Open Access Article
Open Access ArticlePredicting novel targets with Bayesian machine learning by integrating multiple biological signatures†
Xiao
Wei
a,
Tingfei
Zhu
ab,
Hiu Fung
Yip
b,
Xiangzheng
Fu
b,
Dejun
Jiang
a,
Youchao
Deng
a,
Aiping
Lu
b and
Dongsheng
Cao
 *ab
*ab
aXiangya School of Pharmaceutical Sciences, Central South University, Changsha, Hunan 410003, China. E-mail: oriental-cds@163.com
bSchool of Chinese Medicine, Hong Kong Baptist University, Hong Kong, SAR 999077, China
First published on 19th August 2024
Abstract
The identification of targets for candidate molecules is a pivotal stride in the drug development journey, encompassing lead discovery, drug repurposing, and the scrutiny of potential off-target or side effects. Consequently, enhancing the precision of target prediction has significant implications. Moreover, current target prediction methods primarily rely on the principle of ligand-based chemical similarity, lacking the capture of novel compound-target relationships based on ligand high-level characterization similarity. Therefore, in this context, we introduce a pioneering algorithm known as the Fused Multiple Biological Signatures (FMBS) strategy. This approach leverages a Bayesian framework to amalgamate 25 predictable biological space characterizations of molecules to predict novel targets through scaffold hopping, thereby improving target prediction accuracy and providing a versatile tool for a wide range of small-molecule target prediction. When juxtaposed with alternative target prediction methods, FMBS showcases notable efficacy, outperforming traditional descriptors. Through an analysis of scaffold hopping cases, we elucidate how FMBS attains heightened accuracy by assimilating comprehensive and complementary high-dimensional signatures, thereby underscoring its potential in unearthing novel compound-target relationships. The findings underscore that our approach adeptly pinpoints promising candidate targets, thereby expediting drug mechanism exploration through the integration of multiple high-level characterizations.
Introduction
Identifying the targets of candidate molecules is a pivotal step in drug development. It can help confirm targets for natural or synthetic compounds with potential biological activities1,2 to prevent unexpected off-target effects due to incomplete mechanistic studies3 and even uncover new knowledge for existing drugs.4 However, traditional experimental methods for target identification are limited by their resource-intensive and time-consuming characteristics,5 posing significant challenges to the swift progression of drug discovery efforts.To address these limitations, computational target prediction methods have emerged as a valuable adjunct to experimental techniques. They bridge the gaps in target identification by simulating and forecasting potential interactions between compounds and biological targets during the pre-clinical stages of drug development. Among these methods, structure-based target prediction methods are limited by the need for protein crystal structure data and high computational power, with representative methods including reverse molecular docking6 and reverse pharmacophore matching.7 In parallel, ligand-based target prediction strategies have been developed.8 Among them, ligand search methods are favored for their computational efficiency. The theoretical basis of this approach is that small molecules with similar chemical structures or physicochemical properties may interact with the same targets. By comparing the chemical structures or physicochemical properties of query molecules with those of known active target molecules, we can predict other potential targets of the query molecules.
Previous studies have typically conducted similarity searches based on a single level, such as chemical structure or bioactivity characterization. For example, methods, such as the SEA (similarity ensemble approach) server are based on 2D structural similarity9 and SwissTargetPrediction utilizes 2D and 3D descriptors for compound profiling.10 However, structure-based prediction methods are limited in handling scaffold hopping. These methods primarily focus on the intrinsic properties of small molecules and do not account for their interactions with other entities (such as cellular detection), and thus, they are ineffective in driving target prediction guided by biological functions.11 Therefore, to predict novel small molecule-target interactions beyond chemical similarity, some studies incorporate information on advanced biological features into their similarity searches.12 For example, Campillos et al. inferred shared targets between drugs based on side effect similarities of 746 marketed drugs. They validated 13 implicit drug-target relationships through experiments and suggested phenotype information to be a potential indicator for novel molecular interactions.13 Subsequently, Iorio et al. constructed a “drug network” of 1302 nodes (drugs) by computing the similarity of the transcriptional responses containing 6100 genome-wide expression profiles. They also verified an unexpected similarity between cyclin-dependent kinase 2 inhibitors and topoisomerase inhibitors.14
However, relying solely on single-level information for target prediction hinders the comprehensive understanding of compound-target interactions.11 Therefore, an increasing number of studies are incorporating a wider range of phenotype data, which may be complementary to bridging the gap between molecular properties and biological functions, thus providing a more comprehensive understanding of the compound behavior and facilitating the identification of the biological activities of specific targets.15–18 Some studies utilize supervised (such as CMLDR19) and unsupervised machine learning (ML) methods (such as MSSP proposed by Cao et al. based on collaborative filtering recommendation systems20 and RWHNDR proposed by Luo et al.21) to link biological and chemical spaces to improve target prediction. Additionally, researchers from Weill Cornell Medical College combine the structural similarity of molecules with four types of biological activity (including growth inhibition, side effects, bioassays, and gene expression) using Bayesian methods, demonstrating strong target prediction performance.22 These findings suggest that the accuracy of target prediction improves with the emergence of new types of phenotype data. However, these data types focus on several structural or phenotypic information levels that are widely applicable for target prediction. Therefore, there is an urgent need to develop target prediction methods that can capture multidimensional information to identify the interaction between novel small molecules and targets as comprehensively as possible.
Additionally, phenotypic data is obtained through costly biological experiments that result in molecules with multiple types of phenotype data typically representing only a small fraction of chemical space.23,24 Most phenotype-based methods currently cannot predict targets for molecules without phenotypic data, while many types of phenotypic data are complementary.25 Therefore, for many small molecules with incomplete biological activity profiles, the effective utilization of high-level biological information to enhance target prediction is a missing link. It is noteworthy that a method utilizing a Siamese neural network was proposed to predict high-level biological signatures of molecules, which encompassed 25 biological facets covering the entirety of drug development, thus enabling the acquisition of biological signatures for any small molecules.26
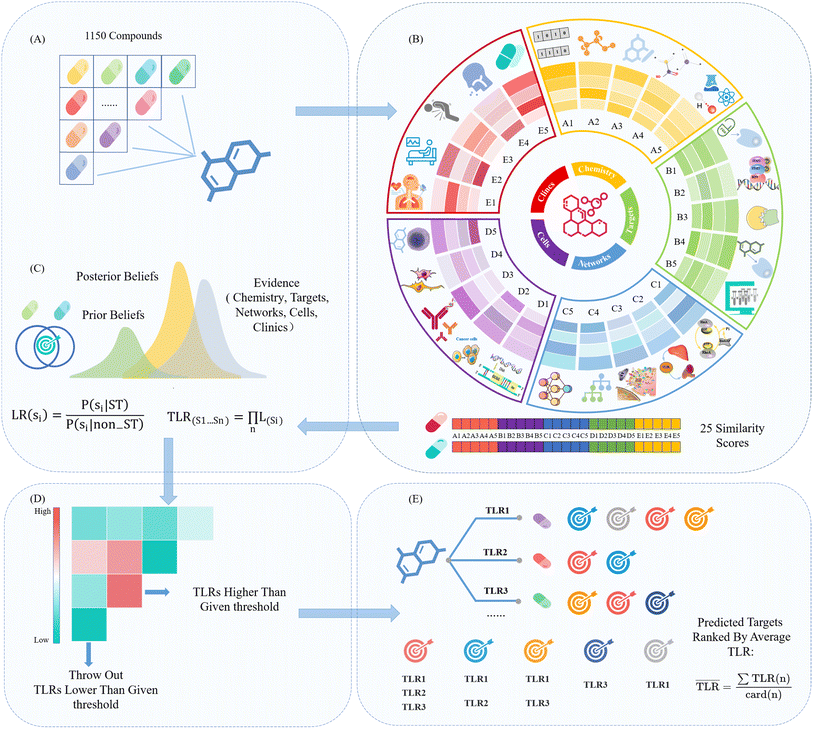
In this study, we propose an approach to address the limitations discussed above: using a Bayesian method with strong interpretability and flexible framework, the similarities of 25 characterizations were combined to predict drug targets more accurately. These 25 characterizations cover various dimensions of the drug discovery process, including molecular targets and biological networks to cellular responses and clinical applications. Subsequently, we validated the predictive performance of this method, including leave-one-out cross-validation and external dataset validation. Compared with other target prediction methods, our model demonstrates equivalent or even better predictive ability in top-k predictions. Finally, through case studies of scaffold hopping, we confirmed that biological signatures reveal drug-target relationships that are difficult to discover solely at the chemical level.
