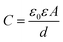Open Access Article
Open Access ArticleHigh-throughput single biomarker identification using droplet nanopore†
Lin-Lin
Zhang
 a,
Cheng-Bing
Zhong
a,
Ting-Jing
Huang
a,
Li-Min
Zhang
b,
Feng
Yan
b and
Yi-Lun
Ying
a,
Cheng-Bing
Zhong
a,
Ting-Jing
Huang
a,
Li-Min
Zhang
b,
Feng
Yan
b and
Yi-Lun
Ying
 *acd
*acd
aMolecular Sensing and Imaging Center, School of Chemistry and Chemical Engineering, Nanjing University, Nanjing 210023, P. R. China. E-mail: yilunying@nju.edu.cn
bSchool of Electronic Science and Engineering, Nanjing University, Nanjing 210023, P. R. China
cState Key Laboratory of Analytical Chemistry for Life Science, Nanjing University, Nanjing 210023, P. R. China
dChemistry and Biomedicine Innovation Center, Nanjing University, Nanjing 210023, P. R. China
First published on 10th April 2024
Abstract
Biomarkers are present in various metabolism processes, demanding precise and meticulous analysis at the single-molecule level for accurate clinical diagnosis. Given the need for high sensitivity, biological nanopore have been applied for single biomarker sensing. However, the detection of low-volume biomarkers poses challenges due to their low concentrations in dilute buffer solutions, as well as difficulty in parallel detection. Here, a droplet nanopore technique is developed for low-volume and high-throughput single biomarker detection at the sub-microliter scale, which shows a 2000-fold volume reduction compared to conventional setups. To prove the concept, this nanopore sensing platform not only enables multichannel recording but also significantly lowers the detection limit for various types of biomarkers such as angiotensin II, to 42 pg. This advancement enables direct biomarker detection at the picogram level. Such a leap forward in detection capability positions this nanopore sensing platform as a promising candidate for point-of-care testing of biomarker at single-molecule level, while substantially minimizing the need for sample dilution.
Introduction
The detection of biomarkers is one of the most common methods for diagnosing disease, forecasting future ailments, and tracking treatment responses.1–4 DNA and peptides have traditionally served as the cornerstone for biomarker tests, indicating various medical conditions.5,6 However, owing to their low-abundance and low-volume levels in biological samples, the accurate detection of biomarkers poses the greatest challenges in providing clinically valuable details about pathological conditions.7–9 Single-molecule biomarker detection technologies provide the capability for sensing with high sensitivity, low sample volume, and high throughput.10–14 Compared to atomic force microscopy, single-molecule fluorescence microscopy and other single-molecule methods, nanopore-based single-molecule electrochemical technology has evolved as an ultrasensitive analytical tool for single biomarker detection, because of its properties including label-free, real-time, low-cost and portability.15–17 The biological nanopore show great possibility for biomarker sensing, which enables qualitative and quantitative analysis of DNA and peptides by identifying and counting characteristic ionic current blockages.18–20 However, for achieving nanopore-based single biomarker identification within a high dynamic range, the measuring systems require (i) low sample consumption with minimum dilution, (ii) successfully repeated formation of lipid bilayer membranes for reproducible nanopore sensing, and (iii) a parallel and integrated multichannel platform for high-throughput biomarker detection.To meet these demands, one feasible approach is to develop a high-throughput and low-sample-volume artificial lipid bilayer formation system.21 Conventionally, the “painting”, “folding” or “vesicle” methods22,23 are used for bilayer formation, which are well-established but challenging to integrate and scale up for high-throughput applications. Current studies make use of microchips to develop platforms for multichannel Black Lipid Membrane (BLM) formation.24,25 The microchip could be integrated with microfluidics for automatic and high-throughput nanopore sensing.26,27 These methods require a relatively substantial sample volume, usually exceeding several hundred microliters.28 In order to further minimize the sample volume, the droplet interface bilayer (DIB) method has been employed in nanopore sensing with micro-to-nano litre volumes.29,30 The DIB method uses two contacted aqueous droplets together within a bulk oil phase in the presence of lipids, allowing for biological nanopore insertion.31 A two-electrode system manipulates a pair of droplets, bringing them closer together while simultaneously measuring the ionic current.32 This technique offers a number of advantages over conventional planar bilayer systems, including low sample volume, good stability, an asymmetric interface, and easy integration for point-of-care use.32,33 To further scale up the system feature, it is challenging to precisely position each droplet to ensure contact with the electrode array for bilayer formation. One potential solution involves incorporating hydrophilic and hydrophobic supports on the microchip to facilitate droplet localization.34–36
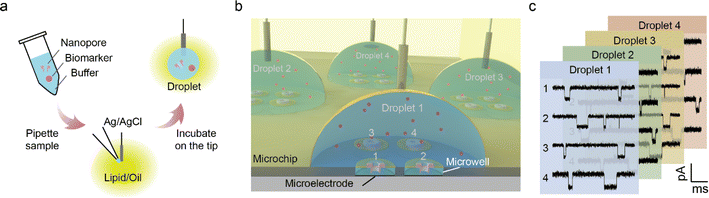
Herein, we developed a droplet nanopore array technique to meet the low-sample-volume, high-throughput and easy-to-maneuver demands. As illustrated in Fig. 1, micro-to-nanolitre droplets, premixed with biological nanopores, analyte and buffer, are pipetted into the oil and lipid phase. The droplet nanopore microchip method achieves high-throughput single biomarker sensing with the limit of detection at the picogram level, which paves the way towards the commercialization of biological nanopores for low-volume clinical biomolecule analysis.
