Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Optimization of a nanoparticle uptake protocol applied to amniotic-derived cells: unlocking the therapeutic potential†
Alessia
Peserico
 *a,
Angelo
Canciello
*a,
Angelo
Canciello
 a,
Giuseppe
Prencipe
a,
Roberto
Gramignoli
b,
Valeria
Melai
c,
Giampiero
Scortichini
c,
Mirella
Bellocci
a,
Giuseppe
Prencipe
a,
Roberto
Gramignoli
b,
Valeria
Melai
c,
Giampiero
Scortichini
c,
Mirella
Bellocci
 c,
Giulia
Capacchietti
a,
Maura
Turriani
a,
Chiara
Di Pancrazio
c,
Paolo
Berardinelli
a,
Valentina
Russo
a,
Mauro
Mattioli
a and
Barbara
Barboni
a
c,
Giulia
Capacchietti
a,
Maura
Turriani
a,
Chiara
Di Pancrazio
c,
Paolo
Berardinelli
a,
Valentina
Russo
a,
Mauro
Mattioli
a and
Barbara
Barboni
a
aDepartment of Bioscience and Technology for Food, Agriculture and Environment, University of Teramo, 64100 Teramo, Italy. E-mail: apeserico@unite.it
bDepartment of Laboratory Medicine, Karolinska Institute, Stockholm, Sweden
cIstituto Zooprofilattico Sperimentale dell'Abruzzo e Molise ‘G. Caporale’, Campo Boario, 64100 Teramo, Italy
First published on 14th August 2024
Abstract
Stem cell-based therapy implementation relies heavily on advancements in cell tracking. The present research has been designed to develop a gold nanorod (AuNR) labeling protocol applied to amniotic epithelial cells (AECs) leveraging the pro-regenerative properties of this placental stem cell source which is widely used for both human and veterinary biomedical regenerative applications, although not yet exploited with tracking technologies. Ovine AECs, in native or induced mesenchymal (mAECs) phenotypes via epithelial–mesenchymal transition (EMT), served as the model. Initially, various uptake methods validated on other sources of mesenchymal stromal cells (MSCs) were assessed on mAECs before optimization for AECs. Furthermore, the protocol was implemented by adopting the biological strategy of MitoCeption to improve endocytosis. The results indicate that the most efficient, affordable, and easy protocol leading to internalization of AuNRs in living mAECs recognized the combination of the one-step uptake condition (cell in suspension), centrifugation-mediated internalization method (G-force) and MitoCeption (mitochondrial isolated from mAECs). This protocol produced labeled vital mAECs within minutes, suitable for preclinical and clinical trials. The optimized protocol has the potential to yield feasible labeled amniotic-derived cells for biomedical purposes: up to 10 million starting from a single amniotic membrane. Similar and even higher efficiency was found when the protocol was applied to ovine and human AECs, thereby demonstrating the transferability of the method to cells of different phenotypes and species-specificity, hence validating its great potential for the development of improved biomedical applications in cell-based therapy and diagnostic imaging.
Introduction
Regenerative medicine holds significant promise for leveraging the therapeutic potential of stem cells. Amniotic-derived cells, with their distinctive characteristics such as widespread availability, absence of ethical concerns, and immune privileges facilitating their use in allo- and xenotransplantation, exhibit broad clinical potential in both medical and veterinary fields, albeit only partially explored.Moreover, the inherent stemness of these cells takes advantage of remarkable plasticity, evident in both in situ and ex situ trans-differentiation processes.1,2 Similar to other stem cell sources, they demonstrate diverse paracrine regulatory functions, including immunomodulatory, antimicrobial, and antifibrotic activities.3–6 Unlike other stem cell sources, these biological attributes are constitutively expressed, defining unique functions of amniotic-derived cells, such as maintaining an immunotolerance barrier between the fetus and the mother.
Additionally, these abilities can be potentiated in response to the surrounding inputs, such as inflammatory mediators. Nevertheless, it has recently been demonstrated that there is an active conversation between them and neighboring cells which can also occur through mitochondrial transfer pathways.7 Based on these premises, amniotic-derived cells have been proposed as candidates for the treatment of severely damaged tissues that cannot be saved even by the most advanced surgical or pharmacological treatments.8,9
Amniotic-derived cells combine the great pro-regenerative actions with the ability of a robust and enduring engraftment into damaged tissues.10–17 This capability enables their substantial contribution to the long-term healing, replacement, and restoration of tissues and organs.18 In addition, epithelial amniotic-derived cells (AECs) as mesenchymal stromal ones (AMSCs) exploit the migratory activity as a mechanism to reach damaged tissues.19 In AECs, the mobility is enhanced because of the activation of the EMT (EMT) process. This activation occurs in response to specific chemoattractant signals released by stromal or resident cells, triggering a recruitment response.20,21 This process, known as “homing”, becomes crucial for the success of tissue regeneration and healing.
Cell mobility also plays a vital role in determining the appropriate injection strategy for transplanting stem cells.22,23 To develop cell therapy systems that can be readily translated into the clinical setting, a precise method for delivering cells into the body still needs to be established. Unfortunately, current protocols suffer from low delivery efficiency, as only a limited number of cells reach the target tissue and remain there after systemic administration.24
Drawing inspiration from stem cell trafficking within the body under physiological and pathological conditions, recent advances have been made in inducing stem cell mobilization and directing patients' stem cells to sites of interest for treating a broad spectrum of diseases.25 This perspective has paved the way for a new paradigm in the field of regenerative medicine, allowing for targeted and personalized management of stem cell-based therapy. Additionally, understanding the mechanisms of cell trafficking represents a challenging biological advancement for standardizing transplant protocols, aiming to maximize the homing abilities of stem cells into injured tissues and establish their long-lasting presence in damaged areas. Achieving these goals would significantly enhance the therapeutic potential of stem cell-based therapies.
A first fundamental prerequisite for achieving these targets is to have a reliable cell tracking system, capable of monitoring the diffusion of stem cells in the host bodies and defining the timing and conditions for the cells to get concentrated in the target zone without affecting their viability and/or their biological functions. In this context, labelling stem cells with nanomaterials working as contrast agents for imaging devices has recently attracted great attention in both the medical and veterinary fields.26,27 Different protocols have been developed so far for labelling mesenchymal stem cells (MSCs) to follow their homing and engraftment,22 monitoring cell survival and viability,28 assessing tissue regeneration,29 optimizing transplantation routes and studying biodistribution30 and understanding their mechanisms of action.31 Importantly, in the veterinary field, tracking models of allogeneic mesenchymal cells have been developed in canines32,33 and equines.34,35
To date, no approaches for tracking amnion-derived cells have been disclosed, despite their significant usage in preclinical and clinical models of stem cell-based treatments.1,2,20,36–39
The current study aimed to optimize a 10 × 41 nm nanoparticle (NP) uptake protocol applied to ovine mesenchymal and epithelial amniotic-derived cells taking the advantage of the capability to manipulate their phenotype in vitro, thereby influencing their immunological and homing characteristics.
Experimental
Ethics statement
No ethical statement is necessary for the use of the ovine-derived amniotic cells since they are obtained from waste tissue obtained from animals slaughtered for feed purposes.Human amniotic epithelial cells derived from discarded human placentae from uncomplicated cesarean resections at 37–42 weeks of gestational age were obtained from Karolinska University Hospital (Stockholm, Sweden) with institutional review board approval and signed informed consent.
Cell isolation and culture
AECs were isolated from the AM of female sheep in mid-pregnancy (3 AM totally) according to Canciello et al.40 AECs were maintained in MEM Eagle Alpha (ECM0850L, Euroclone) supplemented with 10% fetal bovine serum (FBS; #10270-106, Gibco), 100 IU mL−1 penicillin–streptomycin (DE 17-602E, Lonza), 100 IU mL−1 amphotericin B (ECM0009D, Euroclone) and 100 IU mL−1L-glutamine (ECB300D, Euroclone). AECs and mAECs were obtained, respectively, by culturing AECs for three passages in the presence and absence of 25 mM P4 (4 pregnene-3,20-dione, P4; P8783, Sigma-Aldrich) at 38.5 °C and 5% CO2 in the incubator.Human AECs were isolated and collected as reported by Gramignoli et al.41 Briefly, the amnion membrane was mechanically separated from the chorion layer and washed with Ringer's solution (Baxter, Sweden) to remove blood cells. Subsequently, the amnion was immersed in TrypLE solution (Life Tech, Paisley, UK) and incubated for 30 min at 37 °C. The released cells were sedimented by centrifugation at 500 × g for 5 minutes and re-suspended in a cryogenic preservation solution composed by University of Wisconsin solution (Lisieux, France) supplemented with 10% dimethylsulfoxide (DMSO; Sigma-Aldrich, MO, USA). Cells were transferred in controlled freezing containers and stored in the vapor phase in a liquid nitrogen storage tank until uptake experiments. Human AECs were maintained in DMEM Low Glucose (ECM0060, Euroclone) supplemented with 10% fetal bovine serum (FBS; #10270-106, Gibco), 100 IU mL−1 penicillin–streptomycin (DE 17-602E, Lonza), and 100 IU mL−1 amphotericin B (ECM0009D, Euroclone). Human AECs were not passaged before uptake experiements. Cells were kept at 37 °C and 5% CO2 in the incubator for all the experimental procedures.
