubiquinone Q6, Q9 and Q10), as presented in Fig. 9. Ubiquinone Q6 was found in C. bertholletie, C. elegans and C. ramosa, while ubiquinone Q9 was found in C. elegans, C. blakesleeana and C. echinulata. However, C. bertholletie and C. elegans are the only producers of ubiquinone Q10.86 On the other hand, it is worth noting that the bio-transformed compounds are still missing, and further investigations are required.
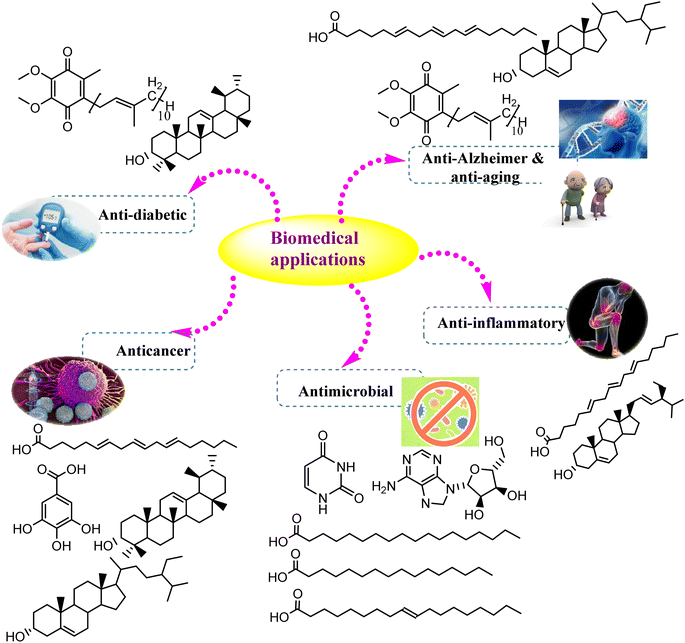
4. Biological, agricultural, environmental, and industrial applications
The secondary metabolites fabricated by endophytic fungi are well defined by their vital role in several applications. As the global population increases, the request for pharmaceutical and agricultural substances has dramatically increased. Hence, Cunninghamella sp. will gain tremendous attention in the future, particularly for the isolation of bioactive compounds.87 The current section will discuss the various biological, industrial, agricultural, and environmental values of previously isolated metabolites, and provide new insights into the recent applications of Cunninghamella sp. and as summarized in Table 4 and Fig. 10.| Antimicrobial activity | ||||||||
|---|---|---|---|---|---|---|---|---|
| Isolated compounds | Cunninghamella sp. | Microorganism | Inhibition zone (mm) | MIC (μg mL−1 | Standard | Inhibition zone (mm) | MIC (μg mL−1) | Ref. |
| Oleic acid | C. blakesleeana | Salmonella typhimurium | 6.5 ± 0 | 700 | Gentamycin | 22.6 ± 1.5 | 01.95 | 65 |
| Staphylococcus aureus | 13.0 ± 0.1 | 250 | Ampicillin | 22.00 ± 1.0 | 01.95 | |||
| C. elegans | 11.0 ± 0.3 | — | Penicillin G | 29.5 ± 0.8 | — | 91 | ||
| Stearic acid | C. blakesleeana | Salmonella typhimurium | 5.9 ± 0.9 | 750 | Gentamycin | 22.6 ± 1.5 | 01.95 | 65 |
| Staphylococcus aureus | 11.0 ± 0.3 | 360 | Ampicillin | 22.00 ± 1.00 | 01.95 | |||
| C. elegans | 15.0 ± 0.5 | — | Streptomycin | 25.0 ± 0.2 | — | 91 | ||
| Palmitic acid | C. blakesleeana | Salmonella typhimurium | 8.0 ± 0.8 | 690 | Gentamycin | 22.6 ± 1.5 | 01.95 | 65 |
| Staphylococcus aureus | 15.0 ± 0.5 | 200 | Ampicillin | 22.00 ± 1.00 | 01.95 | |||
| C. elegans | 13.0 ± 0.1 | — | Penicillin G | 29.5 ± 0.8 | — | 91 | ||
| Adenosine | C. elegans | Staphylococcus aureus | 30.0 ± 0.1 | 20 | Penicillin G | 29.5 ± 0.8 | — | 91 |
| Vancomycin | — | 0.75 | ||||||
| Uridine | C. elegans | Staphylococcus aureus | 11.0 ± 0.1 | 150 | Penicillin G | 29.5 ± 0.8 | — | 91 |
| Vancomycin | — | 0.75 | ||||||
| Gallic acid | C. elegans | Staphylococcus aureus | 5.0 ± 0.5 | 130 | Penicillin G | 29.5 ± 0.8 | — | 91 |
| Gentamicin | — | 0.35 | ||||||
| Uracil | C. elegans | Staphylococcus aureus | 7.0 ± 0.5 | 210 | Penicillin G | 29.5 ± 0.8 | — | 91 |
| Glucose fatty acid esters | C. echinulata | Bacillus subtilis | 14.1 ± 0.5 | — | Eicosapentaenoic acid | 17.0 ± 0.5 | — | 112 |
| Candida albicans | 14.3 ± 0.0 | — | 20.0 ± 0.1 | — | ||||
| Staphylococcus aureus | 14.1 ± 0.5 | — | 17.0 ± 0.2 | — | ||||
| Chitosan | C. elegans | Escherichia coli | 33.8 ± 1 | 0.375 | — | — | — | 90 |
| S. aureus | 33.2 ± 1.3 | 0.375 | — | — | ||||
| C. albicans | 23.5 ± 0.8 | 1.25 | — | — | ||||
| Penicillium expansum | 8.2 ± 0.3 | 2.75 | — | — | ||||
| Anticancer activity | ||||
|---|---|---|---|---|
| Isolated compounds | Cunninghamella sp. | Cancer cell line | IC50 (μM) | Ref. |
| GLA | C. echinulata and C. blakesleeana | HT-29 human colorectal cancer cell line | 255 | 113 |
| GA | C. elegans | SW480 and SW620 colorectal cancer cell lines | 22.39, 11.8 | 98 |
| β-Sitosterol | C. elegans and C. blakesleeana | HCT-116 colon cancer cell | 140 | 99 |
| Anti-inflammatory | |||||
|---|---|---|---|---|---|
| Isolated compounds | Cunninghamella sp. | Cell line/animals | Dose | Inflammatory mediator affected | Ref. |
| Stigmasterol | Salicorn 5 (Cunninghamella sp.) | BEAS-2B human lung epithelial cell line | 20 g mL−1 | IL-13 | 101 |
| Chitosan | C. elegans | Colonic homogenates of colitis mice | 30 mg kg−1 | TNF-α, IL6 and NF-kβ | 104 |
| Anti-Alzheimer and anti-aging | |||||
|---|---|---|---|---|---|
| Isolated compounds | Cunninghamella sp. | Experiment | Dose/IC50 | Enzymes/indicators affected | Ref. |
| GLA | C. echinulata | In silico | 7.6 × 10−5 M | Amyloid cleaving enzyme (BACE1) | 105 |
| C. blakesleeana | |||||
| Ubiquinone Q10 | C. bertholletie | In vitro | 15 μg mL−1 | Senescence-associated secretory phenotype (SASP) indicators (p21, IL-8, CXCL1, and MMP-1) | 106 |
| C. elegans | |||||
| Antidiabetic, antiplatelets and anti-hypercholesteremia | |||||
|---|---|---|---|---|---|
| Isolated compounds | Cunninghamella sp. | Experiment | Dose/IC50 | Biological effect | Ref. |
| α-Amyrin | C. elegans | In vivo | 5 and 10 mg kg−1 | Antidiabetic | 107 |
| C. blakesleeana | Anti-hypercholesteremia | ||||
| Ubiquinone Q10 | C. bertholletie | In vivo | 5 g kg−1 | Antidiabetic | 108 |
| C. elegans | Antioxidant | ||||
| GA | C. elegans | In vitro and in silico | 9.07 μmol L−1 | Antiplatelets | 109 |
| GLA | C. echinulata | In vivo | 2.88 and 7.68 g kg−1 | Anti-hypercholesteremia | 110 |
| C. blakesleeana | |||||
| β-Sitosterol | C. elegans | In vitro | 16 μM | Anti-hypercholesterolemia | 111 |
| Antidiabetic | |||||
| C. blakesleeana | Antioxidant | ||||
4.1. Biological activities
Fungal secondary metabolites are of intense interest due to their considerable biological activities.25–27 A strong relationship has been created between the pharmaceutical and biochemical industries with endophytic fungi, in particular Cunninghamella, due to the abundance of several promising metabolites that have various biological values, like anti-cancer, antioxidants, antidiabetic, anti-inflammatory and antimicrobial values (Table 4). These values offer significant potential for their usage in medicine.88Adenosine was evaluated for its antimicrobial activity, and the results indicated that adenosine was most active against S. aureus with an inhibition zone of 30 ± 0.1 mm at a concentration of 20 μg mL−1 compared to those of streptomycin and penicillin G as standard antibiotics (25 ± 0.2 and 29.5 ± 0.8 mm, respectively).51 Additionally, the topical application of 1 mg mL−1 adenosine to an experimentally excised wound surface sped up the healing process, according to a study on the wound-healing properties of adenosine isolated from C. elegans. Topical application of adenosine showed 76.5% wound correction after 14 days since treatment started, which was close to that of the standard drug latmoxef (100%). Complete wound correction by adenosine occurred after 18 days.51
Moreover, glucose esters of different fatty acids from C. echinulata were synthesized using lipases as biocatalysts. The biological activity assay of glucose fatty acid esters from C. echinulata indicated that they were efficient against Bacillus subtilis, Candida albicans, and Staphylococcus aureus at MIC 40 μg mL−1 with inhibition zones of 14.1, 14.3 and 12.3 mm, respectively. The results revealed the high pathogenic activity of fatty acid esters present in Cunninghamella sp. compared to those founded in U. isabelline. This could be attributed to GLA present in the lipids of C. echinulata, which has been recognized for its antibacterial activity in higher amounts. Furthermore, the glucose fatty acid esters from C. echinulata demonstrated notable insecticidal action against Aedes aegypti larvae with LC50 of 0.54 mg L−1. Additionally, after treatment with 10 g mL−1 of glucose esters, the SKOV-3 ovarian cancer cell line experienced a high proportion of apoptosis (39.2%).92 Furthermore, β-sitosterol exhibits potent antiviral activity against the influenza A virus. A study by Shokry et al. showed that β-sitosterol demonstrated promising antiviral efficacy against A/H1N1 and A/H5N1 strains with IC50 of 0.975 and 0.295 μg mL−1, respectively, compared to zanamivir as a positive control. It has been discovered that β-sitosterol can influence various viral replication processes, including viral adsorption and replication. The significant inhibitory impact of β-sitosterol against the hemagglutinin surface protein and neuraminidase, with docking energies of −6.40 and −29.40 kcal mol−1, was attributed as the mechanism of β-sitosterol's antiviral activity.93
