Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Dual-responsive near-infrared turn-on fluorescent probe for cancer stem cell-specific visualization†
Koji
Miki
 *a,
Masahiro
Oe
a,
Kanae
Suzuki
a,
Koki
Miki
a,
Huiying
Mu
*a,
Masahiro
Oe
a,
Kanae
Suzuki
a,
Koki
Miki
a,
Huiying
Mu
 a,
Yoshimi
Kato
b,
Mayumi
Iwatake
b,
Hiroshi
Yukawa
a,
Yoshimi
Kato
b,
Mayumi
Iwatake
b,
Hiroshi
Yukawa
 bcd,
Yoshinobu
Baba
bc,
Yoshifumi
Ueda
e,
Yasuo
Mori
e and
Kouichi
Ohe
bcd,
Yoshinobu
Baba
bc,
Yoshifumi
Ueda
e,
Yasuo
Mori
e and
Kouichi
Ohe
 *a
*a
aDepartment of Energy and Hydrocarbon Chemistry, Graduate School of Engineering, Kyoto University, Katsura, Nishikyo-ku, Kyoto 615-8510, Japan. E-mail: kojimiki@scl.kyoto-u.ac.jp; ohe@scl.kyoto-u.ac.jp
bInstitute of Nano-Life-Systems, Institutes of Innovation for Future Society, Nagoya University, Furo-cho, Chikusa-ku, Nagoya 464-8603, Japan
cInstitute of Quantum Life Science, Quantum Life and Medical Science Directorate, National Institutes for Quantum Science and Technology (QST), Anagawa 4-9-1, Inage-ku, Chiba 263-8555, Japan
dDepartment of Quantum Life Science, Graduate School of Science, Chiba University, Chiba 265-8522, Japan
eDepartment of Synthetic Chemistry and Biological Chemistry, Graduate School of Engineering, Kyoto University, Katsura, Nishikyo-ku, Kyoto 615-8510, Japan
First published on 10th June 2024
Abstract
Aldehyde dehydrogenase 1A1 (ALDH1A1) stands out as one of the most reliable intracellular biomarkers for stem cells because it is expressed in both cancer stem cells (CSCs) and normal somatic stem cells (NSCs). Although several turn-on fluorescent probes for ALDH1A1 have been developed to visualize CSCs in cancer cells, the discrimination of CSCs from NSCs is difficult. We here report an AND-type dual-responsive fluorescent probe, CHO_βgal, the near-infrared fluorescence of which can be turned on after responding to both ALDH1A1 and β-galactosidase. The AND-type dual responsiveness enables CSCs to be clearly visualized, whereas NSCs are non-emissive in microscopy. CSC-positive metastasis model lungs were successfully discriminated from normal lungs in ex vivo staining experiments using CHO_βgal, whereas the single-input ALDH1A1-responsive probe failed to achieve this discrimination owing to pronounced false-positive fluorescence output from lung NSCs. In tissue slice staining experiments, even in the presence of adjacent normal tissues, the peripheral region-specific localization of CSCs was clear. The versatility of CHO_βgal holds promise not only as a fundamental in vitro research tool for visualizing CSCs but also as a valuable asset in practical tissue staining diagnosis, significantly contributing to the assessment of cancer malignancy.
Introduction
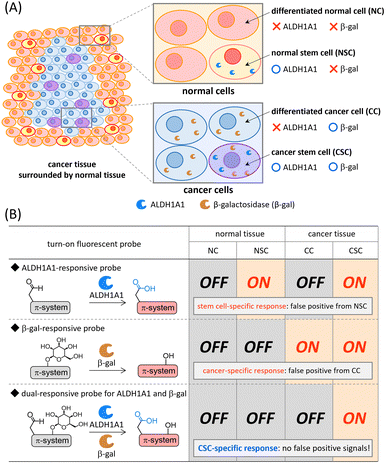
Cancer stem cells (CSCs) have generated great excitement in the research area of cancer because these cells are closely related to cancer initiation, progression, metastasis, and recurrence.1–3 CSCs are a small population of cancer cells that present specific antigens on the surface and/or express specific intracellular enzymes.4 During the past few decades, the CSC concept,1–3 cancer plasticity,5–8 and dormant cancer stem cells6,9,10 have been intensively discussed in the field of oncology in an effort to understand the therapeutic response of cancers. Given that details of cancer progression and metastasis mechanisms remain controversial, the development of reliable molecular imaging probes that can be used both to achieve a deeper understanding of CSCs and as a more efficient imaging agent for rapid and facile cancer malignancy assessment in clinical diagnosis, is highly desired. CSC fluorescence imaging is recognized as one of the most powerful tools because of its technical simplicity, noninvasive nature, and high spatiotemporal resolution.11,12Intracellular enzyme aldehyde dehydrogenase 1A1 (ALDH1A1)13,14 is one of the most promising biomarkers overexpressed in CSCs, and a variety of ALDH1A1-specific fluorescent probes have been developed to date.11 The always-on fluorescent probe ALDEFLUOR15 and analogous derivatives16–18 are frequently used for CSC isolation based on fluorescence-activated cell sorting. Recently, turn-on fluorescent probes,19–24 which are non-emissive before ALDH1A1-mediated transformation, have been developed for both visualization and isolation of CSCs in cancer cells. Some of the near-infrared (NIR) turn-on fluorescent probes were successfully applied to CSC-specific visualization in vivo and ex vivo.20,22 However, ALDH1A1 is known to be an intracellular biomarker of stem cells and it is overexpressed both in CSCs in cancer tissues and normal somatic stem cells (NSCs) in normal tissues (Fig. 1(A)).14,25,26 Hence, the discrimination of CSCs from NSCs is considered to be difficult by using single-input ALDH1A1-responsive probes (Fig. 1(B)). Given that contamination of NSCs is usually unavoidable in surgically excised cancer tissues, a smart probe that becomes emissive in CSCs but remains silent in NSCs is urgently needed for the evaluation of cancer malignancy both in fundamental science and clinical diagnosis/therapy.27