Results and discussion
Design and characterization of microchips
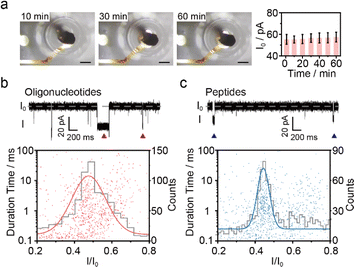
In the first step, the nanopore microchip was fabricated by photolithography, and was designed with separated multi-microwells for supporting multiple bilayers. Each microwell is integrated with an Ag/AgCl microelectrode connecting to an independent amplifier, allowing for multichannel current recording. There are 4 × 4 channels designed to validate the feasibility of the microchip (Fig. S1 and S2†). In brief, a quartz slide (SiO2) was chosen as the substrate because of its excellent properties,37 including a smooth surface for benefiting the bilayer formation and the low loss-conductance for reducing dielectric noise (Fig. S3†). The Ag layer on the top of the Au wire undergoes oxidation, resulting in the formation of irregular and rough Ag/AgCl microelectrodes for providing stable potential (Fig. S4†). To evaluate the stability of bias potential, we measured the open-circuit potential (OCP) of 3 individual Ag/AgCl microelectrodes on a microchip (Fig. 2a and S5†). The OCP values remained relatively stable, with variations of less than 4.0 mV during the continuous recording over 1 hour. Notably, over 3 consecutive days, the microelectrode maintained a stable OCP within the range of 4.8 ± 1.0 mV, demonstrating its long-term ability for electrochemical response. Subsequently, to support bilayers, a hydrophobic passivation layer, SU-8,38,39 was patterned on the microelectrodes to construct microwells with diameters ranging from 50 μm to 150 μm. The contact angle confirmed the hydrophobic nature of SU-8 for lipid adsorption (Fig. S6†), which is consistent with a previous report.40 The depths and profiles of microwells showed uniform fabrication quality (Fig. 2b and S7†). Specifically, the top surface of the microwell exhibited a sufficiently horizontal profile, providing appropriate support for the membrane. Next, the previously reported air bubble method28 was incorporated to verify the ability of the microchip for nanopore sensing. In brief, an air bubble (∼2 μL) was squeezed using a pipette dipped in lipid and manipulated across the microwells to facilitate the spreading of membranes (Fig. S8†). The homemade microchip was connected to an Orbit 16 for multichannel current recording to verify its performance. In our design, the neighbouring microwells have been positioned at a sufficient distance (1.2 mm) to ensure that the membrane formation on each microwell does not interfere with each other. The wild-type (WT) aerolysin (AeL) nanopore sensing of single oligonucleotide denoted as poly(dA)4 (5′-AAAA-3′, dA4), shows good reproducibility as demonstrated by previous studies.41 Here, we incorporated this model system for the verification. As shown in Fig. 2c, the typical blockage current signals of dA4, which was synthesized and HPLC-purified by Sangon Biotech Co., Ltd (Shanghai, China), exhibit no distortion, suggesting low capacitance introduced by the designed microchip. Moreover, the statistical results of blockage current and duration are consistent with a previous report.41 Simultaneous recording from multiple channels and long-time monitoring (>1.5 h) demonstrated the good capability of the microchip for high-throughput nanopore sensing (Fig. S9†). The standard deviation (STD) value of the open-pore current (I0) reveals that microwells with the minimum diameter of 50 μm generate the lowest noise (Fig. 2d), as demonstrated by previous reports.42,43 Thus, in this design, the microwell with a diameter of 50 μm was chosen for further nanopore sensing.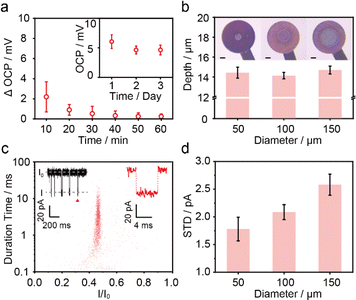 | ||
| Fig. 2 The characterization of microchips. (a) The OCP changes of Ag/AgCl microelectrodes with diameters of 100 μm vs. a Ag/AgCl commercial electrode in 1.0 M Tris–KCl, 1.0 mM EDTA, pH 8.0. Error bars depict the STD of 3 independent measurements. Inset: the OCP of one microelectrode over 3 consecutive days. The data are calculated for each minute with the error indicating daily fluctuations in continuous recording. The raw current trace is shown in Fig. S5.† (b) The optical images and depths of microwells with diameters of 50 μm, 100 μm, and 150 μm. Error bars depict the STD of 3 independent channels within one microchip. The scale bar is 50 μm. (c) Scatter plots and a typical current trace of 1 μM dA4 detected with WT AeL using the air bubble method on the homemade microchip featuring microwells with a diameter of 50 μm. The data were recorded over a 5 min period in the scatter plots (2468 events). Here, I represents the blockade residual current, I0 is the open-pore current, and I/I0 reflects the residual current depth. (d) The STD values of I0 on microwells with diameters of 50 μm, 100 μm and 150 μm. The error bars were calculated from 3 independent experiments. All current data were acquired at +100 mV in 1.0 M KCl, 10.0 mM Tris, and 1.0 mM EDTA at pH 8.0 with 5 kHz filtering and a 100 kHz sampling rate using an Orbit 16 instrument. | ||
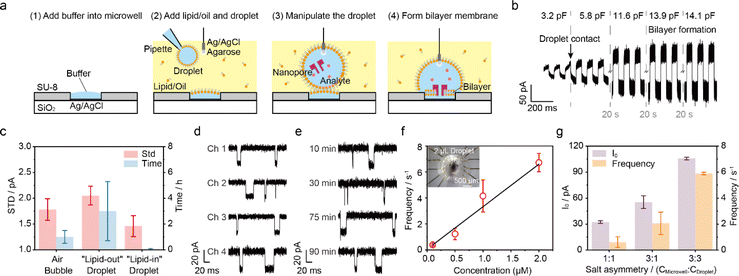
Droplet method for membrane formation
