Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Recombinant silk protein condensates show widely different properties depending on the sample background†
Jennifer
Tersteegen
 a,
Isabell
Tunn
a,
Isabell
Tunn
 a,
Ma
Sand
a,
Ma
Sand
 a,
Teemu
Välisalmi
a,
Teemu
Välisalmi
 a,
Maaria
Malkamäki
a,
Maaria
Malkamäki
 a,
Julie-Anne
Gandier
a,
Julie-Anne
Gandier
 ab,
Grégory
Beaune
ab,
Grégory
Beaune
 c,
Alba
Sanz-Velasco
c,
Alba
Sanz-Velasco
 a,
Eduardo
Anaya-Plaza
a,
Eduardo
Anaya-Plaza
 a and
Markus B.
Linder
a and
Markus B.
Linder
 *a
*a
aDepartment of Bioproducts and Biosystems, Aalto University School of Chemical Engineering, Espoo, Aalto FI-00076, Finland. E-mail: markus.linder@aalto.fi
bHäme University of Applied Sciences HAMK, Hämeenlinna, HAMK FI-13101, Finland
cDepartment of Applied Physics, Aalto University School of Science, Espoo, Aalto FI-00076, Finland
First published on 16th October 2024
Abstract
There is an increasing understanding that condensation is a crucial intermediate step in the assembly of biological materials and for a multitude of cellular processes. To apply and to understand these mechanisms, in vitro biophysical characterisation techniques are central. The formation and biophysical properties of protein condensates depend on a multitude of factors, such as protein concentration, pH, temperature, salt concentration, and presence of other biomolecules as well as protein purification and storage conditions. Here we show how critical the procedures for preparing protein samples for in vitro studies are. We compare two purification methods of the recombinant spider silk protein CBM-AQ12-CBM and study the effect of background molecules, such as DNA, on the formation and properties of the condensates. We characterize the condensates using aggregation induced emitters (AIEs), coalescence studies, and micropipette aspiration. The condensated sample containing background molecules exhibit a lower threshold concentration for condensate formation accompanied by a lower surface tension and longer coalescence time when compared to the pure protein condensates. Furthermore, the partitioning of small AIEs is enhanced in the presence of background molecules. Our results highlight that the purification method and remaining background molecules strongly affect the biophysical properties of spider silk condensates. Using the acquired knowledge about spider silk protein purification we derive guidelines for reproducible condensate formation that will foster the use of spider silk proteins as adhesives or carriers for biomedical applications.
1. Introduction
The need for sustainable and environmentally friendly biomaterials is growing. Protein-based materials are a promising solution to address this need.1,2 At the same time, the role of condensates in biological systems has gained increasing attention since the discovery of membrane-less organelles.3,4 Especially in the field of biological materials, studies on biomolecular condensates have enhanced the understanding of underlying molecular mechanisms.5,6 Among others, condensates can be observed in the formation of mussel byssus threads and the formation of the squid beak.6,7 Condensation, also called coacervation, is also proposed to be an important step in the assembly of fibers from spider silk protein. Already inside the silk glands of spiders, it is assumed that condensation occurs due to increased protein concentration and a change in the pH and salt conditions.8 Likewise, the artificial production of spider silk fibers from recombinantly produced protein has been linked to condensation, which likely needs to occur before fiber pulling.9,10 Recombinant spider silk proteins have been largely studied in the context of materials and biomedical applications.11–14 Condensates are a crucial building block in the assembly process of these artificial biomaterials.9,15,16 This highlights the requirement for reproducible and large-scale protein condensation in the production process of protein-based materials. However, while providing a viable approach to the manufacturing of sustainable and biocompatible materials, the use of biomolecular condensates is challenging.Biomolecular condensates are supramolecular assemblies that result from liquid–liquid phase separation (LLPS). This creates a dense condensate phase, containing a high biomolecule concentration. The surrounding dilute phase contains a low amount of the biomolecule. Weak interactions between the proteins are the driving force for condensation.17,18 Typically, condensates behave in a liquid-like manner, which can be characterized by their tendency to coalesce. Proteins that undergo self-condensation often contain highly repetitive, intrinsically disordered regions (IDRs).19,20 For spider silk proteins in condensates, it has been shown that their secondary structure is rich in α-helices which can transform into β-sheets when fibers are pulled from the solution.9,21 Furthermore, condensates provide a confined space e.g., for adhesives in the mussel's byssus threads.22 At the same time, condensates show an exceptionally low surface tension which seems to be beneficial for substrate infiltration.23,24
Condensates are metastable and known to be highly influenced by sample history and preparation.25,26 For example, pH, salt conditions, and temperature affect the concentration at which LLPS occurs.25,27,28 Additionally, salts (e.g. Na2HPO4) affect the state of condensates by transforming previous liquid-like condensates, that can coalescence, into solid-like condensates.9,27 The significance of these aspects becomes especially evident when comparing biomolecular condensates in vivo and in vitro. Naturally, condensates are influenced by a vast number of other components, that are present inside cells.29,30 For example, RNA and DNA are known to be able to form complex condensates with certain proteins.31,32 By varying RNA and protein ratios, hollow, vesicle-like condensates can be generated.33 However, it is challenging to recreate these conditions when utilizing protein condensates for materials applications. In these cases, starting from a more simplified, pure system followed by a step by step understanding of the influence of specific background molecules is desirable. RNA and DNA are also common background molecules during recombinant protein production.34,35 The purification process of proteins can influence all these variables. By alternating the purification process, different purities and backgrounds can be achieved. However, the use of condensates in large-scale production of biomaterials requires careful balancing of factors such as protein purity, yield, and production costs, as well as factors influencing the biophysical properties of condensates.
This work sheds light on the importance of the purification protocol, and the presence of background molecules for the reproducible condensation of the recombinant spider silk fusion protein CBM-AQ12-CBM. The pure silk protein allows us to engineer a more simplified in vitro model system. The presence of the background molecules in the silk protein resembles crowded environments, similar to those observed in vivo. We qualitatively analyze the effect of background molecules on the condensation of spider silk proteins by concentrating experiments and light microscopy. Utilizing coalescence studies and micropipette aspiration of condensates, we demonstrate that spider silk condensates exhibit different biophysical properties, depending on the presence of background molecules in the protein solution. Background molecules such as DNA and RNA reduce the concentration at which LLPS occurs by 75%. Silk protein condensates that are formed in the presence of background molecules frequently show bursting behavior and have a reduced surface tension, indicating the presence of a shell-like assembly at the condensate's surface. In contrast, we did not observe bursting in the simplified model system of pure silk protein condensates, which exhibit a higher surface tension, indicating the absence of shell-like structures. Furthermore, in the absence of background molecules the silk protein condensates are more selective to the uptake of small molecules such as aggregation induced emitters (AIEs). This work highlights the importance of sample history, purification, and preparation when characterizing the condensation properties of proteins. Our example focuses on silk protein assembly, but similar mechanisms are likely to apply to IDR containing proteins more generally.
