Open Access Article
Open Access ArticleSpectrally separated dual functional fluorescent nanosensors for subcellular lysosomal detection of hypochlorous acid and chloride†
Yunxin
Cui‡
a,
Jianhong
Wu‡
b,
Jingying
Zhai
*b,
Yifu
Wang
a and
Xiaojiang
Xie
 *a
*a
aDepartment of Chemistry, Southern University of Science and Technology, Shenzhen, 518055, China. E-mail: xiexj@sustech.edu.cn
bAcademy for Advanced Interdisciplinary Studies, Southern University of Science and Technology, Shenzhen, 518055, China
First published on 5th January 2024
Abstract
Dynamic monitoring of different chemical species at the subcellular level greatly enriches our understanding of various biochemical processes. Hypochlorous acid (HClO), an indispensable disinfecting weapon in immune responses, is synthesized from chloride ions (Cl−) with the enzyme myeloperoxidase (MPO) in lysosomes. Here, we reported a dual-functional fluorescent nanosensor (NS_Cl&HClO) to simultaneously monitor the subcellular dynamics of HClO and Cl− in lysosomes. Prepared by nanoprecipitation from biocompatible polymers, NS_Cl&HClO contained a Cl− sensitive dye (Q+), a HClO reactive dye (Cy5), and a reference dye (LRed). Spectral separation ensured no cross-response between the two analytes. The nanosensors exhibited a detection limit of ca. 21 nM for HClO and responded to Cl− in a dynamic range of 0 to 170 mM, with excellent selectivity over a range of other reactive species and common biological ions. The dynamics of HClO in the lysosomes of RAW264.7 cells was successfully monitored by the endocytosed NS_Cl&HClO with high contrast, indicating that the proposed nanosensors are very promising for the detection of subcellular Cl− and HClO.
Introduction
The intracellular network contains numerous biochemical reactions in different compartments to maintain the proper functioning and homeostasis of the cell.1–3 Biological pathways often involve a number of different species which differ dramatically in concentrations.1,4,5 For macrophages hunting an exotic species such as a virus or bacterium, the endocytic pathways are extremely important as a part of the innate immune system.6–8 The captured cargo will experience different stages of gradual maturity, and be finally delivered to lysosomes where degradation occurs.9–11 During the past decades, the luminal dynamics of pH, metal ions, anions, and reactive species have drawn particular attention.12–14 For example, the pH is found to gradually decrease from early endosomes to lysosomes for maximum efficiency of various degradative enzymes in the lysosomes.15Fluorescent probes have been widely used to explore different cellular processes because of the high spatial and temporal resolution, high sensitivity, and compatibility with state-of-the-art fluorescence imaging platforms.16–18 Various fluorescent probes were developed with high selectivity to one particular chemical species. However, to better depict the intertwined biological processes, simultaneous monitoring of multiple species is undoubtedly a more desired solution.19–21 Unfortunately, the width of the biological spectral window makes it challenging to combine a large variety of fluorophores without spectral overlap, thus limiting the number of simultaneous detectable targets. In recent years, fluorescent probes aimed at detecting two chemical species have started to emerge in live-cell imaging mainly based on nanosensors and small-molecule synthetic probes.12–14,19–22 For example, the group of Krishnan successfully utilized fluorescently labeled DNA nanoassemblies to monitor the dynamic evolution of pH and hypochlorous acid (HClO) during phagocytosis and revealed that the generation of HClO was highly dependent on the luminal pH.13 Tian and co-workers also reported dual functional fluorescent probes for pH/Ca2+ and pH/Cu2+.23,24 Our group previously reported the simultaneous monitoring of chloride ions (Cl−) and oxygen using dual-functional fluorescent nanosensors in the lysosomes of HeLa cells and found that the luminal chloride concentration decreased during oxygen and glucose deprivation (OGD).25 However, the current pictures only reveal the tip of an iceberg since the chemical environment in the lysosomes remains largely unexplored in numerous processes.
In the lysosomes, Cl− reacts with H2O2 under the catalysis of the enzyme myeloperoxidase (MPO), producing HClO as an extremely potent disinfectant and oxidant.26–28 The level of HClO also reflects the expression of MPO which has been found overexpressed in several chronic diseases including rheumatoid arthritis and atherosclerosis.29–32 Venkatachalam et al. reported that the dysregulation of Cl− in lysosomes could lead to malfunction and weakening of bacteria degradation.33 As HClO is synthesized from Cl−, they are naturally linked and it is highly desirable to develop dual-functional probes targeting these two species.
Herein, we reported a dual-functional probe for Cl− and HClO (abbreviated as NS_Cl&HClO) based on polymeric nanospheres containing fluorescent units responsive to Cl− and HClO, respectively. To our knowledge, it was the first report of a dual-functional fluorescent probe that simultaneously detects Cl− and HClO. NS_Cl&HClO exhibited excellent spectral separation for Cl− and HClO, high sensitivity, and selectivity over a range of anions and reactive species. The nanosensors were delivered to the lysosomes through endocytosis with good biocompatibility. The dynamic changes of Cl− and HClO in the lysosomes were successfully monitored with NS_Cl&HClO in regulating the activity of MPO, the proton pump V-ATPase, and Cl− transporters upon external stimulation.