As shown in Fig. 3a, to facilitate droplet nanopore sensing with low sample volume, the aqueous droplet was added into the oil phase and positioned onto the top of the microwell. In detail, add a buffer solution into microwells (ESI Movie 1†) and fill the top chamber with oil solution instantly to prevent the evaporation of the buffer from microwells. Then, pipette a droplet onto the end of the agarose-coated Ag/AgCl electrode. As the droplet comes into contact with the microwell, the lipid layer thinned spontaneously, followed by bilayer generation within 1 min, with the capacitance response (Fig. 3b). In case of the membranes rupture or unintended multi-nanopore insertion, the membranes could be effortlessly reformed by manipulating the droplet (Fig. S10†). The lipid could be either premixed in the droplet or added into the oil phase, denoted as the “lipid-in” (Fig. S11†) and “lipid-out” (Fig. 3a) method,44 respectively. These two methods were also verified using the commercialized MECA-16 chips due to their mature performance. As a result, the droplet methods generate a comparable current signal of dA4 detection with WT AeL compared to the air bubble method (Fig. S12†). Specifically, the “lipid-out” method provided a long-time stable current trace while the “lipid-in” led to current instantaneous fluctuations in the baseline. But the “lipid-in” droplet method showed a lower STD value at 1.46 ± 0.20 pA, when employing only the short stable baseline value of I0 to evaluate the noise (Fig. 3c). To achieve long-time recording, we used the “lipid-out” droplet method for low-volume nanopore sensing. We pipetted a 2 μL droplet onto MECA-16, which covered all 16 microwells for multichannel recording. The “lipid-out” droplet method exhibited parallel recording of more than 4 channels and continuous recording lasting for more than 1.5 h, extending to even beyond 5 h (Fig. 3d, e, S13 and ESI Movie 2†), while maintaining accuracy and minimizing additional errors from method variations (Fig. S14†). Longer and more consistent droplet nanopore sensing could be further achieved by incorporating improvements such as stabilizing the potential of Ag/AgCl electrodes, suppressing the suspension of membranes, precisely controlling the concentration of nanopore forming proteins for single-channel recording, and optimizing the oil volume to prevent droplet evaporation. Furthermore, a linear relationship between event frequency and analyte concentrations was achieved (Fig. 3f). In a 2 μL droplet, the lowest detectable quality of 0.1 μM dA4 is 254 pg, showing the capability of the droplet method in minimizing sample volumes. The high salt concentration and asymmetric salt concentration can enhance the capture frequency for biomarker blockade.45,46 The lipid bilayer formed by the “lipid-out” droplet method could withstand both high and asymmetric salt concentrations without salt precipitation (Fig. 3g and S15†). This robustness leads to an increased capture frequency, enabling the detection of low-volume biomarkers. The droplet volume in the “lipid-out” method was further reduced from 2 μL to 0.4 μL when employing a homemade microchip with 50 μm microwells (Fig. S16†), thereby concomitantly decreasing the required sample volume. While adopting a 0.4 μL droplet for nanopore sensing, there was no significant evaporation of the droplet due to the presence of oil, as proved with the consistent volumes and I0 (Fig. 4a). As shown in Fig. 4b, 51 pg (0.1 μM) dA4 within a 0.4 μL droplet was successfully detected using the WT AeL-incorporated nanopore microchip.As angiotensin II (Ang II) is a biomarker of influenza and cancer, here, we employed the “lipid-out” droplet method for Ang II analysis. Ang II is involved in the regulation of blood and human metabolism, and implicated in various diseases.47,48 Due to its low quantity in plasma, Ang II detection poses a big challenge.49 The utilization of T232K engineered AeL enables enhanced Ang II detection, characterized by prolonged duration and higher capture efficiency.50 A 0.4 μL droplet containing 42 pg (0.1 μM) Ang II was detected by the T232K AeL nanopores employing homemade microchips featuring microwells of 50 μm diameter (Fig. 4c). It's noteworthy that the sample quality was reduced by over 2000 times compared to the conventional Delrin cup with a volume of 1 mL50 while maintaining the same biomarker concentration.
Conclusions
In conclusion, we developed a droplet nanopore microchip technique which is capable of high-throughput and low-volume single biomarker detection at the picogram level. Utilizing sub-microliter droplets for nanopore sensing, biomarkers such as oligonucleotides and peptides could be effectively identified at qualities below 100 pg, which advanced conventional label-free single biomarker detection methods (Table S1†). This sensing platform facilitates the detection of low-volume samples with minimal buffer solution dilution, enabling integration with instrumentation and high-throughput recording without complex microfluidic systems.Importantly, in our design, each droplet spans multiple microwells for simultaneously multichannel recording, thereby enhancing the limit of detection for low-volume biomarkers. If one nanopore is blocked or its membrane ruptured, the remaining biomarkers could be preferentially analyzed by the adjacent open nanopore instead, increasing the probability of successful detection.51,52 Further improvements include the integration of an automated pico/nano-litre pipette array which could enable parallel droplet deposition onto the microwell array for high-throughput nanopore sensing. Additionally, although we successfully introduced an extra 1 μL solution into the droplet (Fig. S17†), exchanging the solution inside the droplet remains difficult. Using a nanopipette for ultra-low volume sample addition or solution exchange could enhance this technique's efficiency and functionality for detecting multiple samples or analyzing ion channels. Along with the advanced sample purification methods and target capture strategies, the sub-nanomole-level biomarkers would be anticipated to be readily detected even within complex biological samples. This advancement is attributed to the increased frequency of typical events as well as minimum sample dilution.
