Open Access Article
Open Access ArticleSialylation-induced stabilization of dynamic glycoprotein conformations unveiled by time-aligned parallel unfolding and glycan release mass spectrometry†
Yifei
Jia‡
a,
Yichang
Liu‡
 b,
Yamei
Wang‡
a,
Jinyu
Li
b,
Yamei
Wang‡
a,
Jinyu
Li
 *d and
Gongyu
Li
*d and
Gongyu
Li
 *ac
*ac
aTianjin Key Laboratory of Biosensing and Molecular Recognition, Research Center for Analytical Science, Frontiers Science Center for New Organic Matter, College of Chemistry, Nankai University, Tianjin 300071, China. E-mail: ligongyu@nankai.edu.cn
bSchool of Pharmacy, Nantong University, Nantong 226001, Jiangsu, China
cHaihe Laboratory of Sustainable Chemical Transformations, Tianjin 300192, China
dCollege of Chemistry, Fuzhou University, Fuzhou 350108, Fujian, China. E-mail: j.li@fzu.edu.cn
First published on 12th August 2024
Abstract
Sialylation, a critical post-translational modification, regulates glycoprotein structure and function by tuning their molecular heterogeneity. However, characterizing its subtle and dynamic conformational effects at the intact glycoprotein level remains challenging. We introduce a glycoform-resolved unfolding approach based on a high-throughput ion mobility-mass spectrometry (IM-MS) platform. This method integrates high-throughput unfolding with parallel fragmentation, enabling simultaneous analysis of sialylation patterns, stoichiometries, and their impact on conformational stability. Applying this approach to fetuin, we identified distinct sialylation patterns and their differential influence on protein conformation, namely sialylation-induced stabilization during early unfolding and increased flexibility in later unfolding stages. IM-MS-guided molecular dynamics simulations revealed that increased sialylation enhances the initial conformational stability, likely through enhanced electrostatic interactions and hydrogen bonding. These findings highlight the complex interplay between sialylation and protein dynamics and establish glycoform-resolved unfolding IM-MS as a powerful tool for characterizing glycoprotein conformational landscapes.
Introduction
Protein sialylation, the addition of sialic acid to glycoproteins during the final decoration stage of protein bio-synthesis via the endoplasmic reticulum, plays a crucial role in various biological processes and has significant implications for health and disease.1,2 Specifically, sialylation can enhance protein structural stability by protecting them from proteolytic degradation and thermal denaturation.3,4 The negative charges of sialic acids can also influence protein folding states.5 By definition, sialylation is always involved in the interaction systems, including cell–cell recognition, pathogen interactions and many other immune modulations.6–8 For example, sialylation of the Fc region of immunoglobulin G (IgG) can promote anti-inflammatory effects by increasing its affinity for the inhibitory Fcγ receptor (FcγRIIB).9,10 In short, sialylation has been widely involved in diverse scientific and practical application directions, including cancer biology, biomarker development and biotherapeutics discovery. Comprehensive understanding of the chemical composition, environment-responsive molecular patterns and structural dynamics of protein sialylation is central to the development of new diagnostic tools, therapeutic strategies, and improved biopharmaceuticals.However, precisely characterizing the dynamic conformations of glycoproteins and the changes governed by sialylation presents significant technical challenges: (1) microheterogeneity, multiple glycoforms present at each glycosylation site, arising from variations in the number, type (N-acetylneuraminic acid (Neu5Ac) and N-glycolylneuraminic acid (Neu5Gc)), and linkage of sialic acid residues (α2,3vs. α2,6 or α2,8), as well as the underlying glycan structure; (2) sialic acid diversity, comprising a family of over 50 structurally distinct monosaccharides, with further modification by acetylation, methylation, or sulfation; (3) lability of sialic acid, prone to loss during sample preparation and analysis via the acid- and temperature-driven hydrolysis pathway.11,12 The above-mentioned inherent characteristics of sialylation have significantly hindered their accurate characterization. These challenges are exacerbated by limitations in existing analytical techniques, including insufficient isomeric resolution, a shortage of well-characterized sialylated glycoprotein standards and the lack of complementary methods for capturing dynamic conformations in a time-sensitive manner.13,14
Native mass spectrometry (MS) is a powerful tool for analyzing intact protein complexes and glycoproteins.15–17 High-resolution native MS not only decodes glycan heterogeneity, but also elucidates the relationship between structural heterogeneity and glycoprotein interaction.18 For example, Wu et al.19 utilized a native top-down strategy to analyze three glycoproteins with increasing levels of heterogeneity, revealing that glycosylation can regulate glycoprotein assembly. Similarly, integrating native MS and glycoproteomics enabled the characterization of glycosylation patterns on SARS-CoV-2 spike protein and angiotensin-converting enzyme 2 (ACE2), along with the binding stoichiometry of spike-ACE2 complexes mediated by N-glycans.20 However, sialic acid loss during sample preparation, coupled with suppressed ionization efficiency, often necessitates offline enzymatic removal of sialic acids prior to native MS experiments. This approach facilitates reproducible comparisons of compositions and topologies among differentially sialylated isoforms.21,22
Native ion mobility-mass spectrometry (IM-MS), as a powerful structural MS technique, offers conformer-resolving capabilities and enables dynamic conformer manipulation through collision-induced unfolding (CIU).23–25 CIU tracks the stability and unfolding pathway of gas-phase protein ions via stepwise collisional heating, providing insights into dynamic protein conformations and enabling rapid isoform differentiation. For instance, CIU experiments demonstrated a strong correlation between the gas-phase stabilities of IgG1 ions and glycosylation extent.26 Previously, we developed an all-ion unfolding (AIU) strategy to uncover dynamic structural information on intact glycoproteins, effectively sampling representative conformational ensembles in an expedited manner.27 AIU's ability to incorporate all protein charge states, tolerate charge shifts inherent to heterogeneous glycoprotein analysis, and preserve intact protein topology makes it a powerful tool for comprehensive glycoprotein structural characterization.5,28,29 For example, Li et al.5 analyzed the conformational stability of differentially sialylated transferrin by the CIU-IM-MS strategy and confirmed that the conformational stability of Neu5Gc modification was lower than that of Neu5Ac modification on glycoprotein transferrin, while both modifications stabilize its overall conformation.