The antiparasitic efficacy of α-amyrin was in vitro testing against Trypanosoma cruzi. The study showed that the isolated compound was more effective against the amastigote stage than the trypomastigote stage with an IC50 value of 9.08 μg mL−1 compared to that of the reference medication nifurtimox (3.07 μg mL−1). A molecular docking study indicated that α-amyrin has a greater affinity to T. cruzi cysteine synthase (TcCS) with a binding energy of −9.8 kcal mol−1.94
In conclusion, from the tabulated data (Table 4), it was clearly observed that most of the antimicrobial research involved very few species of Cunninghamella, e.g., C. elegans, C. echinulata and C. blakesleeana. In addition, fatty acids exhibited the highest antimicrobial activity compared to other classes. This is consistent with previous literature data, which confirmed the potential antimicrobial activity of fatty acids. However, further investigations are recommended with the incorporation of new active species of Cunninghamella, aiming to isolate new lead compounds.
To maximize the impact of the anticancer efficacy in Cunninghamella sp., a phenolic acid such as gallic acid (GA) was previously isolated from C. elegans.51 Gallic acid has a wide variety of biological activities. Recently, GA has shown strong cytotoxic effects on colon cancer cell lines. Three different types of cell lines, representing various stages of cancer severity, were used to assess GA's cytotoxic effect: colon epithelial cells CRL1790, which represent the non-tumorigenic stage; and colorectal cancer cell lines SW480 and SW620, which represent the primary tumor stage and the aggressive metastatic stage, respectively. The results revealed that GA inhibited the cell growth of SW480 and SW 620 at IC50 of 22.39 ± 2.12 and 11.8 ± 1.5 μM, respectively. Furthermore, GA showed high selectivity toward cancer cells rather than non-cancer cells due to the high IC50 (>100 μM) of GA against CRL1790 cells. Moreover, the results of the cell cycle analysis of the tested cell lines showed that GA induced prominent S and G2/M phases. GA changed the frequency of the cells from 34.2% to 43.8% at the S phase, and 7.7% to 14.5% at the G2/M phase. These results indicate that GA may affect DNA replication, inducing cell cycle arrest in the S and G2/M stages. These findings were supported by an in vivo experimental model, and immunofluorescent analysis of the tumor tissues taken from sacrificed mice showed that GA had a downregulating effect on several G-Quadruplexes (G4)-enriched oncogenes, leading to DNA damage.98
Additionally, β-sitosterol significantly inhibits colon cancer cell growth, which has the ability to suppress HCT-116 cell proliferation. The mechanism was studied, and it was found that β-sitosterol downregulates the gene and protein expression of lymphoid enhancer binding factor (LEF1), which is an oncogenic gene. In addition, it disrupts Wnt/β-catenin pathway transmission in HCT-116 at the same concentration.99 These findings indicated that Cunninghamella may be a valuable source for potential anticancer agents (Table 4).
On the other hand, several applications of chitosan were examined for the treatment of various ailments. In experimental colitis, chitosan was discovered to have anti-inflammatory properties. Using ELISA kits, Jhuundoo et al. discovered that 30 mg kg−1 of chitosan significantly decreased the levels of myeloperoxidase, alkaline phosphatase, TNF-α, IL6 and NF-kβ in the colonic homogenates of colitis mice compared to untreated mice.104
Recently, the impact of ubiquinone Q10 was potentially investigated for anti-ageing activity, and showed significant results. Ubiquinone Q10 biosynthesis decreases with age in different tissues, including the skin. However, it could be modulated by 3-hydroxy-3-methyl-glutaryl-coenzyme A (HMG-CoA) reductase inhibitors such as statins, which resulted in a senescence phenotype. Ubiquinone Q10's impact in the process of skin ageing was studied by Marcheggiani et al. using statin-pretreated cultured human dermal fibroblasts (HDF). The outcomes showed that statin-treated HDF could be prevented from developing senescence and ageing indicators, and could even be saved by ubiquinone Q10 supplementation at a dose of 15 μg mL−1. Along with increasing the extracellular matrix's components including elastin and collagen type 1, it greatly decreased several senescence-associated secretory phenotype (SASP) indicators like p21, IL-8, CXCL1, and MMP-1 (Table 4).106
Another investigation also demonstrated that ubiquinone Q10 has strong anti-inflammatory, antioxidant, and anti-diabetic properties in streptozotocin-induced diabetic rats. The findings showed that diabetic rats treated for 21 days with 5 g kg−1 in rat food experienced a significant decrease in blood glucose, IL-6, malondialdehyde (MDA), and myoglobin levels. These findings indicated that ubiquinone Q10 may have a beneficial effect on diabetes complications.108
In addition, the antiplatelet aggregation activity of GA was verified via different strategies, including surface plasmon resonance (SPR), molecular docking, molecular dynamics simulation with a thrombin inhibition assay. According to the findings, GA can inhibit thrombin with an IC50 of 9.07 μmol L−1, which in turn reduces thrombin-induced platelet aggregation by 35% when compared to cells that have been treated with thrombin. This result was brought on by the thrombin-GA equilibrium system's high binding free energy (−14.6 kcal mol−1).109
Moreover, the anti-hypercholesteremic action of GLA was investigated. It was proved that GLA can lower the body fat content by inducing the activities of liver carnitine palmitoyl transferase and peroxisomal β-oxidation for fatty acids in the liver.110 Sterols from natural sources have various biological activities.51,100 Vasanth et al. recently investigated the anti-adipogenicity of β-sitosterol. It was found that β-sitosterol reduced the viability of 3T3-L1 mouse fibroblast cells in a dose-dependent way by suppressing the cell cycle stages (particularly S and G2/M) with a considerable reduction in the intracellular lipid accumulation and an increase in the glucose uptake. Additionally, the findings demonstrated that β-sitosterol decreased the formation of reactive oxygen species from 91.65% to 42.97% at a concentration of 16 μM (Table 4).111
4.2. Cunninghamella metabolites of agricultural, environmental, and industrial values