AuNRs
Rhodamine (535/575 nm) labelled 10 × 41 nm gold nanorods (AuNRs; CF12-10-808-RHO-FB-DIH-50-1-CS-EP) with a surface plasmon resonance (SPR) 808 nm were obtained from Nanopartz™. AuNRs were managed according to the manifacturer's instructions.Chemicals
The stock solutions of gold (Au) at 100 mg L−1 in 10% (v/v) HCl and rhenium (Re) at 1000 mg L−1 in 2% (v/v) HNO3 were purchased, respectively, from CPAchem Ltd (Bogomilovo, Bulgaria) and Merck KGaA, (Darmstadt, Germany). The nitric and hydrochloric acid used for all dilutions were grade ultrapure, HNO3 ≥60% and HCl ≥30% both supplied by Supelco (Merck KGaA, Darmstadt, Germany). All working solutions, standard, blanks and samples were prepared with type I (resistivity 18.2 MΩ cm) water, obtained from a ELGA LabWater PURELAB Option-Q water purification system (High Wycombe, United Kingdom). Certified reference material NPs SRM 8011 gold nanoparticles (nominal 10 nm diameter) was bought from NIST (Gaithersburg, USA). Argon and helium gases of 99.9995% purity were supplied by Sapio (Monza, Italy).Immunofluorescence
Cells were grown on glass coverslips, treated as indicated in the cell culture section and then fixed with 4% paraformaldehyde (#11699404, VWR) for 10 min and permeabilized with 0.1% (v/v) Triton X-100 (T8787, Sigma-Aldrich) in 1× phosphate buffered saline (PBS; P3813, Sigma-Aldrich) for 10 min. Glass coverslips were then blocked in % (w/v) bovine serum albumin (BSA; A3059, Sigma-Aldrich) in 1× PBS for 1 h at room temperature (RT) and stained overnight at 4 °C with the following primary antibodies![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) anti-cytokeratin-8 (1
anti-cytokeratin-8 (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 200) (ab2530, Abcam) and anti-vimentin (1
200) (ab2530, Abcam) and anti-vimentin (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 200) (M0725, Agilent DAKO). Cy3 (AP124C, Millipore) or Alexa Fluor 488 (ab150113, Abcam) conjugated anti-mouse secondary antibodies were diluted 1
200) (M0725, Agilent DAKO). Cy3 (AP124C, Millipore) or Alexa Fluor 488 (ab150113, Abcam) conjugated anti-mouse secondary antibodies were diluted 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 200 in 1% (w/v) BSA/PBS and incubated on glass coverslips for 40 min at RT. Nuclear counterstaining was obtained with 4′,6-diamidino-2-phenylindole (DAPI; D9542 Sigma-Aldrich) diluted 1
200 in 1% (w/v) BSA/PBS and incubated on glass coverslips for 40 min at RT. Nuclear counterstaining was obtained with 4′,6-diamidino-2-phenylindole (DAPI; D9542 Sigma-Aldrich) diluted 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2000 in 1× PBS. Coverslips were mounted with fluoromount (F4680, Sigma-Aldrich), and cell samples were analyzed using an Axioskop 2 plus incident light fluorescence microscope (Zeiss) equipped with a CCD camera (Axiovision Cam 208color, Zeiss), with a resolution of 1300 × 1030 pixels, configured for fluorescence microscopy, and interfaced to a computer workstation, provided with an interactive and automatic image analyser (Zen 3.4 Blue edition Software, Zeiss). Digital images were acquired using standard filters setup for Cy3, Alexa Fluor 488 or DAPI.
2000 in 1× PBS. Coverslips were mounted with fluoromount (F4680, Sigma-Aldrich), and cell samples were analyzed using an Axioskop 2 plus incident light fluorescence microscope (Zeiss) equipped with a CCD camera (Axiovision Cam 208color, Zeiss), with a resolution of 1300 × 1030 pixels, configured for fluorescence microscopy, and interfaced to a computer workstation, provided with an interactive and automatic image analyser (Zen 3.4 Blue edition Software, Zeiss). Digital images were acquired using standard filters setup for Cy3, Alexa Fluor 488 or DAPI.
Cytotoxicity assay
The CellTiter 96 aqueous one solution cell proliferation assay (MTS; #G3582; Promega) was used to determine the viability of mAECs cells treated with different concentrations of AuNRs. In detail, cells were seeded in 96-well plates at a density of 2 × 104 cells per cm2 and were allowed to attach overnight in a CO2 incubator. Cells were then treated with 0, 1.1, 4.4, and 18 μg mL−1 of AuNRs for 12, 24 and 48 hours. The cells were then washed three times with PBS (P3813, Sigma-Aldrich) to remove the unbound AuNRs. The MTT reagent was added 2 h before the termination of the experiment, and the optical density was acquired at 490 nm. The NP concentration and timeframe were determined based on cell viability below 70%. For long term experiments ovine or human AECs labelled by following the optimized condition for uptake (24 hours one-step with 4.4 μg mL−1 AuNRs + G-force + MitoCeption) were checked for their viability over time (24, 48, 72 and 96 hours) by MTS following the manufacturer's instructions.Internalization of AuNRs
In the two-step method, mAECs adhering as a cell monolayer to the bottom of the support dish were exposed to AuNRs. Specifically, mAECs (2 × 104 cells per cm2) were plated, and after 24 hours, the growth medium was removed, and the cells were cultured with AuNRs dispersed in fresh medium.
The one-step method involved the addition of AuNRs to mAECs as a cell suspension, followed by cell seeding into a low-adherent culture dish (2 × 104 cells per cm2) and subsequent incubation using a cell shaker. A wide concentration range of AuNRs spanning from 0 μg mL−1 to 18 μg mL−1 and at different time points (12, 24 and 48 hours) were tested for evaluating AuNRs uptake performances gathered, respectively, with the one-step and the two-step methods.
For the one-step protocol, AuNRs at a concentration of 1.1, 4.4 μg mL−1 were added to a cell suspension (ovine mesenchymal and epithelial AECs and human epithelial AECs). Cells were then seeded into a low-adherent culture dish (2 × 104 cells per cm2) and incubated using a cell shaker in the incubator for 12, 24 and 48 hours. A control cell plate was also prepared in a similar manner but without the addition of AuNRs (0 μg mL−1 AuNRs).
The protocol used refers to that described by O’Campo and colleagues42 with slight modifications. Briefly, AuNRs, at a concentration of 4.4 μg mL−1, were dispersed in either 6 mL or 1 mL of fresh complete medium and applied to the cell pellet containing 2 × 105 cells for cytofluorimetric studies or 1.2 × 106 cells for ICP-MS analysis.
The samples were then subjected to centrifugal forces (200 × g) for 5 minutes and subsequently seeded in low-adherent culture dishes, achieving a cell density of 2 × 104 cells per cm2. Following a 24 hour incubation at 38.5 °C or 37 °C, the samples were processed for gold quantification using ICP-MS and for the examination of cells that had internalized AuNRs through flow cytometry. A control cell plate was prepared in a similar manner, omitting the addition of AuNRs.
30 minutes before the end of the AuNRs incubation protocol, mitochondria staining was carried out by adding MitoTracker™ Green FM (M7514, Invitrogen) to the cells at a concentration of 20 nM. Subsequently, the cells were incubated at 38.5 °C or 37 °C for 30 minutes following manufacturer's instructions. The cells were then collected by centrifugation at 300 × g and extensively washed (3 times) with DPBS (#D8662; Sigma-Aldrich) before being used for further analyses.
Gold analytical determination with ICP-MS
Once uptake was completed, the cells were collected at the bottom of a tube by centrifugation at 300 × g. Three thorough washes with DPBS (#D8662; Sigma-Aldrich) were performed prior ICP-MS analysis to remove any remaining AuNRs possibly adhered to the cell surface.At the beginning of the analyses, all equipment and glassware were flushed with 1–2% (v/v) HNO3 and then rinsed with type I water. After vigorous mixing, a sample volume (50–100 μl) was solubilized in an adequate quantity of a solution containing 20% (v/v) HNO3 and 2% HCl (v/v) to obtain a sample dilution of 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 100 in HNO3 2% (v/v) and HCl 0.2% (v/v). The sample solutions obtained were analyzed using an inductively coupled plasma mass spectrometer ICP-MS Icap RQ (Thermo Fisher Scientific, Waltham, MA, USA) in KED (kinetic energy discrimination) mode. Instrumental parameters are shown in Table 1.
100 in HNO3 2% (v/v) and HCl 0.2% (v/v). The sample solutions obtained were analyzed using an inductively coupled plasma mass spectrometer ICP-MS Icap RQ (Thermo Fisher Scientific, Waltham, MA, USA) in KED (kinetic energy discrimination) mode. Instrumental parameters are shown in Table 1.
| Spray chamber | Cyclonic (at 2.7 °C) |
| Nebulizer | Micromist at 0.4 mL min−1 |
| Sample/skimmer cone | Platinum |
| Cool gas (L min−1) | 14 |
| Auxiliary gas (L min−1) | 0.8 |
| Plasma power (W) | 1550 |
| Nebulizer gas flow (L min−1) | (0.95–1.15) optimized daily |
| KED mode: collision gas flow (helium) (L min−1) | (4.5–5.5) optimized daily |
The isotopic masses acquired were 197Au, 187Re. Quantitative gold determination was performed using a calibration curve built with increasing addition standard concentrations (0.2, 0.4, 1, 2, 4, 10, 20, 50, 70, 100, 120 μg L−1) in 2% (v/v) HNO3 and 0.2% (v/v) HCl. In all injected solutions (blank, standard and samples), the same amount of Re was added as an internal standard to compensate for the signal fluctuations typical of the ICPMS technique. In each analytical session, the absence of interferents and analytical accuracy were monitored. Therefore, a blank reagent was processed, and the gold concentration was lower than the LOQ value (equal to 0.10 mg L−1) and the recovery of the certified reference material NPs SRM 8011 gold nanoparticles was acceptable in the range (98–103)%.
Quantification of rhodamine positive cells and MitoCeption by flow cytometry
For flow cytometry measurements, following exposure to AuNRs, cells underwent thorough washing with DPBS (#D8662; Sigma-Aldrich) and were subsequently suspended in a fresh complete medium. The flow cytometry analysis was conducted using a CytoFLEX SRT system (Beckman Coulter), where a total of 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 events were analyzed. AuNRs-rhodamine b was excited by a 561 nm yellow laser (Y585 channel), while mitotracker green dye was excited by a 488 nm blue laser (B525 channel). Fluorescence intensity was represented on a standard logarithmic scale. Data analysis and visualization were performed using CytExpert SRT software (version 1.2.10004, Beckman Coulter).
000 events were analyzed. AuNRs-rhodamine b was excited by a 561 nm yellow laser (Y585 channel), while mitotracker green dye was excited by a 488 nm blue laser (B525 channel). Fluorescence intensity was represented on a standard logarithmic scale. Data analysis and visualization were performed using CytExpert SRT software (version 1.2.10004, Beckman Coulter).
Representative flow cytometer plots were shown for each experimental condition. More in detail, the percentage of cells enclosing AuNRs or mitochondria were measured, respectively, by PE intensity (x-axis) or FITC (x-axis) and side scattering (SSC; y-axis) flow cytometry dot plot. First, the percentage of cells containing AuNRs or mitochondria was assigned by cells of higher PE or mitochondria intensity than the threshold intensity (black vertical line). The threshold value was determined by adding the robust SD to the median intensity of the untreated (0 μg mL−1) cells.