Among sialylated glycoproteins, fetuin is an abundant serum glycoprotein that contains both N-linked and O-linked glycosylation sites, displaying extensive molecular heterogeneity.30,31 This inherent complexity, stemming from extensive glycan modifications, conformational flexibility, and intrinsic molecular heterogeneity, has hindered complete structural elucidation of fetuin with native glycans.
To investigate the impact of sialylation on glycoprotein structural dynamics and thermal stability, we selected fetuin as a model system and employed a series of unfolding experiments based on an advanced AIU-based unfolding regime, termed the glycoform-resolved unfolding approach. To correlate sialylation patterns with intact protein dynamics, we designed experiments using a time-aligned parallel (TAP) unfolding and glycan release mode in conjunction with AIU. We performed enzyme-assisted glycoproteomic analyses on human and bovine fetuin samples, including a sialylation-specific enzyme, to profile sialylation and other glycosylation modifications.
To further unveil the molecular details of sialylation effects on the dynamic 3D structure of fetuin, we conducted gas-phase molecular dynamics (MD) simulations32,33 guided by native MS and AIU data, corroborated by classical MD simulations in aqueous solution. Our integrative results demonstrate that terminal sialylation stabilizes fetuin conformation and restricts its dynamic structural fluctuations, likely through modulation of the hydrogen bonding and contact area between glycans and the protein backbone, as well as maintenance of secondary structural elements.
Results and discussion
Fetuin sialylation patterns
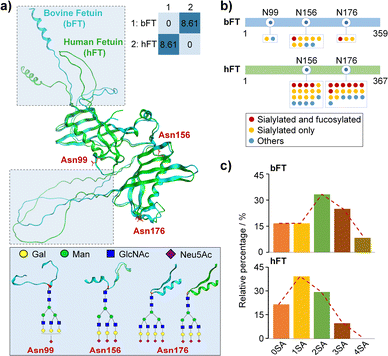
Fetuin is a highly sialylated glycoprotein system with well-characterized glycan profiles and structures.21,30 Bovine (bFT) and human fetuin (hFT) samples display a high sequence conservation showing approximately 65% homology at the amino acid level (Table S1†) and a high structure similarity with a root-mean-square deviation (RMSD) of 0.485 Å after deducting a random coil (Fig. 1a). We carried out EThcD-based bottom-up glycoproteomics experiments to identify and quantify glycosylation patterns, especially for the sialylation level, from different fetuin groups (Tables S2 and S3†).Fig. 1b summarizes the site-specific glycosylation patterns of bFT and hFT, highlighting differences in N-glycan occupancy, structural composition, and microheterogeneity. These results, including differential sialylation levels and stoichiometry, are further corroborated in Fig. 1c. Notably, the di-sialylated glycoform predominates in bFT, while the mono-sialylated form is most abundant in hFT. The unique presence of a tetra-sialylated glycoform in bFT contributes to its higher overall sialylation level compared to hFT. The observed differences in sialylation patterns make hFT and bFT a well-established model system for further interrogation of sialylation-governed protein conformations.
Glycoform-resolved unfolding
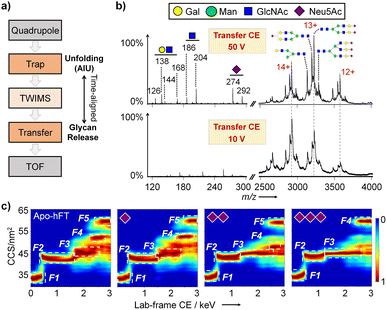
To correlate the intact protein structure with specific glycoforms, we developed a glycoform-resolved unfolding strategy integrating native IM-MS and AIU. Using a Synapt instrument, intact protein ions were first mass-selected in the quadrupole (or not selected for the AIU mode) and activated in the trap cell at optimized energy. The resulting oxonium ions and glycoprotein ions underwent separation in the TWIMS cell, followed by transport through the transfer cell for TAP fragmentation in parallel with trap cell activation (Fig. 2a).34,35 This post-IM fragmentation in the transfer cell facilitates the removal of nonspecific adducts and unannotated glycans, addressing the challenge of limited mass resolving capability due to the inherent molecular heterogeneity of most glycoproteins. Increasing the collision energy (CE) in the transfer cell improved the signal-to-noise (S/N) ratio of protein ions and generated dissociated glycan fragment ions, indicating enhanced glycan resolving power with increased TAP fragmentation energy (Fig. S1 and S2†).However, excessively high CE (100 V and 150 V) led to protein backbone cleavage and increased signal complexity, hindering analyte signal identification. Optimizing the transfer CE to 50 V for hFT analysis resulted in good separation of both saccharide oxonium ions and major proteoforms (charge states 12+, 13+, and 14+). N-linked glycosylation in hFT was confirmed based on the characteristic ion ratio for GlcNAc and GalNAc.36 Notably, the sialoform assignments, determined from glycoform-resolved unfolding experiments under the native MS mode on a Synapt XS instrument (Fig. 2b), were consistent with glycoproteomic analysis (Fig. 1).
