Open Access Article
Open Access ArticleMagneto-controlled electrochemical immunosensing platform to assess the senescence-associated GDF-15 marker in colorectal cancer†
Sandra
Tejerina-Miranda
a,
Víctor
Pérez-Ginés
a,
Rebeca M.
Torrente-Rodríguez
 a,
María
Pedrero
a,
María
Pedrero
 a,
Ana
Montero-Calle
b,
José M.
Pingarrón
a,
Ana
Montero-Calle
b,
José M.
Pingarrón
 a,
Rodrigo
Barderas
*b and
Susana
Campuzano
a,
Rodrigo
Barderas
*b and
Susana
Campuzano
 *a
*a
aDepartamento de Química Analítica, Facultad de CC. Químicas, Universidad Complutense de Madrid, Pza. de las Ciencias 2, 28040-Madrid, Spain. E-mail: susanacr@quim.ucm.es
bChronic Disease Programme, UFIEC, Institute of Health Carlos III, Majadahonda, 28220-Madrid, Spain. E-mail: r.barderasm@isciii.es
First published on 30th January 2024
Abstract
In this study, we report a novel electrochemical immunoplatform using magnetic micro-supports and screen-printed carbon electrodes (SPCEs), overcoming limitations of the methods reported to date, for the rapid and sensitive determination of GDF-15, a molecular marker associated with cellular senescence in aging and cancer development and prognosis. The immunoplatform incorporated a sandwich-type configuration with specific capture and biotinylated-detection antibodies and used a streptavidin–peroxidase (strep–HRP) enzymatic conjugate as label. After magnetic capturing the micro-supports with the sandwich HRP-labeled immunocomplexes onto the surface of a SPCE, the change in cathodic current was quantified in the presence of H2O2 and hydroquinone (HQ), showing a direct correlation with the GDF-15 concentration. The proposed bioplatform exhibited attractive performance characteristics, including a good reproducibility (RSD 4.3%), and a wide linear concentration dynamic range from 140 to 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 pg mL−1 with a low limit of detection (LOD) of 42 pg mL−1 for GDF-15 standards in buffered solutions. The selectivity of the developed method and the storage stability of the capturing immunoconjugates were noteworthy. Indeed, the immunoconjugates showed a reliable performance for over 28 days when stored in a refrigerator. The immunoplatform was applied to a cohort of 19 plasma samples representing the different stages of colorectal cancer (CRC). The method allowed efficient discrimination between healthy individuals and CRC patients, particularly those in advanced stages, within a rapid 75-minute timeframe. The immunoplatform also presents substantial advantages in terms of cost-effectiveness, assay time reduction, and simplicity compared to other available techniques.
000 pg mL−1 with a low limit of detection (LOD) of 42 pg mL−1 for GDF-15 standards in buffered solutions. The selectivity of the developed method and the storage stability of the capturing immunoconjugates were noteworthy. Indeed, the immunoconjugates showed a reliable performance for over 28 days when stored in a refrigerator. The immunoplatform was applied to a cohort of 19 plasma samples representing the different stages of colorectal cancer (CRC). The method allowed efficient discrimination between healthy individuals and CRC patients, particularly those in advanced stages, within a rapid 75-minute timeframe. The immunoplatform also presents substantial advantages in terms of cost-effectiveness, assay time reduction, and simplicity compared to other available techniques.
Introduction
Cellular senescence is a complex state of stable cell cycle arrest in which proliferating cells become unresponsive to growth-promoting stimuli, which typically occurs in response to DNA damage.1,2 Originally thought to be an artifact of cell culture, senescence has emerged as a crucial cellular defence mechanism against stress, playing diverse roles in physiological and pathological processes such as tissue remodelling, injury response, and cancer development.3 One of its hallmarks is the senescence-associated secretory phenotype (SASP), which encompasses a wide range of soluble factors secreted by senescent cells.4 SASP comprises a variety of cytokines, chemokines, growth factors, angiogenic factors, proteases, bioactive lipids, extracellular matrix components, and matrix metalloproteinases,5 and has been shown to be associated with age-related health, mortality from cardiovascular diseases, several types of cancer, and Alzheimer's disease.6,7Focusing on cancer, SASP has a paradoxical role that is still under investigation. On the one hand, it can prevent cell division and promote immune clearance of damaged cells, thus preventing the development of tumors8 but, on the other hand, it can often promote resistance to treatment and cancer recurrence. When non-malignant senescent cells secrete SASP, tumorigenesis of premalignant cells can be driven, counteracting the overall beneficial effects of senescence.6 Consequently, controlling SASP factors emerges as a compelling strategy to manage cancer, with significant potential as biomarkers of senescence and prospects for clinical utility in the detection of cancer and metastasis.7
Colorectal cancer (CRC) is one of the most common – the third most diagnosed cancer in the world – and lethal – the third cause of cancer-related death and the second in men under 50 years.9 Currently, there is a firm conviction that the determination of a specific set of biomarkers can decisively complement traditional methods to provide an earlier and more personalized diagnosis of this neoplasia with the advantages that this represents to improve the survival and quality of life of patients.10