Imaging AuNRs cell internalization with confocal laser scanner analysis
At the end of the incubation period, cells were gently washed three times with DPBS (#D8662; Sigma-Aldrich). Subsequently, the cells were seeded in confocal Petri dishes at a density of 2 × 104 cells per cm2 and allowed the support to be adhered for approximately 3 hours before proceeding with confocal laser scanning analysis.The cells were fixed with 4% paraformaldehyde (#11699404, VWR) for 10 minutes and permeabilized with 0.1% (v/v) Triton X-100 (T8787, Sigma-Aldrich) in 1× phosphate buffered saline (PBS; P3813, Sigma-Aldrich) for 10 minutes. Following this, cell nuclei were stained with DAPI (D9542 Sigma-Aldrich) diluted 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2000 in 1× PBS for 15 minutes.
2000 in 1× PBS for 15 minutes.
In case of phalloidin staining, Alexa Fluor™ 488 phalloidin (green; #A12379) was added at a concentration of 165 nM, incubated for 1 h at RT and washed extensively before confocal laser scanning microscopy (CLSM; Nikon A1R) analysis. CLSM was interfaced to a computer workstation, provided with an interactive and automatic image analyser (NIS-Element AR Analysis v4.40.00 64-bit). Digital images were acquired using a standard laser setup for Cy3 (for AuNRs), FITC 488 nm (for phalloidin) or DAPI (for nuclei).
Isolation of PBMCs
Following the collection of 16 milliliters of fresh peripheral blood at the slaughterhouse, ovine PBMCs were extracted using a density gradient centrifugation method. As directed by the manufacturer, a gradient was constructed using 12 mL of Ficoll-Paque PLUS (#GE17-1440-02, Cytiva). The cells were preserved using the above-discussed methods and kept in liquid nitrogen until they were required for immunological testing.CM production from AECs
CM production was started once the AECs (either ovine or human) reached 24 hours of 4.4 μg mL−1 AuNRs uptake with one-step + G-force + MitoCeption. The cells were cultured 4 hours in serum-free media (ovine AEC with MEM Eagle Alpha, human AECs with DMEM low glucose, both supplemented with 1% penicillin/streptomycin, 1% amphotericin B and 1% L-glutamine) before exposing them to the inflammatory lipopolysaccharide (LPS) stimulus (1 mg ml−1, #L2637; Sigma-Aldrich) for 1 hour.43–45 The cells were then washed twice and cultured in serum-free medium for 24 hours before collecting the conditioned medium (CM). The CM was centrifuged at 300 × g for 10 minutes to remove any remaining cells or debris, and the supernatants were used for the PBMC proliferation assay.PBMCs proliferation assay
The immunomodulatory effect of ovine and human AEC-derived CM was verified by the proliferation rate of PBMCs by using MTS. In detail, PBMCs were seeded in 96-well plates at a concentration of 2 × 105 cells, activated with 10 μg mL−1 of phytohemagglutinin (PHA-L; #00-4977-93; invitrogen) and cultured for 48 h with or without CM. Then, PBMC proliferation was assessed by using the CellTiter 96 aqueous one solution cell proliferation assay (MTS; #G3582; Promega) following the manufacturer's instruction protocol. Data were normalized on PHA-activated PBMCs.qPCR
The total RNA was extracted using a total RNA purification kit (# 17250 Norgen Biotek Corp.) following the manufacturer's instructions. 1 μg of total RNA was retrotranscribed using oligodT primers (#BIO-38029, Bioline) and Tetro reverse transcriptase (#BIO-65050, Bioline), following the manufacturer's instructions. The qPCRs were carried out in triplicate using the SensiFAST SYBR Lo-ROX kit (#BIO-94050, Bioline) on a 7500 fast real-time PCR system (Life Technologies), according to the manufacturer's instructions. The following PCR conditions were used for all the experiments: 95 °C for 10 min, followed by 40 cycles at 95 °C for 10 s and 60 °C for 30 s. Relative quantification was performed by using the ΔΔCt method. GAPDH (glyceraldehyde 3-phosphate dehydrogenase) and YWHAZ (tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein zeta) were selected amongst the housekeeping genes for gene quantification. Gene expression data were the mean ± SD, of the ΔCt values related to the selected housekeeping. Specifically, one of the two housekeeping genes was reported. The expression profiles were similar with both reference genes. The primer sequences are reported in Table 2.| Gene | Forward sequence | Reverse sequence | Annealing temperature (°C) |
|---|---|---|---|
| GAPDH | 5′-TCGGAGTGAACGGATTTGGC-3′ | 5′-CCGTTCTCTGCCTTGACTGT-3′ | 64.4 |
| YWHAZ | 5′-AGACGGAAGGTGCTGAGAAA-3′ | 5′-CGTTGGGGATCAAGAACTTT-3′ | 61.3 |
| Vimentin | 5′-GACCAGCTCACCAACGACA-3′ | 5′-CTCCTCCTGCAACTTCTCCC-3′ | 61.2 |
| Cytokeratin-8 | 5′-CTCAAAGGCCAGGGCTTC-3′ | 5′-CTTGGCCTGGCATCCTTGA-3′ | 61.3 |
Statistical analysis
Data were generated from three independent experiments. Data were plotted and statistically analyzed using the software package GraphPad Prism version 5.0 for Windows. All samples were compared using a one-way ANOVA and Bonferroni post hoc test. Only significant differences (p-value < 0.05) among the samples are indicated in the charts.Results and discussion
NP uptake optimization according to amniotic cell phenotypes
The major objective of the present study is to define whether the currently available protocols validated on mesenchymal stromal cells (MSCs) are effective in amniotic-derived cells. As a further novelty, a biological strategy (mitochondrial transfer, also known as MitoCeption) was tempted to enhance the uptake mechanisms.42Using this setting, the validation of the NP uptake protocol was first performed on ovine mesenchymal amniotic-derived cells (mAECs) resulting from the in vitro transition of epithelial ones (AECs).44 Once the NP uptake protocol was verified and implemented on mAECs, the approach was then extended to the native pool of AECs of both ovine and human origin.
Amniotic-derived cells offer, indeed, the possibility to start from an epithelial phenotype and move towards the mesenchymal one exploiting the physiological pathway that controls epithelial–mesenchymal transition (EMT) in this cell type.
Indeed, amniotic cells with epithelial or mesenchymal phenotype coexist in vivo in the amniotic membrane (AM) where the EMT path is switched on in epithelial labor.46In vitro, AECs undergo spontaneous EMT after isolation from AM, acquiring a mesenchymal phenotype because of expansion in conventional media.47,48 This process can be prevented by progesterone (P4) supplementation by mimicking in culture the hormonal asset of pregnancy as demonstrated recently by our group.44 Crucially, our group proved, in addition, that the amniotic cell phenotype strongly affected the stem cell properties: in particular, to enhance stemness, in vitro44 and in vivo plasticity16,49 and migratory properties.50
Exploiting the effect of P4, for the current research, two different cell typologies (epithelial vs. mesenchymal) have been generated starting from AECs freshly isolated from 3 different fetuses. The phenotype characterization of both the epithelial and mesenchymal cell subpopulations was evaluated after three passages of expansion using specific protein and gene markers. Vimentin was employed as a late mesenchymal marker, while cytokeratin-8 was utilized for epithelial markers (Fig. S1, ESI†).
The interest of the present research towards the AECs, the most studied and used amniotic-derived cells to date,51 is related to their distinctive therapeutic properties and to the research expertise of our research team that widely characterized in vitro and in vivo them over the past years.1,10,16,17,49,52 Furthermore, our interest extends to mesenchymal amniotic-derived cells, benefiting from the collective expertise of various stem cell research groups that occasionally utilize this placental source to isolate fetal mesenchymal stromal cells.
Effect of AuNRs on viability of mAECs
For the present research, gold nanorods (AuNRs) have been selected. Recently, there has been growing interest in a new approach that utilizes gold NPs as a contrast agent for computed tomography (CT) with potential applications in medicine protocols in both medicine26 and veterinary.53 This methodology is gaining attention due to its clinical applicability and low cost-effectiveness. Indeed, research indicates that gold NPs can effectively label various cell types, including stem cells and immune cells, without compromising their therapeutic effectiveness.54 Nevertheless, in vivo experiments have successfully demonstrated noninvasive, quantitative, and longitudinal cell tracking with high sensitivity using this approach using animal models.55–60 ICP-MS approach with flow cytometry and confocal laser scanning analysis.The initial experiments involved the preparation of AuNRs on mAECs to assess their impact on cell viability, employing a validated two-step protocol.26 To employ AuNRs as cell-labeling agents, their cellular absorption and potential toxicity were assessed.
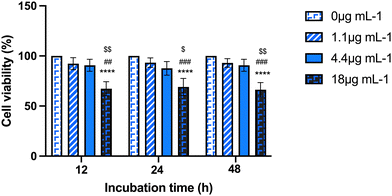
First, a dose–response curve with a range of Au-NRs (from 1.1 to 18 μg mL−1) encompassing the working dose indicated by the manufacturer (4.4 μg mL−1) was screened to assess the influence of AuNRs exposure on mAECs using the MTS assay. Three time points for AuNRs exposure were considered (12, 24, 48 h). As shown in Fig. 1, no cytotoxicity was seen after 12 h, 24 h, or 48 h of AuNRs incubation using the concentration range from 1.1 to 4.4 μg mL−1. In contrast, supplementation of 18 μg mL−1 dramatically reduced mAECs viability roughly 65% regardless of the incubation period. Based on these preliminary data, the experimental set up was then performed by incubating mAECs at both 1.1 and 4.4 μg mL−1.