Experimental section
Materials
Boric acid, sodium chloride, sodium nitrate, sodium fluoride, sodium sulfate, sodium pyrophosphate, sodium bromide, sodium iodide, Pluronic F-127 (average Mn 12![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 600 Da, F127), poly(dimethylsiloxane) bis(3-aminopropyl) terminated (average Mn ∼2500 Da, PDMS-NH2) and lipopolysaccharides (LPS) from pseudomonas aeruginosa were purchased from Sigma-Aldrich. Angeli's salt (GC19428), phorbol myristate acetate (PMA) and the Cell Counting Kit-8 (CCK-8) were purchased from GlpBio Technology USA. Sodium hydroxide, hydrogen peroxide (H2O2, 30%), tert-butyl hydroperoxide (TBHP), sodium bisulfide, sodium sulfite, sodium bicarbonate, sodium nitrite, sodium hypochlorite, S-nitroso-N-acetyl-DL-penicillamine (SNAP), 2, 2′-azobis(2-methylpropionamidine)-dihydrochloride (AAPH), L-cysteine (Cys) and glutathione (GSH) were obtained from Aladdin Chemistry Co. Ltd. Potassium superoxide was purchased from InnoChem Science & Technology Co. Ltd. Beijing. 6-Methoxy-quinoline, cinnamic acid, 5-nitro-2-((3-phenylpropyl)amino)benzoic acid (NPPB) and 4-aminobenzoic acid hydrazide (ABAH) were purchased from Bide Pharmatech. Ltd. Cy5-maleimide (Cy5) was purchased from APExBIO. Lumogen Red (LRed), Bafilomycin A1(BA1), and phosphoric acid were purchased from Macklin Biochemical Co. Ltd., Shanghai. Ultra-filtration tubes (MWCO: 3000) were obtained from Millipore Amicon. LysoTracker Deep Red was obtained from Thermo Fisher Scientific. Tetrahydrofuran (THF), tosyl chloride (TsCl), triethylamine (TEA), acetonitrile (ACN), acetic acid, dichloroethane (DCM) and ethanol (HPLC grade) were purchased from J&K Scientific Ltd. in China. Deionized water was used to prepare aqueous solutions after purification by a Milli-Q Integral 5. Dulbecco's modified Eagle medium (DMEM), fetal bovine serum (FBS), penicillin and streptomycin solution (100×), and sterile phosphate buffered saline (PBS) were purchased from Corning. RAW264.7 cells were purchased from Procell Life Science&Technology Co. Ltd.
600 Da, F127), poly(dimethylsiloxane) bis(3-aminopropyl) terminated (average Mn ∼2500 Da, PDMS-NH2) and lipopolysaccharides (LPS) from pseudomonas aeruginosa were purchased from Sigma-Aldrich. Angeli's salt (GC19428), phorbol myristate acetate (PMA) and the Cell Counting Kit-8 (CCK-8) were purchased from GlpBio Technology USA. Sodium hydroxide, hydrogen peroxide (H2O2, 30%), tert-butyl hydroperoxide (TBHP), sodium bisulfide, sodium sulfite, sodium bicarbonate, sodium nitrite, sodium hypochlorite, S-nitroso-N-acetyl-DL-penicillamine (SNAP), 2, 2′-azobis(2-methylpropionamidine)-dihydrochloride (AAPH), L-cysteine (Cys) and glutathione (GSH) were obtained from Aladdin Chemistry Co. Ltd. Potassium superoxide was purchased from InnoChem Science & Technology Co. Ltd. Beijing. 6-Methoxy-quinoline, cinnamic acid, 5-nitro-2-((3-phenylpropyl)amino)benzoic acid (NPPB) and 4-aminobenzoic acid hydrazide (ABAH) were purchased from Bide Pharmatech. Ltd. Cy5-maleimide (Cy5) was purchased from APExBIO. Lumogen Red (LRed), Bafilomycin A1(BA1), and phosphoric acid were purchased from Macklin Biochemical Co. Ltd., Shanghai. Ultra-filtration tubes (MWCO: 3000) were obtained from Millipore Amicon. LysoTracker Deep Red was obtained from Thermo Fisher Scientific. Tetrahydrofuran (THF), tosyl chloride (TsCl), triethylamine (TEA), acetonitrile (ACN), acetic acid, dichloroethane (DCM) and ethanol (HPLC grade) were purchased from J&K Scientific Ltd. in China. Deionized water was used to prepare aqueous solutions after purification by a Milli-Q Integral 5. Dulbecco's modified Eagle medium (DMEM), fetal bovine serum (FBS), penicillin and streptomycin solution (100×), and sterile phosphate buffered saline (PBS) were purchased from Corning. RAW264.7 cells were purchased from Procell Life Science&Technology Co. Ltd.
