Enhanced in vivo antiviral activity against pseudorabies virus through transforming gallic acid into graphene quantum dots with stimulation of interferon-related immune responses†
Shiyi
Ye
a,
Fei
Su
a,
Junxing
Li
a,
Bin
Yu
a,
Lihua
Xu
a,
Tao
Xiong
c,
Kang
Shao
 *b and
Xiufang
Yuan
*a
*b and
Xiufang
Yuan
*a
aInstitute of Animal Husbandry and Veterinary Science, Zhejiang Academy of Agricultural Sciences, Hangzhou 310021, P. R. China. E-mail: yuanxf@zaas.ac.cn
bCollege of Chemical Engineering, Zhejiang University of Technology, Hangzhou 310014, P. R. China. E-mail: sk1033@zjut.edu.cn
cCollege of Life Science, Yangtze University, Jingzhou 434025, P. R. China
First published on 14th November 2023
Abstract
With the urgent need for antiviral agents, antiviral materials with high biocompatibility and antiviral effects have attracted a lot of attention. In this study, gallic acid, a natural polyphenolic compound, was transformed into biocompatible graphene quantum dots (GAGQDs) which exhibit enhanced antiviral activity against pseudorabies virus (PRV). The as-prepared GAGQDs inhibit PRV proliferation with a 104-fold reduction in viral titers. Investigation of the antiviral mechanism revealed that GAGQDs inhibit the adsorption, invasion and replication of PRV infection. Treatment with GAGQDs regulates the expression levels of interferon-related antiviral proteins, including mitochondrial antiviral-signaling protein (MAVS), signal transducer and activator of transcription 1 (STAT1) and 2′,5′-oligoadenylate synthetase 1 (OAS1), suggesting that GAGQDs can stimulate innate antiviral immune responses, resulting in enhanced antiviral effects. More importantly, GAGQD treatments alleviate clinical symptoms and reduce mortality in PRV-infected mice. Our results reveal the enhanced therapeutic effects of GAGQDs against PRV infection in vitro and in vivo, suggesting the potential of GAGQDs as a promising novel antiviral agent.
1. Introduction
Infectious diseases caused by viruses pose a serious threat to public health security for both humans and animals. Pseudorabies virus (PRV), also known as suid herpesvirus (SuHV-1) or Aujeszky's disease virus belong to alphaherpes viruses along with human herpes simplex virus type 1 and Varicella-zoster virus.1 PRV can infect a wide range of hosts, including mammals, ruminants, carnivores, ungulates and rodents, causing severe clinical symptoms and acute death.2,3 In particular, recent research has reported several cases of PRV infection in humans, suggesting that human beings may also be a potential host of PRV.4–8 Currently, no approved drugs are available for the treatment of pseudorabies in humans or animals. Despite commercial vaccines being widely used in controlling PRV in swine, the battle to control PR has not come to an end due to the mutation and recombination of PRV strains.9,10 Therefore, the development of an efficacious viricide is urgently needed.It is well known that natural compounds are widely accepted for the prevention and treatment of diseases throughout history and have become important resources for therapeutic agents. However, some natural compounds have the defects of poor water solubility or low bioavailability, which limit their application. With the rapid development of nanoscience and nanotechnology, antiviral nanomaterials have become competitive candidates for antiviral agents due to their excellent physicochemical properties and difficulty in causing drug resistance. Studies have revealed that some metallic and carbon-based nanomaterials like silver nanoparticles, graphene oxide, and carbon dots exhibit broad-spectrum antiviral properties.11–17 Among them, graphene quantum dots (GQDs) as a kind of carbon dot (CD) have emerged as very potent antiviral nano-agents due to their low toxicity, extensive precursor sources, low cost, simple synthesis and ease of industrialization.18–22 Basically, GQDs are ultra-small pieces of graphene consisting of one or a few layers (no more than 10) of graphene, and they share many properties with carbon quantum dots and graphene.21,23
Utilizing natural bioactive molecules as precursors for transformation into CDs or GQDs by carbonization is an effective method to overcome the defects of some natural compounds. By regulating suitable carbonization conditions, the as-synthesized CDs or GQDs can not only overcome the defects of precursors and maintain their biological activity, but also obtain the unique physical and chemical properties of nanomaterials. Lin et al. heated curcumin to synthesize carbon quantum dots (Cur-CQDs) which exhibit higher biocompatibility and superior antiviral capabilities against EV71.24 Glycyrrhizic acid was used to synthesize carbon dots which were demonstrated to inhibit the proliferation of porcine reproductive and respiratory syndrome virus and to stimulate interferon production like ISG-60 and ZAP-5 in MARC-145 cells.25 Glycyrrhizic-acid-based carbon dots also exhibit antiviral activity against influenza A virus.26 Although several studies report the antiviral activity of CDs, the specific antiviral mechanisms require further investigation.
In this study, gallic acid (GA), a naturally occurring low molecular weight polyphenolic compound in some fruits and plants, was used as a carbon source because it has several evident pharmacological effects, including anti-inflammatory, anti-microbial, anti-tumour and antioxidant properties; however, it is structurally unstable and has poor water solubility.27,28 As described in Fig. 1, gallic acid derived GQDs (GAGQDs) were synthesized using a simple one-pot hydrothermal method. In vitro assays revealed that the GAGQDs exhibit greatly superior solubility and prominent antiviral activity against PRV compared to GA. Analysis of the antiviral mechanism showed that GAGQDs can suppress the absorption, invasion and replication steps of the PRV proliferation process. In addition, GAGQD treatment can stimulate cells to regulate the expression of some interferon (IFN)-stimulating genes (including OAS1) and some transcription factors (like MAVS, STAT1) in IFN-related signal pathways. We also conducted in vivo studies in mice, which revealed improved survival and protective effects against morbidity in mice treated with GAGQDs. These results suggest that biocompatible GAGQDs may be promising candidates for therapeutic use against PRV infection.
