Open Access Article
Open Access ArticleA dynamic cascade DNA nanocomplex to synergistically disrupt the pyroptosis checkpoint and relieve tumor hypoxia for efficient pyroptosis cancer therapy†
Xiaoni
Wang
a,
Xiyang
Ge
a,
Min
Zhang
a,
Jianghui
Sun
a,
Jin
Ouyang
 b and
Na
Na
b and
Na
Na
 *a
*a
aKey Laboratory of Radiopharmaceuticals, Ministry of Education, College of Chemistry, Beijing Normal University, Beijing 100875, China. E-mail: nana@bnu.edu.cn
bDepartment of Chemistry, College of Arts and Sciences, Beijing Normal University at Zhuhai, Zhuhai City, Guangdong Province 519087, China
First published on 16th April 2024
Abstract
Pyroptosis has attracted widespread concerns in cancer therapy, while the therapeutic efficiency could be significantly restricted by using the crucial pyroptosis checkpoint of autophagy and tumor hypoxia. Herein, a DNA nanocomplex (DNFs@ZnMn), containing cascade DNAzymes, promoter-like ZnO2–Mn nanozymes and photosensitizers, was constructed in one pot through rolling circle amplification reactions to induce pyroptosis through disrupting autophagy. After targeting cancer cells with a high expression of H+ and glutathione, DNFs@ZnMn decomposed to expose DNAzymes and promoter-like ZnO2–Mn nanozymes. Then, sufficient metal ions and O2 were released to promote cascade DNA/RNA cleavage and relieving of tumor hypoxia. The released DNAzyme-1 self-cleaved long DNA strands with Zn2+ as the cofactor and simultaneously exposed DNAzyme-2 to cleave ATG-5 mRNA (with Mn2+ as the cofactor). This cascade DNAzyme-mediated gene regulation process induced downregulation of ATG-5 proteins to disrupt autophagy. Simultaneously, the released ZnO2 donated sufficient H2O2 to generate adequate O2 to relieve tumor hypoxia, obtaining highly cytotoxic 1O2 to trigger pyroptosis. By using dynamic cascade gene silencing to disrupt the pyroptosis checkpoint and synergistic relieving of hypoxia, this DNA nanocomplex significantly weakened cellular resistance to achieve efficient pyroptosis therapy both in vitro and in vivo.
Introduction
Pyroptosis, a programmed proinflammatory cell death by pore formation via cellular swelling with large bubbles, has attracted widespread concern in cancer therapy. Upon formation of inflammasomes, caspase 1 and pro-inflammatory cytokines (IL-1β and IL-18) are activated to cleave gasdermin D, releasing N-terminal domains (GSDMD-N) to execute pyroptosis.1–4 However, the biological application of pyroptosis in cancer therapy is still restricted by the intrinsic adaptive survival pathway of autophagy, a checkpoint to negatively regulate pyroptosis for inducing therapeutic resistance. Actually, autophagy limits the activation of inflammasomes and inflammatory cytokines, and facilitates their degradation.5–7 Specifically, cancer cells usually exhibit higher autophagic activity than normal cells, which further aggravates the degradation to weaken the therapeutic effect of pyroptosis.8–10 Meanwhile, the given reactive oxygen species (ROS) generation is the key upstream step of pyroptosis upon oxidative stress or intracellular osmolarity change, and tumor hypoxia becomes another obstacle of pyroptosis.11–14 However, in response to the oxidative stress caused by the intracellular accumulation of ROS, cancer cells can invoke self-protective autophagy to escape pyroptosis for reducing therapy efficiency.15–18 Consequently, a synergistic strategy for both disrupting the pyroptosis checkpoint of autophagy and relieving tumor hypoxia is desirable for efficient pyroptosis cancer therapy.Based on the advantages of precise initiation and sequence arrangement of DNA, DNA nanocomplexes can be rationally designed and programmed with multiple modules and good adaptability for cancer treatments.19–21 In particular, with inherent negative charges, DNA nanomaterials can potentially reduce nonspecific interactions between DNA nanomaterials and cells for alleviating immunogenicity in cancer therapy.22–25 Specifically, with metal ions as cofactors to activate catalytic activities, DNAzyme can precisely and specifically recognize and cleave DNA/RNA fragments. This facilitated efficient therapy through down-regulating disease-related proteins without disturbing their cellular machinery.26–30 However, DNAzyme-based therapy is still in its infant stages and challenged by the following issues: (1) both DNAzymes and the corresponding aptamers are short-stranded DNA molecules, which normally exhibit inefficient cellular uptake with less stability and nonspecific distribution under physiological conditions.31 (2) The multifunctional DNAzyme-based strategy was normally restricted by insufficient intracellular metal ions and difficult integration of multiple active sites into cascade therapy but without any interference from each other.32 (3) Insufficient functional DNA monotherapy or active sites in one DNA nanocomplex could greatly limit the pyroptosis efficiency,33 which requires the amplification-based strategy for pyroptosis enhancements.
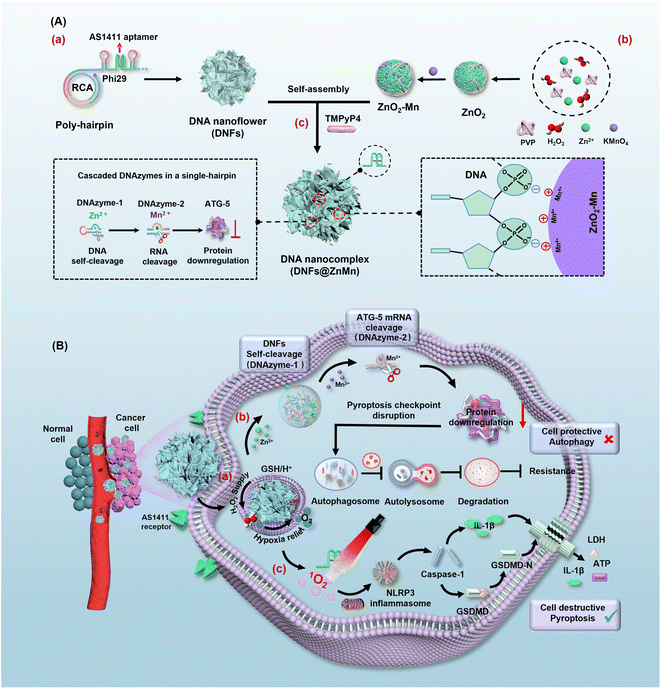
Herein, a multi-functional DNA nanocomplex (DNFs@ZnMn) with cascade DNAzymes, promoter-like ZnO2–Mn nanozymes and intercalated photosensitizers were constructed to dynamically and cascadely disrupt autophagy and relieve tumor hypoxia for enhanced pyroptosis therapy. With rolling circle amplification (RCA) reactions, the expression of functional units of DNAzyme-1, DNAzyme-2 and AS1411 aptamer was exponentially loaded in DNFs@ZnMn for efficient therapy. As shown in Scheme 1A, poly-hairpin containing sequences of the aptamer (AS1411) and cascade DNAzymes (DNAzyme-1 and DNAzyme-2) were designed as the scaffold to intercalate photosensitizers and encapsulate promoter-like ZnO2–Mn particles. Besides, the AS1411 DNA aptamer could form quadruplex structures to facilitate cell specific theranostics upon the intercalation of photosensitizer 5,10,15,20-tetra (1-methylpyridin-4-yl)porphyrin (TMPyP4) and recognition of nucleolin that were overexpressed on the membrane of cancer cells. In blood circulation, the DNA nanocomplex targeted and entered cancer cells by aptamer recognition (Scheme 1B). Thereafter, DNFs@ZnMn was gradually decomposed by consuming H+ and glutathione (GSH) at tumor sites, and released metal ions and DNAzymes to initiate cascade gene therapy for autophagy disruption (Scheme 1B-a). Briefly, with Zn2+ as a cofactor, DNAzyme-1 catalyzes the self-cleavage of ultra long DNA strands to produce downstream therapeutic DNAzyme-2-containing fragments. Afterwards, with Mn2+ as a cofactor, DNAzyme-2 facilitated ATG-5 mRNA cleavage for silencing of the ATG-5 protein. This would reduce the level of cellular autophagy flux for inhibiting the resistance pyroptosis and blocking the response to oxidative stress through autophagy. Simultaneously, the released biocompatible enzyme precursor of ZnO2 donated sufficient H2O2, which was subsequently in situ converted into O2 catalyzed by MnO2 (Scheme 1B-b). The in situ generated O2 would efficiently relieve tumor hypoxia,34–36 which facilitated the following red light-initiated activation of TMPyP4 to catalytically convert O2 into highly cytotoxic ROS of 1O2. Thereafter, NLRP3 inflammasomes were formed to active caspase-1 and pro-inflammatory cytokine IL-1β, and caspase-1 further cleaved gasdermin D to release the N-terminal domain (GSDMD-N) for inducing pyroptosis. Consequently, both IL-1β and GSDMD-N executed pyroptosis via the pore-forming activities.37,38 Besides, ROS is also reported to be crucial for GSDMD oligomerization and pore formation in pyroptosis therapy.39 This ensured the highly efficient pyroptosis cancer therapy by synergistically disrupting the pyroptosis checkpoint and relieving hypoxia. This strategy would ultimately ensure satisfactory and safe antitumor treatment both in vitro and in vivo.