To explore the effects of sialylation on fetuin conformational stability, a CIU experiment was conducted with trap CE from 10 V to 180 V and transfer CE kept at 50 V to maximize the resolving capability of molecular heterogeneity. Charge-separated unfolding fingerprints of bFT and hFT revealed distinct unfolding patterns for different charge states, generally with two to five features observed for charges 12+ to 15+, and corresponding differences in CCS and CIU50 values (Fig. S3 and S4†).
To facilitate intuitive comparison of protein conformations, all-ion unfolding (AIU) fingerprints were generated for each glycoprotein, retaining all conformational information and mitigating the impact of charge state shifts inherent to intact glycoprotein analysis.27 The two fetuin proteins exhibited unique dynamic unfolding characteristics, with conformations clearly distinguished by an RMSD of 26.64% (Fig. S5†). The glycoform-resolving capability of the optimized AIU strategy was further validated by isolating m/z ranges corresponding to specific sialoforms. hFT with different sialoforms displayed distinct unfolding patterns (Fig. 2c). As the number of sialic acids increases, the disparity in CCS values between unfolding intermediates F2 and F3 diminishes gradually. Notably, the tri-sialylated hFT conformation exhibits four features compared to five features among other sialoforms. In contrast, variations in individual sialic acid modifications had negligible effects on bFT conformation, with all sialoforms exhibiting three features (Fig. S6†). Overall, the number of sialic acids is likely linked to the conformational flexibility of glycoprotein fetuin.
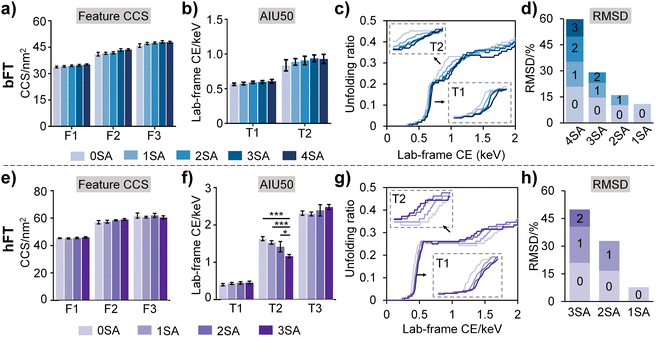
To further elucidate sialylation-induced structural alterations, we quantified a series of conformational parameters, including feature CCS, AIU50 values, and unfolding extents under distinct levels of sialylation as obtained from AIU experiments (Fig. 3). Slight CCS variations were observed across differential sialoforms (Fig. 3a and e), suggesting that sialylation-induced subtle changes in overall conformations are difficult to be effectively resolved by CCS alone. Increasing sialic acid contents led to higher AIU50 values for the first transition (T1) (Fig. 3b and f), implying that sialylation enhances the resistance of glycoprotein to unfolding, presumably through promoting additional electrostatic interactions and hydrogen bonding between sialic acid and protein domains. It is noteworthy that the opposite trend was observed in the second transition (T2) of hFT, speculating that sialylation could promote the conformational flexibility of the protein to some degree (Fig. 3f). This observation is in good accordance with the unfolding feature number–conformational flexibility relationship as shown in Fig. 2c.
Then, unfolding curves were employed to quantify unfolding proportions and provide a dynamic view of sialylation-governed conformational stability and dynamic transitions. The unfolding curves were plotted through monitoring CCS changing percentages against lab-frame collisional energy (Fig. 3c and g). From the unfolding curves, a clear sialylation number-dependent unfolding trend was apparently present as indicated by the gradual increment of unfolding onset energy along with the increase in sialic acid numbers. Consistent with AIU50 results, the shallow slopes of the first transition (T1) in both fetuin proteins with increasing sialic acid contents indicated that sialylation enhances resistance to protein unfolding. Notably, we also observed a uniform sialylation–conformational flexibility relationship from the T2 stage in the unfolding curves (Fig. 3g), in line with data in Fig. 2c and 3f. Furthermore, the sialoform-resolved unfolding strategy amplified the conformational discrepancies between different sialoforms, with RMSDs of 5.9–20.8% for bFT and 7.7–20.9% for hFT (Fig. 3d and h).
Overall, increasing sialylation correlated with enhanced conformational stability, particularly during early unfolding transitions, likely due to increased electrostatic interactions and hydrogen bonding between sialic acid residues and the protein backbone. Interestingly, higher sialylation levels also appeared to promote conformational flexibility in later unfolding stages, suggesting a complex interplay between sialylation and fetuin structural dynamics. These findings highlight the sensitivity of AIU-based approaches for characterizing subtle conformational changes induced by labile glycan modifications.