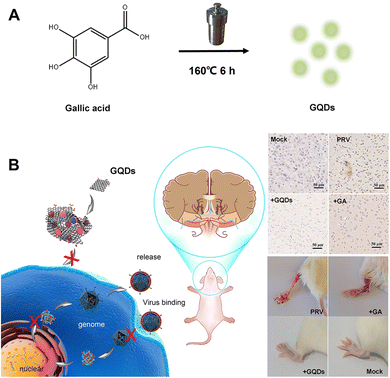 | ||
| Fig. 1 Schematic representation of (A) the one-step synthesis of GAGQDs and (B) their antiviral applications against PRV infection in vitro and in vivo. | ||
2. Experimental
2.1. Cells and viruses
Vero cells were propagated in Dulbecco's Modified Eagle Medium (DMEM, Gibco, USA) supplemented with 10% fetal bovine serum at 37 °C in a 5% CO2 incubator. A viral infection inhibition assay was performed with a PRV variant strain which was isolated from infected pigs immunized with commercial vaccine.2.2. Synthesis of GQDs
GAGQDs were synthesized from gallic acid powder (Aladdin Chemistry Co. Ltd Shanghai, China) with a modified one-pot hydrothermal method. Briefly, 85 mg of gallic acid was dissolved in 10 mL of absolute ethyl alcohol; then the mixture was transferred into a Teflon-lined autoclave and incubated for 6 h at 160 °C. The naturally cooled brownish-yellow supernatant was then transferred to a dialysis membrane (MWCO = 1 kDa) and dialyzed in ultrapure water for 8 h. The ultrapure water was replaced every 2 h during dialysis. The purified GAGQD powder was obtained by lyophilization.2.3. Cytotoxicity assay
The in vitro cytotoxicity of GAGQDs and GA was assessed using Counting Kit-8 (CCK8, Dojindo Molecular Technologies, Inc., Japan) according to the instructions of the product manufacturer. In brief, the Vero cells were seeded in a 96-well tissue culture plate and incubated in 5% CO2 at 37 °C for 24 h. The monolayer cells were incubated with materials at different concentrations diluted with DMEM. Five replicates were made for each concentration. After 72 h of incubation, 10 μL per well CCK8 reagent was added followed by 2 h of incubation at 37 °C. The absorbance at a wavelength of 450 nm was measured using a multifunction microplate reader. The cell viability was calculated as a percentage of the control (untreated) cells.2.4. Antiviral assay
Vero cells were inoculated on 12-well plates and cultured at 37 °C for 24 h. PRV (1000 pfu per well) was preincubated with GAGQDs or GA at different concentrations for 1 h at 37 °C. The confluent monolayer cells were washed twice with serum-free DMEM medium and infected with pretreated PRV. After 1 h of viral adsorption, the supernatant was discarded, and the cells were washed twice to remove the unadsorbed virus. An overlay medium consisting of DMEM, 2% FBS and 1.2% methylcellulose was added to each well. The cells were fixed with 10% formaldehyde after obvious plaques had appeared and stained with crystal violet. The number of plaques was counted.2.5. PRV replication stage analysis
2.6. Western blot
Cells were inoculated into a 6-well plate and cultured to a monolayer. The cells were infected with PRV treated with GAGQDs or GA. When the cells had been cultured for a specific time, the cells were rinsed 3 times with PBS and then treated with RIPA lysis buffer. Sodium dodecyl sulfate (SDS) loading buffer was added into the collected cell lysates and boiled for 10 min to obtain protein samples. Equivalent amounts of proteins from each treated group were loaded and electrophoresed on 12% sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE). Subsequently, the proteins were transferred to nitrocellulose (NC) membranes, followed by blocking for 1 h with 5% skim milk. After washing with TBST 3 times, the NC membrane was incubated with primary antibodies (PRV gE, OAS1, MAVS and so on) overnight at 4 °C. The rinsed membrane was then exposed to the corresponding HRP-conjugated secondary antibodies. The expressed protein signals were collected using an enhanced chemiluminescence instrument. Analysis of signal intensities was performed with ImageJ.2.7. In vivo antiviral efficacy of GAGQDs
All animal experiments were performed in compliance with the guidelines of the Laboratory Animal Center of Zhejiang Academy of Agricultural Sciences and approved by the Experimental Animal Welfare Ethics Committee of Zhejiang Academy of Agricultural Sciences.Forty 6-week-old female Balb/c mice were purchased from SLAC laboratory animal Co. Ltd (Shanghai, China). After one week of acclimation, these mice were randomly divided into 4 groups. The mice were intraperitoneally (i.p.) injected with 100 μL of PBS (mock and PRV controls) or compound (GAGQDs or GA group) at a dose of 50 mg kg−1 body weight. After 24 h, the mice in 3 groups (PRV, GAGQDs and GA group) were intra-plantar infected with PRV (103TCID50). Each mouse was subsequently intragastrically treated with PBS, GAGQDs, or GA daily for 10 consecutive days. The clinical symptoms, mental state, survival rates and body weights of the mice in each group were monitored every day.