Recently, dual-responsive fluorescent probes that can respond to two concurrently generated biological stimuli have been developed.28 Among dual-responsive probes, AND-type probes can suppress false positive signals induced by one of two target stimuli. It is known that the expression level of β-galactosidase is upregulated in a wide range of cancer cells (Fig. 1(A));29,30 therefore, a variety of β-galactosidase-responsive fluorescent probes have been developed for cancer detection.31–34 Despite their cancer-specific identification, the discrimination of CSCs from differentiated cancer cells is considered to be difficult (Fig. 1(B)). Based on this background, we envisioned that a dual-responsive AND-type probe that becomes emissive only after both ALDH1A1- and β-galactosidase-mediated transformations would make specific visualization of CSCs possible (Fig. 1(B)). We herein report a dual-responsive turn-on fluorescent probe CHO_βgal (Fig. 2(A)), in which an NIR fluorescent hemicyanine dye is modified with both ALDH1A1- and β-galactosidase-responsive substrates. In in vitro cell staining using CHO_βgal, CSCs in cancer cells were specifically visualized with high contrast, whereas NSCs in normal cells remained non-emissive. In CHO_βgal staining of excised tissue slices, CSCs in cancer tissues as well as CSCs invading normal tissues were visualized even in the presence of NSCs.
Results and discussion
Probe design
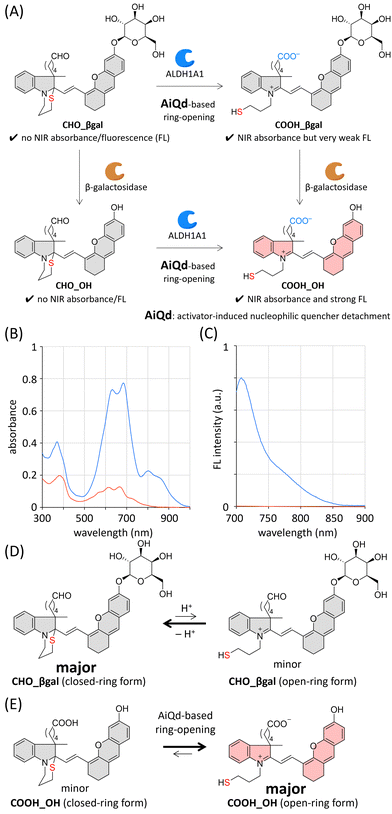
To construct dual-responsive probe CHO_βgal, a member of the hemicyanine dye family, [2-((E)-2-(6-hydroxy-2,3-dihydro-1H-xanthen-4-yl)ethenyl)-3H-indol-1-ium], was selected as a fluorophore, the NIR emission of which is suitable for both in vitro and ex vivo applications.35,36 The NIR emission of the hemicyanine fluorophore is derived from its acceptor–π–donor structure consisting of an electron-accepting iminium group and an electron-donating phenolate moiety. Hence, its fluorescence emission can be quenched by modifying either of these two groups. The ALDH1A1-responsive molecular structure in CHO_βgal consists of an N-substituted benzoindole, an ω-formylbutyl group as an ALDH1A1 substrate, and an ω-mercaptopropyl group as a nucleophilic fluorescence quencher. Before the ALDH1A1-mediated oxidation of a formyl group, the intramolecular cyclization of the nucleophilic mercapto group takes place to form a 1,3-thiazinane ring structure under physiological conditions. Because the electron-accepting iminium group is masked by the mercapto group through this cyclization, it can be expected that CHO_βgal does not show NIR absorbance/fluorescence. After ALDH1A1-mediated transformation, the resulting carboxy group acts as an activator to promote ring-opening of the 1,3-thiazinane to generate an iminium group, thereby recovering the inherent π-extended dye skeleton. This activator-induced nucleophilic quencher detachment (AiQd) mechanism has been developed by us for enzyme-responsive turn-on fluorescent probes.37,38 However, the electron-donating phenolate moiety remains capped with a weakly electron-donating β-galactosyl group after the sequential ALDH1A1-mediated transformation of CHO_βgal and AiQd-based ring-opening; therefore, it can be expected that the plausible product COOH_βgal has NIR absorbance but emits very weak NIR fluorescence. In the case of β-galactosidase-mediated transformation, CHO_βgal can be hydrolysed at the anomeric position of the β-galactosyl group to afford CHO_OH. Given that the 1,3-thiazinane ring is stable under physiological conditions, CHO_OH is assumed to show neither NIR absorbance nor fluorescence. After both ALDH1A1- and β-galactosidase-mediated transformations of CHO_βgal, the plausible product COOH_OH is expected to exhibit strong fluorescence.Based on this design strategy, we prepared CHO_βgal as a dual-responsive AND-type turn-on fluorescent probe together with the plausible products COOH_βgal, CHO_OH, and COOH_OH (Schemes S1–S3 and Fig. S1–S3, ESI†). Because the closed-ring form of CHO_βgal contains two chiral quaternary carbons, it is formed as a mixture of major and minor diastereoisomers. Density functional theory (DFT) calculations support the conclusion that the ω-formylbutyl group and the 2-(6-galactosyl-2,3-dihydro-1H-xanthenyl)ethenyl group are located on the same side in the major diastereoisomer (Fig. S4, ESI†).