However, the droplet method may introduce more background signals (Fig. S18†), which may be because of the concentrated impurity within the confined droplet.53,54 Combining technologies of sample pretreatment, such as protein purification columns, non-specific signals could be decreased efficiently with little sample loss. Furthermore, by employing machine learning algorithms, we can further push the limits of high-sensitivity and high-accuracy biomarker sensing. Therefore, the droplet nanopores indeed pave the way towards the commercialization of biological nanopores for high-throughput, low-volume and portable clinical biomarker analysis by virtue of minimizing the need for extensive dilution.
Experimental section
Microchip fabrication
The SiO2 substrate was cleaned by rinsing with ultrapure water and isopropanol, followed by N2 drying, and heated on a 120 °C hotplate for 10 min. Next, to define the conductor wire, standard lift-off photoresist, ROL-7133 (Kayaku Advanced Materials, Inc., Westborough, MA, USA), served as a lift-off mask for metallization. In order to remove the small amount of residue, O2 plasma cleaning (300 W, 2 min) was implemented to achieve a high aspect ratio55 using a plasma cleaning machine from PVA TePla America Inc (Corona, CA, USA). After defining the wire pattern, 10 nm Cr/150 nm Au (99.999%, Kurt J. Lesker Company®, Jefferson Hills, PA, USA) was deposited by using an electron beam evaporator (EBE) form Kurt J. Lesker Company® (Jefferson Hills, PA, USA). The chip was then soaked in acetone for 10 min to remove the redundant metal. Subsequently, an ROL-7133 photoresist was used again as a lift-off mask for metallization, following by depositing 500 nm Ag (99.999%, Kurt J. Lesker Company®, Jefferson Hills, PA, USA) as the electrode material by EBE. By immersing the chip in a sodium hypochlorite solution (NaClO, available chlorine 34.0 g L−1, Jiangsu Aitefu Co., Ltd, Huai'an, China) for 3 min, a layer of AgCl was formed, ultimately resulting in the fabrication of Ag/AgCl microelectrodes with a diameter of 300 μm. To support a suspended lipid bilayer, the SU-8 2010 photoresist (Kayaku Advanced Materials, Inc., Westborough, MA, USA) was patterned into microwells with diameters of 50–150 μm on the microelectrodes by photolithography using a mask-aligner from SUSS MicroTec Solutions GmbH & Co. KG (Garching, Germany). In detail, a layer of SU-8 2010 was spun onto the chip at 500 rpm for 6 s and 3500 rpm for 30 s, giving a layer with a thickness of about 14 μm. Additionally, use acetone as the edge bead remover (EBR) reagent, to remove any edge bead after spinning the SU-8, if necessary, which is good for close contact between the photomask and resist, resulting in an improved resolution and aspect ratio with a flat surface.56 The reagents and materials we used are all of analytical grade.Microchip characterization
Scanning electron microscopy (SEM) images and element analysis were conducted on a SEM with an energy dispersive X-ray spectrometer (SEM/EDX S-4800) from Hitachi Ltd (Tokyo, Japan) at a voltage of 20.0 kV. The hydrophobicity of SU-8 was measured by using a contact angle meter from Dataphysics Instruments GmbH (Filderstadt, Germany). By dripping 3 μL of ultrapure water onto the surface of the microchip, covered with an SU-8 layer, the contact angle can be measured. The profiles of microelectrodes and microwells were collected by using a step profiler DektakXT from Bruker (Billerica, MA, USA). While testing the microelectrodes, the parameters were set with a range of 6.5 μm, scan duration of 10 s, scan length of 600 μm, and force of 3 mg. While testing the depth of microwells, the range was set at 65.5 μm, scan duration at 10 s, scan length at 300 μm, and force at 2 mg. An optical microscope from Shanghai Zhiqi Industrial Co., Ltd (Shanghai, China) was employed to observe the platform. The OCP measurements were performed on a CHI 852D from Shanghai Chenhua Co., Ltd (Shanghai, China) in 1 M Tris–KCl, 1 mM EDTA, pH 8.0 buffer solution versus a commercial Ag/AgCl electrode with a diameter of 2 mm (Wuhan Brain Link Technology, Wuhan, China) at a sampling interval time of 0.1 s. The OCP of 3 independent Ag/AgCl microelectrodes was measured for 1 hour. As shown in Fig. 2a, the change value of OCP in the first 10 min was calculated from the absolute value of the tenth second and tenth minutes because of the charge effect at the start of measurement (Fig. S5†), and the subsequent OCP change value was calculated from the absolute values of the initial and the final OCP value.Nanopore experiments
The proaerolysin production of WT and T232K was performed in our laboratory and the preparation methods have been reported in previous studies.50,57 The WT and T232K proaerolysin were activated by mixing with trypsin–EDTA (Sigma-Aldrich Co., Ltd, St. Louis, MO, USA), with a mass concentration ratio close to 3![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 50 (v/v) at room temperature for 5 h. Poly(dA)4 was synthesized and HPLC-purified by Sangon Biotech Co., Ltd (Shanghai, China). Angiotensin II (Ang II) was synthesized and HPLC-purified by GL Biochem (Shanghai) Ltd (Shanghai, China). All solutions were prepared using ultrapure water (18.2 MΩ cm at 25 °C) from a Milli-Q system (Billerica, MA, USA).
50 (v/v) at room temperature for 5 h. Poly(dA)4 was synthesized and HPLC-purified by Sangon Biotech Co., Ltd (Shanghai, China). Angiotensin II (Ang II) was synthesized and HPLC-purified by GL Biochem (Shanghai) Ltd (Shanghai, China). All solutions were prepared using ultrapure water (18.2 MΩ cm at 25 °C) from a Milli-Q system (Billerica, MA, USA).