Impact of cellular adhesion on AuNRs uptake
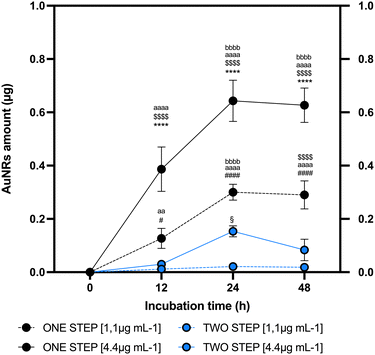
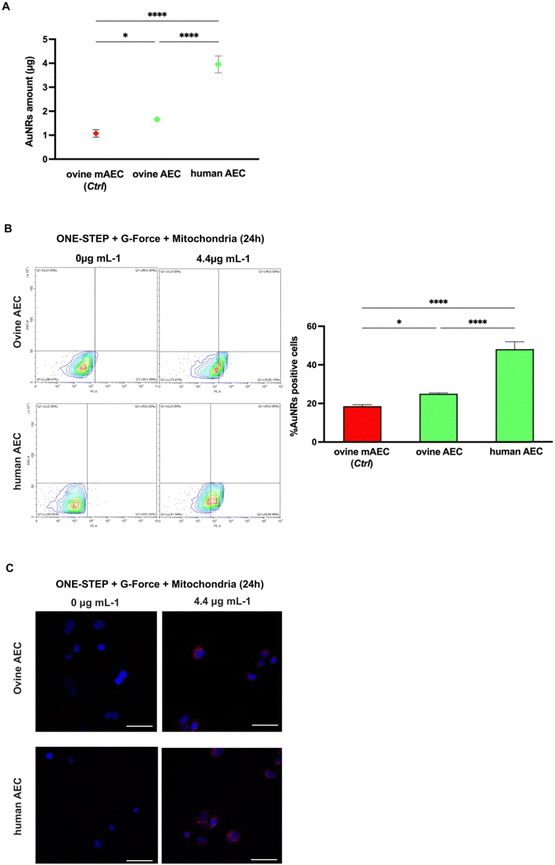
To establish the link between AuNRs uptake protocol and in vitro cell conditions, a comparative analysis was conducted between one-step and two-step approaches. The two-step replicated a method extensively validated in the literature for NP uptake26 where AuNRs are exposed to a cell monolayer where cells adhere to the bottom of the cultural support. On the other hand, the one-step method involves the addition of AuNRs to cells maintained in suspension, using low-adherent culture dish, and shaking. The one-step approach was regarded as promising due to its biological potential and practical advantages. The biological rationale behind employing this protocol is grounded by the evidence demonstrating as NP penetrability through the lipid bilayer is positively influenced by the extension of surface contact area.61 Nevertheless, it may provide the practical advantage of the immediate use of labeled cells without the need for further processing of the sample. The mAECs were exposed to different doses of AuNRs (1.1 and 4.4 μg mL−1) at three time points (12, 24 and 48 hours), utilizing both one-step and two-step approaches. Following each incubation with AuNRs, extensive washing was performed to ensure the removal of any AuNRs potentially attached to the cell surface. Fig. S2 (ESI†) illustrates a representative confocal laser scanner image of mAECs that, following the washing step, show that cells are free of AuNRs on the membrane surface while AuNRs are displayed internally. Subsequently, mAECs were lysed and analyzed for gold content using ICP-MS, a reference analytical technique, consolidated in the last decade for reliable NP-metal quantification.62 As depicted in Fig. 2, both systems reach their maximum uptake at 24 hours. Specifically, the one-step approach performs better, incorporating significantly higher quantities of AuNRs at all evaluated time points. When exposed to 1.1 μg mL−1 AuNRs, an increase of 11, 14, and 12-fold was observed at 12, 24 and 48 hours, respectively, compared to the two-step method. Similarly, exposure to 4.4 μg mL−1 AuNRs resulted in a 12, 19, and 19-fold increase at 12, 24 and 48 hours, respectively, compared to the two-step approach. Additionally, the highest incorporation is observed when cells are exposed to the highest dose of AuNRs (4.4 μg mL−1), showing a peak uptake at 24 h followed by reaching a plateau at longer uptake times (Fig. 2). Consistently, literature evidence demonstrates a similar trend in nanorod uptake rates.63–66 The two-step culture strategy incorporates lower quantities of AuNRs, showing an increase at 24 h only when cells are exposed to a dose of AuNRs equal to 4.4 μg mL−1. Furthermore, extending the uptake times does not reach a plateau; instead, a decrease is observed (Fig. 2).Another critical aspect requiring consideration is the determination of the percentage of cells containing AuNRs acquired at the end of the cell labeling protocol. Evaluating this factor is essential for confirming the efficacy of the proposed cell tracking technique. A rapid and effective method validated in literature for determining the percentage of cells in a specific subset is flow cytometry.26 To date, this analytical technique has been extensively utilized to quantify NP uptake in mammalian cells since the introduction of NP to cells significantly increases side scatter, as documented in several studies.67–72 Importantly, its application for determining the percentage of NP-containing cells has gained significant interest in recent years73 even if the sensitivity in measurements is considerably lower compared to more complex techniques such as ICP-MS-based technologies.74
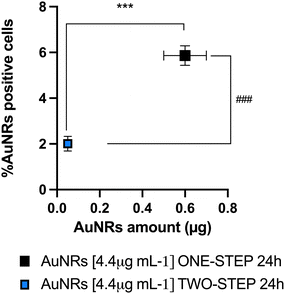
Of note, the applicative advantage of flow cytometry lies in its non-invasiveness. If required, the examined cells can be retrieved using a cell sorter and promptly employed for in vivo cell tracking purposes. A flow cytometric investigation was conducted to assess the percentage of mAECs containing AuNRs using both one and two-step. Notably, this analytical phase focused on the time window between 12 h and 24 h, guided by the results obtained from ICP-MS, which clearly indicated a plateau in AuNRs incorporation at 48 h. As shown in ESI† Fig. S3, a significant increase in the number of labelled cells was detected using the one-step protocol with 4.4 μg mL−1 AuNRs after 24 h of incubation. In more detail, mAECs cultured using the one-step protocol and exposed to 1.1 μg mL−1 and 4.4 μg mL−1 AuNRs for 24 h exhibited a respective 2.2-fold (2.1 ± 0.7% vs. 0.95 ± 0.2%) and 3-fold (5.9 ± 0.4% vs. 2.1 ± 0.3%) increase in positive cells compared to the percentage observed in the two-step approach. Consistent with the results obtained from ICP-MS concerning the internalized AuNRs, flow cytometric analysis exhibited the best performance in terms of cells labeled with AuNRs using the one-step approach at the optimized dose of 4.4 μg mL−1 after 24 hours of exposure. Remarkably, the analysis demonstrated a positive correlation between AuNRs data recorded with ICP-MS (amount of Au internalization) and flow cytometry (rhodamine positive cells) at 4.4 μg mL−1 after 24 h. This latter analysis definitively confirmed the one-step approach as the most efficient protocol for Au uptake and allowed the set-up of the cultural strategy before NP exposure (Fig. 3).
Even though the dynamics of uptake are closely dependent on a multitude of factors related to either NP features and cell type, biological context, and the function of the cells themselves,72,75 It is plausible to hypothesize that the one-step approach exhibits superior uptake capability as first it does not require preliminary steps, such as trypsinization, to prepare the batch of cells for NP labeling. These preliminary steps can be a source of stress and may negatively influence endocytosis dynamics.76
By avoiding potentially harmful procedures such as trypsinization, the one-step approach ensures a more straightforward and less invasive process for NP labeling, leading to more reliable and accurate results. This methodological improvement is particularly advantageous for maintaining the integrity of the cells and the fidelity of the uptake process, thus making the one-step approach more favorable under specific experimental conditions here detailed. Experimentally, suspension cells can also be easier to work with for endocytosis studies because they do not need to be detached from a surface for analysis and/or further applications. This can reduce experimental variability and simplify the protocol potentially offering the practical advantage of having a labelled cell population ready-to-be used for tracking procedures. This avoids additional manipulation, which might potentially cause the loss of the same nanorods from the cells or/and cell viability.
The adoption of combined analytical approaches has yielded robust and highly confident data, providing a precise measure of uptake. Notably, at present, there is no primary method for measuring NP uptake, necessitating the use of orthogonal methods to achieve reliable results77 for78 silica79 NPs. Furthermore, Böhme and colleagues utilized ICP-MS and flow cytometry, as in the present research, to accurately quantify the uptake of Al2O3 NP.80
Cell uptake improvement by combining G-force with mitochondria supplementation
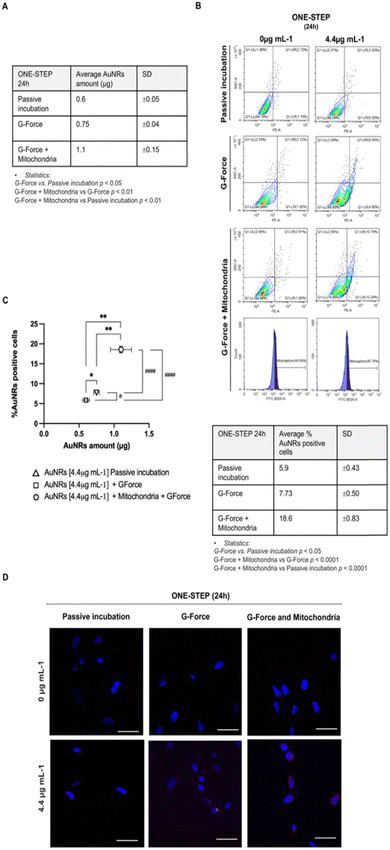
Overall, based on the previous findings, the one-step culture approach using 4.4 μg mL−1 AuNRs after 24 hours of incubation was chosen for further experiments aimed at identifying strategies to enhance cell uptake.First, the addition of AuNRs to mAECs was performed under the influence of G-force, based on literature evidence attesting a positive influence of the centrifugal force (hereinafter referred as G-force) on the internalization efficiency of NP.42 This effect had been attributed to the forced NP deposition on cell plasma membrane. Subsequently, mAECs were incubated with 4.4 μg mL−1 AuNRs for 24 h using the one-step method. Data were then analyzed by comparing the AuNRs uptake performances using ICP-MS and flow cytometry (AuNRs internalized amount and % of AuNRs positive cells, respectively) with or without (referred as passive incubation) G-force. As depicted in Fig. 4, G-force induces a significant increase in both AuNRs amount and cell positivity to AuNRs. More in detail, the AU internalization in mAECs increased 25% (0.75 ± 0.04 vs. 0.6 ± 0.05 μg/106 pellet; Fig. 4A and C) whereas the percentage of positive cells passed from 5.9 to 7.73% (p < 0.05; Fig. 4B and C).
Then, an additional optimization step was tempted. To improve AuNRs uptake, the mechanism of endocytosis was modulated by introducing isolated mitochondria by exploiting a recognized mechanisms of cell-to-cell dialogue (e.g. passage/exchange of organelles).
Importantly, recent evidence showed that internalization of exogenous mitochondria causes the activation of the actin-dependent endocytic pathway, demonstrated even by inhibiting endocytosis function.81–83 Starting from this premise and considering the recent successful attempts demonstrating a novel method to guide the transfer of mitochondria from cell donor to cell recipient, known as MitoCeption,84 freshly isolated mitochondria from mAECs (donor cells) were incorporated into the one-step protocol for AuNRs uptake, utilizing G-force. Data reported in Fig. 4 showed that mitochondria supplementation determines a huge and significant increase in Au amount internalized in mAECs (1.1 ± 0.15 μg, about 2-fold; Fig. 4A and C) and, at the same time, it significantly increases the percentage of positive-rhodamine cells (18.6 ± 0.83%; more than 2.5-fold; Fig. 4B and C).
