Open Access Article
Open Access ArticleSelectivity in the chiral self-assembly of nucleobase-arylazopyrazole photoswitches along DNA templates†
Noemí
Nogal‡
 a,
Santiago
Guisán‡
a,
David
Dellemme
b,
Mathieu
Surin
a,
Santiago
Guisán‡
a,
David
Dellemme
b,
Mathieu
Surin
 *b and
Andrés
de la Escosura
*b and
Andrés
de la Escosura
 *ac
*ac
aDepartament of Organic Chemistry, Universidad Autónoma de Madrid, Campus de Cantoblanco, Madrid 28049, Spain. E-mail: andres.delaescosura@uam.es
bLaboratory for Chemistry of Novel Materials, Center for Innovation in Materials and Polymers, University of Mons – UMONS, 20 Place du Parc, Mons B-7000, Belgium. E-mail: mathieu.surin@umons.ac.be
cInstitute for Advanced Research in Chemistry (IAdChem), Campus de Cantoblanco, Madrid 28049, Spain
First published on 12th March 2024
Abstract
The control of supramolecular DNA assembly through external stimuli such as light represents a promising approach to control bioreactions, and modulate hybridization or delivery processes. Here, we report on the design of nucleobase-containing arylazopyrazole photoswitches that undergo chiral organization upon self-assembly along short DNA templates. Chiroptical spectroscopy shows that the specific nucleobases allow selectivity in the resulting supramolecular DNA complexes, and UV light irradiation triggers partial desorption of the arylazopyrazole photoswitches. Molecular modeling studies reveal the differences of binding modes between the two configurations in the templated assembly. Remarkably, our results show that the photoswitching behaviour controls the self-assembly process along DNA, opening the way to potential applications as nano- and biomaterials.
Introduction
The use of molecular switches and motors in the field of supramolecular chemistry has enabled the development of stimuli-responsive dynamic materials mimicking certain properties of living systems, such as the transport of ions across membranes, multivalent sensing and differentiation of cells.1–5 Recently, light-driven switches and motors have been utilized for fabrication of responsive DNA-based supramolecular assemblies.6–10 Interfacing photo-controllable molecules with programmable DNA structures is expected to provide sophisticated nanomaterials with possible applications in e.g. the modulation of enzymatic activity or photosensitive surfactants capable of controlling the DNA conformation.10–18In this context, azobenzene derivatives are powerful molecular photoswitches for the generation of self-assembled structures with new properties such as smart organic reagents,19 tuneable biomolecules20 and photoactive polymers.21 Recently, novel azoheterocyclic photoswitches have been reported (e.g. arylazopyrazoles),22–25 which show higher thermal stability and facilitate the process of photoisomerization.26,27 Herein we report about two new arylazopyrazole compounds bearing a nucleobase, either thymine or adenine, with the goal to assess their supramolecular organization templated by complementary oligonucleotides (see Fig. 1). This design is motivated by previous works that make use of short DNA single strands such as oligoadenine (dAn) or oligothymine (dTn) to template the self-assembly of molecules bearing a complementary recognition unit.13,28–32 The synthesis of these azo compounds involves the incorporation of a carboxylic acid group at the pyrazole moiety, which enables aqueous solubility as a necessary requirement to be interfaced with the DNA templates.
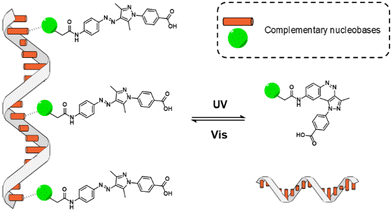 | ||
| Fig. 1 Cartoon of the photoswitchable assemblies formed between oligonucleotide templates and nucleobase-arylazopyrazole derivatives. | ||
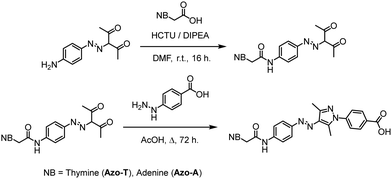
Synthesis of nucleobase-arylazopyrazoles
The synthetic route towards the target arylazopyrazole derivatives Azo-T and Azo-A is shown in Scheme 1. The nucleobase was introduced using the corresponding carboxymethyl derivatives,33,34 through an amide bond formation reaction with (E)-3-((4-aminophenyl)diazenyl)pentane-2,4-dione35 using DMF as solvent and HCTU as coupling reagent.36 The last step consisted in the condensation of 4-hydrazinylbenzoic acid with the 3,5-dimethylisoxazole moiety of the previous nucleobase-functionalized intermediates, to yield the desired products via the Knorr reaction for pyrazole synthesis (procedures details and full characterization of products in the ESI†).37Spectroscopic studies
The self-assembly on DNA templates was assessed by UV-Vis absorption and circular dichroism (CD) spectroscopies, using oligonucleotides with different sequences and lengths, in 50 mM Tris–HCl buffer at pH 7.5 and 3% DMSO to ensure solubility. The stoichiometry of nucleobase-arylazopyrazole compound with respect to the oligonucleotide was 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 in all experiments. Control spectra corresponding to either the oligonucleotides or the azo compounds alone are shown in Fig. S1–S4 (ESI†).