Preparation of F127-Q+ and PDMS-Cy5
F127-Q+: 828 mg of F127 and 88 mg of TsCl were respectively dissolved in 25 mL of DCM, and then 40 μL of TEA was added to the DCM solution. The reactants were stirred at 350 rpm and left at room temperature for 20 h. After the evaporation of DCM, the crude products were further dissolved in 25 mL of ACN, and 170 mg of 6-methoxy-quinoline was directly added to further react at 89 °C for 36 h. The final crude products were purified using dialysis (1500 Da MWCO) against an aqueous solution containing 10% volume of ethanol. Finally, the purified solution was freeze-dried to obtain F127-Q+ as powder for further usage.PDMS-Cy5: 8.6 mg of PDMS-NH2 and 4.5 mg of Cy5-maleimide were dissolved in 8 mL of ethanol, and reacted at room temperature for 35 h. After the evaporation of ethanol, the crude products were repeatedly centrifuged with an Ultra-filtration tube of MWCO at 3000 Da to remove unreacted Cy5-maleimide. Finally, the purified product PDMS-Cy5 was quantified according to the absorption spectrum and stored at 4 °C for further use.
Preparation of NS_Cl&HClO
Typically, 5 mg of F127-Q+, 0.150 mg of PDMS-NH2, a PDMS-Cy5 solution containing 50 nmol of Cy5, and 0.03 mg of LRed were dissolved in 0.5 mL THF. Then, the mixture was injected into 5 mL of deionized water under vortexing at 1000 rpm. THF was then removed via compressed air and the suspension was metered to 5 mL as the stock solution.MALDI-TOF, ESI-MS, and FT-IR characterization of F127-Q+ and PDMS-Cy5
The MALDI-TOF characterization of F127 and F127-Q+ was conducted with cinnamic acid as a matrix via an Autoflex speed LRF (Bruker). PDMS-NH2 and PDMS-Cy5 were characterized through a Q-Exactive electrospray ionization mass spectrometer (ESI-MS) in the positive-ion mode (Thermo Fisher Scientific, USA). In addition, FT-IR spectra of Cy5-maleimide, PDMS-NH2, and PDMS-Cy5 were recorded using a Vertex 80 FTIR purge spectrometer (Bruker).Morphologic characterization of NS_Cl&HClO
The stock solution of the nanosensors was diluted ten times, dropped to a plasma-cleaned copper grid, and left to dry in air. Then, the prepared sample was imaged via transmission electron microscopy (HT-7700, Hitachi) at 100 kV accelerating voltage. The hydrodynamic size of the nanosensors was measured with a Zetasizer Nano ZS analyzer (Malvern Inc.).Fluorescent response to Cl− and HClO
A suspension of NS_Cl&HClO was successively titrated to reach 0, 1, 3, 7, 10, 30, 70, 100, 130, and 170 mM of NaCl in modified Britton–Robinson (MBR) buffer at pH 4.6 and the fluorescence spectra were recorded with excitation at 365 nm on a spectrometer (Fluorolog-3, Horiba Jobin Yvon). A suspension of NS_Cl&HClO containing 1 μM Cy5 in MBR buffer was respectively mixed with NaClO stock solutions to reach 0, 0.1, 0.3, 0.5, 0.8, 1, 2, 5, 8, 10, and 15 μM of HClO, and incubated for 1 h at room temperature. Then, the fluorescence spectra were recorded with excitation at 561 nm and 640 nm, respectively.Selectivity of NS_Cl&HClO over different anions and ROS/RNS/RSS
To evaluate the selectivity to Cl−, 1 mM of different anions (sodium salt) was used. Freshly prepared ROS/RNS were applied to evaluate the selectivity to HClO. The following protocol was used to prepare the reactive species. The hydroxyl radical (·OH) was prepared by mixing 1 mM Fe2+ with 3 mM H2O2. The superoxide anion (O2−) was obtained by dissolving KO2 with DMSO to 1 mM. The alkyl peroxyl radical (ROO·) was generated by the thermolysis of air-saturated aqueous AAPH solution (1 mM) at 310 K. Peroxynitrite (ONOO−) was generated by the reaction between H2O2 (2 mM) and NaNO2 (2 mM) aqueous solution of the same volume. Nitric oxide and nitroxyl (HNO) were derived from 1 mM aqueous solution of SNAP and Angeli's salt, respectively. 1O2 was generated by mixing H2O2 (2 mM) with NaClO solution (2 mM). All measurements were performed after 1 h of incubation at room temperature.Cell viability
RAW264.7 cells were seeded in a 96 well plate (8500 cells/per well) and incubated in DMEM complete medium at 37 °C with 5% CO2 for 24 h. Then, after rinsing with PBS, different concentrations of NS_Cl&HClO were respectively added to the wells with three repeats. After 20 h of incubation, the CCK-8 reagents were added for another 2.5 h of incubation. Finally, the optical density (OD) at 450 nm of each well was read by a microplate reader (Cytation5, BioTek), and the cellular viability was calculated according to the OD values.Live cell imaging
RAW264.7 cells were seeded in a 30 mm glass-bottomed plate (30![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 cells/per plate) and further cultured for 24 h in DMEM complete medium at 37 °C with 5% CO2. To determine cellular localization, NS_Cl&HClO (10 times diluted from the stock) was used to stain RAW264.7 cells for 5 min before 100 nM LysoTracker Deep Red was added to incubate the cells for 1 min. After three times rinsing, the sample was respectively excited with the laser at 405 nm, 561 nm, and 640 nm, and signals from 410–530 nm, 570–630 nm, and 650–700 nm were collected by a laser confocal scanning microscope (CLSM, LSM900, Zeiss).
