Open Access Article
Open Access ArticleMonitoring water harvesting in metal–organic frameworks, one water molecule at a time†
Kelly M.
Hunter
a and
Francesco
Paesani
 *abcd
*abcd
aDepartment of Chemistry and Biochemistry, University of California, La Jolla, San Diego, California 92093, USA
bMaterials Science and Engineering, University of California, La Jolla, San Diego, California 92093, USA
cHalicioğlu Data Science Institute, University of California, La Jolla, San Diego, California 92093, USA
dSan Diego Supercomputer Center, University of California, La Jolla, San Diego, California 92093, USA. E-mail: fpaesani@ucsd.edu
First published on 5th March 2024
Abstract
Metal–organic frameworks (MOFs) have gained prominence as potential materials for atmospheric water harvesting, a vital solution for arid regions and areas experiencing severe water shortages. However, the molecular factors influencing the performance of MOFs in capturing water from the air remain elusive. Among all MOFs, Ni2X2BTDD (X = F, Cl, Br) stands out as a promising water harvester due to its ability to adsorb substantial amounts of water at low relative humidity (RH). Here, we use advanced molecular dynamics simulations carried out with the state-of-the-art MB-pol data-driven many-body potential to monitor water adsorption in the three Ni2X2BTDD variants as a function of RH. Our simulations reveal that the type of halide atom in the three Ni2X2BTDD frameworks significantly influences the corresponding molecular mechanisms of water adsorption: while water molecules form strong hydrogen bonds with the fluoride atoms in Ni2F2BTDD, they tend to form hydrogen bonds with the nitrogen atoms of the triazolate linkers in Ni2Cl2BTDD and Ni2Br2BTDD. Importantly, the large size of the bromide atoms reduces the void volume in the Ni2Br2BTDD pores, which enable water molecules to initiate an extended hydrogen-bond network at lower RH. These findings not only underscore the prospect for precisely tuning structural and chemical modifications of the frameworks to optimize their interaction with water, but also highlight the predictive power of simulations with the MB-pol data-driven many-body potential. By providing a realistic description of water under different thermodynamic conditions and environments, these simulations yield unique, molecular-level insights that can guide the design and optimization of energy-efficient water harvesting materials.
1 Introduction
Lack of access to freshwater is a pressing global challenge,1 with profound impacts ranging from decreased food production2 to long-term public health issues.3,4 With the atmosphere holding approximately 1021 liters of water as either vapor or droplets, atmospheric water harvesting (AWH) represents an appealing solution to provide freshwater to regions grappling with water scarcity.5 AWH boasts several advantages over other technologies: it is not restricted to specific geographical regions as water can be harvested from the atmosphere anywhere in the world, it eliminates the need for transportation, and it does not tap into more limited water resources such as rivers and lakes.4,6 However, for AWH to serve as a reliable source of freshwater, it necessitates sorbent materials endowed with high hydrolytic stability for enduring recycling performance, large porosity and surface area for substantial water vapor uptake, relatively mild regeneration conditions, adsorption isotherms demonstrating a sharp uptake at low relative humidity (RH), and a high deliverable capacity. Metal–organic frameworks (MOFs) hold the promise of fulfilling all these prerequisites.Built from organic linkers and secondary building units (SBUs) composed of metal ions or clusters, MOFs are characterized by large surface areas and a wide range of structural and physical properties that can be tuned for specific applications through multivariate reticular chemistry7,8 and/or post-synthetic approaches.9–11 Several MOFs for potential applications in AWH technologies have been reported in the scientific literature.12–20 Despite some progress, a molecular-level characterization of the water adsorption mechanisms in MOFs, which are key to the design and optimization of frameworks with high adsorption capacities at low RH, remains elusive.21–25 A primary challenge stems from the intricate water–framework interface, which has so far made it difficult to link macroscopic thermodynamic properties, such as isotherms and enthalpies of adsorption, to specific interactions of individual water molecules with the SBUs and organic linkers of the framework as a function of RH. From a simulation perspective, these difficulties are primarily associated with intrinsic limitations of empirical force fields used to model water–framework and water–water interactions, which are unable to correctly reproduce the properties of water across different phases as a function of temperature and pressure.26 Recent advances in molecular dynamics (MD) simulations of water, spurred by the advent of the MB-pol data-driven many-body potential,27–29 have addressed these limitations.30 This progress has paved the way for realistic computational studies, providing an unprecedented level of molecular insight into structural, thermodynamic, dynamical, and spectroscopic properties of water across the entire phase diagram, from gas-phase clusters to liquid water and ice.31–40
In this study, we employ MD simulations using the MB-pol data-driven many-body potential to characterize, at the molecular level, the individual steps associated with water adsorption in the halide-exchanged Ni2X2BTDD MOFs series, where X corresponds to fluoride (F), chloride (Cl), and bromide (Br) atoms. The Ni2X2BTDD MOFs stand out as some of the most promising candidates for AWH applications, owing to their exceptional water uptake capabilities even at low RH values.19 All three Ni2X2BTDD MOFs share the same topology and are synthesized from the same organic linkers, specifically, bis(1H-1,2,3-triazolo[4,5-b],[4′,5′-i])dibenzo[1,4]dioxin, or H2BTDD. They also share the same SBUs with undercoordinated nickel (Ni2+) centers. Their distinction lies in the halide atoms (F, Cl, and Br) that bridge the Ni2+ centers. While the three Ni2X2BTDD frameworks have similar pore sizes, ranging between 22 Å and 23 Å, their different halide functionalization leads to notable variations in water adsorption capacities and framework stabilities. Specifically, Ni2F2BTDD and Ni2Cl2BTDD can adsorb over 100% of their weight in water at 32% RH, while Ni2Br2BTDD can only adsorb 64% of its weight in water, but this occurs below 25% RH, which is one of the lowest RH values for AWH reported for MOFs to date.19 Furthermore, Ni2Cl2BTDD and Ni2Br2BTDD maintain their water uptake capacity for more than 400 adsorption/desorption cycles, while Ni2F2BTDD undergoes partial amorphization and displays a decline in its adsorption capacity after one cycle.19 Given their structural similarities and varying water adsorption capacities, the three Ni2X2BTDD MOFs present an ideal platform to identify specific correlations between water–framework and water–water interactions and characterize the thermodynamics of water adsorption as a function of RH at the molecular level.