2.8. TMT quantitative proteomic analysis
Cells were inoculated into a T75 culture flask and cultured to a monolayer. The cells were divided into 3 groups: treatment with 80 μg mL−1 GAGQDs or GA or DMEM. After treatment for 24 h, the medium was discarded and the cells were washed with precooled PBS 3 times. Then, the cells were placed on ice and 1.5 mL of ice-cold PBS was added. The cells were collected using a cell scraper and transferred to clean tubes. The cell suspensions were centrifuged at 1000 rpm for 10 min, the supernatant was discarded, and the cell pellet was frozen in liquid nitrogen for further proteomic analysis. Each treatment group had 5 biological replicates, three of which were used for Tandem mass tags (TMT) quantitative proteomic analysis, while the other two were used to validate the reliability of the proteomic results.3. Results and discussion
3.1. Characteristics of graphene quantum dots
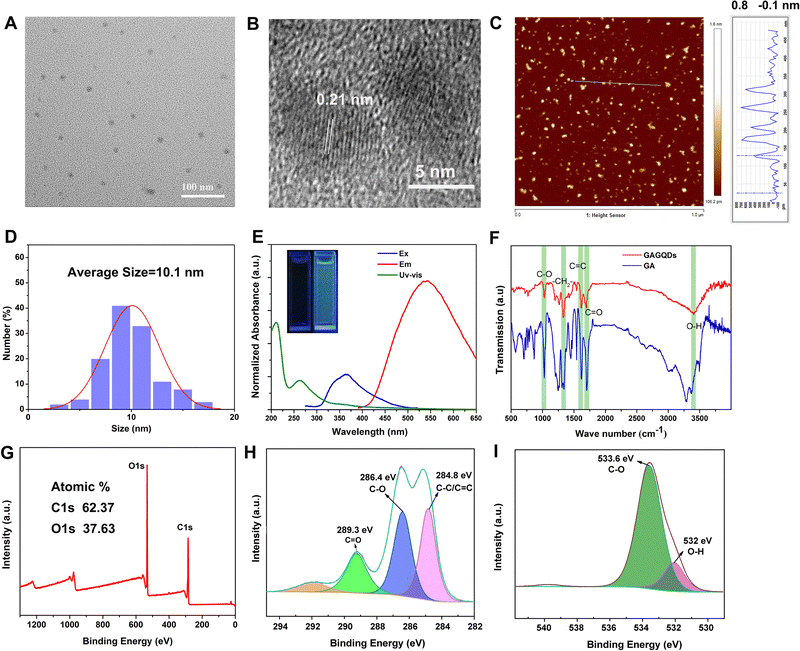
The surface morphology of the GAGQDs was characterized by transmission electron microscopy (Fig. 2A). The as-prepared GAGQDs were uniform, monodispersed spheres with mean diameters of 10.1 nm, from 100 counts (Fig. 2D), suggesting that the reaction process was steady, and no adverse side reaction had occurred. The high-resolution (HR)-TEM image showed that the intracrystalline d-spacing value was 0.21 nm for GAGQDs, corresponding to the (100) plane of graphite,21 suggesting that the carbon core of the GAGQDs formed as sp2 graphene carbon (020 facets) during the carbonization process. The AFM image (Fig. 2C) shows nicely distributed GAGQDs. The AFM line-scan profile showed that the as-prepared GAGQDs are mostly monolayered with a thickness of 0.5–1 nm (Fig. S1, ESI†) because of the rigid molecular structure of gallic acid. As shown in the Raman spectrum (Fig. S2, ESI†), a typical D band relating to local structural defects and disorders at approximately 1390 cm−1 and a G band ascribed to sp2-hybridized carbon atoms in the graphitic layers at approximately 1587 cm−1 were observed, corresponding to the structural features of the GQDs.The morphology and activity of the carbonized dots are closely related to the structure of the precursors and carbonization conditions.29 In this study, after a mild hydrothermal reaction, gallic acid polymerized into single-layered graphene quantum dots instead of carbonized polymer dots, which should be closely related to the benzene ring structure of gallic acid.
The structures of the GAGQDs were further characterized using optical spectroscopies. As shown in Fig. 2E, both GA and GAGQDs showed the same two strong absorbance peaks at about 200 and 270 nm, which were ascribed to the π–π* transition of an aromatic C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C bond. The GAGQDs exhibited very similar spectral properties to GA, further supporting the polyphenolic functional structures being preserved on the GQDs. The absorbance of GAGQDs had a small shoulder at around 350 nm, which is consistent with the n–π* transition of C
C bond. The GAGQDs exhibited very similar spectral properties to GA, further supporting the polyphenolic functional structures being preserved on the GQDs. The absorbance of GAGQDs had a small shoulder at around 350 nm, which is consistent with the n–π* transition of C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O or COOH groups. PL excitation and emission spectra of GAGQDs are shown in Fig. 2E. The PL excitation spectra show pronounced excitation bands at 365 nm, consistent with the features of the absorption spectra. The emission peak of GAGQDs is at 550 nm. The insert in Fig. 2E shows that the GAGQD solutions fluoresce green when excited by directed illumination of 365 nm UV light.
O or COOH groups. PL excitation and emission spectra of GAGQDs are shown in Fig. 2E. The PL excitation spectra show pronounced excitation bands at 365 nm, consistent with the features of the absorption spectra. The emission peak of GAGQDs is at 550 nm. The insert in Fig. 2E shows that the GAGQD solutions fluoresce green when excited by directed illumination of 365 nm UV light.