Materials and methods
Data collection and curation
The original dataset of target prediction was downloaded from the DrugBank (version 5.1.10) database. Proteins were first grouped by protein type (target, enzyme, or transporter) and then subcategorized based on their pharmacological activity. To enhance the accuracy of target prediction, we specifically chose targets marked as pharmacologically active. A series of preparations were applied: (1) targets associated with the human species were retained. (2) Targets that are solely bound to biotech drugs were removed. (3) For each target, small-molecule drugs were retained and biotech drugs, e.g. monoclonal antibodies, were removed. Finally, we obtained the resulting drug-target network containing 688 targets, 1150 drugs, and 3199 associations (Additional file 1†).Twenty-five bioactivity descriptors as compound signatures
The 25 biological descriptors representing different biological spaces are divided into five main aspects (molecule, target, pathway, cell, or clinic), and each was further subdivided into five sub-aspects as follows:(A) At the molecular level, there are five fingerprints, A1–A5: A1 provides 2D structural information of molecules, constructed using a 2048 bit Morgan fingerprint with a bond radius of 2, capturing atom types, bond lengths, and relative positions; A2 utilizes E3FP fingerprints, representing the binary hash of the three best-conformational minima after energy minimization; A3 provides Murcko scaffold information; A4 employs MACCS fingerprints, encoding 166 predefined substructures known to effectively encode molecular structures; A5 offers physicochemical parameters, such as molecular weight, log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) P, refractivity, hydrogen bond donors and acceptors, and alert structures.
P, refractivity, hydrogen bond donors and acceptors, and alert structures.
(B) The target-related information level comprises five fingerprints, B1–B5: B1 provides information on the pharmacological mechanism of molecules; B2 offers metabolic gene information; B3 provides protein structure information; B4 contains binding affinity information from the ChEMBL33 and binding DB; and B5 includes high-throughput screening information from the PubChem database.
(C) The biological network level includes five fingerprints, C1–C5: C1 provides ontology terms associated with small molecules with recognized biological activity; C2 offers metabolic pathway information, mainly focusing on endogenous metabolites; C3 provides information on biological pathways affected by molecular-target interactions; C4 represents biological process information from protein-annotated gene ontology annotations; C5 includes representative protein–protein interaction network information.
(D) The cellular level consists of five fingerprints, D1–D5: D1 provides transcription information of small molecules in different cell lines; D2 contains GI50 data from 60 cancer cell lines; D3 includes screening results of small molecules against approximately 300 yeast mutants, reflecting chemical genetic information; D4 contains data on changes in the cellular morphology induced by small molecules; D5 encompasses cell-based assay data, i.e., primarily growth and proliferation measurements, of small molecules reported in ChEMBL33.
(E) The clinical level includes five fingerprints, E1–E5: E1 includes ATC classification information of drugs; E2 contains drug indication information; E3 provides information on adverse effects of small-molecule drugs; E4 offers disease phenotype information of small-molecule drugs; and E5 includes drug–drug interaction information.
The biological characteristics of 25 dimensions were predicted and completed through Siamese neural networks, which can be applied to any molecule, with each dimension being 128-dimensional. These descriptors were calculated using signatures.26
Integration of molecular signatures using the Bayesian framework
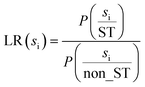 | (1) |
For each biological characterization, the similarity scores were grouped into 20 equally spaced intervals, and LR (si) was computed for each interval. Following this, the exponential functions were independently fitted to the 25 biological levels using Python's ‘predict’ and ‘exp’ functions, which were then utilized to calculate the likelihood values for new pairs of compounds. Ultimately, we chose the Bayesian framework to integrate a variety of information on biological signatures, owing to its high interpretability and the flexibility to accommodate new data types. Consequently, the total likelihood ratio (TLR) 22 was determined as the product of the individual LRs. It was directly proportional to the likelihood of two compounds sharing a target within a given biological characterization:
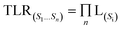 | (2) |
Voting strategy for target prediction
Each query molecule formed compound pairs with all 1150 molecules in the library, and the TLR for each pair was calculated. Compounds with TLR exceeding a given threshold were predicted to share the same target. Each predicted molecule corresponded to one or multiple known targets, and these targets were associated with their corresponding TLR. Subsequently, all known targets of these predicted molecules were compiled to form the predicted target set of the query molecule. Each target molecule in the target set had a corresponding final TLR value. If a protein was targeted by multiple compounds, it had multiple TLRs from different sources. The average TLR was used as the final TLR for that target. Finally, a list of target predictions was obtained by ranking the final TLR values of the targets.Performance evaluation
Our approach involved weighting the predicted target sets for voting to ensure that the Bayesian integration framework effectively distinguishes between the shared and non-shared drug pairs. Therefore, to guarantee the accuracy of the target prediction approach, we initially evaluated the performance of the Bayesian fusion signature framework and then assessed the effectiveness of the target voting strategy.![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2 split. Specifically, each fold of the training set consisted of 528540 drug pairs that were further divided into 9392 ST pairs and 519148 non-ST pairs. Each test set comprised 152135 drug pairs, consisting of 2,348 ST pairs and 129787 non-ST pairs. The probabilities obtained from the training dataset were utilized to calculate the TLR for each drug pair in the test set. Finally, the results from the five test folds were combined to construct an ROC curve and determine the average AUC value.
2 split. Specifically, each fold of the training set consisted of 528540 drug pairs that were further divided into 9392 ST pairs and 519148 non-ST pairs. Each test set comprised 152135 drug pairs, consisting of 2,348 ST pairs and 129787 non-ST pairs. The probabilities obtained from the training dataset were utilized to calculate the TLR for each drug pair in the test set. Finally, the results from the five test folds were combined to construct an ROC curve and determine the average AUC value.