Photophysical properties
Absorption and fluorescence spectra of CHO_βgal and COOH_OH in neutral phosphate buffered solution (PBS, pH 7.4) were measured (Fig. 2(B), (C) and Table S1, ESI†). In the case of CHO_βgal, two broad signals were observed at 300–400 nm and 500–800 nm. As supported by time-dependent DFT calculations (Fig. S5, ESI†), the former absorbance is attributed to the closed-ring form in which the π-system is shortened. Because the intramolecular nucleophilic cyclization of a mercapto group is a reversible reaction, the open-ring form of CHO_βgal was also present (Fig. 2(D)). Therefore, the latter absorbance can be assigned to a hemicyanine dye skeleton in the open-ring form. The molar extinction coefficients (ε, L mol−1 cm−1) of CHO_βgal are 1.2 × 104 at 381 nm, 7.6 × 103 at 614 nm, and 7.3 × 103 at 667 nm. The ε value at 667 nm attributed to the open-ring form is much lower than those of common water-soluble hemicyanine dyes (ε = ∼3 × 104),39 indicating that the closed-ring form of CHO_βgal is a major component in the solution. As expected, almost no NIR emission was detected in the fluorescence spectra. In contrast to CHO_βgal, COOH_OH showed strong absorbance at 600–700 nm (ε = 3.5 × 104 at 681 nm), indicating that the AiQd mechanism is applicable to a hemicyanine skeleton and that the equilibrium of COOH_OH shifts to the open-ring form under neutral conditions (Fig. 2(E)). The broad absorbance around 800 nm can be assigned to the J-aggregate of the hemicyanine dye.40 The fluorescence intensity of COOH_OH at 710 nm was more than 400 times stronger than that of CHO_βgal. We confirmed that both CHO_OH and COOH_βgal were slightly emissive but that the intensities were much weaker than that of COOH_OH (Fig. S6(A), ESI†). Because both the 1,3-thiazinane and phenolate moieties were expected to be pH-responsive, the pH-dependent spectral changes were investigated (Fig. S6–S9, ESI†). Although fluorescence intensity was dependent on pH values, it was confirmed that COOH_OH tends to be much more emissive than the other compounds under physiological conditions. By considering the photophysical properties of the compounds, CHO_βgal is expected to act as an AND-type turn-on fluorescent probe for ALDH1A1 and β-galactosidase.Enzyme responsiveness
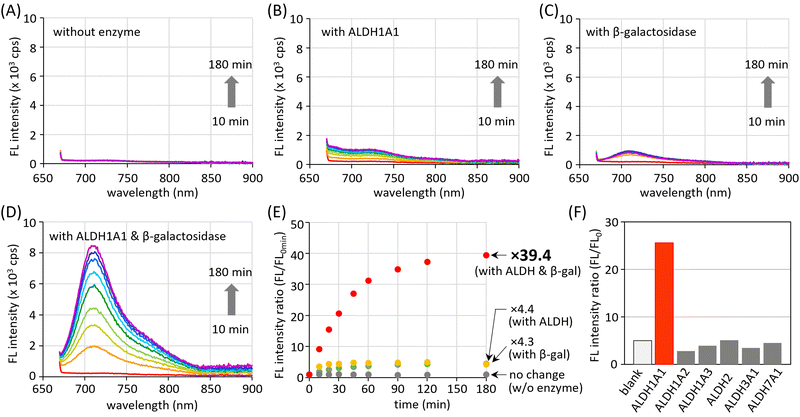
The AND-type dual responsiveness of CHO_βgal to enzymes was examined. In the absence of enzymes, no clear increase in fluorescence intensity was detected (Fig. 3(A)). A buffered aqueous solution of CHO_βgal (20 μM, pH 7.4) was incubated with ALDH1A1 (200 nM) and its fluorescence intensity was monitored. The broad peak at 700–750 nm in the fluorescence spectrum increased gradually during incubation and a peak at 710 nm (excitation: 660 nm) was detected that became 4.4 times stronger after 3 h incubation (Fig. 3(B) and (E)). As described in the previous section, COOH_βgal was slightly emissive because AiQd-based ring-opening shifts the equilibrium to its open-ring form. Hence, the weak NIR fluorescence output after ALDH1A1-mediated transformation is reasonable. When ALDH1A1 was pretreated with its inhibitor disulfiram and incubated with CHO_βgal, the increase in fluorescence intensity was suppressed, explaining the need for a formyl group (Fig. S10, ESI†). When the β-galactosidase-mediated transformation of CHO_βgal was examined, the fluorescence intensity was found to increase 4.3 times after 3 h incubation (Fig. 3(C) and (E)). The formation of weakly emissive CHO_OH was confirmed by liquid chromatography-mass spectroscopy (LC-MS) (Fig. S11, ESI†). The presence of an excess amount of D-galactose, which is a potent capping agent of the β-galactosidase active site, suppressed the increase in the fluorescence intensity (Fig. S12, ESI†). The increase of the fluorescence intensity after treatment of CHO_βgal with both ALDH1A1 and β-galactosidase was then monitored. LC-MS analysis after 1 h incubation confirmed that CHO_βgal was fully consumed and that both COOH_βgal and COOH_OH were generated (Fig. S13, ESI†). The fluorescence intensity was gradually increased to 39.4 times after 3 h incubation (Fig. 3(D) and (E)). These results indicate that the treatment of both ALDH1A1 and β-galactosidase smoothly generates emissive COOH_OH and that there is no significant interference between the two enzymes. Thus, the AND-type turn-on property of CHO_βgal mediated by ALDH1A1 and β-galactosidase was demonstrated. The limits of detection for ALDH1A1 and β-galactosidase were estimated at 1.7 μM and 0.032 U mL−1, respectively, by measuring fluorescence intensities at different concentrations (Fig. S14, ESI†).The stability of CHO_βgal under physiological conditions was examined (Fig. S15, ESI†). The fluorescence intensities after treatment of CHO_βgal with biological thiols such as cysteine, glutathione (GSH), and mercapto-containing protein (bovine serum albumin: BSA) were much weaker than that observed when CHO_βgal was incubated with both enzymes. Treatment of CHO_βgal with either amino acids bearing other functional groups or reactive oxygen species that oxidize a sulfur atom did not lead to fluorescence turn-on. Given that human ALDHs are found in 19 isoforms, the isoform selectivity of CHO_βgal was also examined. To this end, CHO_βgal was treated with several ALDH isoforms in the presence of β-galactosidase. As shown in Fig. 3(F), no significant increase in fluorescence intensity was observed upon incubation with any of the representative ALDH isoforms (ALDH1A2, ALDH1A3, ALDH2, ALDH3A1, or ALDH7A1), indicating the pronounced high selectivity to ALDH1A1.