The bilayer was formed by using either the air bubble or droplet method. To validate the fabricated microchips, the nanopore experiments were carried out by using an Orbit 16 from Nanion Technologies GmbH (Munich, Germany). To validate the droplet nanopore measurements, the commercial MECA-16 microchip (Ionera Technologies GmbH, Freiburg, Germany, available through Nanion Technologies GmbH) and Orbit 16 from Nanion Technologies GmbH (Munich, Germany) were employed. To achieve low sample volume and high-throughput detection, the fabricated microchip was connected to an Orbit 16 from Nanion Technologies GmbH (Munich, Germany). All the current data were sampled at 100 kHz and filtered at 5 kHz with the ranging of 200 pA at the applied voltage of +100 mV, unless otherwise noted. To prepare a Ag/AgCl wire electrode, polish a silver wire (0.25 mm diameter, Goodfellow, UK, 99.99%) with 3000 grit sandpaper, and then electroplate it in 10 wt% hydrochloric acid (HCl, 36 wt%, Nanjing Chemical Reagent Co., Ltd, Nanjing, China) for 10 min to form the Ag/AgCl wire electrode. The Ag/AgCl microelectrodes were under the microwell, while the Ag/AgCl wire inserted into the external chamber. Two Ag/AgCl electrodes were connected to the Orbit 16, allowing the application of a stable voltage and recording of the current between the two Ag/AgCl electrodes. All the nanopore experiments were conducted at 24 ± 3 °C to prevent freezing of oil, as the freezing point of hexadecane is 18 °C.31
Unless otherwise noted, for symmetric salt nanopore sensing, apply a voltage of +100 mV and use the solution of 1.0 M KCl and 10.0 mM Tris with 1.0 mM EDTA at pH 8.0 as the electrolyte buffer. While conducting high or asymmetric salt concentration nanopore sensing at an applied voltage of +60 mV, the solution in microwells consists of 3.0 M KCl and 10 mM Tris with 1.0 mM EDTA at pH 8.0 and the solution in the droplet contains 3.0 M KCl or 1.0 M KCl, both with 10.0 mM Tris and 1.0 mM EDTA at pH 8.0, respectively.
Membrane capacitance measurement
The current noise is dominated by parasitic capacitance, which is typically affected by the capacitance of the lipid bilayer membrane.58 The capacitance (C) of the bilayer membrane is given by:where ε0 is the absolute dielectric constant, ε is the relative dielectric constant of the membrane, A is the area of the membrane, and d is the thickness of the membrane.59
Air bubble method for membrane formation
150 μL of buffer solution was dispensed into the microchip chamber. Once the current overflowed, the electrochemical circuit was connected to the instrument. To prepare a lipid film, 1,2-diphytanoyl-sn-glycero-3-phosphocholine (DPhPC, ≥99%, Avanti Polar Lipids Inc., Alabaster, Al, USA) in chloroform (≥99.5%, Sinopharm Chemical Reagent Co., Ltd, Shanghai, China) was placed in a glass vial, and vacuumed for 2 h to remove the solvent completely. Then 30 mg mL−1 lipid solution was prepared by adding decane (Sigma-Aldrich Co., Ltd, St. Louis, MO, USA) to the DPhPC film. To generate a lipid membrane, a little lipid solution was touched by a pipette tip and placed on the surface of the microwell. An air bubble of 2 μL was squeezed by the pipette, and moved around microwells to spread the lipid membranes evenly.28 After covering the lipid solution on the microwell, the thickness and stability of the membrane were tested by applying a voltage to measure the membrane capacitance and membrane breaking voltage. If the membrane was too thick and unable to break, the pipette was used again to adjust the membrane thickness accordingly. Once a stable bilayer membrane was formed, add the nanopore protein and analyte into the chamber for current recording.“Lipid-out” droplet method for membrane formation
Add the buffer into the microchip and apply air bubble to ensure the conductivity of microelectrode array and commercial Ag/AgCl electrode by monitoring current overflow. Then the buffer solution in the external chamber was removed, leaving only the buffer in microwells. Afterward, an oil bath was prepared by filling the microchip with 100 μL lipid/oil solution instantly. This step was crucial to prevent the evaporation of the solution in the microwells. The lipid/oil solution consisted of 10 mg mL−1 DPhPC in a mixture of 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 (v/v) hexadecane (≥99.8%, Sigma-Aldrich Co., Ltd, St. Louis, MO, USA) and silicone AR 20 (Sigma-Aldrich Co., Ltd, St. Louis, MO, USA) (v/v). Subsequently, an 0.25 mm Ag/AgCl electrode, fixed on a three-axis micro-displacement platform (Zhejiang Star Pneumatic Co., Ltd, Leqing, China), was inserted into the oil chamber for droplet manipulation. But due to the hydrophobicity of the electrode inoil, it's hard to manipulate the aqueous droplet. In order to manipulate the droplet by using the Ag/AgCl electrode effectively, immerse the tip of the electrode with 3 wt% agarose in 1.0 M KCl, 10.0 mM Tris, and 1.0 mM EDTA at pH 8.0 for surface hydrophilic treatment.31 Next, to initiate the formation of a droplet, a 0.4–2 μL pre-prepared solution consisting of the buffer solution, nanopore forming proteins (∼1 μg mL−1), and analyte was pipetted onto the tip of the agarose-Ag/AgCl electrode. The microwells, along with the droplet, were then incubated for a period of 10–20 min, allowing for the self-assembly of lipid monolayers. Then, manipulate the micro-displacement platform to carefully position the droplet onto the microwell for bilayer membrane formation. Finally, apply a voltage of +100 mV for single nanopore insertion and nanopore sensing.