Growth differentiation factor 15 (GDF-15, also known as macrophage inhibitory cytokine 1, MIC-1), belongs to the transforming growth factor β (TGF-β) cytokine superfamily.11 GDF-15 is a widely distributed protein, which plays multiple roles in physiological and pathological conditions.12 Its effects are pleiotropic, encompassing appetite regulation, actions on metabolism, pregnancy, cell survival, immune response, and inflammation.12,13 GDF-15 also plays different roles in the pathophysiology of cardiovascular diseases, autoimmunity, diabetes, and cancer, including CRC. In this sense, GDF-15 is an integral component of SASP that contributes to various cellular processes associated with CRC progression, including inflammation, cell proliferation, apoptosis, and metastasis.14–16 When specifically considering metastasis in CRC, GDF-15 plays an important role as an activator of the TGF-β signalling cascade.17 This intricate and versatile signalling pathway regulates a wide range of cellular processes and holds promise as a target for anticancer drug development in both preclinical and clinical stages.18 Among the processes under the regulation of the TGF-β signalling cascade is metastasis mediated by epithelial-mesenchymal transition (EMT), which is activated by GDF-15.19,20 Therefore, GDF-15 dysregulation within the SASP framework underlines its relevance as a well-established biomarker for detecting and characterizing metastatic CRC.15,21,22
Considering the importance of GDF-15 in assessing the presence and progression of CRC, clinical research strives to develop simple, sensitive, accurate, and cost-effective methods to monitor this biomarker. Traditionally, immunohistochemistry23 and blotting technologies24 have been used for GDF-15 detection, deprived of quantitative ability. Subsequently, Enzyme-Linked Immunosorbent Assay (ELISA) methodologies entailing skilled personnel, relatively long assay durations and bulky plate readers,25–27 and DNA amplification28 have been employed for the determination of GDF-15. Recently, electrochemical immunosensors have been harnessed for GDF-15 quantification, leveraging their unique attributes of sensitivity and selectivity.29–32 However, despite their remarkably low detection limits and demonstrated applicability to the analysis of serum samples for the diagnosis and prognosis of cardiovascular diseases (CVDs), these biosensors required the use of nanomaterials and lengthy manufacturing processes. Furthermore, the reported immunosensors for GDF-15 used glassy carbon electrodes (GCEs), whose use in electrochemical biosensing shows certain well-known limitations, such as susceptibility to fouling, incompatibility with miniaturization and multiplexing, and the requirement of exhaustive electrode surface pretreatment.33–35
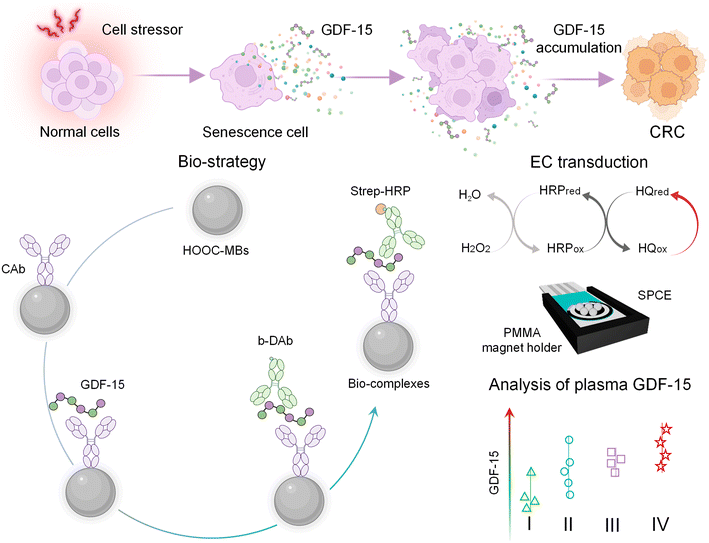
These drawbacks can be overcome by working with screen-printed carbon electrodes (SPCEs) and profiting from the inherent benefits provided by micromagnetic supports in biosensing, derived from their high surface-to-volume ratio, easy manipulation through magnetic fields, and versatility in surface chemistry for bioconjugation and efficient target capture. These benefits lead to innovative biosensing strategies with higher simplicity, improved sensitivity and kinetics, and minimization of matrix effects, thus satisfying the sought-after demands for point-of-care applications.36–40 Accordingly, we report here the first electrochemical immunoplatform involving micromagnetic supports for the determination of GDF-15. This immunoplatform involves a sandwich-type immunoassay, enzymatic labeling with HRP and amperometric transduction at SPCEs, and represents a competitive alternative to other available methodologies for the simple and rapid quantification of GDF-15. Unlike other electrochemical immunoplatforms reported for the determination of GDF-15, all of them using integrated formats and focused on cardiovascular diseases,29–32 the developed immunoplatform was applied to the identification and prognosis of CRC through the analysis of plasma samples.
Experimental section
Apparatus and electrodes
Amperometric measurements were performed at room temperature with a CHI812B potentiostat provided with CHI1140A software. Screen-printed carbon electrodes (SPCEs, DRP110, 4 mm ∅) consisting of a three-electrode cell configuration with a carbon working electrode (WE), a carbon auxiliary electrode (AE) and an Ag pseudo-reference electrode (RE) were used as electrochemical platforms.The SPCEs and their connecting cable (DRP-CAC), were acquired from Metrohm-DropSens. The SPCEs were placed in a homemade polymethylmethacrylate (PMMA) casing with an embedded Nd magnet (∅ 4 × 4 mm, AIMAN GZ), ensuring the capture of the modified microbeads (MBs) on the WE surface. The whole casing was immersed in a 10 mL glass electrochemical cell to carry out the amperometric detection.
A magnetic particle concentrator DynaMag-2 (Cat. No: 12321D, Dynabeads®, Invitrogen™ Thermo Fisher Scientific), a BioSan TS-100 uniform temperature incubator shaker (Thermo), a magnetic stirrer (Inbea), a Crison model Basic 20+ pH-meter, a vortex (Velp Scientifica) and an MPW-65R centrifuge from MPW (Med. Instruments) were also employed.
Reagents and solutions
Carboxylic acid-functionalized MBs (HOOC-MBs, Dynabeads™, Cat. No.: M-270, 2.8 μm ∅, ∼2 × 109 beads mL−1) were purchased from Invitrogen™ Thermo Fisher Scientific. Commercial Blocker™ Casein in PBS (BB) was obtained from Thermo Scientific (Cat. No. 37528). Ethanolamine (ETA), hydroquinone (HQ), hydrogen peroxide (H2O2, 30% v/v), N-(3-dimethyl-aminopropyl)-N′-ethyl-carbodiimide (EDC) and N-hydroxysulfosuccinimide (sulfo-NHS) were acquired from Sigma-Aldrich.Tris-hydroxymethyl-aminomethane-HCl (Tris-HCl), potassium chloride, sodium chloride, sodium di-hydrogen phosphate, di-sodium hydrogen phosphate and 2-(N-morpholino) ethanesulfonic acid (MES) were purchased from Scharlab. All used reagents were of the highest available grade.
Deionized water type I from a Millipore Milli-Q purification system (18.2 MΩ cm) was used for the preparation of all buffer solutions: 0.1 mol L−1 phosphate buffer (PB) solution pH 8.0; 0.1 mol L−1 Tris-HCl pH 7.2; 0.025 mol L−1 MES buffer pH 5.0; 0.01 mol L−1 phosphate buffer saline solution (PBS) pH 7.4 and 0.05 mol L−1 PB solution pH 6.0. For the activation and the blocking step of the HOOC-MBs an EDC/sulfo-NHS mixture solution (50 mg mL−1 each) prepared in MES buffer pH 5.0 and a 1.0 M ethanolamine (ETA) solution prepared in 100 mM PB buffer pH 8.0 were used, respectively. For the amperometric measurements, solutions of 100 mM HQ and 100 mM H2O2 were freshly prepared in 50 mM PB pH 6.0 just before their use.