Results and discussion
FMBS: a fused multiple biological signatures strategy for predicting targets
We designed a strategy called fused multiple biological signatures (FMBS) to predict compound targets. This strategy is based on the Bayesian framework and utilizes multiple high-level biological characterization resources. The underlying concept of the method is that similar compounds share similar targets, where the degree of similarity was determined here by integrating 25 biological characterizations within a Bayesian framework. Specifically, when given a set of compounds with known targets, we computed pairwise similarity scores of compounds for each level (total of 25 levels) and used the proportion of pairs with at least one shared target (ST pairs), among all pairs as the prior probability. The likelihood scores were derived using the prior probability, enabling the inference of the likelihood that new compound pairs are shared. Ultimately, we calculated a total likelihood ratio, which was directly proportional to the likelihood of compound pairs being shared by integrating the distinct likelihood ratio across 25 dimensions within the Bayesian framework. Subsequently, a voting strategy was used to rank the sets of shared compound targets, thereby enriching potential correct targets (Fig. 1).Analysis of compound-target data and 25 bioactivity signatures
We retrieved 3199 drug-target interactions (DTIs) for FMBS from the DrugBank (version 5.1.10) database to estimate accurate and high-quality compound-target relations. The extracted interactions involved a total of 1150 drugs and 688 targets. Among the 1150 drugs, the number of associated targets ranged from 1 to 30, with a mean of 2.78. Among the 688 targets, the number of associated drugs ranged from 1 to 51, with a mean of 4.65. The 1150 drugs generated a total of 660![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 675 drug pairs, including 11
675 drug pairs, including 11![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 740 ST pairs and 648
740 ST pairs and 648![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 935 non-ST pairs.
935 non-ST pairs.
We employed newly proposed biological signatures for compound representation. The biological signatures are categorized into five levels according to the increasing complexity of the drug discovery pr. These levels span from the interaction between a small molecule (A, chemistry) and its associated protein receptors (B, targets) to the activated biological pathways (C, networks) and the resulting observable phenotypic changes (D, cells), ultimately extending to the clinical levels (E, clinics). Following this, each level (A–E) was further divided into five sublevels (1–5), revealing finer characteristics of molecules in the biological space (see the Materials and methods for details of 25 biological signatures). It is noteworthy that since these 25 characterizations can be predicted, they can be applied to all molecules.
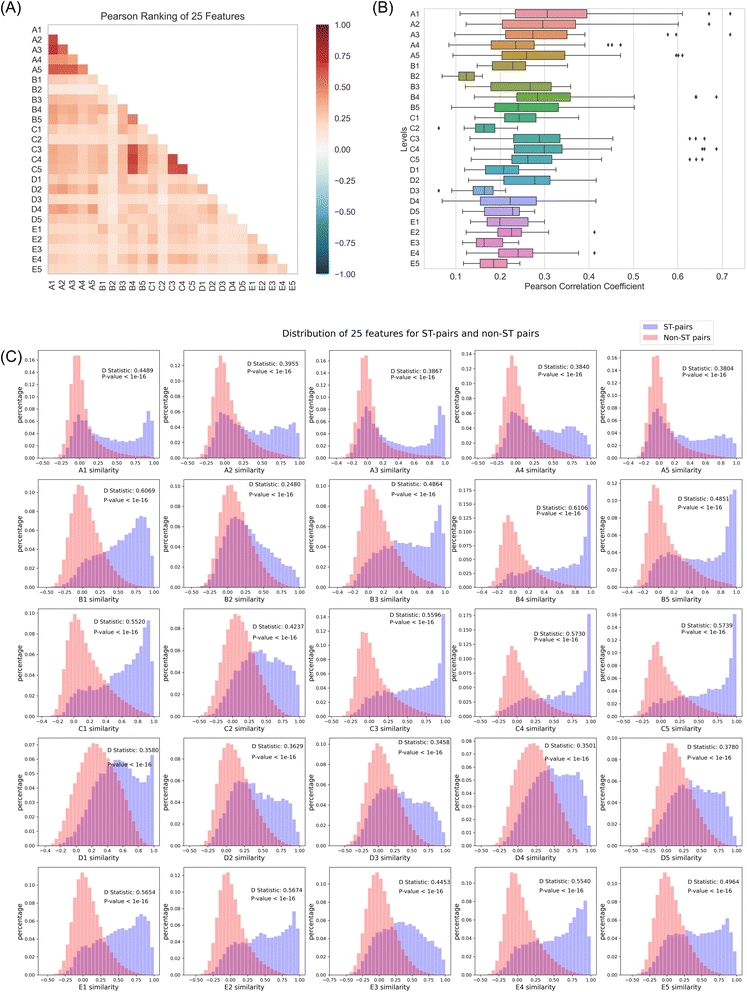
In order to explore the correlation among the 25 characterizations, we initially computed the similarity scores of 660![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 675 drug pairs for each of the 25 levels, separately. Subsequently, the Pearson correlation coefficient was utilized as a standard to assess the correlation between the similarities across the 25 levels. The results (Fig. 2A) indicated that the overall correlation among the 25 levels was relatively low. It is well-established that descriptors with more similar information exhibit higher correlation, and vice versa. Therefore, it is not difficult to understand that the correlation between the A levels and the C levels (mainly C3, C4, and C5) is relatively high, as they are closely and directly related to structure and biological pathway information, respectively. Additionally, the B4 level compiles information on targets with explicit mechanisms, while the C3, C4, and C5 levels assemble biological signals triggered by compounds, which also reflect part of the mechanistic information. Therefore, the relatively higher correlation between B4 and C3, C4, and C5 is reasonable. Even so, the correlation coefficients of most of the levels are below 0.5, and the similarity of the highly correlated levels mentioned above does not exceed 0.7 (Fig. 2B). Therefore, it can be considered that each level provides new information related to compounds to varying degrees, suggesting that integrating these complementary, high-dimensional signatures can provide more comprehensive information.
675 drug pairs for each of the 25 levels, separately. Subsequently, the Pearson correlation coefficient was utilized as a standard to assess the correlation between the similarities across the 25 levels. The results (Fig. 2A) indicated that the overall correlation among the 25 levels was relatively low. It is well-established that descriptors with more similar information exhibit higher correlation, and vice versa. Therefore, it is not difficult to understand that the correlation between the A levels and the C levels (mainly C3, C4, and C5) is relatively high, as they are closely and directly related to structure and biological pathway information, respectively. Additionally, the B4 level compiles information on targets with explicit mechanisms, while the C3, C4, and C5 levels assemble biological signals triggered by compounds, which also reflect part of the mechanistic information. Therefore, the relatively higher correlation between B4 and C3, C4, and C5 is reasonable. Even so, the correlation coefficients of most of the levels are below 0.5, and the similarity of the highly correlated levels mentioned above does not exceed 0.7 (Fig. 2B). Therefore, it can be considered that each level provides new information related to compounds to varying degrees, suggesting that integrating these complementary, high-dimensional signatures can provide more comprehensive information.
