Open Access Article
Open Access ArticleStimuli-instructed sequential morphological transformations for molecular imaging
Peiyao
Chen
a,
Liling
Meng
a,
Tuotuo
Zhang
b and
Yao
Sun
 *b
*b
aKey Laboratory of Fermentation Engineering (Ministry of Education), National “111” Center for Cellular Regulation and Molecular Pharmaceutics, Hubei Key Laboratory of Industrial Microbiology, School of Food and Biological Engineering, Hubei University of Technology, Wuhan 430068, China
bNational Key Laboratory of Green Pesticide, College of Chemistry, Central China Normal University, Wuhan 430079, China. E-mail: sunyaogbasp@mail.ccnu.edu.cn
First published on 10th February 2024
Abstract
Stimuli-instructed sequential morphological transformations hold great promise for molecular imaging. During these processes, probes exhibit a spatially–temporally responsive cascade self-assembly/disassembly or tandem self-assembly transition toward dual stimuli along with sequential imaging signal switching, thus providing multiple types of information for precise and sensitive imaging or efficient theranostics of diseases. In this critical review, we provide an overview of dual stimuli-instructed cascade self-assembly and disassembly or tandem self-assembly of probes for molecular imaging and focus on the design strategies and potential applications of these probes. Finally, we discuss the current challenges and highlight perspective ideas in this field. We anticipate that this review will provide useful information on stimuli-instructed sequential morphological transformations and expand their applications to other fields.
1. Introduction
Molecular imaging is an emerging medical imaging science subfield, which is broadly defined as the characterization and measurement of biological processes at the cellular and molecular level in living subjects.1–4 Molecular imaging techniques commonly used in academic research and clinical disease diagnosis include positron emission tomography (PET), computed tomography (CT), magnetic resonance imaging (MRI), and optical imaging,5,6 permitting noninvasive imaging of disease-related biomarkers in real time. Imaging probes or contrast agents are one of the key components in molecular imaging, which generate signals upon interaction with the target of interest. Currently, “active probes” exhibiting “always on” imaging signals are commonly used in molecular imaging. Generally, an extended time delay is required to eliminate off-target active probes to produce a sufficient signal-to-noise ratio (SNR) for imaging, leading to a substantial loss of sensitivity even while the probe is highly specific.7 In contrast, “activatable probes” with “turn-off/on” signals at target sites display improved sensitivity and specificity for real-time imaging of target biomolecules in vivo.8–11Till now, numerous stimuli have been employed to develop activatable probes, including exogenous stimuli (e.g., ultrasound, light, heat, magnetic field) or endogenous stimuli (e.g., enzymes, acidity, glutathione (GSH), reactive oxygen species (ROS)).12,13 Since many disease-related biomarkers are also detectable in normal tissues or plasma, a single parameter to stimulate probe activation for disease diagnosis might cause “false-positive” results.14,15 Many efforts have been made to develop multiple stimuli-activated probes for molecular imaging.16,17 Previously, Fan and co-workers reported “dual lock-and-key”-controlled nanoprobes, which could be activated in the presence of both hyaluronidase and thiols, for ultrahigh specific fluorescence imaging of tumors in vivo.18 Compared with “single-lock-and-key”-controlled nanoprobes, these dual lock-and-key-controlled nanoprobes possessed lower background, higher tumor-to-background ratio and tumor-to-liver ratio, achieving tumor imaging with high specificity.
In the meantime, a lot of stimuli-activated probes actualize signal enrichments or changes through morphological transformations during the response processes, such as self-assembly/disassembly or tandem self-assembly of nanostructures. For example, 68Ga- or 18F-labeled small-molecule radiotracers, which were subjected to stimuli-instructed self-assembly to form nanoparticles and retained inside tumor cells, achieved enhanced PET imaging of tumors.19,20 Self-assembly/disassembly of fluorophores characterized by aggregation-caused quenching (ACQ) or aggregation-induced emission (AIE) effect could turn “on/off” signals for activated fluorescence imaging.21,22 Wang and co-workers verified that cyanine dyes that formed an undefined structure in a loose column showed excellent fluorescence properties, while H-aggregated cyanine dyes formed in a P helical column tended to enhance the photothermal conversion efficiency of the dyes.23 A variety of systems have been developed to control the assembly/disassembly of imaging motifs.24–26 Peptides, derived from natural proteins or artificial manufacture, can self-assemble to form multiple supramolecular structures including nanoparticles, nanofibers, and nanotubes through non-covalent interactions such as hydrogen bonding, hydrophobic interactions, aromatic interactions, and so on. In addition, physical (e.g., co-assembly) or chemical (e.g., chemical integration) modifications of peptide molecules are relatively easy to carry out, leading to controllable imaging properties.27 Considering that earlier reviews have summarized stimuli-triggered morphological transformation of peptide-based probes,28–30 in this review, we focus mainly on dual stimuli-instructed cascade self-assembly and disassembly or tandem self-assembly of peptide-based probes for molecular imaging (Fig. 1), which enables precise and sensitive imaging or efficient theranostics and provides multiple types of information about diseases. We discuss the mechanisms and potential applications of these probes and provide some challenges and perspective ideas in this field. We anticipate that this critical review will promote the generation of versatile designs for stimuli-instructed sequential morphological transformations and expand their applications to other fields.
2. Cascade self-assembly and disassembly
Normally, stimuli-instructed cascade self-assembly and disassembly of nanofibers or nanoparticles would lead to the cascade switch of signals (e.g., fluorescence, photoacoustic (PA), or 19F MRI signals). In addition, some researchers attempted to produce bioluminescence substrates to turn “on” signals through these processes. In this section, we have summarized probes that displayed dual stimuli-instructed cascade self-assembly and disassembly in Table 1 and discussed the design strategies in detail.| Probe | Initial morphologya | Stimulus #1a | Intermediate morphologya | Stimulus #2a | Final morphologya | Potential application | Ref. |
|---|---|---|---|---|---|---|---|
| a The probe presenting initial morphology was first transformed into intermediate morphology triggered by stimulus #1, which was subsequently transformed into final morphology upon the interaction with stimulus #2. | |||||||
| PEA-NBD-Yp | Molecules | ALP | Nanofibers | GSH | Molecules | Trace sequential assembly and disassembly of nanofibers in living cells | 34 |
| 1 | Molecules | GSH | Nanoparticles | Furin | Molecules | Furin imaging with enhanced sensitivity | 35 |
| P-CyPt | Molecules | ALP | Nanoparticles | GSH | Molecules | Tumor imaging and drug release monitoring | 36 |
| 2 | Molecules | GSH | Nanoparticles | FAAH | Molecules | Long-term imaging of FAAH activity | 37 |
| 3-1 | Molecules | GSH | Nanoparticles | Lgmn | Molecules | 19F MR imaging of Lgmn activity in tumors of zebrafish at a low dosage | 45 |
| 4 | Nanoaggregates | eDHFR | Probe/protein complexes | TMP | Nanoaggregates | Sense the protein activity in a reversible manner | 47 |
Optical imaging, such as fluorescence and bioluminescence, has the advantages of non-ionizing radiation, high spatiotemporal resolution, and high sensitivity.31–33 To date, sequential stimuli have been skillfully used in manipulating the aggregation state of a dye to regulate its optical performance. In 2021, Liang and co-workers reported a 7-nitro-1,2,3-benzoxadiazole (NBD)-labeled fluorescent probe PEA-NBD-Yp to trace sequential alkaline phosphatase (ALP)-triggered assembly and GSH-triggered disassembly of nanofibers in living cells.34 Upon extracellular ALP-participated dephosphorylation of the phosphate group, the nonfluorescent PEA-NBD-Yp (Off1) was converted to PEA-NBD-Y, which self-assembled into fluorescent nanofibers (On1) on the cell outer membrane. Then, the nanofibers were translocated into cells to light up the cells (On2), and the monomer in the nanofibers (i.e., PEA-NBD-Y) subsequently underwent intracellular GSH-mediated reduction of the disulfide bond to produce NBD-Y, leading to the disassembly of nanofibers to turn “off” the fluorescence (Off2) (Fig. 2a). Time-course fluorescence imaging of HeLa cells incubated with PEA-NBD-Yp provided clear indications of the Off1–On1–On2–Off2 processes (Fig. 2b), demonstrating the ability of PEA-NBD-Yp to trace sequential assembly and disassembly of nanofibers in living cells.