The standard and the antibody pair used for the determination of Human GDF-15 were those provided in the commercial ELISA kit Human GDF-15 DuoSet ELISA (Cat. No. DY957, from R&D Systems) containing a Recombinant Human GDF-15 Standard, a Mouse Anti-Human GDF-15 Capture Antibody (CAb) and a Biotinylated Goat Anti-Human GDF-15 Detection Antibody (b-DAb). Streptavidin–HRP (strep–HRP) conjugate from Roche (Cat. No. 11089153001) was employed as enzymatic tracer.
IgG from human serum (hIgG, Cat. No. I2511), albumin from human serum (HSA, Cat. No. A1653), human haemoglobin (Hb, Cat. No. H7379), all from Sigma-Aldrich, and human TNFα (Cat. No. DY210) from R&D Systems, were assessed as potential interferents.
Sandwich immunoassay implementation on MBs
MBs modification was performed using 1.5 mL microcentrifuge tubes employing successive incubation and washing steps. Incubation steps were carried out using 25 μL of the corresponding solution in a thermoshaker at 25 °C under constant stirring (950 rpm). On the other hand, washing steps were done with 50 μL of the required solution. After each step, the microcentrifuge tube with the MBs suspension was placed in the magnetic separator for 2 min with the subsequent removal of the supernatant.For each measurement, a 3 μL-aliquot of the HOOC-MBs suspension was placed in a 1.5 mL microcentrifuge tube and washed twice with MES buffer for 10 min. Subsequently, the carboxylic groups on the MBs surface were activated by incubation with freshly prepared EDC/sulfo-NHS solution for 35 min. After activation of the carboxylic groups, two washing steps were performed with MES buffer.
Thereafter, the activated HOOC-MBs were incubated for 45 min in a 25 μg mL−1 CAb solution prepared in MES buffer. The CAb-MBs were washed twice with the same buffer and incubated in 1.0 M ETA solution for 60 min to deactivate the remaining groups after CAb immobilisation. Then, the modified MBs were washed with 0.1 M Tris-HCl buffer (pH 7.2) and twice with 0.01 M PBS pH 7.4. The CAb-MBs were used for the measurements at the time of preparation or stored (in filtered PBS, at 4 °C) for later use.
For the determination of GDF-15, HRP-labeled sandwich immunocomplexes were formed upon three successive incubation steps: in the standard target protein solution (in PBS pH 7.4) for 15 min; then in a 0.5 μg mL−1 solution (in BB) of b-DAb for 30 min; and in a 1/1000 strep–HRP solution (in BB) for 30 min.
The resulting magnetic bioconjugates were washed twice with BB and re-suspended in 50 μL of PB pH 6.0 to perform the amperometric measurements.
Amperometric measurements
First, the SPCE was placed in the Nd-embedded-PMMA casing. Then, 50 μL of the MBs suspension were disposed on the surface of the WE of the SPCE (a new one was used for each determination).The SPCE/PMMA casing set was connected to the potentiostat through the cable and immersed into a measuring cell containing 10 mL of PB buffer pH 6.0 and 100 μL of freshly prepared 0.1 mol L−1 HQ solution. Amperometric detection was made by applying continuous stirring and a constant potential of −0.20 V vs. the Ag pseudo-reference electrode.
Once the background current was stabilized, 50 μL of a freshly prepared 0.1 mol L−1 H2O2 were added to the electrochemical cell producing a cathodic current variation due to the HQ-mediated enzymatic reduction of H2O2. The signals given in the manuscript were calculated as the difference between the steady-state current (after H2O2 addition) and the background current (before H2O2 addition) and are the mean values resulting from three replicates. The error bars were calculated as the standard deviation (SD) of these replicates.
Analysis of plasma samples
Plasma samples from patients with CRC and healthy individuals were provided by the biobank of the San Carlos Clinical Hospital after approval by the Ethical Review Committee (CEI PI 45). These samples, stored at −80 °C, were handled and used anonymously accomplishing all the ethical issues. Experiments were performed in agreement with relevant guidelines and regulations of Hospital Universitario Clínico San Carlos, Instituto de Salud Carlos III and Universidad Complutense de Madrid. All individuals provided written informed consent for the use of their biological samples for research purposes, adhering to ethical principles outlined by Spain (LOPD 15/1999) and the European Union Fundamental Rights of the EU (2000/C364/01).Although no matrix effect was observed when analysing 25 times diluted plasma samples, considering the low content of the target protein, the determination was performed employing standard additions calibration plots for GDF-15 with concentrations between 0 and 5000 pg mL−1.