2. Results & discussion
Protocol for reproducible silk protein production and condensation
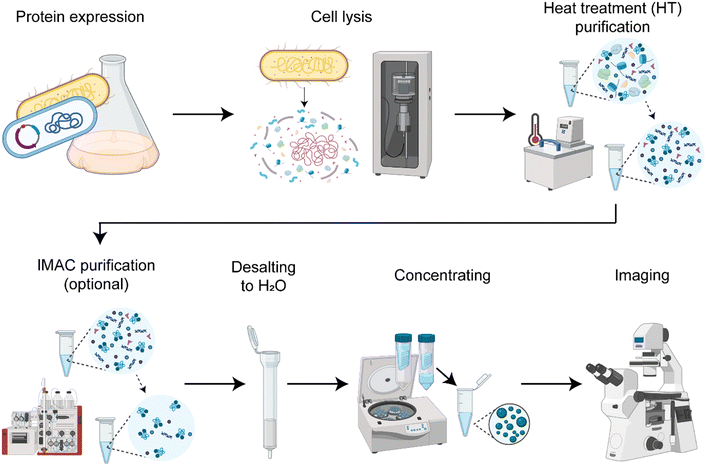
We established a protocol to produce the spider silk protein CBM-AQ12-CBM (Fig. S1, ESI†) with reproducible concentration and properties (Fig. 1). Consistent production results enable controlled condensation. In short, the protocol involves recombinant protein production in E. coli, cell lysis by sonication (Fig. S2, ESI†), protein purification and desalting to deionized water, and a final concentrating step with centrifugal concentrators. Depending on the purification method used, we either have background molecules present imitating a crowded in vivo environment or create a simplified in vitro model system. The purification method developed here always involves a heat treatment for 30 min at 70 °C. The silk protein CBM-AQ12-CBM is stable at this temperature and does not denature. Thus, heat treatment is a fast and easy purification step, especially for large amounts of silk protein. However, after heat treatment, background molecules, such as DNA, RNA and other heat stable proteins are present in the protein solution. After heat treatment and desalting additional His-tag immobilized metal affinity chromatography (IMAC) can be carried out to receive a pure spider silk protein solution.Quantification of background molecules in silk protein solution
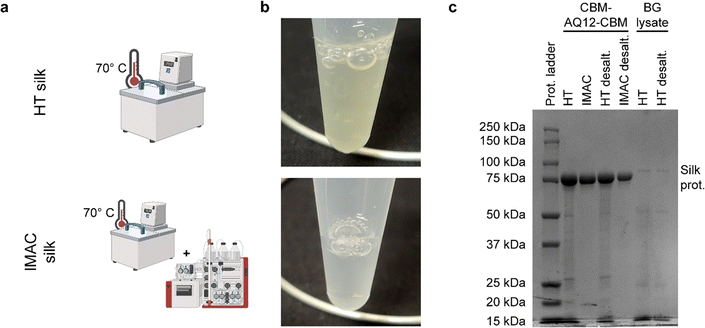
Heat treatment (HT) purification for 30 min at 70 °C results in a slightly yellowish-colored heat treated protein solution (called HT silk), while HT followed by IMAC purification leads to a clear, transparent protein solution (called IMAC silk) (Fig. 2(a) and (b)). Sodium dodecyl sulfate–polyacrylamide gel (SDS–PAGE) of purified protein samples before and after desalting shows a strong band around 85 kDa corresponding to the CBM-AQ12-CBM spider silk protein (Fig. 2(c)). Several faint bands indicating the presence of smaller proteins are visible in the HT sample and are not removed by desalting. In contrast, no additional bands are visible after IMAC. Furthermore, a control, free from silk protein, was prepared to obtain the components in the purification background (called BG lysate). This sample also shows multiple faint bands, indicating the presence of a variety of proteins.The type of background molecules, as well as the yield of silk proteins, were further investigated by amino acid analysis and DNA/RNA extraction. Amino acid analysis of HT silk, IMAC silk, and BG lysate was performed to determine the concentration of CBM-AQ12-CBM and to estimate the total protein amount, including other heat stable proteins. For this, the concentration of CBM-AQ12-CBM was calculated based on the protein sequence and the number of amino acids measured during amino acid analysis. This value was compared to the total amount of amino acids to determine the amount of protein in the BG lysate. However, due to the degradation of specific amino acids (e.g., cysteine and methionine) during the hydrolysis process required for amino acid analysis, this is only an estimation. A whole genome extraction of the differently purified protein samples was carried out with and without RNase to estimate the amount of RNA and DNA present in the samples.
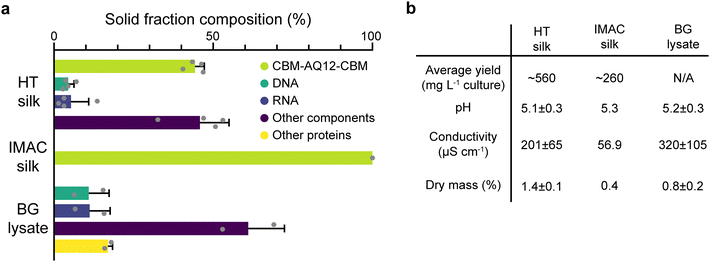
HT silk shows the largest solid fraction with a dry mass of 1.4 ± 0.1% (mean ± standard error of the mean, n = 4), while IMAC silk has a dry mass of 0.4%. The dry mass of BG lysate is 0.8 ± 0.2% (n = 2) (Fig. 3(b)). HT silk contains a significant number of various background molecules which include DNA, RNA, and other components (Fig. 3(a)). Other components might be, e.g., sugars. The fraction of these background molecules is estimated to be around 4.5 ± 1.8% of DNA, 5.3 ± 5.6% of RNA, and 45.8 ± 9.2% of other components, respectively (n = 4). It was not possible to identify other heat stable proteins in the HT silk based on the composition of amino acids given by the amino acid analysis. However, SDS–PAGE gives an estimation of their size and relative amount (Fig. 2(c)). This becomes more evident from the solid fraction composition of the BG lysate which consists of around 10.9 ± 6.4% DNA, 11.2 ± 6.4% RNA, 16.9 ± 1.4% other heat stable proteins, and 61 ± 11.4% other components (n = 2). No background components of any kind were detected in the IMAC silk. In addition to the presence of background molecules, HT silk and IMAC silk differ in their average yield of silk protein with 560 and 260 mg L−1 of culture, respectively. The pH shows no significant variation between the different samples and is around 5.1–5.3 after the final desalting step. The conductivity is highest in the BG lysate with 320 ± 105 μS cm−1 (n = 2) and lowest in IMAC silk with 56.9 μS cm−1 (n = 1). The conductivity of HT silk is around 201 ± 65 μS cm−1 (n = 4).
When comparing the dry mass composition of HT silk and the IMAC silk it becomes clear that there is a multitude of various background molecules present in HT silk, such as DNA and RNA. Both are known to affect protein condensation and might also interact with the spider silk protein. The composition of the background becomes clearer from the dry mass composition of the BG lysate which essentially shows the background molecule composition without the spider silk protein. The complex composition of the background hinders the prediction of possible interactions between the background molecules and the silk protein. In contrast, the absence of detectable background molecules in IMAC silk eases the understanding of molecular interaction between silk proteins without the influence of other molecules and thus creates a simplified model system. Reducing the complexity of the system, batch-to-batch variation and unwanted molecular interactions enables reproducible protein condensation.