000 cells/per plate) and further cultured for 24 h in DMEM complete medium at 37 °C with 5% CO2. To determine cellular localization, NS_Cl&HClO (10 times diluted from the stock) was used to stain RAW264.7 cells for 5 min before 100 nM LysoTracker Deep Red was added to incubate the cells for 1 min. After three times rinsing, the sample was respectively excited with the laser at 405 nm, 561 nm, and 640 nm, and signals from 410–530 nm, 570–630 nm, and 650–700 nm were collected by a laser confocal scanning microscope (CLSM, LSM900, Zeiss).
To monitor the dynamics of lysosomal Cl− and HClO, RAW264.7 cells were stained with NS_Cl&HClO for 5 min, and after three times rinsing with PBS, the sample was directly imaged with different time-lapses. The cells were also imaged after challenging with 7 μg mL−1 LPS and then 3 μg mL−1 PMA. For the BA1 and NPPB groups, RAW264.7 cells were first incubated with 200 nM BA1 or 100 μM NPPB for 1 h and then stained with NS_Cl&HClO for 5 min. Next, the samples were imaged in the basal medium containing 200 nM BA1 or 100 μM NPPB. For the ABAH group, RAW264.7 cells were incubated with 100 μM ABAH. All the samples were respectively excited at 405 nm, 561 nm, and 640 nm, and signals were collected from the range of 410–530 nm (blue channel), 570–630 nm (orange channel), and 650–700 nm (red channel), respectively.
Results and discussion
Working principle and preparation of NS_Cl&HClO
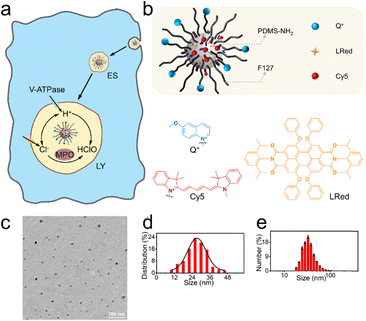
Fig. 1a shows the endocytosis of the proposed nanosensors (NS_Cl&HClO) by a macrophage. After a few early stages (ES), the nanosensors are expected to be delivered to the lysosomes where they will show fluorescence response to Cl− and HClO simultaneously. The lysosomes are highly dynamic organelles enclosing various enzymes for degradation. It is known that V-ATPase on the plasma membrane is employed to pump H+ into the lysosomal lumen to keep the luminal pH around 4 to 5.15 The acidic environment is crucial for the proper functioning of lysosomal enzymes including MPO, which converts HClO from Cl−.The design of the dual-functional nanosensors (NS_Cl&HClO) had the following features: 1) the fluorescence bands of Q+ (emission peak around 448 nm) and Cy5 (emission peak around 670 nm) were well separated to avoid spectral cross-response; 2) the Cl− sensitive probe Q+ located on the surface of the nanospheres ensured high sensitivity to Cl−; 3) the hydrophobic core of PDMS allowed the addition of lipophilic reference dye LRed and enabled ratiometric measurements of HClO. In other words, fluorescence resonance energy transfer (FRET) could occur between the reference dye LRed and Cy5 due to spectral overlap. Also, LRed was not sensitive to Cl− and HClO (shown in Fig. S1†) which ensured that it was very suitable to act as a reference dye. In addition, the polymer matrix also enhanced the selectivity to HClO through an anion-exchange-type mechanism according to our previous findings.34
Through nanoprecipitation, NS_Cl&HClO was self-assembled from a poly(dimethylsiloxane) (PDMS) derivative, a modified amphiphilic triblock copolymer Pluronic F-127 (F127), and a chemically inert fluorescent reference dye LRed (Fig. 1b). The Cl−-sensitive fluorophore, 6-methoxy-quinolinium (Q+), was chemically attached to the distal end of F127. The HClO-sensitive fluorophore, Cy5, was attached to the amine terminal of PDMS-NH2 through the Michael addition between amine and maleic amide. Fig. S2† shows a slight increase of m/z for F127-Q+ compared with the reactant F127. The successful formation of PDMS-Cy5 was confirmed with mass spectroscopy (Fig. S3†). From the Fourier transform infrared (FT-IR) spectrum (Fig. S4†) of PDMS-Cy5, the stretching of the C–H bond of the maleic amide at 697 cm−1 also disappeared which further confirmed the successful ligation. The obtained nanosensors exhibited an average diameter of 27.4 nm from transmission electron microscopy (TEM, Fig. 1c and d) and the average hydrodynamic diameter was determined by dynamic light scattering (DLS) to be 32.7 ± 1.1 nm (Fig. 1e). Nanoparticles of similar size were reported to be readily taken up by various cells through endocytic pathways without resorting to further surface modification.35
Fluorescent response of NS_Cl&HClO
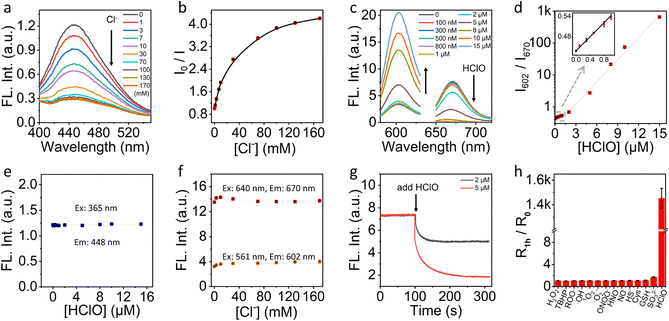