Recently, the industrial and environmental applications of naturally occurring compounds have gained tremendous interest to produce green and sustainable substances. Chitosan is one of the most crucial compounds to be widely applied in different sectors, including food and beverages, wastewater treatment, biopharmaceutics, cosmetics and toiletries, and agricultural uses.114 The biological activity of chitosan is significantly dependent on its molecular weight and molecular weight distribution (100 < Mw < 500 kDa).115As a bio-protector and substitute for traditional fertilizers, chitosan from the Cunninghamella fungi can be used. Chitosan's efficiency against tomato wilt caused by Ralstonia solanacearum bacterium was compared to conventional fertilizers in a study. According to the findings, plants exposed to conventional fertilizers (NPKF) began to exhibit severe disease symptoms one week after being infected with R. solanacearum, and all the plants perished two weeks later. However, plants treated with chitosan developed improved plant traits and bacterial disease resistance.116 Moreover, chitosan isolated from C. elegans revealed a suitable inhibition activity of 81.7% against mycelial growth of Scytalidium lignicola, which causes potential decrease in cassava production all over the world.117
In another study, chitosan from C. elegans was found to have potent fungicidal activity against Fusarium oxysporum f. sp. tracheiphilum, which is a pathogenic fungus responsible for one of the most frequent diseases in cowpea (Vigna unguiculata L.) crops. This has great socioeconomic importance, especially in Brazil, because it represents a popular dietary source of protein, carbohydrates, and iron, and can be used for animal feed and for the recovery of soil fertility as green manure. This pathogen causes a disease known as Fusarium wilt, which can lead to a reduction in plant growth, chlorosis, wilting and premature leaf fall, all of which almost inevitably lead to the death of the afflicted plants. The results indicated that higher concentrations of fungal chitosan (4.0–6.0 mg mL−1) were responsible for the lowest Fusarium wilt disease severity index in cowpea plants. This was because these higher chitosan concentrations directly induced catalase and peroxidase activity in plants, which in turn controlled the reactive oxygen species equilibrium for plant resistance, and led to a significant decrease in the disease severity in cowpea. These results are crucial for establishing sustainable agriculture and avoiding the usage of pesticides.118
Additionally, fungal chitosan has an inhibitory effect against Botrytis cinerea and Penicillium expansum, which can deteriorate the fruit crop of table grape after harvesting. Chitosan can inhibit the mycelial growth of B. cinerea and P. expansum at MIC of 15 mg mL−1 at 80.4% and 85.7%, respectively. Moreover, chitosan can inhibit spore germination of the two previous fungi at 98.2% and 94.3%, respectively, at the same MIC.119 Consequently, chitosan could be used effectively for extending the shelf life of foods and keeping them safe for a long time. As a bio-preservative in processed fish sausages made from Nile tilapia (Oreochromic niloticus), fungal chitosan also plays a positive role in the food sector. The results showed that fish sausages treated with 1.5% chitosan significantly reduced the microbiological load of coliforms, yeasts, molds, E. coli, and S. aureus, while maintaining the sensory quality of the sausages for a 28 days storage period at 4 °C. Mean ratings for the odor, taste, color, and texture of the chitosan-treated samples from the panelists were 93.2%, 88.3%, 92.1%, and 89.8%, respectively. The control samples' comparable scores, on the other hand, had mean values of 71.4%, 64.2%, 87.9%, and 81.1%, respectively.120
The environmental value of fungal chitosan and chitin was represented by the highest affinity of chitosan and chitin from C. elegans for copper and iron adsorption, respectively.121 Chitin is the parent compound of chitosan that is found in several organisms, forming the exoskeletons of crustaceans, mollusks, insects, algae and the cell wall of fungi.122,123 Additionally, chitosan was demonstrated to be a powerful metal adsorbent for zinc and lead ions, and its adsorption capacity significantly increased as the metal concentration increased.90 This affinity made them potential agents for heavy metal bioremediation in polluted environments, as shown in Fig. 11.
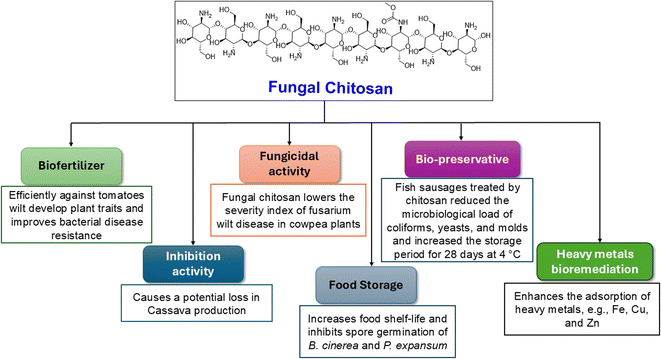 | ||
| Fig. 11 Agricultural, environmental, and industrial values of chitosan isolated from Cunninghamella species. | ||
5. Different enzymes from Cunninghamella species
The swift advancements of biotechnology and discoveries in the field of enzymology have triggered great curiosity toward enzyme preparations. The Cunninghamella genus possesses diverse biosynthetic machinery, which include crucial genes and enzymes utilized in the biotransformation and modification of different organic compounds.124 The enzymatic system in Cunninghamella can be divided into two classes based on their responsibilities and relation with mammalian systems.125 It includes enzymes responsible for biotransformation or xenobiotic metabolism, while the second category includes enzymes with different functions and is incorporated in industrial usage.125 Currently, enzymes derived from Cunninghamella sp. are involved in a variety of applications encompassing industry, agriculture, medicine, and bioremediation.5.1. Cunninghamella enzymes of biological values
Cunninghamella sp. have several enzymes comparable to the human enzyme system such as CYP-450 enzymes, which can metabolize a variety of medicines and transform them into biologically active forms.126,127 In that context, Cunninghamella has the capacity to catabolize xenobiotic chemicals into products that can then be derivatized by additional chemical reactions to create metabolites with unique biological functions. The gene encoding for CYP 5313D1 is thought to play a role in the metabolism of xenobiotics such as the non-steroidal anti-inflammatory medicine flurbiprofen, according to in silico analysis and expression studies. The CYP 5313D1 gene was obtained from C. elegans. When it was heterologously overexpressed in yeast (Pichia pastoris), a recombinant strain that metabolized flurbiprofen to 4′ hydroxy flurbiprofen was identified using GC-MS analysis. This metabolite is the same one produced by C. elegans when flurbiprofen is used as a substrate.26 It is possible to derivatize this hydroxylated metabolite utilizing the Mannich base reaction to create a variety of compounds that work as multifunctional drugs with strong anti-platelet aggregation, anti-neuroinflammatory, and antioxidant properties.128 In addition, a recent study investigated the impact of CYP-450 reductase enzyme derived from C. elegans against the nitro-reduction of flutamide.129 The results induced the potential conversion of flutamide as an anticancer drug to the nitro-reduced metabolite, which is similar to that produced from the same substrate in human NADPH: CYP-450 reductase shown in Fig. 12.129 The nitro reductase effect was prolonged to other substances like nilutamide and environmental pollutants, e.g., 1,3-dinitronaphthalene and 1-nitronaphthalene. Comparative studies with cell lysates of recombinant yeast showed the high sensitivity of reductases toward oxygen.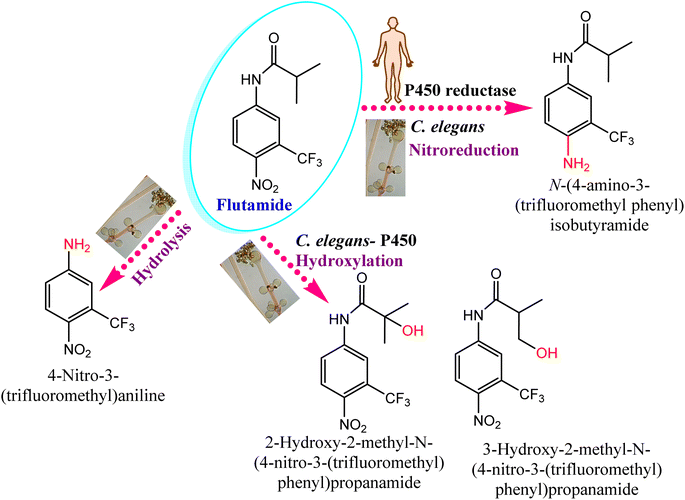 | ||
| Fig. 12 Effect of the CYP-450 reductase enzyme on flutamide biotransformation. Flutamide biotransformed by Cunninghamella elegans produces the same metabolites as those present in humans. | ||
Following the action of the CYP-450 enzyme in the same species, the metabolic breakdown of propiconazole was recently investigated.130 The enzyme initially performs hydroxylation and oxidation reactions of propyl groups in phase I metabolism. Five metabolites were accumulated after 3 days of post-treatment, and indicated that 98% of propiconazole was approximately degraded.130 Interestingly, the formed metabolites were comparable to previously identified compounds from other natural sources, e.g., animals, plants and soil, as presented in Fig. 13.