In vitro CSC visualization
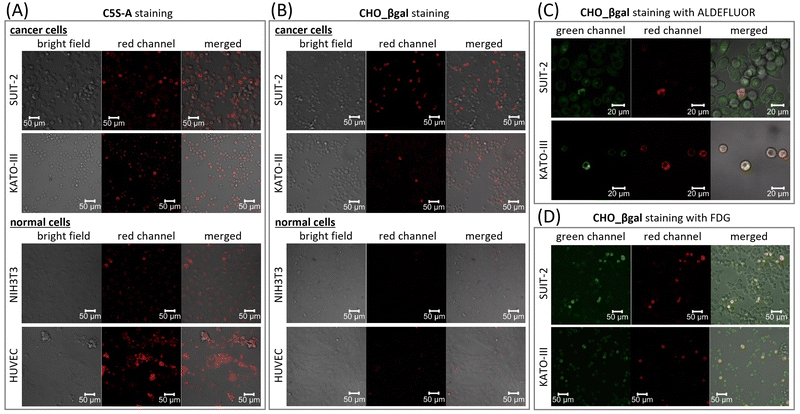
To verify the dual responsiveness of CHO_βgalin vitro, human pancreatic carcinoma cell line SUIT-2 and human gastric signet ring carcinoma cell line KATO-III, both of which are known to contain CSCs with high ALDH1A1 activity, were treated with CHO_βgal (Fig. 4 and Fig. S18, ESI†).30,41,42 Mouse embryonic fibroblast cell line NIH3T3 and human umbilical vein endothelial cell line HUVEC were chosen as representative β-galactosidase-negative normal cells for control experiments. To demonstrate the superiority of dual responsiveness, single-input ALDH1A1-responsive fluorescent probe C5S-A22 was used as a control molecule. Based on the ALDEFLUOR assay,15 we verified that NIH3T3 and HUVEC cells contain 11% and 33% ALDH1-positive cells, respectively (Fig. S22, ESI†). After treatment of SUIT-2 cells with C5S-A, a small population of brightly emissive cells that could be assigned to CSCs was visualized (Fig. 4(A) and Fig. S18(A), ESI†). In addition, it was confirmed that another cancer cell KATO-III as well as normal cells NIH3T3 and HUVEC contained brightly emissive ALDH1A1-positive cells. These results clearly indicate that single-input probe C5S-A is not suitable for discriminating between CSCs and NSCs.The high cell viability of both cancer and normal cells was confirmed after 24 h incubation with CHO_βgal (0.01–100 μM), indicating that CHO_βgal has low cytotoxicity at a concentration of <20 μM (Fig. S16, ESI†). We next examined the turn-on nature of CHO_βgal in both cancer and normal cells. Cancer cell staining using CHO_βgal resulted in brightly emissive cells being observed together with a large number of non-emissive cells (Fig. 4(B) and Fig. S18(B), ESI†). Notably, no washing step was necessary to obtain high-contrast visualization because of the turn-on property of CHO_βgal. Furthermore, the signal/noise ratios (FCSC/FCC) of CHO_βgal, which were obtained by dividing the fluorescence intensities of CSCs (FCSC) by those of differentiated cancer cells (FCC), were 11.4 ± 2.0 for SUIT-2 and 15.4 ± 3.5 for KATO-III, which are much higher than those of C5S-A (FCSC/FCC = 4.4 ± 0.9 for SUIT-2 and 3.6 ± 1.3 for KATO-III) (Fig. S17, ESI†). The high signal/noise ratios suggest that CHO_βgal is a suitable probe for high contrast CSC visualization. It is noted that more than 10 μM of CHO_βgal should be used for clear staining because of lower fluorescence intensity of hemicyanine dyes compared with cyanine dyes. As shown in the enzymatic transformation experiments, conversion of CHO_βgal catalysed by either ALDH1A1- or β-galactosidase alone did not yield a strong fluorescence output; therefore, the strong fluorescence was obtained only through conversion of CHO_βgal catalysed by both enzymes. To confirm that the expression levels of ALDH1A1 and β-galactosidase are both upregulated in CSCs, co-staining experiments were conducted using CHO_βgal with green-fluorescent ALDEFLUOR or fluorescein di-β-D-galactopyranoside (FDG),43,44 which is a commercial green-fluorescent β-galactosidase-specific single-input probe (Fig. 4(C), (D) and Fig. S19, ESI†). In low-magnification mode, ALDEFLUOR staining did not clearly distinguish ALDH1-positive cells of SUIT-2 from ALDH1-negative cells because of its always-on nature. On the other hand, in higher magnification mode, slightly brighter cells were identified in the green channel and the cells were also emissive in the deep-red channel (Fig. 4(C) and Fig. S19(A), ESI†). In the case of KATO-III, brighter cells were identified in the green channel even in low magnification mode and the stained cells were the same as those seen in the deep-red channel (Fig. S20, ESI†). This is probably because the ALDH1A1 expression level in CSCs of KATO-III is higher than that of SUIT-2.30 In co-staining experiments using CHO_βgal