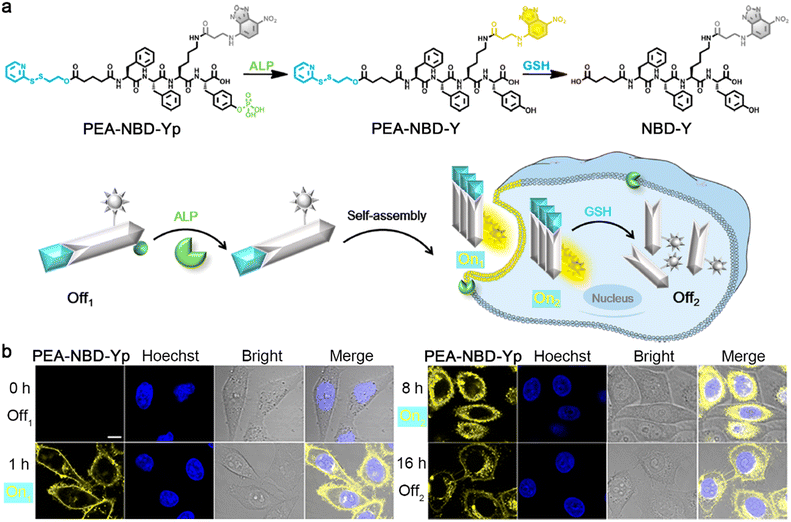 | ||
| Fig. 2 (a) Chemical structures and schematic illustration of the fluorescence changes during extracellular ALP-instructed self-assembly and intracellular GSH-responsive disassembly of PEA-NBD-Yp. (b) Fluorescence images of HeLa cells incubated with PEA-NBD-Yp for different times. Scale bar: 10 μm. Reproduced with permission from ref. 34. Copyright 2021, American Chemical Society. | ||
Besides the above-mentioned morphological transformations, sequential stimuli were also employed to control the formation and destruction of nanoparticles for molecular imaging. Previously, Liang and co-workers reported a “smart” dual quenching strategy for furin imaging with enhanced sensitivity.35 Specifically, a single quenched small-molecule precursor 1-P based on fluorescence resonance energy transfer (FRET) was developed by linking fluorescein isothiocyanate (FITC) to its dark quencher 4-((4-(dimethylamino)phenyl)azo)benzoic acid (DABCYL) through a furin cleavable peptide sequence. Upon the cleavage of furin, the fluorophore FITC was separated from its quencher to turn on the fluorescence. Furthermore, 1-P was artfully attached to a 2-cyanobenzothiazole (CBT)–cysteine (Cys) condensation system to obtain probe 1 with a single quench fluorescence property. In particular, 1 was subjected to intracellular GSH-instructed reduction of the disulfide bond to trigger CBT–Cys condensation reaction to yield a cyclic dimer (1-D), which self-assembled into dual quenched nanoparticles (1-NPs) to further lower the fluorescence intensity due to the ACQ effect. Later, a free FITC-labelled peptide sequence was released from the nanoparticles under the cleavage of furin to turn “on” the fluorescence with a high SNR because of the destruction of both FRET and ACQ effects (Fig. 3a). To support these hypotheses, 1 or 1-P was applied for fluorescence imaging of living cells. As shown in Fig. 3b, the cells coincubated with 1 and Cys(StBu)-Lys-CBT, which was used to avoid the interference of intracellular abundant cysteine to guarantee the formation of nanoparticles inside cells, exhibited a 25-fold enhancement of FITC fluorescence intensity compared to those cells preincubated with furin inhibitor II (Inh), whereas the cells treated with single quenched 1-P showed only a 4-fold enhancement of fluorescence intensity compared to the cells pretreated with furin inhibitor II. These cell imaging results confirmed the ability of this dual quenching strategy-based probe 1 to improve the SNR of fluorescence imaging.
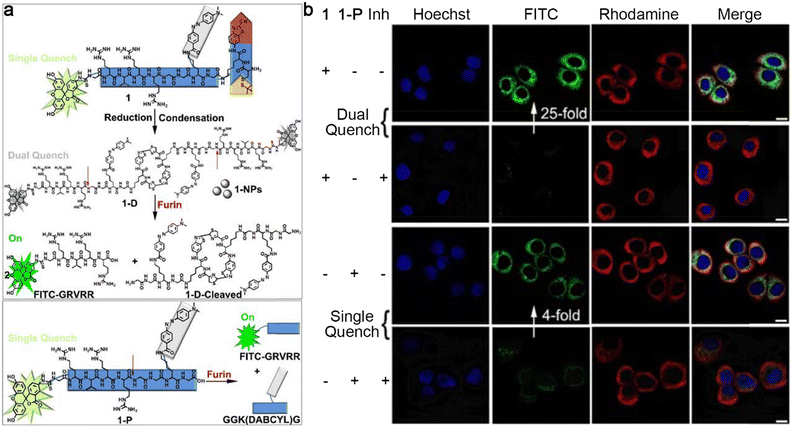 | ||
| Fig. 3 (a) Chemical structures and schematic illustrations of dual quenched small-molecule probe 1 or single quenched control probe 1-P for the imaging of furin. (b) Confocal laser scanning microscopy images of MDA-MB-468 cells treated with 1 together with Cys(StBu)-Lys-CBT (top row), pretreated with furin inhibitor II, then 1 together with Cys(StBu)-Lys-CBT (top middle row), 1-P (bottom middle row), pretreated with furin inhibitor II then 1-P (bottom row). The nuclei were stained with Hoechst 33342 (blue). Green fluorescence was from FITC in 1 or 1-P. Red fluorescence was immunofluorescence staining of furin. Scale bar: 10 μm. Reproduced with permission from ref. 35. Copyright 2018, American Chemical Society. | ||
In 2023, Ye and co-workers reported a sequential stimuli-responsive cisplatin prodrug (P-CyPt) for tumor theranostics.36 Briefly, an ALP-cleavable phosphate group caged merocyanine fluorophore was linked with a cisplatin prodrug (Pt(IV)) through a D-dipeptide (D-Phe-D-Phe) to construct P-CyPt. When incubated with tumor cells, P-CyPt was first subjected to ALP-triggered dephosphorylation to generate CyPt. During this process, the fluorescence emission at 710 nm and PA signal at 700 nm of merocyanine fluorophore were switched “on”. At the same time, CyPt was self-assembled into nanoparticles (PtIVNPs) in the physiological environment with a turned-on PA signal at 750 nm and further anchored on the cell membranes to enhance the retention and endocytosis of the prodrug. After being translocated into tumor cells, PtIVNPs were subjected to intracellular abundant GSH-triggered reduction to release anti-cancer drug cisplatin and finally disassemble into Cy-COOH. The disassembly of the nanoparticles further enhanced the NIR fluorescence intensity at 710 nm due to the elimination of the ACQ effect and turned “off” the PA signal at 750 nm (Fig. 4a). After intravenous (i.v.) injection into tumor-bearing mice, small-molecule prodrug P-CyPt could penetrate deeply into tumor tissues and be dephosphorylated by the membrane-bound ALP to generate PtIVNPs and anchor on the cell membranes, which promoted the cell uptake and prolonged the retention of the drug in the tumor region. Within tumor cells, PtIVNPs were disassembled by the reduction of endogenous GSH to release cisplatin and deplete GSH to effectively kill tumor cells (Fig. 4b). In particular, these processes were accompanied by the changes in NIR fluorescence and PA signals, permitting in vivo tumor imaging with high sensitivity and spatial-resolution, and drug release monitoring in real-time. Transmission electron microscopy (TEM) analysis clearly showed the assembly and disassembly processes of P-CyPt sequentially interacting with ALP and GSH (Fig. 4c). Subsequently, fluorescence imaging of subcutaneous HeLa tumor-bearing mice i.v. injected with P-CyPt demonstrated strong fluorescence signals at 1 h in tumor regions, which were maintained for 4 h. The tumor of the mice injected with PtIVNPs also displayed bright fluorescence at 1 h, but the signals decreased quickly (Fig. 4d). Moreover, PA signals from the tumor of P-CyPt-treated mice at 700 nm reached the maximum at 2 h and remained strong within 4 h, while the signal at 750 nm gradually declined to the baseline after it reached the maximum value at 2 h. However, the mice injected with PtIVNPs showed weak PA signals at 700 or 750 nm in tumor regions (Fig. 4e). It was inferred from these imaging results that P-CyPt could be transformed into PtIVNPs in tumor regions at 2 h, which were disassembled after 4 h. Meanwhile, P-CyPt was further applied for imaging-guided therapy of orthotopic liver tumors. On the whole, this sequential ALP- and GSH-triggered self-assembly and disassembly strategy provided an efficient drug delivery method with tumor imaging and drug release monitoring properties, which might assume the role of a general drug delivery platform for cancer theranostics.