2 Methods
2.1 Force fields and potential energy functions
Each MOF structure was first optimized using periodic density functional theory (DFT) calculations carried out with the Vienna Ab initio Simulation Package (VASP),41–44 using the PBE exchange–correlation functional45 with Grimme's D3 dispersion correction46 in combination with a projector-augmented wave (PAW) treatment47,48 with a 700 eV kinetic energy cutoff. A 2 × 2 × 4 k-point grid was used, and forces were converged to a tolerance of 0.05 eV Å−1. Following periodic optimization, a cluster surrounding the Ni2+ metal center was extracted for each MOF (see ESI† for details). All bonded parameters involving the Ni2+ metal center were fit using a genetic algorithm, and all bonded parameters for the organic linker were taken from the General Amber Force Field (GAFF).49 Atomic point charges were calculated for the clusters using the charge model 5 (CM5) charge scheme.50 The Lennard-Jones (LJ) parameters were taken from the Universal Force Field (UFF)51 for the Ni, F, Cl, and Br atoms, and the LJ parameters for the organic linker atoms (H, C, N, O) were taken from GAFF.49 Force field parameters are given in Tables S1–S15 in the ESI.†The MB-pol data-driven many-body potential was used to simulate water.27–29 MB-pol has been shown to correctly predict the properties of water from the gas to the condensed phase for structural, dynamical, and thermodynamic properties.31 The many-body dipole moment (MB-μ) was used to calculate the dipole moment of water for the IR spectra.33 Local mode and local monomer (LM) calculations52–54 from ref. 55 were utilized to add the Fermi resonance contribution to the IR spectra.
Nonbonded interactions between water and the MOF were represented through electrostatic and van der Waals (VDW) interactions. The Ni–OW, X–OW (X = F, Cl, Br), and X–HW interactions were fit to a Buckingham potential using a genetic algorithm (see ESI† for details). Cross interactions between water and the remaining MOF atoms were described through Lennard-Jones (LJ) parameters. The LJ parameters were derived from Lorentz–Berthelot mixing rules using the LJ parameters of the TIP4P/2005 water model,56 which is the closest point-charge model to MB-pol.57
2.2 Molecular dynamics simulations
Classical molecular dynamics (MD) simulations were performed using an in-house code based on the DL_POLY_2 simulation package,58 which was modified to include MB-pol water.27–29 Simulations for each halide MOF were carried out for a system containing 1 × 1 × 3 unit cells under periodic boundary conditions. The initial configurations for each MD simulation of the different halide MOFs were generated from Packmol.59 Each configuration started with 54 water molecules (1H2O/Ni2+), and 54 water molecules were added to each subsequent simulation (+1H2O/Ni2+) up to the point of pore saturation on the adsorption isotherm (12H2O/Ni2+ for Ni2F2BTDD, 13H2O/Ni2+ for Ni2Cl2BTDD, and 11H2O/Ni2+ for Ni2Br2BTDD).19 MD simulations were first performed utilizing a constant number of molecules, pressure, and temperature in the isobaric–isothermal ensemble (NPT = constant number of atoms, pressure, and temperature). The system was heated to 1000 K for 10 ps at 1.0 atm, cooled to 500 K for 20 ps at 1.0 atm, and cooled to 300 K for 20 ps at 1.0 atm, all in the NPT ensemble, to randomize the initial distribution of water molecules. MD simulations were then performed for 1 ns at 300 K and 1.0 atm in the NPT ensemble to equilibrate the system, calculate structural information, and obtain the average box dimensions. The average box dimensions listed in Tables S16–S18 in the ESI† were used for all subsequent simulations at each loading. The angles of each simulation box were 90°, 90°, and 120° along the x, y, and z dimensions, respectively. Following the initial equilibration, the system was simulated with a constant number of molecules, volume, and temperature in the canonical ensemble (NVT = constant number of atoms, volume, and temperature) for 100 ps at 300 K followed by a constant number of molecules, volume, and energy in the microcanonical ensemble (NVE = constant number of atoms, volume, and energy) for 50 ps to obtain dynamical information, including the IR spectra, where the temperature remained constant around 300 K. Subsequent configurations were heated to 1000 K for 10 ps in the NVT ensemble to randomize the distribution of water, cooled to 300 K for 10 ps in the NVT ensemble, and then run for 50 ps in the NVE ensemble where the temperature remained constant around 300 K. 19 of these additional trajectories were run at each loading for each halide, giving a total of 20 independent MD simulations in which all dynamical properties were averaged over. The entropy was calculated as a function of RH using the two-phase thermodynamic (2PT) model.60 While approximate like any other molecular model of entropy, the 2PT model provides an intuitive approach to calculating the entropy of water depending on the local environment. Using the 2PT model, MB-pol predicts a value of 71.9 ± 0.6 J mol−1 K−1 for the entropy of liquid water at ambient conditions, which is in good agreement with the experimental value of 69.95 ± 0.03 J mol−1 K−1.All MD simulations were performed with a time step of 0.2 fs, and the equations of motion were propagated according to the velocity-Verlet algorithm. The temperature was maintained at 300 K by a Nosé–Hoover chain of four thermostats.61 The NPT ensemble was generated according to the algorithm described in ref. 62. Short-range interactions were truncated at an atom–atom distance of 9.0 Å, and the electrostatics were calculated using the Ewald sum.63
The molecular models used in the MD simulations, which were conducted using in-house software based on the DL_POLY_2 simulation package, are available upon request from the authors in a format compatible with the MBX interface64,65 for LAMMPS.66
3 Results
3.1 Thermodynamics of water adsorption