with FDG, CHO_βgal-positive cells were included in FDG-positive cells (Fig. 4(D), and Fig. S19(B), ESI†). Surprisingly, fluorescence from CHO_βgal-negative cells was observed in the green channel, but the intensity was significantly weaker than fluorescence from CHO_βgal-positive cells. The co-staining results strongly suggest that the β-galactosidase activity is upregulated in ALDH1A1-positive CSCs in both SUIT-2 and KATO-III. The use of CHO_βgal confirmed that FDG is an effective probe for detecting ALDH1A1-positive CSCs in specific cell lines. Pre-treatment of cells with either the ALDH1A1 inhibitor disulfiram or β-galactosidase competitive substrate D-galactose successfully suppressed the fluorescence turn-on of CHO_βgal (Fig. S21, ESI†). No positive images were observed with NIH3T3 and HUVEC (Fig. 4(B) and Fig. S18(B), ESI†), indicating that CHO_βgal can be used to successfully distinguish CSCs from NSCs in vitro. Flow cytometry analysis of HUVEC cells also supports that CHO_βgal remains silent in NSCs (Fig. S23, ESI†). These results suggest that CHO_βgal staining has the potential to selectively visualize/isolate CSCs from tissues containing both normal and cancer cells harvested from patients. Furthermore, through co-staining experiments using commercial single-input probes, we found that the β-galactosidase expression levels in CSCs are clearly higher than those in differentiated cancer cells in specific cell lines. More importantly, these results suggest that the new design of dual-responsive probes for ALDH1A1 and other biological analytes will find new biomarkers that are specifically expressed in CSCs.
Ex vivo CSC visualization
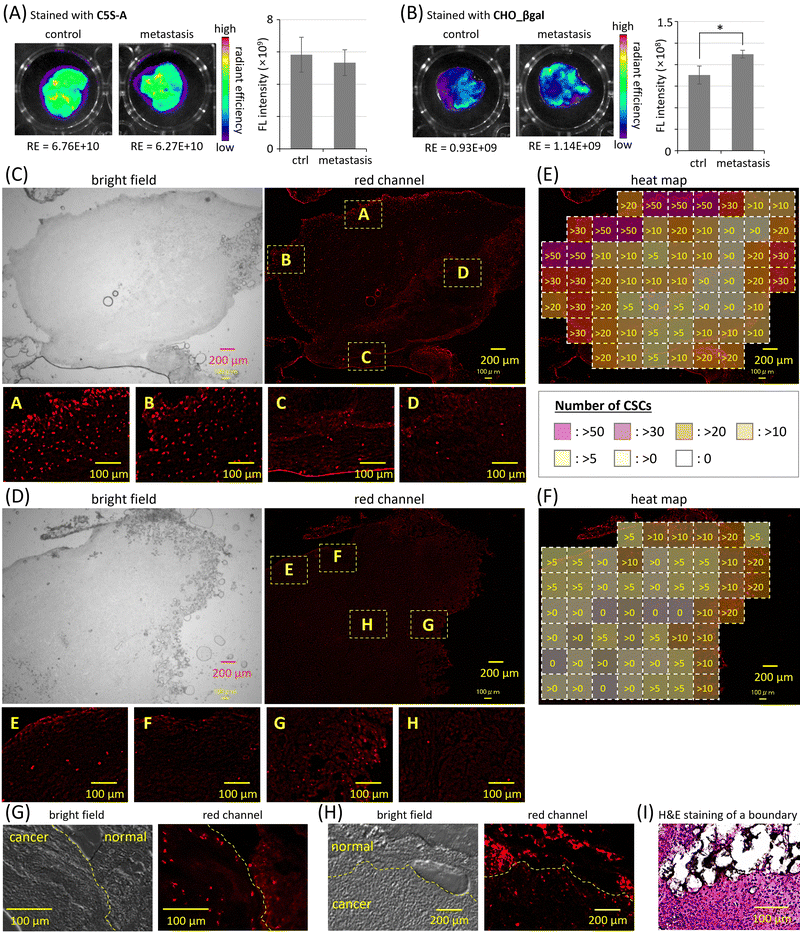
In cancer metastasis, cancer cells break off from the original cancer tissues and spread to other organs; therefore, cancer metastasis is often associated with adverse prognoses. The invasion of CSCs through the basal membrane into blood vessels is proposed as one of the most plausible mechanisms of cancer metastasis initiation.1–3 The intravenous injection of cancer cells into mice is a facile method to prepare model mice bearing metastatic cancer in the lungs,45 in which the CSC population increases. The excised lungs of nude mice five days after intravenous injection of SUIT-2 cells were stained by using single-input probe C5S-A and dual-responsive probe CHO_βgal (Fig. 5 and Fig. S24, ESI†). The excised lungs of nude mice without injection of SUIT-2 cells were also stained for control experiments. In the case of C5S-A, strong fluorescence was observed in the lungs of both metastasis model mice and normal mice (Fig. 5(A) and Fig. S24(A), ESI†); there was no significant difference between these two groups. The strong fluorescence is considered to be caused by ALDH1A1 expressed in NSCs of lung tissues. In contrast, in the case of CHO_βgal, much weaker fluorescence was observed in both the control and metastasis lungs, indicating that CHO_βgal successfully suppresses the false-positive signals from NSCs (Fig. 5(B) and Fig. S24(B), ESI†). A significant difference between the two groups was confirmed; however, the difference in fluorescence intensities was not large. As shown in Fig. 3(E), the fluorescence intensity increments of CHO_βgal after the treatment either with only ALDH1A1 or with both ALDH1A1 and β-galactosidase were 4.4 times and 39.4 times, respectively. The number of NSCs in metastasis model lungs is expected to be much greater than that of CSCs. Although the difference in fluorescence intensity increments was clear in the enzyme treatment experiments, the fluorescence after conversion of CHO_βgal by ALDH1A1 in NSCs pushed up the background fluorescence, resulting in the small difference in fluorescence intensity between the metastasis model lungs and normal lungs.Finally, by staining excised slices of both