Results and discussion
This work reports the first electrochemical immunoassay involving micromagnetic supports and SPCEs for the determination of GDF-15. The bioplatform entails efficient capture of the target protein, GDF-15, through a specific antibody (CAb) onto the surface of HOOC-MBs. Subsequently, GDF-15 is sandwiched using a biotinylated detector antibody (b-DAb) and further enzymatically labeled with a streptavidin–horseradish peroxidase (strep–HRP) conjugate. Upon magnetic capture of the MBs bearing the immunoconjugates onto the surface of the SPCEs, electrochemical detection is performed by monitoring the cathodic current measured at −0.20 V vs. the Ag pseudo-reference electrode utilizing the H2O2/HQ system. The design of the immunoplatform and the reactions involved in the amperometric transduction are illustrated in Scheme 1.Optimization of experimental variables
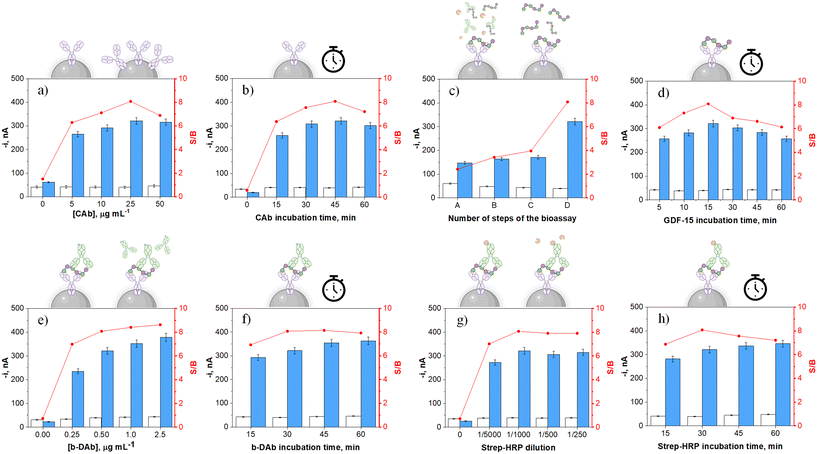
In seeking to optimize the effectiveness of our proposed immunodetection strategy, a comprehensive evaluation of each operational parameter, which directly impacts its proper functioning, was undertaken. The selection criterion for this evaluation was based on attaining the utmost signal-to-blank ratio (S/B) under two conditions: the absence (B) and the presence (S) of 1000 pg mL−1 GDF-15 standard. A graphical display of the evaluated variables, their corresponding ranges, and the selected values are shown and summarized in Fig. 1 and Table 1, respectively. Other experimental parameters, such as the quantity of HOOC-MBs utilized per determination and all the factors associated with the amperometric transduction, were optimized in previous studies.41,42| Variable | Studied range | Selected value | |
|---|---|---|---|
| Number of steps | 1–3 | 3 | |
| Concentrations, μg mL−1*/dilution** | CAb* | 0.0–50.0 | 25.0 |
| b-DAb* | 0.0–2.5 | 0.5 | |
| Strep–HRP** | 0–1/250 | 1/1000 | |
| Incubation times, min | CAb | 0–60 | 45 |
| GDF-15 | 5–60 | 15 | |
| b-DAb | 15–60 | 30 | |
| Strep–HRP | 15–60 | 30 | |
Fig. 1a and b show higher S/B ratios for MBs incubated in a 25 μg mL−1 CAb solution for 45 min. The decreases observed for larger concentrations and longer incubation times were attributed to smaller S responses, probably due to hindered target recognition by CAb crowding.43 Furthermore, as multiple steps are involved in forming sandwich immunocomplexes on the surface of CAb-MBs, it is important to check how the used protocol (i.e., the number of involved steps) influences the measurements. Four different protocols, all starting from the CAb-MBs preparation and involving different 30 min incubation steps using single or mixture solutions of the bioreagents employed for the formation of the HRP-labeled sandwich immunocomplexes, have been evaluated as detailed in Table 2.
| Protocol | Successive incubation steps (30 min each) | Assay time, min | ||
|---|---|---|---|---|
| GDF-15 | b-DAb | Strep–HRP | ||
| A | 1 | 30 | ||
| B | 1 | 2 | 60 | |
| C | 1 | 2 | 60 | |
| D | 1 | 2 | 3 | 90 |
As Fig. 1c shows, the specific signal in the presence of GDF-15 achieved a larger value with the 3-step protocol (D) involving the sequential incubation of CAb-MBs with the target protein, b-DAb, and the enzymatic tracer strep–HRP solutions, respectively. Conversely, the incubation of CAb-MBs immunoconjugates with mixtures of GDF-15 and b-DAb (protocol B), b-DAb and strep–HRP (protocol C), or GDF-15, b-DAb, and strep–HRP (protocol A) resulted in lower S responses, suggesting poorer GDF-15 capture and/or labeling efficiencies. Therefore, 15 min was selected as the incubation time of CAb-MBs with the target antigen (Fig. 1d). In addition, according to Fig. 1e, the S/B ratio exhibited an ascending trend with increasing b-DAb concentration. Nevertheless, since this trend is not pronounced, 0.5 μg mL−1 b-DAb was chosen for further work to avoid an unnecessary cost increase per determination.
Fig. 1g shows as a larger S/B ratio was obtained with a 1/1000 dilution of the strep–HRP conjugate. It is important to note that bars 0 in Fig. 1a, e and g confirmed the need of using the three bioreagents to carry out the assay which took place through the formation of sandwich-type immune complexes labeled with HRP. These results also confirmed the effectiveness of the blocking step after CAb immobilization on MBs, which effectively prevented nonspecific adsorption of b-DAb or strep–HRP in the absence of GDF-15.
Considering the incubation times for b-DAb and strep–HRP conjugate (Fig. 1f and h), it was seen that 30 min incubations were adequate for GDF-15 sandwiching and enzymatic labeling, respectively, since incubation times did not significantly improve the S/B ratios. Using the optimized conditions, it is important to highlight that, counting from the deactivated CAb-MBs, the determination of GDF-15 can be completed in as short time as 75 min.
Analytical and operational characteristics
Under the optimized working conditions summarized in Table 1, a calibration plot was constructed for GDF-15 standards in buffered solutions. As it can be seen in Fig. 2, a linear calibration plot (r = 0.996) was obtained over the 140–10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 pg mL−1 GDF-15 concentration range, fitting the equation – i, nA = (0.236 ± 0.005) nA mL pg−1 [GDF-15] + (56 ± 21) nA. The limit of detection (LOD) and quantification (LOQ) values were estimated using, respectively, the 3 × sb/slope and 10 × sb/slope criteria (sb: standard deviation of ten amperometric measurements in the absence of GDF-15).44–46 The obtained values were 42.0 and 140.0 pg mL−1, respectively.
000 pg mL−1 GDF-15 concentration range, fitting the equation – i, nA = (0.236 ± 0.005) nA mL pg−1 [GDF-15] + (56 ± 21) nA. The limit of detection (LOD) and quantification (LOQ) values were estimated using, respectively, the 3 × sb/slope and 10 × sb/slope criteria (sb: standard deviation of ten amperometric measurements in the absence of GDF-15).44–46 The obtained values were 42.0 and 140.0 pg mL−1, respectively.