Fig. 1 presents the experimental adsorption isotherms for the three Ni2X2BTDD MOFs measured as a function of RH in ref. 19, along with the theoretical values of the adsorption enthalpy and entropy calculated for water in the Ni2X2BTDD from MD simulations carried out with the MB-pol data-driven many-body potential. The enthalpy of adsorption directly reports on the strength and interplay of water–framework and water–water interactions. The balance between these interactions is crucial for determining the onset RH values for water adsorption and pore filling, which in turn dictates the overall performance of a MOF for AWH applications. Ni2F2BTDD displays the most negative enthalpy of adsorption at all RH values, which directly correlates with fluoride providing the strongest interactions with water among the halide atoms.67–70 The enthalpy of adsorption of Ni2Cl2BTDD and Ni2Br2BTDD does not, however, follow the order of chloride–water and bromide–water interactions, suggesting differences in the modes of adsorption between the two frameworks. The differences in the physico-chemical properties of the fluoride, chloride, and bromide ions, their interactions with water, and adsorption modes manifest in distinct pore-filling onsets (Fig. 1A). Specifically, Ni2Br2BTDD fills its pores at a lower RH value (24% RH), compared to Ni2F2BTDD and Ni2Cl2BTDD (32% RH), which is mirrored by steep increases calculated for the corresponding enthalpies of adsorption at the same RH values (Fig. 1B). The increase in enthalpy of adsorption predicted by the MD simulations as the RH increases indicates that water–water interactions, which become the dominant interactions at high RH values, are relatively weaker than water–framework interactions. The predicted larger enthalpy of adsorption for Ni2F2BTDD, indicative of stronger water–framework interactions, aligns with its observed partial amorphization and reduced water adsorption capacity in successive adsorption/desorption cycles.19 | ||
| Fig. 1 (A) Experimental adsorption isotherms of the three Ni2X2BTDD MOFs reported in ref. 19. (B and C) Adsorption enthalpy and entropy, respectively, calculated for water in the Ni2X2BTDD MOFs as a function of relative humidity from MD simulations carried out with the MB-pol data-driven many-body potential of water. | ||
The entropy of water calculated from the MD simulations displays the opposite trend compared to the enthalpy of adsorption, with water in Ni2Cl2BTDD and Ni2F2BTDD having the highest and lowest entropy up to the pore-filling step, respectively (Fig. 1C). Since at low RH values, water in the MOF pores exhibit gas-like behavior modulated by water–framework interactions, the entropy of water adsorbed in all three Ni2X2BTDD MOFs remains appreciably larger than the entropy of liquid water up to the pore-filling step.23,71 At the RH values corresponding to the pore-filling steps, the entropy of adsorption displays a sudden drop for all three MOFs and approaches the value of liquid water as the RH increases. The variation of the entropy of adsorption as a function of RH effectively mirrors the variation of the enthalpy of adsorption, indicating that as the pores get filled, the adsorbed water molecules acquire bulk-like behavior.
3.2 Early stages of water adsorption
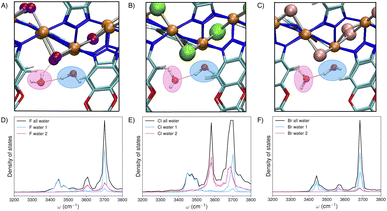
The OH vibrations of water molecules are especially sensitive to their surrounding environment.72 Monitoring the evolution of the infrared (IR) spectra of water adsorbed in the three Ni2X2BTDD MOFs in the OH-stretch frequency range thus provides a direct probe into the development of hydrogen-bonded networks inside the pores and offers molecular-level insights into the adsorption mechanisms. At low RH, the experimental IR spectra (Fig. 2) display four distinct features at ∼3700 cm−1, ∼3500 cm−1, ∼3300 cm−1, and ∼3200 cm−1. This contrasts with the broad, featureless OH-stretch band seen in the IR spectrum of liquid water.33 Using the simulated IR spectra for comparison, the highest frequency peak at ∼3700 cm−1 can be attributed to stretch vibrations of OH bonds not involved in hydrogen bonding. The position and width of this high-frequency peak are reminiscent of the free-OH peak in the sum-frequency generation spectrum of the air/water interface.35 Our theoretical analysis suggests that the feature at ∼3200 cm−1, often linked to ice-like hydrogen-bonded structures, actually arises from Fermi resonances.73 These resonances result from couplings between the OH-stretch fundamental vibrations and the HOH bending overtones.55 The other two peaks indicate distinct local hydrogen-bonding environments that develop inside the MOF pores as water adsorption increases with RH. The discrepancies between the experimental and calculated IR spectra observed in Fig. 2 can be attributed to various factors. These include the omission of higher-body framework–water interactions and nuclear quantum effects in the MD simulations. Previous studies have shown that not accounting for nuclear quantum effects in MD simulations of vibrational spectra of water generally leads to spectral features that are narrower and exhibit a blueshift when compared to experimental results.74 | ||
| Fig. 2 Experimental IR spectra of water from ref. 19 for Ni2F2BTDD at 10% RH (A), Ni2Cl2BTDD at 5% RH (B), and Ni2Br2BTDD at 5% RH (C). (D) IR spectrum of water in Ni2F2BTDD at 9.2% RH (light red shaded area) obtained by combining the IR spectrum calculated from the MD simulations (light red solid curve) with the Fermi resonance contributions (dark red solid curve). (E) IR spectrum of water in Ni2Cl2BTDD at 4.3% RH (light green shaded area) obtained by combining the IR spectrum calculated from the MD simulations (light green solid curve) with the Fermi resonance contributions (dark green solid curve). (F) IR spectrum of water in Ni2Br2BTDD at 5.4% RH (light blue shaded area) obtained by combining the IR spectrum calculated from the MD simulations (light blue solid curve) with the Fermi resonance contributions (dark blue solid curve). The simulated IR spectra red-shifted by 215 cm−1 (Ni2F2BTDD and Ni2Cl2BTDD) or 219 cm−1 (Ni2Br2BTDD) to align the position of the free-OH peak with the corresponding experimental peak, and the HOH bending overtones are blue-shifted by 10 cm−1, accordingly. The main features of the experimental and simulated IR spectra are indicated by dashed lines. | ||
MD simulations, performed by incrementally adding water molecules, reveal that the initial water molecules entering the pores of the three Ni2X2BTDD MOFs bind to one of the open Ni2+ metal centers, as depicted by the water molecule inside the blue area in Fig. 3A–C. The spatial distributions of these Ni-bound water molecules, however, vary depending on the specific halide ion present in the framework, giving rise to characteristic peaks in the corresponding IR spectra (Fig. 3D–F).