1 in all experiments. Control spectra corresponding to either the oligonucleotides or the azo compounds alone are shown in Fig. S1–S4 (ESI†).
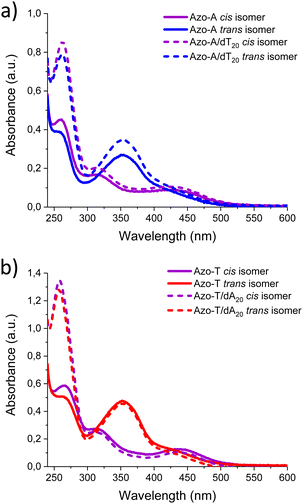
Fig. 2 shows the effect of photoisomerisation on the UV-Vis absorption spectra for Azo-A and Azo-T (20 μM) in presence and/or absence of dT20 and dA20 (2 μM). The absorption band at 260 nm doubles in intensity due to the presence of oligonucleotides in the mixture. In addition, in the upper panel of the figure, corresponding to Azo-A samples, the π → π* transition band of the arylazopyrazole trans isomer shows a considerable increase in absorption after irradiating for 15 min with blue light. When the samples are irradiated with UV light for 5 min, a weaker band appears in the visible region, with a maximum wavelength around 430 nm, corresponding to the n → π* transition of the arylazopyrazole cis isomer.
We investigated how the type of nucleobase in the oligonucleotide template influences the chiroptical response in the spectral region corresponding to the π → π* transition band of the arylazopyrazole trans isomer (Fig. 3a and b). DNA templates such as dT20 and dA20 were employed as model sequences to induce the self-assembly with Azo-A and Azo-T, respectively. As commonly observed with other types of supramolecular assemblies in aqueous media,38–40 the addition of salt promotes the non-covalent interactions between both components. For Azo-A, an induced circular dichroism (ICD) in the spectral region where it absorbs appears with dT20 at both 1 and 5 M NaCl (blue curves in Fig. 3a and b), meaning that Azo-A adopts a chiral assembly when interacting with the template. The ICD signal intensity increases with the medium ionic strength. For Azo-T (red curves in Fig. 3a and b), there is ICD signal only at the highest salt concentration, indicating that a higher ionic strength is required for the formation of a supramolecular complex with dA20. The most important observation from these results, however, is that the sign of the CD band is determined by the canonical base present in the oligonucleotide: a positive Cotton effect with maximum at 360 nm is observed for dA20, in contrast to the negative one found at 340 nm for dT20. This effect was not observed when the azo compound with the opposite nucleobase was used (Azo-A with dA20 and Azo-T with dT20; see Fig. S5 and S6), proving that complementarity between the bases is necessary for self-assembly to occur.