Fourier transform infrared (FTIR) spectroscopy was performed to examine the functional groups of GAGQDs and GA (Fig. 2F). Both GAGQDs and GA revealed O–H stretching vibrations (3407 cm−1, 3369 cm−1), C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O stretching vibrations (1691 cm−1, 1701 cm−1), C
O stretching vibrations (1691 cm−1, 1701 cm−1), C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C stretching vibrations (1617 cm−1, 1618 cm−1), –CH2– bending vibrations (1330 cm−1, 1339 cm−1) and C–O stretching vibrations (1028 cm−1, 1026 cm−1), which indicate that the as-formed GAGQDs still retain abundant −OH and −COOH surface functional groups after high-temperature carbonization.
C stretching vibrations (1617 cm−1, 1618 cm−1), –CH2– bending vibrations (1330 cm−1, 1339 cm−1) and C–O stretching vibrations (1028 cm−1, 1026 cm−1), which indicate that the as-formed GAGQDs still retain abundant −OH and −COOH surface functional groups after high-temperature carbonization.
The elemental constituents and functional groups of GAGQDs were characterized by X-ray photoelectron spectroscopy (XPS). As shown in Fig. 2G, two peaks of C1s and O1s could obviously be recognized at 290 and 529 eV. The elemental analysis revealed that GAGQDs contained 62.37% carbon and 37.63% oxygen. C1s spectra can be decomposed into 3 peaks, corresponding to different functional groups. The peak at 284.8 eV was assigned to C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C/C–C, while C–O was at 286.4 eV and C
C/C–C, while C–O was at 286.4 eV and C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O was at 289.3 eV. Compared to the spectra of the parental compound GA, most characteristic functional group peaks were retained during the carbonization process, indicating that the as-synthetic GAGQDs preserve most functional groups and fragment molecules presumably along with their biological activities.
O was at 289.3 eV. Compared to the spectra of the parental compound GA, most characteristic functional group peaks were retained during the carbonization process, indicating that the as-synthetic GAGQDs preserve most functional groups and fragment molecules presumably along with their biological activities.
3.2. GAGQDs exhibit potent antiviral activity against PRV
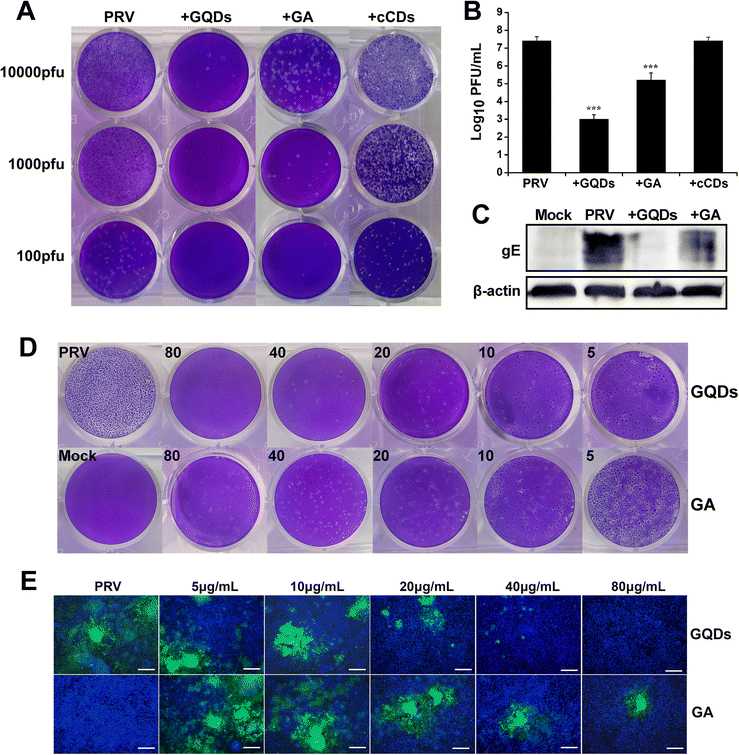
Cytotoxicity experiments were performed to assess the biocompatibility of GAGQDs in vitro. Briefly, Vero cells were cultured separately with GAGQDs or gallic acid at different concentrations ranging from 5 to 320 μg mL−1 for 48 h. As shown in Fig. S3 (ESI†), the cell viability was about 80% when treated with 160 μg mL−1 GAGQDs, while the viability was 45.6% when treated with gallic acid of the same concentration. These results revealed significant improvement in biocompatibility through transformation into GAGQDs. Furthermore, the cell morphology was not affected when the concentration of the materials (both GAGQDs and GA) was 80 μg mL−1 (Fig. S4, ESI†). Thus, 80 μg mL−1 was used in subsequent experiments.Gallic acid has been reported to have antiviral activity.30,31 Gallic acid based GQDs showed better stability and biocompatibility, but their antiviral effect on PRV was unclear. To evaluate the antiviral potentials of GAGQDs, Vero cells were infected with PRV in the presence GAGQDs, GA and traditional CDs synthesized from sodium citrate (cCDs). As shown in Fig. 3A, virus incubation with GAGQDs (80 μg mL−1), followed by infection of cells exhibited much better antiviral effects than parental GA. The addition of GAGQDs significantly reduced PRV titers with a maximum reduction of approximately 104-fold by plaque assay (Fig. 3B). However, cCDs showed no inhibitory effect against PRV at the same concentration, which may be attributed to citric acid and ethylenediamine: the raw materials of the cCDs showed no antiviral activity. These results indicate that the antiviral activity of carbon quantum dots or GQDs is closely related to their synthetic precursors. The expression levels of PRV gE protein was further investigated to confirm the influence of GAGQDs on PRV proliferation. Western blot analysis revealed a remarkable decrease in the expression levels of PRV gE protein under treatment with GAGQDs (Fig. 3C).