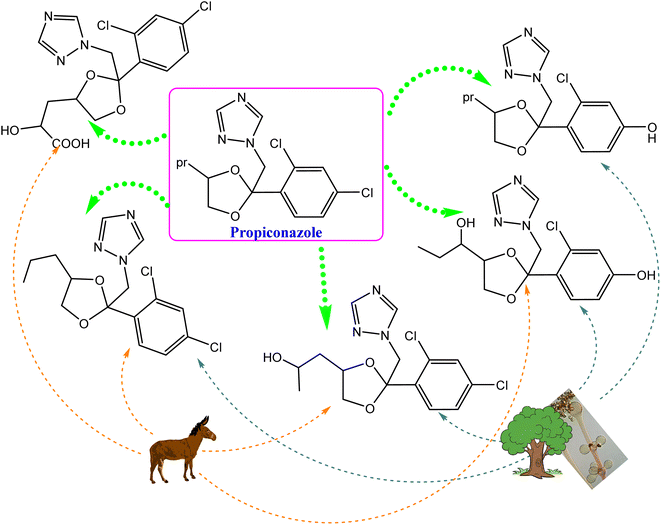 | ||
| Fig. 13 Comparison of the propiconazole breakdown and metabolic pathways between Cunninghamella elegans and other environmental sources, e.g., soil, plants, and animals. | ||
Another study involved C. blakesleeana for improving the low oral bioavailability of paeoniflorin.131 Paeoniflorin is a glycoside compound isolated from Paeonia lactiflora, and has various pharmacological effects, including diabetes mellitus-associated macrovascular complications,132 neuroprotective, antidepressant,133 anti-inflammatory,134 anti-Parkinson,135,136 and anti-Alzheimer effects.137,138 Using a comparison of mass spectroscopy and transcriptomics, the gene (G6046) for a paeoniflorin-converting enzyme was extracted from C. blakesleeana. It was investigated whether optimizing the conditions would promote the highest enzyme activity. When the enzyme activity for paeoniflorin metabolism was tested in vitro, benzoic acid and other benzoate substances were produced, which may make them easier to absorb into the bloodstream, pass through the blood–brain barrier, and enter the central nervous system, where they can exert the pharmacological effects previously mentioned with the greatest ease.139
Moreover, Cunninghamella can produce the catalase enzyme, which is considered a potent antioxidant defense enzyme.140 Exogenous catalase generated from microorganisms like Cunninghamella can be added to the diet as an exogenous supplement to boost immunity against problems brought on by oxidative stress. Exogenous catalase from microorganisms was added to the meal in an in vivo experiment to see if it could reduce the damage that lipopolysaccharides (LPs) caused to the intestinal mucosa of weaned pigs. The findings demonstrated that feeding pigs a diet supplemented with 2000 mg kg−1 of exogenous catalase for 35 days helped to reduce the negative effects that LPs had on the intestinal mucosa. This was done by increasing the amount of catalase and super oxide dismutase in the intestines, and lowering the levels of malondialdehyde and H2O2 in the blood. The amounts of proinflammatory cytokines, such as tumor necrosis factor alpha (TNF-α) and interleukin 6 (IL-6), are also reduced by exogenous catalase supplementation of the food by 11.82% and 15%, respectively. Additionally, compared to pigs merely receiving LP treatment, exogenous catalase in the meal enhanced secretory immunoglobulin A content by 18.14%.141
5.2. Cunninghamella enzymes of agricultural, environmental, and industrial values
Many Cunninghamella sp., such as C. echinulata and C. SL2 species, were proved to produce cellulase and xylanase enzymes using cellulose and xylan degradability in vitro methods.142–144 Thiep et al.145 studied the use of these enzymes from Cunninghamella SL2 sp. as biofertilizers to improve the soil for tea and Arabica coffee plants. According to the findings, Cunninghamella SL2 sp. exhibited significant enzymatic activity for cellulose and xylan with a clear zone of 22.5 and 24.4 mm, respectively. According to the study, these enzymes can break down organic materials and be employed as biofertilizers, as shown in Fig. 14. In comparison to the control plants (45.7 cm per plant, 5.6 g per plant, and 68.9 g per plant), they enhanced the plant height, root weight, and plant weight of coffee plants to 55.8 cm per plant, 9.2 g per plant, and 100.9 g per plant, respectively. Additionally, compared to the control (37.5 cm per plant, 2.3 g per plant, and 8.5 g per plant), these enzymes enhanced the plant height, root weight, and plant weight of tea plants to 54 cm per plant, 5.9 g per plant, and 22.3 g per plant, respectively. As a result, Cunninghamella sp.’ cellulase and xylanase enzymes can be utilized as biofertilizers to enhance the characteristics of the soil and the growth of plants.145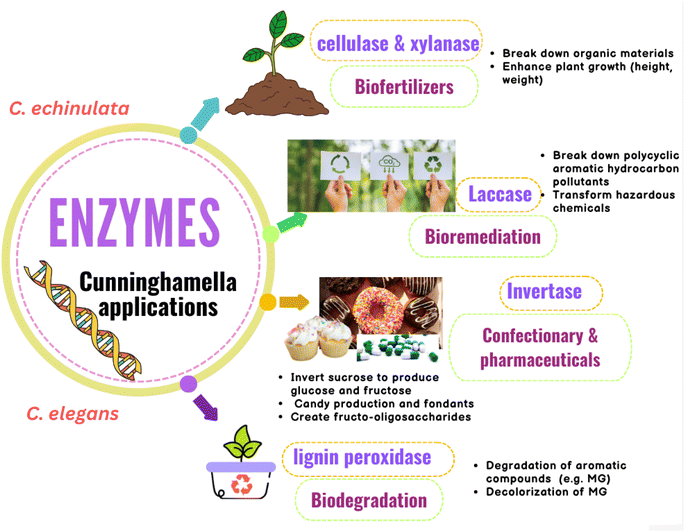 | ||
| Fig. 14 Diagram representing the agricultural, environmental and industrial applications of Cunninghamella-derived enzymes with their impacts. | ||
Cunninghamella produces laccase, a key lignin degradation enzyme that can break down polycyclic aromatic hydrocarbon pollutants (PAHs). Thus, it can be used in bioremediation strategies and has been found to be able to transform a wide range of hazardous chemicals, as tabulated in (Table 5).146 These hydrocarbons are continuously increasing due to industrial expansion, posing numerous risks to people, including the development of cancer and toxicity. According to a study to assess the laccase enzyme activity of C. echinulata, it has a strong ability to degrade three-ring PAHs (anthracene and phenanthrene) as well as phenolic compounds, with breakdown percentages ranging from 96.03% to 99.98% at various levels of PAHs (1%, 2%, 3%, 4% and 5%). High PAH concentrations were observed to boost the activity of the laccase enzyme. In addition, Cunninghamella might use PAHs as a source of carbon for growth.147
| Cunninghamella sp. | Enzyme | Application | Ref. |
|---|---|---|---|
| Cunninghamella sp. | Cytochrome P450 (CYPs) | Antioxidant, metabolism of xenobiotics, anti-platelets aggregation, anti-neuroinflammatory | 26 and 128 |
| C. elegans | Cytochrome P450 reductase | Nitroreductase activity | 129 |
| C. blakesleeana | Catalase | Antioxidant | 140 |
| C. echinulata and C. SL2 | Cellulase and xylanase | Biofertilizers | 142–144 |
| C. echinulata | Laccase | Bioremediation | 146 |
| C. elegans | Lignin peroxidase | Biodegradation of lignin-related aromatic compounds, e.g., dye malachite green (MG) | 149 |
| C. echinulata | Invertase | Confectionary & pharmaceutical industry | 157 and 158 |
Additionally, it was shown that the lignin peroxidase enzyme is essential for the biodegradation of lignin-related aromatic compounds, including the dye malachite green (MG), which is an N-methylated diamino triphenyl methane dye.148 It can be used to dye cotton, silk, wool, jute, leather, and pottery. MG is extremely harmful to mammals, especially humans.149 Cunninghamella sp. are considered important sources for lignin peroxidase enzymes.150 Roushdy et al. studied the lignin peroxidase effect from C. elegans in the decolorization of MG. The findings demonstrated that in the presence of 5 mL of a C. elegans cell-free extract containing lignin peroxidase enzyme, 100% decolorization of MG was seen at doses of 10, 20, and 50 mg L−1 of MG. Additionally, the results showed that static conditions were better when compared to shaking conditions because in aerobic conditions, oxygen and dye competed for the decreased electron carriers.151
Cunninghamella sp. was found to have the ability of invertase enzyme production.152 Invertase is a hydrolyzing enzyme that is capable of breaking down the α-1,4 glycosidic linkage between D-glucose and D-fructose of sucrose.153 Due to the colored byproducts produced by acid hydrolysis processes, such as hydroxy methyl furfural, which is harmful to humans, enzyme hydrolysis of sucrose is preferable to acid hydrolysis.154,155 A study demonstrated that C. echinulata produces invertase enzyme at a high level.152 The conditions were ideal for optimum enzyme synthesis, and the fungus produced a lot of enzymes in culture media supplemented with apple peel. Due to its capacity to invert sucrose to produce a glucose and fructose mixture known as invert syrup, which is sweeter than sucrose due to the high sweetness of fructose, the invertase enzyme has a wide range of industrial applications. These applications comprise the confectionary industry and pharmaceutical industry, and include the formulation of drugs, digestive tablets, and cough syrups, as shown in Fig. 14.156 Due to its hygroscopic character, it can also be employed as a humectant in the production of candy and fondants. Invertase can also create fructo-oligosaccharides, which are great for diabetic patients because they have a lower calorie content while maintaining a similar sweetness.157,158
6. Conclusion
Endophytic fungi are increasingly recognized as a significant source of useful metabolites, which can be distinct derivatives that are either more potent or have identical potency to those of their host plants. Among the most well-known endophytes is Cunninghamella, which is attributed to its abundance of isolated metabolites and its capacity to biotransform a wide range of substrates into more active forms with a variety of biological effects, such as antimicrobial, anticancer, anti-inflammatory, anti-Alzheimer, antiaging, antidiabetic, antiplatelet, and antihypercholesteremia effects. Under the deep investigation for rediscovering the chemical profile of Cunninghamella, it was found to be superior in the biodiversity of FA production. However, none of the elucidated flavonoids have been reported yet, while a few alkaloids, steroids, polyphenols and polysaccharides were isolated. In contrast, Cunninghamella was distinguished in the biotransformation process. It revealed successful results in producing significant and rare metabolites, including steroids, alkaloids and flavonoids when compared to other classes like FAs, polyphenols and quinones, which have yet to be verified via biotransformation. These results inspired researchers to further study the chemistry of the Cunninghamella species, with the aim of answering these contradictions to discover new lead compounds. The current study rediscovered the potential applications of Cunninghamella sp. and associated metabolites in the context of different biological, agricultural, industrial and environmental disciplines. In addition to being a significant source of valuable compounds, Cunninghamella is also thought to be a valuable source of important enzymes, such as lignin peroxidase, catalase, cellulase, xylanase, laccase, and CYP 450, which have a variety of uses in industry, agriculture, medicine, and the environment. However, the use of Cunninghamella as a host candidate for heterologous expression of valuable proteins has not been attempted. Future work is recommended to focus on exploiting Cunninghamella sp. in more complicated biotechnology applications other than the perspective and retrospective studies used in drug discovery.Data availability
No data were used to create the information presented in the present research.Conflicts of interest
The authors declare no conflict of interest.References
- L. L. Xu, Y. Y. Li, T. Han, Q. Y. Zhang, Q. L. Ming, K. Rahman and L. P. Qin, Appl. Microbiol. Biotechnol., 2013, 97, 7617–7625 CrossRef PubMed.