Background molecules lower the critical concentration for LLPS of silk proteins
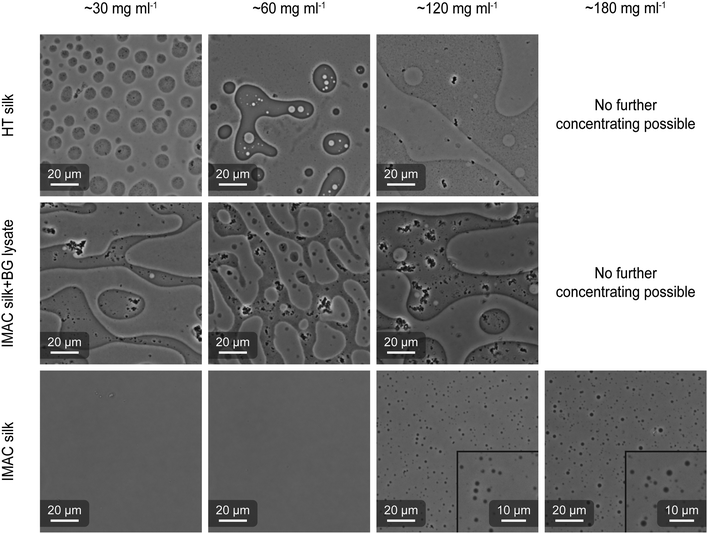
HT silk and IMAC silk were prepared as described above (see the methods section for details). A third sample was prepared by mixing IMAC silk with BG lysate (called IMAC silk + BG lysate). Each sample was concentrated in centrifugal concentrators (30 kDa cutoff) containing 6 ml of 2 mg ml−1 protein solution. The concentrating progress was regularly monitored. When a desired final concentration (final volume) was reached, the concentrated protein solution was collected and imaged using optical microscopy (Fig. 4). For the HT silk and IMAC silk + BG lysate, LLPS occurred at a protein concentration of 30 mg ml−1. When further concentrating these samples to 60 and 120 mg ml−1, the coexistence of condensates in dilute phase (normal phase) and regions with dense phase volume fractions higher than 50% (inverted phase) was observed in the same sample. It was not possible to concentrate these samples to 180 mg ml−1. The IMAC silk showed no presence of condensates at protein concentrations of 30 and 60 mg ml−1, but at 120 mg ml−1. In contrast to the first two samples, IMAC silk was possible to be concentrated to 180 mg ml−1, resulting in LLPS. Thus, the presence of background molecules reduces the critical concentration for LLPS by 75%. In all samples coalescence of condensates was observed, indicating liquid-like properties. Upon dilution of condensated samples we observe that condensates disappear, indicating that condensate formation is reversible. The presence of an inverted phase was identified by the partition of silk protein precipitates, which form due to the metastability of the system, into the dense phase, as well as by adding free green fluorescent protein (eGFP) which partitions into the dilute phase (Fig. S3, ESI†).Scanning electron microscopy (SEM) shows filamentous structures originating from silk proteins
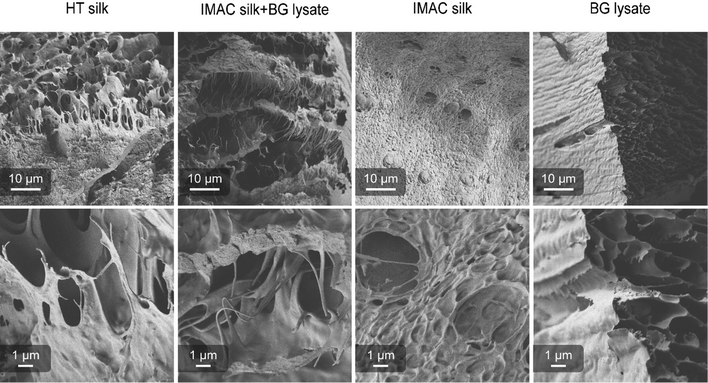
Differences in the supramolecular interactions of the silk protein in condensated samples and additional interactions with the background molecules were investigated using SEM. Concentrated samples of HT silk, IMAC silk + BG lysate, IMAC silk and only BG lysate were vitrified with liquid ethane (−180 °C) and freeze dried. All samples, except BG lysate, contained condensates. Freeze dried samples were cracked, transferred to carbon tape coated stubs and sputter coated with 6 nm Au/Pd. The consistency of the samples after freeze-drying varied between the different samples. Samples containing silk protein required a force to break into pieces, while the BG lysate behaved like a powder. Samples were imaged with the Sigma VP scanning electron microscope (Zeiss) using a SE2 detector (Fig. 5). The BG lysate sample shows a spongy structure that can arise during sample preparation. However, in contrast to the BG lysate, all samples containing silk protein show thin filament-like structures. These are most prominent in HT silk and IMAC silk + BG lysate. Additionally, a layered structure is very prominent in IMAC silk + BG lysate. The filamentous structures are observed to stretch between the layers. Structures that might correlate to condensates were only observed in the IMAC silk. The filamentous structures likely originate from the silk protein and might be the reason for the stronger material properties that prevent the freeze-dried samples from breaking down into a powder. Similar layered structures, as observed especially in the IMAC silk + BG lysate, were previously described for hydrogels of the silk protein eADF4.36 However, in this case the effect of cross-linking was investigated and the layered structure was assumed to be a result of the chemical sample fixation which differs from the vitrification done in this study.Background molecules affect the uptake of AIEs by silk condensates
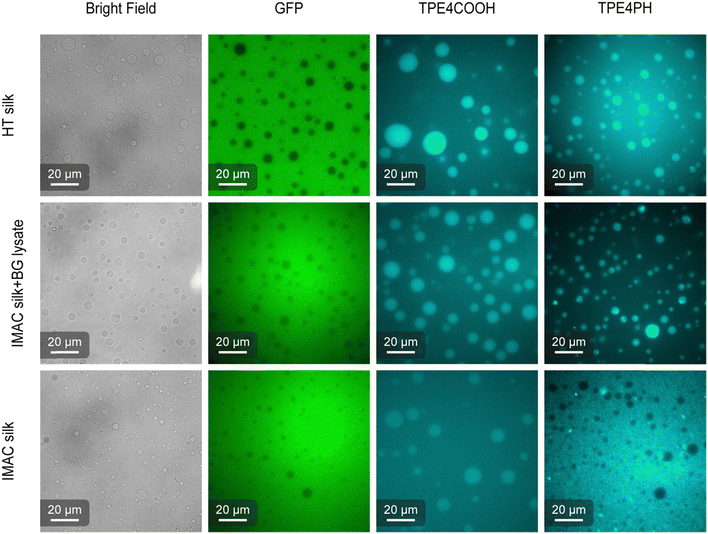
AIEs are small organic molecules that show increased photoemission in an aggregated state, triggered by the restriction of molecular vibrations and rotations in the dense environment of the condensates.37,38 The partition of two different AIEs, 4,4′,4′′,4′′′-(ethene-1,1,2,2-tetrayl)tetrabenzoic acid (TPE4COOH) and 4,4′,4′′,4′′′-(ethene-1,1,2,2-tetrayltetrakis(benzene-4,1-diyl))tetrakis(1-hexylpyridin-1-ium) bromine (TPE4PH) (Fig. S4, ESI†), was studied to investigate differences in the biophysical properties of the condensates (Fig. 6).39 Free eGFP was used to verify the identity of the dilute phase in the sample. TPE4COOH partitions preferably into the condensates for all three types of condensates. However, fluorescence is still detectable also in the dilute phase. The AIE fluorescence emission contrast between the dense and dilute phase is higher for the HT silk and the IMAC silk + BG lysate compared to the IMAC silk. TPE4PH only partitions into the condensates of samples that contain background molecules i.e., HT silk and IMAC silk + BG lysate. TPE4PH stays in the dilute phase for IMAC silk. Thus, the selective uptake of small molecules, such as AIEs, by the spider silk condensates studied here depends on the presence of background molecules. In the case of TPE4COOH the presence of these molecules increases partitioning into the condensate. The partitioning effect is even stronger for TPE4PH which only enters into the condensates in the presence of background molecules. The increased uptake of other molecules into condensates as well as the decrease of the condensation threshold have been previously described for crowding agents.40 Therefore, the background molecules present in the HT silk and the IMAC silk + BG lysate might act as crowding agents. We further discuss this hypothesis in the last paragraph of this section.Micropipette aspiration shows that the presence of background molecules affects the surface tension of condensates
Micropipette aspiration was used to determine the biophysical properties of the IMAC silk condensates. The biophysical properties of the HT silk condensates were determined using micropipette aspiration as described before.24 The IMAC silk condensates were aspirated with 20 Pa applied pressure ΔP with a micrometer-sized glass pipette (Fig. 7(a) and Video S1, ESI†) followed by the retraction of the tongue when ΔP = 0 Pa (Fig. 7(b) and Video S1, ESI†). The aspiration and retraction length were measured (Fig. 7(c)) and plotted against time, to obtain the aspiration rate![[L with combining dot above]](https://www.rsc.org/images/entities/i_char_004c_0307.gif) a and retraction rate
a and retraction rate ![[L with combining dot above]](https://www.rsc.org/images/entities/i_char_004c_0307.gif) r from the slope of the corresponding curves (Fig. 7(d) and (e)). The aspiration and retraction rates were used to calculate the bulk viscosity ηb and the critical pressure ΔPc of aspiration (see eqn (2) and (4) in the methods section). The bulk viscosity ηb of the IMAC condensates was 5.4 ± 0.7 Pa s (mean ± standard error of the mean, n = 9) (Fig. 7(f) and Table S1, ESI†), while the effective bulk viscosity ηeff (eqn (5)) of the HT silk condensates was only 2.2 ± 0.4 Pa s (n = 17) (original data published previously in Tunn et al.24). The surface tension of the IMAC silk condensates was obtained from the Young Laplace law (eqn (1)). The mean surface tension γ of the IMAC silk condensates was 52.2 ± 3.2 μN m−1 (n = 9). The surface tension of the HT silk condensates was more than two times lower (19.1 ± 2.2 μN m−1 (n = 17)) than the surface tension of the IMAC silk condensates.24 Interestingly, some of the HT silk condensates burst upon aspiration. Therefore, we assume that the HT silk condensates form a shell-like assembly at the interface between the dense and dilute phase with a surface viscosity ηs of 53.2 ± 9.3 μN s m−1 (n = 17).24 We did not observe any bursting of the IMAC silk condensates during micropipette aspiration or under any other experimental conditions tested. Therefore, we assume that the IMAC silk condensates do not have a shell-like assembly and report the viscosity of the bulk condensate ηb. The reduced surface tension of HT silk condensates can be explained by the assembly of background molecules on the interface. In this case the background molecules would act as a surfactant. Furthermore, the assembly on the interface could explain the bursting of condensates observed with HT silk.