Moreover, the antiviral activity of GAGQDs was concentration dependent. The inhibition of PRV plaque formation was enhanced with an increase in GAGQD concentration (Fig. 3D). GAGQDs showed anti-PRV activity at a low concentration of 5 μg mL−1. To further confirm the result, an inhibitory assay was performed using recombinant PRV expressing green fluorescent protein (GFP-PRV). Compared with GA treatment, the number of PRV-infected cells was significantly decreased in the presence of GAGQDs (Fig. 3E), which was consistent with the plaque-reduction assay. These results further demonstrated the efficient antiviral activity of GAGQDs against PRV.
3.3. GAGQDs inhibit PRV adsorption, invasion and replication
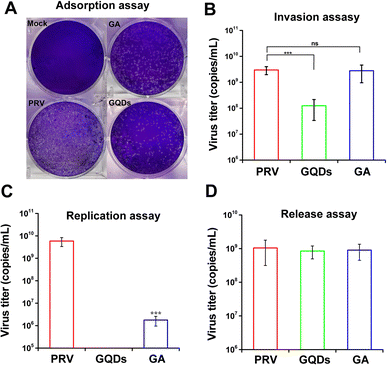
In order to identify the potential antiviral mechanism of GAGQDs, the effects of GAGQDs on each stage of viral proliferation were analyzed. In an adsorption assay, treatment with GAGQDs significantly reduced the number of plaques of PRV (Fig. 4A), indicating that GAGQDs can inhibit PRV adsorption. Moreover, GA also exhibits adsorb inhibition activity, but is less effective than GAGQDs. The influence of GAGQDs and GA on the invasion process of PRV was evaluated by real-time PCR. As shown in Fig. 4B, GAGQDs reduced the virus titer of PRV by about 10-fold, while the GA-treated group showed no invasion inhibition. In the replication assay, PRV replication was almost completely abolished in the GAGQD-treated group, while GA treatment also decreased the infectious virus titer compared to the control group (Fig. 4C). These results indicate that both GAGQDs and GA inhibit PRV replication, but GAGQDs show more effective replication inhibition than GA. Moreover, whether the GAGQDs influence the release stage of PRV was analyzed. As shown in Fig. 4D, no significant difference was observed between GAGQDs, GA and the control group, indicating that GAGQDs and GA had no inhibitory effect on the viral release of PRV. In summary, GAGQDs suppress PRV infection by targeting the stages of absorption, invasion and replication, while the raw material GA inhibits only the absorption and replication processes.3.4. GAGQDs regulate the expression of antiviral genes MAVS and OAS1 in Vero cells
To further understand the antiviral mechanism of GAGQDs, the intracellular proteins levels were explored by Tandem mass tag (TMT)-based quantitative proteomic analysis. A total of 6373 proteins were identified, among which 588 were upregulated and 455 downregulated in the GAGQD-treated group while 621 were upregulated and 415 downregulated in the GA-treated group. The overview of differentially expressed cellular proteins and quantified proteins are summarized by a dot-plot (Fig. S6, ESI†).To validate the proteomic results, 3 typical differentially expressed proteins were selected for western blot analysis. As shown in Fig. 5B, the expressions of mitochondrial antiviral-signaling protein (MAVS) and 2′,5′-oligoadenylate synthetase 1 (OAS1) were upregulated in GAGQD-treated cells, while these proteins were not significantly changed in GA-treated cells. In GAGQD-treated cells, the expression of STAT1 (signal transducer and activator of transcription 1) was downregulated, but not significantly altered in GA-treated cells. ImageJ software was used to convert the gray values to the quantitative results. As shown in Fig. 5B, the ratios identified by the western blot assay were consistent with those from TMT-based proteomic analysis. GAGQD treatment regulated the MAPK signaling pathway (Fig. 5A and Fig. S8, ESI†) but not GA treatment (Fig. S9, ESI†), suggesting that GAGQDs inhibit PRV infection by regulating the expression levels of the transcription factors relevant to the MAPK pathway.
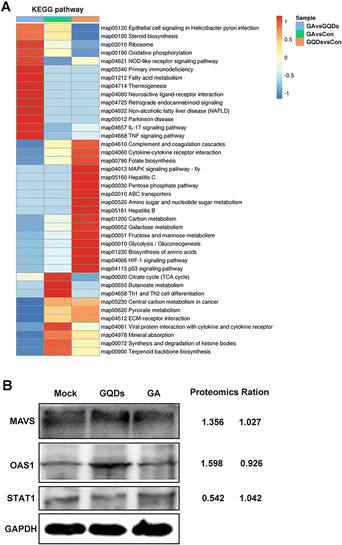 | ||
| Fig. 5 (A) Effect of GAGQDs on the KEGG pathway in Vero cells. (B) Proteomic data from western blot analysis. | ||
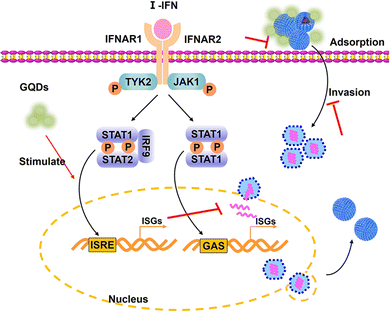
Combined with previous results, the GAGQDs inhibit PRV infection by multisite inhibition mechanisms. GAGQDs can regulate innate cellular immunity related signaling pathways and stimulate the expression of ISGs to achieve antiviral properties. MAVS is a key adapter protein of RIG-I/MDA5 antiviral signaling and IFN production, which have been widely demonstrated to assist the host in defending against viral infection.32,33 Studies have shown that viruses can promote the degradation or cleavage of MAVS to escape the host's innate antiviral immune response. For example, SARS-CoV-2 membrane glycoprotein M was found to attenuate the innate antiviral response by inhibiting the aggregation of MAVS and its recruitment of downstream molecules like TRAF3, TBK1, and IRF3.34 OAS1, an important interferon-stimulated gene was upregulated in GAGQD-treated cells. The OAS1/RNaseL signaling pathway is closely related to resistance to numerous viral infections.35 In humans and animals, OAS1 has been demonstrated to have antiviral activities against numerous viruses, such as severe acute respiratory syndrome coronavirus 2,36 West Nile virus,37 hepatitis C virus,38 and porcine reproductive and respiratory syndrome virus.39 In this study, GAGQDs showed a more efficient anti-PRV effect than GA, which may be attributed to MAVS and OAS1 being upregulated in GAGQDs-treated cells rather than in GA-treated cells.
