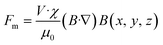Open Access Article
Open Access ArticlePortable microfluidic immunoassay platform for the detection of inflammatory protein biomarkers†
Gihoon
Choi
 a,
Betty B.
Mangadu
a,
Yooli K.
Light
b and
Robert J.
Meagher
a,
Betty B.
Mangadu
a,
Yooli K.
Light
b and
Robert J.
Meagher
 *b
*b
aSandia National Laboratories, Biotechnology & Bioengineering Dept., Livermore, CA, USA
bSandia National Laboratories, Systems Biology Dept., Livermore, CA, USA. E-mail: rmeaghe@sandia.gov
First published on 8th March 2024
Abstract
Cytokines and acute-phase proteins are promising biomarkers for inflammatory disease. Despite its potential, early diagnosis based on these biomarkers remains challenging without technology enabling highly sensitive protein detection immediately after sample collection, because of the low abundance and short half-life of these proteins in bodily fluids. Enzyme-linked immunosorbent assay (ELISA) is a gold-standard method for such protein analysis, but it often requires labor-intensive and time-consuming sample handling and as well as a bulky benchtop platereader, limiting its utility in the clinical site. We developed a portable microfluidic immunoassay device capable of sensitive, quantitative, and high-throughput protein detection at point-of-need. The portable microfluidic system performs eight magnetic bead-based sandwich immunoassays from raw samples in 40 min. An innovative bead actuation strategy was incorporated into the system to automate multiple sample handling steps with minimal user intervention. The device enables quantitative protein analysis with picomolar sensitivity, as demonstrated using human samples spiked with interleukin-6 and C-reactive protein. The affinity-based assays are highly specific to the target without cross-reactivity. Therefore, we envision the reported device offering ultrasensitive and field-deployable immunoassay tests for timely and accurate clinical diagnosis.
1 Introduction
Detection of specific protein biomarkers is an essential clinical procedure for the diagnosis of various diseases. For example, cytokines and acute-phase proteins are key inflammatory biomarkers for the diagnosis of numerous life-threatening diseases due to their dynamic nature in the immune response to infections (e.g., sepsis,1 AIDS,2 SARS-CoV-23,4) and other pathological conditions (e.g., cancer,5 Parkinson's disease,6 diabetes7). Abnormal levels of such protein biomarkers are strongly correlated with the onset and progression of infection, making them valuable tools for early detection and monitoring of diseases.1,3–7 More specifically, elevated concentrations of Interleukin-6 (IL-6, 1 ng mL−1) and C-reactive protein (CRP, 5000 ng mL−1) have been frequently reported in patients with severe inflammatory conditions, and can be associated with mortality in many clinical cases.1,8 Whereas, lower concentrations (5–25 pg mL−1 IL-6; 3000 ng mL−1 CRP) are typically present in healthy clinical specimens. The practical challenges for detection of cytokines are the low traceable change in concentration and short half-lives in bodily fluid such as serum (e.g., TNF-α: 18.2 min).9–11 The use of panels comprising multiple biomarkers showed improved specificity.1,10 Thus, the ability to detect multiple inflammatory biomarkers is of interest for diagnostic accuracy. Therefore, rapid, sensitive, quantitative, and multiplexed detection of such biomarkers is urgently needed for timely and precise diagnosis, enabling prompt administration of appropriate therapies.5Enzyme-linked immunosorbent assay (ELISA) is a gold standard technique for sensitive and quantitative detection of proteins, in some cases enabling detection at femtomolar concentrations.12 However, complicated sample handling, long turnaround time (∼3 h), and bulky benchtop plate washers and scanners limit its use at the point-of-need.13 Thus, clinical samples are typically transported to the nearest central laboratory for immunoassay tests. Such sample transportation can pose a considerable delay and increased risk of sample degradation, compromising test accuracy.9 Lateral flow strips (or paper-based ELISA) are commonly used for rapid, low-cost, and portable immunoassay tests. However, their sensitivity highly depends on the types of the sample matrix, and their qualitative readout is inadequate for biomarkers that require quantitative analysis for a correct decision. To improve the sensitivity, signal amplification methods were coupled with conventional ELISA such as nanozymes,14 plasmonic labels,15 and nucleic acid amplifications.16–18 Despite the promising potential, nanozyme face limitations related to substrate selectivity and catalytic efficiency.19 Additionally, the reproducibility of plasmonic labels is compromised due to their susceptibility to variations in solution conditions, such as pH, temperature, and ionic strength.19 The application of nucleic acid amplifications introduces challenges such as increased assay complexity and extended assay time, particularly during the amplification step. These considerations must be carefully addressed to be implemented in the point-of-care setting. Alternatively, the microfluidic approach is well suited for on-site clinical diagnosis because of its inherent attributes of portability, low sample/reagent volumes, and automated processing.20 To this end, extensive research efforts have been made over the past decade to translate laboratory-quality immunoassays into a microfluidic format.21–24
Microfluidic immunoassays frequently involve use of solid-phase particles, so-called bead-based ELISA, because a high surface-to-volume ratio promotes antigen–antibody binding. The use of beads significantly reduces the assay time while improving the sensitivity, making bead-based assays ideal for sequential immunoassay procedures (i.e., streamlined incubation, washing, and reaction).13,25 However, implementing bead-based immunoassays in microfluidics requires strategic actuation of beads and/or liquid droplets in a controllable manner,26 which can be challenging and complicated.
Numerous microfluidic immunoassay platforms have been reported in the past, and they can be categorized by the bead actuation methods such as flow-based microfluidic, digital ELISA (e.g., droplet and microwell array), centrifugal, and miniaturized robotic platforms (e.g., electrowetting and electromagnetic arrays). A flow-based microfluidic immunoassay platform in which reagent inflows are sequentially merged with bead-containing droplets via T-junction, and a magnet is used to collect the beads from droplets for optical measurement.21 Despite its automated capability, tubing and infusion pumps are not ideal for portability.