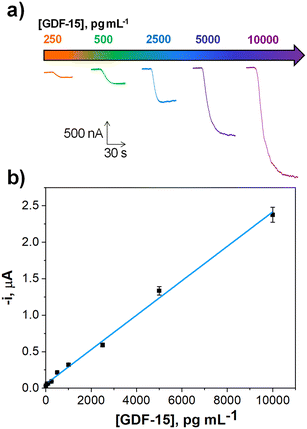 | ||
| Fig. 2 Actual amperometric traces recorded at the indicated concentrations (a) and calibration graph (b) constructed for the determination of GDF-15 with the developed immunoplatform. | ||
It is important to highlight the low LOD achieved by the bioplatform, considerably lower than the cut-off values established for GDF-15 in serum samples to identify CRC patients (around 1000 pg mL−1).14,47
The reproducibility of the measurements and the storage stability of the CAb-MBs were evaluated. The responses for 1000 pg mL−1 GDF-15 measured with ten different bioplatforms prepared the same day, gave a relative standard deviation (RSD) value of 4.3%. This result highlights both the reproducibility of the methodology used to implement the sandwich-type immunoassay on the MBs, and that of the amperometric measurements. Furthermore, the amperometric responses provided in the absence and in the presence of 2500 pg mL−1 GDF-15 by immunoplatforms constructed using the stored CAb-MBs (at 4 °C resuspended in filtered PBS) were compared. The results obtained (Fig. S1 in the ESI†) showed that the as prepared immunoplatforms gave similar S/B values for at least 28 days after the CAb-MBs preparation.
Table S1 (in the ESI†) summarizes the most relevant characteristics of other electrochemical immunosensors recently reported in the literature for the determination of GDF-15.
As it can be seen, the reported immunosensors29–32 claimed lower LOD values than that achieved with the developed immunoplatform. However, all of them involved complex, long, and laborious protocols for their preparation and used nanomaterials. Therefore, although the developed immunoplatform, provides a slightly lower detectability, but, nevertheless, able to respond to the demands of the clinic, it is attractive in terms of simplicity, preparation and testing time and use of disposable substrates, thus exhibiting advantages for its potential future implementation in POC and/or multiplexing devices. It is also important to note that all the reported immunosensors were applied for cardiovascular diseases, thus leaving unexplored the possibility of using GDF-15 in the diagnosis and prognosis of other diseases of high prevalence and mortality such as CRC.
On the other hand, the comparison of the linear ranges of the bioplatform and those specified in the commercial ELISA kit that uses the same immunoreagents (DY957 DuoSet ELISA, R&D Systems, 7.8–500 pg mL−1), and taking into account that the bioplatform uses a standard/sample volume 4 times lower (25 vs. 100 μL) both technologies offer similar sensitivity but the electrochemical biotool is attractive for point-of-care applications and even home testing, due to the requirement of cost-effectiveness, simple and portable instrumentation.
Selectivity
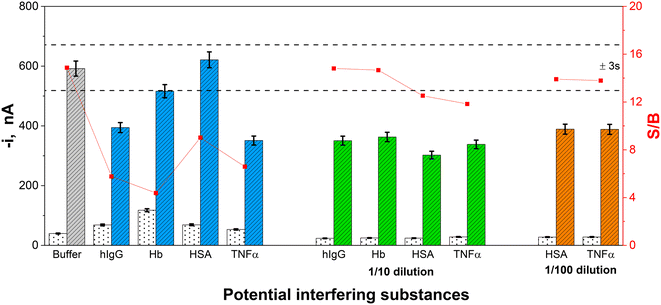
To glimpse whether there would be interferences when applying the developed immunoplatform to real samples, the effect of the following proteins found in human serum, hIgG, HSA, Hb and TNFα, was evaluated. Therefore, the effect of diluted solutions of these potential interfering proteins (at the concentration levels reported in healthy individuals and indicated in Fig. 3 caption) on the amperometric signals recorded in the absence and in the presence of 2500 pg mL−1 GDF-15, was checked.Furthermore, the supplier company of the employed immunoreagents (DY957 DuoSet ELISA, R&D Systems) certified no cross-reactivity for recombinant human GDF-11, for recombinant mouse GDF-5, GDF-6, GDF-7, GDF-8 and Pro-GDF-8, all of them prepared at a 50 ng mL−1 concentration.
As can be seen in Fig. 3, the four tested proteins interfered with the determination of GDF-15 at the largest concentrations tested.
The interference of hIgG, Hb and HSA is widely described in the literature; hIgG interference can be attributed both to its nonspecific adsorption on the surface of MBs48 and to the presence of circulating human antibodies which can react with mouse-expressed antibodies, as it is the CAb involved in the developed immunoplatform, leading to inaccurate results.49 The interference observed for Hb, which significantly affects the amperometric response in the absence of GDF-15, can be attributed to its peroxidase activity which has been widely reported to cause false positives.50 The interference from HSA, especially when used at concentrations equal to or larger than 5 mg mL−1, has been mainly attributed to the presence of IgG in low-purity HSA lots.49,51 The interference observed for TNFα is not so well described in the literature as that for the other tested proteins. This interference can be attributed to interaction with the target proteins or to nonspecific recognition by the used antibodies. A hypothesis that may justify this result is the tendency of TNFα to form clusters that can somehow trap the target protein, preventing its recognition.52
As expected, an appropriate dilution of these four non-target proteins (10 times for hIgG and Hb, and 100 times for HSA and TNFα) provoked minimization of their interference.
Analysis of plasma samples
The designed immunoplatform was used for the analysis of 19 plasma samples from healthy individuals (4) and from CRC patients (15) diagnosed at different stages of the disease (I–IV).The possible existence of a matrix effect was evaluated by comparing the slope values of the calibration plot constructed for GDF-15 in buffer solution with those prepared in 25-fold diluted solutions of representative plasma samples from a healthy subject and a stage IV CRC patient.