PRV achieves immune escape by regulating the cellular antiviral immune response and inflammatory responses mediated by the JAK/STAT and MAPK pathways.40 A previous study showed that PRV infection in PK-15 cells could significantly upregulate the expression levels of c-Jun and STAT1,41 which are associated with the MAPK pathway. The expression level of STAT1 was significantly reduced upon GAGQD treatment in this study. GAGQD treatment can stimulate cells to regulate the expression of some IFN-stimulating genes (including OAS1) and some transcription factors (like MAVS, STAT1) in IFN-related signal pathways, which play an important role in limiting viral infection. Combined with the proteomic results, GAGQDs can regulate the innate cellular immunity related signaling pathways and thus exhibit antiviral properties.
3.5. GAGQDs inhibit PRV infection in vivo
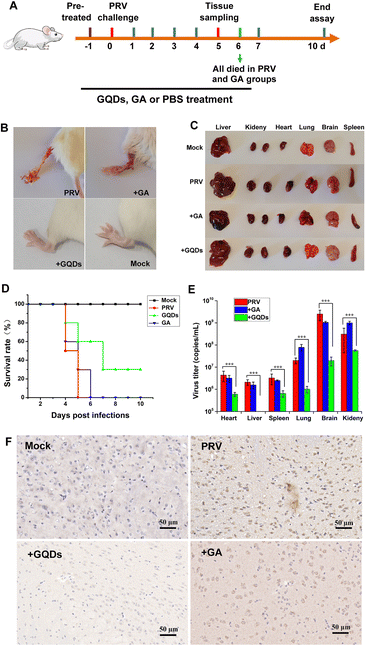
Considering the potent antiviral efficacy of GAGQDs on cells, we proceeded to verify and compare the anti-PRV activity of GAGQDs and GA in an animal model (Fig. 6A). Seven-week-old female Balb/c mice were intra-plantar infected with PRV. Each mouse was subsequently treated daily with PBS, GAGQDs, or GA via intragastric administration. Four days after PRV infection, all the mice in the PRV-infected group developed severe neurological symptoms, like abnormal excitement, itching, and gnawing hindlimbs (the inoculum site, Fig. 6B), and then died. In the GAGQD-treated group (50 mg kg−1), fewer mice developed symptoms, and the onset of symptoms was about 24 h later than in the PRV-infected group or GA-treated group. It was found that the PRV-infected group and the GA-treated group had obvious infarct in the spleen or congestion in the liver, but there were no significant differences between the GAGQD-treated group and the control group (Fig. 6C). All the mice in the PRV-infected group died 5 d after infection, and all the mice in the GA-treated group died 6 d after infection. The remaining mice were kept until the 10th day. Three mice in the GAGQD-treated group still survived without any symptoms. Infected mice treated with GAGQDs exhibited significant anti-PRV efficacy, featuring a higher survival rate (3/10) and milder clinical appearances, while all the mice in the GA-treated group died (Fig. 6D).Furthermore, in the GAGQD-treated group, no viruses were detected in the organs (brain, lung, heart, liver, spleen and kidney) of non-symptomatic mice, while symptomatic mice showed a much lower virus load than the PRV or GA-treated groups (Fig. 6E). Moreover, the results of brain immunohistochemistry targeting the PRV gE protein showed that a PRV-positive signal was found in mice from the PRV-infected group and GA-treated group, but not in the GAGQD-treated group. These results clearly demonstrate that GAGQDs are an effective antiviral agent for PRV infection in vivo.
Conclusions
In the present study, we synthesized graphene quantum dots from gallic acid using a simple one-pot hydrothermal method. The as-synthesized GAGQDs were employed as a high-performance inhibitor against PRV infection in vitro and in vivo. Compared to natural GA, GAGQDs exhibit greatly superior solubility and prominent antiviral activity against PRV. In addition, GAGQDs can suppress the absorption, invasion and replication steps of the PRV proliferation process. We also conducted in vivo studies in mice, which revealed that treatment with GAGQDs significantly improved survival rates and protective effects against morbidity in mice. Based on proteomic data, we found that GAGQDs can stimulate cells to modulate the expression of some transcription factors in interferon (IFN)-related signal pathways (Fig. 7). These results prove that GAGQDs are directly involved in innate immune regulation to achieve an antiviral effect. In summary, we used gallic acid as a carbon source to synthesize GAGQDs with enhanced water dispersibility, higher biocompatibility and improved antiviral properties, which may be promising candidates for therapeutic use against PRV infection.Author contributions
Shiyi Ye: conceptualization, methodology, investigation, visualization and writing the manuscript; Fei Su: methodology, data curation; Bin Yu: Resources; Junxing Li: methodology and investigation; Lihua Xu: methodology and investigation; Tao Xiong: methodology and resources; Kang Shao: conceptualization, data curation, writing – review & editing; Xiufang Yuan: project administration, supervision, conceptualization, funding acquisition. All authors have given approval to the final version of the manuscript.Conflicts of interest
There are no conflicts to declare.Acknowledgements
This work was financially supported by the Key R&D Program of Zhejiang Province (Grant No. 2022C02024) and Zhejiang Provincial Natural Science Foundation of China (Grant No. LTGN23C180002).References
- M. Vallbracht, M. Backovic, B. G. Klupp, F. A. Rey and T. C. Mettenleiter, Adv. Virus Res., 2019, 104, 225–281 CrossRef CAS PubMed.