To demonstrate that mitochondrial transfer facilitated the enhanced internalization of AuNRs through MitoCeption, the organelles were pre-labelled with the fluorophore Mitotracker green. The MitoCeption was then quantified using flow cytometry.
As reported in Fig. 4B, mitochondria transfer occurred by involving 45.63 ± 0.6% of mAECs (Ctrl: 0 μg mL−1 AuNRs) and 45.74 ± 1% of cells exposed to 4.4 μg mL−1. Significantly, MitoCeption synergizes with the mechanism of NP micropinocytosis. In fact, nearly all cells tested positive for AuNRs (83.10 ± 1%) exhibited incorporated mitochondria (Fig. S4, ESI†).
Confocal imaging was finally employed to further validate the enhancement in cell uptake achieved by combining G-force with MitoCeption. As depicted in Fig. 4D confocal laser scanning microscopy provided clear information consistent with the improved uptake data discussed earlier.
AuNRs uptake standardization applied to AECs
Finally, the protocol optimized for mAECs was extended to the native source of ovine AECs (see Fig. S1, ESI†) and then translated to human AECs. Human AECs have been extensively researched and utilized.20 In contrast to ovine AECs, isolated from animals at mid-gestation, human AECs are term cells obtained from the amniotic membrane. Biologically, they have a distinct background; the ovine AECs used in this study are isolated at a stage where they actively support pregnancy, whereas the human one adopted for the present research are cells that have fulfilled their biological role. Nonetheless, they remain a significant source of fetal stem cells due to their broad pro-regenerative properties.To this purpose, ovine AECs and human AECs were exposed to AuNRs uptake using the optimized protocol. More in detail, the previous source of isolated mitochondria collected from ovine-derived mAECs was supplemented to one-step cultured cells exposed to G force treatment.
ICP-MS analysis confirmed greater internalization in AECs compared to mAECs, regardless of species origin. More in detail, ovine AECs and human AECs internalized 1.5 (1.7 ± 0.1 vs. 1.1 ± 0.15 μg/106 pellet) and 3.6 (4.0 ± 0.4 vs. 1.1 ± 0.15 μg/106 pellet) times, respectively, more that ovine mAECs (Fig. 5A).
The result of improved uptake ability of the cells with the native epithelial phenotype was also confirmed by flow cytometry (Fig. 5B) showing a 1.3 (25.0 ± 0.5 vs. 18.6 ± 0.83%) and 2.6 (48.1 ± 3.8 vs. 18.6 ± 0.83%) fold increase in the percentage of rhodamine-positive ovine and human AECs, respectively. Confocal analysis corroborated these data (Fig. 5C). Notably, the human derived AECs showed a very high uptake efficiency that could be dependent of their species specificity (lipid composition of plasma membrane, endocytosis/pynocitosis function, etc.) or from the different lifetime of the two amniotic stem cell sources (late human vs. middle stage ovine).
Considering that the objective of labeling these cells is their use in regenerative medicine protocols, it is essential to evaluate the long-term effects of nanoparticle incorporation, ensuring the preservation of cellular and biological functionalities.
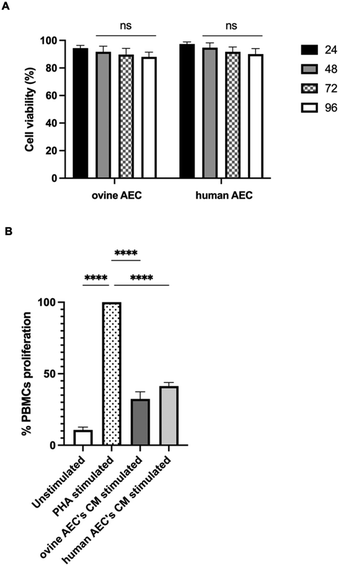
To this end, the long-term effect of NP incorporation on cellular functionality was monitored by MTS assay in both ovine and human AECs labeled with the optimized labeling condition, with the observation endpoint set at 96 hours. AuNRs labeling remained stable over time, with an unchanged percentage of AuNR-positive cells detected by flow cytometry compared to the 24-hour time point (Fig. S5, ESI†). Notably, cell viability data indicate a perfect preservation of cellular functionality over time (Fig. 6A).
Also, biological properties of AECs bearing AuNRs were evaluated. Accordingly, our recent studies described how AECs exhibit significant immunomodulatory properties. These properties are expressed through the paracrine release of specific immunomodulatory molecules that regulate the inflammatory microenvironment, promoting tissue regeneration.50,85 Specifically, it has been observed that conditioned media (CM) from this cell source, whether from ovine or human AECs, play a crucial role in modulating immune cell activities by suppressing the proliferation of different immune cells, including peripheral blood mononuclear cells (PBMCs).50,85–87 This response was significantly heightened when the cells were exposed to inflammatory-like stimuli.85,86
The immunomodulatory influence of CM derived from LPS-stimulated ovine and human AECs was tested using the PBMCs activation test. As shown in Fig. 6B, CM inhibited the PHA-induced proliferation of PBMCs by approximately 70% (p < 0.0001 vs. PHA) and ∼60% (p < 0.0001 vs. PHA) for ovine and human AECs, respectively. These data confirm that the native attitudes of these cells are preserved upon AuNRs incorporation.
Overall, the present results proved the efficiency of the combo protocol involving physical (G-force) and biological (MitoCeption) strategies in improving AuNRs incorporation.
Furthermore, the optimized uptake protocol can be effectively utilized with amniotic-derived cells, enabling the generation of a batch of labeled cells ranging from 2 × 105 to 5 × 105, depending on the AECs phenotype and the species of origin used for AnNRs incorporation, starting from an initial concentration of 1 × 106.
The availability of this quantity of cells does not impose a restriction on AECs, as each amniotic membrane can yield 100 to 500 million viable cells in the ovine placenta,40 and 80 to 300 million viable cells in a full-term human placenta.41
Our research group has optimized the recovery protocol of ovine AECs from the AM at middle gestational stage (fetus of 25–30 mm in size), allowing over 20 millions of cells to be to obtained from 10 cm2 of AM.40 This practical aspect is quite relevant in order to document the feasibility of the proposed protocol in generating a realistic amount of autologous immune-privileged labelled cells ready to be used for biomedical applications in medicine and veterinary. The protocol formulation seems to exceed the intracellular AuNR concentration necessary to be visualized with CT as reported as demonstrated by Kim and colleagues, who achieved CT detection with at least 2 × 104 cells60 labeled with Au using a passive incubation protocol.
Of relevance, the present results were obtained by requiring the use of a small initial AuNR concentrations (4.4 μg mL−1) if compared with other tracking protocols that started from initial Au NP doses ranging from μg mL−1 up to mg mL−1.56,60,64,65,88,89
Finally, the optimized protocol proposed in this research appears to significantly enhance AU internalization efficiency, even at the single-cell level. Premising that our group did not undertake single-cell quantification, as previously documented by Wang and colleagues,63 our results indirectly affirmed an internalized Au amount of 4.0 μg in a population of 0.48 × 106 labeled cells (equivalent to 48.1% out of 1 × 106 cells in human AECs). This yielded a single-cell incorporation of approximately 8.3 pg per cell, originating from a 4.4 μg mL−1 of AuNRs solution. The amount of incorporated gold exceeds the yield levels reported in the literature concerning AuNRs, particularly considering that the nanoparticle exposure concentration in this study was maintained at lower levels than those documented in previous studies.64,65
This high yield data coupled with a low exposure concentration is highly relevant from a biological standpoint, as it is preferable for avoiding changes in cellular functionality and cytotoxicity.90 This becomes particularly critical, as in our case, for regenerative medicine applications, necessitating the development of systems that not only track cells but also require them to function at the site of repair.
Conclusions
In this study, an optimized protocol for the uptake of gold NPs (AuNRs) by amniotic-derived cells, leveraging cell suspension (one-step), G-force and MitoCeption, was developed. This new uptake method significantly improved AuNRs incorporation in ovine amniotic-derived cells independently of cell phenotype (epithelial and mesenchymal) and in human epithelial cells, enhancing their application for regenerative medicine and biomedical imaging.The ability to greatly internalize NPs without compromising cell vitality or therapeutic properties opens new avenues in both human and veterinary medicine, particularly in non-invasive tracking of cell therapies and enhancing diagnostic precision with AuNRs as CT contrast agents. Future research will focus on standardizing this labeling protocol for clinical use, by assessing long-term effects in preclinical models to ensure safety and stem cell retention of their native regenerative potential. By establishing a new cell labeling protocol for cellular tracking, this study also unveils key insights into the mechanisms of cellular communication setting the stage for advanced studies to unravel the mechanisms behind NPs uptake and mitochondrial transfer, potentially revolutionizing personalized stem cell-based strategies of repair.
Author contributions
Alessia Peserico: validation, formal analysis, investigation, data curation, writing – original draft. Angelo Canciello: investigation, resources, visualization. Giuseppe Prencipe: investigation, resources, visualization. Roberto Gramignoli: resources. Valeria Melai: investigation. Giampiero Scortichini: resources. Mirella Bellocci: investigation. Giulia Capacchietti: investigation. Maura Turriani: resources. Chiara Di Pancrazio: resources. Paolo Beradinelli: supervision. Valentina Russo: supervision. Mauro Mattioli: conceptualization, methodology, supervision. Barbara Barboni: conceptualization, methodology, supervision, writing – review & editing, project administration, funding acquisition.Data availability
Any additional information is available from the corresponding author upon request.Conflicts of interest
There are no conflicts to declare.Acknowledgements
This research was funded by Fondazione Tercas, Project Title: “New frontiers in stem cell homing” 2021. The authors gratefully acknowledge the financial support from Next Generation EU with “PNRR - MISURA B4 – Centri di ricerca per l'innovazione” and the Italian Ministry of Education, Universities and Research (MUR) with “DM 1274/2021 art. 1, comma 1, lett. c”. The authors also thank Fabiana Verni for her valuable contribution to retrieving and transporting the biological material.Notes and references
- B. Barboni, V. Russo, P. Berardinelli, A. Mauro, L. Valbonetti and H. Sanyal, et al., Placental Stem Cells from Domestic Animals: Translational Potential and Clinical Relevance, Cell Transplant., 2018, 27(1), 93–116 CAS . Available from: https://pubmed.ncbi.nlm.nih.gov/29562773/.
- A. Umezawa, A. Hasegawa, M. Inoue, A. Tanuma-Takahashi, K. Kajiwara and H. Makino, et al., Amnion-derived cells as a reliable resource for next-generation regenerative medicine, Placenta, 2019, 84, 50–56 CrossRef CAS PubMed . Available from: https://pubmed.ncbi.nlm.nih.gov/31272680/.