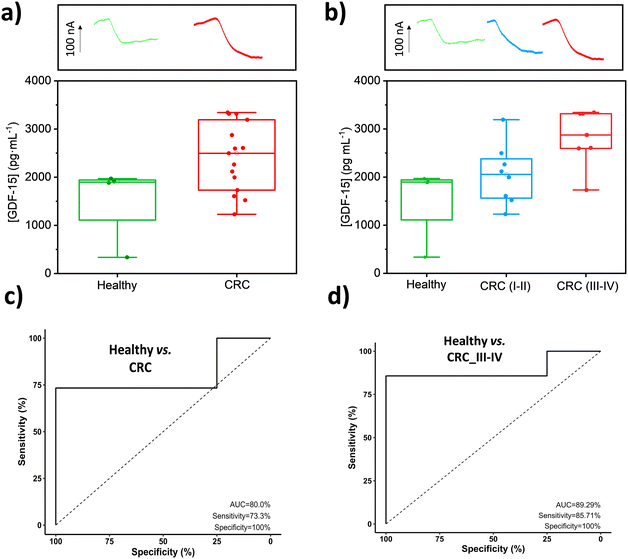
No statistically significant differences were found between the slope values of the calibration plots constructed with GDF-15 standards prepared in buffered solutions ((0.24 ± 0.01) nA mL pg−1) and in 25-fold diluted plasma samples from healthy individuals ((0.25 ± 0.01) nA mL pg−1) and CRC patients ((0.24 ± 0.01) nA mL pg−1), texp values of 0.219 and 0.012, respectively, which are lower than ttab = 4.303 (n = 3, α = 0.05).53 Therefore, under the mentioned conditions, no matrix effect was apparent. However, the low target protein content in the diluted samples led us to carry out the determination by applying the standard additions method through the supplementation of 25-diluted plasma samples with increasing concentrations of GDF-15, from 0 to 5000 pg mL−1. The obtained results are displayed in Fig. 4, where the concentrations found for healthy individuals (healthy group), for CRC patients (CRC group) and for CRC patients at early stages of the disease (CRC (I and II) group) and at advanced stages of the disease (CRC (III and IV) group), are compared.
These results confirmed larger GDF-15 plasmatic concentrations in CRC patients (range 1229–3342 pg mL−1, mean 2413.7 pg mL−1) compared to healthy subjects (range 337–1971 pg mL−1, mean 1525.6 pg mL−1) and, within the former, larger at advanced stages (III–IV: 1732–3342 pg mL−1, mean 2826.2 pg mL−1) than at early stages (I–II: 1229–3193 pg mL−1, mean 2052.8 pg mL−1). This agrees with the gradual increase reported in the serological concentration of GDF-15 with the subsequent stages of CRC advancement.54,55 These results are perfectly in line with the valuable potential of GDF-15 for CRC detection and prognosis.14,17,20,21,22,47,56–60 Indeed, it has been reported that GDF-15 levels in serum gradually increase in the process of conversion from adenomatous polyps to colorectal carcinoma14,59 and that CRC patients with an increased serum level of GDF-15 had a 2.09-fold higher risk of death11 and worse cancer-specific survival (CSS).21 In our results, this would correspond to the CRC plasma samples at advanced stages (1.85-fold higher GDF-15 plasma values than healthy individuals). It is also important to note that the concentration ranges found with the bioplatform in the explored cohort agree with those reported in the literature for healthy subjects: (427–851 pg mL−1),61 (167.9–1203.9 pg mL−1),14 (33.9–2398.9 pg mL−1)47 and CRC patients: (908–3152 pg mL−1),61 (240.1–3713.4 pg mL−1)14 and (112.0–5178 pg mL−1).47
In addition, a more exhaustive analysis of the results provided by the developed bioplatform using ROC curves (Fig. 4c and d, Table 3) shows its potential to discriminate between healthy individuals and CRC patients, and particularly between healthy subjects and CRC patients with advanced stages (III and IV), yielding the plasma cut-off values of GDF-15 that are also included in the Table and allow these discriminations. These results agree with the role of GDF-15 as a negative prognostic marker in CRC whose elevated plasma levels correlate with increased risk of recurrence and reduced overall survival.44,55
| Parameter | Healthy vs. CRC | Healthy vs. CRC III and IV |
|---|---|---|
| AUC (%) | 80.0 | 89.3 |
| Sensitivity (%) | 73.3 | 85.7 |
| Specificity (%) | 100.0 | 100.0 |
| Cut-off, pg mL−1 | 1983.5 | 2283.0 |
In addition, the accuracy of the results provided by the bioplatform in the analysis of plasma samples was evaluated by performing recovery studies. A representative sample of each pool (healthy, CRC I, CRC II, CRC III, and CRC IV) was spiked with 1000 pg mL−1 GDF-15 standard. The obtained results, using the same protocol as for non-supplemented samples and upon the subtraction of the endogenous content found in each sample from the overall concentration determined after sample spiking, provided recovery values between 100 and 106% for the 5 types of samples analyzed, thus confirming the accuracy of the methodology.
Conclusions
This article reports the first electrochemical immunoplatform for the determination of GDF-15 employing HRP-labeled sandwich immunocomplexes linked to the surface of MBs captured on SPCEs for amperometric transduction.The developed bioplatform exhibits analytical (LOD considerably lower than the cut-off value established in serum to diagnose several types of cancer) and operational characteristics compatible with its clinical application. The developed bioplatform has been tested for 19 plasma samples of healthy individuals and CRC patients at different stages of the disease. The results show its potential for a minimally invasive diagnosis and prognosis in only 75 min and using a minimum amount of sample (1 μL plasma/determination).
This new biotool represents an attractive alternative to conventional technologies (blotting and ELISA), as well as to other recently described electrochemical immunosensors, for the simple, rapid, and point-of-need determination of GDF-15. These features, together with the demonstrated applicability and compatibility with integration in multiplexing devices, offer it as a tool to help understanding GDF-15 regulation and expression in metastatic colon cancer and therefore providing more reliable diagnoses and prognoses of this neoplasm. Furthermore, considering that recent literature reports that GDF-15 promoted 5-Fluorouracil (5-FU, the conventional chemotherapeutic in CRC treatment) resistance, its determination can contribute to both precision medicine and therapy.60,62
Conflicts of interest
The authors declare no conflict of interest.Acknowledgements
The financial support of Grant PID2019-103899RB-I00 funded by MCIN/AEI/10.13039/501100011033, EU's Horizon 2020 funding programme (UCM's Specific Research Fund FEI-EU-22-08) and PI20CIII/00019 and PI23CIII/00027 grants from the AES-ISCIII Program co-founded by FEDER funds are gratefully acknowledged.References
- F. d'Adda di Fagagna, Nat. Rev. Cancer, 2008, 8(7), 512–522 CrossRef PubMed.
- R. Shmulevich and V. Krizhanovsky, Antioxid. Redox Signaling, 2021, 34(4), 324–334 CrossRef CAS PubMed.
- D. Paramos-De-Carvalho, A. Jacinto and L. Saúde, eLife, 2021, 10, e72449 CrossRef CAS PubMed.
- N. Ohtani, Inflammation Regener., 2022, 42, 11 CrossRef CAS PubMed.