- Z. Bo and X. Li, Viruses, 2022, 14, 1003 CrossRef CAS PubMed.
- L. Tan, J. Yao, Y. Yang, W. Luo, X. Yuan, L. Yang and A. Wang, Virol. Sin., 2021, 36, 588–607 CrossRef PubMed.
- J. Ai, S. Weng, Q. Cheng, P. Cui, Y. Li, H. Wu, Y. Zhu, B. Xu and W. Zhang, Emerging Infect. Dis., 2018, 24, 1087–1090 CrossRef.
- H. Yang, H. Han, H. Wang, Y. Cui, H. Liu and S. Ding, Front. Neurol., 2019, 10, 534 CrossRef PubMed.
- L. Zheng, X. Liu, D. Yuan, R. Li, J. Lu, X. Li, K. Tian and E. Dai, Transboundary Emerging Dis., 2019, 66, 2562–2565 CrossRef PubMed.
- Q. Liu, X. Wang, C. Xie, S. Ding, H. Yang, S. Guo, J. Li, L. Qin, F. Ban, D. Wang, C. Wang, L. Feng, H. Ma, B. Wu, L. Zhang, C. Dong, L. Xing, J. Zhang, H. Chen, R. Yan, X. Wang and W. Li, Clin. Infect. Dis., 2021, 73, e3690–e3700 CrossRef CAS.
- J. Ou, S. Cai, F. Zheng, G. Lu and G. Zhang, J. Infect., 2020, 80, 578–606 CrossRef.
- Z. Bo, Y. Miao, R. Xi, X. Gao, D. Miao, H. Chen, Y. S. Jung, Y. Qian and J. Dai, Transboundary Emerging Dis., 2021, 68, 1454–1464 CrossRef CAS PubMed.
- G. Wang, R. Chen, P. Huang, J. Hong, J. Cao, Q. Wu, W. Zheng, L. Lin, Q. Han, Y. Chen and N. Xia, Antiviral Res., 2021, 186, 105014 CrossRef CAS PubMed.
- X. C. Xu, J. Zhang, S. Liu, C. H. Wang, H. Y. Wang, H. H. Fan, Y. G. Tong, H. Y. Liu and D. S. Zhou, Small Struct., 2022, 3, 2200021 CrossRef CAS.
- A. Mushtaq, M. Z. Iqbal and X. D. Kong, J. Mater. Chem. B, 2022, 10, 5323–5343 RSC.
- E. Sharifi, S. Yousefiasl, M. Trovato, R. Sartorius, Y. Esmaeili, H. Goodarzi, M. Ghomi, A. Bigham, F. D. Moghaddam, M. Heidarifard, S. Pourmotabed, E. Nazarzadeh Zare, A. C. Paiva-Santos, N. Rabiee, X. Wang and F. R. Tay, J. Nanobiotechnol., 2023, 21, 199 CrossRef CAS PubMed.
- X. H. Pan, Y. P. Zhang, Y. M. Zhao, S. Q. Yao, C. X. Guan, L. Q. Wang and L. Y. Chen, Arch. Virol., 2022, 167, 1619–1636 CrossRef CAS PubMed.
- Z. A. Ratan, F. R. Mashrur, A. P. Chhoan, S. M. Shahriar, M. F. Haidere, N. J. Runa, S. Kim, D. H. Kweon, H. Hosseinzadeh and J. Y. Cho, Pharmaceutics, 2021, 13, 2034 CrossRef CAS PubMed.
- S. Ye, K. Shao, Z. Li, N. Guo, Y. Zuo, Q. Li, Z. Lu, L. Chen, Q. He and H. Han, ACS Appl. Mater. Interfaces, 2015, 7, 21571–21579 CrossRef CAS.
- D. J. H. Tng and J. G. H. Low, Antiviral Res., 2023, 210, 105488 CrossRef CAS.
- A. Serrano-Aroca, K. Takayama, A. Tunon-Molina, M. Seyran, S. S. Hassan, P. P. Choudhury, V. N. Uversky, K. Lundstrom, P. Adadi, G. Palu, A. A. A. Aljabali, G. Chauhan, R. Kandimalla, M. M. Tambuwala, A. Lal, T. M. Abd El-Aziz, S. Sherchan, D. Barh, E. M. Redwan, N. G. Bazan, Y. K. Mishra, B. D. Uhal and A. Brufsky, ACS Nano, 2021, 15, 8069–8086 CrossRef CAS PubMed.
- Y. X. Xue, C. C. Liu, G. Andrews, J. Y. Wang and Y. Ge, Nano Convergence, 2022, 9, 15 CrossRef CAS PubMed.
- P. Innocenzi and L. Stagi, Chem. Sci., 2020, 11, 6606–6622 RSC.