1 (v/v) hexadecane (≥99.8%, Sigma-Aldrich Co., Ltd, St. Louis, MO, USA) and silicone AR 20 (Sigma-Aldrich Co., Ltd, St. Louis, MO, USA) (v/v). Subsequently, an 0.25 mm Ag/AgCl electrode, fixed on a three-axis micro-displacement platform (Zhejiang Star Pneumatic Co., Ltd, Leqing, China), was inserted into the oil chamber for droplet manipulation. But due to the hydrophobicity of the electrode inoil, it's hard to manipulate the aqueous droplet. In order to manipulate the droplet by using the Ag/AgCl electrode effectively, immerse the tip of the electrode with 3 wt% agarose in 1.0 M KCl, 10.0 mM Tris, and 1.0 mM EDTA at pH 8.0 for surface hydrophilic treatment.31 Next, to initiate the formation of a droplet, a 0.4–2 μL pre-prepared solution consisting of the buffer solution, nanopore forming proteins (∼1 μg mL−1), and analyte was pipetted onto the tip of the agarose-Ag/AgCl electrode. The microwells, along with the droplet, were then incubated for a period of 10–20 min, allowing for the self-assembly of lipid monolayers. Then, manipulate the micro-displacement platform to carefully position the droplet onto the microwell for bilayer membrane formation. Finally, apply a voltage of +100 mV for single nanopore insertion and nanopore sensing.
“Lipid-in” droplet method for membrane formation
First, prepare 250–500 μg mL−1 liposome solution by mixing DPhPC in 1.0 M KCl, 10.0 mM Tris, and 1.0 mM EDTA at pH 8.0. The solution mixture was vortexed for 1 min and then extruded through a 220 nm filter (Sigma-Aldrich Co., Ltd, St. Louis, MO, USA) to form liposomes. The liposome solution is added to the microchip, followed by the removal of any excess solution. Next, the top chamber of the microchip was filled with 100 μL of a 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 (v/v) mixture of hexadecane and silicone AR 20. A 2 μL droplet of the pre-prepared liposome solution which contained nanopores and analyte, was pipetted onto the tip of the agarose-Ag/AgCl electrode. After approximately 5 min of incubation, the droplet was carefully moved onto the microwell, allowing the formation of bilayer membranes.
1 (v/v) mixture of hexadecane and silicone AR 20. A 2 μL droplet of the pre-prepared liposome solution which contained nanopores and analyte, was pipetted onto the tip of the agarose-Ag/AgCl electrode. After approximately 5 min of incubation, the droplet was carefully moved onto the microwell, allowing the formation of bilayer membranes.
Data processing software
All the current data were analyzed with MOSAIC 1.3 software.60 Usually, the scatter plots were divided into translocation events and collision events, according to previous studies.50 For further analysis, the collision events that were normally shorter than 0.1 ms were excluded. The current trace was visualized by using Clampfit 10.2 (Molecular Devices LLC, San Jose, CA, USA) and the analyzed results were shown using OriginLab 8.0 (OriginLab Corporation, Northampton, MA, USA). The extracted blockades were analyzed statistically by using Gaussian fittings to the histogram of the residual current depth (I/I0). The capture frequency of the blockades was calculated by using f = 1/τon, where τon represents the interval time. The τon was analyzed statistically by using Gaussian fitting to the histogram of the common logarithm.Data availability
All the experimental data related to this study are available from the corresponding author upon reasonable request.Author contributions
Y.-L. Y. conceived the original idea of the research, and L.-M. Z. and F. Y. supervised the project. L.-L. Z. and C.-B. Z. designed the platform. L.-L. Z. and T.-J. H. conducted the experiments. L.-L. Z. processed the data and wrote the manuscript. All authors discussed the results and commented on the manuscript.Conflicts of interest
There are no conflicts to declare.Acknowledgements
This research was supported by the National Key R&D Program of China (2022YFA1304604), National Natural Science Foundation of China (22027806 and 22090054), and the Sino-German Center Call for the Mobility Programme (M-0134). The authors thank Prof. Yi-Tao Long for the instrumentations support and helpful discussion. The authors thank Dr Pan-Ke Zhang for microchip fabrication training and help. The authors also thank Dr Meng-Yin Li, Jun-Ge Li, and Yan Gao for aerolysin protein production and helpful discussion, and Dr Si-Min Lu for the discussion of electrochemical measurements.References
- G. Jung, E. Hernandez-Illan, L. Moreira, F. Balaguer and A. Goel, Nat. Rev. Gastroenterol. Hepatol., 2020, 17, 111–130 CrossRef PubMed.
- O. Hansson, Nat. Med., 2021, 27, 954–963 CrossRef CAS PubMed.
- Y. Chang, Y. Chen, Y. Shao, B. Li, Y. Wu, W. Zhang, Y. Zhou, Z. Yu, L. Lu, X. Wang and G. Guo, Biosens. Bioelectron., 2021, 175, 112915 CrossRef CAS PubMed.
- X. Yang, F. Zhang, X. Jin, Y. Jiao, X. Zhang, Y. Liu, C. Amatore and W. Huang, Proc. Natl. Acad. Sci. U. S. A., 2023, 120, e2219994120 CrossRef CAS PubMed.
- S. V. S. Bratman, Y. C. Yang, M. A. J. Iafolla, Z. Liu, A. R. Hansen, P. L. Bedard, S. Lheureux, A. Spreafico, A. A. Razak, S. Shchegrova, M. Louie, P. Billings, B. Zimmermann, H. Sethi, A. Aleshin, D. Torti, K. Marsh, J. Eagles, I. Cirlan, Y. Hanna, D. L. Clouthier, S. C. Lien, P. S. Ohashi, W. Xu, L. L. Siu and T. J. Pugh, Nat. Cancer, 2020, 1, 873–881 CrossRef CAS PubMed.
- X. Zhang, Z. Zhang, W. Diao, C. Zhou, Y. Song, R. Wang, X. Luo and G. Liu, TrAC, Trends Anal. Chem., 2023, 159, 116904 CrossRef CAS.
- P. Chen, X. Jiang, K. Huang, P. Hu, X. Li, L. Wei, W. Liu, L. Wei, C. Tao, B. Ying, X. Wei and J. Geng, ACS Appl. Mater. Interfaces, 2019, 11, 36476–36484 CrossRef CAS PubMed.