normal and cancer tissue, we evaluated the CSC staining ability of CHO_βgal. The normal and cancer tissues as well as the boundary region between these two tissues were identified by using hematoxylin and eosin (H&E) staining (Fig. 5(I) and Fig. S32, ESI†). No strong fluorescence was observed from adipose tissue slices, which were used as a normal tissue model with low β-galactosidase expression level (Fig. S25, ESI†). In cancer tissue slices, which were prepared by the intraperitoneal injection of SUIT-2 cells into femoral subcutaneous adipose tissues of mice, much stronger fluorescence was detected from the entire tissue slice together with many bright spots (Fig. 5(C) and Fig. S26, ESI†). When cancer tissue slices were treated with a mixture of CHO_βgal and ALDH1A1 inhibitor disulfiram, the number of bright spots decreased significantly (Fig. 5(D) and Fig. S27, ESI†). Similar bright spots were observed in the cancer tissue slices stained by C5S-A; however, these spots were difficult to be discriminated because of the high background signal of C5S-A (Fig. S28, ESI†). The number of bright spots was also decreased under co-treatment of C5S-A with disulfiram (Fig. S29, ESI†). These results clearly indicate that the bright spots observed in both CHO_βgal and C5S-A staining arise from fluorescence from CSCs. The heat maps of the numbers of CSCs detected in Fig. 5(C) and (D) are summarized in Fig. 5(E) and (F), respectively. Interestingly, CSCs were localized in the peripheral region of cancer tissues, whereas almost no CSCs were detected in the core of the tissues. Considering that CSCs are closely related to cancer progression and metastasis,46 it is reasonable to conclude that active CSCs are specifically localized at the peripheral region of cancer tissues. It is known that cancer tissues contain invasive leader cells, the so-called “leading front”, having proliferative, invasive, and metastatic abilities compared with other trailing follower cells.47 As shown in Fig. 5(E), it is obvious that the density of CSCs was higher at the left upper side, but lower at the lower side. This result suggests that CHO_βgal staining can point out the location of the active “leading front” in malignant cancer tissues.
Because CSCs in cancer tissue slices were successfully visualized, the discrimination of CSCs from NSCs in tissue slices was next examined. When a boundary region between cancer and adipose tissues was excised and the slices were stained with CHO_βgal (Fig. 5(G) and Fig. S30, ESI†), many bright spots were observed. It was confirmed that the ALDH1A1 inhibitor disulfiram successfully suppressed the fluorescence turn-on in the sliced tissues (Fig. S31, ESI†). The many bright spots that were assignable to CSCs were predominantly observed on the side of the cancer tissue near the boundary region. This observation is consistent with the distribution of stained cells in cancer tissue slices shown in Fig. 5(C). In the case of tissue slice staining using single-input probe C5S-A, many bright spots were observed in both cancer and adipose tissues (Fig. 5(H) and Fig. S28, ESI†) and the emission could be suppressed by ALDH1A1 inhibitor disulfiram (Fig. S29, ESI†). These results clearly indicate that the single-input probe could respond to ALDH1A1 in both CSCs and NSCs, being not suitable for CSC-specific visualization. In contrast, in the case of CHO_βgal staining, the CSC-specific visualization at the boundary region was not influenced by the presence of NSCs. These results strongly suggest that CHO_βgal staining will be applicable to practical cancer malignancy assessment using excised tissue slices that contain both cancer and normal tissues. Some bright spots were observed on the side of adipose tissues near the boundary region, probably indicating that CHO_βgal staining can reveal CSCs invading normal tissues.