- C.-T. Hsieh, S. Gu, Y. A. Gandomi, C.-C. Fu, P.-Y. Sung, R.-S. Juang and C.-C. Chen, J. Colloid Interface Sci., 2023, 630, 1–10 CrossRef CAS PubMed.
- D. Iannazzo, A. Pistone, S. Ferro, L. De Luca, A. M. Monforte, R. Romeo, M. R. Buemi and C. Pannecouque, Bioconjugate Chem., 2018, 29, 3084–3093 CrossRef CAS PubMed.
- C. Hadad, J. M. González-Domínguez, S. Armelloni, D. Mattinzoli, M. Ikehata, A. Istif, A. Ostric, F. Cellesi, C. M. Alfieri, P. Messa, B. Ballesteros and T. Da Ros, Nano Res., 2021, 14, 674–683 CrossRef CAS.
- C. Lin, L. Chang, H. Chu, H.-J. Lin, P. Chang, R. Y. L. Wang, B. Unnikrishnan, J. Mao, S. Chen and C. Huang, Small, 2019, 15, 1902641 CrossRef CAS PubMed.
- T. Tong, H. Hu, J. Zhou, S. Deng, X. Zhang, W. Tang, L. Fang, S. Xiao and J. Liang, Small, 2020, 16, 1906206 CrossRef CAS PubMed.
- C. Li, P. Han, H. Mao, C. Lv, K. Huang and M. Jin, ACS Appl. Mater. Interfaces, 2023, 15, 10441–10451 CrossRef CAS.
- J. Bai, Y. Zhang, C. Tang, Y. Hou, X. Ai, X. Chen, Y. Zhang, X. Wang and X. Meng, Biomed. Pharmacother., 2021, 133, 110985 CrossRef CAS PubMed.
- S. Lu, L. Liu, H. Wang, W. Zhao, Z. Li, Z. Qu, J. Li, T. Sun, T. Wang and G. Sui, Biomater. Sci., 2019, 7, 3258–3265 RSC.
- C. Xia, S. Zhu, T. Feng, M. Yang and B. Yang, Adv. Sci., 2019, 6, 1901316 CrossRef CAS.
- J.-H. Lee, M. Oh, J. H. Seok, S. Kim, D. B. Lee, G. Bae, H.-I. Bae, S. Y. Bae, Y.-M. Hong and S.-O. Kwon, Viruses, 2016, 8, 157 CrossRef PubMed.
- N. A. A. Zahrani, R. M. El-Shishtawy and A. M. Asiri, Eur. J. Med. Chem., 2020, 204, 112609 CrossRef PubMed.
- Z. Ren, T. Ding, Z. Zuo, Z. Xu, J. Deng and Z. Wei, Front. Immunol., 2020, 11, 1030 CrossRef CAS.
- Y. Q. Chen, Y. H. Shi, J. Wu and N. Qi, Front. Microbiol., 2021, 12, 744348 CrossRef PubMed.
- Y. Z. Fu, S. Y. Wang, Z. Q. Zheng, Y. Huang, W. W. Li, Z. S. Xu and Y. Y. Wang, Cell Mol. Immunol., 2021, 18, 613–620 CrossRef CAS.
- H. Kristiansen, C. A. Scherer, M. McVean, S. P. Iadonato, S. Vends, K. Thavachelvam, T. B. Steffensen, K. A. Horan, T. Kuri, F. Weber, S. R. Paludan and R. Hartmann, J. Virol., 2010, 84, 11898–11904 CrossRef CAS PubMed.
- S. R. Zhou, G. Butler-Laporte, T. Nakanishi, D. R. Morrison, J. Afilalo, M. Afilalo, L. Laurent, M. Pietzner, N. Kerrison, K. Q. Zhao, E. Brunet-Ratnasingham, D. Henry, N. Kimchi, Z. Afrasiabi, N. Rezk, M. Bouab, L. Petitjean, C. Guzman, X. Q. Xue, C. Tselios, B. Vulesevic, O. Adeleye, T. Abdullah, N. Almamlouk, Y. H. Chen, M. Chasse, M. Durand, C. Paterson, J. Normark, R. Frithiof, M. Lipcsey, M. Hultstrom, C. M. T. Greenwood, H. Zeberg, C. Langenberg, E. Thysell, M. Pollak, V. Mooser, V. Forgetta, D. E. Kaufmann and J. B. Richards, Nat. Med., 2021, 27, 659–667 CrossRef CAS.
- H. T. Tag-El-Din-Hassan, N. Sasaki, K. Moritoh, D. Torigoe, A. Maeda and T. Agui, Jpn. J. Vet. Res., 2012, 60, 95–103 Search PubMed.
- N. G. B. El Din, M. A. Anany, R. M. Dawood, M. K. Ibrahim, R. El-Shenawy, Y. S. El Abd and M. K. El Awady, Viral Immunol., 2015, 28, 509–516 CrossRef.
- J. H. Zhao, N. Feng, Z. H. Li, P. Wang, Z. Y. Qi, W. Liang, X. Zhou, X. W. Xu and B. Liu, Antiviral Res., 2016, 132, 268–273 CrossRef CAS PubMed.
- X. Li, S. Chen, L. Zhang, G. Niu, X. Zhang, L. Yang, W. Ji and L. Ren, Int. J. Mol. Sci., 2022, 23, 4469 CrossRef CAS.
- X. Song, H. Hu, Z. Hu, Y. Zhang, H. Wan, Z. Yin, L. Li, X. Liang, X. Zhao and L. Yin, Front. Microbiol., 2022, 13, 985108 CrossRef.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d3tb01844j |
| This journal is © The Royal Society of Chemistry 2024 |