Results and discussion
Preparation and characterization of DNFs@ZnMn
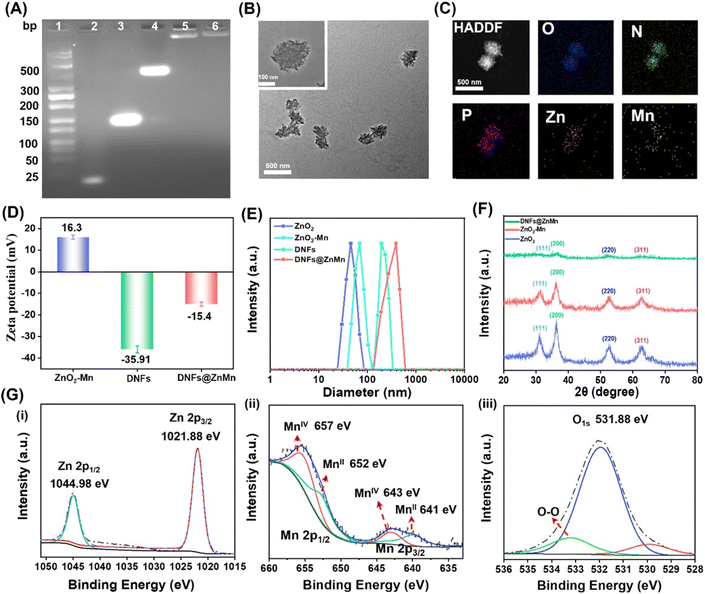
To obtain DNFs@ZnMn, the DNA nanoflower of a sterically RCA-caged DNA nanocomplex with ultra long single DNA strands was first prepared via RCA technology (Scheme 1A-a). The poly-hairpin DNA template was designed to contain the multi-functional complementary strands corresponding to the AS1411 aptamer, cascade DNAzyme-1 for self-cleavage and DNAzyme-2 for cleaving mRNAs. This facilitated the amplification of long single-stranded DNA in the presence of primers. Simultaneously (Scheme 1A-b), the cascade nanozymes of ZnO2–Mn were obtained via reduction of KMnO4 by the PVP-modified ZnO2 (obtained by a reaction between Zn(OAc)2 and H2O2). Thereafter, the DNFs@ZnMn was finally obtained upon the adsorption of the ZnO2–Mn nanozyme and porphyrin photosensitizer TMPyP4 onto DNA nanoflowers (Scheme 1A-c).To confirm the successful synthesis of DNFs@ZnMn, a series of characterization studies have been carried out. As demonstrated by poly-acrylamide gel electrophoresis (PAGE) analysis, the step-by-step products, including primers, ssDNA, a circular DNA template of RCA (Circ-DNA), DNA nanoflowers and DNFs@ZnMn, were confirmed based on bands with different migration performances (Fig. 1A). The morphology characterization studies were further performed by transmission electron microscope (TEM) analysis, which revealed the presence of 300 nm DNA nanoflowers (Fig. S1†) and DNFs@ZnMn (Fig. 1B). This morphology was convenient to load smaller sized ZnO2–Mn nanozymes on DNA nanoflowers (Fig. S2 and S3†). Besides, the successful synthesis of DNFs@ZnMn was further confirmed by using the element mapping images (Fig. 1C), which demonstrated the even distribution of Zn and Mn species (from ZnO2–Mn nanozymes) in DNFs@ZnMn.
The chemical characterization studies were further carried out to examine the preparation of DNFs@ZnMn. Initially, the surface charge changes during the synthesis were examined. As demonstrated by the zeta-potential analysis (Fig. 1D), DNFs@ZnMn (−15.4 mV) was obtained upon the electrostatic interaction between positively charged ZnO2–Mn nanozymes (+16.3 mV) and negatively charged DNA nanoflowers (−35.91 mV). In addition, dynamic light scattering (DLS) analysis showed the similar sizes of DNA nanoflowers and DNFs@ZnMn, which verified the loading of smaller ZnO2–Mn nanozymes into larger DNA nanoflowers (Fig. 1E). Furthermore, the presence of peroxide groups (O22−) can be confirmed by a strong Raman band of O–O tensile vibration (at 844 cm−1, Fig. S4†). In addition, similar crystal structures of ZnO2 nanoparticles, ZnO2–Mn nanozymes and DNFs@ZnMn were exhibited in X-ray diffraction (XRD) patterns (Fig. 1F), which could be due to the weak interactions for loading nanozymes onto DNA nanoflowers. Moreover, the chemical composition states of different nanomaterials were examined by X-ray photoelectron spectroscopy (XPS). Similar peaks of Zn 2p, Mn 2p, and O 1s for both ZnO2–Mn nanozymes (Fig. S5†) and DNFs@ZnMn (Fig. 1G) were exhibited, which indicated no obvious effect of MnO2 on the crystal structure of ZnO2. Besides, the surface-absorbed oxygen and peroxide oxygen (O22−) were also confirmed in DNFs@ZnMn by using the peaks at 532.0 eV and 530.0 eV, respectively. In addition, the successful loading of photo-sensitizer TMPyP4 in DNFs@ZnMn was also confirmed by using UV-vis absorption spectra (Fig. S6†). Moreover, the good stability of DNFs@ZnMn in a biological environment was further verified by the absence of an obvious fragment in images of agarose gel electrophoresis (Fig. S7†). Besides, DNFs@ZnMn exhibited the satisfactory long-term stability (Fig. S8†), which ensured the following biological applications. Consequently, DNFs@ZnMn was confirmed to be successfully prepared, which exhibited satisfactory stability in biological environments for the subsequent intracellular pyroptosis therapy.