- Y. Dai, Y. Wu, G. Liu and J. J. Gooding, Angew. Chem., Int. Ed., 2020, 59, 20754–20766 CrossRef CAS PubMed.
- T. N. Tiambeng, D. S. Roberts, K. A. Brown, Y. Zhu, B. Chen, Z. Wu, S. D. Mitchell, T. M. Guardado-Alvarez, S. Jin and Y. Ge, Nat. Commun., 2020, 11, 3903 CrossRef CAS PubMed.
- H. Li, Y. Huang, G. Hou, A. Xiao, P. Chen, H. Liang, Y. Huang, X. Zhao, L. Liang, X. Feng and B. Guan, Sci. Adv., 2019, 5, eaax4659 CrossRef CAS PubMed.
- M. Raveendran, A. J. Lee, R. Sharma, C. Walti and P. Actis, Nat. Commun., 2020, 11, 4384 CrossRef CAS PubMed.
- Y. Ying, Z. Hu, S. Zhang, Y. Qing, A. Fragasso, G. Maglia, A. Meller, H. Bayley, C. Dekker and Y. Long, Nat. Nanotechnol., 2022, 17, 1136–1146 CrossRef CAS PubMed.
- V. Fedyuk, N. Erez, N. Furth, O. Beresh, E. Andreishcheva, A. Shinde, D. Jones, B. B. Zakai, Y. Mavor, T. Peretz, A. Hubert, J. E. Cohen, A. Salah, M. Temper, A. Grinshpun, M. Maoz, A. Zick, G. Ron and E. Shema, Nat. Biotechnol., 2023, 41, 212–221 CrossRef CAS PubMed.
- X. Chang, Y. Huo, C. Zhao, W. Sun, Z. Song, Z. Qi, J. Wang, C. Jia and X. Guo, Adv. Sens. Res., 2023, 2, 2200084 CrossRef.
- D. Kwak, J. Kim, M. Lee, K. Ryu and S. Chi, Anal. Chem., 2020, 92, 14303–14308 CrossRef CAS PubMed.
- R. Wu, Y. Wang, J. Yu, H. Li, C. Yu, H. Wang, M. Wang and B. Li, Angew. Chem., Int. Ed., 2023, 62, e202304891 CrossRef CAS PubMed.
- Y. Qiao, J. J. Hu, Y. Hu, C. Duan, W. Jiang, Q. Ma, Y. Hong, W. H. Huang, F. Xia and X. Lou, Angew. Chem., Int. Ed., 2023, 62, e202309671 CrossRef CAS PubMed.
- C. Cao, Y. Ying, Z. Hu, D. Liao, H. Tian and Y. Long, Nat. Nanotechnol., 2016, 11, 713–718 CrossRef CAS PubMed.
- C. Cao, D. Liao, J. Yu, H. Tian and Y. Long, Nat. Protoc., 2017, 12, 1901–1911 CrossRef CAS PubMed.
- X. Wu, M. Li, S. Yang, J. Jiang, Y. Ying, P. R. Chen and Y. Long, Angew. Chem., Int. Ed., 2023, 62, e202300582 CrossRef CAS PubMed.
- E. B. Stephenson, J. L. Korner and K. S. Elvira, Nat. Chem., 2022, 14, 862–870 CrossRef CAS PubMed.
- S. Oiki, in Patch Clamp Techniques: From Beginning to Advanced Protocols, ed. Y. Okada, Springer, Tokyo, 2012, pp. 229–275 Search PubMed.
- P. Kongsuphol, K. B. Fang and Z. Ding, Sens. Actuators, B, 2013, 185, 530–542 CrossRef CAS.
- G. Baaken, M. Sondermann, C. Schlemmer, J. Ruhe and J. C. Behrends, Lab Chip, 2008, 8, 938–944 RSC.
- K. Ogishi, T. Osaki, Y. Morimoto and S. Takeuchi, IEEE MEMS, 2022, pp. 381–383 Search PubMed.
- T. Osaki, Y. Watanabe, R. Kawano, H. Sasaki and S. Takeuchi, J. Microelectromech. Syst., 2011, 20, 797–799 CAS.
- H. Wakebe, T. Fukushima and T. Tanaka, Electron. Commun. Jpn., 2022, 105, e12343 CrossRef.
- E. Zaitseva, A. Obergrussberger, C. Weichbrodt, M. Boukhet, F. Bernhard, C. Hein, G. Baaken, N. Fertig and J. C. Behrends, in Patch Clamp Electrophysiology: Methods and Protocols, ed. M. Dallas and D. Bell, Springer, Humana, New York, 2021, vol. 2188, pp. 67–92 Search PubMed.
- A. Alcinesio, O. J. Meacock, R. G. Allan, C. Monico, V. Restrepo Schild, I. Cazimoglu, M. T. Cornall, R. Krishna Kumar and H. Bayley, Nat. Commun., 2020, 11, 2105 CrossRef CAS PubMed.
- F. G. Downs, D. J. Lunn, M. J. Booth, J. B. Sauer, W. J. Ramsay, R. G. Klemperer, C. J. Hawker and H. Bayley, Nat. Chem., 2020, 12, 363–371 CrossRef CAS PubMed.
- S. Leptihn, O. K. Castell, B. Cronin, E. H. Lee, L. C. Gross, D. P. Marshall, J. R. Thompson, M. Holden and M. I. Wallace, Nat. Protoc., 2013, 8, 1048–1057 CrossRef CAS PubMed.
- R. Strutt, J. W. Hindley, J. Gregg, P. J. Booth, J. D. Harling, R. V. Law, M. S. Friddin and O. Ces, Chem. Sci., 2021, 12, 2138–2145 RSC.