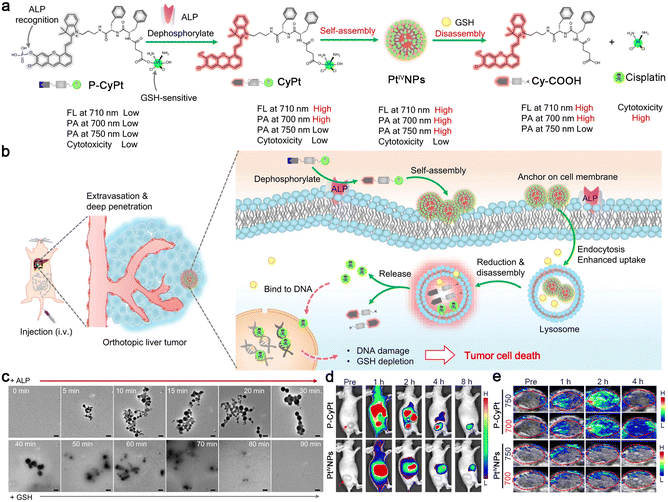 | ||
| Fig. 4 (a) Proposed mechanism of ALP-triggered self-assembly of P-CyPt to form PtIVNPs and GSH-triggered disassembly to release cisplatin for tumor therapy. The changes in NIR fluorescence and PA signals reflected the self-assembly and disassembly processes of P-CyPt, which could be used for imaging-guided precise chemotherapy of tumors. (b) Schematic illustration of the mechanism of P-CyPt with NIR fluorescence and PA dual modalities for imaging-guided therapy of tumors. (c) TEM images of P-CyPt incubated with ALP (0–30 min), followed by incubation with GSH (30–90 min). Scale bars: 200 nm. NIR fluorescence (d) and PA (e) images of HeLa tumors of the mice i.v. injected with P-CyPt or PtIVNPs for different times. Reproduced with permission from ref. 36. Copyright 2023, Springer Nature. | ||
Different from the above strategy to control the performance of a dye, Liang and co-workers developed a latent bioluminescence probe 2, which was susceptible to sequential stimuli-controlled self-assembly and disassembly to produce bioluminescence substrates for long-term imaging of fatty acid amide hydrolase (FAAH) activity.37 As illustrated in Fig. 5a, 2 was imbibed by cells and reacted with intracellular GSH to yield reduction product 2-Red. Subsequently, 2-Red underwent CBT–Cys cycloaddition to form amphiphilic cyclic dimer 2-Dimer, which immediately self-assembled into nanoparticles (2-NPs) and were retained in the cells. Upon interaction with FAAH, 2-NPs were disassembled and the bioluminescence substrates Lys-Luc or NH2-Luc were slow-released to achieve persistent bioluminescence imaging. Long-term bioluminescence imaging of mice xenografted with firefly luciferase-transfected MDA-MB-231 tumors showed that the mice intraperitoneally (i.p.) injected with 2 demonstrated bright bioluminescence signals in tumor regions for at least 16 h. However, the mice treated with FAAH inhibitor URB597 and 2 exhibited much weaker bioluminescence intensities in tumor regions, indicating that FAAH was essential to emanate persistent bioluminescence. Furthermore, the tumors of FAAH substrates AMA- and Lys-Lus- or cleavage product NH2-Luc-treated mice displayed strong bioluminescence signals but decreased quickly (Fig. 5b). These imaging results manifested the ability of 2 for persistent imaging of FAAH activity. In addition, the same group reported two different configurations of probes, which could be reduced by GSH to yield linear luciferin oligomers and self-assembled into nanoparticles, then hydrolyzed by intracellular proteases to produce NH2-Luc for bioluminescence generation.38
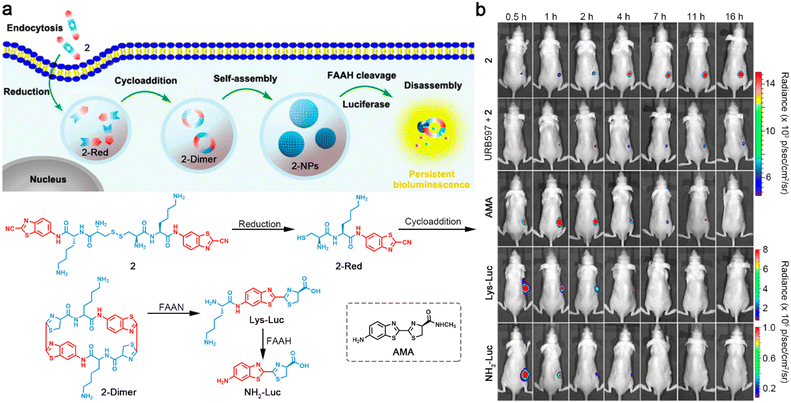 | ||
| Fig. 5 (a) Schematic illustration of intracellular reduction-induced cycloaddition and self-assembly of 2 to construct cyclic D-luciferin-based 2-NPs, and FAAH-instructed disassembly to yield Lys-Luc and NH2-Luc for persistent bioluminescence imaging of FAAH. Chemical structure of a control compound AMA. (b) Bioluminescence images of mice bearing firefly luciferase-transfected MDA-MB-231 tumors after i.p. injection of 2, URB597 with 2, AMA, Lys-Luc, or NH2-Luc for different times. Reproduced with permission from ref. 37. Copyright 2016, American Chemical Society. | ||
MRI is one of the most commonly used imaging techniques in clinics, which is characterized by noninvasive and nonradiative properties, excellent temporal and spatial resolution, and unlimited tissue penetration depth.39,40 Conventional MRI detects 1H nuclei, abundant in water and fat in the human body. There have been several 1H MRI contrast agents reported, which could self-assemble into nanostructures in target sites to enhance the imaging contrast.41,42 At the same time, 19F MRI is a promising imaging modality in various clinical applications. 19F MRI holds comparable sensitivity to 1H MRI. Endogenous 19F nuclear magnetic resonance (NMR) signal is hardly detectable in animal bodies, leading to low background signals.43 Previous studies have shown that fluorinated aggregates could broaden and attenuate 19F NMR signals, which recover on the disassembly.44–46 For example, Liang and co-workers reported a small-molecule 19F MRI contrast agent 3-1 and a control probe 3-2.45 As demonstrated in Fig. 6a, after entering cells, probe 3-1 was reduced by plentiful intracellular GSH to trigger the CBT–Cys condensation reaction and self-assemble into nanoparticles (3-1-NPs) with switched-off 19F NMR signals. Soon afterward, 3-1-NPs were disassembled by legumain (Lgmn) to turn the 19F NMR signals “on”. On the other hand, the control probe 3-2 could self-assemble into nanoaggregates (3-2-NPs) inside cells upon the interaction with GSH and Lgmn for 19F MR “turn-off” imaging. TEM images of 3-1 treated with GSH and Lgmn sequentially clearly depicted the assembly and disassembly of the nanoparticles, while the same treatment of 3-2 resulted only in the formation of the nanoparticles (Fig. 6b). Additionally, strong 19F MRI signals were observed from the tumor regions of the zebrafish injected with 3-1. In contrast, no obvious 19F MRI signals could be detected from healthy zebrafish receiving an injection of the same dose of 3-1. Moreover, a much higher dose of 3-2 was required to light up the whole bodies of healthy zebrafish or nontumor sites of tumor-bearing zebrafish (Fig. 6c). This “smart” cascade self-assembly and disassembly strategy made it possible for 19F MR imaging of Lgmn activity in tumors of zebrafish at a low dosage.