Examination of cascade enzyme-like activities for synergistic mRNA cleavage and relieving of tumor hypoxia
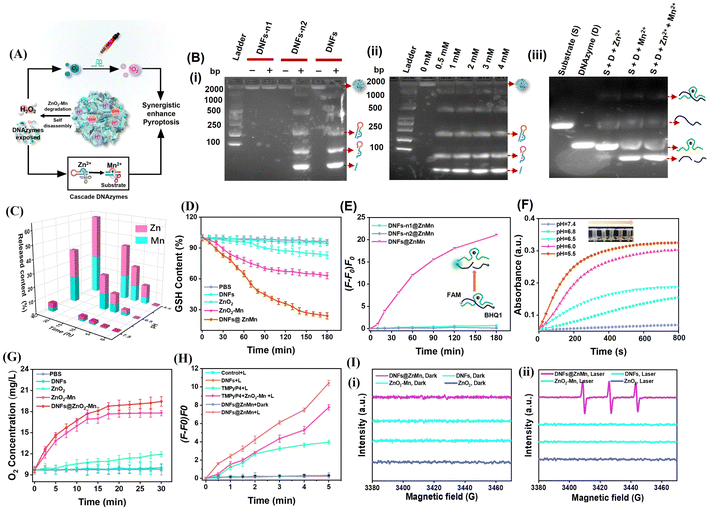
As illustrated in Fig. 2A-i, upon the self-disassembly of DNFs@ZnMn at tumor sites with a high expression of GSH and H+, the ZnO2–Mn nanozyme was exposed to release Zn2+ (cofactor of DNAzyme-1) and Mn2+ (cofactor of DNAzyme-2). Consequently, in the presence of Zn2+, ultra-long DNA chains of DNA nanoflowers were cleaved by DNAzyme-1. This simultaneously resulted in the exposure of DNAzyme-2 because the 5′- and 3′-termini of DNAzyme-2 were designed to be elongated with DNAzyme-1. Therefore, cascade mRNA cleavage was initiated by the exposed DNAzyme-2 with Mn2+ as the cofactor, which would contribute to tumor-specific gene silencing for autophagy disruption. Simultaneously (Fig. 2A-ii), the released ZnO2 initiated the relieving of tumor hypoxia upon MnO2-catalytic generation of sufficient O2 from H2O2 (obtained from ZnO2). Thereafter, upon light-irradiation, adequate 1O2 was generated (with TMPyP4 as the photosensitizer) to be synergistic with autophagy disruption (Fig. 2A-i), for enhancing the subsequent pyroptosis therapy.To examine the cascade enzyme-like activities for synergistic mRNA cleavage and relieving of tumor hypoxia, a series of experiments have been carried out and the comparison of corresponding species was employed. Initially, the role of the Zn2+ cofactor in the cleavage of DNA nanoflowers by DNAzyme-1 was confirmed by PAGE analysis (Fig. 2B-i), which only exhibited significant bands of DNA lysates in the presence of both Zn2+ and DNAzyme-1. In addition, DNA lysates increased with increasing Zn2+ concentration (Fig. 2B-ii), and the released Zn2+ from ZnO2–Mn nanozyme efficiently triggered DNAzyeme-1 for mRNA substrate cleavage as free Zn2+ did (Fig. S9†). Furthermore, mRNA substrate cleavage was highly specific only with Mn2+ as the cofactor of DNAzyeme-2, and the presence of Zn2+ and other ions would have no effect on the cleavage (Fig. 2B-iii and S10†). Based on the pH-dependent disassembly of DNFs@ZnMn shown by TEM and DLS (Fig. S11†), the size of the nanomaterials decreased and backbone structures were significantly disrupted after 24 h of treatment at pH = 5.5. The release of Zn2+ and Mn2+ at different pH values was also examined by inductively coupled plasma optical emission spectrometry (ICP-OES) tests. As resulted (Fig. 2C), DNFs@ZnMn was relatively stable under natural conditions (pH = 7.4), while Zn2+/Mn2+ expression increased with increasing acidic levels due to the acidic stimulation of the ZnO2 component. Besides, GSH was also consumed during the decomposition of DNFs@ZnMn under acidic conditions, demonstrated by the significant depletion of added GSH in the reaction systems (Fig. 2D). Therefore, efficient decomposition of of DNFs@ZnMn upon GSH/H+ stimulation at tumor sites was confirmed, which facilitated the subsequent tumor-specific therapy.
To examine the cleavage performance of the mRNA substrate by DNFs@ZnMn, ATG-5 mRNA was labelled with a fluorophore (FAM)/quencher (BHQ1) at its 3′- and 5′-ends, respectively. Thus, the cleavage of mRNA can be indicated by the increased FL signals. In the experiments, mRNAs were incubated with DNA nanoflowers (DNFs) and the ones without DNAzyme-1 (DNFs-n1) or without DNAzyme-2 (DNFs-n2) for the comparison. As a result (Fig. 2E and S12†), only a dramatic increase of FL signals was recorded after incubating mRNAs with DNFs@ZnMn, while no significant response was observed for the groups without one or both DNAzymes. This further confirmed the cascade cleavage of the mRNA by DNAzyme-1 and DNAzyme-2, which facilitated the highly specific gene silencing therapy.
DNFs@ZnMn was expected to function as a CAT-like nanozyme to generate H2O2 for decomposing H2O2 into O2 at the tumor sites, devoted to relieving hypoxia for efficient ROS generation. To examine the performance of cascade relieving of tumor hypoxia by DNFs@ZnMn (illustrated in Fig. 2A-ii), the generation of both H2O2 and O2 was evaluated. First, H2O2 generation from ZnO2 was monitored by incubating DNFs@ZnMn with Ti(SO4)2, which formed yellow precipitates to exhibit 412 nm-absorption under acidic conditions. The standardized linear relationship between absorption of Ti(SO4)2 and H2O2 concentration is exhibited in Fig. S13.† As shown in Fig. 2F, the UV-vis absorption at 412 nm increased with the increase in acidity (from pH 7.4 to pH 5.5), and the highest absorption was recorded at pH 5.5. This indicated the pH-initiated DNFs@ZnMn disassembly for generating H2O2. Thereafter, adequate O2 was generated upon the reaction between H2O2 and MnO2, along with obtaining Mn2+. This was further confirmed by the time-dependent and H2O2-dependent generation of dissolved O2 in the presence of MnO2 (DNFs@ZnMn and ZnO2–Mn nanozyme) (Fig. 2G and S14A†), which was also facilitated under acidic conditions (Fig. S14B†). Therefore, the relieving of tumor hypoxia was realized by the cascade generation of adequate O2 upon the disassembly of DNFs@ZnMn under acidic conditions.
Besides, the generation of 1O2 under light-irradiation was further evaluated with SOSG as a singlet oxygen probe, exhibiting FL signals in the presence of 1O2. As a result (Fig. 2H and S15†), upon light-irradiation (660 nm, 0.2 W cm−2, 10 min), the highest amount of 1O2 was generated in the DNFs@ZnMn group. In addition, the intensity of the signal of 1O2 enhanced with increasing irradiation time, which was assessed by the 1,3-diphenylisobenzofuran (DPBF) probe (Fig. S16†), while a lower amount of 1O2 was recorded for the groups in the absence of one or more factors (Zn, Mn, TMPyP4, and light-irradiation). This further confirmed the feasibility of ROS generation (1O2) upon the cascade supply of O2 by the present TME-irradiated decomposition of DNFs@ZnMn. It should be noted that no ROS species of ·OH was observed, as demonstrated by the characteristic absorption at 652 nm with 3,3′,5,5′-tetramethylbenzidine (TMB) as the probe (Fig. S17†). This was in accordance with the electron spin resonance (ESR) spectra, exhibiting 1O2 peaks (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1
1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 triplet signals) (Fig. 2I) and the absence of an ·OH peak (1
1 triplet signals) (Fig. 2I) and the absence of an ·OH peak (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2
2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2
2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 signals) (Fig. S18A†) after light-irradiation. In addition, the 1O2 signals increased with increasing acidity (Fig. S18B†), which was in accordance with the evaluation of the 1O2 amount at different pH values (Fig. S19†). This further confirmed the efficient tumor-specific obtaining of 1O2 for efficient therapy under acidic conditions of TME. Consequently, the cascade enzyme-like activities for synergistic mRNA cleavage and relieving of tumor hypoxia were confirmed, facilitating the subsequent intracellular therapy upon pyroptosis.