A droplet-based digital ELISA platform performed compartmentalized bead-based immunoassays in millions of carrier droplets and successfully achieved single-molecule sensitivity.27 However, the scalability of the test throughput is constrained by the limited optics and color-coded bead options. Besides, prolonged incubation time (3 h) is necessary to form immunocomplexes, possibly due to the absence of a mixing unit. A standalone digital ELISA platform28 using a microwell array-embedded disc dispenses the bead-containing solution by applying vacuum pressure at outlets and settling the beads into femtoliter microwells by gravity. The instrument has a built-in shaker mechanism for thorough reagent mixing, a robotic pipette for reagent/sample transferring, and twenty-four individual test arrays on a single microfluidic disc for high-throughput measurement within a reasonable assay time (1 h). However, the bulky floor-standing instrument with a large footprint is not compatible with portability.
Centrifugal microfluidic platforms have been used to drive the beads for a streamlined immunoassay process without peripheral devices.22,29–31 However, centrifugal force can only direct the beads in one direction. Therefore, a complicated sample sequence that requires multi-directional bead operation (e.g., agitation) is challenging. Moreover, limited bead operation in the microfluidic chip can increase reliance on off-chip steps (e.g., off-chip incubation).22,31
Electrowetting-based digital microfluidics (DMF) enables flexible discrete droplet actuation (move, split, and merge) on a hydrophobic surface.23,32,33 This method holds great promise for adapting magnetic bead-based immunoassay. However, active magnetic bead actuation for resuspension and agitation is challenging to implement in a droplet, especially with a stationary magnetic trap. In addition, the adsorption of proteins and detergents causes biofouling or cross-contamination, affecting surface wettability and creating challenges for reproducibility with complex sample matrices.
As opposed to controlling droplets against stationary beads, active magnetic actuation on beads can alleviate such problems. For example, Chiou et al. developed a system that electromagnetically actuates magnetic beads in stationary droplets.34 However, the system is only compatible with certain types of beads (e.g., large size, high susceptibility) because small electromagnets cannot provide sufficient magnetic fields to induce strong magnetic forces on small beads. The necessity to use large beads is at odds with the benefit of a high surface-to-volume ratio to maximize sensitivity. Despite numerous approaches reported in scientific literature, integrating reliable, robust, and automated immunoassays is still a common hurdle for point-of-care microfluidic devices.
We present a portable microfluidic immunoassay device with autonomous sample preparation capability for sensitive protein detection, intended for field-deployable use. The system consists of a mobile analyzer and a compact microfluidic chip. The platform integrates electromechanical and optical subsystems, and the microfluidic chip contains eight separate testing units spaced for 8-channel pipette compatibility. An embedded programmable planar electromagnetic microactuator array actuates a permanent magnet, which effectively amplifies the weak magnetic field from the microactuator array. Thus, magnetic beads can be reliably manipulated against the stationary droplets by strong magnetic interaction, enabling an autonomous sample process. The system facilitates a complete immunoassay process (incubation, washing, and chemiluminescent signal amplification) with a 40 min total processing time for eight raw samples. We incorporated a one-pot incubation assay in a microfluidic chip to reduce sample steps and assay time. The device achieved a sub-picomolar detection sensitivity (IL-6: 10 pg mL−1; and CRP: 100 pg mL−1) spiked into model sample matrices such as human serum and fetal bovine serum. This level of sensitivity is sufficient to identify abnormal physiological disorders at an early stage.35–38 The ability to quantify inflammation response markers should facilitate monitoring of the immune status of patients during disease progression to inform adjustment of therapies. We envision that this microfluidic immunoassay device will serve as a versatile tool for timely and accurate diagnosis in clinical settings.
2 Materials and methods
2.1 Instrumentation
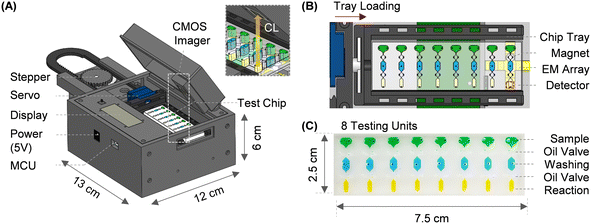
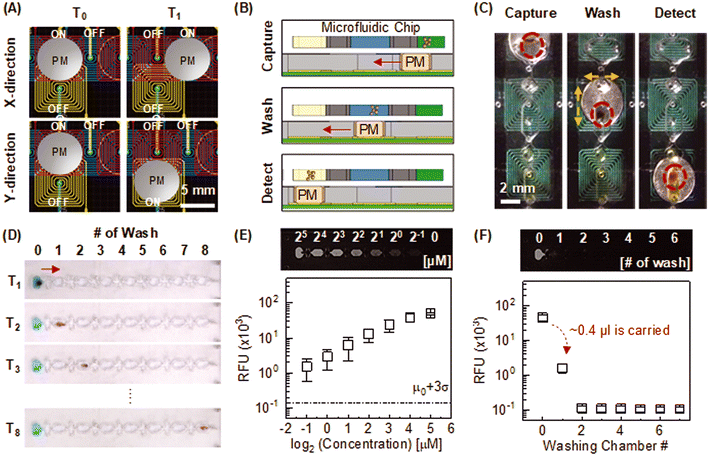
Fig. 1A shows a system overview of the microfluidic immunoassay system. The platform enclosure was designed in SolidWorks, 3D printed, and assembled in the lab. The fully integrated system has a footprint of 11 × 12 × 8 cm and contains optical and electromechanical subsystems (Fig. S1†). Optical subsystem: a low-noise complementary metal oxide semiconductor (CMOS) imager (ULS24, Anitoa Systems) is incorporated into the system to measure the chemiluminescent signal. The image sensor surface is directly in contact with the microfluidic channel to maximize the collection of blue emission light (peak wavelength: 425 nm). In this optical configuration, the sensing area (3.6 × 3.6 mm) sufficiently covers the detection zone of the individual testing unit without an optical lens or filter. We attach mylar reflective film on top of the permanent magnet underneath the detection zone during the chemiluminescent measurement to further increase the light collection. Electromechanical subsystems: integrated electromechanical subsystems include linear, rotary, and electromagnetic actuators. For a linear actuator, a rack-and-pinion mechanism converts the rotary motion of the stepper motor into a linear motion using in-house fabricated rack/pinion gears. A unipolar stepper motor enables the precision positioning of the microfluidic chip. On the right edge of the rack, a high-torque servomotor is installed as a rotary joint to rotate the microfluidic chip (Fig. 1B). A custom-printed planar electromagnetic microactuator array is used to actuate a neodymium magnet (grade: N52) that further manipulates the magnetic beads in a microfluidic channel in a programmed manner. Each rectangular-shaped planar coil has a copper trace with a width of 170 μm, a thickness of 36 μm, and a pitch of 170 μm. Adjacent planar coils are partially overlapped in different printed circuit board (PCB) layers with a 4.5 mm gap. It is noteworthy that magnetic force follows an inverse square relationship with distance.39 Therefore, a neodymium magnet close to the coil center can be actuated by a small electromagnetic field with low power consumption (<2.5 W). To transport a neodymium magnet from one location to another, a localized electromagnetic field is generated from the coil one at a time by sequentially switching the transistor. A customized PCB with an embedded microcontroller unit (MCU) operates all integrated electronic components. The entire system is powered by a USB connection or a DC power adapter (5 V, 500 mA).2.2 Microfluidic chip