We first characterized the fluorescence response of NS_Cl&HClO to Cl− and HClO at pH 4.6 to mimic the lysosome pH, respectively. Fig. 2a shows that the emission intensity at 448 nm gradually decreases with increasing Cl− concentration under 365 nm excitation. Quantification according to the dynamic quenching model resulted in the calibration curve in Fig. 2b, which fitted well with the two-site Stern–Volmer equation. The response indicated different accessibility of Q+ to Cl− which was also reported in previous chloride nanosensors based on dynamic quenching.36,37As shown in Fig. 2c, with excitation at 640 nm, the emission of Cy5 at 670 nm was highly sensitive to HClO which agreed with a previous report on the oxidation of Cy5 by HClO.38 With increasing HClO concentration, the decrease of emission at 670 nm was accompanied by an increase of the emission of LRed at 602 nm (excited at 561 nm). This ratiometric response was caused by the spectral overlap between the emission of LRed and the excitation of Cy5. From the ratiometric response (Fig. 2d), the detection limit was estimated to be ca. 21 nM according to three times the standard deviation over the sensitivity. Amazingly, with just 15 μM HClO, the fluorescence signal changed over 1400-fold. This excellent sensitivity underpinned the high contrast in further fluorescence cell imaging.
To evaluate if there was a cross-response between Cl− and HClO, we evaluated the fluorescence intensity of NS_Cl&HClO at 448 nm at different HClO concentrations. As shown in Fig. 2e, the presence of HClO caused no interference to the fluorescence of the Q+ moiety. Similarly, as shown in Fig. 2f, the emissions at both 602 and 670 nm were insensitive to the concentration change of Cl−.
Fig. 2g shows the kinetic monitoring of the emission intensity at 670 nm when 2 or 5 μM HClO was added to the sample of NS_Cl&HClO. The response time (t95%) was determined to be 78 s which was close to several reported HClO probes.13,39 Moreover, NS_Cl&HClO exhibited excellent selectivity for biological applications. Fig. 2h shows the selectivity to HClO over different reactive oxygen species (ROS), reactive nitrogen species (RNS), and reactive sulfur species (RSS). The concentrations used for different species were specified in the figure caption and were comparable or much higher than common concentrations found in cells. The selectivity to Cl− was also evaluated (Fig. S5†). In agreement with previous reports, Q+ exhibited a good selectivity to halides over a range of other anions.36,40 Surprisingly, the selectivity to HClO over a range of ROS and RNS was improved compared with free Cy5 in water,38 which was ascribed to the polymeric barrier and anion exchange nature of the nanosensor. A similar observation was reported in our previous work where the selectivity to sulfite was enhanced in polymeric nanosensors.35
Toxicity and colocalization
The cell toxicity was examined before further cellular experiments. As shown in Fig. S6,† using 160 mg L−1 of NS_Cl&HClO (calculated by total mass) in the culture media showed negligible influence on the viability of RAW264.7 cells. Up to 70% of viability was still observed when 1600 mg L−1 of NS_Cl&HClO was supplemented. The excellent compatibility was ascribed to the use of the biocompatible matrix materials (F127 and PDMS).41 Next, the location of the nanosensors in cells was studied with the commercial tracker LysoTracker Deep Red. As shown in Fig. S7,† the fluorescence from the nanosensors and the tracker exhibited a good overlap with a Pearson's correlation coefficient of ca. 0.9. Therefore, the nanosensors were promising for measurements in the lysosomes.Cell imaging with chemical stimulation
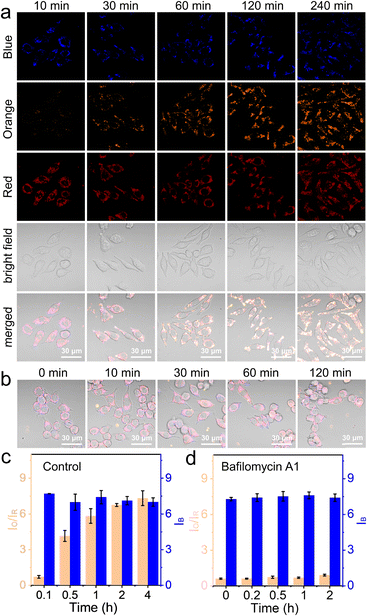
In this work, NS_Cl&HClO was used in confocal laser scanning microscopy (CLSM) to study the lysosomal dynamics of Cl− and HClO during different chemical stimulations. First, as shown in Fig. 3a, RAW264.7 macrophages incubated with NS_Cl&HClO were imaged in a normal DMEM culture medium. While the blue channel corresponded to the emission of Q+ for Cl−, the orange and red channels corresponded to the emission of LRed and Cy5, respectively. Notably, from ca. 10 min to 4 h after the nanosensors were taken up by RAW264.7, the blue channel showed no significant change, but the red channel gradually darkened and the orange channel significantly brightened up. As shown in Fig. 3c, the ratio of orange over red, Io/IR, increased over 9-fold, indicating an increasing level of HClO generation. The results indicated that NS_Cl&HClO could be used to detect the endogenous HClO in RAW264.7 which was reported to be very challenging because of the low expression of MPO.8Lipopolysaccharides (LPS), endotoxins from the cell wall of Gram-negative bacteria, are known to activate the inflammatory responses of macrophages. Together with phorbol myristate acetate (PMA), a protein kinase C activator, they have previously been used to increase the expression of MPO in macrophages.42 Here, we also stimulated the RAW264.7 cells with LPS and PMA (Fig. S8a†). In this case, NS_Cl&HClO indeed exhibited more change in IO/IR (more than 49-fold, Fig. S8b†). The blue channel also exhibited a small degree of oscillation. However, there was no conclusive trend based on the acquired data so far.