r from the slope of the corresponding curves (Fig. 7(d) and (e)). The aspiration and retraction rates were used to calculate the bulk viscosity ηb and the critical pressure ΔPc of aspiration (see eqn (2) and (4) in the methods section). The bulk viscosity ηb of the IMAC condensates was 5.4 ± 0.7 Pa s (mean ± standard error of the mean, n = 9) (Fig. 7(f) and Table S1, ESI†), while the effective bulk viscosity ηeff (eqn (5)) of the HT silk condensates was only 2.2 ± 0.4 Pa s (n = 17) (original data published previously in Tunn et al.24). The surface tension of the IMAC silk condensates was obtained from the Young Laplace law (eqn (1)). The mean surface tension γ of the IMAC silk condensates was 52.2 ± 3.2 μN m−1 (n = 9). The surface tension of the HT silk condensates was more than two times lower (19.1 ± 2.2 μN m−1 (n = 17)) than the surface tension of the IMAC silk condensates.24 Interestingly, some of the HT silk condensates burst upon aspiration. Therefore, we assume that the HT silk condensates form a shell-like assembly at the interface between the dense and dilute phase with a surface viscosity ηs of 53.2 ± 9.3 μN s m−1 (n = 17).24 We did not observe any bursting of the IMAC silk condensates during micropipette aspiration or under any other experimental conditions tested. Therefore, we assume that the IMAC silk condensates do not have a shell-like assembly and report the viscosity of the bulk condensate ηb. The reduced surface tension of HT silk condensates can be explained by the assembly of background molecules on the interface. In this case the background molecules would act as a surfactant. Furthermore, the assembly on the interface could explain the bursting of condensates observed with HT silk.
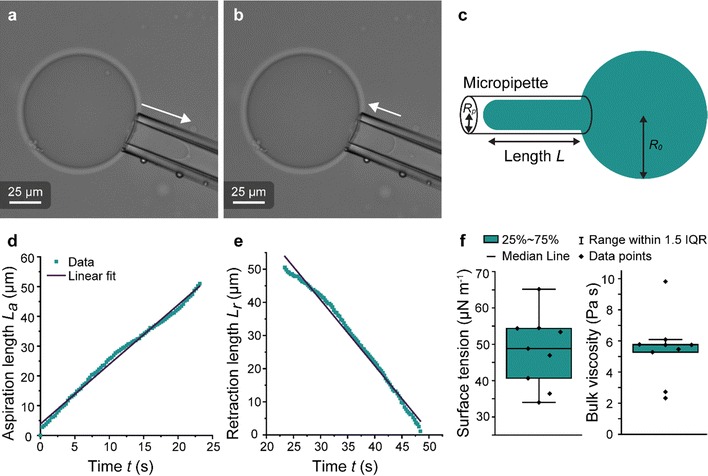 | ||
Fig. 7 Micropipette aspiration of IMAC silk condensates. (a) Aspiration of a condensate. Aspiration direction marked with the arrow. (b) Retraction of the tongue at ΔP = 0 Pa. Arrow points at the retraction direction. (c) Scheme of the micropipette aspiration with pipette radius Rp, the condensate radius R0 and the length L of the aspirated or retracting condensate, respectively. (d) Aspiration curve with linear fit La = ![[L with combining dot above]](https://www.rsc.org/images/entities/i_char_004c_0307.gif) at + n of a condensate. (e) Retraction curve with linear fit: Lr = at + n of a condensate. (e) Retraction curve with linear fit: Lr = ![[L with combining dot above]](https://www.rsc.org/images/entities/i_char_004c_0307.gif) rt + n′ of a condensate. (f) Box plots of the surface tension (obtained from eqn (1)) and the bulk viscosity (obtained from eqn (2)). IQR = inter quartile range. Number of datasets: 9. rt + n′ of a condensate. (f) Box plots of the surface tension (obtained from eqn (1)) and the bulk viscosity (obtained from eqn (2)). IQR = inter quartile range. Number of datasets: 9. | ||
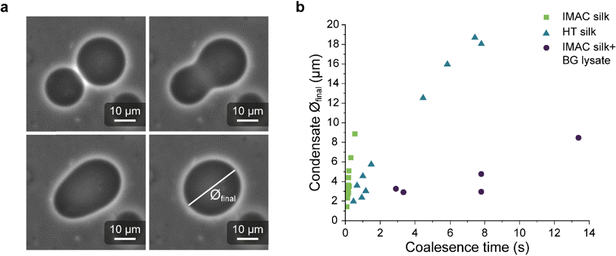
The coalescence time of condensates is affected by background molecules
Light microscopy videos were acquired to study the coalescence of IMAC silk condensates, HT silk condensates and IMAC silk + BG lysate condensates (Videos S2–S4, ESI†). The final condensate diameter after the coalescence and the total time of the coalescence event were determined with ZEN microscopy software (Zeiss) (Fig. 8(a)). The final condensate diameter was plotted against the coalescence time (Fig. 8(b)). Observed IMAC silk condensates and IMAC silk + BG lysate condensates were generally smaller than those from HT silk. The coalescence time increased with the condensate size in all samples. Coalescence of the observed IMAC silk condensates is fastest with less than 1 s. Coalescence of the observed HT silk condensates was slower compared to IMAC silk condensates with a similar size range. In contrast, IMAC silk + BG lysate condensates show a much slower coalescence time of up to 14 s for condensates in the same size range.Various aspects can explain the increased coalescence time in the presence of background molecules. One possibility is the higher total amount of macromolecules in samples with background such as HT silk and IMAC silk + BG lysate that can lead to increased viscosity of the dilute phase. The change in the aspect ratio over time was fitted using an exponential decay function (see details in the methods section) to obtain the characteristic relaxation time scale of the coalescence of the condensates (Fig. S5a–c, ESI†). The characteristic relaxation time was then plotted against the characteristic length to obtain the inverse capillary velocity (ICV) of the condensates (Fig. S5d, ESI†). The ICV was 0.01 s μm−1 for IMAC silk (n = 11), 0.07 s μm−1 for HT silk (n = 10) and 0.31 s μm−1 for IMAC silk+ BG lysate (n = 5). Using the surface tension values obtained from micropipette aspiration (HT silk and IMAC silk) the bulk viscosity ηb,c of the condensates can be determined from the ICV. The ηb,c of the IMAC silk was 0.5 Pa s and 1.4 Pa s for the HT silk condensates. These bulk viscosities are 2–10× lower than the bulk viscosities obtained from micropipette aspiration with 5.4 Pa s for IMAC silk and 2.2 Pa s for HT silk condensates. The difference of the viscosity obtained from the aspiration data from that of the coalescence data may be due to viscous dissipation, the geometry of the micropipette setup and the ICV analysis, which does not take the viscosity of the dilute phase into account.