Then, the Kolmogorov–Smirnov test was conducted on similarity scores of the ST pairs and non-ST pairs based on 25 levels to evaluate the discriminative capability of the 25 characterizations. The relevant D statistic served as the evaluation metric. In Fig. 2C, it was observed that all characterizations could significantly differentiate between the ST and non-ST pairs (P-value < 2 × 10−16). As anticipated, the information conveyed by levels, B1 and B4, was the most relevant in separating ST pairs from all pairs, thus exhibiting the highest discriminative ability among all evaluated characterizations (D = 0.61). Following closely were the C-related levels, especially level C4 (D = 0.57), which revolved around biological processes in gene ontology annotations and have previously been demonstrated to facilitate the discovery of new drug-target relationships or mechanisms.27 Subsequently, levels E1, E2, and E4 contain higher-level biological features related to clinical information, with D values of approximately 0.56. This result supports the principle that drug targets are directly linked to clinical phenotypes, such as indications and side effects. Surprisingly, characterizations commonly used in target prediction, such as structure similarity (A levels) and gene expression profile similarity (D levels), did not achieve the expected discriminative effects, with corresponding D values around 0.4 and 0.36, respectively. For structural similarity, the phenomenon of activity cliffs in compounds is difficult to address through structural similarity alone. Moreover, the biological activity of many compounds depends not only on their structure but also on their dynamic behavior, metabolic processes, and other factors. Additionally, the signatures at level A were obtained through further dimensionality reduction of chemical fingerprints, which may have resulted in some loss of information. These factors could contribute to the low discriminative power of structural similarity. Therefore, integrating these high-level biological layers to complement each other and capture more information about the interactions between drug targets may improve the discriminative ability. For gene expression profile similarity, the complexity of gene expression profiles within biological systems (with high noise levels and variability among individuals) and the inherent redundancy of gene data may contribute to achieving the expected discriminative effect.
Overall, through the above exploration, we found that each characterization may describe different aspects of drugs and possess potential for target prediction. Therefore, we opted to keep all levels and proposed a hypothesis: combining these 25 characterizations can enhance target prediction ability.
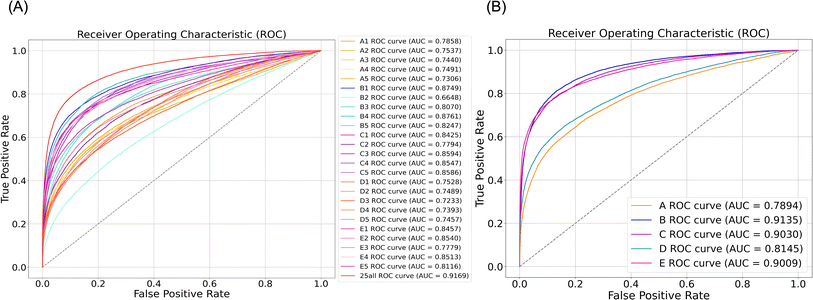
FMBS increases the accuracy of distinguishing ST and non-ST pairs
To confirm the hypothesis, we adopted a Bayesian framework to integrate 25 biological characterizations and evaluated its performance in distinguishing ST pairs from all drug pairs using 5-fold cross-validation. The total likelihood ratio (TLR) for each testing drug pair with known targets was calculated by multiplying its individual likelihood ratio (LR) at each level. As depicted in Fig. 3A, integrating all characterizations within the framework resulted in superior performance compared to any single characterization, achieving an average area under the curve (AUC) of up to 0.92. Moreover, it is noteworthy that, for each main level (A, B, C, D, E), the discriminative capability surpassed that of the corresponding 5 sub-levels (see Fig. 3). Additionally, the AUC for the discriminative capability of the method, which integrated 25 characterizations, was moderately higher than that of any individual main level. This suggests that the discriminative capability increases with the incorporation of additional dimensional information. Spontaneously, this preliminary evidence supported our hypothesis that the integration of multiple types of information may improve target prediction performance by enhancing accuracy in distinguishing between ST and non-ST pairs.We observed the ability of each level to distinguish between the ST and non-ST pairs to further investigate their respective contributions to the Bayesian prediction framework. The results indicated that each level exhibited a discriminatory ability, with the AUC ranging from 0.72 to 0.87, as illustrated in Fig. 3A. The performance of each characterization was ranked from high to low as follows (the top 8 characterizations): B4 > B1 > C3 > C5 > C4 > E2 > E4 > C1. Meanwhile, Fig. 3B demonstrated that the discriminatory ability that was achieved by integrating the results of five different levels using the Bayesian framework was highly consistent with the results of the K–S distribution test. Therefore, both individual levels and the five main levels demonstrated that the prediction accuracies of the B, C, and E levels (B: 0.91; C: 0.90; E: 0.90), representing target, network, and clinical information, were higher. In contrast, the discriminatory abilities of the A and D levels (A: 0.79; D: 0.81), representing chemical structure and cellular information, were relatively lower. This suggested to researchers that when computational or data constraints prevented the simultaneous use of information from all levels, priority could be given to information closely related to the B, C, and E levels to ensure the accuracy of target prediction.
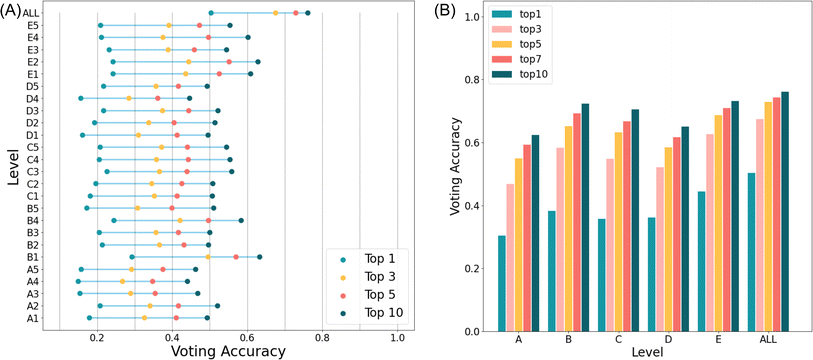
Evaluation of the target prediction performance after finishing target voting
To obtain the target prediction list, we conducted target voting on the predicted ST drug pairs. The best prediction accuracy was obtained after implementing the voting strategy using the leave-one-out method under different TLRs (refer to “Materials and methods”); the corresponding TLR of the best prediction accuracy was utilized as the default threshold for subsequent experiments, such as external validation set evaluation. As anticipated, the voting results aligned with the outcomes of distinguishing between shared and non-shared drug pairs, indicating that as the number of integrated information levels increased, the predictive capability for target identification progressively improved (see ESI Fig. 2†). The fusion of 25 information levels exhibited superior target prediction accuracy compared to that of the five levels, which, in turn, surpassed that of a single level. Meanwhile, Fig. 4A illustrates that the target prediction accuracy significantly increased after integrating 25 levels compared to individual levels. These findings emphasized that each level may capture varying degrees of information regarding new drug-target interactions. Interestingly, when distinguishing between shared and non-shared drug pairs, level ‘B’ demonstrated the highest performance. However, the voting results suggested that information from level ‘E’ contributed to more accurate final target predictions, which approached the outcomes obtained from the fusion of all levels. This implied that clinical information held greater relevance for target prediction, which is consistent with the understanding that targets are closely associated with clinical aspects (Fig. 4B).The method generated 1150 TLRs between the drug pairs for each compound to be predicted. Subsequently, a voting process was employed to calculate the average likelihood value for each protein, resulting in a ranked list of target predictions arranged from highest to lowest likelihood values. Undoubtedly, increasing the number of selected targets would also enhance the accuracy. However, considering the actual experimental costs, we calculated the accuracy for the top-k targets (k = 1–10) and used them to assess the accuracy of the target predictions. As anticipated, the integration of information from 25 levels led to a substantial enhancement in target prediction accuracy, with the actual target recall rate of our method gradually increasing as k ranged from 1 to 10. Following fusion, the prediction accuracy of the top 10, represented by a single feature, rose from around 50% to over 70% (Fig. 4A). In comparison to the accuracy of the top 1, which was represented by a single feature and did not exceed 30%, the accuracy of the top 1 represented by fusion was approximately 50%. This implied that by employing our method for target prediction, roughly half of the drugs could achieve accurate target predictions at the top 1, which narrowed the scope of targets for subsequent testing and decreased experimental expenses. Of note, the target prediction accuracy exhibited a substantial increase from the top 1 to the top 3, reaching up to 20%. However, the growth rate slowed notably beyond the top 5–10. Therefore, this suggested that in practical usage of the model, opting for the top 5 struck a relative balance between cost and accuracy.