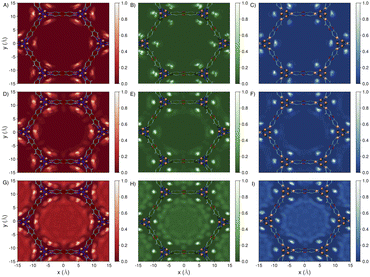
Fig. 4A–C shows that while the Ni-bound water molecules localize in a rounded three-dimensional region perpendicular to the Ni2+ metal center in Ni2Cl2BTDD and Ni2Br2BTDD, they are more delocalized along the F–Ni–F axis in Ni2F2BTDD. These differences become more apparent from the analyses of the radial distribution functions (RDFs) describing spatial correlations between the oxygen (OW) and hydrogen (HW) atoms of the Ni-bound water molecule and the Ni2+, halide, and nitrogen (n = n1 and n2) atoms of the framework (see Fig. S1, S2 and Tables S1–S15 in the ESI† for the complete lists of atom labels and force field parameters). Specifically, the OW–Ni RDFs in Fig. 4D show that the average distance of the Ni-bound water molecules from the Ni2+ open sites is effectively the same in all three MOFs. In contrast, the HW–X RDFs in Fig. 4E show that the average distance between the H atoms of the Ni-bound water molecules and the halide atoms of the frameworks is significantly different among the three MOFs, indicating that the Ni-bound water molecules establish hydrogen bonds with the fluoride atoms in Ni2F2BTDD but not with the chloride and bromide atoms in Ni2Cl2BTDD and Ni2Br2BTDD. This different spatial arrangement can be explained by considering the larger electronegativity and smaller size of fluoride compared to chloride and bromide, which lead to significantly stronger fluoride–water interactions.67 The trend is reversed in the HW–n RDFs shown in Fig. 4F, with the H atoms of the Ni-bound water molecules approaching the N atoms of the triazolate linkers more closely in Ni2Cl2BTDD and Ni2Br2BTDD than in Ni2F2BTDD. The Ni-bound water molecules act as anchor points, initiating the formation of extended hydrogen-bonding networks by donating a hydrogen bond to a second water molecule, hereafter referred to as the H-bonded molecule (shown inside the pink area in Fig. 3A–C).
 | ||
| Fig. 4 (A–C) Spatial distributions of one water molecule adsorbed in Ni2F2BTDD, Ni2Cl2BTDD, and Ni2Br2BTDD, respectively. Red isosurfaces correspond to water oxygen atoms, and white isosurfaces correspond to water hydrogen atoms. Color scheme of atoms in panels A through C: Ni = orange, F = purple, Cl = green, Br = pink, N = blue, C = cyan, O = red, H = white. (D–F) Radial distribution functions (RDFs) describing spatial correlations for the OW–Ni (the first peak of the RDF is magnified in the inset), HW–X, and HW–n atom pairs, respectively, calculated for water in Ni2F2BTDD (red curves), Ni2Cl2BTDD (green curves), and Ni2Br2BTDD (blue curves). Refer to Fig. S1 and S2 in the ESI† for the complete list of atom labels. | ||
Analysis of the MD trajectories and associated vibrational density of states (Fig. 3D–F) reveals that the Ni-bound water molecules in all three MOFs exhibit a free-OH, contributing to the peak at ∼3700 cm−1. The second OH bond of the Ni-bound water molecules instead engages in hydrogen bonding with another water molecule, contributing to the peak at ∼3450 cm−1. The H-bonded water molecules also exhibit free-OH bonds that point toward the center of the pore and contribute to the peak at 3700 cm−1. The second OH bond of the H-bonded molecule instead interacts with the framework, forming weak hydrogen bonds with neighboring fluoride atoms in Ni2F2BTDD, and triazolate groups in Ni2Cl2BTDD and Ni2Br2BTDD, contributing to the peak at ∼3550–3600 cm−1. The IR spectra of water adsorbed in the three MOFs at low RH can thus be assigned to hydrogen-bonded water dimers that are anchored to the open Ni2+ centers. As the RH increases, the four distinct peaks grow into one broad and featureless band similar to that observed in the IR spectrum of liquid water, indicating the emergence of a continuum of hydrogen-bond topologies (Fig. S4 in the ESI†).