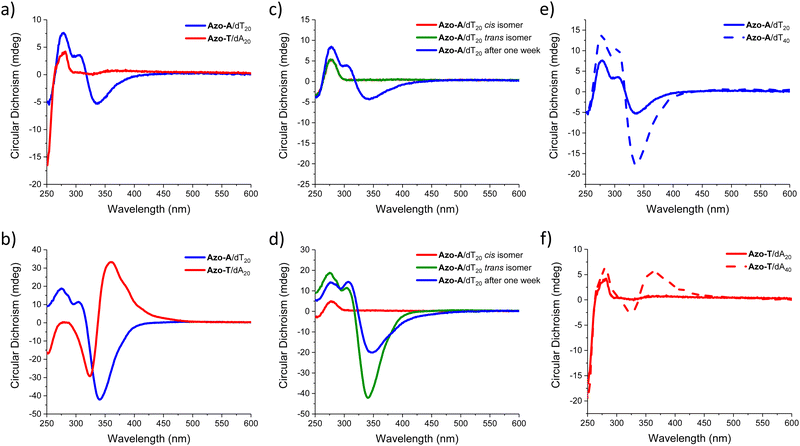 | ||
| Fig. 3 (a and b) CD spectra of oligonucleotide/nucleobase-arylazopyrazole assemblies at 20 °C and (a) 1 M or (b) 5 M NaCl (the corresponding UV-Vis spectra are shown in Fig. S10, ESI†). (c and d) CD spectra of Azo-A/dT20 assemblies at 20 °C and (c) 1 M or (d) 5 M NaCl, recorded after the initial preparation, irradiation with 365 nm UV light, 465 nm blue light, and after one week. (e and f) CD spectra showing the effect of oligonucleotide templates with a different number of bases at 20 °C and NaCl 1M, for (e) Azo-A/dT20 and Azo-A/dT40; and for (f) Azo-T/dA20 and Azo-T/dA40. | ||
After showing the self-assembly of trans arylazopyrazoles on DNA templates, we investigated the effect of UV light on the assemblies, as it switches the chromophore from the trans to the cis configuration. Fig. S7–S9 (ESI†) show the 1H-NMR and UV-Vis monitoring of the photoswitching behaviour of Azo-A and Azo-T in DMSO, revealing trans/cis ratios in the two possible photostationary states of 18![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 82 and 66
82 and 66![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 34 for the former (as determined by NMR in 16 mM concentration), and of 4
34 for the former (as determined by NMR in 16 mM concentration), and of 4![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 96 and 65
96 and 65![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 35 for the latter. Fig. S10 (ESI†) presents, on the other hand, the photoisomerization of all the supramolecular assemblies studied in aqueous medium, monitored through UV-Vis spectroscopy and CD. With regards to the mixed assemblies, Fig. 3c and d represents the reversible photoswitching behaviour of the Azo-A/dT20 system when irradiated with UV light of 365 nm and then with blue light of 465 nm, following the process by CD at two NaCl concentrations (1 M and 5 M). For both conditions, the ICD band completely vanishes upon irradiation with UV light, as a consequence of the establishment of the photostationary state with predominance of the cis isomer, which may indicate partial desorption of Azo-A upon irradiation.9 At low ionic strength (Fig. 3c), the signal is not recovered upon irradiation with blue light (Fig. S11, ESI†), and it takes a week for partially recovering the initial chiroptical response, as a consequence of its reassembly. For the solutions with a high salt concentration (5 M, Fig. 3d), the CD signal recovery after irradiation with blue light is much more efficient, doubling its intensity with respect to the initial trans isomer. This ICD is maintained over time, yet with a partial decrease in intensity after one week (Fig. S12, ESI†). For the Azo-T/dA20 system, the same experiment was performed. At a lower concentration of NaCl (1 M), no sign of self-assembly is observed, even a long time after irradiation with blue light (Fig. S13, ESI†). In contrast, when the ionic strength of the medium is high (5 M NaCl), supramolecular self-assembly is indicated by the ICD signatures, which vanish upon irradiation with UV light. Upon irradiation of the samples with blue light, the ICD signal increases significantly with respect to the initial one, but it decays to half of the intensity after one week (Fig. S14, ESI†).
35 for the latter. Fig. S10 (ESI†) presents, on the other hand, the photoisomerization of all the supramolecular assemblies studied in aqueous medium, monitored through UV-Vis spectroscopy and CD. With regards to the mixed assemblies, Fig. 3c and d represents the reversible photoswitching behaviour of the Azo-A/dT20 system when irradiated with UV light of 365 nm and then with blue light of 465 nm, following the process by CD at two NaCl concentrations (1 M and 5 M). For both conditions, the ICD band completely vanishes upon irradiation with UV light, as a consequence of the establishment of the photostationary state with predominance of the cis isomer, which may indicate partial desorption of Azo-A upon irradiation.9 At low ionic strength (Fig. 3c), the signal is not recovered upon irradiation with blue light (Fig. S11, ESI†), and it takes a week for partially recovering the initial chiroptical response, as a consequence of its reassembly. For the solutions with a high salt concentration (5 M, Fig. 3d), the CD signal recovery after irradiation with blue light is much more efficient, doubling its intensity with respect to the initial trans isomer. This ICD is maintained over time, yet with a partial decrease in intensity after one week (Fig. S12, ESI†). For the Azo-T/dA20 system, the same experiment was performed. At a lower concentration of NaCl (1 M), no sign of self-assembly is observed, even a long time after irradiation with blue light (Fig. S13, ESI†). In contrast, when the ionic strength of the medium is high (5 M NaCl), supramolecular self-assembly is indicated by the ICD signatures, which vanish upon irradiation with UV light. Upon irradiation of the samples with blue light, the ICD signal increases significantly with respect to the initial one, but it decays to half of the intensity after one week (Fig. S14, ESI†).