- A. Papait, A. R. Silini, M. Gazouli, R. Malvicini, M. Muraca and L. O’Driscoll, et al., Perinatal derivatives: How to best validate their immunomodulatory functions, Front. Bioeng. Biotechnol., 2022, 10, 981061 CrossRef PubMed . Available from: https://pubmed.ncbi.nlm.nih.gov/36185431/.
- M. Magatti, E. Vertua, A. Cargnoni, A. Silini and O. Parolini, The Immunomodulatory Properties of Amniotic Cells: The Two Sides of the Coin, Cell Transplant., 2018, 27(1), 31–44 Search PubMed . Available from: https://pubmed.ncbi.nlm.nih.gov/29562786/.
- T. Miki, Stem cell characteristics and the therapeutic potential of amniotic epithelial cells, Am. J. Reprod. Immunol., 2018, 80(4), e13003 CrossRef PubMed . Available from: https://pubmed.ncbi.nlm.nih.gov/29956869/.
- C. H. Wassmer and E. Berishvili, Immunomodulatory Properties of Amniotic Membrane Derivatives and Their Potential in Regenerative Medicine, Curr. Diabetes Rep., 2020, 20(8), 31 CrossRef PubMed . Available from: https://pubmed.ncbi.nlm.nih.gov/32519069/.
- I. Taiko, C. Takano, M. Nomoto, S. Hayashida, K. Kanemaru and T. Miki, Selection of red fluorescent protein for genetic labeling of mitochondria and intercellular transfer of viable mitochondria, Sci. Rep., 2022, 12(1), 19841 CrossRef CAS PubMed . Available from: https://pubmed.ncbi.nlm.nih.gov/36400807/.
- D. M. Hoang, P. T. Pham, T. Q. Bach, A. T. L. Ngo, Q. T. Nguyen and T. T. K. Phan, et al., Stem cell-based therapy for human diseases, Signal Transduction Targeted Ther., 2022, 7(1), 272 CrossRef PubMed . Available from: https://pubmed.ncbi.nlm.nih.gov/35933430/.
- Y. Jin, S. Li, Q. Yu, T. Chen and D. Liu, Application of stem cells in regeneration medicine, MedComm, 2023, 4(4), e291 CrossRef CAS PubMed . Available from: https://pubmed.ncbi.nlm.nih.gov/37337579/.
- A. Peserico, B. Barboni, V. Russo, D. Nardinocchi, M. Turriani and C. Cimini, et al., AEC and AFMSC Transplantation Preserves Fertility of Experimentally Induced Rat Varicocele by Expressing Differential Regenerative Mechanisms, Int. J. Mol. Sci., 2023, 24(10), 8737 CrossRef CAS PubMed . Available from: https://pubmed.ncbi.nlm.nih.gov/37240083/.
- A. Muttini, L. Valbonetti, M. Abate, A. Colosimo, V. Curini and A. Mauro, et al., Ovine amniotic epithelial cells: in vitro characterization and transplantation into equine superficial digital flexor tendon spontaneous defects, Res. Vet. Sci., 2013, 94(1), 158–169 CrossRef CAS PubMed . Available from: https://pubmed.ncbi.nlm.nih.gov/22954787/.
- A. Colosimo, V. Curini, V. Russo, A. Mauro, N. Bernabò and M. Marchisio, et al., Characterization, GFP gene Nucleofection, and allotransplantation in injured tendons of ovine amniotic fluid-derived stem cells, Cell Transplant., 2013, 22(1), 99–117 CAS . Available from: https://pubmed.ncbi.nlm.nih.gov/22507078/.
- B. Barboni, C. Mangano, L. Valbonetti, G. Marruchella, P. Berardinelli and A. Martelli, et al., Synthetic bone substitute engineered with amniotic epithelial cells enhances bone regeneration after maxillary sinus augmentation, PLoS One, 2013, 8(5), e63256 CrossRef CAS PubMed . Available from: https://pubmed.ncbi.nlm.nih.gov/23696804/.
- C. Mangano, B. Barboni, L. Valbonetti, P. Berardinelli, A. Martelli and A. Muttini, et al., In Vivo Behavior of a Custom-Made 3D Synthetic Bone Substitute in Sinus Augmentation Procedures in Sheep, J. Oral Implantol., 2015, 41(3), 241–251 CrossRef PubMed . Available from: https://pubmed.ncbi.nlm.nih.gov/23829685/.
- P. Berardinelli, L. Valbonetti, A. Muttini, A. Martelli, R. Peli and V. Zizzari, et al., Role of amniotic fluid mesenchymal cells engineered on MgHA/collagen-based scaffold allotransplanted on an experimental animal study of sinus augmentation, Clin. Oral Invest., 2013, 17(7), 1661–1675 CrossRef PubMed . Available from: https://pubmed.ncbi.nlm.nih.gov/23064983/.
- V. Russo, A. Mauro, A. Peserico, O. Di Giacinto, M. El Khatib and M. R. Citeroni, et al., Tendon Healing Response Is Dependent on Epithelial-Mesenchymal-Tendon Transition State of Amniotic Epithelial Stem Cells, Biomedicines, 2022, 10(5), 1177 CrossRef CAS PubMed . Available from: https://pubmed.ncbi.nlm.nih.gov/35625913/.
- B. Barboni, V. Russo, V. Curini, A. Mauro, A. Martelli and A. Muttini, et al., Achilles tendon regeneration can be improved by amniotic epithelial cell allotransplantation, Cell Transplant., 2012, 21(11), 2377–2395 CAS . Available from: https://pubmed.ncbi.nlm.nih.gov/22507232/.
- L. A. Fortier and M. Lintz, Clinically, bone marrow aspirate concentrate, and micronized adipose tissue are equally efficacious in treatment of knee pain, but their benefits have not been directly correlated to retention of the respective stem cell niche Basic Science of Resident St, Oper. Tech. Sports Med., 2020, 28(4), 150776, DOI:10.1016/j.otsm.2020.150776.
- Z. Wu, G. Hui, Y. Lu, T. Liu, Q. Huang and L. Guo, Human amniotic epithelial cells express specific markers of nerve cells and migrate along the nerve fibers in the corpus callosum, Neural Regener. Res., 2012, 7(1), 41–45 Search PubMed . Available from: https://pubmed.ncbi.nlm.nih.gov/25806057/.
- Q. Zhang and D. Lai, Application of human amniotic epithelial cells in regenerative medicine: a systematic review, Stem Cell Res. Ther., 2020, 11(1), 439 CrossRef PubMed . Available from: https://pubmed.ncbi.nlm.nih.gov/33059766/.
- A. R. Nejad, A. A. Hamidieh, M. A. Amirkhani and M. M. Sisakht, Update review on five top clinical applications of human amniotic membrane in regenerative medicine, Placenta, 2021, 103, 104–119 CrossRef CAS PubMed . Available from: https://pubmed.ncbi.nlm.nih.gov/33120046/.
- J. L. Liesveld, N. Sharma and O. S. Aljitawi, Stem cell homing: From physiology to therapeutics, Stem Cells, 2020, 38(10), 1241–1253 CrossRef PubMed . Available from: https://pubmed.ncbi.nlm.nih.gov/32526037/.
- Y. Yin, X. Li, X. T. He, R. X. Wu, H. H. Sun and F. M. Chen, Leveraging Stem Cell Homing for Therapeutic Regeneration, J. Dent. Res., 2017, 96(6), 601–609 CrossRef CAS PubMed . Available from: https://pubmed.ncbi.nlm.nih.gov/28414563/.
- A. De Becker and I. Van Riet, Homing and migration of mesenchymal stromal cells: How to improve the efficacy of cell therapy?, World J. Stem Cells, 2016, 8(3), 73–87 CrossRef PubMed . Available from: https://pubmed.ncbi.nlm.nih.gov/27022438/.
- F. D. Miller and D. R. Kaplan, Mobilizing endogenous stem cells for repair and regeneration: are we there yet?, Cell Stem Cell, 2012, 10(6), 650–652 CrossRef CAS PubMed . Available from: https://pubmed.ncbi.nlm.nih.gov/22704501/.
- A. Peserico, C. Di Berardino, V. Russo, G. Capacchietti, O. Di Giacinto and A. Canciello, et al., Nanotechnology-Assisted Cell Tracking, Nanomaterials, 2022, 12(9), 1414 CrossRef CAS PubMed . Available from: https://pubmed.ncbi.nlm.nih.gov/35564123/.
- M. B. Gugjoo, Therapeutic Applications of Mesenchymal Stem Cells in Veterinary Medicine, Springer Nat. ISBN: 978:428.
- A. K. Srivastava and J. W. M. Bulte, Seeing stem cells at work in vivo, Stem Cell Rev. Rep., 2014, 10(1), 127–144 CrossRef PubMed . Available from: https://pubmed.ncbi.nlm.nih.gov/23975604/.
- L. Accomasso, C. Gallina, V. Turinetto and C. Giachino, Stem Cell Tracking with Nanoparticles for Regenerative Medicine Purposes: An Overview, Stem Cells Int., 2016, 7920358 CrossRef PubMed . Available from: https://pubmed.ncbi.nlm.nih.gov/26839568/.
- M. Sanchez-Diaz, M. I. Quiñones-Vico, R. S. de la Torre, T. Montero-Vílchez, A. Sierra-Sánchez and A. Molina-Leyva, et al., Biodistribution of Mesenchymal Stromal Cells after Administration in Animal Models and Humans: A Systematic Review, J. Clin. Med., 2021, 10(13), 2925 CrossRef PubMed . Available from: https://pubmed.ncbi.nlm.nih.gov/34210026/.
- J. W. M. Bulte and H. E. Daldrup-Link, Clinical Tracking of Cell Transfer and Cell Transplantation: Trials and Tribulations, Radiology, 2018, 289(3), 604–615 CrossRef PubMed . Available from: https://pubmed.ncbi.nlm.nih.gov/30299232/.
- J. H. Yoo, C. Park, D. I. Jung, C. Y. Lim, B. T. Kang and J. H. Kim, et al., In vivo cell tracking of canine allogenic mesenchymal stem cells administration via renal arterial catheterization and physiopathological effects on the kidney in two healthy dogs, J. Vet. Med. Sci., 2011, 73(2), 269–274 CrossRef PubMed . Available from: https://pubmed.ncbi.nlm.nih.gov/20953134/.