- R. Kumari and P. Jat, Front. Cell Dev. Biol., 2021, 9, 645593 CrossRef PubMed.
- M. Takasugi, Y. Yoshida, E. Hara and N. Ohtani, FEBS J., 2022, 290(5), 1348–1361 CrossRef PubMed.
- A. Domen, C. Deben, J. Verswyvel, T. Flieswasser, H. Prenen, M. Peeters, F. Lardon and A. Wouters, J. Exp. Clin. Cancer Res., 2022, 41, 360 CrossRef CAS PubMed.
- J. Yang, M. Liu, D. Hong, M. Zeng and X. Zhang, Front. Cell Dev. Biol., 2021, 9, 722205 CrossRef PubMed.
- R. L. Siegel, N. Sandeep Wagle, A. Cercek, R. A. Smith and A. Jemal, Ca-Cancer J. Clin., 2023, 73, 233–254 CrossRef PubMed.
- X. Hua, M. Kratz, R. C. Malen, J. Y. Dai, S. Lindström, Y. Zheng and P. A. Newcomb, Br. J. Cancer, 2021, 125(6), 806–815 CrossRef CAS PubMed.
- L. N. Lawton, M. F. Bonaldo, P. C. Jelenc, L. Qiu, S. A. Baumes, R. A. Marcelino, G. M. de Jesus, S. Wellington, J. A. Knowles, D. Warburton, S. Brown and M. B. Soares, Gene, 1997, 203, 17–26 CrossRef CAS PubMed.
- A. Assadi, A. Zahabi and R. A. Hart, Pflugers Arch., 2020, 472, 1535–1546 CrossRef CAS PubMed.
- P. Iglesias, R. A. Silvestre and J. J. Díez, Endocrine, 2023, 81, 419–431 CrossRef CAS PubMed.
- C. Li, X. Wang, J. I. Casal, J. Wang, P. Li, W. Zhang, E. Xu, M. Lai and H. Zhang, J. Cell. Mol. Med., 2016, 20(8), 1420–1426 CrossRef CAS PubMed.
- M. Vocka, D. Langer, J. Petryl, M. Kalousová, T. Zima, T. Hanuš and L. B. Petruzelka, J. Clin. Oncol., 2016, 34(15_suppl), e15098 Search PubMed.
- S. J. Baek and T. E. Eling, Pharmacol. Ther., 2019, 198, 46–58 CrossRef CAS PubMed.
- C. Li, J. Wang, J. Kong, J. Tang, Y. Wu, E. Xu, H. Zhang and M. Lai, Oncotarget, 2015, 7(1), 860–872 CrossRef PubMed.
- A. B. Baba, B. Rah, G. M. Bhat, I. Mushtaq, S. Parveen, R. Hassan, M. H. Zargar and D. Afroze, Front. Pharmacol., 2022, 13, 791272 CrossRef CAS PubMed.
- J. Xu, S. Lamouille and R. Derynck, Cell Res., 2009, 19(2), 156–172 CrossRef CAS PubMed.
- Y. Zhang, X. Wang, M. Zhang, Z. Zhang, L. Jiang and L. Li, Biotechnology, 2018, 46(sup2), 652–658 CAS.
- Y. Wang, T. Jiang, M. Jiang and G. Shuijing, BMC Cancer, 2019, 19, 177 CrossRef PubMed.
- J. Robotycka, S. Mielcarska, M. Dawidowicz, A. Kula, B. Ochman, T. Niebudek, B. Strzałkowska, D. Waniczek and E. Świętochowska, Med. Res. J., 2022, 7(3), 208–214 CrossRef.
- K. Souček, A. Malenovská, Z. Kahounová, J. Remšík, Z. Holubcová, T. Soukup, D. Kurfürstová, J. Bouchal, T. Suchánková, E. Slabáková and A. Hampl, J. Assist. Reprod. Genet., 2018, 35(8), 1407–1417 CrossRef PubMed.
- P. J. Emmerson, K. L. Duffin, S. Chintharlapalli and X. Wu, Front. Physiol., 2018, 9, 1712 CrossRef PubMed.
- J. Trovik, H. B. Salvesen, T. Cuppens, F. Amant and A. C. Staff, Int. J. Gynecol. Cancer, 2014, 24(2), 252–259 CrossRef PubMed.
- Y. Zhang, W. Jiang, L. Wang and K. Lingappan, Toxicol. Appl. Pharmacol., 2017, 332, 8–14 CrossRef CAS PubMed.
- K. Liu, B. Liu, G. A. Wittert, C. Thompson, A. T. Hutchison and L. K. Heilbronn, Obes. Res. Clin. Pract., 2023, 17(1), 91–93 CrossRef PubMed.
- T. Ebai, F. M. S. De Oliveira, L. Löf, L. Wik, C. Schweiger, A. Larsson, U. Keilholtz, J. Haybaeck, U. Landegren and M. Kamali-Moghaddam, Clin. Chem., 2017, 63(9), 1497–1505 CrossRef CAS PubMed.
- M. Chen, L. Zhao, D. Wu, S. Tu, C. Chen, H. Guo and Y. Xu, Anal. Chim. Acta, 2022, 1223, 340194 CrossRef CAS PubMed.
- Y. Jiao, Z. Huang, M. Chen, X. Zhou, H. Lu, B. Wang and X. Dai, Anal. Methods, 2022, 14, 1420–1429 RSC.
- C. Chen, J. Kang, S. Wang, S. Chen, H. Guo and M. Chen, Microchim. Acta, 2023, 190, 92 CrossRef CAS PubMed.
- M. Chen, Y. Jiao, C. Chen, Y. Wang, H. Lu and X. Dai, Microchem. J., 2023, 193, 109150 CrossRef CAS.
- A. M. Abdel-Aziz, H. H. Hassan and I. H. A. Badr, Anal. Chem., 2020, 92(11), 7947–7954 CrossRef CAS PubMed.
- E. Sramkova, T. Bystron and K. Bouzek, Electrochim. Acta, 2021, 379, 138177 CrossRef CAS.
- M. Devi, M. Vomero, E. Fuhrer, E. Castagnola, C. Gueli, S. Nimbalkar, M. Hirabayashi, S. Kassegne, T. Stieglitz and S. Sharma, J. Neural Eng., 2021, 18(4), 041007 CrossRef PubMed.