A microfluidic chip is constructed by casting polydimethylsiloxane (PDMS) into a 3D-printed reusable master mold (Fig. S1†). A negative mold is printed with a commercial resin-based stereolithography 3D printer (Form 3, Formlabs). After printing, the mold is sonicated with isopropyl alcohol, and then post-cured in a UV box at 60 °C for 30 min. The mold surface is silane-modified (1H,1H,2H,2H-perfluorodecyltrichlorosilane, CAS 102488-49-3, Sigma-Aldrich) following oxygen plasma treatment to prevent PDMS from sticking to the mold surface and uncured resin residues from interfering with PDMS curing.40,41 To fabricate the PDMS device, curing and elastomer agents are thoroughly mixed in 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 10 (m/m), degassed in a vacuum desiccator, then cured at 65 °C for 2 hours. Reagent-loading holes are punched on a PDMS stamp and irreversibly bonded to a glass slide immediately after oxygen plasma treatment (RF power at 20 W for 1.5 min). We baked the device at 80–90 °C for an hour to enhance strong covalent bonding between PDMS and glass substrate.
10 (m/m), degassed in a vacuum desiccator, then cured at 65 °C for 2 hours. Reagent-loading holes are punched on a PDMS stamp and irreversibly bonded to a glass slide immediately after oxygen plasma treatment (RF power at 20 W for 1.5 min). We baked the device at 80–90 °C for an hour to enhance strong covalent bonding between PDMS and glass substrate.
The compact reagent microfluidic chip (25 × 70 mm) has eight spatially isolated testing units spaced for standard 8-channel pipette compatibility (Fig. 1C). Each testing unit is comprised of sample, washing, and reaction chambers. These three chambers hold 15 μl of sample diluent with 25 μg of functionalized magnetic beads, 15 μl of washing buffer (PBS with 0.5% TWEEN-20), and 8 μl of chemiluminescent substrate (SuperSignal™ ELISA Femto Substrate, 37075, ThermoFisher Scientific). In addition, two oil valves are filled with 10 μl of mineral oil with 0.1% ABIL EM-90 (Evonik) to separate the chambers. All reagents required for magnetic bead-based sandwich immunoassay and mineral oils for valving are pre-loaded into the chip using an 8-channel pipette before the test (Fig. S2B† for detailed loading instructions).
2.3 Workflow
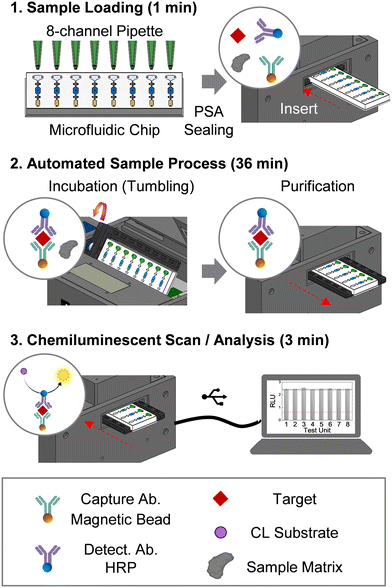
The testing workflow comprises sample loading, automated sample preparation, and chemiluminescent (CL) measurement/analysis (Fig. 2). Briefly, 15 μL of human serum or other test sample is loaded into each sample chamber using an 8-channel pipette, then sealed with clear pressure-sensitive adhesive (PSA). Once a prepared microfluidic chip is inserted, the platform facilitates the automated sample process (i.e., incubation and washing) (Fig. 2). During the incubation, the microfluidic chip tumbles to keep the beads in motion for efficient binding. The incubation was performed at room temperature. To maintain uniformity in incubation time among the eight tests, magnetic beads from each testing unit are moved into the washing chambers as soon as the incubation step is completed. Bead washing is achieved by using the embedded programmable planar microactuator to move the beads in a back-and-forth motion in the washing chamber. After washing, the CL signal from each testing unit is measured individually using the actuator to move the beads into the detection chamber, and the resulting chemiluminescent signal is analyzed using a custom-developed program.2.4 Magnetic bead-based protein detection assays
The integrated magnetic bead-based sandwich immunoassay has three components: capture beads, detector antibody, and target protein samples. Capture bead preparation: 30 μg of target-specific polyclonal antibody [IL-6 antibody: P620 (Invitrogen); CRP antibody: AF1707 (R&D Systems)] is covalently coupled to 1 mg of epoxy-activated superparamagnetic bead surface using a commercially available antibody-coupling kit (14311D, Invitrogen). After overnight incubation at room temperature, antibody-coupled beads are thoroughly washed three times with purification buffer to remove the unbound antibody excess and then blocked with SuperBlock buffer (37518, Thermo Scientific) to prevent non-specific binding. The final stock concentration of the capture bead is set to 10 mg mL−1. Horseradish peroxidase (HRP)-labeled detector preparation: monoclonal antibody [IL-6 antibody: P620 (Invitrogen); CRP antibody: MAB17072 (R&D Systems)] is labeled using an HRP conjugation kit (ab102890, Abcam). The final concentration of the HRP-labeled detector is adjusted to 1 μg mL−1 with phosphate buffer saline (PBS) with 0.05% Tween-20. Target protein sample preparation: target protein (IL-6 or CRP) is spiked into PBS, human serum, or fetal bovine serum (FBS, Gibco) to validate the assay in different sample matrix contexts. Pooled human serum from anonymous donors was purchased from Innovative Research (Novi, MI, USA). The use of de-identified pooled specimens was reviewed by the Institutional Review Board of Sandia National Laboratories and determined to be exempt according to United States federal regulations as outlined in the code of federal regulations 10 CFR 745.101(b).2.5 The principle behind the permanent magnet actuation using planar micro-coil array