- J. Zhao, T. Shan, Y. Mou and L. Zhou, Rev. Med. Chem., 2011, 11, 159–168 CrossRef CAS PubMed.
- E. Z. Attia, B. A. Khalifa, G. M. Shaban, W. M. Abdelraheem, M. Mustafa, U. R. Abdelmohsen and M. H. El-Katatny, S. Afr. J. Bot., 2022, 151, 218–227 CrossRef CAS.
- A. Gupta, V. Meshram, M. Gupta, S. Goyal, K. A. Qureshi, M. Jaremko and K. K. Shukla, Biomolecules, 2023, 13, 1038 CrossRef CAS PubMed.
- A. Khiralla, R. Spina, M. Varbanov, S. Philippot, P. Lemiere, S. Slezack-Deschaumes, P. André, I. Mohamed, S. M. Yagi and D. Laurain-Mattar, Microorganisms, 2020, 8, 1–20 CrossRef PubMed.
- H. Widjajanti, Muharni, E. Nurnawati and V. Tripuspita, in IOP Conference Series: Earth and Environmental Science, IOP Publishing Ltd, 2022, vol. 976 Search PubMed.
- Y. Teimoori-Boghsani, A. Ganjeali, T. Cernava, H. Müller, J. Asili and G. Berg, Front. Microbiol., 2020, 10, 3013 CrossRef PubMed.
- K. Aini, H. Widjajanti, A. Setiawan and A. R. Kurniawati, Biodiversitas, 2022, 23, 521–532 Search PubMed.
- A. A. da Silva, J. C. Polonio, A. M. Bulla, A. D. Polli, J. C. Castro, L. C. Soares, V. A. de Oliveira-Junior, V. E. P. Vicentini, A. J. B. de Oliveira, J. E. Gonçalves, R. A. C. Gonçalves, J. L. Azevedo, B. A. de Abreu-Filho and J. A. Pamphile, Nat. Prod. Res., 2021, 36, 3158–3162 CrossRef PubMed.
- N. Kapoor, A. Ntemafack, R. Chouhan and S. G. Gandhi, Arch. Phytopathol. Plant Prot., 2022, 55, 454–473 CrossRef CAS.
- L. Tedersoo, S. Sánchez-Ramírez, U. Kõljalg, M. Bahram, M. Döring, D. Schigel, T. May, M. Ryberg and K. Abarenkov, Fungal Diversity, 2018, 90, 135–159 CrossRef.
- D. J. McLaughlin, E. G. McLaughlin and P. A. Lemke, The Mycota. Vol. VII. Part A. Systematics and Evolution, Springer-Verlag, Berlin, 2001 Search PubMed.
- J. Guo, H. Wang, D. Liu, J. N. Zhang, Y. H. Zhao, T. X. Liu and Z. H. Xin, Mycol. Prog., 2015, 14, 1–8 CrossRef.
- R. Zheng and G. Chen, Mycotaxon, 2001, 80, 1–75 Search PubMed.
- J. Yu, G. Walther, A. D. Van Diepeningen, A. H. G. G. Van Den Ende, R. Y. Li, T. A. A. Moussa, O. A. Almaghrabi and G. S. De Hoog, Med. Mycol., 2015, 53, 99–106 CrossRef CAS PubMed.
- Z. Y. Zhang, Y. X. Zhao, X. Shen, W. H. Chen, Y. F. Han, J. Huang and Z. Q. Liang, Phytotaxa, 2020, 455, 31–39 CrossRef.
- O. Navanukroh, A. Jitmuang, M. Chayakulkeeree and P. Ngamskulrungroj, Transplant Infect. Dis., 2014, 16, 658–665 CrossRef CAS PubMed.
- J. García-Rodríguez, I. Quiles-Melero, K. Humala-Barbier, A. Monzon and M. Cuenca-Estrella, Mycoses, 2012, 55, 463–465 CrossRef PubMed.
- E. Alvarez, D. A. Sutton, J. Cano, A. W. Fothergill, A. Stchigel, M. G. Rinaldi and J. Guarro, J. Clin. Microbiol., 2009, 47, 1650–1656 CrossRef CAS PubMed.
- V. Hallur, H. Prakash, M. Sable, C. Preetam, P. Purushotham, R. Senapati, S. A. Shankarnarayan, N. D. Bag and S. M. Rudramurthy, J. Fungi, 2021, 7, 670 CrossRef CAS PubMed.
- H. Prakash and A. Chakrabarti, Microorganisms, 2021, 9, 1–12 CrossRef PubMed.
- W. Jeong, C. Keighley, R. Wolfe, W. L. Lee, M. A. Slavin, D. C. M. Kong and S. C. A. Chen, Clin. Microbiol. Infect., 2019, 25, 26–34 CrossRef CAS PubMed.
- F. A. Alasmary, A. S. Awaad, S. M. Alqahtani, R. M. El-Meligy, D. A. Abdullah and S. I. Alqasoumi, Saudi Pharm. J., 2020, 28, 1197–1202 CrossRef CAS PubMed.
- W. Chen, M. K. Lee, C. Jefcoate, S. C. Kim, F. Chen and J. H. Yu, Genome Biol. Evol., 2014, 6, 1620–1634 CrossRef PubMed.
- D. Zhang, Y. Yang, J. E. A. Leakey and C. E. Cerniglia, FEMS Microbiol. Lett., 1996, 138, 221–226 CrossRef CAS PubMed.
- W. Palmer-Brown, R. Miranda-CasoLuengo, K. H. Wolfe, K. P. Byrne and C. D. Murphy, Sci. Rep., 2019, 9, 9240 CrossRef PubMed.
- S. Mehrotra and V. Goyal, Genome Biol. Evol., 2014, 12, 164–171 Search PubMed.
- O. Asperger, H. Steinbrenner, A. Lehmann, M. Petsch and H. Griengl, Appl. Microbiol. Biotechnol., 1999, 51, 516–522 CrossRef CAS.
- R.-F. Wang, W.-W. Cao, A. A. Khan and C. E. Cerniglia, FEMS Microbiol. Lett., 2000, 188, 55–61 CrossRef CAS PubMed.
- E. Cinteza, A. Nicolescu, T. Ciomartan, L. C. Gavriliu, C. Voicu, A. Carabas, M. Popescu and I. Margarint, Diagnostics, 2022, 12, 657 CrossRef CAS PubMed.
- K. Lemmer, H. Losert, V. Rickerts, G. Just-Nübling, A. Sander, M. L. Kerkmann and K. Tintelnot, Mykosen, Suppl., 2002, 45, 31–36 CrossRef CAS PubMed.