In conclusion, this study validated our hypothesis that with the increase in the variety of integrated characterization types, the accuracy of target prediction also improved. Specifically, augmenting conventional chemical structure models with additional advanced characterization information significantly enhanced the prediction of molecular-protein binding interactions. Therefore, our target prediction method was highly competitive.
Comparison with alternative approaches
To further validate the excellent target prediction capability of FMBS, we compared it with other methods. Firstly, to ensure the fairness and reliability of model evaluation, we randomly selected 791 compounds from ChEMBL33 with Ki/IC50 values less than 10 μM in 2022 and 2021 as an external validation set. To avoid potential bias, these compounds did not appear in our modeling dataset. After preprocessing, our external validation dataset ultimately comprised 791 compounds and 181 proteins, representing 934 compound-target interactions. The accuracy defined in this study was then used to compare our FMBS with other methods, and the comparative results are presented in Table 1. Detailed information on validation data can be found in Additional file 1.†| Method | Top 1 | Top 3 | Top 5 | Top 7 | Top 10 | |
|---|---|---|---|---|---|---|
| PPB2 | DNN | 15.78 | 30.05 | 36.36 | 40.03 | 44.57 |
| NN(ECFP4) | 19.82 | 37.75 | 47.22 | 51.14 | 54.92 | |
| NN(MQN)_NB(ECFP4) | 15.03 | 31.57 | 38.89 | 43.94 | 49.12 | |
| NN(XFP) | 11.24 | 24.62 | 30.43 | 35.48 | 39.39 | |
| NN(XFP)_NB(ECFP4) | 16.16 | 34.72 | 41.67 | 46.47 | 51.01 | |
| NN_NB(ECFP4) | 09.22 | 25.13 | 35.48 | 42.17 | 48.86 | |
| NB(ECFP4) | 10.46 | 23.54 | 29.08 | 34.92 | 40.92 | |
| NN_MQN | 10.48 | 20.08 | 23.11 | 26.52 | 30.81 | |
| SEA | 20.57 | 34.41 | 42.56 | 46.44 | 50.97 | |
| FMBS | 32.45 | 52.27 | 62.25 | 70.08 | 78.41 | |
The comparison results demonstrate that FMBS outperformed other methods in terms of recall rate for top-k predictions, including the popular PPB2 method NN(XFP)_NB(ECFP4) (top 10 Recall: 51.01% vs. 78.41%). Additionally, when compared with SEA, it was found that among 773 molecules, 50.97% of the molecules had at least one known target predicted within the top 10 predictions of SEA (with an additional 19 molecules not yielding prediction results). Among the 791 molecules, 78.41% of the molecules had at least one known target predicted within the top 10 predictions of our method. Of particular note is that our method correctly predicted 32.45% of ligands for the top 1 prediction, whereas SEA achieved only 20.57%. Since SEA is acknowledged for its high target prediction recall rate,28 it is particularly encouraging that our method surpasses SEA in the recall rate. Overall, these remarkable results highlight the effectiveness of our method as a robust tool to predict potential targets that may strongly interact with compounds. As previously mentioned, we attributed the performance of FMBS to inferring relationships between new drug targets using novel types of information, thereby enhancing target prediction accuracy through the integration of diverse information types. In subsequent experiments, we further investigated this by conducting a case study on scaffold-hopping.
Analysis of target prediction caused by scaffold-hopping
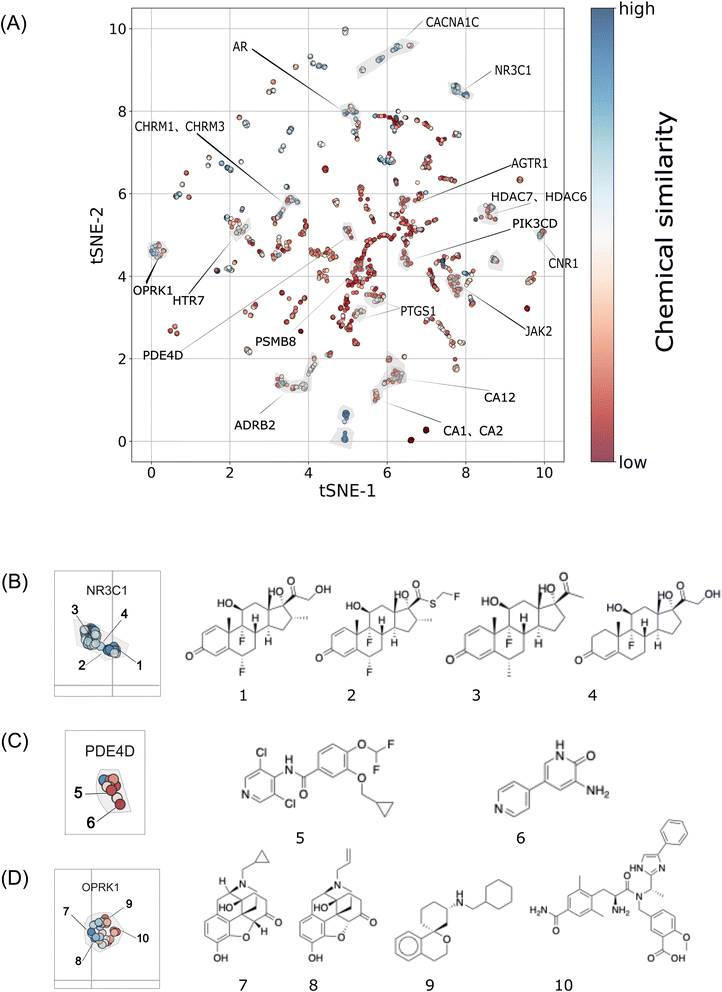
To demonstrate that the biological fingerprints employed in this study contained high-level biological information beyond chemical structures, and thus, possessed scaffold-hopping capabilities to enhance target prediction accuracy, we further analyzed the biological and chemical similarities of active molecules for specific targets to investigate the practical effectiveness of 25 biological descriptors in target prediction. Specifically, we explored the clustering distribution of all molecules in the test set and validation set at the biological level (all 25 biological signatures) to observe the biological similarity between molecules. We color-coded the molecules based on their chemical similarity while highlighting molecules corresponding to specific proteins. From the results in Fig. 5A, it was observed that according to biological fingerprint clustering, most ligands of the same target were clustered in the same region, indicating that biological fingerprints could indeed reflect the biological functions of compounds, further demonstrating the rationality of using biological fingerprints for target prediction. Additionally, it was observed that some molecules, such as the red cluster in Fig. 5A (ligands of proteins, such as AGTR1, PTGS1, CA1, CA2, JAK2, etc.), were more similar at the biological level, but they had lower chemical similarity, indicating that these molecules could improve the ability of target prediction through our method by overcoming the scaffold-hopping phenomenon. Similarly, in the blue cluster in Fig. 5A, ligands of proteins, such as CACNA1C, AR, CHRM1, CHRM3, NR3C1, etc., included molecules that were similar at both the biological and chemical levels, which meant that it was difficult for these proteins to achieve scaffold-hopping when searching for ligands. Next, specific case analyses were conducted for different situations.Some clustering attempts group molecules with similar chemical structures together, indicating their proximity in both biological and chemical spaces. In such cases, a more comprehensive target space can be predicted through searches based on either chemical structure similarity or biological descriptor similarity. For example, targeting NR3C1, 25 biological descriptors cause ligands of NR3C1 to cluster (see Fig. 5B). These molecules all share cyclopentane as their core structure, exhibiting high chemical similarity. Specifically, there is a Morgan fingerprint similarity of 0.76 between compounds 3 and 4 and a similarity of 0.5 between compounds 4 and 6.