We implemented a multilayer planar electromagnetic microactuator as an energy-efficient method to facilitate long-range transport and manipulation of a permanent magnet for actuating magnetic beads. Control of the permanent neodymium magnet using a planar spiral electromagnetic coil was previously reported.42,43 A rectangular spiral electromagnetic coil generates the highest magnetic field strength and gradient at the innermost turn of the coil with applying current, thus inducing magnetic interaction with a permanent magnet. The magnetic force exerted on the neodymium magnet can be described in equations.39,43
Using a planar coil array to drive a permanent magnet has several benefits. The permanent magnet itself is incapable of programmed control, while the planar electromagnet itself is too weak to directly actuate magnetic beads in channels. The partially-overlapped double-layer planar coil concept has been previously reported to manipulate magnetic beads in a microfluidic device. However, the weak electromagnetic field generated from the planar coil is insufficient to directly actuate magnetic beads without topographical assistance (i.e., it works only with specific fluidic channel geometry).34 Besides, relying on the coils alone works only for bead products with high magnetic susceptibility, and the magnetic properties are material and size dependent. The permanent magnet is more suitable for bead actuation due to its relatively high magnetic susceptibility. The combined permanent magnet and planar electromagnet approach mitigates such limitations and allows programmable bead manipulation in a microfluidic chip. A similar mechanism was recently integrated for portable nucleic acid testing device,44 but only 1D bead motion was available. Multilayer planar electromagnetic microactuator enables 2D bead motion (X-, Y-, and diagonal), enabling a greater variety of programmable bead motions.
2.6 Data analysis
Raw images from the CMOS image sensor are captured from eight testing units to obtain the relative luminescence unit (RLU). To achieve optimal detection sensitivity, the exposure time of the image sensor setting is tuned for two different protein detection assays. Microfluidic chips are typically configured to run seven experimental tests and one negative control (i.e., CL substrate + plain magnetic beads). Using a custom-developed MATLAB program, 12 × 12 pixel values are extracted, averaged, and then subtracted from the RLU obtained from the negative control. The detection limit is defined as the lowest detectable analyte concentration with a 3× standard deviation (σ) higher than the mean signal background (μ0).3 Results and discussion
3.1 Validation of imaging-based chemiluminescence sensing for immunoassay tests
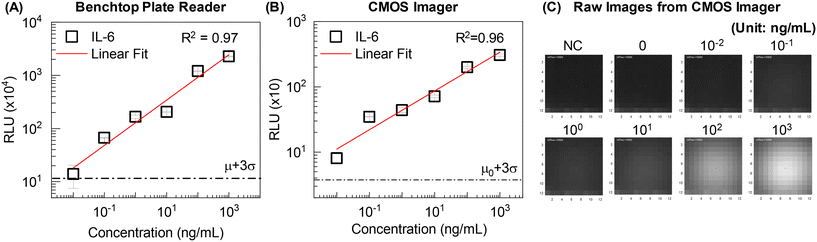
For comprehensive protein diagnostics, sensitive and quantitative detection is a prerequisite. To evaluate the system-integrated chemiluminescence sensing, we benchmarked with the same assays processed manually and read using a benchtop commercial plate reader (Enspire). The magnetic bead-based sandwich immunoassay was performed in a tube with 10-fold serially diluted IL-6 as a model target. The plate reader and integrated CMOS imager were each used to measure CL signals from matched samples (same concentration and preparation steps). The standard curves showed linear trends, with a 10 pg mL−1 detection limit, for the benchtop plate reader and CMOS image sensor (Fig. 3). The small standard deviation indicates excellent reproducibility and limit of quantification (LOQ). These results confirmed that the integrated imaging-based sensing mode has quantitative ability and sensitivity comparable to the benchtop instrument for the IL-6 detection assay (Fig. 3A and B). Fig. 3C shows raw images obtained from CMOS sensors corresponding to each IL-6 concentration. The exported RLU values from Fig. 3B showed good agreement with gray-scale images (Fig. 3C). The lens-free configuration of the CMOS image sensor minimized the optical path loss (close contact with the sensing window), enabling detection sensitivity comparable to the benchtop instrument where a photomultiplier tube (PMT) is usually embedded. While a PMT has superior gain (106) and signal-to-noise ratio (∼50 dB),45 the larger footprint, high voltage requirements, and fragility are less attractive for portability. The low-noise, low-power, and small-sized CMOS image sensor showed detection capability similar to a PMT with IL-6 detection assay. This implies immunoassay sensitivity in this case is limited by the affinity reagents, rather than by the choice of sensor. We confirmed from the validation that integrated image-based chemiluminescence sensing is amenable to achieving highly sensitive protein analysis.3.2 Automated sample preparation
One of the major challenges for field deployment of immunoassay devices is the complexity of sample handling steps. For rapid diagnostic tests (RDTs), quick assay time (≤15 min) with simpler sample preparation is often achieved at the expense of sensitivity. Conversely, highly sensitive sandwich ELISA requires several time-intensive incubations and laborious wash steps. This is performed manually by laboratory personnel or automatically using benchtop sample preparation instruments in the clinical laboratory, but this is often not practical in a low-resource setting. For field deployment of ELISA-like immunoassay tests, a portable device with automated sample preparation capability is highly desired.For automated sample preparation, we embedded a programmable planar electromagnetic actuator into the microfluidic immunoassay device (Fig. 4A). Magnetic beads were actuated against the stationary reagent droplets in the fluidic channel/chamber by controlled neodymium magnet motion on electromagnet micro-actuators (Fig. 4B). The integrated sample preparation consists of incubation, washing, and CL reaction steps (Fig. 4B and C). During the incubation step, functionalized beads specifically capture the complex of the target protein and HRP-conjugated detection antibody in the sample chamber (Fig. 2– step 2: left circle panel). After the incubation, the neodymium magnet drags the target-captured magnetic beads to the washing chamber. For washing (Fig. 4B and C), magnetic beads were agitated in the X and Y directions to eliminate potential reagent carryovers (i.e., sample matrix, unbound analyte and HRP-labeled detector antibodies) from the sample chamber (Fig. 2 – step 2: right circle panel). Thoroughly washed beads were then transferred to the reaction chamber for CL measurement (Fig. 2 – step 3: circle panel and 4B and C).