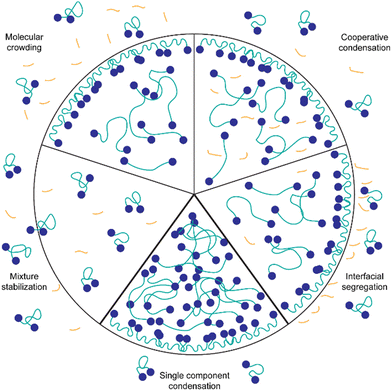
Possible interaction mechanisms of the background molecules with the silk-protein
The presented results indicate that the background molecules have a strong effect on the protein concentration required for condensation, the partitioning of dye molecules and the biophysical properties of the spider silk condensates. However, through Fourier-transform-infrared spectroscopy (FTIR) of IMAC silk and HT silk it is not possible to detect a change in protein secondary structure (Fig. S6, ESI†). Fig. 9 illustrates possible mechanisms of the interaction of background molecules with condensate forming proteins in general. Probable mechanisms relevant for the system studied here are discussed in the following paragraphs.Background molecules can affect protein condensation in various ways. One common phenomenon is molecular crowding. Crowding agents such as dextran and polyethylene glycol (PEG) are known to induce LLPS by increasing the effective concentration of the protein.41 This results in LLPS occurring at lower protein concentrations. It is possible that some components of the background molecules act as crowding agents. Evidence pointing towards the crowding hypothesis is that uptake of TPE4PH into the HT silk and the IMAC silk + BG lysate condensates is enhanced compared to IMAC silk condensates where it is excluded. It has been shown previously that crowding agents can promote the uptake of molecules into condensates.40 However, due to the complex composition it is difficult to predict which molecules present in the background could have a crowding effect.
Another possible interaction between background molecules and condensating proteins is cooperative condensation, i.e. complex coacervation.42 In particular, DNA and RNA are known to form complexes with intracellular proteins that contain RNA or DNA binding domains.33,43–45 Generally, negatively charged RNA or DNA can interact with positive charges of proteins to form complex condensates. The spider silk protein CBM-AQ12-CBM has a mainly neutral AQ12 middle part, and slightly negatively charged CBM terminal domains at pH values higher than the isoelectric point. This indicates a low probability of complex condensation with negatively charged DNA and RNA. However, other weak molecular interactions such as H-bonding and hydrophobic interactions between DNA/RNA and the intrinsically disordered middle part could occur.46 Cooperative effects could also result in a lower concentration needed for LLPS since the number of possible interactions and the concentration of macromolecules participating in the condensation is increased.
Furthermore, the presence of background molecules affects the biophysical properties of the condensates, such as surface tension and viscosity. Micropipette aspiration revealed that the surface tension is two times higher for the IMAC silk condensates compared to the HT silk that contains background molecules. The lower surface tension of the HT silk condensates might contribute to the condensation at lower protein concentrations observed for the HT silk. In the HT silk the background molecules could act in a similar way as surfactant and assemble at the interface of condensates (interfacial segregation). Additionally, if a surface tension reduction takes place by background molecules assembling at the interface, these molecules are likely to interact with the condensating proteins at the surface of the condensate. This could reduce the fluctuations of the proteins at the interface leading to the formation of a thin protein shell. The existence of a thin protein shell might also explain the longer fusion times observed for the HT silk condensates as well as the bursting of the HT silk condensates studied by Tunn et al.24 The formation of condensates with a shell layer has been previously found for several intracellular complex condensates containing proteins and DNA/RNA.38 However, the presence of the background molecules also increases the overall viscosity of the system, which could lead to increased fusion times of the HT silk and the IMAC silk + BG lysate condensates.
Another possible action mechanism of the background molecules is the stabilization of the mixture of these molecules and the proteins. For example, background molecules could screen charges on the surface of proteins and thus inhibit molecular interactions between the proteins. This would reduce the likelihood of LLPS occuring.47 Since we observe condensation of the spider silk protein at lower protein concentrations in the presence of background molecules, mixture stabilization seems unlikely in our system.
We assume that any kind of fragmented DNA or RNA could have a similar effect on the protein condensation as we observe with HT silk and IMAC silk + BG lysate. This is due to our sample preparation involving cell lysis with a lysis buffer that includes DNase, which likely results in HT silk and BG lysate mainly containing fragmented DNA. Additionally, RNases present in the environment will likely cut the remaining RNA in the HT silk and BG lysate into fragments. We further tested this by adding ssDNA from salmon testes (deoxyribonucleic acid sodium salt from salmon testes, Sigma-Aldrich) to the IMAC silk (Fig. S7, ESI†). Sonicated and thus fragmented DNA enhances condensation of IMAC silk while long DNA that was not sonicated shows no effect. However, adding sonicated ssDNA to IMAC silk seems to easily gel the condensates.
In the case of the IMAC silk that does not contain any detectable background molecules, we assume that single component condensation takes place. The IMAC silk condensates formed are simple condensates containing only CBM-AQ12-CBM. The concentration of the IMAC silk needed to form condensates is four times higher than for the HT silk that contains background molecules. The results of viscosity measurements of the IMAC silk varies 10-fold between the coalescence (0.5 Pa s) and micropipette techniques (5.4 Pa s) but less than 2-fold for the HT silk condensates (1.4 and 2.2 Pa s). A lower sample viscosity could explain the fast coalescence of the IMAC silk condensates determined by coalescence analysis. In contrast, a higher viscosity of the IMAC silk could be explained by the overall higher silk protein concentration. The viscosity of the HT silk condensates is influenced by the presence of background molecules.
3. Conclusions
The formation of condensates of recombinant silk proteins was strongly affected by the presence of background co-solutes. Several properties of the condensates—such as surface tension, viscosity, uptake of dyes—were also strongly affected. The presence of background molecules creates a complex environment where individual contributions are difficult to pinpoint. We find several implications of this strong context dependency. One is practical—when producing biosynthetic materials, the assembly steps become difficult to control if there are variations in parameters that are unknown, posing a risk, for example to batch to batch variations. We cannot expect any system to be 100% pure, so the effect of co-solutes will always be present, just the extent of their effect is varied. On the other hand, in natural systems materials such as silks are not a single component, and we should expect that interactions between components affect strongly how materials are formed.48 We should therefore expect that designs for biosynthetic materials could benefit from involving the use of multicomponent systems. Furthermore, silk condensates are a promising material in various biomedical applications. Condensates in general are possible candidates for drug delivery systems, bio-adhesives, and for tissue engineering and repair.49–53 Acquiring a deeper understanding of mechanisms taking place at the condensate interface is crucial for applications. Additionally, silk proteins have high biocompatibility and low cytotoxicity.54–56 Here condensation was achieved more easily with background components, and as we demonstrated earlier proteins assembled in this way can form highly functional materials.9,11 A deeper understanding of the interactions involved will be needed.In our interpretation, the background molecules might act as a molecular crowder, as components partition into the condensates (cooperative condensation) or form an interfacial layer on the surface of the protein condensate (interfacial segregation). Combinations of these are also possible. Further research will focus on determining the identity of the background molecules affecting the condensate formation in the HT sample and their action mechanisms. IMAC silk as a simplified model system will allow investigation of the influence of various macromolecules on LLPS and condensate properties in a controlled way.57 This will enable us to decipher conditions that benefit the condensate properties for material formation.