Having studied the influence of light on the photoreversible self-assembly of oligonucleotide templates with nucleobase-arylazopyrazole compounds, we evaluated the effect of the number of nucleobases within the template on the ICD signal for the trans isomer of the two possible complementary assemblies (i.e., Azo-A/dTn and Azo-T/dAn). In particular, we compared the effectiveness of dT20 and dT40 as templates at 1 M NaCl. For both systems (Fig. 3e and f), the ICD signal intensity increases with the oligonucleotide length. For Azo-T (Fig. 3f), the use of dT40 as template actually leads to a significant CD response under salinity conditions that do not allow its chiral self-assembly with dT20, showing the ICD signal maximum within the π → π* absorption region of the Azo-Ttrans isomer.
Taking the conditions that best promote supramolecular self-assembly (5 M NaCl), we assessed the selectivity of dT20 and dA20 when competing for the same arylazopyrazole. The samples contained 1 μM of each oligonucleotide (dT20 + dA20) and 20 μM of either Azo-A or Azo-T. These were subjected to an irradiation cycle, first with UV light to disrupt previously existing aggregates, thus facilitating that the three species present in the medium come from a molecularly dissolved state, followed by blue light to isomerise the azo compound back to the trans form. As it can be observed in Fig. 4, for both samples the sign of the Cotton effect is induced by the oligonucleotide that is complementary to the added azo compound: positive for Azo-T (at 360 nm, red curve) because it interacts preferentially with dA20; negative for Azo-A (at 340 nm, blue curve) due to its favourable association with dT20. These results point to a certain degree of selectivity in the binding of the studied nucleobase-arylazopyrazoles to the complementary oligonucleotide. Besides, the interaction between Azo-T and dA20 leads to a much more intense chiroptical response than the opposite counterparts. In order to shed light on the interactions and binding modes between these azo compounds and their complementary template, as well as the impact of the trans or cis configuration, molecular dynamics (MD) simulations were then carried out.
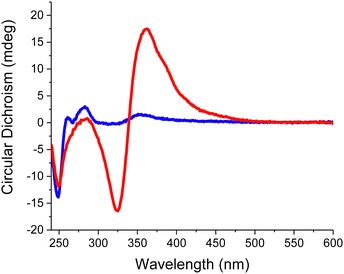 | ||
| Fig. 4 CD spectra of the assemblies obtained between a mixture of dT20 and dA20 (at 1 μM each) with either 20 μM Azo-A (blue) or Azo-T (red). | ||
Molecular dynamics simulations
MD simulations were performed with the AMBER package on assemblies of 10 arylazopyrazole molecules with a complementary oligonucleotide template of 20 bases, thus reproducing the experimental stoichiometry.41 Both arylazopyrazole derivatives were described with a reparametrized version of the GAFF 2 force-field.42 Details on the reparametrization of the force-field and on the simulation protocol are given in the ESI.† Two systems were simulated: The Azo-A/dT20 and Azo-T/dA20 supramolecular complexes, with separated runs for the trans and cis isomers. Two replicas were performed for each configuration of the system, which gives a total of 8 simulations. The complexes were solvated in water boxes, using the 4-point OPC model, with NaCl concentration as in experiments (5 M).43 In all cases, the azo compounds were preorganised along the template (Fig. S18, ESI†). The H-bonds between the complementary bases were constrained during 250 ns, to avoid disassembly of the complex during its equilibration. After this step, the simulations were extended for at least 1 μs without any restraints.In all simulations, the number of H-bonds between nucleobases decreases as soon as the constraints are removed (Fig. S19, ESI†). This shows that the azo derivatives are not “glued” to the template and are still dynamic, which can lead to reorganisations along the DNA strand. However, some bases remain H-bonded on the microsecond timescale, as shown in the snapshots of Fig. 5 and evidenced by heatmaps of H-bonds (Fig. S20, ESI†). These interactions are strongly reinforced by the stacking of the arylazopyrazole units, stabilised by π-type interactions, whose number remains quite stable during the whole simulation (Fig. S21, ESI†). In general, the number of hydrogen bonds and aromatic interactions is higher for the molecules in the trans configuration than for the ones in cis. MD simulations indicate that the assembly is driven by the stacking between the conjugated molecular regions, whereas H-bonds with the template help to order the stacks of azo compounds, which is in line with the ICD signal. The dihedral angle between the nucleobase of the azo compound