To investigate reagent carryover during the magnetic bead transfer, we performed a sequential washing test in the microfluidic chip. Eight washing chambers are connected in series but separated by the oil valves (Fig. 4D). A mixture of fluorescence dye and magnetic beads was loaded in the first chamber. Since carryover volume is linearly correlated with the amount of magnetic beads for the water–oil surface tension valve,46 the number of beads (25 μg) was set to be identical to the microfluidic immunoassay. The sequential washing was automatically facilitated as magnetic beads transferred from chamber 1 through 8 in series. After the washing, endpoint fluorescence intensity was measured at each washing chamber. We noticed a substantial increase in the signal background of the first washing chamber, implying a carryover of about 0.4 μL from the preceding chamber (Fig. 4E and F). However, the baseline was unchanged from the second washing chamber, indicating that a single washing step was sufficient for removal of reagent carryover.
One of the main challenges for magnet-involved bead actuation lies in the resuspension of clumped beads into a bulk solution after driving beads. This can lead to unreliable washing since partially clumped beads are only locally exposed to the bulk washing buffer along the bead actuation path (see ESI† text and Fig. S2 for detailed 1D and 2D washing comparison). In cases where bead actuation is limited to unidirectional or bidirectional motion, it is suggested to adopt a narrow and elongated chamber shape along the bead actuation path to maximize the effective washing volume. However, such geometric requirements can impose constraints on the flexibility of microfluidic chamber (or channel) design. The key benefit of the EM-based actuation lies in its ability to induce multi-directional bead motion, enabling beads to sample a larger volume of wash buffer within a compact space. Besides, it is more scalable than linear actuators and servo motors. Therefore, EM-based actuation offer better adaptability for translating more complicated magnetic bead-based bioassays, requiring multiple unit operations, such as a series of incubation and washing, into the microfluidic format.
3.3 Uniformity of eight testing units
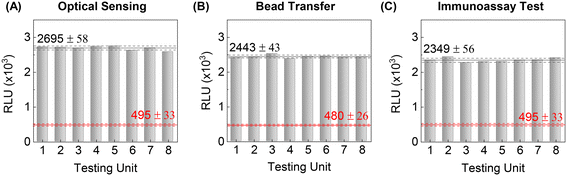
Unit-to-unit consistency among eight tests is an essential prerequisite for quantitative comparison. The test consistency for the microfluidic immunoassay platform relies on uniform optical sensing and reproducible sample preparation. We first evaluated the CL sensing uniformity among eight testing units by directly introducing HRP-labelled magnetic bead aliquots into each reaction chamber without performing any microfluidic sample preparation steps. A permanent magnet was placed underneath the sensing zone to form a magnetic bead clump. Each testing unit was scanned individually for the CL measurement, using the rack-and-pinion mechanism to translate the chip past the detector. As expected, the quantitative results among eight testing units showed excellent optical uniformity with small variation (Fig. 5A, black-dotted lines), implying good optical alignment between the CMOS sensor and the final position of the bead cluster in the reaction chamber.Next, we evaluated the optical uniformity while incorporating the automated sample washing sequence. HRP-labelled magnetic bead aliquots were introduced in each sample chamber and transferred toward the reaction chamber through oil valves. As shown in Fig. 5B, quantitative uniformity is still valid with an automated sample process, indicating consistent and reliable magnetic bead transfers from the sample to the reaction chamber with negligible bead loss at the water–oil interface. To minimize bead loss between chambers, we designed a narrow funnel-shaped channel between chambers and valves to assist in forming the magnetic bead cluster and maximizing the magnetic force acting on the beads for efficient capture (Fig. S2†). Note that the mass of the magnetic bead cluster is proportional to the magnetic force.47 This magnetic pulling force on the bead cluster generates the small break-away bead droplet as opposed to the capillary force (i.e., bead holding force) at the water–oil interface. Adding 0.1% ABIL EM-90 surfactant in mineral oil lowers the interfacial tension at the aqueous-to-oil interface, to facilitate splitting of the bead droplet from the mother droplet; thus, magnetic beads can reliably transport from chamber to chamber without significant loss.
Lastly, we performed a microfluidic IL-6 detection assay to see if the uniformity among testing units is maintained while performing the entire immunoassay process, including incubation and washing. An identical concentration of purified IL-6 aliquot was loaded into each of the eight testing units, and then processed by the platform following the programmed sequence. The eight test results showed excellent consistency with minimal variation (Fig. 5C, black-dotted lines). This demonstrates a robust reagent mixing and washing throughout the microfluidic immunoassay process. While manual sandwich immunoassay on microplates (e.g., ELISA) is prone to human errors due to iterative liquid exchanging steps, the integrated autonomous sample process mitigates such errors and allows reliable eight tests on the mobile immunoassay platform.