1 signals) (Fig. S18A†) after light-irradiation. In addition, the 1O2 signals increased with increasing acidity (Fig. S18B†), which was in accordance with the evaluation of the 1O2 amount at different pH values (Fig. S19†). This further confirmed the efficient tumor-specific obtaining of 1O2 for efficient therapy under acidic conditions of TME. Consequently, the cascade enzyme-like activities for synergistic mRNA cleavage and relieving of tumor hypoxia were confirmed, facilitating the subsequent intracellular therapy upon pyroptosis.
Evaluations of intracellular delivery of DNFs@ZnMn
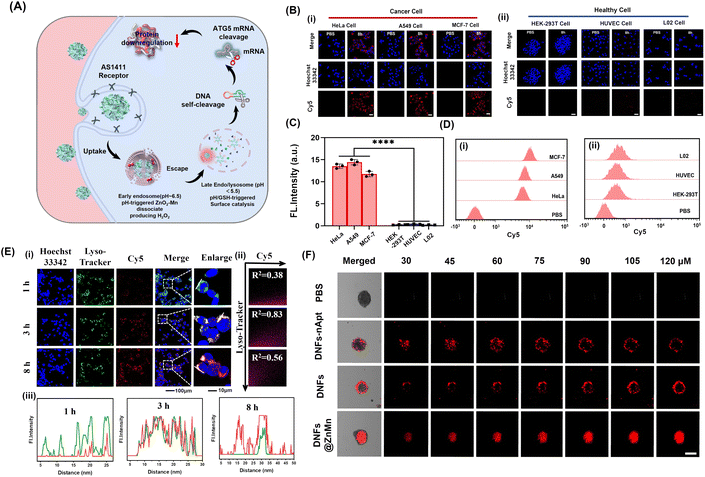
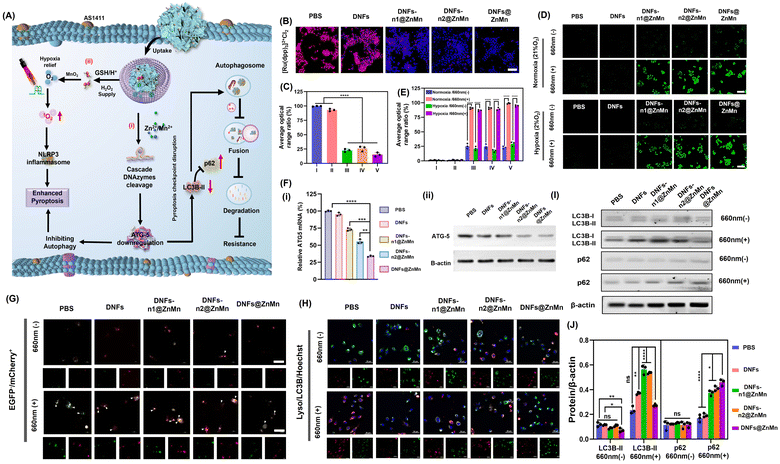
For improving tumor-specific efficacy and reducing side effects, the uptake efficiency and tumor-specific delivery of DNFs@ZnMn into cancer cells was crucial. As shown in Fig. 3A, with the AS1411 receptor loaded, DNFs@ZnMn targeted tumor cells, which initiated the internalization pathways including the internalization into early endosomes (∼pH 6.5) followed by the endosome/lysosome (pH < 5.5). Thereafter, upon GSH/H+ stimulation, DNFs@ZnMn was decomposed for cascade gene silencing of mRNA, including cleavage of ultra-long DNA chains and the subsequent mRNA cleavage. To evaluate the tumor-specific cellular uptake, Cy5 was labelled on DNFs@ZnMn for cell imaging by confocal laser scanning microscopy (CLSM) and cytometry analysis. As a result, the significant red signals of Cy5 were observed in groups of cancer cells with high nucleolin expression (Fig. 3B). In addition, the cellular uptake efficiency of the nanocomplex with a nucleolin-targeting AS1411 aptamer (DNFs@ZnMn) was significantly higher than the efficiency of that without an aptamer (DNFs–nApt@ZnMn), increasing upon the incubation (Fig. S20†). This was in accordance with the comparison of FL intensities in cancer cells and normal cells (HEK-293T, HUVEC, L02 cells, low nucleolin expression), demonstrating the specific targeting of cancer cells with high nucleolin expression by the AS1411 aptamer (Fig. 3C and D). The specific cellular uptake would endow DNFs with high therapeutic efficacy and minimized off-target toxicity. The cellular uptake pathway of DNFs@ZnMn was studied by treating HeLa cells with specific endocytic inhibitors at different temperatures. As a result (Fig. S21†), the cellular uptake relied on clathrin/lipid-raft mediated endocytosis and micropinocytosis due to the significant uptake inhibition by their inhibitors. This indicated that DNFs@ZnMn was first internalized into early endosomes (∼pH 6.5), followed by entering into the late endosome/lysosome (pH < 5.5). In addition, the inhabitation at a low temperature of 4 °C indicated that the uptake was an energy-dependent process.To further examine the intracellular subcellular localization of DNFs@ZnMn in HeLa cells, the co-localization of lysosomes and DNFs@ZnMn (with Cy5 labelled) was carried out. As indicated by Lysotracker (green) and Cy5 (red), the red and green fluorescence signals displayed a strong overlap in HeLa cells, and the Pearson correlation coefficient (PCC) was 0.97 at 3 h (Fig. 3E), while at 8 h, the red signals increased obviously along with the separation of the two signals (PCC was 0.56), which indicated the escape of DNFs@ZnMn from endosomes to cytoplasm. However, for the DNA nanoflowers without nanozymes, PCC remained at relatively high values, indicating their entrapping in endosomes without nanozymes (Fig. S22†). This further confirmed the generation of ROS species by ZnO2–Mn nanozyme in acidic endosome–lysosome environments, which facilitated breaking of the lysosome membrane for endo-lysosomal escape. Moreover, HeLa cell-based multicellular spheroids (MCSs) were constructed to assess tumor infiltration properties, which demonstrated the high tumor penetration at deep levels in the DNFs@ZnMn group, only with both AS1411 aptamer and ZnO2–Mn nanozymes loaded (Fig. 3F). Consequently, the tumor-specific uptake of DNFs@ZnMn was confirmed, which initiated intracellular decomposition to release multiple reagents for the subsequent cascade and synergistic applications.
Thereafter, the tumor-specific intracellular decomposition of DNFs@ZnMn was initiated by the high-concentration of H+/GSH, achieving the disassembly and structural collapse of ZnO2–Mn, and thereby promote the release of DNAzyme cofactors of Zn2+ and Mn2+. This was further verified by evaluating the intracellular contents of Zn2+ and Mn2+ by inductively coupled plasma mass spectrometry (ICP-MS) analysis. As a result (Fig. S23†), a significant increase in both ions in cancer cells was recorded after incubation with HeLa cells for 24 h, which would be sufficient for cascade activation of two DNAzymes. Besides, the intracellular Zn2+ was tracked by using a blue fluorescent probe (zincquin ethyl ester) in HeLa cell imaging, which exhibited increased FL signals upon the incubation (Fig. S24 and S25†). Consequently, the metal ions were confirmed to be efficiently released into cancer cells for the subsequent therapy. Therefore, DNFs@ZnMn was efficiently and specifically delivered into tumor sites and the corresponding reagents were in situ released for the subsequent tumor-specific therapy.