3.3 Pore filling
At a loading of 1H2O/Ni2+, which corresponds to ∼1–5% RH depending on the specific Ni2X2BTDD MOF, Fig. 5A–C show that the water molecules inside the pores of all three MOFs localize near the open Ni2+ sites, following the same structural arrangements identified in Fig. 4A–C, with the water density in Ni2F2BTDD being more elongated due to relatively stronger interactions with the fluoride atoms of the framework. As the RH increases to 3H2O/Ni2+ (Fig. 5D–F), the water molecules localized near different Ni2+ sites begin to connect to each other through the formation of bridging hydrogen bonds, as revealed by the increased water density near the organic linkers, which is particularly evident in Ni2F2BTDD and Ni2Cl2BTDD. It is important to note that a loading of 3H2O/Ni2+ corresponds to different RH values in the three MOFs due to differences in the thermodynamics of the adsorption process, as manifested in the different trends in the corresponding isotherms, enthalpies, and entropies of adsorption. Specifically, a loading of 3H2O/Ni2+ is equivalent to 32.1% RH in Ni2F2BTDD and 31.7% RH in Ni2Cl2BTDD, which occur in correspondence of the pore filling step in the adsorption isotherms of both MOFs. In contrast, a loading of 3H2O/Ni2+ is below the onset of pore filling in Ni2Br2BTDD, corresponding to 24% RH. It is thus apparent that pore filling in the three Ni2X2BTDD MOFs occurs after the water molecules are able to form connected hydrogen-bonded chains that elongate along the pore channels and provide additional hydrogen-bonding sites. The different RH values needed for pore filling in the three MOFs is due to the different sizes of the fluoride, chloride, and bromide atoms that decorate the pore surface of the three frameworks, as originally suggested in ref. 19. In particular, due to their larger sizes, the bromide atoms of Ni2Br2BTDD protrude into the pore more than the fluoride and chloride atoms in the analogous MOFs. This implies that a relatively larger number of water molecules (i.e., higher RH) is needed to encircle the bromide atoms in order to form fully connected hydrogen-bonded chains that can then bridge with water molecules localized near the adjacent Ni2+ sites (Fig. 5D–F).Fig. 5G–I show well-defined density patterns of water inside the pores at the maximum water loading in each of the three MOFs. Due to stronger fluoride–water interactions, the first layer of water molecules in Ni2F2BTDD is able to template the hydrogen-bonding structure of the additional water molecules down to the center of the pore. This templating effect becomes progressively weaker from Ni2F2BTDD to Ni2Br2BTDD, following the same trend as the underlying halide–water interactions.68 Importantly, while water in Ni2Br2BTDD has the highest density near the Ni2+ sites, areas of little to no water density also exist right next to these regions. These areas of low water density are a direct consequence of the bromide atoms protruding further into the pores than the other two halide atoms, effectively hindering the formation of well-connected hydrogen-bonding networks that manifest in a relatively more disordered distribution of water molecules inside the pores (Fig. 5I).
Fig. 6 shows the distributions of different hydrogen-bonding topologies, classified in terms of the average number of hydrogen bonds that each water molecule donates (nD) and accepts (mA), determined from MD simulations at the maximum water loading in each of the three Ni2X2BTDD MOFs. In Ni2F2BTDD, a larger fraction of water molecules donate a hydrogen bond to the fluoride atoms of the framework, while still accepting hydrogen bonds from the neighboring water molecules. On the other hand, water molecules in Ni2Cl2BTDD and Ni2Br2BTDD, on average, donate a relatively larger number of hydrogen bonds to the N atoms of the triazolate linkers. The lifetime of the H2O–X and H2O–N hydrogen bonds also vary, reflecting the strength of the underlying interactions. Specifically, the hydrogen-bond correlation functions, which directly report on the probability that a hydrogen bond remains intact for a given time,75 indicate that water has the longest hydrogen-bond lifetime in Ni2Br2BTDD when interacting with the N atom (Table 1 and Fig. S9 in the ESI†). Long hydrogen-bond lifetimes are also calculated for the hydrogen bonds established by the water molecules with the fluoride atoms in Ni2F2BTDD and the N atoms in Ni2Cl2BTDD. The differences in hydrogen-bond lifetimes are rationalized by considering that the larger size of the bromide ions effectively push the water molecules toward the framework where they establish strong hydrogen bonds with the N atoms of the triazolate linkers. The distinct hydrogen-bonding arrangements developed by the water molecules in the pores of the three MOFs also explain the larger enthalpy of adsorption of water exhibited by Ni2Br2BTDD compared to Ni2Cl2BTDD.