3.4 Detection of protein biomarkers: sensitivity and specificity
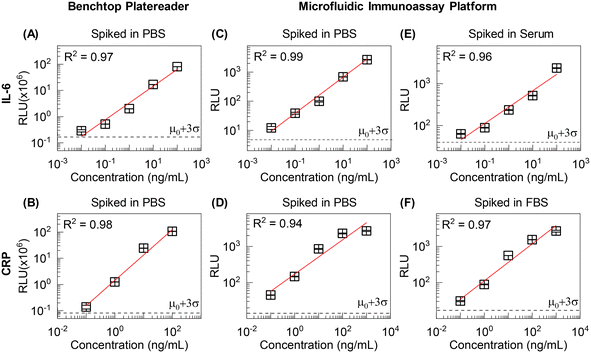
Highly sensitive detection of specific protein biomarkers from raw clinical specimens is an essential requirement for accurate clinical measurements. For laboratory-based ELISA procedures, femtomolar detection sensitivity can be achieved with sequential sample steps where multiple incubation and washing are necessary for analyte capture, blocking, and detector binding. Sequential sample steps complicate the entire test workflow, and mass transfer limitations in microtiter plate wells significantly increase assay time (up to 3 h), making standard ELISA impractical for POC diagnostics. To this end, we considered a one-pot incubation where capture, analyte, and detector reagents are all assembled simultaneously in the sample chamber.To evaluate the detection capability of the device, we performed multiple sensitivity tests with model targets IL-6 and CRP using a benchtop platereader and microfluidic device. We designed parallel experiments to validate i) a one-pot incubation assay, ii) the assay compatibility with a microfluidic device, and iii) the sample matrix effect. Various known concentrations of protein targets (IL-6: 0.01–100 ng mL−1; CRP: 0.1–1000 ng mL−1) were 10-fold serially diluted with PBS and human serum (for IL-6) or FBS (for CRP) to represent sample matrices of differing complexity. The reason to use FBS for CRP sensitivity validation is because the concentration of CRP in normal human serum is well above the detection limit. Although FBS is not a perfect model for human serum, it models high abundance of proteins that could interfere nonspecifically with immunoassay. For all experiments, the measurement was repeated three times for each sample at different concentrations to ensure the reproducibility of the test. Fig. 6 shows the quantitative results from IL-6 and CRP standards at various conditions (e.g., manual microtiter assay versus automated microfluidic assay and simple versus complex sample matrices). A good linearity (R2 = 0.94–0.99) with a wide quantifiable range (≥5 orders of magnitude) demonstrates the quantitative ability of microfluidic devices with one-pot incubation (Fig. 6C and D). This dynamic range is comparable with the results obtained from the commercial platereader (Fig. 6A and B). In Fig. 6B, the signal saturation observed in the highest concentration sample (103 ng mL−1) was primarily attributed to the limited dynamic range of the benchtop instrument readout. Thus, linear regression was performed using unsaturated sample data. The device showed high analytical sensitivity (IL-6: 10 pg mL−1; CRP: 100 pg mL−1) in all sample matrices tested (Fig. 6). This level of sensitivity is comparable to a benchtop plate reader12 (Fig. 6A–D) and sufficient to detect low levels of inflammatory biomarkers in bodily fluids (pM range).9–11,35–38 We did not observe any indication of non-specific binding, such as elevated signal background or considerable variation. For optimal detection sensitivity, we set the exposure time of the CMOS imager to 3 s and 10 s for CRP and IL-6 detection assays, respectively. The extended exposure time benefits from increasing the overall chemiluminescent signal. However, noise remains a limiting factor for achieving higher analytical sensitivity. The sensitivity was not significantly improved with exposure time higher than 3 s for the CRP detection assay and 10 s for IL-6 detection assay. Note binding kinetics and affinity for the capture antibody, target analyte, and detector antibody are also limiting factor for detection sensitivity, in addition to instrumental characteristics. Therefore, optimal sensor settings should be determined for different detection assays.
We further investigated whether integrated sample washing enhanced device performance in the presence of the sample matrix. Without the on-chip sample washing, an order of magnitude higher detection limit (i.e., 480 pg mL−1 IL-6) and decreased signal-to-background ratios were observed for PBS and serum spiked samples, indicating the clear presence of the sample matrix effect (Fig. S3†). Although a high background may imply non-specific binding due to ionic or hydrophobic interaction with serum proteins,48 we believe the high background originated from the small amount of unbound HRP-labeled antibodies carried over to the reaction chamber along with bead pellet since the increased background was observed even without the serum matrices (Fig. S3A†). For viscous serum-containing samples, such unintended carry-over can be more significant than the sample spiked in PBS (Fig. S3B†). However, the incorporated washing step mitigates such sample matrix hindrance and enables sensitive protein detection (Fig. 6C and E).
Lastly, to demonstrate the specificity of the device, we performed a cross-reactivity test using IL-6 and CRP samples (Fig. S4†). To ensure the constant signal background, a microfluidic device was prepared to perform triplicates of positive and negative samples and two internal negative controls (i.e., plain beads). As expected, the qualitative results identify the corresponding target protein biomarkers without cross-reactivity (Fig. S4†). Moreover, no false positive results with human sera containing various antibody kinds further confirm the excellent specificity (Fig. 6E and F). Although we only tested two types of protein biomarkers for sensitivity and specificity validation, the eight testing units can be configured differently for high-throughput detection of numerous protein biomarkers.