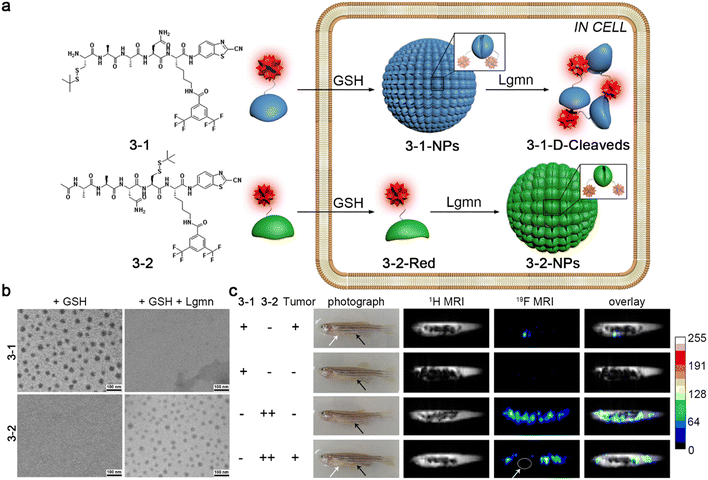 | ||
| Fig. 6 (a) Chemical structures of probe 3-1 and 3-2. Schematic illustration of GSH- and Lgmn-instructed sequential self-assembly and disassembly of probe 3-1 with “off” and “on” 19F NMR signals and GSH- and Lgmn-controlled self-assembly of probe 3-2 to turn “off” 19F NMR signals. (b) TEM images of probe 3-1 or 3-2 treated with GSH or GSH and Lgmn sequentially. (c) 1H MR and 19F MR images of HEK 293T tumor-bearing zebrafish or healthy zebrafish injected with probe 3-1 or 3-2. The white or black arrows showed the tumor sites or the injection sites, respectively. Reproduced with permission from ref. 45. Copyright 2015, American Chemical Society. | ||
Unlike the abovementioned small-molecule probes, which first self-assembled into nanostructures and then disassembled to release free imaging motifs for imaging, Hamachi and co-workers reported unique protein-responsive nanoaggregates with reversible self-assembly/disassembly properties for intracellular protein sensing (Fig. 7a).47 Briefly, this reported probe 4 was constituted by a folate receptor or dihydrofolate reductase (eDHFR) targeting ligand methotrexate (MTX) and a phenylalanine (Phe)-decorated tetramethylrhodamine (TMR) fluorophore connected by a linker (Fig. 7b). In the physiological environment, 4 could self-assemble into spherical or oval less fluorescent nanoaggregates. After entering cells through endocytosis, the nanoaggregates could bind to the target protein selectively and reversibly, turning its fluorescence “on” and then “off” (Fig. 7a). When probe 4 was incubated with the green fluorescent protein (GFP)-fused eDHFR stably overexpressing HeLa cells (HeLa-DG cells), strong red fluorescence from TMR in 4 was observed and colocalized well with the green fluorescence from the GFP without washing, indicating that probe 4 could efficiently bind to eDHFR and turn “on” the fluorescence (Fig. 7c). The follow-up addition of the eDHFR inhibitor trimethoprim (TMP) resulted in an acute decrease of the TMR fluorescence intensity inside cells (Fig. 7d), reflecting the ability of probe 4 to sense the protein activity reversibly.
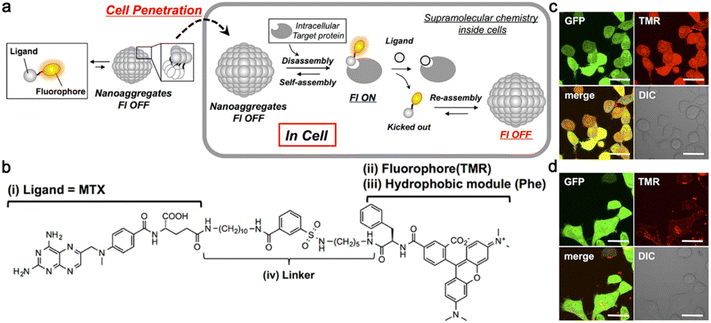 | ||
| Fig. 7 (a) Schematic illustration of reversible self-assembly/disassembly nanoaggregates for intracellular protein sensing. (b) Chemical structure of probe 4 for eDHFR imaging. Probe 4 contained four parts: (i) a folate receptor or eDHFR targeting ligand MTX, (ii) a fluorophore TMR, (iii) a hydrophobic module Phe, and (iv) a linker. Confocal laser scanning microscopy images of HeLa-DG cells treated with 4 for 12 h (c) and then treated with TMP (d) without any washing. Scale bar: 20 μm. Reproduced with permission from ref. 47. Copyright 2014, American Chemical Society. | ||
3. Tandem self-assembly
Probes with stimuli-instructed tandem self-assembly performance usually exhibit a molecule-to-nanoparticle-to-nanofiber transition with controllable fluorescence signals. Besides, other morphological transformations, including nanoparticle-to-nanofiber-to-nanofiber and nanoparticle-to-nanoparticle-to-nanofiber transitions are also employed to regulate fluorescence or PA signals. The following section will focus on the nanostructure transformations of probes and their potential biomedical applications (Table 2).| Probe | Initial morphologya | Stimulus #1a | Intermediate morphologya | Stimulus #2a | Final morphologya | Potential application | Ref. |
|---|---|---|---|---|---|---|---|
| a The probe presenting initial morphology was first transformed into intermediate morphology triggered by stimulus #1, which was subsequently transformed into final morphology upon the interaction with stimulus #2. | |||||||
| TPE-1p | Molecules | ALP | Nanoparticles | ChT-L | Nanofibers | Monitor ChT-L | 48 |
| 5-1 | Molecules | ALP | Nanoparticles | GSH | Nanofibers | Cancer diagnostics and treatment | 49 |
| 6-1 | Molecules | ALP | Nanofibers | Reductase | Nanofibers | Selectively inhibit lung cancer cells | 50 |
| DEVD-DLPA@C3 | Nanoparticles | Caspase-3 | Moniliform nanofibers | mtROS | Smooth nanofibers | Self-amplified cooperative antitumor therapy | 51 |
| PKK-S-PEG | Nanoparticles | GSH | Nanoparticles | Laser irradiation | Nanofibers | Accelerate the formation of nanomaterials at tumor sites | 53 |
In 2023, Zhong and co-workers developed a cascade-activated probe to monitor the chymotrypsin-like (ChT-L) activity of proteasomes.48 As depicted in Fig. 8a, the AIE luminogen (AIEgen)–peptide conjugate TPE-1p was composed of three parts: an AIEgen tetraphenyl ethylene (TPE) used as the imaging motif; a Tyr(H2PO3)-Tyr dipeptide as a substrate for ALP, and its dephosphorylation product Tyr-Tyr could be recognized by ChT-L; an Asp-Asp dipeptide to improve the solubility of the probe in the physiological environment. The presence of the phosphate group could effectively avoid the direct recognition of ChT-L toward TPE-1p, ensuring the morphological transformation process for ChT-L activity of proteasome monitoring with a high SNR. TPE-1p emitted a weak fluorescence in the physiological environment. When incubated with ALP- and ChT-L-overexpressing cancer cells, TPE-1p was first dephosphorylated by membrane-bound ALP to yield TPE-1, which tended to partially assemble into nanoparticles with slightly increased fluorescence emission. After entering cells, TPE-1 was degraded by ChT-L to produce TPE-Y and reassembled into nanofibers, greatly improving the fluorescence intensity (Fig. 8b). Fluorescence images of ALP- and ChT-L-overexpressing HeLa cells incubated with TPE-1p showed bright blue fluorescence, while the pretreatment of ALP inhibitor L-Phe and ChT-L inhibitor bortezomib (Btz) would significantly weaken the fluorescence. Interestingly, pretreatment with L-Phe only also led to a similar degree of fluorescence decrease to both ALP and ChT-L inhibited cells, yet Btz-pretreated cells still exhibited a weak fluorescence, indicating that the fluorescence of TPE-1p was sequentially activated by ALP and ChT-L (Fig. 8c).