After the above control experiments, we evaluated RAW264.7 under the stimulation of Bafilomycin A1 (BA1), a specific inhibitor of V-ATPase.43 The blockage of the proton-pumps was shown in the literature to increase the pH in lysosomes. The question here was whether this would influence the generation of HClO with MPO and whether the nanosensor NS_Cl&HClO was capable of detecting any signal change.
As shown in Fig. 3b and S9,† during the 2 h of BA1 stimulation, the orange channel remained dark and the intensity of the red channel was not significantly decreased. Fig. 3d shows that the value of IO/IR remained small throughout the experiment (ca. 0.5-fold increase). This meant much less HClO was generated compared to that in the control experiment. On the other hand, the intensity of the blue channel remained unchanged, indicating that stimulation with BA1 did not cause too much change in the chloride ion concentration.
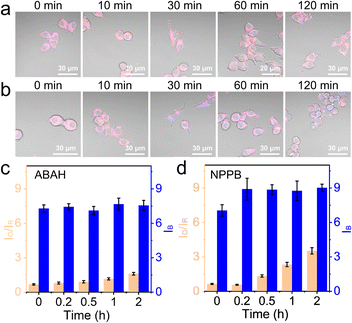
The influence of BA1 was rather universal because an increase in lysosomal pH impacted the whole compartment that functioned the best at acidic pH. To be more specific, 100 μM ABAH, which was reported to irreversibly inactivate MPO,44 was used to culture the cells for 1 h before imaging with CLSM. As shown in Fig. 4a, c, and S10,† the change of IO/IR was very small during the first 1 h, and slightly increased by 1.4-fold from 0 h to 2 h. This confirmed that ABAH was indeed able to inhibit MPO but the effect was not as much as BA1.
Here, the experiments with BA1 and ABAH did not cause too much intensity change in the blue channel. The chloride ion concentration in the lysosomes was reported in several previous works to be over 80 mM.25,45 Therefore, the consumption of Cl− during the production of HClO was negligible. However, it was unclear whether a change of luminal Cl− would impact the generation of HClO. With NS_Cl&HClO, we were able to monitor the cells under the chemical stimulation of NPPB, which inhibited broad-spectrum anion transporters in cells including those for Cl−.46 When RAW264.7 cells were treated with NPPB (Fig. 4b and S11†), the intensity of the blue channel increased at 10 min, indicating a reduced Cl− concentration. At the same time, the generation of HClO was also clearly delayed by decreased Cl− with the value of IO/IR increasing to only 4.5-fold after 2 h (Fig. 4d). These results showed that a decrease of luminal Cl− concentration in the lysosomes would decrease the level of HClO, which was synthesized from Cl−. However, by comparing the value of IO/IR, the effect was again not as significant as BA1 and with time the suppression was alleviated. Further experiments for quantitative correlation were continued in the lab.