Conclusions
In summary, we have developed the ALDH1A1- and β-galactosidase-responsive turn-on fluorescent probe CHO_βgal and applied it to in vitro and ex vivo CSC visualization. CHO_βgal was successfully synthesized by decorating hemicyanine dye with ALDH1A1 and β-galactosidase-responsive substrates. Both ALDH1A1- and β-galactosidase-mediated transformations gave the emissive COOH_OH from CHO_βgal, but neither of the two independent transformations yielded strong fluorescence. In in vitro confocal laser scanning microscopy observations, CHO_βgal clearly and selectively lit up CSCs in the presence of differentiated cancer cells. Thanks to the AND-type dual responsiveness of CHO_βgal, false-positive signals from NSCs were suppressed, which has not been attained with single-input ALDH1A1-responsive probes. As shown in ex vivo imaging of excised lungs, CHO_βgal can effectively avoid false-positive assignments, discriminating metastasis model lungs from normal lungs. The tissue slice staining using CHO_βgal elucidated that active CSCs localize at the peripheral region in cancer tissues and it was able to point out the invasive “leading front” in malignant cancer tissues. Furthermore, CSC-specific visualization in tissue slices was not disturbed by NSCs. To our knowledge, this is the first example of visualization of ALDH1A1-positive CSCs in surgically excised tissue slices containing NSCs. Hence, we believe that the CHO_βgal staining will become not only a reliable and facile diagnostic method for detecting CSC-positive cancer tissues and evaluating cancer malignancy, but also a powerful and promising tool for pursuing the origin of CSCs and cancer plasticity.Author contributions
Koji M. and M. O. conceived and designed the study. Koji M., M. O., K. S., and Koki M. synthesized and characterized the compounds. Koji M., M. O., K. S., and H. M. performed in vitro cell experiments. Koji M. performed theoretical calculation. Koji M., M. O., Y. K., M. I, H. Y., Y. B., Y. U., and Y. M. performed ex vivo metastasis model lung staining. Y. K., M. I, H. Y., and Y. B. performed ex vivo tissue slice staining and H&E staining. K. O. directed the project. The manuscript was written by Koji M. and K. O.Conflicts of interest
There are no conflicts to declare.Acknowledgements
This work was supported by JSPS KAKENHI Grant Numbers 20H02811, 21J15733, 21H00424, 22K19027, and 24H00841. K. M. appreciates the financial support from the Asahi Glass Foundation and Hoansha Foundation. This research was partially supported by the Advanced Research Infrastructure for Materials and Nanotechnology in Japan (ARIM, Nagoya University) of MEXT. The authors wish to acknowledge the Division for Medical Research Engineering, Nagoya University Graduate School of Medicine, for the usage of CM 3050 S and BZ-9000. Computation time was provided by the SuperComputer System, Institute for Chemical Research, Kyoto University. We acknowledged Prof. Hiroshi Nonaka and Ms Mengchu Wang in Kyoto University for flow cytometry analysis.References
- E. Batlle and H. Clevers, Nat. Med., 2017, 23, 1124–1134 Search PubMed.
- B. Beck and C. Blanpain, Nat. Rev. Cancer, 2013, 13, 727–738 Search PubMed.
- Q. Gao, Y. Zhan, L. Sun and W. Zhu, Stem Cell Rev. Rep., 2023, 19, 2141–2154 Search PubMed.
- L. Walcher, A.-K. Kistenmacher, H. Suo, R. Kitte, S. Dluczek, A. Strauβ, A.-R. Blaudszun, T. Yevsa, S. Fricke and U. Kossatz-Boehlert, Front. Immunol., 2020, 11, 1280 Search PubMed.
- N. M. Warrier, N. Kelkar, C. T. Johnson, T. Govindarajan, V. Prabhu and P. Kumar, Eur. J. Cell Biol., 2023, 102, 151321 Search PubMed.
- R. Paul, J. F. Dorsey and Y. Fan, Pharmcol. Ther., 2022, 231, 107985 Search PubMed.
- P. B. Gupta, I. Pastushenko, A. Skibinski, C. Blanpain and C. Kuperwasser, Cell Stem Cell, 2019, 24, 65–78 Search PubMed.
- C. E. Meacham and S. J. Morrison, Nature, 2013, 501, 328–337 Search PubMed.
- M. L. De Angelis, F. Francescangeli, F. La Torre and A. Zeuner, Front. Oncol., 2019, 9, 626 Search PubMed.
- G. L. Ismaeel, A. H. Abdul-Hussein, H. M. Qasim, N. K. Abed, A. T. Jalil, A. A. Suleiman and S. H. Dilfy, Gene Rep., 2023, 30, 101717 Search PubMed.
- J. Han, M. Won, J. H. Kim, E. Jung, K. Min, P. Jangili and J. S. Kim, Chem. Soc. Rev., 2020, 49, 7856–7878 Search PubMed.
- H. Jariyal, C. Gupta, V. S. Bhat, J. R. Wagh and A. Srivastava, Stem Cell Rev. Rep., 2019, 15, 755–773 Search PubMed.
- X. Xu, S. Chai, P. Wang, C. Zhang, Y. Yang, Y. Yang and K. Wang, Cancer Lett., 2015, 369, 50–57 Search PubMed.
- H. Tomita, K. Tanaka, T. Tanaka and A. Hara, Oncotarget, 2016, 7, 11018–11032 Search PubMed.
- R. W. Storms, A. P. Trujillo, J. B. Springer, L. Shah, O. M. Colvin, S. M. Ludeman and C. Smith, Proc. Natl. Acad. Sci. U. S. A., 1999, 96, 9118–9123 Search PubMed.
- I. Min, H. Wang, R. C. Mease, Y. Byun, X. Yang, J. Wang, S. D. Leach and M. G. Pomper, Nat. Commun., 2014, 5, 3662 Search PubMed.
- A. Yagishita, T. Ueno, H. Esumi, H. Saya, K. Kaneko, K. Tsuchihara and Y. Urano, Bioconjugate Chem., 2017, 28, 302–306 Search PubMed.
- A. Yagishita, T. Ueno, K. Tsuchihara and Y. Urano, Bioconjugate Chem., 2021, 32, 234–238 Search PubMed.
- S. Maity, C. M. Sadlowski, J.-M. G. Lin, C.-H. Chen, L.-H. Peng, E.-S. Lee, G. K. Vegesna, C. Lee, S. H. Kim, D. Mochly-Rosen, S. Kumar and N. Murthy, Chem. Sci., 2017, 8, 7143–7151 Search PubMed.