- C. E. G. Hoskin, V. R. Schild, J. Vinals and H. Bayley, Nat. Chem., 2022, 14, 650–657 CrossRef CAS PubMed.
- R. Watanabe, T. Komatsu, Y. Urano and H. Noji, Lab Chip, 2018, 18, 2849–2853 RSC.
- Z. Dong and Q. Fang, TrAC, Trends Anal. Chem., 2020, 124, 115812 CrossRef CAS.
- Y. Jin, W. Xu, H. Zhang, R. Li, J. Sun, S. Yang, M. Liu, H. Mao and Z. Wang, Proc. Natl. Acad. Sci. U. S. A., 2022, 119, e2105459119 CrossRef CAS PubMed.
- A. Fragasso, S. Schmid and C. Dekker, ACS Nano, 2020, 14, 1338–1349 CrossRef CAS PubMed.
- J. Cama, H. Bajaj, S. Pagliara, T. Maier, Y. Braun, M. Winterhalter and U. F. Keyser, J. Am. Chem. Soc., 2015, 137, 13836–13843 CrossRef CAS PubMed.
- J. de Novais Schianti, F. do Nascimento, J. Cordoba Ramirez, M. Machida, L. H. Gabrielli, H. E. Hernandez-Figueroa and S. Moshkalev, J. Vac. Sci. Technol., A, 2018, 36, 021403 CrossRef.
- I. Czolkos, Y. Erkan, P. Dommersnes, A. Jesorka and O. Orwar, Nano Lett., 2007, 7, 1980–1984 CrossRef CAS PubMed.
- L. Zhang, C. Zhong, J. Li, H. Niu, Y. Ying and Y. Long, J. Electroanal. Chem., 2022, 915, 116266 CrossRef CAS.
- M. Mayer, J. K. Krienel, M. T. Tosteson and G. M. Whitesides, Biophys. J., 2003, 85, 2684–2695 CrossRef CAS PubMed.
- E. Stava, C. Siyoung, K. Hyun-Seok and R. H. Blick, IEEE Trans. NanoBiosci., 2012, 11, 169–175 Search PubMed.
- W. L. Hwang, M. Chen, B. Cronin, M. A. Holden and H. Bayley, J. Am. Chem. Soc., 2008, 130, 5878–5879 CrossRef CAS PubMed.
- I. C. Nova, I. M. Derrington, J. M. Craig, M. T. Noakes, B. I. Tickman, K. Doering, H. Higinbotham, A. H. Laszlo and J. H. Gundlach, PloS One, 2017, 12, e0181599 CrossRef PubMed.
- L. S. Lastra, Y. M. N. D. Y. Bandara, M. Nguyen, N. Farajpour and K. J. Freedman, Nat. Commun., 2022, 13, 2186 CrossRef CAS PubMed.
- S. Cicalese, K. Okuno and S. Eguchi, J. Cardiovasc. Pharmacol., 2021, 77, 43–48 CrossRef CAS PubMed.
- T. C. Santiago, L. Parra, J. V. Nani, T. M. Fidalgo, N. J. Bradshaw and M. A. F. Hayashi, J. Neurochem., 2023, 166, 138–155 CrossRef CAS PubMed.
- F. Chen, Z. Cheng, Y. Peng, Z. Wang, C. Huang, D. Liu, B. Wang, B. Pan and W. Guo, J. Chromatogr. B, 2021, 1179, 122740 CrossRef CAS PubMed.
- J. Jiang, M. Li, X. Wu, Y. Ying, H. Han and Y. Long, Nat. Chem., 2023, 15, 578–586 CrossRef CAS PubMed.
- K. Chuah, Y. Wu, S. R. C. Vivekchand, K. Gaus, P. J. Reece, A. P. Micolich and J. J. Gooding, Nat. Commun., 2019, 10, 2109 CrossRef PubMed.
- P. Cadinu, M. Kang, B. P. Nadappuram, A. P. Ivanov and J. B. Edel, Nano Lett., 2020, 20, 2012–2019 CrossRef CAS PubMed.
- M. A. Bakshloo, J. J. Kasianowicz, M. Pastoriza-Gallego, J. Mathe, R. Daniel, F. Piguet and A. Oukhaled, J. Am. Chem. Soc., 2022, 144, 2716–2725 CrossRef PubMed.
- M. A. Bakshloo, S. Yahiaoui, H. Ouldali, M. Pastoriza-Gallego, F. Piguet and A. Oukhaled, Proteomics, 2022, 22, 2100056 CrossRef PubMed.
- Y. Huaa, W. P. King and C. L. Henderson, Microelectron. Eng., 2008, 85, 934–936 CrossRef.
- H. Lee, K. Lee, B. Ahn, J. Xu, L. Xu and K. W. Oh, J. Microelectromech. Syst., 2011, 21, 125006 Search PubMed.
- Y. Wang, C. Cao, Y. Ying, S. Li, M. Wang, J. Huang and Y. Long, ACS Sens., 2018, 3, 779–783 CrossRef CAS PubMed.
- A. J. W. Hartel, S. Shekar, P. Ong, I. Schroeder, G. Thiel and K. L. Shepard, Anal. Chim. Acta, 2019, 1061, 13–27 CrossRef CAS PubMed.
- K. Luo, K. Jeong, J. Oh, S. Choi, T. Jeon and Y. Kim, RSC Adv., 2017, 7, 49858–49862 RSC.
- A. Balijepalli, J. Ettedgui, A. T. Cornio, J. W. F. Robertson, K. P. Cheung, J. J. Kasianowicz and C. Vaz, ACS Nano, 2014, 8, 1547–1553 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available: Supplementary figures and supplementary movies. See DOI: https://doi.org/10.1039/d3sc06795e |
| This journal is © The Royal Society of Chemistry 2024 |