| Hydrogen-bond type | τ HB (ps) |
|---|---|
| H2O–F in Ni2F2BTDD | 77 |
| H2O–N in Ni2F2BTDD | 20 |
| H2O–Cl in Ni2Cl2BTDD | 12 |
| H2O–N in Ni2Cl2BTDD | 78 |
| H2O–Br in Ni2Br2BTDD | 24 |
| H2O–N in Ni2Br2BTDD | >100 |
4 Conclusions
Exchanging the halide atoms in Ni2X2BTDD leads to three MOFs with different properties and water adsorption capacities. Ni2F2BTDD and Ni2Cl2BTDD adsorb over 100% of their weight in water at 32% RH, while Ni2Br2BTDD adsorbs 64% of its weight in water below 25% RH, one of the best water uptake values reported to date.19 By providing a realistic representation of water adsorption in all three MOFs, our MD simulations carried out with the MB-pol data-driven many-body potential provide unique, molecular-level insights into the underlying interactions and thermodynamic driving forces.Theoretical analyses of the simulated IR spectra allows for monitoring the evolution of extended hydrogen-bond networks as a function of RH. At low RH values, the main spectral features are assigned to free OH stretches, water–framework hydrogen bonds, water–water hydrogen bonds, and Fermi resonances, while a continuum of hydrogen-bonding environments exist at higher RH values, resembling the hydrogen-bond network of liquid water. All three MOFs exhibit higher enthalpies of water adsorption at low RH values, indicating stronger water–framework interactions than water–water interactions. Furthermore, Ni2F2BTDD exhibits the largest enthalpy of adsorption at all RH values, which directly correlates with relatively stronger fluoride–water interactions. These strong interactions are likely responsible for the partial amorphization and decrease in water uptake capacity of Ni2F2BTDD after the first adsorption cycle. On the other hand, the distinct physico-chemical properties of the chloride and bromide atoms play a central role in modulating water adsorption in Ni2Cl2BTDD and Ni2Br2BTDD. Specifically, the water adsorption capacity is greater in Ni2Cl2BTDD than Ni2Br2BTDD because of the smaller atomic size of Cl, resulting in a larger void space within the pores. At low RH values, the water molecules saturate the undercoordinated Ni2+ centers of both Ni2Cl2BTDD and Ni2Br2BTDD frameworks before establishing hydrogen bonds with the N atoms of the triazolate linkers. Due to the large size of the bromide ions, the water molecules in Ni2Br2BTDD protrude into the pore to the greatest extent, which favors the development of extended hydrogen-bonding networks and the onset of pore filling at the lowest RH value compared to Ni2F2BTDD and Ni2Cl2BTDD. Overall, Ni2Cl2BTDD exhibits the lowest enthalpy of adsorption, which is a critical parameter for AWH applications determining the energy input required to desorb water from the MOF pores. Importantly, the entropy of adsorption calculated for water in the three MOFs exhibits the opposite trend to the enthalpy of adsorption, with higher values for water in Ni2Cl2BTDD at low RH and a progressive decrease toward the value for liquid water at high RH. The distinct trends in thermodynamic properties, spatial arrangements, and mobility of the water molecules in the three Ni2X2BTDD MOFs directly correlate with the lifetime of the underlying hydrogen bonds, which are particularly long for water hydrogen-bonded to the N atoms of the triazolate linkers in Ni2Br2BTDD.
Besides providing molecular-level insights into how structural and chemical modifications of topologically identical frameworks modulate the water adsorption capacities of MOFs, the in-depth view into the structure, thermodynamics, and dynamics of water confined in Ni2X2BTDD gained from MD simulations with the MB-pol data-driven many-body potential highlight the key role of computer simulations with realistic models in guiding the design and optimization of energy-efficient water harvesting materials.
Data availability
All molecular dynamics trajectories and computer codes used in the analyses presented in this study are available from the authors upon request.Author contributions
F. P. conceived the research. K. M. H. and F. P. designed the research. K. M. H. performed the simulations. K. M. H. and F. P. analyzed and discussed the results, and wrote the paper. F. P. acquired funding and administered the project.Conflicts of interest
There are no conflicts to declare.Acknowledgements
This research was supported by the National Science Foundation through award no. 2311260. All simulations were carried out on Expanse at the San Diego Supercomputer Center (SDSC) through allocation CHE230052 from the Advanced Cyberinfrastructure Coordination Ecosystem: Services & Support (ACCESS) program, which is supported by NSF grants no. 2138259, 2138286, 2138307, 2137603, and 2138296, as well as Triton Shared Computing Cluster (TSCC) at SDSC.Notes and references
- M. M. Mekonnen and A. Y. Hoekstra, Sci. Adv., 2016, 2, e1500323 CrossRef PubMed.
- A. Dinar, A. Tieu and H. Huynh, Global Food Secur., 2019, 23, 212–226 CrossRef.
- M. Salehi, Environ. Int., 2022, 158, 106936 CrossRef PubMed.
- Y. Tu, R. Wang, Y. Zhang and J. Wang, Joule, 2018, 2, 1452–1475 CrossRef CAS.
- K. Yang, T. Pan, Q. Lei, X. Dong, Q. Cheng and Y. Han, Environ. Sci. Technol., 2021, 55, 6542–6560 CrossRef CAS PubMed.
- X. Zhou, H. Lu, F. Zhao and G. Yu, ACS Mater. Lett., 2020, 2, 671–684 CrossRef CAS.
- Z. Ji, T. Li and O. M. Yaghi, Science, 2020, 369, 674–680 CrossRef CAS PubMed.
- S. Canossa, Z. Ji, C. Gropp, Z. Rong, E. Ploetz, S. Wuttke and O. M. Yaghi, Nat. Rev. Mater., 2023, 8, 331–340 CrossRef.
- Z. Wang and S. M. Cohen, Chem. Soc. Rev., 2009, 38, 1315–1329 RSC.
- K. K. Tanabe and S. M. Cohen, Chem. Soc. Rev., 2011, 40, 498–519 RSC.
- M. Kalaj and S. M. Cohen, ACS Cent. Sci., 2020, 6, 1046–1057 CrossRef CAS PubMed.
- H. Furukawa, F. Gándara, Y.-B. Zhang, J. Jiang, W. L. Queen, M. R. Hudson and O. M. Yaghi, J. Am. Chem. Soc., 2014, 136, 4369–4381 CrossRef CAS PubMed.
- A. Cadiau, J. S. Lee, D. Damasceno Borges, P. Fabry, T. Devic, M. T. Wharmby, C. Martineau, D. Foucher, F. Taulelle, C.-H. Jun, Y. K. Hwang, N. Stock, M. F. De Lange, F. Kapteijn, J. Gascon, G. Maurin, J.-S. Chang and C. Serre, Adv. Mater., 2015, 27, 4775–4780 CrossRef CAS PubMed.
- D. Fröhlich, E. Pantatosaki, P. D. Kolokathis, K. Markey, H. Reinsch, M. Baumgartner, M. A. van der Veen, D. E. De Vos, N. Stock, G. K. Papadopoulos, S. K. Henninger and C. Janiakothers, J. Mater. Chem. A, 2016, 4, 11859–11869 RSC.