Intracellular examinations of synergistic relieving of tumor hypoxia and autophagy disruption
The synergistic pathways for relieving of tumor hypoxia and autophagy disruption are crucial for the present efficient apoptosis treatments. As shown in Fig. 4A, after the tumor-specific uptake and decomposition of DNFs@ZnMn upon GSH/H+ simulation in tumor microenvironments, multiple reagents were released for the two synergistic processes. On the one hand, the ZnO2–Mn nanozymes initiated the cascade cleavage of ATG-5 mRNA to downregulate the ATG-5 protein, which impaired cytoprotective autophagy protein to inhibit autophagy (Fig. 4A-i). On the other hand, adequate 1O2 was generated upon the cascade reliving of tumor hypoxia and photocatalysis reactions, which initiated the subsequent pyroptosis via the NLRP3-caspase 1-GSDMD pathway (Fig. 4A-ii). Therefore, the inhibition of autophagy, the reliving of tumor hypoxia and the synergistic performance between each other were further examined in cancer cells.For the intracellular examinations of the synergistic inhibition of autophagy and reliving of tumor hypoxia, HeLa cells were imaged after incubation with different species. Initially, the content of O2 was evaluated with a red fluorescence indicator of [Ru(dpp)3]2+Cl2. As a result (Fig. 4B and C), on comparison with the DNA nanoflowers without nanozymes, a significant amount of O2 was recorded in the groups with nanozymes (DNFs-n1@ZnMn and DNFs-n2@ZnMn) and the highest O2 content was observed in DNFs@ZnMn (Fig. 4C). This could be generated from the highest content of H2O2 after treatment by DNFs@ZnMn (Fig. S26†). Thus, the relieving of tumor hypoxia by ZnO2–Mn nanozymes was confirmed, and was even synergistically enhanced in the presence of DNAzyme-1 and DNAzyme-2 for autophagy inhibition. This was also in accordance with the generation of ROS species upon light-irradiation at 660 nm, with singlet oxygen sensor green (SOSG) as the probe (Fig. 4D and E). More significantly, a similar expression of ROS was exhibited in both normal (21% O2) and hypoxic (2% O2) environments, which further confirms the role of reliving tumor hypoxia by DNFs@ZnMn, while the ROS levels significantly increased for the DNFs@ZnMn group after light-irradiation, which indicated effective reliving of tumor hypoxia for efficient ROS generations. In fact, this intracellular generation of ROS species was tumor specific with minimized off-target toxicity, showing no obvious signals in the normal cells of HUVEC (Fig. S27†). Besides, intracellular GSH depletion was also confirmed during the incubation of HeLa cells with DNFs@ZnMn (Fig. S28†), which not only simulated the cancer-specific release of multiple reagents, but also accelerated the accumulation of intracellular toxic ROS for enhanced therapy. Consequently, the synergistic reliving of tumor hypoxia and the generation of toxic ROS species were confirmed in cancer cells, which facilitated the tumor-specific therapy.
Considering that the ROS species contribute to membrane dysfunctions structurally and functionally, the membrane dysfunctions were therefore examined to evaluate therapy performance of DNFs@ZnMn. Given that the mitochondria are the main sites of ROS production and the main ROS targets, mitochondrial depolarization upon dysfunctions was initially evaluated by using the membrane potential (Δψ) using a JC-1 probe. As a result, the most significant decrease in Δψ (with the increase in the green/red ratio) was recorded in the DNFs@ZnMn group under light irradiation (Fig. S29†), demonstrating the efficient mitochondrial damage. This was also in accordance with the performance of mitochondrial permeability transition pore (mPTP) activation, which was evaluated by the calcein-AM loading/CoCl2 quenching strategy (Fig. S30†). Furthermore, the endosomal membrane disruption by DNFs@ZnMn was also examined, which could be due to massive peroxidation of membrane lipids by ROS. In the experiments, the integrity of endosomes was evaluated by using an acridine orange (AO) indicator, which exhibited decreased red signals upon endosomal membrane disruption. As a result (Fig. S31†), the most significant decrease in red signals was recorded for HeLa cells treated with DNFs@ZnMn under light irradiation. This was attributed to lysosomal membrane permeabilization (LMP), which was induced by lysosomal dysfunction and therefore released protons into the cytoplasm for intracellular acidification.40 According to the quantification of BCEF-AM fluorescence with different intracellular pH values in cell imaging (Fig. S32†), the significantly decreased intracellular pH values were further confirmed after DNFs@ZnMn treatments (Fig. S33†). In addition, without light irradiation, no obvious pH decrease was observed, further confirming the activation of PDT under light irradiation. Besides, the highest LPO level was recorded in HeLa cells treated with DNFs@ZnMn (Fig. S34†). Consequently, upon reliving of tumor hypoxia, DNFs@ZnMn triggered ROS-mediated severe damage to mitochondria and endosomal membranes for the subsequent cancer therapy.
In fact, the synergistic relieving of tumor hypoxia for ROS-based therapy and autophagy disruption upon downregulation of ATG-5 mRNA and proteins were further examined by quantitative reverse transcription-polymerase chain reaction (qPCR) and western blot (WB) analysis. As indicated, only the obviously decreased expression of ATG-5 mRNA was observed in the group of DNFs@ZnMn (Fig. 4F-i), while no significant downregulation was recorded without nanozymes (DNFs, DNFs-n1, and DNFs-n2). This was also in accordance with the western blot (WB) analysis (Fig. 4F-ii and S35†). Therefore, the cascaded and synergistic downregulation of ATG-5 was confirmed for autophagy inhibition.
Subsequently, the autophagy level was characterized by cell imaging of autophagy markers (autophagosomes and autolysosomes) with mCherry+/EGFP− as the probe, which can indicate the accumulation of autolysosomes for enhanced autophagic flux by yellow puncta. As a result (Fig. 4G), the PBS and DNF groups exhibited obvious yellow EGFP−/mCherry+ signals (autophagosomes), indicating an increased accumulation of autophagosomes after light irradiation, while after DNFs@ZnMn treatment, autophagosome formation was significantly inhibited after light irradiation, suggesting the blockage of autophagy. This confirmed the inhabitation of autophagy upon the synergistic ROS-based procedures and cascade ATG-5 disruption, boosting the therapeutic applications of pyroptosis. Besides, the autophagy protein marker of LC3B (microtubule-associated protein 1 light chain 3) was also imaged, which exhibited a significant decrease in the colocalization of the accumulated autophagosome (LC3B) and lysosome in the DNFs@ZnMn group with light irradiation (Fig. 4H). These results indicated that DNFs@ZnMn has great potential to disrupt the pyroptosis checkpoint for efficient pyroptotic cancer therapy.
The immunoblotting analysis was also carried out to evaluate the autophagy level by the expression of autophagy protein markers LC3 (including two forms of LC3-I and LC3-II) and p62. During autophagy, the cytosolic form of LC3-I could be converted to the active membrane-bound LC3-II, which indicated autophagosome formation and autophagy initiation by the increased LC3-II/LC3-I ratio.41 Simultaneously, the formed autophagosome will be internalized and digested by lysosomes and degraded p62 protein (an autophagy substrate). Consequently, the autophagy inhibition could be indicated by the significant increase in the LC3-II/LC3-I ratio and the increased p62 expression, which was further evaluated by the western blotting analysis. As a result (Fig. 4I, J and S36†), both the decrease in LC3-II/LC3-I and up-regulated p62 expression were recorded in the DNFs@ZnMn group with light irradiation, confirming the downregulation of autophagy. These indicated that the negative regulation caused by protective autophagy could be inhibited to boost the potential therapeutic pyroptosis therapy. Consequently, the synergistic relieving of tumor hypoxia and autophagy disruption can be achieved by DNFs@ZnMn under light irradiation, which facilitated the subsequent efficient pyroptosis cancer therapy.