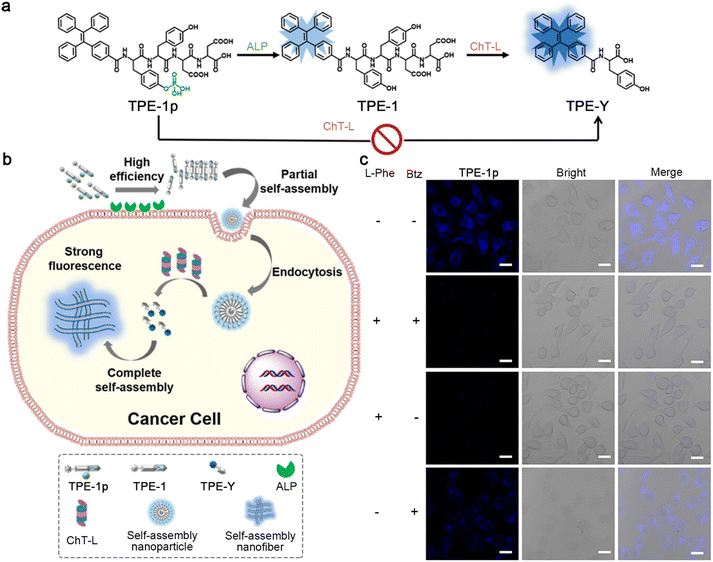 | ||
| Fig. 8 (a) Chemical structures of TPE-1p, its ALP hydrolysis product TPE-1, and the subsequent ChT-L cleavage product TPE-Y. ChT-L cannot recognize the cleavage site in TPE-1p directly to produce TPE-Y. (b) Schematic illustration of cascade activation of TPE-1p for the monitoring of the ChT-L activity of proteasomes. (c) Fluorescence images of HeLa cells preincubated with (or without) L-Phe and/or Btz and then with TPE-1p. Scale bar: 20 μm. Reproduced with permission from ref. 48. Copyright 2018, American Chemical Society. | ||
In addition, Yang and co-workers reported a tandem molecular self-assembly system, which might improve performance in cancer diagnostics and treatment.49 In this system, a tumor-targeting peptide derivative 5-1 was designed, which was first responsive to extracellular ALP to yield 5-2 and then to intracellular GSH to produce 5-3 (Fig. 9a). When incubated with ALP- and GSH-overexpressing liver cancer cells (e.g., HepG2 and QGY7703), 5-1 was dephosphorylated to form nanoparticles outside cells, promoting cell uptake through endocytosis. After targeting and entering the cells, the nanoparticles were reduced to transform into nanofibers, which prolonged the retention time inside cells (Fig. 9b). The nanofibers formed inside cells might alter the viscosity of cancer cells or interact with important intracellular proteins, inducing cancer cell death. Certainly, yellow fluorescent dots were observed around HepG2 cells incubated with 5-1 for 0.5 h. When extending the incubation time to 4 h, yellow fluorescent fibers inside the cells were observed, clearly indicating the nanoparticle-to-nanofiber transformation. Dense nanofibers in QGY7703 cells were also observed when incubated with 5-1 for 4 h (Fig. 9c).
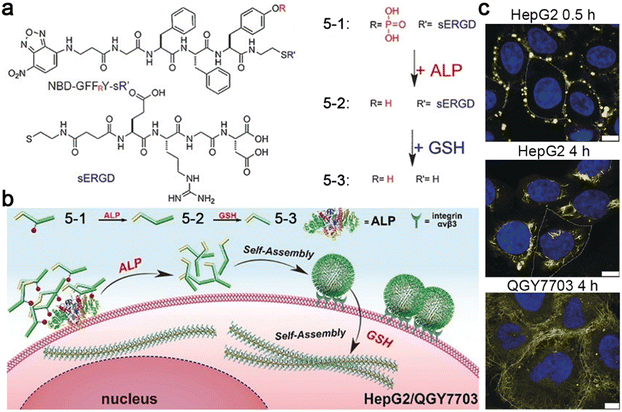 | ||
| Fig. 9 (a) Chemical structures of 5-1, 5-2, and 5-3. (b) Proposed mechanism of ALP- and GSH-controlled tandem self-assembly of 5-1 incubated with liver cancer cells. (c) Confocal laser scanning microscopy images of HepG2 or QGY7703 cells treated with 5-1 for different times. The nuclei were stained with Hoechst 33342 (blue). Yellow fluorescence was from NBD in 5-1. The cell boundary is delineated using a white dashed line. Scale bar: 10 μm. Reproduced with permission from ref. 49. Copyright 2018, Wiley-VCH. | ||
Later, the same group developed a small-molecule probe 6-1 based on ALP- and reductase-controlled tandem self-assembly to selectively inhibit lung cancer cells.50 As illustrated in Fig. 10a, 6-1 was first dephosphorylated by membrane-bound ALP to produce 6-2 and self-assembled into nanofibers. The as-formed nanofibers then interacted with αvβ3 integrin on the cell membranes and were endocytosed by lung cancer cells. The azo group in 6-2 could accelerate lysosomal escape and mitochondrial accumulation, and was reduced by reductase in the mitochondrial membrane to yield 6-3, which re-assembled into nanofibers in the mitochondrial membrane. The in situ formation of the nanofibers would disrupt the mitochondrial membrane to release cytochrome c (Cyt C) and produce ROS, which increased the endoplasmic reticulum (ER) stress and activated the unfolded protein response (UPR), resulting in cancer cell death. Confocal laser scanning microscopy images of A549 lung cancer cells incubated with 6-1 showed that the NBD peptide first gathered in the lysosomes at the 1 h time point and escaped from the lysosomes within 4 h and accumulated in the mitochondria at the 8 h time point (Fig. 10b).
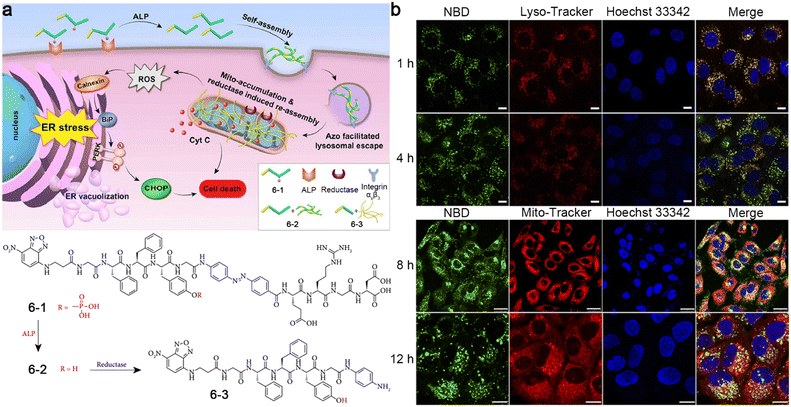 | ||
| Fig. 10 (a) Proposed mechanism of ALP- and reductase-controlled tandem self-assembly of 6-1 to induce ER stress, leading to cell death. Chemical structures of 6-1, 6-2, and 6-3. (b) Confocal laser scanning microscopy images of A549 cells incubated with 6-1 for different times. Green fluorescence was from NBD in 6-1. The lysosomes or mitochondria were stained with Lyso-Tracker at 1 h and 4 h, or Mito-Tracker at 8 h and 12 h, respectively (red). The nuclei were stained with Hoechst 33342 (blue). Scale bar: 25 μm in 1 h, 4 h, and 8 h time points, 10 μm in 12 h time point. Reproduced with permission from ref. 50. Copyright 2019, Exclusive Licensee Science and Technology Review Publishing House. | ||
In 2022, Zhu and co-workers reported a sequential stimuli-instructed nanoparticle-to-nanofiber transition and the following polymerization inside cells for self-amplified cooperative antitumor therapy.51 A diacetylene-decorated lipidated peptide amphiphile (DEVD-DLPA) was developed, which could target integrin and be cleaved by caspase-3. Then, a mitochondria-targeting photosensitizer (C3) was encapsulated by DEVD-DLPA to construct DEVD-DLPA@C3 nanoparticles (Fig. 11a). Generally, DEVD-DLPA@C3 nanoparticles could transform into moniliform nanofibers by the cleavage of caspase-3 to release free C3. In the presence of ROS, the diacetylene groups in the nanofibers were crosslinked to yield polymerized diacetylene (PDA) emitting red fluorescence,52 leading to the reassembly of the nanofiber to form dense nanofibers (smooth fibers) (Fig. 11b). As originally conceived, DEVD-DLPA@C3 nanoparticles could be efficiently internalized by tumor cells through actively targeting integrin for photodynamic therapy (PDT). Upon laser irradiation, ROS were generated to induce the apoptosis of the tumor cells and the activation of caspase-3. In turn, the nanoparticles could transform into nanofibers by the activated caspase-3, and free C3 was released and retained on the mitochondrial membrane. Consequently, another laser irradiation further spawned mitochondrial dysfunction and mitochondrial ROS (mtROS) burst, triggering in situ polymerization for self-amplified circulation of mtROS to improve therapeutic efficacy significantly (Fig. 11c). To study the intracellular polymerization of the diacetylene groups, confocal laser scanning microscopy was employed to monitor the PDA signals. As shown in Fig. 11d, bright red fluorescence was observed from 4T1 cells treated with DEVD-DLPA@C3 nanoparticles plus twice laser irradiation, while no obvious fluorescence signal from PDA was detected when cells were treated with DEVD-DLPA@C3 without laser irradiation. In the control experiments, no red fluorescence or very weak fluorescence was observed from the cells treated with DEVD-C12@C3 nanoparticles (without diacetylene groups), EEVE-DLPA@C3 nanoparticles (cannot be recognized by caspase-3), DEVD-DLPA nanoparticles (without photosensitizer C3) or EEVE-DLPA nanoparticles, suggesting that polymerization of the nanofibers could be initiated mainly by ROS produced during the PDT process.