Of note, there are also some clusters corresponding to sets of diverse compounds, meaning that they are close in biological space but exhibit significant differences in chemical structure. Therefore, relying solely on chemical structure similarity for target prediction is limited, often leading to blind spots attributed to scaffold-hopping. However, biological fingerprints can fill this gap. For example, regarding the PDE4D target (see Fig. 5C), its ligand compounds are clustered in the same biological space, but they have significantly different core structures. For instance, compounds 1 and 2 have a Morgan fingerprint Tanimoto similarity of only 0.1. These novel potential active molecules with unique scaffolds are often overlooked by methods relying on chemical structure similarity searches. Therefore, this is also a key reason why our approach, integrating multiple high-level descriptors, can uncover newer compound-target associations, thereby enhancing target prediction accuracy.
The majority of clusters contain mixed components, comprising both chemically relevant and dissimilar subgroups. For instance, in Fig. 5D for the OPRK1 target, ligands include compounds 7 and 8 with a similarity of 0.75, as well as compounds 9 and 10 with a similarity of 0.086. Our strategy can improve target prediction accuracy for these molecules as well.
Application of FMBS and case study
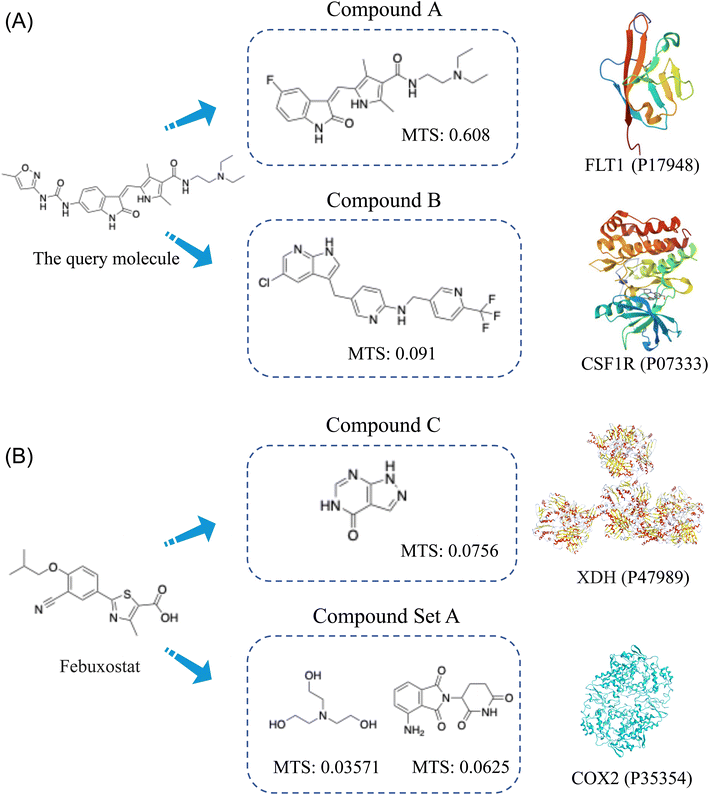
To further validate the predictive capability of FMBS to identify novel compound-target pairs through scaffold-hopping, we applied our FMBS method and SEA websites to predict targets for some molecules from ChEMBL33 (which were not included in our modeling dataset). We found that some new compound-target interactions could be predicted through our method, but they were difficult to identify through methods based on chemical similarity. The query molecule in Fig. 6A is a typical molecule selected for in-depth exploration. Both the FMBS method and SEA predicted the target FLT1 for this query molecule as a case study. Our method formed compound pairs that target this query molecule by pairing it with compound B (with a high Morgan fingerprint Tanimoto similarity of up to 0.608), indicating that this query molecule could be predicted through chemical similarity. Additionally, our method also worked via compound B-targeted CSF1R29 (has been validated) with a chemical similarity of only 0.091 between the query molecule and compound B. This target was not predicted by SEA, suggesting that it might have been predicted through biological information similarity. This result is consistent with the conclusions drawn from the case analysis of scaffold-hopping. Further elucidation demonstrates that our method is capable of capturing targets with ligands of high chemical similarity, as well as those of low chemical similarity through high-level biological information, thus integrating more comprehensive information to enhance target prediction accuracy.Additionally, we conducted predictions on several drugs by listing their corresponding drug targets explicitly documented in DrugBank,30 along with the targets predicted by our method and supported by literature validation in Table 2. Febuxostat (DB04854), a notable example, was initially annotated in DrugBank30 as targeting xanthine dehydrogenase/oxidase for the treatment of chronic hyperuricemia and gout.31 However, subsequent research demonstrated that Febuxostat could mitigate paraquat-induced pulmonary toxicity by upregulating COX2 expression, thereby elucidating a novel mechanism for its ameliorative effects.32 Moreover, earlier experiments also reported similar inhibitory effects of Febuxostat on COX2 activity in a diabetic renal injury model.33 Remarkably, FMBS successfully predicted its target as COX2. Interestingly, XDH ranks in the top1 in the prediction results of FMBS, while COX2 ranks top2. From Fig. 6B, it can be observed that FMBS targets XDH through compound C, and it targets COX2 through molecules in a compound set A. The structures of these compounds differ significantly from Febuxostat itself, with a Morgan fingerprint Tanimoto similarity of less than 0.08, indicating that this target prediction is achieved by capturing biological-level information. This further exemplifies how FMBS can effectively facilitate the prediction of potential new drug targets, thereby offering valuable insights for drug repurposing and adverse effect discovery in pharmaceutical research.