4 Conclusion
We developed a microfluidic immunoassay device for sensitive and high-throughput detection of proteins at POC. The system consists of a mobile analyzer and ready-to-use microfluidic devices. Within 40 min, the system automatically prepares eight samples for CL measurement from raw human serum. The excellent unit-to-unit consistency indicated robust eight-plexed sample preparation with minimal bead loss and high optical uniformity. Additionally, the linear standard curve showed the quantitative ability of the platform. The device achieved sub-picomolar detection sensitivity with IL-6 and CRP-spiked human serum and FBS, respectively. Finally, we confirmed that integrated sample washing reduced the non-specific signal originating from the sample matrix effect, thus improving the detection sensitivity. Although we only tested the IL-6 and CPR detection assays for device validation, it can be adapted to sensitively and specifically detect various proteins by conjugating capture antibodies for the target of interest on magnetic beads. The eight-sample device has potential usage for versatile protein analysis, such as antibody screening and affinity assay development. In addition, the reported device will enable highly sensitive immunoassays for accurate and timely diagnosis at point-of-need.Author contributions
G. C. was responsible for the design and integration of the platform. B. B. M., and Y. K. L. contributed to the development and optimization of the bead-based sandwich immunoassay. G. C. and B. B. M. performed all the experiments and carried out data analysis. R. J. M. conceived the study, designed experiment, supervised work. All authors contributed to writing, reviewing, and editing the manuscript.Conflicts of interest
G Choi and RJ Meagher are co-inventors on patents or patent applications related to the technology described in this article. All authors declare that they have no affiliations with or involvement in any organization or entity with any financial interests in the subject matter or materials discussed in this manuscript.Acknowledgements
The reported work is funded by Sandia National Laboratories Laboratory Directed Research and Development (LDRD) Program. This article is authored by an employee of National Technology & Engineering Solutions of Sandia, LLC under Contract No. DE-NA0003525 with the U.S. Department of Energy (DOE). The employee owns all right, title and interest in and to the article and is solely responsible for its contents. The United States Government retains and the publisher, by accepting the article for publication, acknowledges that the United States Government retains a non-exclusive, paid-up, irrevocable, world-wide license to publish or reproduce the published form of this article or allow others to do so, for United States Government purposes. The DOE will provide public access to these results of federally sponsored research in accordance with the DOE Public Access Plan https://www.energy.gov/downloads/doe-public-access-plan.References
- H. Dollner, L. Vatten and R. Austgulen, J. Clin. Epidemiol., 2001, 54, 1251–1257 CrossRef CAS PubMed.
- A. H. Borges, J. L. O'Connor, A. N. Phillips, F. F. Ronsholt, S. Pett, M. J. Vjecha, M. A. French, J. D. Lundgren, S. Insight, E. S. Groups and the S. S. C., J. Infect. Dis., 2015, 212, 585–595 CrossRef CAS PubMed.
- F. Liu, L. Li, M. Xu, J. Wu, D. Luo, Y. Zhu, B. Li, X. Song and X. Zhou, J. Clin. Virol., 2020, 127, 104370 CrossRef CAS PubMed.
- J. J. Guirao, C. M. Cabrera, N. Jimenez, L. Rincon and J. M. Urra, Mol. Immunol., 2020, 128, 64–68 CrossRef CAS PubMed.
- J. F. Rusling, C. V. Kumar, J. S. Gutkind and V. Patel, Analyst, 2010, 135, 2496–2511 RSC.
- M. Dufek, I. Rektorova, V. Thon, J. Lokaj and I. Rektor, J. Parkinson's Dis., 2015, 2015, 898192 Search PubMed.
- A. D. Pradhan, J. E. Manson, N. Rifai, J. E. Buring and P. M. Ridker, JAMA, 2001, 286, 327–334 CrossRef CAS PubMed.
- T. Niehues, LymphoSign Journal, 2018, 5, 35–47 CrossRef.
- C. Liu, D. Chu, K. Kalantar-Zadeh, J. George, H. A. Young and G. Liu, Adv. Sci., 2021, 8, e2004433 CrossRef PubMed.
- G. Liu, C. Jiang, X. Lin and Y. Yang, View, 2021, 2, 20210003 CrossRef CAS PubMed.
- T. L. Whiteside, Clin. Diagn. Lab. Immunol., 1994, 1, 257 CAS.
- S. Zhang, A. Garcia-D'Angeli, J. P. Brennan and Q. Huo, Analyst, 2014, 139, 439–445 RSC.
- A. I. Barbosa and N. M. Reis, Analyst, 2017, 142, 858–882 RSC.
- S. Oh, J. Kim, V. T. Tran, D. K. Lee, S. R. Ahmed, J. C. Hong, J. Lee, E. Y. Park and J. Lee, ACS Appl. Mater. Interfaces, 2018, 10, 12534–12543 CrossRef CAS PubMed.
- R. De La Rica and M. M. Stevens, Nat. Protoc., 2013, 8, 1759–1764 CrossRef PubMed.
- M. You, P. Peng, Z. Xue, H. Tong, W. He, P. Mao, Q. Liu, C. Yao and F. Xu, Analyst, 2021, 146, 2871–2877 RSC.
- Q. Guo, J.-J. Han, S. Shan, D.-F. Liu, S.-S. Wu, Y.-H. Xiong and W.-H. Lai, Biosens. Bioelectron., 2016, 86, 990–995 CrossRef CAS PubMed.
- H. Ravan and R. Yazdanparast, Anal. Chim. Acta, 2012, 733, 64–70 CrossRef CAS PubMed.
- P. Peng, C. Liu, Z. Li, Z. Xue, P. Mao, J. Hu, F. Xu, C. Yao and M. You, TrAC, Trends Anal. Chem., 2022, 152, 116605 CrossRef CAS.
- Y. Shi, P. Ye, K. Yang, J. Meng, J. Guo, Z. Pan, Q. Bayin and W. Zhao, J. Healthc. Eng., 2021, 2021, 2959843 Search PubMed.
- L. Metzler, U. Rehbein, J. N. Schonberg, T. Brandstetter, K. Thedieck and J. Ruhe, Anal. Chem., 2020, 92, 10283–10290 CrossRef CAS PubMed.
- C.-Y. Koh, U. Y. Schaff, M. E. Piccini, L. H. Stanker, L. W. Cheng, E. Ravichandran, B.-R. Singh, G. J. Sommer and A. K. Singh, Anal. Chem., 2015, 87, 922–928 CrossRef CAS PubMed.