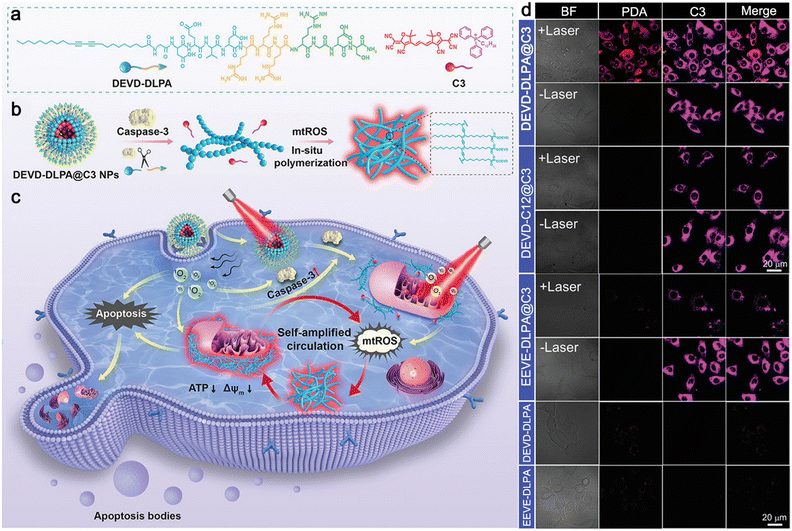 | ||
| Fig. 11 (a) Chemical structures of DEVD-DLPA and C3. (b) Proposed mechanism of caspase-3-instructed nanoparticle-to-nanofiber morphological transformation and mtROS-instructed in situ polymerization of DEVD-DLPA@C3 nanoparticles. (c) Schematic illustration of PDT-induced intracellular polymerization for self-amplified cooperative antitumor therapy. (d) Confocal laser scanning microscopy images of 4T1 cells incubated with different nanoparticles and irradiated with 655 nm laser for 5 min twice or not. Red fluorescence was from PDA formed in the dense nanofibers. Pink fluorescence was from C3 in the nanoparticles. Reproduced with permission from ref. 51. Copyright 2022, Wiley-VCH. | ||
Additionally, Wang and co-workers reported a photothermal-promoted morphology transformation (PMT) strategy to promote the accretion of drugs at tumor sites.53 To be specific, a polymer–peptide conjugate PKK-S-PEG was constructed of four parts: a cytotoxic peptide [KLAKLAK]2 to target and destroy mitochondria, a hydrogen-bond unit KLVFF originated from amyloid β-protein, a hydrophilic polyethylene glycol (mPEG) linked by a disulfide bond, and photothermal/PA molecule purpurin-18 (P18). In the physiological environment, PKK-S-PEG could self-assemble into nanoparticles, where PEG was the key to maintain its nanostructure. After the internalization of the nanoparticles through endocytosis, the disulfide bonds of PKK-S-PEG were reduced by intracellular GSH, and the PEG shell was stripped to produce PKK nanoparticles with minor morphology changes. Driven by the hydrogen bonding of KLVFF, the nanoparticles were slowly transformed into nanofibers on the mitochondrial membrane for tumor therapy, accompanied by the red shift of the PA signal. Interestingly, this morphology transformation could be accelerated by the risen temperature generated from P18 under 655 nm laser irradiation, which lowered the Gibbs free energy of the reassembly process, thus improving the tumor therapeutic effect (Fig. 12a). TEM imaging of PKK nanoparticles in phosphate-buffered saline (PBS) showed that it took 240 min for the nanoparticles to completely transform into nanofibers spontaneously, while only 60 min was needed when the solution was irradiated with a 655 nm laser to raise the temperature to 42 °C (Fig. 12b). PA imaging of tumors further confirm the feasibility of the PMT strategy. HeLa tumor-bearing mice were i.v. injected with PKK-S-PEG and the tumor sites were irradiated (or not) with a 655 nm laser every two hours six times post-injection. PA images are shown in Fig. 12c. Compared with the tumors without laser irradiation, the PA signals of tumors from the mice treated with PKK-S-PEG and laser irradiation at 730 nm were significantly increased, while those at 685 nm were weakened, suggesting that laser irradiation could accelerate the nanoparticle-to-nanofiber transition process.
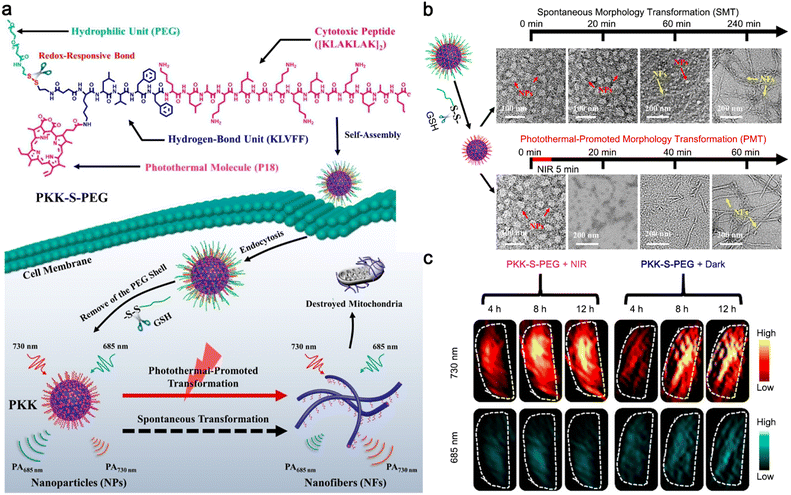 | ||
| Fig. 12 (a) Schematic illustration of photothermal-promoted nanoparticle-to-nanofiber transformation to destroy mitochondria in tumor cells, along with the red-shift of PA signal. (b) Time-course TEM images of PKK-S-PEG incubated with GSH and then irradiated with 655 nm laser or not. (c) PA imaging at 730 and 685 nm of tumors from mice treated with PKK-S-PEG and then irradiated with 655 nm laser or not. Reproduced with permission from ref. 53. Copyright 2020, American Chemical Society. | ||
4. Conclusions and perspective
In this review, we summarized the stimuli-instructed sequential morphological transformations of probes for molecular imaging. Dual stimuli-instructed cascade self-assembly and disassembly or tandem self-assembly was discussed in detail, including dual stimuli-instructed molecule-to-nanofiber/nanoparticle-to-molecule transition, molecule-to-nanoparticle-to-nanofiber transition and so on, inducing the fluorescence, PA, bioluminescence or 19F MRI signal changes for precise and sensitive imaging or efficient theranostics of diseases. Stimuli-instructed cascade self-assembly and disassembly of probes might improve the SNR35 and reduce the dosage for imaging.45 In addition, the disassembly of nanoaggregates might contribute to the elimination of probes in living bodies. However, the rapid elimination of imaging motifs might lower the signal intensities, interfering with the outputs caused by the disassembly of probes in turn. As for tandem self-assembly strategies, they might be advantageous in the development of theranostic systems.49–51 However, the imaging motifs have fewer options because they are required to show switchable imaging properties along with aggregation states (e.g., nanoparticles or nanofibers). Choosing appropriate self-assembly motifs, stimuli, and imaging motifs to construct probes is important for the successful demonstration of dual stimuli-instructed sequential morphological transformations in the physiological environment with switchable imaging signals. Despite some encouraging results that have been obtained in this field, large challenges remain to be addressed before clinical translation in the future.First of all, the imaging modalities adopted in this field are confined to fluorescence, PA, bioluminescence, and 19F MRI due to the lack of versatile imaging motifs with controllable signals changed with the morphology of probes, which greatly limits the development of this field. For example, to the best of our knowledge, stimuli-instructed sequential morphological transformations for PET imaging were barely reported. The possible reason might be that PET signals are almost unchanged whether the radiotracers are assembled into (or disassembled from) nanostructures. Fluorescence and PA seem to be the most promising modalities to fabricate sequential stimuli-responsive probes. Some dyes packed in different states displayed distinct fluorescence or PA signals.23,53 Previous studies have shown that a few fluorophores (e.g., Cy5.5) fabricated into nanostructures exhibited a stimulus-responsive disassembly to turn “on” fluorescence in vivo.21,54 However, the intracellular formation of Cy5.5 nanoparticles still emitted bright fluorescence from cells.55,56 These phenomena might be caused by the loose packing of Cy5.5 in the nanoparticles, which brought about little fluorescence quenching due to the ACQ effect and mass fluorophores retained inside cells to light up the cells. Thus, these fluorophores are not suitable for constructing cascade self-assembly and disassembly strategy-based probes because of the subtle changes in imaging signals. Fluorophores with red-shifted or blue-shifted fluorescence emission in the aggregate state (e.g., coumarin and pyrene)57–59 might be utilized as substitutes. However, their emission wavelengths are generally located in the visible region, which might suffer from poor depth penetration and low SNR for in vivo imaging. Another promising alternative is Fe3O4 nanoparticles, demonstrating aggregation-dependent interlocked variations of longitudinal and transversal relaxivities (R1 and R2) for MR imaging,60 which remain to be explored.