and the arylazopyrazole moiety is strongly restrained for the stacked molecules, in contrast to what happens for Azo-A or Azo-T alone (Fig. S24, ESI†). Important aromatic interactions between conjugated molecules assembled on a DNA strand have been observed previously, for different systems.44,45 The molecules that bind efficiently to the oligonucleotide can serve as “anchors” for the stacking of other units, which may remain in the organised stacks even without being themselves H-bonded to the template. The linearity of the trans configuration allows the conjugated regions to stack to their neighbours and maximise the interactions, while there is more disorder with the cis configuration. This disorder is also due to the greater flexibility of the cis isomers, as shown by RMSD measurements, which increase from trans to cis. The stacking mode of the azo derivatives in their trans configuration may also explain the important role played by the salt concentration. Within the stacks, where the nucleobases are directed towards the template, the carboxylate groups at the other end of the molecules become close to each other. A high concentration of salt helps to decrease the electrostatic repulsion and, therefore, to stabilise the complexes. Radial distribution functions illustrate well the high concentration of Na+ ions close to the carboxylate moieties, for all systems (Fig. S25, ESI†). Interestingly, in one simulation of the Azo-A/dT20 complex, the cis isomers maintained their H-bonds with the template on the microsecond timescale. This suggests that such isomers can also bind efficiently to the oligonucleotide, implying that the disappearance of the ICD signal observed after photoisomerisation may not only be caused by a dissociation of the supramolecular complex, but also by the loss of organisation of the stacked arylazopyrazole units. Fig. 5 illustrates clearly the disordered and dense packing of the molecules in cis, shown on the right, in comparison to the well-aligned trans isomers, shown on the left. These simulations indicate that even the binding to the template does not improve the stacking interactions of the cis isomers. Furthermore, except in this particular case, dissociation of the DNA strand is much more frequent for the cis azo derivative, in our simulations.
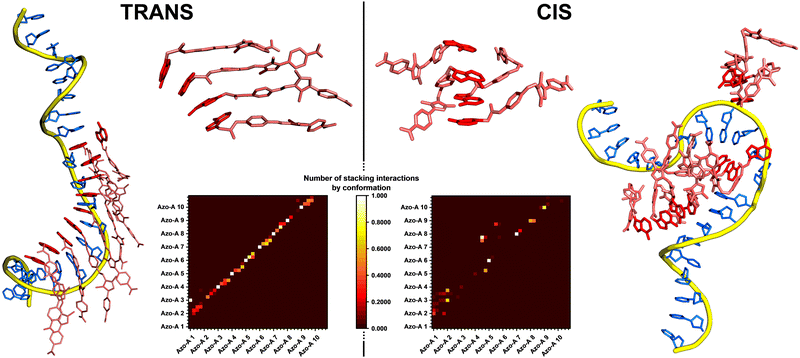 | ||
| Fig. 5 MD snapshots illustrating the Azo-A/dT20 supramolecular complex after 1.25 μs (1 μs without restraints), with the azo compound in the trans (left) or cis (right) configuration. A zoom on 4 molecules (top center) shows the organisation of the stacking for each isomer. These snapshots show important differences between the two configurations. The DNA backbone is shown as a yellow tube, its nucleobases are coloured in blue, the Azo-A molecules are represented as pink sticks and their nucleobases are coloured in red. Heatmaps of the stacking interactions between the molecules, calculated for the last 500 ns of the simulation, are shown (bottom center). 40 residues are represented on the x and y axes, one for each aromatic cycle (4 per azo molecule, the molecules being numbered from Azo-A 1 to Azo-A 10). At the crossing of a pair of two residues is located a coloured square, whose colour indicates the number of stacking interactions detected between this pair. The trans isomer shows a “diagonal pattern”, meaning that the molecules are stacked consecutively (Azo-A 1 interacts with Azo-A 2, Azo-A 2 interacts with Azo-A 3, and so on). The cis isomer shows more dispersed interactions, with less organisation. More details and the maps for all systems are given in ESI.† | ||
A structural study was also carried out to better understand the organisation of the azo compound units in their trans configuration within the stacks. Their conjugated part being quite linear, we can measure the angle between two consecutive molecules to evaluate the rotation between two arylazopyrazole moieties (more details are given in ESI†). The average angle between the conjugated regions of two stacked molecules is 23 ± 10°. The distributions of angles are fairly homogeneous in the 0–40° range, which indicates that some mobility is allowed (Fig. S26 and S27, ESI†). In comparison, the angle between two consecutive nucleobases in double-stranded DNA in its B-form is ca. 34°. This shows that the rotation of the azo units does not follow perfectly the rotation of the bases in the oligonucleotide, which could explain, in part, why only some H-bonds are maintained along the template, given the predominance of π-stacking interactions.