- V. Hallur, H. Prakash, M. Sable, C. Preetam, P. Purushotham, R. Senapati, S. A. Shankarnarayan, N. D. Bag and S. M. Rudramurthy, J. Fungi, 2021, 7, 670 CrossRef CAS PubMed.
- R.-F. Wang, W.-W. Cao, A. A. Khan and C. E. Cerniglia, FEMS Microbiol. Lett., 2000, 188, 55–61 CrossRef CAS PubMed.
- E. Cinteza, A. Nicolescu, T. Ciomartan, L. C. Gavriliu, C. Voicu, A. Carabas, M. Popescu and I. Margarint, Diagnostics, 2022, 12, 657 CrossRef CAS PubMed.
- E. P. Feofilova and L. S. Kuznetsova, Appl. Biochem. Microbiol., 2008, 44, 613–618 CrossRef CAS.
- E.-M. Zhou, X.-A. Chen, M.-M. Zhou, L.-Y. Xu, D. Wang, H.-P. Shen and W.-Q. Xu, Infect., Genet. Evol., 2024, 120, 105575 CrossRef CAS PubMed.
- Z. Da-Fang, S. Lu, L. Lei and H.-H. Huang, Acta Pharmacol. Sin., 2003, 24(5), 442–447 Search PubMed.
- X.-Y. Liu, H. Huang and R.-Y. Zheng, Mycotaxon, 2001, LXXX, 77–95 Search PubMed.
- D. G. M. El Menoufy Hassaan and W. A. Elkhateeb, in Fungi Bioactive Metabolites: Integration of Pharmaceutical Applications, ed. S. S. Deshmukh Sunil Kumar and J. A. Takahashi, Springer Nature Singapore, Singapore, 2024, pp. 743–762 Search PubMed.
- J. Wu, G. Du, J. Zhou and J. Chen, J. Biotechnol., 2014, 188, 72–80 CrossRef CAS PubMed.
- J. F. Wang, S. S. Liu, Z. Q. Song, T. C. Xu, C. S. Liu, Y. G. Hou, R. Huang and S. H. Wu, Molecules, 2020, 25, 5112 CrossRef CAS PubMed.
- H. Cao, X. Chen, A. R. Jassbi and J. Xiao, Biotechnol. Adv., 2015, 33, 214–223 CrossRef CAS PubMed.
- X. L. Xin, G. J. Fan, Z. Sun, N. Zhang, Y. Li, R. Lan, L. Chen and P. P. Dong, J. Mol. Catal. B:Enzym., 2015, 122, 141–146 CrossRef CAS.
- Y. Q. Shi, X. L. Xin, H. C. Zhang, B. J. Zhang, C. Y. Wang, J. Hou, Q. P. Yuan, S. Deng, Y. Tian and X. C. Ma, J. Asian Nat. Prod. Res., 2012, 14, 906–912 CrossRef CAS PubMed.
- Y. Q. Shi, X. L. Xin, Q. P. Yuan, C. Y. Wang, B. J. Zhang, J. Hou, Y. Tian, S. Deng, S. S. Huang and X. C. Ma, J. Asian Nat. Prod. Res., 2012, 14, 1002–1007 CrossRef CAS PubMed.
- A. A. Ghadai, A. A. E. S. Kamillia and S. I. Abdel Rahim, Afr. J. Pharm. Pharmacol., 2016, 10, 411–418 CrossRef.
- E. A. Abourashed, J. R. Mikell and I. A. Khan, Bioorg. Med. Chem., 2012, 20, 2784–2788 CrossRef CAS PubMed.
- H. Inoue, H. Kumagai, M. Osono, M. Matsufujia, T. Sameshimaa, N. Kawamuraa, T. Someno, M. Ishizuka and T. Takeuchi, J. Antibiot., 2003, 56, 209–213 CrossRef CAS PubMed.
- T. Someno, H. Inoue, H. Kumagai, M. Ishizuka and T. Takeuchi, J. Antibiot., 2003, 56, 214–218 CrossRef CAS PubMed.
- D. D. Baker and K. A. Alvi, Curr. Opin. Biotechnol., 2004, 15, 576–583 CrossRef CAS PubMed.
- A. S. Awaad, T. A. Alhamed, M. Dj, S. Ga, N. A. Al-Jaber, M. R. Al-Outhman, Z. Me, R. M. El-Meligy and A. M. Alafeefy, Life Sci. J., 2014, 11, 1097–8135 Search PubMed.
- M. Phaya, S. Chalom, K. Ingkaninan, K. Ounnunkad, N. Chandet, S. G. Pyne and P. Mungkornasawakul, Artif. Cells, Nanomed., Biotechnol., 2021, 49, 166–172 CrossRef CAS PubMed.
- Y. F. Lü, K. Y. Chen, H. L. Li, Y. H. Pei, R. H. Liu and W. D. Zhang, Helv. Chim. Acta, 2008, 91, 819–824 CrossRef.
- L. L. Chu, L. Q. My and H. N. Quang, in Fungal Secondary Metabolites, ed. K. A. Abd-Elsalam and H. I. Mohamed, Elsevier, 2024, pp. 73–90 Search PubMed.
- J. Carvalho, L. Paixão, M. Dolabela, P. Marinho and A. Marinho, Acta Amazonica, 2016, 46, 69–72 CrossRef CAS.
- S. Saha, T. Nath Dhar and C. Sengupta, Czech J. Food Sci., 2013, 31(2), 194–202 Search PubMed.
- S. K. Sukrutha, Z. Adamechova, K. Rachana, J. Savitha and M. Certik, J. Am. Oil Chem. Soc., 2014, 91, 1507–1513 CrossRef CAS.
- S. Fakas, S. Papanikolaou, M. Galiotou-Panayotou, M. Komaitis and G. Aggelis, Appl. Microbiol. Biotechnol., 2006, 73, 676–683 CrossRef CAS PubMed.
- S. Bellou, A. Moustogianni, A. Makri and G. Aggelis, Appl. Biochem. Biotechnol., 2012, 166, 146–158 CrossRef CAS PubMed.
- A. B. Al-Hawash, S. Li, X. Zhang, X. Zhang and F. Ma, Food Biosci., 2018, 21, 1–7 CrossRef CAS.
- M. Gharieb, M. Badawy and G. ElReesh, J. Exp. Biol., 2022, 18, 259 Search PubMed.
- W. B. Suleiman and A. H. Hashem, Res. J. Pharm., Biol. Chem. Sci., 2018, 9, 764–774 CAS.
- Y. Zhao, H. Wang, T. Liu and Z. Xin, Eur. Food Res. Technol., 2014, 238, 621–633 CrossRef CAS.
- F. A. Alasmary, A. S. Awaad, S. M. Alqahtani, R. M. El-Meligy, D. A. Abdullah and S. I. Alqasoumi, Saudi Pharm. J., 2020, 28, 1197–1202 CrossRef CAS PubMed.
- F. A. Alasmary, A. S. Awaad, S. M. Alqahtani, R. M. El-Meligy, D. A. Abdullah and S. I. Alqasoumi, Saudi Pharm. J., 2020, 28, 1197–1202 CrossRef CAS PubMed.
- L. Pierre Luhata and T. Usuki, Bioorg. Med. Chem. Lett., 2021, 48, 128248 CrossRef CAS PubMed.
- T. D. Viet, T. D. Xuan and L. H. Anh, Molecules, 2021, 26, 7248 CrossRef CAS PubMed.
- X. Wang, M. Zhang, Y. Zhao, H. Wang, T. Liu and Z. Xin, Food Chem., 2013, 141, 2066–2074 CrossRef CAS PubMed.
- A.-R. S. Ibrahim and A. E. Ragab, Nat. Prod. Res., 2023, 37, 3722–3726 CrossRef CAS PubMed.
- A. E. Ragab, A. R. S. Ibrahim and F. Léon, Rec. Nat. Prod., 2020, 14, 292–296 CrossRef CAS.
- B. Von Der Haar And and H. Schrempf, J. Bacteriol., 1995, 177(1), 152–155 CrossRef PubMed.
- A. E. Ragab, A. R. S. Ibrahim and F. Léon, Rec. Nat. Prod., 2020, 14, 292–296 CrossRef CAS.
- A. R. S. Ibrahim, K. M. Elokely, D. Ferreira and A. E. Ragab, Molecules, 2018, 23, 970 CrossRef PubMed.
- M. S. Ahmad, S. Zafar, M. Bibi, S. Bano, A. T. Wahab, A. U. Rahman and M. Iqbal Choudhary, Steroids, 2014, 82, 53–59 CrossRef CAS PubMed.
- M. Siddiqui, M. S. Ahmad, A. T. Wahab, S. Yousuf, N. Fatima, N. N. Shaikh, A. U. Rahman and M. I. Choudhary, PLoS One, 2017, 12, e0171476 CrossRef PubMed.