Evaluation of cell apoptosis performance
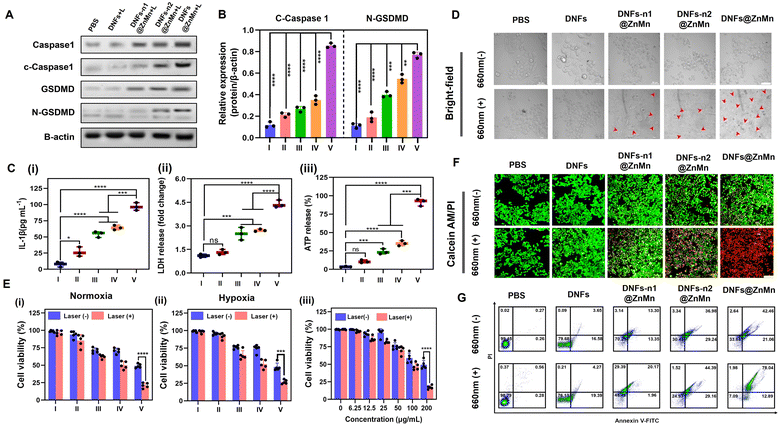
Upon the aforementioned cascade and synergistic relieving of tumor hypoxia (to enhance ROS generation) and autophagy disruption, efficient pyroptosis would be initiated. Via attacking ROS on the subcellular membrane, pyroptosis-inducing factors were released for the subsequent pyroptosis of tumor cells.42–45 Pyroptosis could be employed via the NLRP3-caspase 1-GSDMD pathway, including the formation of NLRP3 inflammasomes to active caspase-1. Thereafter, both pro-inflammatory cytokines IL-1β (Pro-1L-1β) and gasdermin D (GSDMD) were cleaved to release the N-terminal domain (GSDMD-N) and 1L-1β for executing pyroptosis via the pore-forming activities. Accordingly, pyroptosis was confirmed by the western blotting analysis, based on the significantly increased expression of cleaved-caspase 1 (c-caspase 1) and N-GSDMD after the treatments with DNFs@ZnMn (Fig. 5A, B and S37†). In addition, as the hallmark of pyroptosis upon cell rupture, the extracellular accumulation of inflammatory cytokine IL-1β, lactic dehydrogenase (LDH) and adenosine triphosphate (ATP) were evaluated with laser irradiation. As a result (Fig. 5C–E), the most significant increase of these extracellular contents was recorded in the group of DNFs@ZnMn, revealing the high pyroptosis efficiency. Consequently, pyroptosis was confirmed by the detection of the corresponding markers.Furthermore, the cell experiments were also carried out for evaluating apoptosis. As shown in Fig. 5F, a typical pyroptotic cell morphology was observed for the HeLa cells treated with DNFs@ZnMn. This further confirmed the efficient apoptosis by DNFs@ZnMn, which was also in accordance with the cell viability evaluations. As shown in Fig. 5G-i, the lowest cell viability resulted for HeLa cells treated with DNFs@ZnMn under light irradiation, displaying 79.5% photo-toxicity. More significantly, under hypoxic conditions (2% O2), a similar photo-toxicity was still exhibited (Fig. 5G-ii), which further confirmed the reliving of hypoxia for efficient therapy by DNFs@ZnMn. Furthermore, the cytotoxicity of cells by this apoptosis-based therapy was dose-dependent, which indicated the important roles of DNFs@ZnMn in efficient therapy (Fig. 5G-iii). In addition, the highest cytotoxicity was recorded at pH 5.5, which confirmed the selective therapy under TME conditions (Fig. S38†). Besides, the live/dead staining assays also confirmed the efficient therapy by the significant increase in red signals (indicating dead cells) in the DNFs@ZnMn group under light irradiation. This indicated that the blocking of protective autophagy significantly improved the therapeutic effect (Fig. 5H), while relatively low red signals were recorded in DNFs-n1@ZnMn (without DNAzyme-1) and DNFs-n2@ZnMn (without DNAzyme-2) groups, which further confirmed the cascade gene silencing of ATG-5 mRNA for apoptosis therapy. This was also in accordance with the annexin V-FITC/propidium iodide (PI) apoptosis analysis by flow cytometry (Fig. 5I). Besides, no obvious apoptosis was observed for normal HUVEC cells treated with DNFs@ZnMn (Fig. S39†), while a significant decrease in cell migration rate was exhibited after DNFs@ZnMn treatment (Fig. S40†). This further confirmed the satisfactory therapy performance of DNFs@ZnMn. Consequently, the cascade and synergistic relieving of tumor hypoxia and disrupting self-protection of cancer cells upon blocking the pyroptosis checkpoint were confirmed, thereby boosting the efficient pyroptosis cancer therapy.
In vivo examinations
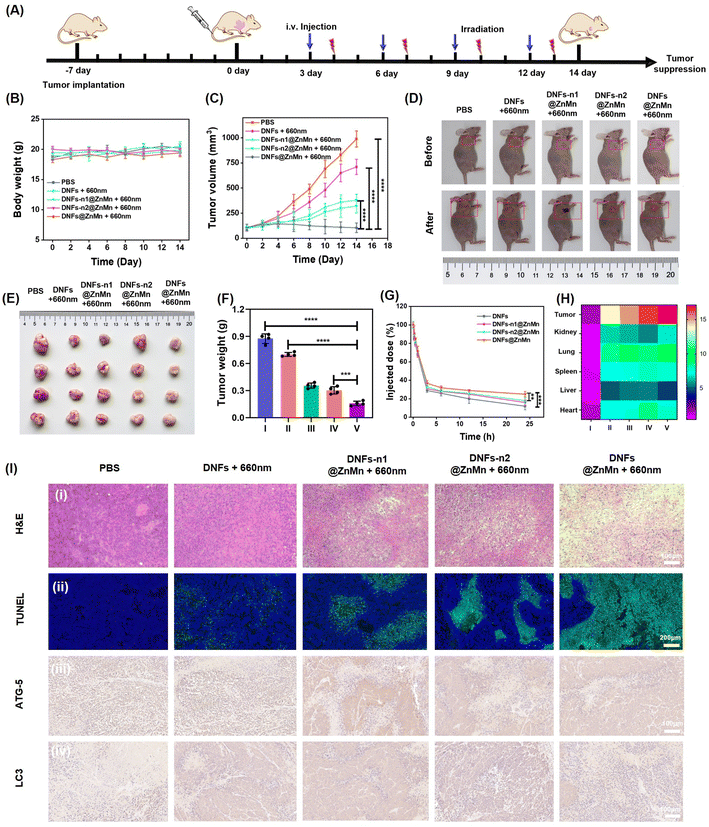
The in vivo therapeutic performance of the present DNFs@ZnMn was evaluated. First, the good biocompatibility of the DNF nanocomplex was confirmed by a hemolytic assay (Fig. S41†), which indicated no significant damage in erythrocytes without obvious adsorption of the released heme. This was in accordance with minimal damage to the kidney and liver (Fig. S42†), as indicated by the evaluation of long-term toxicity by hematology analysis and blood biochemical tests. Subsequently, the in vivo therapeutic performance was assessed with HeLa tumor-bearing mice as the models. As shown in Fig. 6A, different DNF nanocomplexes were intravenously injected every 2 days through the tail vein, and the tumor volumes and body weights were monitored every 2 days for 14 days. As indicated, the body weight was not affected by all the DNF nanocomplex groups (Fig. 6B), while the most significant tumor inhibition was only exhibited upon DNFs@ZnMn treatments under light-irradiation (Fig. 6C). This was further confirmed by the pictures of the tumors from each group (Fig. 6D). Besides, the tumors from the mice were excised and weighed after 14 days, and the most significant tumor suppression of 70.4% occurred in the DNFs@ZnMn group (Fig. 6E and F). Consequently, the efficient inhibition of tumor growth was confirmed, which was achieved by successful autophagy blockage to inhibit the self-protection of cancer cells with low side-effects.Besides, the in vivo delivery and tumor-specific targeting were also evaluated by the comparison of different DNF species with or without ZnO2–Mn functionalized. As shown by a pharmacokinetic study (Fig. 6G), without functionalization, the blood circulation half-time (t1/2) of DNA nanoflowers (DNFs) was relatively short (1.38 h). This was attributed to the negatively charged surface and the stable magnesium pyrophosphate skeleton of DNFs. While with the ZnO2–Mn and DNAzymes loaded (DNFs@ZnMn), the half-time increased to 2.05 h, due to the more compact nanostructure for increasing stability in blood circulation. Furthermore, the tumor-specific targeting was evaluated by imaging of different tissues from the mice treated with Cy5-labelled nanomaterials. As a result (Fig. 6H, S43 and S44†), DNFs@ZnMn exhibited the most significant accumulation at tumor tissues compared to other groups.