- A. J. Rieth, S. Yang, E. N. Wang and M. Dincă, ACS Cent. Sci., 2017, 3, 668–672 CrossRef CAS PubMed.
- M. Gao, J. Wang, Z. Rong, Q. Shi and J. Dong, RSC Adv., 2018, 8, 39627–39634 RSC.
- S. M. T. Abtab, D. Alezi, P. M. Bhatt, A. Shkurenko, Y. Belmabkhout, H. Aggarwal, Ł. J. Weseliński, N. Alsadun, U. Samin, M. N. Hedhili and M. Eddaoudi, Chem, 2018, 4, 94–105 Search PubMed.
- N. Hanikel, M. S. Prévot, F. Fathieh, E. A. Kapustin, H. Lyu, H. Wang, N. J. Diercks, T. G. Glover and O. M. Yaghi, ACS Cent. Sci., 2019, 5, 1699–1706 CrossRef CAS PubMed.
- A. J. Rieth, A. M. Wright, G. Skorupskii, J. L. Mancuso, C. H. Hendon and M. Dincă, J. Am. Chem. Soc., 2019, 141, 13858–13866 CrossRef CAS PubMed.
- Z. Chen, P. Li, X. Zhang, P. Li, M. C. Wasson, T. Islamoglu, J. F. Stoddart and O. K. Farha, J. Am. Chem. Soc., 2019, 141, 2900–2905 CrossRef CAS PubMed.
- J. Canivet, A. Fateeva, Y. Guo, B. Coasne and D. Farrusseng, Chem. Soc. Rev., 2014, 43, 5594–5617 RSC.
- N. C. Burtch, H. Jasuja and K. S. Walton, Chem. Rev., 2014, 114, 10575–10612 CrossRef CAS PubMed.
- A. J. Rieth, K. M. Hunter, M. Dincă and F. Paesani, Nat. Commun., 2019, 10, 4771 CrossRef PubMed.
- X. Liu, X. Wang and F. Kapteijn, Chem. Rev., 2020, 120, 8303–8377 CrossRef CAS PubMed.
- N. Hanikel, X. Pei, S. Chheda, H. Lyu, W. Jeong, J. Sauer, L. Gagliardi and O. M. Yaghi, Science, 2021, 374, 454–459 CrossRef CAS PubMed.
- G. A. Cisneros, K. T. Wikfeldt, L. Ojamäe, J. Lu, Y. Xu, H. Torabifard, A. P. Bartók, G. Csányi, V. Molinero and F. Paesani, Chem. Rev., 2016, 116, 7501–7528 CrossRef CAS PubMed.
- V. Babin, C. Leforestier and F. Paesani, J. Chem. Theory Comput., 2013, 9, 5395–5403 CrossRef CAS PubMed.
- V. Babin, G. R. Medders and F. Paesani, J. Chem. Theory Comput., 2014, 10, 1599–1607 CrossRef CAS PubMed.
- G. R. Medders, V. Babin and F. Paesani, J. Chem. Theory Comput., 2014, 10, 2906–2910 CrossRef CAS PubMed.
- F. Paesani, Acc. Chem. Res., 2016, 49, 1844–1851 CrossRef CAS PubMed.
- S. K. Reddy, S. C. Straight, P. Bajaj, C. Huy Pham, M. Riera, D. R. Moberg, M. A. Morales, C. Knight, A. W. Götz and F. Paesani, J. Chem. Phys., 2016, 145, 194504 CrossRef PubMed.
- S. E. Brown, A. W. Götz, X. Cheng, R. P. Steele, V. A. Mandelshtam and F. Paesani, J. Am. Chem. Soc., 2017, 139, 7082–7088 CrossRef CAS PubMed.
- G. R. Medders and F. Paesani, J. Chem. Theory Comput., 2015, 11, 1145–1154 CrossRef CAS PubMed.
- S. K. Reddy, D. R. Moberg, S. C. Straight and F. Paesani, J. Chem. Phys., 2017, 147, 244504 CrossRef PubMed.
- G. R. Medders and F. Paesani, J. Am. Chem. Soc., 2016, 138, 3912–3919 CrossRef CAS PubMed.
- D. R. Moberg, S. C. Straight and F. Paesani, J. Phys. Chem. B, 2018, 122, 4356–4365 CrossRef CAS PubMed.
- D. R. Moberg, S. C. Straight, C. Knight and F. Paesani, J. Phys. Chem. Lett., 2017, 8, 2579–2583 CrossRef CAS PubMed.
- D. R. Moberg, D. Becker, C. W. Dierking, F. Zurheide, B. Bandow, U. Buck, A. Hudait, V. Molinero, F. Paesani and T. Zeuch, Proc. Natl. Acad. Sci. U. S. A., 2019, 116, 24413–24419 CrossRef CAS PubMed.
- T. E. Gartner III, K. M. Hunter, E. Lambros, A. Caruso, M. Riera, G. R. Medders, A. Z. Panagiotopoulos, P. G. Debenedetti and F. Paesani, J. Phys. Chem. Lett., 2022, 13, 3652–3658 CrossRef CAS PubMed.
- S. L. Bore and F. Paesani, Nat. Commun., 2023, 14, 3349 CrossRef CAS PubMed.
- G. Kresse and J. Hafner, Phys. Rev. B: Condens. Matter Mater. Phys., 1993, 47, 558 CrossRef CAS PubMed.
- G. Kresse and J. Hafner, Phys. Rev. B: Condens. Matter Mater. Phys., 1994, 49, 14251 CrossRef CAS PubMed.
- G. Kresse and J. Furthmüller, Comput. Mater. Sci., 1996, 6, 15–50 CrossRef CAS.