- E. Baydoun, A. Wahab, N. Shoaib, M. S. Ahmad, R. Abdel-Massih, C. Smith, N. Naveed and M. I. Choudhary, Steroids, 2016, 115, 56–61 CrossRef CAS PubMed.
- M. I. Choudhary, N. T. Khan, S. G. Musharraf, S. Anjum and A. U. Rahman, Steroids, 2007, 72, 923–929 CrossRef CAS PubMed.
- K. Lisowska, J. Długoński, J. P. Freeman and C. E. Cerniglia, Chemosphere, 2006, 64, 1499–1506 CrossRef CAS PubMed.
- R. Farooq, N. Hussain, S. Yousuf, A. T. Wahab, M. S. Ahmad, A. U. Rahman and M. I. Choudhary, RSC Adv., 2018, 8, 21985–21992 RSC.
- L. R. R. Berger, M. B. de Araújo, D. P. da Costa, M. A. B. de Lima, J. W. L. de Almeida and E. V. de Medeiros, Int. J. Biol. Macromol., 2020, 161, 101–108 CrossRef CAS PubMed.
- M. M. Jaworska and E. Konieczna, Appl. Microbiol. Biotechnol., 2001, 56, 220–224 CrossRef CAS PubMed.
- M. Wada, Y. Nishimura, Y. Watanabe, T. Takita and S. Innami, Biosci., Biotechnol., Biochem., 1997, 61, 1206–1208 CrossRef CAS PubMed.
- Z. X. Yuan, Z. R. Zhang, D. Zhu, X. Sun, T. Gong, J. Liu and C. T. Luan, Mol. Pharm., 2009, 6, 305–314 CrossRef CAS PubMed.
- W. S. Paiva, F. E. de Souza Neto, M. F. Queiroz, L. A. N. C. Batista, H. A. O. Rocha and A. C. de Lima Batista, Molecules, 2021, 27, 171 CrossRef PubMed.
- M. M. Müller, R. Kantola and V. Kitunen, Mycol. Res., 1994, 98, 593–603 CrossRef.
- R. K. Shiosaki, K. Okada, N. Buarque De Gusmão, P. Nigam, P. S. Falcão, N. Henrique Da Silva, K. Fukushima, M. Miyaji, M. De Campos-Takaki, D. Galba and M. Campos Takaki, Rev. Iberoam. Micol., 2001, 18, 123–127 CAS.
- K. L. Rana, D. Kour, T. Kaur, R. Devi, C. Negi, A. N. Yadav, N. Yadav, K. Singh and A. K. Saxena, in Microbial Endophytes: Functional Biology and Applications, Elsevier, 2020, pp. 273–305 Search PubMed.
- V. K. Singh and A. Kumar, Symbiosis, 2023, 90, 111–125 CrossRef PubMed.
- N. H. Park, J. S. Choi, S. Y. Hwang, Y. C. Kim, Y. K. Hong, K. K. Cho and I. S. Choi, Bot. Stud., 2013, 54, 1–9 CrossRef PubMed.
- A. A. Tayel, M. M. Gharieb, H. R. Zaki and N. M. Elguindy, Int. J. Biol. Macromol., 2016, 83, 277–281 CrossRef CAS PubMed.
- A. S. Awaad, T. A. Alhamed, M. Dj, S. Ga, N. A. Al-Jaber, M. R. Al-Outhman, Z. Me, R. M. El-Meligy and A. M. Alafeefy, Life Sci. J., 2014, 11, 261–268 Search PubMed.
- H. A. El-Baz, A. M. Elazzazy, T. S. Saleh, M. Dourou, J. A. Mahyoub, M. N. Baeshen, H. R. Madian and G. Aggelis, Appl. Sci., 2021, 11, 2700 CrossRef CAS.
- S. Shokry, A. Hegazy, A. M. Abbas, I. Mostafa, I. H. Eissa, A. M. Metwaly, G. Yahya, A. M. El-Shazly, K. M. Aboshanab and A. Mostafa, Vaccines, 2023, 11, 228 CrossRef CAS PubMed.
- D. Pardo-Rodriguez, A. Cifuentes-López, J. Bravo-Espejo, I. Romero, J. Robles, C. Cuervo, S. M. Mejía and J. Tellez, Trop. Med. Infect. Dis., 2023, 8, 263 CrossRef PubMed.
- F. S. Kenny, S. E. Pinder, I. O. Ellis, J. M. W. Gee, R. I. Nicholson, R. P. Bryce and J. F. R. Robertson, Int. J. Cancer, 2000, 85, 643–648 CrossRef CAS PubMed.
- G. Kalampounias, C. Gardeli, S. Alexis, E. Anagnostopoulou, T. Androutsopoulou, P. Dritsas, G. Aggelis, S. Papanikolaou and P. Katsoris, J. Fungi, 2024, 10, 130 CrossRef CAS PubMed.
- K. J. A. Kairemo, A. P. Jekunen, E. T. Korppi-Tommola and S. O. Pyrhönen, Pancreas, 1998, 16, 105–106 CrossRef CAS PubMed.
- V. Sanchez-Martin, M. D. C. Plaza-Calonge, A. Soriano-Lerma, M. Ortiz-Gonzalez, A. Linde-Rodriguez, V. Perez-Carrasco, I. Ramirez-Macias, M. Cuadros, J. Gutierrez-Fernandez, J. Murciano-Calles, J. C. Rodríguez-Manzaneque, M. Soriano and J. A. Garcia-Salcedo, Cancers, 2022, 14, 2648 CrossRef CAS PubMed.
- S. Gu, F. Liu, X. Xie, M. Ding, Z. Wang, X. Xing, T. Xiao and X. Sun, Cell Signal, 2023, 104, 110585 CrossRef CAS PubMed.
- Y. Zhao, H. Wang, T. Liu and Z. Xin, Eur. Food Res. Technol., 2014, 238, 621–633 CrossRef CAS.
- J. Zhang, C. Zhang, L. Miao, Z. Meng, N. Gu and G. Song, Pharm. Biol., 2023, 61, 449–458 CrossRef CAS PubMed.
- L. J. Leventhal, E. G. Boyce and R. B. Zurier, Ann. Intern. Med., 1993, 119, 867–873 CrossRef CAS PubMed.
- S. Sergeant, E. Rahbar and F. H. Chilton, Eur. J. Pharmacol., 2016, 785, 77–86 CrossRef CAS PubMed.
- H. D. Jhundoo, T. Siefen, A. Liang, C. Schmidt, J. Lokhnauth, A. Béduneau, Y. Pellequer, C. C. Larsen and A. Lamprecht, Pharmaceutics, 2020, 12, 1–16 CrossRef PubMed.
- K. Youn, J. Lee, E. Y. Yun, C. T. Ho, M. V. Karwe, W. S. Jeong and M. Jun, J. Funct. Foods, 2014, 10, 187–191 CrossRef CAS.
- F. Marcheggiani, S. Kordes, I. Cirilli, P. Orlando, S. Silvestri, A. Vogelsang, N. Möller, T. Blatt, J. M. Weise, E. Damiani and L. Tiano, Free Radical Biol. Med., 2021, 165, 282–288 CrossRef CAS PubMed.
- O. Sajjad Alsawad, Z. A. Al-Mayyahi, N. A. Khudhair and L. Hossein Jawid, Syst. Rev. Pharm., 2020, 11, 736–746 Search PubMed.
- Y. J. Jbrael, B. K. Hamad and I. Al-Haitham, J. Pure Appl. Sci., 2023, 36, 85–104 Search PubMed.
- Y. Zhang, X. Wang, B. Lu, Y. Gao, Y. Zhang, Y. Li, H. Niu, L. Fan, Z. Pang and Y. Qiao, Chin. Herb. Med., 2022, 14, 303–309 CAS.
- R. Takada, M. Saitoh and T. Mori, J. Nutr., 1994, 124, 469–474 CrossRef CAS PubMed.
- K. Vasanth, G. C. Minakshi, K. Velu, T. Priya, R. M. Kumar, I. Kaliappan and G. P. Dubey, J. Food Biochem., 2022, 46, e14170 CrossRef CAS PubMed.
- H. A. El-Baz, A. M. Elazzazy, T. S. Saleh, M. Dourou, J. A. Mahyoub, M. N. Baeshen, H. R. Madian and G. Aggelis, Appl. Sci., 2021, 11, 2700 CrossRef CAS.
- M. J. González-Fernández, I. Ortea and J. L. Guil-Guerrero, Toxicol. Res., 2020, 9, 474–483 CrossRef PubMed.
- S. Aichayawanich and M. Saengprapaitip, in IOP Conference Series: Earth and Environmental Science, Institute of Physics Publishing, 2019, vol. 301 Search PubMed.
- M. R. Kasaai, J. Arul and G. Charlet, Sci. World J., 2013, 2013, 508540 CrossRef PubMed.