- L. Coudron, M. B. McDonnell, I. Munro, D. K. McCluskey, I. D. Johnston, C. K. Tan and M. C. Tracey, Biosens. Bioelectron., 2019, 128, 52–60 CrossRef CAS PubMed.
- K. Wu, X. He, J. Wang, T. Pan, R. He, F. Kong, Z. Cao, F. Ju, Z. Huang and L. Nie, Front. Bioeng. Biotechnol., 2022, 10, 1112327 CrossRef PubMed.
- V. Mani, B. V. Chikkaveeraiah and J. F. Rusling, Expert Opin. Med. Diagn., 2011, 5, 381–391 CrossRef CAS PubMed.
- Y. Zhang and N. T. Nguyen, Lab Chip, 2017, 17, 994–1008 RSC.
- V. Yelleswarapu, J. R. Buser, M. Haber, J. Baron, E. Inapuri and D. Issadore, Proc. Natl. Acad. Sci. U. S. A., 2019, 116, 4489–4495 CrossRef CAS PubMed.
- D. H. Wilson, D. M. Rissin, C. W. Kan, D. R. Fournier, T. Piech, T. G. Campbell, R. E. Meyer, M. W. Fishburn, C. Cabrera, P. P. Patel, E. Frew, Y. Chen, L. Chang, E. P. Ferrell, V. von Einem, W. McGuigan, M. Reinhardt, H. Sayer, C. Vielsack and D. C. Duffy, J. Lab. Autom., 2016, 21, 533–547 CrossRef CAS PubMed.
- G. Czilwik, S. Vashist, V. Klein, A. Buderer, G. Roth, F. Von Stetten, R. Zengerle and D. Mark, RSC Adv., 2015, 5, 61906–61912 RSC.
- Y. Zhao, G. Czilwik, V. Klein, K. Mitsakakis, R. Zengerle and N. Paust, Lab Chip, 2017, 17, 1666–1677 RSC.
- D. I. Walsh, 3rd, G. J. Sommer, U. Y. Schaff, P. S. Hahn, G. J. Jaffe and S. K. Murthy, Lab Chip, 2014, 14, 2673–2680 RSC.
- Y. Wang, Q. Ruan, Z. C. Lei, S. C. Lin, Z. Zhu, L. Zhou and C. Yang, Anal. Chem., 2018, 90, 5224–5231 CrossRef CAS PubMed.
- A. H. C. Ng, R. Fobel, C. Fobel, J. Lamanna, D. G. Rackus, A. Summers, C. Dixon, M. D. M. Dryden, C. Lam, M. Ho, N. S. Mufti, V. Lee, M. A. M. Asri, E. A. Sykes, M. D. Chamberlain, R. Joseph, M. Ope, H. M. Scobie, A. Knipes, P. A. Rota, N. Marano, P. M. Chege, M. Njuguna, R. Nzunza, N. Kisangau, J. Kiogora, M. Karuingi, J. W. Burton, P. Borus, E. Lam and A. R. Wheeler, Sci. Transl. Med., 2018, 10, eaar6076 CrossRef PubMed.
- C. H. Chiou, D. J. Shin, Y. Zhang and T. H. Wang, Biosens. Bioelectron., 2013, 50, 91–99 CrossRef CAS PubMed.
- J. Song, D. W. Park, S. Moon, H.-J. Cho, J. H. Park, H. Seok and W. S. Choi, BMC Infect. Dis., 2019, 19, 1–11 CrossRef CAS PubMed.
- A. Ortqvist, J. Hedlund, B. Wretlind, A. Carlstrom and M. Kalin, Scand. J. Infect. Dis., 1995, 27, 457–462 CrossRef CAS PubMed.
- M. El-Shabrawy, M. E. Alsadik, M. El-Shafei, A. A. Abdelmoaty, A. S. Alazzouni, M. M. Esawy and M. A. Shabana, Egypt. J. Bronchol., 2021, 15, 1–7 CrossRef.
- M. A. Zimmerman, C. H. Selzman, C. Cothren, A. C. Sorensen, C. D. Raeburn and A. H. Harken, Arch. Surg., 2003, 138, 220–224 CrossRef PubMed.
- A. Beyzavi and N.-T. Nguyen, J. Micromech. Microeng., 2008, 18, 095018 CrossRef.
- G. Comina, A. Suska and D. Filippini, Lab Chip, 2014, 14, 424–430 RSC.
- S. Razavi Bazaz, N. Kashaninejad, S. Azadi, K. Patel, M. Asadnia, D. Jin and M. Ebrahimi Warkiani, Adv. Mater. Technol., 2019, 4, 1900425 CrossRef CAS.
- Y. Kantaros, B. V. Johnson, S. Chowdhury, D. J. Cappelleri and M. M. Zavlanos, IEEE T. Robot., 2018, 34, 1472–1489 Search PubMed.
- S. Chowdhury, W. Jing and D. J. Cappelleri, Micromachines, 2015, 7, 3 CrossRef PubMed.
- T. Liu, G. Choi, Z. Tang, A. Kshirsagar, A. J. Politza and W. Guan, Biosens. Bioelectron., 2022, 209, 114255 CrossRef CAS PubMed.
- S. M. Delgado, D. J. Kinahan, F. S. Sandoval, L. A. Julius, N. A. Kilcawley, J. Ducree and D. Mager, Lab Chip, 2016, 16, 4002–4011 RSC.
- N. M. Adams, A. E. Creecy, C. E. Majors, B. A. Wariso, P. A. Short, D. W. Wright and F. R. Haselton, Biomicrofluidics, 2013, 7, 14104 CrossRef PubMed.
- Z. Long, A. M. Shetty, M. J. Solomon and R. G. Larson, Lab Chip, 2009, 9, 1567–1575 RSC.
- I. Buchwalow, V. Samoilova, W. Boecker and M. Tiemann, Sci. Rep., 2011, 1, 28 CrossRef PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d3sd00258f |
| This journal is © The Royal Society of Chemistry 2024 |