| Drug ID | Drug name | Target in DrugBank | Our predicted targets (have been validated) |
|---|---|---|---|
| DB04854 | Febuxostat | XDH | COX2 (ref. 32 and 33) |
| DB01076 | Atorvastatin | HMGCR | ABCB1 (ref. 34) |
| DB13873 | Fenofibric acid | PPARA | PPARD, PPARG35,36 |
| DB01203 | Nadolol | ADRB1 | ADRB2 (ref. 37 and 38) |
| DB00682 | Warfarin | VKORC1 | ABCC2, PDE10A39 |
Overall, these cases validate the improved predictive capability of our method through scaffold-hopping, which is accomplished by integrating diverse biological signatures. Furthermore, they demonstrate our method's potential to forecast new targets, expedite drug repurposing, and investigate potential mechanisms of drug side effects.
Conclusion
In summary, this study introduces a novel target prediction approach that integrates 25 informative, high-dimensional, and universal molecular characterizations. These characterizations encompass comprehensive information on molecules, such as their structural characteristics and pathways to clinical drug stages. In contrast to conventional general descriptors, such as chemical descriptors, which often lead to blind spots in target prediction due to the challenges in capturing scaffold transitions, our method integrates multiple high-dimensional descriptors to improve performance by capturing more compound-target relationships. Additionally, commonly used biological characterizations can only characterize a small fraction of molecules, resulting in the weak generalization ability of multi-level target prediction models. However, the characterizations adopted in our approach are predictable, thus enabling the prediction of targets for any given molecule. Nevertheless, it is important to acknowledge the limitations of this study. Although our method can theoretically be applied to all molecules, the accuracy may vary due to factors, such as the reliability of the prediction of molecular characterization. At the same time, different characterizations contribute differently to the task and selecting strongly correlated characterizations according to the task can save computational resources. These findings provide valuable guidance for future research, suggesting the exploration of using other multimodal computational methods to enhance target prediction performance and adopting other methods beyond similarity searching to improve model accuracy. Furthermore, when the modeling dataset is based on compounds rather than drugs, the applicability of the model is broadened, and its accuracy is improved. Overall, this study provides valuable insights into efficiently identifying new targets and drug repurposing in the entire field using the Bayesian framework and new high-dimensional characterizations. Improving the accuracy of target prediction is of significant practical importance to save drug development costs and uncover drug mechanisms.Data availability
All the datasets supporting the conclusion of this article are available in the Additional files. The code implementing the FMBS has been deposited in GitHub (https://github.com/wei-xiao-ya/FMBS).†Author contributions
CDS and WX designed the study. WX collected the data, built and tested the models, prepared the diagrams and figures, and wrote the manuscript. ZTF contributed to valuable discussions. YHF assisted in building models related to Bayesian integration. WX, ZTF, YHF, and CDS reviewed and revised the manuscript. FXZ, JDJ, DYC, and LAP helped check and improve the manuscript. All authors have read and approved the final version of the manuscript.Conflicts of interest
The authors declare no conflict of interest.Acknowledgements
This work was supported by the National Key Research and Development Program of China [2021YFF1201400], National Natural Science Foundation of China [22173118, 22220102001], Hunan Provincial Science Fund for Distinguished Young Scholars [2021JJ10068], the Science and Technology Innovation Program of Hunan Province [2021RC4011], The Natural Science Foundation of Hunan Province [2022JJ80104], and The 2020 Guangdong Provincial Science and Technology Innovation Strategy Special Fund [2020B1212030006, Guangdong-Hong Kong-Macau Joint Lab]. Funding for open access charge: HKBU Strategic Development Fund project [SDF19-0402-P02]. We acknowledge Haikun Xu, and the High-Performance Computing Center of Central South University for their support. The study was approved by the university's review board.References
- M. Williams, Target validation, Curr. Opin. Pharmacol., 2003, 3(5), 571–577 CrossRef CAS PubMed
.
- A. L. Hopkins, Network pharmacology: the next paradigm in drug discovery, Nat. Chem. Biol., 2008, 4(11), 682–690 CrossRef CAS PubMed
.
- J. N. Chan, C. Nislow and A. Emili, Recent advances and method development for drug target identification, Trends Pharmacol. Sci., 2010, 31(2), 82–88 CrossRef CAS PubMed
.
- B. Reisberg, R. Doody and A. Stöffler,
et al., Memantine in moderate-to-severe Alzheimer's disease, N. Engl. J. Med., 2003, 348(14), 1333–1341 CrossRef CAS
.
- M. Floris, S. Olla and D. Schlessinger,
et al., Genetic-Driven Druggable Target Identification and Validation, Trends Genet., 2018, 34(7), 558–570 CrossRef CAS PubMed
.
- P. S. Kharkar, S. Warrier and R. S. Gaud, Reverse docking: a powerful tool for drug repositioning and drug rescue, Future Med. Chem., 2014, 6(3), 333–342 CrossRef CAS PubMed
.
- D. Rognan, Structure-Based Approaches to Target Fishing and Ligand Profiling, Mol. Inf., 2010, 29(3), 176–187 CrossRef CAS PubMed
.
- H. Ding, I. Takigawa and H. Mamitsuka,
et al., Similarity-based machine learning methods for predicting drug-target interactions: a brief review, Briefings Bioinf., 2014, 15(5), 734–747 CrossRef
.
- M. J. Keiser, B. L. Roth and B. N. Armbruster,
et al., Relating protein pharmacology by ligand chemistry, Nat. Biotechnol., 2007, 25(2), 197–206 CrossRef CAS PubMed
.
- D. Gfeller, A. Grosdidier and M. Wirth,
et al., SwissTargetPrediction: a web server for target prediction of bioactive small molecules, Nucleic Acids Res., 2014, 42(Web Server issue), W32–W38 CrossRef CAS PubMed
.
- A. F. Fliri, W. T. Loging and P. F. Thadeio,
et al., Biological spectra analysis: Linking biological activity profiles to molecular structure, Proc. Natl. Acad. Sci. U. S. A., 2005, 102(2), 261–266 CrossRef CAS PubMed
.
- A. M. Wassermann, E. Lounkine and L. Urban,
et al., A screening pattern recognition method finds new and divergent targets for drugs and natural products, ACS Chem. Biol., 2014, 9(7), 1622–1631 CrossRef CAS PubMed
.
- M. Campillos, M. Kuhn and A. C. Gavin,
et al., Drug target identification using side-effect similarity, Science, 2008, 321(5886), 263–266 CrossRef CAS PubMed
.