Molecular mechanism of sialylation effects uncovered by IM-MS-guided MD simulation
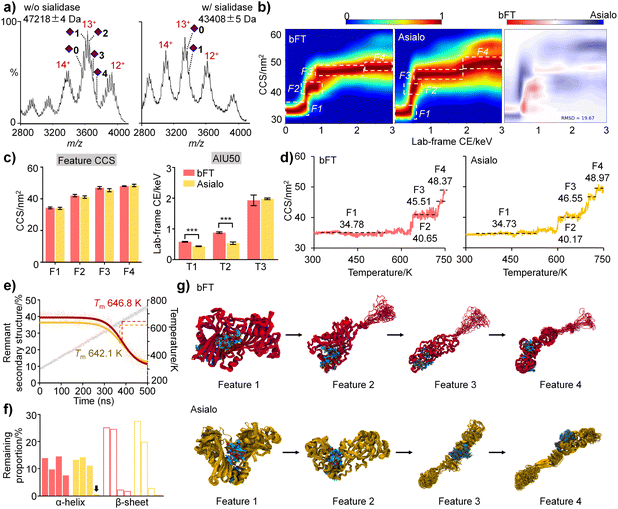
To clarify the interplay between sialylation and protein structure, bFT was subjected to sialidase digestion. Both native MS (Fig. 4a) and proteomics (Table S4†) confirmed substantial removal of sialic acids. Consistent with previous findings, sialylation influenced the unfolding pathway. Fully sialylated bFT exhibited a distinct intermediate state (F2), while the asialo form was more susceptible to conformational transitions, resulting in a significant conformational difference (RMSD = 19.67%) (Fig. 4b). Complete sialylation correlated with larger CCS values and significantly higher AIU50 values, further supporting a stabilizing effect of sialylation on protein structure (Fig. 4c).We next applied IM-MS-guided molecular dynamics (MD) simulations with a linear temperature gradient from 300 to 750 K to further elucidate dynamic conformational changes influenced by sialylation at Asn99 and Asn156 in the gas phase. MD results for both fetuin protein sialoforms exhibited four unfolding features with acceptable CCS errors, mirroring the experimental observations (Fig. 4d). The melting temperature (Tm) of the sialylated isoform (646.8 K) was higher than that of asialo fetuin (642.1 K), indicating that sialylation helps maintain protein conformation (Fig. 4e).
The starting structures for the gas-phase MD simulations were obtained from MD simulations in aqueous solution. Comparison of the representative MD structures of bFT and asialo in aqueous solution (Fig. S7†) indicates that the sialylation groups made contact with a loop on the surface of the protein (residues 255–298). This induced the formation of folded secondary structures within the contact surface. In contrast, this loop in asialo form lacked constraints and was unable to form any folded secondary structures. As the simulation temperature increases, the thermal motion of the amino acids intensifies, leading to weakened interactions between residues. Consequently, the hydrogen bonds connecting the backbones become unstable, resulting in the collapse of the secondary structure, which leads to a decrease in the proportion of remnant secondary structure. Analysis of α-helix and β-sheet content across unfolding intermediates revealed that sialic acids favor the proper folding and stability of secondary structures in bFT (Fig. 4f). Representative unfolding features during heating (Fig. 4g) suggest that sialylated glycans can envelop and compact the protein structure.
To further investigate the relevance of sialylation-induced stability in an aqueous environment, MD simulations were performed with explicit water. It was observed that glycans tend to adhere to protein surface in solution (Fig. S8†). Sialylated glycans exhibited a larger contact area with bFT (1873.7 ± 28.8 Å2) compared to asialylated glycans (1554.6 ± 26.4 Å2). Additionally, sialylated glycans formed an average of 14.4 ± 3.1 hydrogen bonds with bFT, while asialylated glycans formed 9.8 ± 3.0. This indicated that sialic acid facilitates interactions between glycans and the protein. These interactions might assist proteins in resisting unfolding during heating activation.
Elevated temperature weakened these interactions, decreasing hydrogen bond numbers by 9.8% and 32.5% for bFT and asialo-bFT, respectively (Fig. S8a†). Interestingly, the contact area between glycans and protein increased with temperature elevating, with a greater change observed for bFT compared to asialo-bFT (Fig. S8b†). Furthermore, consistent with gas-phase simulations, bFT exhibited a higher melting temperature Tm than asialo-bFT in aqueous solution (Fig. S9†). Taken together, sialylated glycans may mitigate thermal unfolding of bFT by effectively coating the protein. We suspect that terminal sialylation stabilizes fetuin conformation and restricts its dynamic structural fluctuations, likely through modulation of hydrogen bonding and contact area between glycans and the protein backbone, as well as maintenance of secondary structural elements.
Conclusions
In conclusion, this study demonstrates the power of integrating IM-MS with MD simulations to unravel the intricate relationship between protein sialylation and conformational stability. By employing a glycoform-resolved unfolding strategy, we revealed that increasing sialylation enhances the structural stability of fetuin proteins, particularly during early unfolding transitions. This stabilizing effect is likely mediated by increased electrostatic interactions and hydrogen bonding between sialic acid residues and the protein backbone. Interestingly, higher sialylation levels also appear to promote conformational flexibility in later unfolding stages, suggesting a complex interplay between glycosylation and protein dynamics.MD simulations, guided by IM-MS results, provided valuable insights into the molecular mechanisms underlying sialylation-induced stabilization. In the gas phase, sialylated glycans were found to envelop and compact the protein structure, favoring the proper folding and stability of secondary structural elements. Simulations in aqueous environments further corroborated these findings, revealing that sialic acids facilitate interactions between glycans and the protein surface, potentially mitigating structural perturbations induced by elevated temperatures.
The consistency between experimental and computational results highlights the potential of glycoform-resolved unfolding IM-MS as a rapid and cost-effective tool for predicting protein stability in solution. This integrated approach offers a promising avenue for elucidating the structure–function relationships of glycoproteins and may have broad implications for the rational design of glycoengineered therapeutics with enhanced stability and efficacy.