- W. Lai, D. Tang, J. Zhuang, G. Chen and H. Yang, Anal. Chem., 2014, 86, 5061–5068 CrossRef CAS PubMed.
- Y. Lin, Q. Zhou, J. Li, J. Shu, Z. Qiu, Y. Lin and D. Tang, Anal. Chem., 2016, 88, 1030–1038 CrossRef CAS PubMed.
- B. A. Otieno, C. E. Krause and J. F. Rusling, Methods Enzymol., 2016, 571, 135–150 CAS.
- S. Fortunati, C. Giliberti, M. Giannetto, A. Bertucci, S. Capodaglio, E. Ricciardi, P. Giacomini, V. Bianchi, A. Boni, I. De Munari, R. Corradini and M. Careri, Biosens. Bioelectron.: X, 2023, 15, 100404 CAS.
- S. Fortunati, M. Giannetto, C. Giliberti, M. Mattarozzi, A. Bertucci and M. Careri, Analysis Sensing, 2023, e202300062 CrossRef.
- M. Eguílaz, M. Moreno-Guzmán, S. Campuzano, A. González-Cortés, P. Yáñez-Sedeño and J. M. Pingarrón, Biosens. Bioelectron., 2010, 26(2), 517–522 CrossRef PubMed.
- F. Conzuelo, M. Gamella, S. Campuzano, D. G. Pinacho, A. Reviejo, M. P. Marco and J. M. Pingarrón, Biosens. Bioelectron., 2012, 36(1), 81–88 CrossRef CAS PubMed.
- D. M. Kim, X. T. Yao, R. Vanam and M. S. Marlow, mAbs, 2019, 11(7), 1319–1330 CrossRef CAS PubMed.
- K. Hasebe and J. Osteryoung, Anal. Chem., 1975, 47, 2412 CrossRef CAS PubMed.
- L. H. Keith, W. Crummett, J. Deegan Jr., R. A. Libby, J. K. Taylor and G. Wentler, Anal. Chem., 1983, 55, 2210–2218 CrossRef CAS.
- P. Borman and D. Elder, Q2(R1) validation of analytical procedures: text and methodology, ICH quality guidelines: an implementation guide, ed. A. Teasdale, D. Elder and R. W. Nims, Wiley, Hoboken, 2017, pp. 127–166 Search PubMed.
- X. Wang, Z. Yang, H. Tian, Y. Li, M. Li, W. Zhao, C. Zhang, T. Wang, J. Liu, A. Zhang, D. Shen, C. Zheng, J. Qi, D. Zhao, J. Shi, L. Jin, J. Rao and W. Zhang, Oncotarget, 2017, 8(15), 24892–24901 CrossRef PubMed.
- B. Arévalo, V. Serafín, M. Garranzo-Asensio, R. Barderas, P. Yáñez-Sedeño, S. Campuzano and J. M. Pingarrón, Biosens. Bioelectron.: X, 2023, 13, 100325 Search PubMed.
- S. E. F. Melanson, M. J. Tanasijevic and P. Jarolim, Circulation, 2007, 116, e501–e504 CrossRef PubMed.
- D. V. Grigorieva, I. V. Gorodki, A. V. Sokolov, O. V. Kosmachevskaya, A. F. Topunov, I. V. Buko, E. E. Konstantinova, S. N. Cherenkevich and O. M. Panasenko, Bull. Exp. Biol. Med., 2013, 155, 118–121 CrossRef CAS PubMed.
- B. Arévalo, M. Blázquez-García, A. Valverde, V. Serafín, A. Montero-Calle, G. Solís-Fernández, R. Barderas, P. Yáñez-Sedeño, S. Campuzano and J. M. Pingarrón, Bioelectrochemistry, 2022, 146, 108157 CrossRef PubMed.
- E. S. Vanamee and D. L. Faustman, Front. Immunol., 2023, 14, 1225704 CrossRef CAS PubMed.
- J. M. Andrade and M. G. Estévez-Pérez, Anal. Chim. Acta, 2014, 838, 1–12 CrossRef CAS PubMed.
- U. Wallin, B. Glimelius, K. Jirström, S. Darmanis, R. Y. Nong, F. Pontén, C. Johansson, L. Påhlman and H. Birgisson, Br. J. Cancer, 2011, 104, 1619–1627 CrossRef CAS PubMed.
- K. Jakubowska, A. Pryczynicz, V. Dymicka-Piekarska, D. Cepowicz, D. Jagodzińska, Ł. Lewczuk, A. Lebelt, M. Ozimkiewicz, P. Kiszło and K. Guzińska-Ustymowicz, Prog. Health Sci., 2016, 6(1), 40–48 CrossRef CAS.
- D. A. Brown, R. L. Ward, P. Buckhaults, T. Liu, K. E. Romans, N. J. Hawkins, A. R. Bauskin, K. W. Kinzler, B. Vogelstein and S. N. Breit, Clin. Cancer Res., 2003, 9, 2642–2650 CAS.
- R. Barderas, M. Mendes, S. Torres, R. A. Bartolomé, M. López-Lucendog, R. Villar-Vázquez, A. Peláez-García, E. Fuente, F. Bonilla and J. I. Casal, Mol. Cell. Proteomics, 2013, 12, 1602–1620 CrossRef CAS PubMed.
- M. Vocka, D. Langer, V. Fryba, J. Petrtyl, T. Hanus, M. Kalousova, T. Zima and L. Petruzelka, Cancer Biomarkers, 2018, 21(4), 869–874 CAS.
- I. F. Kamel, H. M. Elsadek, A. Mokhtar Ahmad and A. I. Elagrody, Egypt. J. Hosp. Med., 2021, 85(2), 3639–3644 CrossRef.
- C. Lungulescu, D. Sur, Ś. Răileanu, Ś. M. Dumitru, E. A. Dumitrescu and C. Virgil Lungulescu, J. Med. Rad. Onc., 2022, II(1), 1–8 Search PubMed.
- H. Xue, B. Lü, J. Zhang, M. Wu, Q. Huang, Q. Wu, H. Sheng, D. Wu, J. Hu and M. Lai, J. Proteome Res., 2010, 9, 545–555 CrossRef CAS PubMed.
- H. Zheng, S. Yu, C. Zhu, T. Guo, F. Liu and Y. Xu, Exp. Cell Res., 2021, 398, 112394 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d3sd00311f |
| This journal is © The Royal Society of Chemistry 2024 |