An interesting question will be to understand how widely our results for silk-IDRs can apply to other types of IDRs.58 The formation of biomolecular condensates through IDRs is of wide interest in biological research.59,60 As a methodology to understand their functions, comparing them in vivo and in vitro is an essential approach.61,62 Not only should we think that background components in in vitro studies affect properties of IDRs, but also that unavoidably the in vivo environments have a strong presence of background.63 The evaluation of this background should be included in any study on the condensation of IDRs.
4. Methods
Protein expression and purification
The silk fusion protein CBM-AQ12-CBM (85 kDa) was used in this study. This protein has a triblock structure, with an AQ12 block flanked on both sides by a cellulose binding module (CBM).9 The AQ12 block is an engineered silk protein sequence made up of 12 repeats derived from the major ampulla gland silk fibroin 3 from Araneus diadematus.64 At the C-terminus of the silk-protein a polyhistidine (6× His) has been added, to facilitate affinity purification. The cloning procedure has been described previously.9 The silk protein was expressed in E. coli BL21 (DE3) (ThermoFisher Scientific) with a pEt-28a (+) (kanR) (Novagen) vector at 30 °C using EnPresso media (EnPresso B 500, EnPresso GmbH) in accordance with the protocol of the manufacturer. Full-baffled plastic flasks were used for the expression (Tunair). 24 h after the induction, the cells were harvested (11![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 949 rcf for 15 min) and resuspended in 50 mM tris/HCl lysis buffer (pH 7.4) containing 100 mM NaCl, 3 mM MgCl2, 1 mg ml−1 lysozyme (EC 3.2.1.17, Merck), 20 μg ml−1 DNase I (EC 3.1.21.1, Merck) and protease inhibitor tablet (1 tablet/50 ml buffer, SIGMAFAST™ Protease Inhibitor Cocktail Tablets, EDTA-Free, Merck). For resuspension, 3 ml of the lysis buffer was added for every gram of the pellet. After 1 h incubation at room temperature (RT) the cells were lysed by sonication (40% amplitude, 3 × 1 min with 2 s pulse time) using a Q500 sonicator (Qsonica) with 1/2” sonicator tip 4406 (Ramcon). The majority of non-target proteins was removed through heat treatment purification (30 min, 70 °C) and centrifugation (2 × 15 min at 3200 rcf, RT).
949 rcf for 15 min) and resuspended in 50 mM tris/HCl lysis buffer (pH 7.4) containing 100 mM NaCl, 3 mM MgCl2, 1 mg ml−1 lysozyme (EC 3.2.1.17, Merck), 20 μg ml−1 DNase I (EC 3.1.21.1, Merck) and protease inhibitor tablet (1 tablet/50 ml buffer, SIGMAFAST™ Protease Inhibitor Cocktail Tablets, EDTA-Free, Merck). For resuspension, 3 ml of the lysis buffer was added for every gram of the pellet. After 1 h incubation at room temperature (RT) the cells were lysed by sonication (40% amplitude, 3 × 1 min with 2 s pulse time) using a Q500 sonicator (Qsonica) with 1/2” sonicator tip 4406 (Ramcon). The majority of non-target proteins was removed through heat treatment purification (30 min, 70 °C) and centrifugation (2 × 15 min at 3200 rcf, RT).
E. coli cells containing an empty pEt-28a (+) (kanR) (Novagen) vector instead of the vector coding for the silk protein, were used to prepare the BG lysate as a control. The same expression and lysis procedures were performed on the control cells.
Part of the HT silk was further purified by His-tag immobilized metal affinity chromatography (IMAC) (ÄKTA-pure, Cytiva) using HisTrap FF columns (Cytiva) and binding buffer (500 mM NaCl, 20 mM Imidazole, pH 7.4) and elution buffer (500 mM NaCl, 500 mM Imidazole, pH 7.4). All silk protein and the control BG lysate were desalted using Econo-Pac 10DG columns (Bio-Rad) into deionized water. The whole volume of one production batch for each sample was mixed in plastic tubes to achieve consistent protein concentration and solution composition. Conductivity (Jenway 4520 conductivity meter) and pH (pH-indicator strips 4.0–7.0, Merck) were measured for all samples. The protein solution was stored at −80 °C after flash freezing with liquid nitrogen.
SDS-PAGE
SDS-PAGE electrophoresis was performed for samples obtained at different stages of the expression and purification. Pre-cast 10% Mini-Protean TGX gels (Bio-Rad) and Precision Plus Protein Standard Dual Color (Bio-Rad) were used. SDS-PAGE was performed at constant 110 V ∼60 min and staining was done with Coomassie brilliant blue R-250. The destaining solution contained 48% acetic acid and 40% ethanol. The gel was imaged with a Gel Doc XR+ Gel Documentation System (Bio-Rad).Amino acid analysis
Amino acid analysis was carried out to determine the concentration of CBM-AQ12-CBM as well as the total amount of protein. The analysis was done as previously described.11,24 In short, samples were hydrolyzed in 6 M HCl (Merck), containing 0.1% phenol (Sigma-Aldrich). L-Norleucine (Sigma-Aldrich) was added to all samples as an internal standard. After acid evaporation, samples were resuspended in a citric acid buffer (Sykam GmbH). The samples were measured with S433 amino acid analyzer (Sykam GmbH) using a 570 nm and 440 nm UV detector. The retention time of individual amino acids was determined with an external standard (Sykam GmbH). The concentration of CBM-AQ12-CBM was quantified with the internal standard and the proteins sequence. Specifically, alanine, glycine, glutamine, and glutamic acid were used for concentration determination, due to these amino acids being most abundant in CBM-AQ12-CBM. The total protein amount was determined by taking all the detected amino acids into account. However, since certain amino acids degrade during hydrolysis, this only provides an estimation of the total protein amount.DNA/RNA quantification
DNA and RNA present as the background in protein solutions and BG lysate were quantified with the Wizard Genomic DNA Purification Kit (Promega). Since the samples were already lysed and purified no additional lysis was performed. Instead, 600 μl of the sample were added to a microcentrifuge tube. For each sample (HT silk, IMAC silk, BG lysate) two tubes were prepared. In one tube of each sample, 3 μl of RNase solution were added and the tubes were inverted 2–5 times. After 15 min incubation at 37 °C, 200 μl of protein precipitation buffer was added to each microcentrifuge tube. Tubes were vortexed for 20 s followed by a 5 min incubation on ice and centrifuging for 3 min at 16![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 rcf. The supernatant was transferred to clean tubes containing 600 μl of RT isopropanol and the tubes were gently inverted a few times. Samples were centrifuged for 2 min at 16
000 rcf. The supernatant was transferred to clean tubes containing 600 μl of RT isopropanol and the tubes were gently inverted a few times. Samples were centrifuged for 2 min at 16![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 rcf and the pellet was washed with RT 70% ethanol. The pellet was air dried for 15 min and samples were rehydrated in 40 μl deionized water overnight at 8 °C. Absorption was measured from 200 nm to 350 nm with an EON microplate reader (Biotek). Absorbance at 260 nm was used to calculate DNA and RNA concentrations. Samples prepared with RNase were used to calculate the amount of DNA while samples without RNase were used to calculate the DNA + RNA amount. To determine the amount of RNA, the amount of DNA was subtracted from the DNA + RNA amount. Samples only containing DNA were calculated with an extinction coefficient of 50, while samples containing a mixture of DNA and RNA were calculated with an extinction coefficient of 45.