Fetuin exhibits pleiotropic functionality, impacting processes such as bone mineralization, insulin resistance, and immune modulation.37–39 Notably, fetuin can attenuate insulin signalling via binding to the ectodomain of the insulin receptor β-subunit, a process potentially regulated by sialylation. Preliminary experiments (Fig. S10†) revealed that sialylated fetuin significantly reduced glucose uptake compared to its asialo form in HepG2 cells. This suggests that sialylation may enhance fetuin's interaction with the insulin receptor, hindering GLUT4 transporter exocytosis and downstream glucose metabolism. While a detailed investigation of the biological ramifications and molecular underpinnings of fetuin sialylation falls outside the scope of this study, it presents a compelling avenue for future research.
Experimental
Materials
Bovine fetuin (FETB15-N-1, 95%) was obtained from Alpha Diagnostic Intl. Inc. Human fetuin (SRP3283, 95%) and neuraminidase from C. perfringens (sialidase, N2133) were obtained from Sigma-Aldrich (St. Louis, MO). Trypsin was purchased from Beijing Shengxia Proteins Scientific Ltd (Beijing, China).In vitro de-sialylation of bovine fetuin
Bovine fetuin (bFT) protein was de-sialylated with sialidase as previously described.5 Briefly, the incubation mixture with a total volume of 250 μL consisted of 100 μL bFT (1 mg mL−1), 25 μL bovine serum albumin (BSA, 0.5 mg mL−1), 25 μL sialidase (1 U mL−1), and 100 μL sodium acetate (100 mM, pH 5.0). The samples were incubated overnight at 37 °C while shaking.Sample preparation for native IM-MS
Proteins with sialylation were buffer-exchanged with 100 mM ammonium acetate (pH 2.0) two times using an Amicon Ultra-0.5 Centrifugal Filter (10 kDa, MWCO, Millipore) to protonate the negative charge of sialic acids. The samples were then dialyzed in 100 mM ammonium acetate (pH 7.0) overnight and buffer-exchanged with the same buffer three times before analysis. Proteins treated with sialidase were directly buffer-exchanged to 100 mM ammonium acetate (pH 7.0). All the samples were stored at 4 °C until analysis.Native IM-MS and AIU experiments
In the CIU workflows, data acquisition typically begins with a preliminary MS1 scan to observe the protein signals of interest, followed by quadrupole ion filtering to select the desired charge state. Similar to previous reports,5,28,29 for the AIU workflow, precursor ion selection is omitted and all ions in a wide mass range are subjected to collisional unfolding, in a nontargeted activation mode. This workflow critically relies on the integration of structural information from all observed charge states in one parameter, CCSacc. This accumulated data is then ready for further visualization and quantitative analysis. AIU fingerprints were obtained on a Synapt XS IM-MS instrument (Waters Corp, Manchester, UK). Briefly, all protein samples were diluted to 10 μM and loaded into homemade borosilicate needles. The capillary voltage was maintained at 1.0–1.5 kV through a platinum wire inserted into the spray needle solution, with the sampling cones set to 50. The source temperature was set to 70 °C. The IM T-wave ion guide was operated at 3 mbar with wave height and wave velocity values of 40 V and 550 m s−1, respectively. Mass spectra and drift time distributions were obtained for the ions at multiple trap collision energies in steps of 10 V from 10 V to 180 V. To facilitate the CCSacc data integration, it is necessary to convert the trap voltages into lab-frame collision energy (CE) values, with the same protocols as previously reported.40 To improve the resolution of glycoprotein detection, the post-IM fragmentation in the transfer cell was optimized from 10 V to 150 V. Detailed procedures for AIU-CCSacc were kept the same as those in our recent publications.27,29IM-MS-guided MD simulations
To elucidate dynamic conformational changes influenced by sialylation, we established a novel workflow of IM-MS-guided classic MD simulations with a linear temperature gradient from 300 to 750 K, modified from a recent report of IM-MS-guided MD simulation on peptide systems.41 In this study, IM-MS results provide two indispensable aspects for the MD simulations: (1) IM-MS experiments provide charge state and unfolding pathway information to guide MD simulations; (2) MD structures only with a certain CCS error range matching the IM-MS experimental results were used to perform further comparative analysis and reproducing the unfolding behavior. In other words, IM-MS experimental results have been used to guide the construction of the simulation models and the selection of the simulation method. Therefore, we constructed different glycosylation models based on the 3D structure of the proteins and experimentally observed charge state distributions. The linear temperature gradient method was employed to reproduce the protein unfolding process. More details on MD can be found in the ESI.†IM-MS data analysis
The TOF-MS was typically operated over the m/z range of 100–8000. CCS calibration curves were generated using a previously described protocol, and using literature CCS values with nitrogen (N2) derived for use with the Synapt instrument platform.42,43 Specifically, cytochrome C from horse heart, β-lactoglobulin, bovine serum albumin, and concanavalin A were selected as protein standards and prepared at 0.5 mg mL−1 for CCS calibration. To generate the CIU fingerprints, only the data at m/z values corresponding to the selected charge state of the precursor ions were selected for analysis. As previously reported,44 once the amount of the parent ion was less than five percent of the total signal, the CIU fingerprinting experiments were terminated. The data were normalized at each voltage through dividing the intensities of ions at each drift time by the maximum ion intensity observed at that voltage. CIUSuite2 (version 2.2) is a series of Python modules for the generation and manipulation of CIU fingerprints developed by the Ruotolo Research Group at the University of Michigan.45 CIU fingerprinting, subsequent CIU50 and RMSD calculations, and feature analysis were carried out using the most recent version of CIUSuite software.Data availability
The data supporting this article have been included as part of the ESI.†Author contributions
G. L. designed the project. Y. J. and Y. W. collected MS data. Y. L. and J. L. performed MD simulations. Y. J. drafted the manuscript. All authors were involved in the data analysis, wrote the paper and approved the final version of this manuscript.Conflicts of interest
There are no conflicts to declare.Acknowledgements
We acknowledge the instrumental assistance for glycoproteomic characterization from Dr Yuan Liu and Prof. Dr Lingjun Li (UW-Madison). This work is financially supported by the Fundamental Research Funds for the Central Universities (Nankai University, 63241610) and the National Natural Science Foundation of China (22104064, 22293030, 22293032, 22173020).Notes and references
- H. Černocká, L. Římánková and V. Ostatná, Fetuin and asialofetuin at charged surfaces: influence of sialic acid presence, J. Electroanal. Chem., 2021, 902, 115801 CrossRef.