In sum, MD simulations shed some light on the differences between the cis and trans isomers on the supramolecular organization along the DNA template. However, the proposed conformations and assemblies do not allow us to interpret the differences of CD signals, in particular of their opposite signs for assemblies of Azo-T/dA20vs. Azo-A/dT20 (Fig. 3b). This would require an extensive quantum-chemical study to calculate the different contributions to the CD signals,46 as a complementary perspective to this work.
Conclusions
We have designed a new type of arylazopyrazole photoswitch, functionalized with nucleobases, which can be interfaced with DNA templates. Upon templated self-assembly, the nucleobase-arylazopyrazole derivatives undergo chiral organization in the trans form, and the self-assembly is (partially) disrupted when isomerized with UV light, as a result of the cis configuration. As revealed by MD simulations, cooperation of H-bond and π–π stacking interactions, with a predominance of the latter, is key to induce and maintain the dynamical order of the resulting assemblies. While azo compounds in the trans configuration are well stabilized in stacks along DNA templates, the assemblies with a cis configurations of the molecular photoswitch result in a loss of organization with partially unbound templates. Interestingly, the Cotton effect in the ICD signals depends on the template sequence (i.e. with adenine or thymine moieties), and we have observed a selective binding of the complementary nucleobase-arylazopyrazole in competition experiments between both azo compounds. Overall, our results suggest a possible strategy to achieve photoreversible chiral assemblies of arylazopyrazoles by using base-pairing interactions with oligonucleotides. Possible improvements of the compounds design and the exploration of functional roles for these photoswitches will be carried out in the near-future, with the aim to open research avenues towards potential applications as nano- and biomaterials.Author contributions
N. N. performed the experiments; S. G synthesised the compounds; D. D. performed the calculations; M. S. and A. d. l. E. conceived the project; N. N., M. S. and A. d. l. E. designed and analyzed the experiments; N. N., D. D., M. S. and A. d. l. E. wrote and edited the manuscript.Conflicts of interest
There are no conflicts to declare.Acknowledgements
This work was supported by the Fonds de la Recherche Scientifique – FNRS and by the French Community of Belgium within the framework of a FRIA grant for D. D. M. S. is a FNRS research director and acknowledges FNRS-FWO for EOS Grant no. 30650939. Computational resources are provided by the Consortium des Équipements de Calcul Intensif (CÉCI), funded by FNRS (Grant No. U.G.018.18) and Wallonia Region, and we acknowledge EuroCC Belgium, for awarding this project access to the LUMI supercomputer, owned by the EuroHPC Joint Undertaking, hosted by CSC (Finland) and the LUMI consortium. In addition, this research was funded by the Spanish Agencia Estatal de Investigación (AEI, project PID2020-119306GB-100) and the European Union (CLASSY project, Horizon 2020 Research and Innovation Program, Grant Agreement No. 862081).Notes and references
- J. Volaric, W. Szymanski, N. A. Simeth and B. L. Feringa, Chem. Soc. Rev., 2021, 50, 12377–12449 RSC.
- W. Z. Wang, L. B. Huang, S. P. Zheng, E. Moulin, O. Gavat, M. Barboiu and N. Giuseppone, J. Am. Chem. Soc., 2021, 143, 15653–15660 CrossRef CAS PubMed.
- Q. Zhou, J. Chen, Y. Luan, P. A. Vainikka, S. Thallmair, S. J. Marrink, B. L. Feringa and P. van Rijn, Sci. Adv., 2020, 6, eaay2756 CrossRef CAS PubMed.
- M. D. Pluth and K. N. Raymond, Chem. Soc. Rev., 2007, 36, 161–171 RSC.
- S. J. Loeb, Chem. Soc. Rev., 2007, 36, 226–235 RSC.