000 rcf and the pellet was washed with RT 70% ethanol. The pellet was air dried for 15 min and samples were rehydrated in 40 μl deionized water overnight at 8 °C. Absorption was measured from 200 nm to 350 nm with an EON microplate reader (Biotek). Absorbance at 260 nm was used to calculate DNA and RNA concentrations. Samples prepared with RNase were used to calculate the amount of DNA while samples without RNase were used to calculate the DNA + RNA amount. To determine the amount of RNA, the amount of DNA was subtracted from the DNA + RNA amount. Samples only containing DNA were calculated with an extinction coefficient of 50, while samples containing a mixture of DNA and RNA were calculated with an extinction coefficient of 45.
Dry mass
The dry mass of protein solutions and BG lysate was determined after freeze-drying (Christ Alpha 2–4) and weighing the residual pellet.Concentrating
HT and IMAC CBM-AQ12-CBM, as well as BG lysate, were thawed and centrifuged at RT for 15 min at 3200 rcf. Three samples were prepared, 2 mg ml−1 HT silk, diluted with deionized water, 2 mg ml−1 IMAC silk, diluted with deionized water, and 2 mg ml−1 IMAC silk, diluted with BG lysate (ratio 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1). For each sample, four centrifugal concentrators with a 30 kDa cutoff (Vivaspin, Sartorius) were each filled with 6 ml of the prepared protein solution. Samples were concentrated in RT at 1200 rcf until the desired protein concentrations of 30, 60, 120, and 180 mg ml−1 were achieved for each sample. The remaining volume of the concentrated protein solution was used as an estimate of the protein concentration before collection of the sample from the concentrator tube. The final concentration of the samples was determined based on the weight of the collected solution assuming a density of 1 g cm−3. Concentrated samples were used for further studies on the same day as they were concentrated.
1). For each sample, four centrifugal concentrators with a 30 kDa cutoff (Vivaspin, Sartorius) were each filled with 6 ml of the prepared protein solution. Samples were concentrated in RT at 1200 rcf until the desired protein concentrations of 30, 60, 120, and 180 mg ml−1 were achieved for each sample. The remaining volume of the concentrated protein solution was used as an estimate of the protein concentration before collection of the sample from the concentrator tube. The final concentration of the samples was determined based on the weight of the collected solution assuming a density of 1 g cm−3. Concentrated samples were used for further studies on the same day as they were concentrated.
Imaging to study the effect of protein concentration on LLPS of silk proteins
Samples were imaged with an AxioVert.A1 inverted light microscope (Zeiss) after concentration and collection of the sample. Before pipetting the samples onto the glass slides, the samples were gently mixed with a pipette tip by stirring. 3 μl of sample were imaged between two coverslips. Imaging was done with a 40×/0.6 Ph2 objective and phase 2 contrast ring. The presence of an inverted phase was studied by mixing 4.5 μl of concentrated protein solution with 0.5 μl free eGFP (Filter set 38, Zeiss). Free eGFP partitions into the dilute phase but not into the dense phase.Imaging to study the partition behavior of aggregation induced emitters in silk protein condensates
Condensated samples (4.5 μl) (prepared as previously described) were mixed with either 4,4′,4′′,4′′′-(1,1,2,2-ethenetetrayl)tetrabenzoic acid (TPE4COOH) or 4,4′,4′′,4′′′-(ethene-1,1,2,2-tetrayltetrakis(benzene-4,1-diyl))tetrakis(1-hexylpyridin-1-ium) bromine (TPE4PH) (0.5 μl) so that the final AIE concentration was 55 μM or 40 μM, respectively. The presence of the correct phase was verified by the addition of free eGFP. Images were taken with a Nikon Eclipse Ti inverted microscope (Nikon), ORCA-flash 4.0 LT digital camera (Hamamatsu, Japan), 60×/1.4 oil immersion objective lens, 1.5× tube lens and micromanager software (version 2.0.0). Bright field images were taken for all samples. Samples containing free eGFP were imaged with 5% 470 nm light (LDI Laser Diode Illuminator, 89 North) and emission light was collected between 485–535 nm. Samples containing TPE4COOH were excited with 50% 405 nm light (LDI Laser Diode Illuminator, 89 North) and the emitted light was detected from 410–460 nm. Samples containing TPE4PH were imaged with 20% 445 nm (LDI Laser Diode Illuminator, 89 North) with emission light collection in the range of 450–505 nm. Data analyzation was carried out in Fiji (ImageJ 1.54f).Micropipette aspiration
Micropipette aspiration of the IMAC silk condensates was performed in ca. 2 mm high measurement chambers made by spacing 2 coverslips with 6 layers of parafilm. The chamber was filled with 5–10 μl condensate solution and sealed with a small amount of oil (Immersol 518F, Carl Zeiss) to prevent evaporation. The experiments were conducted at room temperature using pulled (PN-31, Narishige) borosilicate capillaries (WPI, 1 mm/0.5 mm outer/inner diameter). The radius of the pipettes Rp is typically 10–20 μm. To enable horizontal insertion of the micropipette into the observation chamber the pipettes were bent with a microforge (MF-900, Narishige). A piezo electric pressure controller (OB1 Mk3, Elveflow) connected to the micropipette was used to fill the micropipette with water before the experiment and control the pressure during experiments. Before the aspiration of the condensates a small amount of dilute phase was aspirated, and the zero pressure of the micropipette was determined. At first the micropipette was brought into contact with a condensate of radius R0. The aspiration was conducted by applying a constant negative suction pressure ΔP. After releasing the pressure (ΔP = 0 Pa) the relaxation of the condensate was recorded. Videos of the aspiration and retraction with a frame rate of about 10 fps were acquired in bright field mode using a Nikon Eclipse Ti inverted microscope (Nikon) equipped with an ORCA-flash 4.0 LT digital camera (Hamamatsu, Japan). The microscope was operated using micromanager (version 2.0.0). The data was analyzed in Fiji (ImageJ 1.54f) with the manual tracking plugin to measure the aspiration and retraction length. The measure function of Fiji was used to determine the diameter of the micropipette and the condensate. Linear fits and calculations were performed in Microsoft Excel. The surface tension γ of the condensate was obtained from the critical pressure ΔPc using the Young–Laplace law:1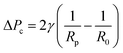 | (1) |
![[L with combining dot above]](https://www.rsc.org/images/entities/i_char_004c_0307.gif) a (eqn (2)).
a (eqn (2)).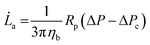 | (2) |
![[L with combining dot above]](https://www.rsc.org/images/entities/i_char_004c_0307.gif) r is defined as
r is defined as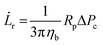 | (3) |
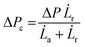 | (4) |
For the full derivation of all equations please see Guevorkian et al.2 The values reported in the main text are the mean ± standard error of the mean (SEM) with the number of datasets in brackets. All data are presented in Table S1 (ESI†).