- C. Anorma, J. Hedhli, T. E. Bearrood, N. W. Pino, S. H. Gardner, H. Inaba, P. Zhang, Y. Li, D. Feng, S. E. Dibrell, K. A. Kilian, L. W. Dobrucki, T. M. Fan and J. Chan, ACS Cent. Sci., 2018, 4, 1045–1055 Search PubMed.
- T. E. Bearrood, G. Aguirre-Figueroa and J. Chan, Bioconjugate Chem., 2020, 31, 224–228 Search PubMed.
- M. Oe, K. Miki, Y. Ueda, Y. Mori, A. Okamoto, Y. Funakoshi, H. Minami and K. Ohe, ACS Sens., 2021, 6, 3320–3329 Search PubMed.
- M. Oe, K. Suzuki, K. Miki, H. Mu and K. Ohe, ChemPlusChem, 2022, 87, e202200319 Search PubMed.
- S. Li, W. Tang and X. Duan, New J. Chem., 2023, 47, 545–549 Search PubMed.
- I. Ma and A. L. Allan, Stem Cell Rev. Rep., 2011, 7, 292–306 Search PubMed.
- G. Vassalli, Stem Cell Inter., 2019, 2019, 3904645 Search PubMed.
- F. Rossi, H. Noren, R. Jove, V. Beljanski and K.-H. Grinnemo, Stem Cell Res. Ther., 2020, 11, 489 Search PubMed.
- J. L. Kolanowski, F. Liu and E. J. New, Chem. Soc. Rev., 2018, 47, 195–208 Search PubMed.
- D. H. Juers, B. W. Matthews and R. E. Huber, Protein Sci., 2012, 21, 1792–1807 Search PubMed.
- A. Tsherniak, F. Vazquez, P. G. Montgomery, B. A. Weir, G. Kryukov, G. S. Cowley, S. Gill, W. F. Harrington, S. Pantel, J. M. Krill-Burger, R. M. Meyers, L. Ali, A. Goodale, Y. Lee, G. Jiang, J. Hsiao, W. F. J. Gerath, S. Howell, E. Merkel, M. Ghandi, L. A. Garraway, D. E. Root, T. R. Golub, J. S. Boehm and W. C. Hahn, Cell, 2017, 170, 564–576 Search PubMed.
- J. Zhang, P. Cheng and K. Pu, Bioconjugate Chem., 2019, 30, 2089–2101 Search PubMed.
- Y. Yao, Y. Zhang, C. Yan, W.-H. Zhu and Z. Guo, Chem. Sci., 2021, 12, 9885–9894 Search PubMed.
- M. Li, M. Yang and W.-H. Zhu, Mater. Chem. Front., 2021, 5, 763–774 Search PubMed.
- L. Li, F. Jia, Y. Li and Y. Peng, RSC Adv., 2024, 14, 3010–3023 Search PubMed.
- H. Li, H. Kim, F. Xu, J. Han, Q. Yao, J. Wang, K. Pu, X. Peng and J. Yoon, Chem. Soc. Rev., 2022, 51, 1795–1835 Search PubMed.
- Z. Zeng, S. S. Liew, X. Wei and K. Pu, Angew. Chem., Int. Ed., 2021, 60, 26454–26475 Search PubMed.
- M. Oe, K. Miki and K. Ohe, Org. Biomol. Chem., 2020, 18, 8620–8624 Search PubMed.
- M. Oe, K. Miki, A. Masuda, K. Nogita and K. Ohe, Chem. Commun., 2022, 58, 1510–1513 Search PubMed.
- X.-P. Fam, J. Huang, T.-B. Ren, L. Yuan and X.-B. Zhang, Anal. Chem., 2023, 95, 1566–1573 Search PubMed.
- Z. Li, P.-Z. Liang, L. Xu, S.-S. Zhang, K. Li, Q. Wu, S.-F. Lou, T.-B. Ren, L. Yuan and S.-B. Zhang, Nat. Commun., 2023, 14, 1843 Search PubMed.
- Y. Hoshino, J. Nishida, Y. Katsuno, D. Koinuma, T. Aoki, N. Kokudo, K. Miyazono and S. Ehata, Am. J. Pathol., 2015, 185, 1457–1470 Search PubMed.
- Y. Katsuno, S. Ehata, M. Yashiro, K. Yanagihara, K. Hirakawa and K. Miyazono, J. Pathol., 2012, 228, 391–404 Search PubMed.
- B. Rotman, J. A. Zderic and M. Edelstein, Proc. Natl. Acad. Sci. U. S. A., 1963, 50, 1–6 Search PubMed.
- F. Debacq-Chainiaux, J. D. Erusalimsky, J. Campisi and O. Toussaint, Nat. Protoc., 2009, 4, 1798–1806 Search PubMed.
- N. Kitamura, T. Iwamura, S. Taniguchi, H. Yamanari, M. A. Kawano, K. Hollingworth and T. Setoguchi, Clin. Exp. Metastasis, 2001, 18, 561–571 Search PubMed.
- Q. Li, Z. Guo, G. Li, Y. Zhang, X. Liu, B. Li, J. Wang and X. Li, Cancer Cell Int., 2023, 23, 305 Search PubMed.
- J.-s Wu, J. Jiang, B.-j Chen, K. Wang, Y.-l Tang and X.-h Liang, Transl. Oncol., 2021, 14, 100899 Search PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d4tb00897a |
| This journal is © The Royal Society of Chemistry 2024 |