- F. Ricken, A. D. Can, S. Gräber, M. Häusler and W. Jahnen-Dechent, Post-translational modifications glycosylation and phosphorylation of the major hepatic plasma protein fetuin-A are associated with CNS inflammation in children, PLoS One, 2022, 17(10), e0268592 CrossRef CAS PubMed.
- T. S. Raju, J. B. Briggs, S. M. Chamow, M. E. Winkler and A. J. Jones, Glycoengineering of therapeutic glycoproteins: in vitro galactosylation and sialylation of glycoproteins with terminal N-acetylglucosamine and galactose residues, Biochemistry, 2001, 40(30), 8868–8876 CrossRef CAS PubMed.
- Q. Zhou and H. Qiu, The Mechanistic Impact of N-Glycosylation on Stability, Pharmacokinetics, and Immunogenicity of Therapeutic Proteins, J. Pharm. Sci., 2019, 108(4), 1366–1377 CrossRef CAS PubMed.
- G. Li, A. Phetsanthad, M. Ma, Q. Yu, A. Nair, Z. Zheng, F. Ma, K. DeLaney, S. Hong and L. Li, Native Ion Mobility-Mass Spectrometry-Enabled Fast Structural Interrogation of Labile Protein Surface Modifications at the Intact Protein Level, Anal. Chem., 2022, 94(4), 2142–2153 CrossRef CAS PubMed.
- Y. Yao, G. Kim, S. Shafer, Z. Chen, S. Kubo, Y. Ji, J. Luo, W. Yang, S. P. Perner, C. Kanellopoulou, A. Y. Park, P. Jiang, J. Li, S. Baris, E. K. Aydiner, D. Ertem, D. J. Mulder, N. Warner, A. M. Griffiths, C. Topf-Olivestone, M. Kori, L. Werner, J. Ouahed, M. Field, C. Liu, B. Schwarz, C. M. Bosio, S. Ganesan, J. Song, H. Urlaub, T. Oellerich, S. A. Malaker, L. Zheng, C. R. Bertozzi, Y. Zhang, H. Matthews, W. Montgomery, H. Y. Shih, J. Jiang, M. Jones, A. Baras, A. Shuldiner, C. Gonzaga-Jauregui, S. B. Snapper, A. M. Muise, D. S. Shouval, A. Ozen, K. T. Pan, C. Wu and M. J. Lenardo, Mucus sialylation determines intestinal host-commensal homeostasis, Cell, 2022, 185(7), 1172–1188 CrossRef CAS PubMed.
- C. Dobie and D. Skropeta, Insights into the role of sialylation in cancer progression and metastasis, Br. J. Cancer, 2021, 124(1), 76–90 CrossRef CAS PubMed.
- R. Vattepu, S. L. Sneed and R. M. Anthony, Sialylation as an Important Regulator of Antibody Function, Front. Immunol., 2022, 13, 818736 CrossRef CAS PubMed.
- Y. Kaneko, F. Nimmerjahn and J. V. Ravetch, Anti-inflammatory activity of immunoglobulin G resulting from Fc sialylation, Science, 2006, 313(5787), 670–673 CrossRef CAS PubMed.
- T. S. Raju and S. E. Lang, Diversity in structure and functions of antibody sialylation in the Fc, Curr. Opin. Biotechnol., 2014, 30, 147–152 CrossRef CAS PubMed.
- S. Yang, L. Zhang, S. Thomas, Y. Hu, S. Li, J. Cipollo and H. Zhang, Modification of Sialic Acids on Solid Phase: Accurate Characterization of Protein Sialylation, Anal. Chem., 2017, 89(12), 6330–6335 CrossRef CAS PubMed.
- N. de Haan, S. Yang, J. Cipollo and M. Wuhrer, Glycomics studies using sialic acid derivatization and mass spectrometry, Nat. Rev. Chem., 2020, 4(5), 229–242 CrossRef CAS PubMed.
- T. Nishikaze, H. Tsumoto, S. Sekiya, S. Iwamoto, Y. Miura and K. Tanaka, Differentiation of Sialyl Linkage Isomers by One-Pot Sialic Acid Derivatization for Mass Spectrometry-Based Glycan Profiling, Anal. Chem., 2017, 89(4), 2353–2360 CrossRef CAS PubMed.
- D. J. Harvey, Fragmentation of negative ions from carbohydrates: part 1. Use of nitrate and other anionic adducts for the production of negative ion electrospray spectra from N-linked carbohydrates, J. Am. Soc. Mass Spectrom., 2005, 16(5), 622–630 CrossRef CAS PubMed.
- Y. Yang, F. Liu, V. Franc, L. A. Halim, H. Schellekens and A. J. Heck, Hybrid mass spectrometry approaches in glycoprotein analysis and their usage in scoring biosimilarity, Nat. Commun., 2016, 7, 13397 CrossRef CAS PubMed.