- H. Asanuma, T. Toda, K. Murayama, X. Liang and H. Kashida, J. Am. Chem. Soc., 2010, 132, 14702–14703 CrossRef CAS PubMed.
- Y. Kamiya, T. Takagi, H. Ooi, H. Ito, X. Liang and H. Asanuma, ACS Synth. Biol., 2015, 4, 365–370 CrossRef CAS PubMed.
- N. A. Simeth, S. Kobayashi, P. Kobauri, S. Crespi, W. Szymanski, K. Nakatani, C. Dohno and B. L. Feringa, Chem. Sci., 2021, 12, 9207–9220 RSC.
- J. Rubio-Magnieto, T. A. Phan, M. Fossépré, V. Matot, J. Knoops, T. Jarrosson, P. Dumy, F. Serein-Spirau, C. Niebel, S. Ulrich and M. Surin, Chem. – Eur. J., 2018, 24, 706–714 CrossRef CAS PubMed.
- F. A. Aldaye, A. L. Palmer and H. F. Sleiman, Science, 2008, 321, 1795–1799 CrossRef CAS PubMed.
- A. Diguet, N. K. Mani, M. Geoffroy, M. Sollogoub and D. Baigl, Chem. – Eur. J., 2010, 16, 11890–11896 CrossRef CAS PubMed.
- N. C. Seeman, Nature, 2003, 421, 427–431 CrossRef PubMed.
- M. Surin and S. Ulrich, ChemistryOpen, 2020, 9, 480–498 CrossRef CAS.
- A. Sikder, C. Esen and R. K. O’Reilly, Acc. Chem. Res., 2022, 55, 1609–1619 CrossRef CAS PubMed.
- S. Lu, J. Shen, C. Fan, Q. Li and X. Yang, Avd. Sci., 2021, 8, 2100328 CAS.
- D. Y. Tam, X. Zhuang, S. W. Wong and P. K. Lo, Small, 2019, 15, 1805481 CrossRef PubMed.
- S. Samai, D. J. Bradley, T. L. Y. Choi, Y. Yan and D. S. Ginger, J. Phys. Chem. C, 2017, 121, 6997–7004 CrossRef CAS.
- N. A. Simeth, P. Mendoza, V. R. A. Dubach, M. C. A. Stuart, J. W. Smith, T. Kudernac, W. R. Browne and B. L. Feringa, Chem. Sci., 2022, 13, 3263–3272 RSC.
- L. Osorio-Planes, C. Rodríguez-Escrich and M. A. Pericàs, Org. Lett., 2014, 16, 1704 CrossRef CAS PubMed.
- T. Shimoboji, E. Larenas, T. Fowler, S. Kulkarni, A. S. Hoffman and P. S. Stayton, Proc. Natl. Acad. Sci. U. S. A., 2002, 99, 16592–16596 CrossRef CAS PubMed.
- Y. Yu, M. Nakano and T. Ikeda, Nature, 2003, 425, 145 CrossRef CAS PubMed.
- T. Wendler, C. Schütt, C. Näther and R. Herges, J. Org. Chem., 2012, 77, 3284–3287 CrossRef CAS PubMed.
- J. Garcia-Amorós, M. C. R. Castro, P. Coelho, M. M. M. Raposo and D. Velasco, Chem. Commun., 2013, 49, 11427–11429 RSC.
- J. Calbo, C. E. Weston, A. J. P. White, H. S. Rzepa, J. Contreras-García and M. J. Fuchter, J. Am. Chem. Soc., 2017, 139, 1261–1274 CrossRef CAS PubMed.
- C. E. Weston, R. D. Richardson, P. R. Haycock, A. J. P. White and M. J. Fuchter, J. Am. Chem. Soc., 2014, 136, 11878–11881 CrossRef CAS PubMed.
- L. Stricker, E. C. Fritz, M. Peterlechner, N. L. Doltsinis and B. J. Ravoo, J. Am. Chem. Soc., 2016, 138, 4547–4554 CrossRef CAS PubMed.
- M. A. Gerkman, R. S. L. Gibson, J. Calbo, Y. Shi, M. J. Fuchter and G. G. D. Han, J. Am. Chem. Soc., 2020, 142, 8688–8695 CrossRef PubMed.
- R. Iwaura, Y. Kikkawa, M. Ohnishi-Kameyama and T. Shimizu, Org. Biomol. Chem., 2007, 5, 3450–3455 RSC.