Conclusions
Dual-functional fluorescent nanosensors targeting two different chemical species were advantageous for the dynamic monitoring of intracellular biological reactions. Since HClO was produced from Cl− in lysosomes, we developed a dual-functional fluorescent nanosensor, NS_Cl&HClO, targeting lysosomal Cl− and HClO simultaneously. This probe was prepared with nanoprecipitation using the polymer-bound small molecular fluorescent probes Cy5 and Q+ for HClO and Cl−, respectively. A reference dye LRed was added to produce ratiometric fluorescence response, acting as the energy transfer donor of Cy5. The spectral separation between Q+, LRed, and Cy5 ensured no cross-response between Cl− and HClO. The matrix materials PDMS and F127 ensured good biocompatibility as evaluated with the commercial CCK-8 assay.NS_Cl&HClO showed an excellent detection limit (ca. 21 nM), rapid response (t95% = 78 s at 5 μM), and enhanced selectivity for HClO over a range of biological ions and reactive species. The sensitivity to Cl− was also suited to monitor endolysosomal chloride ions which was typically between 0.01 and 0.1 M. The endolysosomal dynamics of Cl− and HClO were imaged simultaneously with NS_Cl&HClO under different chemical stimulations. Notably, NS_Cl&HClO enabled high-contrast ratiometric fluorescence imaging of the generation of HClO in RAW264.7 under normal conditions. Stimulation with BA1 to block the V-ATPase was found to significantly reduce the production of HClO with less impact on Cl−. Inhibition of the enzyme MPO by ABAH also reduced the level of HClO as monitored under CLSM. Lastly, when NPPB, an inhibitor of broad-spectrum anion transporters, was applied to RAW264.7, a decrease of the luminal Cl− concentration was detected by NS_Cl&HClO in the blue channel, and the generation of HClO was found to be suppressed at the same time. Overall, the proposed dual-functional fluorescent nanosensor and preliminary cellular experiments formed a unique platform for investigating and correlating subcellular Cl− and HClO simultaneously.
Author contributions
Y. Cui and J. Wu: investigation, methodology and validation – original draft. Y. Wang: investigation. J. Zhai: supervision, funding acquisition, writing – review & editing. X. Xie: conceptualization, writing – review & editing and funding acquisition.Conflicts of interest
There are no conflicts to declare.Acknowledgements
The authors acknowledge financial support from the Department of Science and Technology of Guangdong Province (2021A1515010330), and the Shenzhen Science and Technology Program (No. 202110293000007). The authors acknowledge the assistance of TEM imaging from SUSTech Core Research Facilities.Notes and references
- H. Xu, E. Martinoia and I. Szabo, Cell Calcium, 2015, 58, 1–10 CrossRef PubMed.
- B. A. Schröder, C. Wrocklage, A. Hasilik and P. Saftig, Proteomics, 2010, 10, 4053–4076 CrossRef PubMed.
- R. Zenobi, Science, 2013, 342, 1243259 CrossRef CAS PubMed.
- T. Stauber and T. J. Jentsch, Annu. Rev. Physiol., 2013, 75, 453–477 Search PubMed.
- C. C. Scott and J. Gruenberg, BioEssays, 2011, 33, 103–110 CrossRef CAS PubMed.
- W. L. Lee, R. E. Harrison and S. Grinstein, Microbes Infect., 2003, 5, 1299–1306 Search PubMed.
- A. Di, M. E. Brown, L. V. Deriy, C. Li, F. L. Szeto, Y. Chen, P. Huang, J. Tong, A. P. Naren, V. Bindokas, H. C. Palfrey and D. J. Nelson, Nat. Cell Biol., 2006, 8, 933–944 Search PubMed.
- A. Strzepa, K. A. Pritchard and B. N. Dittel, Cell. Immunol., 2017, 317, 1–8 CrossRef CAS PubMed.
- R. Perera and R. Zoncu, Annu. Rev. Cell Dev. Biol., 2016, 32, 223–253 CrossRef CAS PubMed.
- C. Winterbourn and A. Kettle, Antioxid. Redox Signaling, 2012, 18, 642–660 CrossRef PubMed.
- O. V. Vieira, R. J. Botelho and S. Grinstein, Biochem. J., 2002, 366, 689–704 CrossRef CAS PubMed.
- K. Leung, K. Chakraborty, A. Saminathan and Y. Krishnan, Nat. Nanotechnol., 2019, 14, 176–183 CrossRef CAS PubMed.
- S. Thekkan, M. S. Jani, C. Cui, K. Dan, G. Zhou, L. Becker and Y. Krishnan, Nat. Chem. Biol., 2019, 15, 1165–1172 CrossRef CAS PubMed.
- N. Narayanaswamy, K. Chakraborty, A. Saminathan, E. Zeichner, K. Leung, J. Devany and Y. Krishnan, Nat. Methods, 2019, 16, 95–102 CrossRef CAS PubMed.
- J. R. Casey, S. Grinstein and J. Orlowski, Nat. Rev. Mol. Cell Biol., 2010, 11, 50–61 CrossRef CAS PubMed.
- M. Fernández-Suárez and A. Y. Ting, Nat. Rev. Mol. Cell Biol., 2008, 9, 929–943 CrossRef PubMed.
- E. Specht, E. Braselmann and A. Palmer, Annu. Rev. Physiol., 2017, 79, 93–117 CrossRef CAS PubMed.
- X. Li, X. Gao, W. Shi and H. Ma, Chem. Rev., 2014, 114, 590–659 CrossRef CAS PubMed.
- L. He, B. Dong, Y. Liu and W. Lin, Chem. Soc. Rev., 2016, 45, 6449–6461 RSC.
- J. L. Kolanowski, F. Liu and E. J. New, Chem. Soc. Rev., 2018, 47, 195–208 RSC.
- Y. Yue, F. Huo, F. Cheng, X. Zhu, T. Mafireyi, R. M. Strongin and C. Yin, Chem. Soc. Rev., 2019, 48, 4155–4177 RSC.