Finally, the anti-tumor activity and biosafety of DNFs@ZnMn were evaluated using hematoxylin and eosin (H&E) and terminal deoxynucleotidyl transferase dUTP nick end labeling (TUNEL) staining. As exhibited by imaging of both tumor tissues (Fig. 6I-i and ii) and tissues of other major organs (Fig. S45†), the most obvious unclear condensation and extensive apoptosis were observed in the DNFs@ZnMn group under light irradiation. This confirmed the therapeutic performance of DNFs@ZnMn, in accordance with the expression evaluation of protein markers by immunohistochemistry staining. As indicated in Fig. 6I-iii, the significantly decreased expression of ATG-5 protein resulted in tumor tissues treated with DNFs@ZnMn, which further confirmed the efficient silencing of ATG-5 for autophagy inhibition. In fact, efficient autophagy inhibition was also demonstrated by the significantly decreased expression of LC3 protein in the tumor tissues treated with DNFs@ZnMn. This was also in accordance with the data in western blotting analysis of HeLa cells (Fig. 4I). Consequently, the cascade autophagy inhibition was confirmed by the in vivo experiments, which was synergistic to relieving of tumor hypoxia for efficient pyroptosis cancer therapy.
Conclusion
A DNA nanocomplex with cascade DNAzymes, promoter-like Zn–Mn nanozymes and intercalated photosensitizers was successfully designed for efficient pyroptosis therapy by dynamic cascade disrupting autophagy and relieving of tumor hypoxia. Upon enrichment at tumor sites with a high expression of H+ and GSH, DNFs@ZnMn was gradually decomposed. This induced the release of cascade DNAzymes and ZnO2–Mn nanozyme, which provided cofactors of Zn2+ and Mn2+ to achieve cascade gene therapy of autophagy disruption. Simultaneously, the tumor hypoxia can be efficiently relieved by decomposition of of ZnO2 to generate H2O2 and then obtaining adequate O2 catalyzed by MnO2. This facilitated the subsequent obtaining of adequate cytotoxic 1O2 for efficient pyroptosis therapy. Upon RCA reactions for obtaining long-stranded DNFs@ZnMn, efficient cellular uptake with good stability and tumor specific therapy was obtained. In addition, the multiple metal active sites were integrated for dynamic cascade autophagy inhibition and relieving of tumor hypoxia without any interference from each other. Additionally, the high performance of pyroptosis therapy could be ensured by the sufficient multifunctional active sites based on the RCA-based synthesis strategy. It should be noted that this is an effort towards synergistic pyroptosis therapy upon dynamic cascade disruption autophagy and relieving tumor hypoxia in complex biological systems. The therapeutic efficacy should be further improved upon further studies. These could involve not only a more precise design to achieve efficient synergistic therapy (such as functional sequence-templated assembly with versatile inorganic materials), but also comprehensive mechanism examinations of enhanced pyroptosis therapy upon autophagy disruption. This work will motivate the future design of therapeutic regimens for more biocompatible and controllable cancer treatments based on high-performance pyroptosis therapy.Data availability
All relevant data is presented in the manuscript and ESI.† Raw data is available upon request by email to the corresponding author.Author contributions
X. Wang conceived the experiments, wrote the manuscript, and interpreted the data. X. Ge, M. Zhang and J. Sun supported figure preparations. J. Ouyang supported the characterizations. N. Na administered and supervised the project and acquired the funding.Conflicts of interest
There are no conflicts to declare.Acknowledgements
We gratefully acknowledge the financial support provided by the National Natural Science Foundation of China (No. 22274012), the Fundamental Research Funds for the Central Universities (No. 2233300007) and the Key Project of Science and Technology Plan of Beijing Education Commission (No. KZ20231002807).Notes and references
- P. Broz, P. Pelegrin and F. Shao, Nat. Rev. Immunol., 2020, 20, 143–157 CrossRef CAS PubMed.
- J. Shi, Y. Zhao, K. Wang, X. Shi, Y. Wang, H. Huang, Y. Zhuang, T. Cai, F. Wang and F. Shao, Nature, 2015, 526, 660–665 CrossRef CAS PubMed.
- X. Liu, Z. Zhang, J. Ruan, Y. Pan, V. G. Magupalli, H. Wu and J. Lieberman, Nature, 2016, 535, 153–158 CrossRef CAS PubMed.
- J. Ding, K. Wang, W. Liu, Y. She, Q. Sun, J. Shi, H. Sun, D. C. Wang and F. Shao, Nature, 2016, 535, 111–116 CrossRef CAS PubMed.
- V. G. Magupalli, R. Negro, Y. Tian, A. V. Hauenstein, G. Di Caprio, W. Skillern, Q. Deng, P. Orning, H. B. Alam, Z. Maliga, H. Sharif, J. J. Hu, C. L. Evavold, J. C. Kagan, F. I. Schmidt, K. A. Fitzgerald, T. Kirchhausen, Y. Q. Li and H. Wu, Science, 2020, 369, eaas8995 CrossRef CAS PubMed.
- V. A. Rathinam, S. K. Vanaja and K. A. Fitzgerald, Nat. Immunol., 2012, 13, 333–342 CrossRef CAS PubMed.
- T. Saitoh, N. Fujita, M. H. Jang, S. Uematsu, B. G. Yang, T. Satoh, H. Omori, T. Noda, N. Yamamoto, M. Komatsu, K. Tanaka, T. Kawai, T. Tsujimura, O. Takeuchi, T. Yoshimori and S. Akira, Nature, 2008, 456, 264–268 CrossRef CAS PubMed.
- Y. Huang, X. You, L. Wang, G. Zhang, S. Gui, Y. Jin, R. Zhao and D. Zhang, Angew. Chem., Int. Ed., 2020, 59, 10042–10051 CrossRef CAS PubMed.
- P. Pellegrini, A. Strambi, C. Zipoli, M. Hagg-Olofsson, M. Buoncervello, S. Linder and A. De Milito, Autophagy, 2014, 10, 562–571 CrossRef CAS PubMed.
- F. Xu, X. Li, X. Huang, J. Pan, Y. Wang and S. Zhou, Sci. Adv., 2020, 6, eabb8725 CrossRef CAS PubMed.
- C. S. Shi, K. Shenderov, N. N. Huang, J. Kabat, M. Abu-Asab, K. A. Fitzgerald, A. Sher and J. H. Kehrl, Nat. Immunol., 2012, 13, 255–263 CrossRef CAS PubMed.
- C. Q. Qiao, Z. Yang, X. L. Liu, R. L. Zhang, Y. Q. Xia, L. X. Wang, Z. Chen, Q. Jia, R. H. Wang, Y. Yang and Z. L. Wang, Nano Lett., 2022, 22, 8250–8257 CrossRef CAS PubMed.