- G. Wang, R. N. de Jong, E. T. J. van den Bremer, P. W. H. I. Parren and A. J. R. Heck, Enhancing Accuracy in Molecular Weight Determination of Highly Heterogeneously Glycosylated Proteins by Native Tandem Mass Spectrometry, Anal. Chem., 2017, 89(9), 4793–4797 CrossRef CAS PubMed.
- K. Gupta, J. Li, I. Liko, J. Gault, C. Bechara, D. Wu, J. T. S. Hopper, K. Giles, J. L. P. Benesch and C. V. Robinson, Identifying key membrane protein lipid interactions using mass spectrometry, Nat. Protoc., 2018, 13(5), 1106–1120 CrossRef CAS PubMed.
- D. Wu and C. V. Robinson, Understanding glycoprotein structural heterogeneity and interactions: insights from native mass spectrometry, Curr. Opin. Struct. Biol., 2022, 74, 102351 CrossRef CAS PubMed.
- D. Wu and C. V. Robinson, Native Top-Down Mass Spectrometry Reveals a Role for Interfacial Glycans on Therapeutic Cytokine and Hormone Assemblies, Angew. Chem., Int. Ed. Engl., 2022, 61(49), e202213170 CrossRef CAS PubMed.
- T. J. El-Baba, C. A. Lutomski, S. A. Burnap, J. R. Bolla, L. A. Baker, A. J. Baldwin, W. B. Struwe and C. V. Robinson, Uncovering the Role of N-Glycan Occupancy on the Cooperative Assembly of Spike and Angiotensin Converting Enzyme 2 Complexes: Insights from Glycoengineering and Native Mass Spectrometry, J. Am. Chem. Soc., 2023, 145(14), 8021–8032 CrossRef CAS PubMed.
- S. Chen, D. Wu, C. V. Robinson and W. B. Struwe, Native Mass Spectrometry Meets Glycomics: Resolving Structural Detail and Occupancy of Glycans on Intact Glycoproteins, Anal. Chem., 2021, 93(30), 10435–10443 CrossRef CAS PubMed.
- H. Y. Yen, I. Liko, J. Gault, D. Wu, W. B. Struwe and C. V. Robinson, Correlating Glycoforms of DC-SIGN with Stability Using a Combination of Enzymatic Digestion and Ion Mobility Mass Spectrometry, Angew. Chem., Int. Ed. Engl., 2020, 59(36), 15560–15564 CrossRef CAS PubMed.
- V. Gabelica, A. A. Shvartsburg, C. Afonso, P. Barran, J. L. P. Benesch, C. Bleiholder, M. T. Bowers, A. Bilbao, M. F. Bush, J. L. Campbell, I. D. G. Campuzano, T. Causon, B. H. Clowers, C. S. Creaser, E. De Pauw, J. Far, F. Fernandez-Lima, J. C. Fjeldsted, K. Giles, M. Groessl, C. J. Hogan Jr, S. Hann, H. I. Kim, R. T. Kurulugama, J. C. May, J. A. McLean, K. Pagel, K. Richardson, M. E. Ridgeway, F. Rosu, F. Sobott, K. Thalassinos, S. J. Valentine and T. Wyttenbach, Recommendations for reporting ion mobility mass spectrometry measurements, Mass Spectrom. Rev., 2019, 38(3), 291–320 CrossRef CAS PubMed.
- Y. Tian, L. Han, A. C. Buckner and B. T. Ruotolo, Collision Induced Unfolding of Intact Antibodies: Rapid Characterization of Disulfide Bonding Patterns, Glycosylation, and Structures, Anal. Chem., 2015, 87(22), 11509–11515 CrossRef CAS PubMed.
- S. M. Dixit, D. A. Polasky and B. T. Ruotolo, Collision induced unfolding of isolated proteins in the gas phase: past, present, and future, Curr. Opin. Chem. Biol., 2018, 42, 93–100 CrossRef CAS PubMed.
- Y. Tian and B. Ruotolo, Collision Induced Unfolding Detects Subtle Differences in Intact Antibody Glycoforms and Associated Fragments, Int. J. Mass Spectrom., 2017, 425, 1–9 Search PubMed.
- A. Phetsanthad, G. Li, C. K. Jeon, B. T. Ruotolo and L. Li, Comparing Selected-Ion Collision Induced Unfolding with All Ion Unfolding Methods for Comprehensive Protein Conformational Characterization, J. Am. Soc. Mass Spectrom., 2022, 33(6), 944–951 CrossRef CAS PubMed.
- R. Zhao, N. Liu, Z. Zheng and G. Li, Enhanced Stability Differentiation of Therapeutic Polyclonal Antibodies with All Ion Unfolding-Ion Mobility-Mass Spectrometry, J. Am. Soc. Mass Spectrom., 2023, 34(10), 2289–2295 CrossRef CAS PubMed.
- Z. Zheng, M. Ma, Y. Jia, Y. Cui, R. Zhao, S. Li, C. Wenthur, L. Li and G. Li, Expedited Evaluation of Conformational Stability-Heterogeneity Associations for Crude Polyclonal Antibodies in Response to Conjugate Vaccines, Anal. Chem., 2023, 95(29), 10895–10902 CrossRef CAS PubMed.
- Y.-H. Lin, V. Franc and A. J. R. Heck, Similar Albeit Not the Same: In-Depth Analysis of Proteoforms of Human Serum, Bovine Serum, and Recombinant Human Fetuin, J. Proteome Res., 2018, 17(8), 2861–2869 CrossRef CAS PubMed.