- Y. Huang, Y. Zhang, F. Huo, J. Chao, F. Cheng and C. Yin, J. Am. Chem. Soc., 2020, 142, 18706–18714 CrossRef CAS PubMed.
- Z. Liu, H. Pei, L. Zhang and Y. Tian, ACS Nano, 2018, 12, 12357–12368 CrossRef CAS PubMed.
- Y. Han, C. Ding, J. Zhou and Y. Tian, Anal. Chem., 2015, 87, 5333–5339 CrossRef CAS PubMed.
- J. Li, J. Zhai, Y. Wang, W. Yang and X. Xie, Chem. Commun., 2020, 56, 14980–14983 RSC.
- Z. M. Prokopowicz, F. Arce, R. Biedroń, C. L. L. Chiang, M. Ciszek, D. R. Katz, M. Nowakowska, S. Zapotoczny, J. Marcinkiewicz and B. M. Chain, J. Immunol., 2010, 184, 824–835 Search PubMed.
- Y. Adachi, A. L. Kindzelskii, A. R. Petty, J.-B. Huang, N. Maeda, S. Yotsumoto, Y. Aratani, N. Ohno and H. R. Petty, J. Immunol., 2006, 176, 5033–5040 CrossRef CAS PubMed.
- T. Henrique de Araujo, S. S. Okada, E. E. B. Ghosn, N. N. Taniwaki, M. R. Rodrigues, S. Rogerio de Almeida, R. A. Mortara, M. Russo, A. Campa and R. C. Albuquerque, Cell. Immunol., 2013, 281, 27–30 CrossRef CAS PubMed.
- M. Rahmati, A. Mobasheri and M. Mozafari, Bone, 2016, 85, 81–90 CrossRef CAS PubMed.
- M. J. Steinbeck, L. J. Nesti, P. F. Sharkey and J. Parvizi, J. Orthop. Res., 2007, 25, 1128–1135 CrossRef CAS PubMed.
- A. Daugherty, J. L. Dunn, D. L. Rateri and J. W. Heinecke, J. Clin. Invest., 1994, 94, 437–444 CrossRef CAS PubMed.
- Y. W. Yap, M. Whiteman and N. S. Cheung, Cell. Signalling, 2007, 19, 45219–45228 CrossRef PubMed.
- C. O. Wong, S. Gregory, H. Hu, Y. Chao, V. E. Sepúlveda, Y. He, D. Li-Kroeger, W. E. Goldman, H. J. Bellen and K. Venkatachalam, Cell Host Microbe, 2017, 21, 719–730 CrossRef CAS PubMed.
- J. Li, X. Ma, W. Yang, C. Guo, J. Zhai and X. Xie, Anal. Chem., 2021, 93, 11758–11764 CrossRef CAS PubMed.
- F. Zhao, Y. Zhao, Y. Liu, X. Chang, C. Chen and Y. Zhao, Small, 2011, 7, 1322–1337 CrossRef CAS PubMed.
- D. G. Chris, Meas. Sci. Technol., 2001, 12, 53–88 CrossRef.
- L. Baù, F. Selvestrel, M. Arduini, I. Zamparo, C. Lodovichi and F. Mancin, Org. Lett., 2012, 14, 2984–2987 CrossRef PubMed.
- D. Oushiki, H. Kojima, T. Terai, M. Arita, K. Hanaoka, Y. Urano and T. Nagano, J. Am. Chem. Soc., 2010, 132, 2795–2801 CrossRef CAS PubMed.
- R. Zhang, B. Song and J. Yuan, TrAC, Trends Anal. Chem., 2018, 99, 1–33 CrossRef CAS.
- S. Jayaraman and A. S. Verkman, Biophys. Chem., 2000, 85, 49–57 CrossRef CAS PubMed.
- N. Kamaly, B. Yameen, J. Wu and O. C. Farokhzad, Chem. Rev., 2016, 116, 2602–2663 CrossRef CAS PubMed.
- S. E. Gomez-Mejiba, Z. Zhai, M. S. Gimenez, M. T. Ashby, J. Chilakapati, K. Kitchin, R. P. Mason and D. C. Ramirez, J. Biol. Chem., 2010, 285, 20062–20071 CrossRef CAS PubMed.
- S. Kissing, P. Saftig and A. Haas, Int. J. Med. Microbiol., 2018, 308, 58–67 Search PubMed.
- A. J. Kettle, C. A. Gedye and C. C. Winterbourn, Biochem. J., 1997, 321, 503–508 CrossRef CAS PubMed.
- S. Saha, V. Prakash, S. Halder, K. Chakraborty and Y. Krishnan, Nat. Nanotechnol., 2015, 10, 645–651 CrossRef CAS PubMed.
- M. L. Aiken, R. G. Painter, Y. Zhou and G. Wang, Free Radical Biol. Med., 2012, 53, 2308–2317 CrossRef CAS PubMed.
Footnotes |
| † Electronic supplementary information (ESI) available: Additional characterization of materials, spectroscopic and cellular studies. See DOI: https://doi.org/10.1039/d3sd00275f |
| ‡ Equal contribution. |
| This journal is © The Royal Society of Chemistry 2024 |