- G. Kresse and J. Furthmüller, Phys. Rev. B: Condens. Matter Mater. Phys., 1996, 54, 11169 CrossRef CAS PubMed.
- J. P. Perdew, K. Burke and M. Ernzerhof, Phys. Rev. Lett., 1996, 77, 3865 CrossRef CAS PubMed.
- S. Grimme, J. Antony, S. Ehrlich and H. Krieg, J. Chem. Phys., 2010, 132, 154104 CrossRef PubMed.
- P. E. Blöchl, Phys. Rev. B: Condens. Matter Mater. Phys., 1994, 50, 17953 CrossRef PubMed.
- G. Kresse and D. Joubert, Phys. Rev. B: Condens. Matter Mater. Phys., 1999, 59, 1758 CrossRef CAS.
- J. Wang, R. M. Wolf, J. W. Caldwell, P. A. Kollman and D. A. Case, J. Comput. Chem., 2004, 25, 1157–1174 CrossRef CAS PubMed.
- A. V. Marenich, S. V. Jerome, C. J. Cramer and D. G. Truhlar, J. Chem. Theory Comput., 2012, 8, 527–541 CrossRef CAS PubMed.
- A. K. Rappé, C. J. Casewit, K. Colwell, W. A. Goddard III and W. M. Skiff, J. Am. Chem. Soc., 1992, 114, 10024–10035 CrossRef.
- Y. Wang and J. M. Bowman, J. Chem. Phys., 2011, 134, 154510 CrossRef PubMed.
- X. Cheng and R. P. Steele, J. Chem. Phys., 2014, 141, 104105 CrossRef PubMed.
- X. Cheng, J. J. Talbot and R. P. Steele, J. Chem. Phys., 2016, 145, 124112 CrossRef PubMed.
- K. M. Hunter, F. A. Shakib and F. Paesani, J. Phys. Chem. B, 2018, 122, 10754–10761 CrossRef CAS PubMed.
- J. L. Abascal and C. Vega, J. Chem. Phys., 2005, 123, 234505 CrossRef CAS PubMed.
- A. Hudait, D. R. Moberg, Y. Qiu, N. Odendahl, F. Paesani and V. Molinero, Proc. Natl. Acad. Sci. U. S. A., 2018, 115, 8266–8271 CrossRef CAS PubMed.
- W. Smith and T. Forester, J. Mol. Graphics, 1996, 14, 136–141 CrossRef CAS PubMed.
- L. Martínez, R. Andrade, E. G. Birgin and J. M. Martínez, J. Comput. Chem., 2009, 30, 2157–2164 CrossRef PubMed.
- S.-T. Lin, P. K. Maiti and W. A. Goddard III, J. Phys. Chem. B, 2010, 114, 8191–8198 CrossRef CAS PubMed.
- M. E. Tuckerman, Statistical Mechanics: Theory and Molecular Simulation, Oxford University Press, 2010 Search PubMed.
- G. J. Martyna, A. Hughes and M. E. Tuckerman, J. Chem. Phys., 1999, 110, 3275–3290 CrossRef CAS.
- A. R. Leach, Molecular Modelling: Principles and Applications, Pearson Education Limited, Essex, UK, 2nd edn, 2001 Search PubMed.
- M. Riera, C. Knight, E. F. Bull-Vulpe, X. Zhu, H. Agnew, D. G. A. Smith, A. C. Simmonett and F. Paesani, J. Chem. Phys., 2023, 159, 054802 CrossRef CAS PubMed.
- MBX v1.0, 2021, http://paesanigroup.ucsd.edu/software/mbx.html Search PubMed.
- A. P. Thompson, H. M. Aktulga, R. Berger, D. S. Bolintineanu, W. M. Brown, P. S. Crozier, P. J. Veld, A. Kohlmeyer, S. G. Moore, T. D. Nguyen, R. Shan, M. J. Stevens, J. Tranchida, C. Trott and S. J. Plimpton, Comput. Phys. Commun., 2022, 271, 108171 CrossRef CAS.
- P. Bajaj, A. W. Gotz and F. Paesani, J. Chem. Theory Comput., 2016, 12, 2698–2705 CrossRef CAS PubMed.
- F. Paesani, P. Bajaj and M. Riera, Adv. Phys.: X, 2019, 4, 1631212 CAS.
- P. Bajaj, X.-G. Wang, T. Carrington Jr and F. Paesani, J. Chem. Phys., 2017, 148, 102321 CrossRef PubMed.
- P. Bajaj, M. Riera, J. K. Lin, Y. E. Mendoza Montijo, J. Gazca and F. Paesani, J. Phys. Chem. A, 2019, 123, 2843–2852 CrossRef CAS PubMed.
- K. M. Hunter, J. C. Wagner, M. Kalaj, S. M. Cohen, W. Xiong and F. Paesani, J. Phys. Chem. C, 2021, 125, 12451–12460 CrossRef CAS.
- H. Bakker and J. Skinner, Chem. Rev., 2010, 110, 1498–1517 CrossRef CAS PubMed.
- A. A. Kananenka and J. Skinner, J. Chem. Phys., 2018, 148, 244107 CrossRef PubMed.
- M. Rossi, H. Liu, F. Paesani, J. Bowman and M. Ceriotti, J. Chem. Phys., 2014, 141, 181101 CrossRef PubMed.
- A. Luzar and D. Chandler, Nature, 1996, 379, 55–57 CrossRef CAS.
Footnote |
| † Electronic supplementary information (ESI) available: Details about the molecular models used in the MD simulations along with the complete list of force field parameters used to describe the Ni2X2BTDD framework. See DOI: https://doi.org/10.1039/d3sc06162k |
| This journal is © The Royal Society of Chemistry 2024 |