- I. A. Bhat, T. B. Sivanarayanan, A. Somal, S. Pandey, M. K. Bharti and B. S. K. Panda, et al., An allogenic therapeutic strategy for canine spinal cord injury using mesenchymal stem cells, J. Cell. Physiol., 2019, 234(3), 2705–2718 CrossRef CAS PubMed . Available from: https://pubmed.ncbi.nlm.nih.gov/30132873/.
- J. Burk, D. Berner, W. Brehm, A. Hillmann, C. Horstmeier and C. Josten, et al., Long-Term Cell Tracking Following Local Injection of Mesenchymal Stromal Cells in the Equine Model of Induced Tendon Disease, Cell Transplant., 2016, 25(12), 2199–2211 Search PubMed . Available from: https://pubmed.ncbi.nlm.nih.gov/27392888/.
- C. Horstmeier, A. B. Ahrberg, D. Berner, J. Burk, C. Gittel and A. Hillmann, et al., In Vivo Magic Angle Magnetic Resonance Imaging for Cell Tracking in Equine Low-Field MRI, Stem Cells Int., 2019, 5670106 Search PubMed . Available from: https://pubmed.ncbi.nlm.nih.gov/31933650/.
- Q. W. Liu, Q. M. Huang, H. Y. Wu, G. S. L. Zuo, H. C. Gu and K. Y. Deng, et al., Characteristics and Therapeutic Potential of Human Amnion-Derived Stem Cells, Int. J. Mol. Sci., 2021, 22(2), 1–33 Search PubMed . Available from: https://pubmed.ncbi.nlm.nih.gov/33478081/.
- O. Parolini and M. Caruso, Review: Preclinical studies on placenta-derived cells and amniotic membrane: an update, Placenta, 2011, 32(Suppl 2), S186–S195 CrossRef PubMed . Available from: https://pubmed.ncbi.nlm.nih.gov/21251712/.
- E. Antoniadou and A. L. David, Placental stem cells, Best Pract. Res. Clin. Obstet. Gynaecol., 2016, 31, 13–29 CrossRef PubMed . Available from: https://pubmed.ncbi.nlm.nih.gov/26547389/.
- C. Qiu, Z. Ge, W. Cui, L. Yu and J. Li, Human Amniotic Epithelial Stem Cells: A Promising Seed Cell for Clinical Applications, Int. J. Mol. Sci., 2020, 21(20), 1–26 Search PubMed . Available from: https://pubmed.ncbi.nlm.nih.gov/33086620/.
- A. Canciello, A. Cerverò-Varona, M. Turriani, V. Russo and B. Barboni, Amniotic Membrane and Amniotic Epithelial Cell Culture, Methods Mol. Biol., 2024, 2749, 135–149 CrossRef CAS PubMed . Available from: https://pubmed.ncbi.nlm.nih.gov/38133781/.
- R. Gramignoli, R. C. Srinivasan, K. Kannisto and S. C. Strom, Isolation of Human Amnion Epithelial Cells According to Current Good Manufacturing Procedures, Curr. Protoc. Stem Cell Biol., 2016, 37, 1E.10.1–1E.10.13 CrossRef PubMed . Available from: https://pubmed.ncbi.nlm.nih.gov/27171794/.
- S. M. Ocampo, V. Rodriguez, L. De La Cueva, G. Salas, J. L. Carrascosa and M. Josefa Rodriguez, et al., g-force induced giant efficiency of nanoparticles internalization into living cells, Sci. Rep., 2015, 5, 15160 CrossRef CAS PubMed . Available from: https://pubmed.ncbi.nlm.nih.gov/26477718/.
- P. Flores-Espinosa, M. Pineda-Torres, R. Vega-Sánchez, G. Estrada-Gutiérrez, A. Espejel-Nuñez and A. Flores-Pliego, et al., Progesterone elicits an inhibitory effect upon LPS-induced innate immune response in pre-labor human amniotic epithelium, Am. J. Reprod. Immunol., 2014, 71(1), 61–72 CrossRef CAS PubMed . Available from: https://pubmed.ncbi.nlm.nih.gov/24128422/.
- A. Canciello, V. Russo, P. Berardinelli, N. Bernabò, A. Muttini and M. Mattioli, et al., Progesterone prevents epithelial-mesenchymal transition of ovine amniotic epithelial cells and enhances their immunomodulatory properties, Sci. Rep., 2017, 7(1), 3761 CrossRef PubMed.
- A. Canciello, G. Teti, E. Mazzotti, M. Falconi, V. Russo and A. Giordano, et al., Progesterone Prolongs Viability and Anti-inflammatory Functions of Explanted Preterm Ovine Amniotic Membrane, Front. Bioeng. Biotechnol., 2020, 8, 135 CrossRef PubMed.
- L. S. Richardson, R. N. Taylor and R. Menon, Reversible EMT and MET mediate amnion remodeling during pregnancy and labor, Sci. Signaling, 2020, 13(618), eaay1486 CrossRef CAS PubMed . Available from: https://pubmed.ncbi.nlm.nih.gov/32047115/.
- A. Alcaraz, A. Mrowiec, C. L. Insausti, E. M. García-Vizcaíno, C. Ruiz-Canada and M. C. López-Martínez, et al., Autocrine TGF-β induces epithelial to mesenchymal transition in human amniotic epithelial cells, Cell Transplant., 2013, 22(8), 1351–1367 Search PubMed . Available from: https://pubmed.ncbi.nlm.nih.gov/23031712/.
- A. Lange-Consiglio, G. Accogli, F. Cremonesi and S. Desantis, Cell Surface Glycan Changes in the Spontaneous Epithelial–Mesenchymal Transition of Equine Amniotic Multipotent Progenitor Cells, Cells Tissues Organs, 2014, 200(3–4), 212–226 CrossRef CAS PubMed . Available from: https://pubmed.ncbi.nlm.nih.gov/26337136/.
- B. Barboni, V. Russo, V. Gatta, N. Bernabò, P. Berardinelli and A. Mauro, et al., Therapeutic potential of hAECs for early Achilles tendon defect repair through regeneration, J. Tissue Eng. Regener. Med., 2018, 12(3), e1594–e1608 CAS . Available from: https://pubmed.ncbi.nlm.nih.gov/29024514/.
- A. Cerverò-Varona, A. Canciello, A. Peserico, A. A. Haidar Montes, M. R. Citeroni and A. Mauro, et al., Graphene oxide accelerates TGFβ-mediated epithelial–mesenchymal transition and stimulates pro-inflammatory immune response in amniotic epithelial cells, Mater. Today Bio, 2023, 22, 100758 CrossRef PubMed . Available from: https://pubmed.ncbi.nlm.nih.gov/37600353/.
- I. Fathi and T. Miki, Human Amniotic Epithelial Cells Secretome: Components, Bioactivity, and Challenges, Front. Med., 2022, 8, 763141 CrossRef PubMed . Available from: https://pubmed.ncbi.nlm.nih.gov/35083233/.
- L. Greco, V. Russo, C. Rapino, C. Di Germanio, F. Fezza and N. Bernabò, et al., Characterization of Endocannabinoid System and Interleukin Profiles in Ovine AEC: Cannabinoid Receptors Type-1 and Type-2 as Key Effectors of Pro-Inflammatory Response, Cells, 2020, 9(4), 1008 CrossRef CAS PubMed . Available from: https://pubmed.ncbi.nlm.nih.gov/32325674/.
- D. Yitbarek and G. G. Dagnaw, Application of Advanced Imaging Modalities in Veterinary Medicine: A Review, Vet. Med., 2022, 13, 117–130 Search PubMed . Available from: https://pubmed.ncbi.nlm.nih.gov/35669942/.
- R. Meir and R. Popovtzer, Cell tracking using gold nanoparticles and computed tomography imaging, Wiley Interdiscip. Rev.: Nanomed. Nanobiotechnol., 2018, 10(2), e1480 Search PubMed . Available from: https://pubmed.ncbi.nlm.nih.gov/28544497/.
- R. Meir, K. Shamalov, O. Betzer, M. Motiei, M. Horovitz-Fried and R. Yehuda, et al., Nanomedicine for Cancer Immunotherapy: Tracking Cancer-Specific T-Cells in Vivo with Gold Nanoparticles and CT Imaging, ACS Nano, 2015, 9(6), 6363–6372 CrossRef CAS PubMed . Available from: https://pubmed.ncbi.nlm.nih.gov/26039633/.
- O. Betzer, R. Meir, T. Dreifuss, K. Shamalov, M. Motiei and A. Shwartz, et al., In-vitro Optimization of Nanoparticle-Cell Labeling Protocols for In-vivo Cell Tracking Applications, Sci. Rep., 2015, 5, 15400 CrossRef CAS PubMed . Available from: https://pubmed.ncbi.nlm.nih.gov/26507853/.
- R. Meir, O. Betzer, M. Motiei, N. Kronfeld, C. Brodie and R. Popovtzer, Design principles for noninvasive, longitudinal and quantitative cell tracking with nanoparticle-based CT imaging, Nanomedicine, 2017, 13(2), 421–429 CrossRef CAS PubMed . Available from: https://pubmed.ncbi.nlm.nih.gov/27720990/.
- O. Betzer, A. Shwartz, M. Motiei, G. Kazimirsky, I. Gispan and E. Damti, et al., Nanoparticle-based CT imaging technique for longitudinal and quantitative stem cell tracking within the brain: application in neuropsychiatric disorders, ACS Nano, 2014, 8(9), 9274–9285 CrossRef CAS PubMed . Available from: https://pubmed.ncbi.nlm.nih.gov/25133802/.
- P. Chhour, P. C. Naha, S. M. O’Neill, H. I. Litt, M. P. Reilly and V. A. Ferrari, et al., Labeling monocytes with gold nanoparticles to track their recruitment in atherosclerosis with computed tomography, Biomaterials, 2016, 87, 93–103 CrossRef CAS PubMed . Available from: https://pubmed.ncbi.nlm.nih.gov/26914700/.
- T. Kim, N. Lee, D. R. Arifin, I. Shats, M. Janowski and P. Walczak, et al., In Vivo Micro-CT Imaging of Human Mesenchymal Stem Cells Labeled with Gold-Poly-L-Lysine Nanocomplexes, Adv. Funct. Mater., 2017, 27(3), 1604213 CrossRef PubMed . Available from: https://pubmed.ncbi.nlm.nih.gov/28713230/.
- X. Zhang, G. Ma and W. Wei, Simulation of nanoparticles interacting with a cell membrane: probing the structural basis and potential biomedical application, NPG Asia Mater., 2021, 13, 52, DOI:10.1038/s41427-021-00320-0.