Furthermore, spatially–temporally controllable stimuli responses are the key to realize sequential morphological transformations, which are still confronted with challenges. Normally, spatially controllable stimuli-responsive probes exploit the difference in cell environments. For example, the combination of membrane-bound ALP and intracellular GSH is a common strategy to trigger sequential morphological transformations.34,36,49 However, skilled designs of the probes are required to restrict internalization before ALP cleavage. As for temporally controllable probes, which usually utilize the reaction rates between stimuli and probes, it is impossible to eliminate reactions at the same time. A possible solution to this problem is to trigger the upregulation of some molecules (e.g., ROS)51 or biomarkers (e.g., caspase-3)61 through the probe itself, to induce the following morphological transformation of the probe in turn. However, attention should be attached to the interference from endogenous ROS and the time window of caspase-3 activation. In addition, the employment of exogenous stimuli (e.g., ultrasound, light, heat, magnetic field) to trigger the morphological transformations is a brilliant settlement to achieve spatial–temporal control. The ingenious design of the molecule to control the reaction sequence is also a good method to solve this problem.62
Last but not least, the uncontrollability of the morphological transformations in the complex environment of living bodies63,64 might be a huge obstacle for clinical translations. The selection of stable self-assembly motifs and selectively responsive motifs with strong anti-interference to ensure the nanostructure formation should be made to address this concern. Moreover, the development of powerful tools such as bio-TEM to visualize the morphological transformations inside cells might be helpful in screening potential probes.
In summary, stimuli-instructed sequential morphological transformations hold great promise for precise and sensitive imaging or efficient theranostics of diseases, and great efforts are still needed to develop other systems to solve the existing problems in this field. We anticipate that this review will provide useful information on stimuli-instructed sequential morphological transformations and expand their applications to other fields.
Abbreviations
| PET | Positron emission tomography |
| CT | Computed tomography |
| MRI | Magnetic resonance imaging |
| SNR | Signal-to-noise ratio |
| GSH | Glutathione |
| ROS | Reactive oxygen species |
| ACQ | Aggregation-caused quenching |
| AIE | Aggregation-induced emission |
| PA | Photoacoustic |
| NBD | 7-Nitro-1,2,3-benzoxadiazole |
| ALP | Alkaline phosphatase |
| FRET | Fluorescence resonance energy transfer |
| FITC | Fluorescein isothiocyanate |
| DABCYL | 4-((4-(Dimethylamino)phenyl)azo)benzoic acid |
| CBT | 2-Cyanobenzothiazole |
| Cys | Cysteine |
| i.v. injection | Intravenous injection |
| TEM | Transmission electron microscopy |
| FAAH | Fatty acid amide hydrolase |
| i.p. injection | Intraperitoneal injection |
| NMR | Nuclear magnetic resonance |
| Lgmn | Legumain |
| eDHFR | Dihydrofolate reductase |
| MTX | Methotrexate |
| TMR | Tetramethylrhodamine |
| GFP | Green fluorescent protein |
| TMP | Trimethoprim |
| ChT-L | Chymotrypsin-like |
| AIEgens | AIE luminogens |
| TPE | Tetraphenyl ethylene |
| Btz | Bortezomib |
| Cyt C | Cytochrome c |
| ER | Endoplasmic reticulum |
| UPR | Unfolded protein response |
| PDA | Polymerized diacetylene |
| PDT | Photodynamic therapy |
| mtROS | Mitochondrial ROS |
| PMT | Photothermal-promoted morphology transformation |
| mPEG | Polyethylene glycol |
| P18 | Purpurin-18 |
| PBS | Phosphate-buffered saline |
| R 1 | Longitudinal relaxivity |
| R 2 | Transversal relaxivity |
Author contributions
The manuscript was written through the contributions of all authors. All authors have approved the final version of the manuscript. Peiyao Chen: conceptualization, investigation, writing – original draft, review & editing, funding acquisition. Liling Meng: investigation, writing – original draft. Tuotuo Zhang: writing – review & editing. Yao Sun: supervision, conceptualization, writing – review & editing.Conflicts of interest
There are no conflicts to declare.Acknowledgements
This work was supported by the National Natural Science Foundation of China (Grant 22304049).References
- R. Weissleder and U. Mahmood, Radiology, 2001, 219, 316–333 CrossRef CAS PubMed.
- J. K. Willmann, N. van Bruggen, L. M. Dinkelborg and S. S. Gambhir, Nat. Rev. Drug Discovery, 2008, 7, 591–607 CrossRef CAS PubMed.
- S. Achilefu, Chem. Rev., 2010, 110, 2575–2578 CrossRef CAS PubMed.
- P. Cheng and K. Pu, Nat. Rev. Mater., 2021, 6, 1095–1113 CrossRef CAS.
- R. Weissleder and M. J. Pittet, Nature, 2008, 452, 580–589 CrossRef CAS PubMed.
- M. Baker, Nature, 2010, 463, 977–979 CrossRef CAS PubMed.
- H. Kobayashi, M. R. Longmire, M. Ogawa and P. L. Choyke, Chem. Soc. Rev., 2011, 40, 4626–4648 RSC.
- Z. Zhang, P. Chen and Y. Sun, Molecules, 2023, 28, 5360 CrossRef CAS PubMed.
- J. Li, F. Cheng, H. Huang, L. Li and J.-J. Zhu, Chem. Soc. Rev., 2015, 44, 7855–7880 RSC.
- Y. Wang, Y. Hu and D. Ye, Angew. Chem., Int. Ed., 2022, 61, e202209512 CrossRef CAS PubMed.
- X. Wu, R. Wang, N. Kwon, H. Ma and J. Yoon, Chem. Soc. Rev., 2022, 51, 450–463 RSC.
- Y. Lu, A. A. Aimetti, R. Langer and Z. Gu, Nat. Rev. Mater., 2016, 2, 16075 CrossRef.
- Z. Liu, G. Liang and W. Zhan, Chem. Res. Chin. Univ., 2021, 37, 889–899 CrossRef CAS.
- I. Takashima, R. Kawagoe, I. Hamachi and A. Ojida, Chem. – Eur. J., 2015, 21, 2038–2044 CrossRef CAS PubMed.
- K. Ren, Y. Liu, J. Wu, Y. Zhang, J. Zhu, M. Yang and H. Ju, Nat. Commun., 2016, 7, 13580 CrossRef CAS PubMed.
- M. P. Nikitin, V. O. Shipunova, S. M. Deyev and P. I. Nikitin, Nat. Nanotechnol., 2014, 9, 716–722 CrossRef CAS PubMed.
- M. You, L. Peng, N. Shao, L. Zhang, L. Qiu, C. Cui and W. Tan, J. Am. Chem. Soc., 2014, 136, 1256–1259 CrossRef CAS PubMed.
- Y. Tang, Y. Li, X. Hu, H. Zhao, Y. Ji, L. Chen, W. Hu, W. Zhang, X. Li, X. Lu, W. Huang and Q. Fan, Adv. Mater., 2018, 30, 1801140 CrossRef PubMed.
- P. Chen, H. Wang, H. Wu, P. Zou, C. Wang, X. Liu, Y. Pan, Y. Liu and G. Liang, Anal. Chem., 2021, 93, 6329–6334 CrossRef CAS PubMed.
- J. Lin, D. Gao, S. Wang, G. Lv, X. Wang, C. Lu, Y. Peng and L. Qiu, J. Am. Chem. Soc., 2022, 144, 7667–7675 CrossRef CAS PubMed.
- L. Xu, N. Liu, W. Zhan, Y. Deng, Z. Chen, X. Liu, G. Gao, Q. Chen, Z. Liu and G. Liang, ACS Nano, 2022, 16, 19328–19334 CrossRef CAS PubMed.
- Z. Gao, H. Gao, D. Zheng, T. Xu, Y. Chen, C. Liang, L. Wang, D. Ding and Z. Yang, Sci. China: Chem., 2020, 63, 398–403 CrossRef CAS.
- R. Zheng, J. Yang, M. Mamuti, D.-Y. Hou, H.-W. An, Y. Zhao and H. Wang, Angew. Chem., Int. Ed., 2021, 60, 7809–7819 CrossRef CAS PubMed.