- P. G. A. Janssen, S. Jabbari-Farouji, M. Surin, X. Vila, J. C. Gielen, T. F. A. de Greef, M. R. J. Vos, P. H. H. Bomans, N. A. J. M. Sommerdijk, P. C. M. Christianen, P. Leclère, R. Lazzaroni, P. van der Schoot, E. W. Meijer and A. P. H. J. Schenning, J. Am. Chem. Soc., 2009, 131, 1222–1231 CrossRef CAS.
- R. Hofsäβ, P. Ensslen and H. A. Wagenknecht, Chem. Commun., 2019, 55, 1330–1333 RSC.
- M. Surin, Polym. Chem., 2016, 7, 4137–4150 RSC.
- F. J. Rizzuto, C. M. Platnich, X. Luo, Y. Shen, M. D. Dore, C. Lachance-Brais, A. Guarné, G. Cosa and H. F. Sleiman, Nat. Chem., 2021, 13, 843–849 CrossRef CAS PubMed.
- P. Kumar, A. Srivastava, C. Sah, S. Devi and S. Venkataramani, Chem. – Eur. J., 2019, 25, 11924–11932 CrossRef CAS.
- S. Vela-Gallego, Z. Pardo-Botero, C. Moya and A. de la Escosura, Chem. Sci., 2022, 13, 10715–10724 RSC.
- M. E. Mercurio, S. Tomassi, M. Gaglione, R. Russo, A. Chambery, S. Lama, P. Stiuso, S. Cosconati, E. Novellino, S. Di Maro and A. Messere, J. Org. Chem., 2016, 81, 11612–11625 CrossRef CAS PubMed.
- S. Morales-Reina, C. Giri, M. Leclercq, S. Vela-Gallego, I. de la Torre, J. R. Castón, M. Surin and A. de la Escosura, Chem. – Eur. J., 2020, 26, 1082–1090 CrossRef CAS PubMed.
- L. Knorr, Ber. Dtsch. Chem. Ges., 1883, 16, 2597–2599 CrossRef.
- J. Mikkilä, E. Anaya-Plaza, V. Liljeström, J. R. Caston, T. Torres, A. de la Escosura and M. A. Kostiainen, ACS Nano, 2016, 10, 1565–1571 CrossRef PubMed.
- S. Yang, G. Schaeffer, E. Mattia, O. Markovitch, K. Liu, A. S. Hussain, J. Ottelé, A. Sood and S. Otto, Angew. Chem., Int. Ed., 2021, 60, 11344–11349 CrossRef CAS PubMed.
- S. Yang, Y. Geiger, M. Geerts, M. J. Eleveld, A. Kiani and S. Otto, J. Am. Chem. Soc., 2023, 145, 16889–16898 CrossRef CAS PubMed.
- D. A. Case, T. E. Cheatham 3rd, T. Darden, H. Gohlke, R. Luo, K. M. Merz Jr, A. Onufriev, C. Simmerling, B. Wang and R. J. Woods, J. Comput. Chem., 2005, 16, 1668–1688 CrossRef PubMed.
- J. Wang, R. M. Wolf, J. W. Caldwell, P. A. Kollman and D. A. Case, J. Comput. Chem., 2004, 25, 1157–1174 CrossRef CAS.
- S. Izadi, R. Anandakrishnan and A. V. Onufriev, J. Phys. Chem. Lett., 2014, 5, 3863–3871 CrossRef CAS PubMed.
- M. Surin, P. G. A. Janssen, R. Lazzaroni, P. Leclère, E. W. Meijer and A. P. H. J. Schenning, Adv. Mater., 2009, 21, 1126–1130 CrossRef CAS.
- J. Rubio-Magnieto, M. Kumar, P. Brocorens, J. Idé, S. J. George, R. Lazzaroni and M. Surin, Chem. Commun., 2016, 52, 13873–13876 RSC.
- N. H. List, J. Knoops, J. Rubio-Magnieto, J. Idé, D. Beljonne, P. Norman, M. Surin and M. Linares, J. Am. Chem. Soc., 2017, 139, 14947–14953 CrossRef PubMed.
Footnotes |
| † Electronic supplementary information (ESI) available: The synthesis and characterization of compounds, spectroscopy studies of the DNA-templated assemblies and MD simulations. See DOI: https://doi.org/10.1039/d4tb00041b |
| ‡ N. N. and S. G. contributed equally to this work. |
| This journal is © The Royal Society of Chemistry 2024 |