- B. Meermann and V. Nischwitz, ICP-MS for the Analysis at the Nanoscale—A Tutorial Review, J. Anal. At. Spectrom., 2018, 33, 1432–1468 RSC.
- H. Wang, B. Chen, M. He, X. Li, P. Chen and B. Hu, Study on uptake of gold nanoparticles by single cells using droplet microfluidic chip-inductively coupled plasma mass spectrometry, Talanta, 2019, 200, 398–407 CrossRef CAS PubMed . Available from: https://pubmed.ncbi.nlm.nih.gov/31036201/.
- B. E. White, M. K. White, Z. A. Nima Alsudani, F. Watanabe, A. S. Biris and N. Ali, Cellular Uptake of Gold Nanorods in Breast Cancer Cell Lines, Nanomaterials, 2022, 12(6), 937 CrossRef CAS PubMed . Available from: https://pubmed.ncbi.nlm.nih.gov/35335749/.
- Y. Xiao, W. Xu, Y. Komohara, Y. Fujiwara, H. Hirose and S. Futaki, et al., Effect of Surface Modifications on Cellular Uptake of Gold Nanorods in Human Primary Cells and Established Cell Lines, ACS Omega, 2020, 5(50), 32744–32752 CrossRef CAS PubMed . Available from: https://pubmed.ncbi.nlm.nih.gov/33376912/.
- R. Agarwal, V. Singh, P. Jurney, L. Shi, S. V. Sreenivasan and K. Roy, Mammalian cells preferentially internalize hydrogel nanodiscs over nanorods and use shape-specific uptake mechanisms, Proc. Natl. Acad. Sci. U. S. A., 2013, 110(43), 17247–17252 CrossRef CAS PubMed . Available from: https://pubmed.ncbi.nlm.nih.gov/24101456/.
- J. Park, M. K. Ha, N. Yang and T. H. Yoon, Flow Cytometry-Based Quantification of Cellular Au Nanoparticles, Anal. Chem., 2017, 89(4), 2449–2456 CrossRef CAS PubMed . Available from: https://pubmed.ncbi.nlm.nih.gov/28192941/.
- S. Y. Choi, N. Yang, S. K. Jeon and T. H. Yoon, Semi-quantitative estimation of cellular SiO2 nanoparticles using flow cytometry combined with X-ray fluorescence measurements, Cytometry, Part A, 2014, 85(9), 771–780 CrossRef PubMed . Available from: https://pubmed.ncbi.nlm.nih.gov/24980896/.
- N. Bohmer, A. Rippl, S. May, A. Walter, M. B. Heo and M. Kwak, et al., Interference of engineered nanomaterials in flow cytometry: A case study, Colloids Surf., B, 2018, 172, 635–645 CrossRef CAS PubMed . Available from: https://pubmed.ncbi.nlm.nih.gov/30243217/.
- X. Zhao and Y. Ibuki, Evaluating the toxicity of silver nanoparticles by detecting phosphorylation of histone H3 in combination with flow cytometry side-scattered light, Environ. Sci. Technol., 2015, 49(8), 5003–5012 CrossRef CAS PubMed . Available from: https://pubmed.ncbi.nlm.nih.gov/25815977/.
- R. M. Zucker and W. K. Boyes, Detection of Silver and TiO2 Nanoparticles in Cells by Flow Cytometry, Methods Mol. Biol., 2020, 2118, 415–436 CrossRef CAS PubMed . Available from: https://pubmed.ncbi.nlm.nih.gov/32152995/.
- Q. Xia, J. Huang, Q. Feng, X. Chen, X. Liu and X. Li, et al., Size- and cell type-dependent cellular uptake, cytotoxicity and in vivo distribution of gold nanoparticles, Int. J. Nanomed., 2019, 14, 6957–6970 CrossRef CAS PubMed . Available from: https://pubmed.ncbi.nlm.nih.gov/32021157/.
- H. R. Shin, M. Kwak, T. G. Lee and J. Y. Lee, Quantifying the level of nanoparticle uptake in mammalian cells using flow cytometry, Nanoscale, 2020, 12(29), 15743–15751 RSC . Available from: https://pubmed.ncbi.nlm.nih.gov/32677657/.
- F. Laborda, E. Bolea and J. Jiménez-Lamana, Single particle inductively coupled plasma mass spectrometry: a powerful tool for nanoanalysis, Anal. Chem., 2014, 86(5), 2270–2278 CrossRef CAS PubMed . Available from: https://pubmed.ncbi.nlm.nih.gov/24308527/.
- P. Rees, J. W. Wills, M. R. Brown, C. M. Barnes and H. D. Summers, The origin of heterogeneous nanoparticle uptake by cells, Nat. Commun., 2019, 10, 2341 CrossRef PubMed . Available from: https://pubmed.ncbi.nlm.nih.gov/31138801/.
- T. Serdiuk, S. Alekseev, V. Lysenko, V. Skryshevsky and A. Géloën, Trypsinization-dependent cell labeling with fluorescent nanoparticles, Nanoscale Res. Lett., 2014, 9(1), 568 CrossRef PubMed . Available from: https://pubmed.ncbi.nlm.nih.gov/25328505/.
- B. Drasler, D. Vanhecke, L. Rodriguez-Lorenzo, A. Petri-Fink and B. Rothen-Rutishauser, Quantifying nanoparticle cellular uptake: which method is best?, Nanomedicine, 2017, 12(10), 1095–1099 CrossRef CAS PubMed . Available from: https://pubmed.ncbi.nlm.nih.gov/28447906/.
- C. Gottstein, G. Wu, B. J. Wong and J. A. Zasadzinski, Precise quantification of nanoparticle internalization, ACS Nano, 2013, 7(6), 4933–4945 CrossRef CAS PubMed . Available from: https://pubmed.ncbi.nlm.nih.gov/23706031/.
- S. Vranic, N. Boggetto, V. Contremoulins, S. Mornet, N. Reinhardt and F. Marano, et al., Deciphering the mechanisms of cellular uptake of engineered nanoparticles by accurate evaluation of internalization using imaging flow cytometry, Part. Fibre Toxicol., 2013, 10, 2 CrossRef CAS PubMed . Available from: https://pubmed.ncbi.nlm.nih.gov/23388071/.
- S. Böhme, H. J. Stärk, T. Meißner, A. Springer, T. Reemtsma and D. Kühnel, et al., Quantification of Al2O3 nanoparticles in human cell lines applying inductively coupled plasma mass spectrometry (neb-ICP-MS, LA-ICP-MS) and flow cytometry-based methods, J. Nanopart. Res., 2014, 16(9), 2592 CrossRef PubMed . Available from: https://pubmed.ncbi.nlm.nih.gov/25285033/.
- E. Katrangi, G. D’Souza, S. V. Boddapati, M. Kulawiec, K. K. Singh and B. Bigger, et al., Xenogenic transfer of isolated murine mitochondria into human rho0 cells can improve respiratory function, Rejuvenation Res., 2007, 10(4), 561–570 CrossRef PubMed . Available from: https://pubmed.ncbi.nlm.nih.gov/18069915/.
- K. A. Sinclair, S. T. Yerkovich, P. M. A. Hopkins and D. C. Chambers, Characterization of intercellular communication and mitochondrial donation by mesenchymal stromal cells derived from the human lung, Stem Cell Res. Ther., 2016, 7(1), 91 CrossRef PubMed . Available from: https://pubmed.ncbi.nlm.nih.gov/27406134/.
- M. Jackson and A. Krasnodembskaya, Analysis of Mitochondrial Transfer in Direct Co-cultures of Human Monocyte-derived Macrophages (MDM) and Mesenchymal Stem Cells (MSC), Bio-Protoc., 2017, 7(9), e2255 Search PubMed . Available from: https://pubmed.ncbi.nlm.nih.gov/28534038/.
- A. Caicedo, V. Fritz, J. M. Brondello, M. Ayala, I. Dennemont and N. Abdellaoui, et al., MitoCeption as a new tool to assess the effects of mesenchymal stem/stromal cell mitochondria on cancer cell metabolism and function, Sci. Rep., 2015, 5, 9073 CrossRef CAS PubMed . Available from: https://pubmed.ncbi.nlm.nih.gov/25766410/.
- V. Di Lollo, A. Canciello, M. Orsini, N. Bernabò, M. Ancora and M. Di Federico, et al., Transcriptomic and computational analysis identified LPA metabolism, KLHL14 and KCNE3 as novel regulators of Epithelial-Mesenchymal Transition, Sci. Rep., 2020, 10(1), 4180 CrossRef CAS PubMed . Available from: https://pubmed.ncbi.nlm.nih.gov/32144311/.
- D. Rossi, S. Pianta, M. Magatti, P. Sedlmayr and O. Parolini, Characterization of the Conditioned Medium from Amniotic Membrane Cells: Prostaglandins as Key Effectors of Its Immunomodulatory Activity, PLoS One, 2012, 7(10), e46956 CrossRef CAS PubMed . Available from:/pmc/articles/PMC3468614/.
- H. Li, J. Y. Niederkorn, S. Neelam, E. Mayhew, R. A. Word and J. P. McCulley, et al., Immunosuppressive factors secreted by human amniotic epithelial cells, Invest. Ophthalmol. Visual Sci., 2005, 46(3), 900–907 CrossRef PubMed . Available from: https://pubmed.ncbi.nlm.nih.gov/15728546/.
- G. W. Marquart, J. Stoddard, K. Kinnison, F. Zhou, R. Hugo, R. Ryals, S. Shubert, T. J. McGill and M. R. Mackiewicz, Increasing the Efficacy of Gold Nanorod Uptake in Stem Cell-Derived Therapeutic Cells: Implications for Stem Cell Labeling and Optical Coherence Tomography Imaging, ACS Appl. Nano Mater., 2022, 5(5), 6995–7008 CrossRef CAS.
- A. Silvestri, V. Zambelli, A. M. Ferretti, D. Salerno, G. Bellani and L. Polito, Design of functionalized gold nanoparticle probes for computed tomography imaging, Contrast Media Mol. Imaging, 2016, 11(5), 405–414 CrossRef CAS PubMed . Available from: https://pubmed.ncbi.nlm.nih.gov/27377033/.
- R. Chandrasekaran, T. Madheswaran, N. Tharmalingam, R. J. Bose, H. Park and D. H. Ha, Labeling and tracking cells with gold nanoparticles, Drug Discovery Today, 2021, 26(1), 94–105 CrossRef CAS PubMed . Available from: https://pubmed.ncbi.nlm.nih.gov/33130336/.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d4tb00607k |
| This journal is © The Royal Society of Chemistry 2024 |