- S. Wu, P. Wang, J. Qin, Y. Pei and Y. Wang, Adv. Funct. Mater., 2021, 31, 2102160 CrossRef CAS.
- C. Chung, W. Seo, P. Silwal and E. K. Jo, J. Hematol. Oncol., 2020, 13, 100 CrossRef PubMed.
- R. Jin, L. Liu, W. Zhu, D. Li, L. Yang, J. Duan, Z. Cai, Y. Nie, Y. Zhang, Q. Gong, B. Song, L. P. Wen, J. M. Anderson and H. Ai, Biomaterials, 2019, 203, 23–30 CrossRef CAS PubMed.
- P. Nunes, T. Ernandez, I. Roth, X. Qiao, D. Strebel, R. Bouley, A. Charollais, P. Ramadori, M. Foti, P. Meda, E. Féraille, D. Brown and U. Hasler, Autophagy, 2013, 9, 550–567 CrossRef CAS PubMed.
- T. Lopez-Hernandez, D. Puchkov, E. Krause, T. Maritzen and V. Haucke, Nat. Cell Biol., 2020, 22, 815–827 CrossRef CAS PubMed.
- X. Guo, F. Li, C. Liu, Y. Zhu, N. Xiao, Z. Gu, D. Luo, J. Jiang and D. Yang, Angew. Chem., Int. Ed., 2020, 59, 20651–20658 CrossRef CAS PubMed.
- J.-L. Xie, T.-J. Xie, Y.-J. Luo, K. Mao, C.-Z. Huang, Y.-F. Li and S.-J. Zhen, Chin. Chem. Lett., 2024, 35, 109137 CrossRef CAS.
- Y. Dong, C. Yao, Y. Zhu, L. Yang, D. Luo and D. Yang, Chem. Rev., 2020, 120(17), 9420–9481 CrossRef CAS PubMed.
- H. J. Vaughan, J. J. Green and S. Y. Tzeng, Adv. Mater., 2020, 32, 1901081 CrossRef CAS PubMed.
- A. Lacroix and H. F. Sleiman, ACS Nano, 2021, 15, 3631–3645 CrossRef CAS PubMed.
- F. Praetorius, B. Kick, K. L. Behler, M. N. Honemann, D. Weuster-Botz and H. Dietz, Nature, 2017, 552, 84–87 CrossRef CAS PubMed.
- Y. Dong, C. Yao, Y. Zhu, L. Yang, D. Luo and D. Yang, Chem. Rev., 2020, 120, 9420–9481 CrossRef CAS PubMed.
- H. Fan, X. Zhang and Y. Lu, Sci. China: Chem., 2017, 60, 591–601 CrossRef CAS.
- Z. Wu, H. Fan, N. S. R. Satyavolu, W. Wang, R. Lake, J. H. Jiang and Y. Lu, Angew. Chem., Int. Ed., 2017, 56, 8721–8725 CrossRef CAS PubMed.
- H. Wang, Q. Wu, X. Gong, K. Ma, X. Liu and F. Wang, Angew. Chem., Int. Ed., 2020, 59, 5965–5971 CrossRef PubMed.
- S. Khan, B. Burciu, C. D. M. Filipe, Y. F. Li, K. Dellinger and T. F. Didar, ACS Nano, 2021, 15, 13943–13969 CrossRef CAS PubMed.
- C. Gu, X. L. Liu, L. Luo, J. Q. Chen, X. Zhou, G. H. Chen, X. Huang, L. Yu, Q. Chen, Y. Yang and Y. Yang, Angew. Chem., Int. Ed., 2023, e202307020 CAS.
- Q. Mou, Y. Ma, F. Ding, X. Gao, D. Yan, X. Zhu and C. Zhang, J. Am. Chem. Soc., 2019, 141, 6955–6966 CrossRef CAS PubMed.
- D. Luo, X. Lin, Y. Zhao, J. L. Hu, F. Y. Mo, G. G. Song, Z. Q. Zou, F. Wang and X. Q. Liu, Chem. Sci., 2022, 13, 5155 RSC.
- C. Yao, H. D. Qi, X. M. Jia, Y. W. Xu, Z. B. Tong, Z. Gu and D. Y. Yang, Angew. Chem., Int. Ed., 2022, 61, e202113619 CrossRef CAS PubMed.
- Y. Xiao, T. Zhang, X. B. Ma, Q. C. Yang, L. L. Yang, S. C. Yang, M. Y. Liang, Z. G. Xu and Z. J. Sun, Adv. Sci., 2021, 8, 2101840 CrossRef CAS PubMed.
- D. Luo, X. Lin, Y. Zhao, J. L. Hu, F. Y. Mo, G. G. Song, Z. Q. Zou, F. Wang and X. Q. Liu, Chem. Sci., 2022, 13, 5155 RSC.
- W. T. Zhang, S. H. Li, X. N. Liu, C. Y. Yang, N. Hu, L. N. Dou, B. X. Zhao, Q. Y. Zhang, Y. R. Suo and J. L. Wang, Adv. Funct. Mater., 2018, 28, 1706375 CrossRef.
- Q. Y Liu, L. Y. Shi, Y. Liao, X. S. Cao, X. Q. Liu, Y. X. Yu, Z. F. Wang, X. H. Lu and J. W. Wang, Adv. Sci., 2022, 9, 2200005 CrossRef PubMed.
- X. Y. Xu, H. H. Fan, Y. Yang, S. K. Yao, W. H. Yu, Z. Guo and W. H. Tan, Angew. Chem., Int. Ed., 2023, 62, e2023030 Search PubMed.
- W. Li, S. L. Yin, Y. Shen, H. Y. Li, L. Yuan and X. B. Zhang, J. Am. Chem. Soc., 2023, 145, 3736–3747 CrossRef CAS PubMed.
- W. T. Zhang, Z. W. Liu, J. W. Zhu, Z. Q. Liu, Y. Zhang, G. Qin, J. S. Ren and X. G. Qu, J. Am. Chem. Soc., 2023, 145, 16658–16668 CrossRef CAS PubMed.
- X. B. Ma, W. Su, M. J. Ye, Y. Gao, W. Qiu, M. Y. Liang, P. Xue, Y. J. Kang, Z. J. Sun and Z. G. Xu, Nano Today, 2023, 48, 101727 CrossRef CAS.
- X. Z. Zhou, R. Rong, G. H. Liang, H. B. Li, M. L. You, Z. Zeng, H. H. Xiao, D. Ji and X. B. Xia, Nano Today, 2022, 47, 101668 CrossRef CAS.
- Y. Chen, W. Li, S. Kwon, Y. X. Wang, Z. T. Li and Q. Y. Hu, J. Am. Chem. Soc., 2023, 145, 9815–9824 CrossRef CAS PubMed.
- G. H Wang, B. Li, H. Tian, L. S. Xie, J. Yan, W. Sang, J. Li, Z. Zhang, W. X. Li and Y. L. Dai, Adv. Funct. Mater., 2023, 33, 2213425 CrossRef.
- P. Lu, X. J. Liu, X. Chu, F. L. Wang and J. H. Jiang, Chem. Sci., 2023, 14, 2562–2571 RSC.
- Y. Liang, P. P Lei, R. An, P. Y. Du, S. Y. Liu, Y. Wei and H. J. Zhang, Angew. Chem., Int. Ed., 2024, 63, e20231730 Search PubMed.
Footnote |
| † Electronic supplementary information (ESI) available: Experimental section; additional data for material characterization, including the data of fluorescence spectra and TEM, detailed DNA sequence, cell imaging, colocalization experiments, flow cytometric analyses, ex vivo imaging of organs and tumor tissues, and H&E staining of major tissue sections. See DOI: https://doi.org/10.1039/d4sc01147c |
| This journal is © The Royal Society of Chemistry 2024 |