- S. Yang, Y. Wang, Q. Wang, F. Li and D. Ling, Chem. Biomed. Imaging, 2023, 1, 340–355 CrossRef CAS PubMed.
- S. Wang, K. Zhou, X. Lyu, H. Li, Z. Qiu, Z. Zhao and B. Z. Tang, Chem. Biomed. Imaging, 2023, 1, 509–521 CrossRef CAS.
- J. Gao, J. Zhan and Z. Yang, Adv. Mater., 2020, 32, 1805798 CrossRef CAS PubMed.
- J. Zhang, Y. Wang, B. J. Rodriguez, R. Yang, B. Yu, D. Mei, J. Li, K. Tao and E. Gazit, Chem. Soc. Rev., 2022, 51, 6936–6947 RSC.
- Q. Jiang, X. Liu, G. Liang and X. Sun, Nanoscale, 2021, 13, 15142–15150 RSC.
- W. Huang, Y. Yang and D. Ye, Chin. J. Chem., 2023, 41, 2382–2399 CrossRef CAS.
- Y. Xu, Q. Jiang, X. Sun and G. Liang, Analysis Sensing, 2024, 4, e202300039 CrossRef CAS.
- Y. Jiang and K. Pu, Chem. Rev., 2021, 121, 13086–13131 CrossRef CAS PubMed.
- X. Huang, J. Song, B. C. Yung, X. Huang, Y. Xiong and X. Chen, Chem. Soc. Rev., 2018, 47, 2873–2920 RSC.
- Y. Duan Kenry and B. Liu, Adv. Mater., 2018, 30, 1802394 CrossRef PubMed.
- M. Zhang, C. Wang, C. Yang, H. Wu, H. Xu and G. Liang, Anal. Chem., 2021, 93, 5665–5669 CrossRef CAS PubMed.
- Z. Hai, J. Wu, D. Saimi, Y. Ni, R. Zhou and G. Liang, Anal. Chem., 2018, 90, 1520–1524 CrossRef CAS PubMed.
- X. Wen, R. Zhang, Y. Hu, L. Wu, H. Bai, D. Song, Y. Wang, R. An, J. Weng, S. Zhang, R. Wang, L. Qiu, J. Lin, G. Gao, H. Liu, Z. Guo and D. Ye, Nat. Commun., 2023, 14, 800 CrossRef CAS PubMed.
- Y. Yuan, F. Wang, W. Tang, Z. Ding, L. Wang, L. Liang, Z. Zheng, H. Zhang and G. Liang, ACS Nano, 2016, 10, 7147–7153 CrossRef CAS PubMed.
- Z. Zheng, G. Li, C. Wu, M. Zhang, Y. Zhao and G. Liang, Chem. Commun., 2017, 53, 3567–3570 RSC.
- Z. Zhou, L. Yang, J. Gao and X. Chen, Adv. Mater., 2019, 31, 1804567 CrossRef PubMed.
- N. Vogt, Nat. Methods, 2017, 14, 111 CrossRef CAS.
- H.-D. Xu, X. Cheng, X. Sun, P. Chen, W. Zhan, X. Liu, X. Wang, B. Hu and G. Liang, Nano Lett., 2023, 23, 6178–6183 CrossRef CAS PubMed.
- L. Dong, J. Qian, Z. Hai, J. Xu, W. Du, K. Zhong and G. Liang, Anal. Chem., 2017, 89, 6922–6925 CrossRef CAS PubMed.
- I. Tirotta, V. Dichiarante, C. Pigliacelli, G. Cavallo, G. Terraneo, F. B. Bombelli, P. Metrangolo and G. Resnati, Chem. Rev., 2015, 115, 1106–1129 CrossRef CAS PubMed.
- Y. Takaoka, T. Sakamoto, S. Tsukiji, M. Narazaki, T. Matsuda, H. Tochio, M. Shirakawa and I. Hamachi, Nat. Chem., 2009, 1, 557–561 CrossRef CAS PubMed.
- Y. Yuan, S. Ge, H. Sun, X. Dong, H. Zhao, L. An, J. Zhang, J. Wang, B. Hu and G. Liang, ACS Nano, 2015, 9, 5117–5124 CrossRef CAS PubMed.
- Z. Zheng, H. Sun, C. Hu, G. Li, X. Liu, P. Chen, Y. Cui, J. Liu, J. Wang and G. Liang, Anal. Chem., 2016, 88, 3363–3368 CrossRef CAS PubMed.
- T. Yoshii, K. Mizusawa, Y. Takaoka and I. Hamachi, J. Am. Chem. Soc., 2014, 136, 16635–16642 CrossRef CAS PubMed.
- Q. Jiao, Y. Zheng, S. Pei, X. Luo, X. Wu, K. Xu and W. Zhong, Anal. Chem., 2023, 95, 9097–9106 CrossRef CAS PubMed.
- J. Zhan, Y. Cai, S. He, L. Wang and Z. Yang, Angew. Chem., Int. Ed., 2018, 57, 1813–1816 CrossRef CAS PubMed.
- D. Zheng, Y. Chen, S. Ai, R. Zhang, Z. Gao, C. Liang, L. Cao, Y. Chen, Z. Hong, Y. Shi, L. Wang, X. Li and Z. Yang, Research, 2019, 2019, 4803624 CAS.
- T. Ma, R. Chen, N. Lv, Y. Li, Z.-R. Yang, H. Qin, Z. a. Li, H. Jiang and J. Zhu, Small, 2022, 18, 2204759 CrossRef CAS PubMed.
- H. Jiang, X.-Y. Hu, S. Schlesiger, M. Li, E. Zellermann, S. K. Knauer and C. Schmuck, Angew. Chem., Int. Ed., 2017, 56, 14526–14530 CrossRef CAS PubMed.
- X.-H. Zhang, D.-B. Cheng, L. Ji, H.-W. An, D. Wang, Z.-X. Yang, H. Chen, Z.-Y. Qiao and H. Wang, Nano Lett., 2020, 20, 1286–1295 CrossRef CAS PubMed.
- L. Xu, W. Zhan, Y. Deng, X. Liu, G. Gao, X. Sun and G. Liang, Adv. Healthcare Mater., 2022, 11, 2200453 CrossRef CAS PubMed.
- D. Ye, A. J. Shuhendler, L. Cui, L. Tong, S. S. Tee, G. Tikhomirov, D. W. Felsher and J. Rao, Nat. Chem., 2014, 6, 519–526 CrossRef CAS PubMed.
- Z. Chen, M. Chen, Y. Cheng, T. Kowada, J. Xie, X. Zheng and J. Rao, Angew. Chem., Int. Ed., 2020, 59, 3272–3279 CrossRef CAS PubMed.
- C. Wu, C. Wang, T. Zhang, G. Gao, M. Wei, Y. Chen, X. Li, F. Wang and G. Liang, Adv. Healthcare Mater., 2022, 11, 2101346 CrossRef CAS PubMed.
- Y. Zhong, J. Zhan, G. Xu, Y. Chen, Q. Qin, X. Liao, S. Ma, Z. Yang and Y. Cai, Angew. Chem., Int. Ed., 2021, 60, 8121–8129 CrossRef CAS PubMed.
- L. Ye, Q. Yao, F. Xu, L. He, J. Ding, R. Xiao, L. Ding and B. Luo, J. Biomater. Appl., 2022, 36, 1064–1075 CrossRef CAS PubMed.
- P. Zhang, J. Zeng, Y. Li, C. Yang, J. Meng, Y. Hou and M. Gao, Angew. Chem., Int. Ed., 2021, 60, 8130–8138 CrossRef CAS PubMed.
- Y. Wang, W. Du, T. Zhang, Y. Zhu, Y. Ni, C. Wang, F. M. Sierra Raya, L. Zou, L. Wang and G. Liang, ACS Nano, 2020, 14, 9585–9593 CrossRef CAS PubMed.
- X. Sun, T. Xia, L. Xu, W. Zhan and G. Liang, Anal. Chem., 2023, 95, 5839–5842 CrossRef CAS PubMed.
- H. Lin, Y. Chen and J. Shi, Chem. Soc. Rev., 2018, 47, 1938–1958 RSC.
- Z. Zhang, Y. Hu, Y. Chen, Z. Chen, Y. Zhu, M. Chen, J. Xia, Y. Sun and W. Xu, Front. Oncol., 2022, 12, 1024789 CrossRef CAS PubMed.
| This journal is © The Royal Society of Chemistry 2024 |