- S. O. Wagner, L. C. Iwanne, R. S. O. Jessica, C. B. S. L. Maria, M. S. A. Thatiana, P. S. Newton and V. N. S. Emmanuella, Afr. J. Agric. Res., 2017, 12, 42–49 CrossRef.
- L. R. R. Berger, M. B. de Araújo, D. P. da Costa, M. A. B. de Lima, J. W. L. de Almeida and E. V. de Medeiros, Int. J. Biol. Macromol., 2020, 161, 101–108 CrossRef CAS PubMed.
- L. R. R. Berger, N. P. Stamford, L. G. Willadino, D. Laranjeira, M. A. B. de Lima, S. M. M. Malheiros, W. J. de Oliveira and T. C. M. Stamford, Biol. Control, 2016, 92, 45–54 CrossRef CAS.
- C. E. V. de Oliveira, M. Magnani, C. V. de Sales, A. L. de Souza Pontes, G. M. Campos-Takaki, T. C. M. Stamford and E. L. de Souza, Int. J. Food Microbiol., 2014, 171, 54–61 CrossRef PubMed.
- A. A. Tayel, Int. J. Biol. Macromol., 2016, 93, 41–46 CrossRef CAS PubMed.
- L. De Oliveira Franco, R. De Cássia, C. Maia, A. Lúcia, F. Porto Arminda, S. Messias, K. Fukushima Galba and M. De Campos-Takaki, Braz. J. Microbiol., 2004, 35, 243–247 CrossRef.
- S. Crognale, C. Russo, M. Petruccioli and A. D’annibale, Fermentation, 2022, 8, 76 CrossRef CAS.
- A. Pellis, G. M. Guebitz and G. S. Nyanhongo, Gels, 2022, 8, 393 CrossRef CAS PubMed.
- C. D. Murphy, Biotechnol. Lett., 2015, 37, 19–28 CrossRef CAS PubMed.
- A.-R. S. Ibrahim, M. K. Mansour, M. M. A. Ahmed, R. Ulber and A. Zayed, Bioorg. Chem., 2023, 140, 106801 CrossRef CAS PubMed.
- M. D. Gonçalves, F. Tomiotto-Pellissier, R. L. N. de Matos, J. P. Assolini, B. T. da Silva Bortoleti, V. M. Concato and N. S. Arakawa, Curr. Drug Metab., 2021, 22, 1035–1064 CrossRef PubMed.
- J. Zhan, H. Guo, J. Dai, Y. Zhang and D. Guo, Tetrahedron Lett., 2002, 43, 4519–4521 CrossRef CAS.
- H. Liu, X. Qiang, Q. Song, W. Li, Y. He, C. Ye, Z. Tan and Y. Deng, Bioorg. Med. Chem., 2019, 27, 991–1001 CrossRef CAS PubMed.
- M. F. Khan and C. D. Murphy, Biochem. Biophys. Rep., 2022, 29, 101209 CAS.
- Z. Zhai, M. Meng, Z. Zhang, J. Kim and Y. Zhu, Arch. Microbiol., 2024, 206, 356 CrossRef CAS PubMed.
- J. B. Yu, Z. X. Zhao, R. Peng, L. Bin Pan, J. Fu, S. R. Ma, P. Han, L. Cong, Z. W. Zhang, L. X. Sun, J. D. Jiang and Y. Wang, Front. Pharmacol, 2019, 10, 268 CrossRef CAS PubMed.
- J. S. Wang, Y. Huang, S. Zhang, H. J. Yin, L. Zhang, Y. H. Zhang, Y. W. Song and D. D. Li, Oxid. Med. Cell. Longevity, 2019, 1, 5647219 Search PubMed.
- J. Cheng, M. Chen, H.-Q. Wan, X.-Q. Chen, C.-F. Li, J.-X. Zhu, Q. Liu, G.-H. Xu and L.-T. Yi, J. Ethnopharmacol., 2021, 274, 114046 CrossRef CAS PubMed.
- C. Chen, P. Du and J. Wang, Mol. Med. Rep., 2015, 12, 3937–3943 CrossRef CAS PubMed.
- X. S. Gu, F. Wang, C. Y. Zhang, C. J. Mao, J. Yang, Y. P. Yang, S. Liu, L. F. Hu and C. F. Liu, Neurochem. Res., 2016, 41, 2923–2936 CrossRef CAS PubMed.
- M. Zheng, C. Liu, Y. Fan, P. Yan, D. Shi and Y. Zhang, Neuropharmacology, 2017, 116, 412–420 CrossRef CAS PubMed.
- K. Wang, L. Zhu, X. Zhu, K. Zhang, B. Huang, J. Zhang, Y. Zhang, L. Zhu, B. Zhou and F. Zhou, Cell. Mol. Neurobiol., 2014, 34, 227–234 CrossRef CAS PubMed.
- Z. Lan, L. Chen, Q. Fu, W. Ji, S. Wang, Z. Liang, R. Qu, L. Kong and S. Ma, Brain Res., 2013, 1498, 9–19 CrossRef CAS PubMed.
- Y. Ye, H. Pei, X. Cao, X. Liu, Z. Li, B. Wang, Y. Pan and J. Zheng, Molecules, 2023, 28, 1289 CrossRef CAS PubMed.
- A. A. Forist, J. Am. Pharm. Assoc., 1958, 47, 185–187 CrossRef CAS.
- J. Chen, F. Li, W. Yang, S. Jiang and Y. Li, Microbiol. Spectrum, 2021, 9, e00654 CAS.
- S. Banakar, S. Jiao, T. Basaiah, T. Banjagere, S. P. Banakar and K. J. Thirumalesh, Int. J. Curr. Res., 2012, 4, 76–82 Search PubMed.
- F. Bhadra, A. Gupta, M. Vasundhara and M. S. Reddy, 3 Biotech, 2022, 12, 86 CrossRef PubMed.
- S. C. B. Gopinath, P. Anbu and A. Hilda, Mycoscience, 2005, 46, 119–126 CrossRef CAS.
- N. V. Thiep, K. Soytong, N. O. Thi Kim, P. Huy Quang and P. Hai Yen, Int. J. Agric. Technol., 2019, 15(5), 797–806 CAS.
- M. Alexander, Biodegradation and Bioremediation, Gulf Professional Publishing, 1999 Search PubMed.
- E. M. El-Morsy, E. E. Hafez, M. T. Mohesien and G. M. Abogadallah, Sci. J. Damietta Fac. Sci., 2012, 1, 65–75 Search PubMed.
- R. A. Schnick, Prog. Fish-Cult., 1988, 50, 190–196 CrossRef.
- G. Parshetti, S. Kalme, G. Saratale and S. Govindwar, Acta Chim. Slov., 2006, 53, 492–498 CAS.
- M. Roushdy and E. I. El-Agamy, J. Am. Sci., 2011, 7, 6–13 Search PubMed.
- M. Roushdy, N Y Sci. J., 2011, 4, 80–88 Search PubMed.
- L. M. Rasbold, V. M. Delai, C. M. da Cruz Kerber, M. R. Simões, P. R. Heinen, J. L. da Conceição Silva, R. de Cássia Garcia Simão, M. K. Kadowaki and A. Maller, J. Appl. Microbiol., 2022, 132, 2832–2843 CrossRef CAS PubMed.
- L. E. Trujillo Toledo, D. Martinez García, E. Pérez Cruz, L. M. Rivera Intriago, J. N. Pérez and J. M. Pais Chanfrau, Enzymes Food Biotechnol., 2019, 451–469 Search PubMed.
- M. N. de Almeida, V. M. Guimarães, D. L. Falkoski, B. R. de Camargo, G. C. Fontes-Sant’ana, G. P. Maitan-Alfenas and S. T. de Rezende, J. Food Biochem., 2018, 42, e12551 CrossRef.
- D. Martínez, C. Menéndez, F. M. Echemendia, E. R. Pérez, L. E. Trujillo, A. Sobrino, R. Ramírez, Y. Quintero and L. Hernández, Microb. Cell Fact., 2014, 13, 1–9 CrossRef PubMed.
- U. Andjelković, A. Milutinović-Nikolić, N. Jović-Jovičić, P. Banković, T. Bajt, Z. Mojović, Z. Vujčić and D. Jovanović, Food Chem., 2015, 168, 262–269 CrossRef PubMed.
- T. Chand Bhalla, Bansuli, N. Thakur, Savitri and N. Thakur, LWT, 2017, 77, 178–185 CrossRef CAS.
- D. Linde, B. Rodríguez-Colinas, M. Estévez, A. Poveda, F. J. Plou and M. Fernández Lobato, Bioresour. Technol., 2012, 109, 123–130 CrossRef CAS PubMed.
Footnote |
| † Both authors contributed equally to this work. |
| This journal is © The Royal Society of Chemistry 2024 |