- F. Iorio, R. Bosotti and E. Scacheri,
et al., Discovery of drug mode of action and drug repositioning from transcriptional responses, Proc. Natl. Acad. Sci. U. S. A., 2010, 107(33), 14621–14626 CrossRef CAS PubMed
.
- F. Cheng, W. Li and Z. Wu,
et al., Prediction of polypharmacological profiles of drugs by the integration of chemical, side effect, and therapeutic space, J. Chem. Inf. Model., 2013, 53(4), 753–762 CrossRef CAS PubMed
.
- G. Wu, J. Liu and C. Wang, Predicting drug-disease interactions by semi-supervised graph cut algorithm and three-layer data integration, BMC Med. Genomics, 2017, 10(Suppl 5), 79 CrossRef PubMed
.
- O. Laufkötter, N. Sturm and J. Bajorath,
et al., Combining structural and bioactivity-based fingerprints improves prediction performance and scaffold hopping capability, J. Cheminf., 2019, 11(1), 54 Search PubMed
.
- A. M. Wassermann, E. Lounkine and M. Glick, Bioturbo similarity searching: combining chemical and biological similarity to discover structurally diverse bioactive molecules, J. Chem. Inf. Model., 2013, 53(3), 692–703 CrossRef CAS
.
- H. Luo, J. Wang and C. Yan,
et al., A Novel Drug Repositioning Approach Based on Collaborative Metric Learning, IEEE/ACM Trans. Comput. Biol. Bioinf., 2021, 18(2), 463–471 CAS
.
- D. S. Cao, S. L. Jiang and Y. D. Guan,
et al., A multi-scale systems pharmacology approach uncovers the anti-cancer molecular mechanism of Ixabepilone, Eur. J. Med. Chem., 2020, 199, 112421 CrossRef CAS PubMed
.
- H. Luo, J. Wang and M. Li,
et al., Computational Drug Repositioning with Random Walk on a Heterogeneous Network, IEEE/ACM Trans. Comput. Biol. Bioinf., 2019, 16(6), 1890–1900 CAS
.
- N. S. Madhukar, P. K. Khade and L. Huang,
et al., A Bayesian machine learning approach for drug target identification using diverse data types, Nat. Commun., 2019, 10(1), 5221 CrossRef
.
- T. Sterling and J. J. Irwin, ZINC 15–Ligand Discovery for Everyone, J. Chem. Inf. Model., 2015, 55(11), 2324–2337 CrossRef CAS
.
- J. L. Reymond, The chemical space project, Acc. Chem. Res., 2015, 48(3), 722–730 CrossRef CAS PubMed
.
- A. M. Wassermann, E. Lounkine and J. W. Davies,
et al., The opportunities of mining historical and collective data in drug discovery, Drug Discovery Today, 2015, 20(4), 422–434 CrossRef PubMed
.
- M. Bertoni, M. Duran-Frigola and I. M. P. Badia,
et al., Bioactivity descriptors for uncharacterized chemical compounds, Nat. Commun., 2021, 12(1), 3932 CrossRef CAS PubMed
.
- F. Guo, X. Tang and W. Zhang,
et al., Exploration of the mechanism of traditional Chinese medicine by AI approach using unsupervised machine learning for cellular functional similarity of compounds in heterogeneous networks, XiaoErFuPi granules as an example, Pharmacol. Res., 2020, 160, 105077 CrossRef CAS PubMed
.
- K. Y. Ji, C. Liu and Z. Q. Liu,
et al., Comprehensive assessment of nine target prediction web services: which should we choose for target fishing?, Briefings Bioinf., 2023, 24(2), bbad014 CrossRef PubMed
.
- Q. Lv, X. Pan and D. Wang,
et al., Discovery of (Z)-1-(3-((1H-Pyrrol-2-yl)methylene)-2-oxoindolin-6-yl)-3-(isoxazol-3-yl)urea Derivatives as Novel and Orally Highly Effective CSF-1R Inhibitors for Potential Colorectal Cancer Immunotherapy, J. Med. Chem., 2021, 64(23), 17184–17208 CrossRef CAS PubMed
.
- D. S. Wishart, Y. D. Feunang and A. C. Guo,
et al., DrugBank 5.0: a major update to the DrugBank database for 2018, Nucleic Acids Res., 2018, 46(D1), D1074–d82 CrossRef CAS
.
- M. Hu and B. Tomlinson, Febuxostat in the management of hyperuricemia and chronic gout: a review, Ther. Clin. Risk Manage., 2008, 4(6), 1209–1220 CAS
.
- M. A. E. Ahmed, E. M. El Morsy and A. A. E. Ahmed, Protective effects of febuxostat against paraquat-induced lung toxicity in rats: Impact on RAGE/PI3K/Akt pathway and downstream inflammatory cascades, Life Sci., 2019, 221, 56–64 CrossRef CAS PubMed
.
- H. J. Lee, K. H. Jeong and Y. G. Kim,
et al., Febuxostat ameliorates diabetic renal injury in a streptozotocin-induced diabetic rat model, Am. J. Nephrol., 2014, 40(1), 56–63 CrossRef CAS PubMed
.
- M. L. Becker, L. E. Visser and R. H. Van Schaik,
et al., Influence of genetic variation in CYP3A4 and ABCB1 on dose decrease or switching during simvastatin and atorvastatin therapy, Pharmacoepidemiol. Drug Saf., 2010, 19(1), 75–81 CrossRef CAS PubMed
.
- I. Inoue, F. Itoh and S. Aoyagi,
et al., Fibrate and statin synergistically increase the transcriptional activities of PPARalpha/RXRalpha and decrease the transactivation of NFkappaB, Biochem. Biophys. Res. Commun., 2002, 290(1), 131–139 CrossRef CAS PubMed
.
- Y. Rival, A. Stennevin and L. Puech,
et al., Human adipocyte fatty acid-binding protein (aP2) gene promoter-driven reporter assay discriminates nonlipogenic peroxisome proliferator-activated receptor gamma ligands, J. Pharmacol. Exp. Ther., 2004, 311(2), 467–475 CrossRef CAS
.
- I. Ozakca, E. Arioglu and S. Guner,
et al., Role of beta-3-adrenoceptor in catecholamine-induced relaxations in gastric fundus from control and diabetic rats, Pharmacology, 2007, 80(4), 227–238 CrossRef CAS PubMed
.
- J. W. Wisler, S. M. Dewire and E. J. Whalen,
et al., A unique mechanism of beta-blocker action: carvedilol stimulates beta-arrestin signaling, Proc. Natl. Acad. Sci. U. S. A., 2007, 104(42), 16657–16662 CrossRef CAS PubMed
.
- L. Granadeiro, R. P. Dirks and J. B. Ortiz-Delgado,
et al., Warfarin-exposed zebrafish embryos resembles human warfarin embryopathy in a dose and developmental-time dependent manner - From molecular mechanisms to environmental concerns, Ecotoxicol. Environ. Saf., 2019, 181, 559–571 CrossRef CAS PubMed
.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d4sc03580a |
| This journal is © The Royal Society of Chemistry 2024 |