- M. Windwarder and F. Altmann, Site-specific analysis of the O-glycosylation of bovine fetuin by electron-transfer dissociation mass spectrometry, J. Proteonomics, 2014, 108, 258–268 CrossRef CAS PubMed.
- J. Li, W. Lyu, G. Rossetti, A. Konijnenberg, A. Natalello, E. Ippoliti, M. Orozco, F. Sobott, R. Grandori and P. Carloni, Proton Dynamics in Protein Mass Spectrometry, J. Phys. Chem. Lett., 2017, 8(6), 1105–1112 CrossRef CAS PubMed.
- T. Meyer, V. Gabelica, H. Grubmüller and M. Orozco, Proteins in the gas phase, Wiley Interdiscip. Rev.: Comput. Mol. Sci., 2013, 3(4), 408–425 CAS.
- C. W. Damen, W. Chen, A. B. Chakraborty, M. van Oosterhout, J. R. Mazzeo, J. C. Gebler, J. H. Schellens, H. Rosing and J. H. Beijnen, Electrospray ionization quadrupole ion-mobility time-of-flight mass spectrometry as a tool to distinguish the lot-to-lot heterogeneity in N-glycosylation profile of the therapeutic monoclonal antibody trastuzumab, J. Am. Soc. Mass Spectrom., 2009, 20(11), 2021–2033 CrossRef CAS PubMed.
- J. Castro-Perez, T. P. Roddy, N. M. M. Nibbering, V. Shah, D. G. McLaren, S. Previs, A. B. Attygalle, K. Herath, Z. Chen, S.-P. Wang, L. Mitnaul, B. K. Hubbard, R. J. Vreeken, D. G. Johns and T. Hankemeier, Localization of Fatty Acyl and Double Bond Positions in Phosphatidylcholines Using a Dual Stage CID Fragmentation Coupled with Ion Mobility Mass Spectrometry, J. Am. Soc. Mass Spectrom., 2011, 22(9), 1552–1567 CrossRef CAS PubMed.
- A. Halim, U. Westerlind, C. Pett, M. Schorlemer, U. Ruetschi, G. Brinkmalm, C. Sihlbom, J. Lengqvist, G. Larson and J. Nilsson, Assignment of saccharide identities through analysis of oxonium ion fragmentation profiles in LC-MS/MS of glycopeptides, J. Proteome Res., 2014, 13(12), 6024–6032 CrossRef CAS PubMed.
- A. S. Goustin, N. Derar and A. B. Abou-Samra, Ahsg-fetuin blocks the metabolic arm of insulin action through its interaction with the 95-kD beta-subunit of the insulin receptor, Cell. Signal., 2013, 25(4), 981–988 CrossRef CAS PubMed.
- J. F. Trepanowski, J. Mey and K. A. Varady, Fetuin-A: a novel link between obesity and related complications, Int. J. Obes., 2015, 39(5), 734–741 CrossRef CAS PubMed.
- D. Leto and A. R. Saltiel, Regulation of glucose transport by insulin: traffic control of GLUT4, Nat. Rev. Mol. Cell Biol., 2012, 13(6), 383–396 CrossRef CAS PubMed.
- M. T. Donor, S. O. Shepherd and J. S. Prell, Rapid Determination of Activation Energies for Gas-Phase Protein Unfolding and Dissociation in a Q-IM-ToF Mass Spectrometer, J. Am. Soc. Mass Spectrom., 2020, 31(3), 602–610 CrossRef CAS PubMed.
- G. Li, C. K. Jeon, M. Ma, Y. Jia, Z. Zheng, D. G. Delafield, G. Lu, E. V. Romanova, J. V. Sweedler, B. T. Ruotolo and L. Li, Site-specific chirality-conferred structural compaction differentially mediates the cytotoxicity of Abeta42, Chem. Sci., 2023, 14(22), 5936–5944 RSC.
- M. F. Bush, Z. Hall, K. Giles, J. Hoyes, C. V. Robinson and B. T. Ruotolo, Collision Cross Sections of Proteins and Their Complexes: A Calibration Framework and Database for Gas-Phase Structural Biology, Anal. Chem., 2010, 82(22), 9557–9565 CrossRef CAS PubMed.
- K. Richardson, D. Langridge, S. M. Dixit and B. T. Ruotolo, An Improved Calibration Approach for Traveling Wave Ion Mobility Spectrometry: Robust, High-Precision Collision Cross Sections, Anal. Chem., 2021, 93(7), 3542–3550 CrossRef CAS PubMed.
- G. Li, K. DeLaney and L. Li, Molecular basis for chirality-regulated Abeta self-assembly and receptor recognition revealed by ion mobility-mass spectrometry, Nat. Commun., 2019, 10(1), 5038 CrossRef PubMed.
- J. D. Eschweiler, J. N. Rabuck-Gibbons, Y. Tian and B. T. Ruotolo, CIUSuite: A Quantitative Analysis Package for Collision Induced Unfolding Measurements of Gas-Phase Protein Ions, Anal. Chem., 2015, 87(22), 11516–11522 CrossRef CAS PubMed.
Footnotes |
| † Electronic supplementary information (ESI) available: Additional experimental details, Fig. S1 to S10, and Tables S1 to S4. See DOI: https://doi.org/10.1039/d4sc03672g |
| ‡ Y. J., Y. L. and Y. W. contributed equally to this work. |
| This journal is © The Royal Society of Chemistry 2024 |