For comparison with the biophysical properties of the HT silk condensates the data published in Tunn et al. were used.24,65 The surface tension of the HT silk condensates was calculated before. The effective viscosity ηeff of the HT condensates is
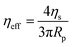 | (5) |
Analysis of coalescence of silk protein condensates
The samples were prepared and concentrated until LLPS occurred, as previously described. 5 μl of sample were imaged in a chamber, constructed of 2 coverslips and a 50 μm thick spacer tape (iSpacer, SunJinLab), to prevent evaporation. Videos of fusion events were taken with an Axio Observer.Z1 (Zeiss) inverted light microscope (20×/0.5 Ph2 objective, 1.6× tube lens, phase 2 contrast) and Prime BSI sCMOS camera (Photometris). The frame rate was approximately 54 fps. The final condensate diameter after coalescence and coalescence time were determined with ZEN microscopy software (Zen lite 3.9, Zeiss). For further analysis the coalescing condensates were approximated by an ellipse. The aspect ratio (AR = llong/lshort) of the ellipse over the coalescence time was determined as described previously and fitted with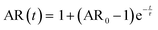 | (6) |
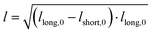 ). The inverse capillary velocity (ICV) was obtained from the slope of a linear fit to the data:
). The inverse capillary velocity (ICV) was obtained from the slope of a linear fit to the data: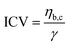 | (7) |
The surface tension γ obtained from micropipette aspiration allowed us to calculate the viscosity ηb,c of the condensates.
Scanning electron microscopy
Samples of HT silk, IMAC silk, and IMAC silk + BG lysate were concentrated until LLPS occurred, as previously described. Additionally, a control free of silk protein was prepared by mixing BG lysate with water, and concentrated similar to IMAC silk + BG lysate. A small amount of sample was transferred into a microcentrifuge tube and vitrified in liquid ethane (−180 °C), followed by freeze-drying (Christ Alpha 2–4). Freeze-dried samples were fractured and transferred to stubs coated with carbon tape. The samples were sputter coated with 6 nm Au/Pd and imaged with a Sigma VP scanning electron microscope (Zeiss), SE2 detector at 1.5 kV.Author contributions
Design and conceptualization of the study: J. T., M. B. L., J.-A. G., A. S.-V. (AIEs), E. A.-P. (AIEs). Experimental part: J. T. (protein expression and purification, concentrating and imaging, amino acid analysis, quantification of background molecules, SEM, coalescence studies, data analysis), I. T. (micropipette aspiration, AIEs), M. S. (protein expression and purification), T. V. (amino acid analysis, SEM and data analysis), M. M. (plasmid for BG lysate), G. B. (micropipette aspiration), A. S.-V. (synthesis of AIEs). Writing – original draft: J. T., I. T. (micropipette aspiration and parts of discussion), M. S. (parts of methods). Writing – finalizing: M. B. L.Data availability
All newly generated data are presented in the manuscript and the ESI.† The raw data are available on Zenodo.org: DOI: https://doi.org/10.5281/zenodo.12529262.Conflicts of interest
The authors declare no conflicts of interest.Acknowledgements
This work was funded by Novo Nordisk Fonden (NNF20OC0061306), the Research Council of Finland (Projects 346105, 364199, 341057, and 346632), the Center of Excellence Program (2022–2029) in Life-Inspired Hybrid Materials (LIBER), and the Wihuri foundation (JT). The table of content figure, as well as Fig. 1 and 2, was created with BioRender.com. We thank Françoise Brochard-Wyart for help with the data analysis for condensate properties. We thank Yin Yin for providing eGFP. We thank Alberto Scacchi, Adam Harmat and Dmitry Tolmachev, and Maria Sammalkorpi for discussions on the hypothesis for mechanisms. The authors acknowledge the provision of facilities and technical support by OtaNano – Nanomicroscopy Center (Aalto-NMC) and the Bioeconomy Infrastructure at Aalto University.References
- N. H. C. S. Silva, C. Vilela, I. M. Marrucho, C. S. R. Freire, C. Pascoal Neto and A. J. D. Silvestre, J. Mater. Chem. B, 2014, 2, 3715 RSC
.
- C. Lendel and N. Solin, RSC Adv., 2021, 11, 39188–39215 RSC
.
- S. Alberti and A. A. Hyman, Nat. Rev. Mol. Cell Biol., 2021, 22, 196–213 CrossRef CAS PubMed
.
- A. S. Lyon, W. B. Peeples and M. K. Rosen, Nat. Rev. Mol. Cell Biol., 2021, 22, 215–235 CrossRef CAS PubMed
.
- A. Miserez, J. Yu and P. Mohammadi, Chem. Rev., 2023, 123, 2049–2111 CrossRef CAS PubMed
.
- Y. Sun, Z. W. Lim, Q. Guo, J. Yu and A. Miserez, MRS Bull., 2020, 45, 1039–1047 CrossRef
.
- Y. Tan, S. Hoon, P. A. Guerette, W. Wei, A. Ghadban, C. Hao, A. Miserez and J. H. Waite, Nat. Chem. Biol., 2015, 11, 488–495 CrossRef CAS PubMed
.
- M. Heim, D. Keerl and T. Scheibel, Angew. Chem., Int. Ed., 2009, 48, 3584–3596 CrossRef CAS PubMed
.
- P. Mohammadi, A. S. Aranko, L. Lemetti, Z. Cenev, Q. Zhou, S. Virtanen, C. P. Landowski, M. Penttilä, W. J. Fischer, W. Wagermaier and M. B. Linder, Commun. Biol., 2018, 1, 86 CrossRef PubMed
.
- T. Välisalmi, H. Bettahar, Q. Zhou and M. B. Linder, Int. J. Biol. Macromol., 2023, 250, 126161 CrossRef PubMed
.
- L. Lemetti, J. Tersteegen, J. Sammaljärvi, A. S. Aranko and M. B. Linder, ACS Sustainable Chem. Eng., 2022, 10, 552–561 CrossRef CAS
.
- T. Välisalmi, N. Roas-Escalona, K. Meinander, P. Mohammadi and M. B. Linder, Langmuir, 2023, 39, 4370–4381 CrossRef PubMed
.
- K. Schacht, T. Jüngst, M. Schweinlin, A. Ewald, J. Groll and T. Scheibel, Angew. Chem., Int. Ed., 2015, 54, 2816–2820 CrossRef CAS PubMed
.
- T. U. Esser, V. T. Trossmann, S. Lentz, F. B. Engel and T. Scheibel, Mater. Today Bio, 2021, 11, 100114 CrossRef CAS PubMed
.
- B. Gabryelczyk, F.-E. Sammalisto, J.-A. Gandier, J. Feng, G. Beaune, J. V. I. Timonen and M. B. Linder, Mater. Today Bio, 2022, 17, 100492 CrossRef CAS PubMed
.
- A. D. Malay, T. Suzuki, T. Katashima, N. Kono, K. Arakawa and K. Numata, Sci. Adv., 2020, 6, eabb6030 CrossRef CAS PubMed
.
- S. F. Banani, H. O. Lee, A. A. Hyman and M. K. Rosen, Nat. Rev. Mol. Cell Biol., 2017, 18, 285–298 CrossRef CAS PubMed
.
- M. Abbas, W. P. Lipiński, J. Wang and E. Spruijt, Chem. Soc. Rev., 2021, 50, 3690–3705 RSC
.
- Y.-H. Lin, J. D. Forman-Kay and H. S. Chan, Biochemistry, 2018, 57, 2499–2508 CrossRef CAS PubMed
.
- V. N. Uversky, I. M. Kuznetsova, K. K. Turoverov and B. Zaslavsky, FEBS Lett., 2015, 589, 15–22 CrossRef CAS PubMed
.
- Y. Shen, F. S. Ruggeri, D. Vigolo, A. Kamada, S. Qamar, A. Levin, C. Iserman, S. Alberti, P. S. George-Hyslop and T. P. J. Knowles, Nat. Nanotechnol., 2020, 15, 841–847 CrossRef CAS PubMed
.
- J. Wang and T. Scheibel, Biomacromolecules, 2018, 19, 3612–3619 CrossRef CAS PubMed
.
- E. Spruijt, J. Sprakel, M. A. Cohen Stuart and J. Van Der Gucht, Soft Matter, 2010, 6, 172–178 RSC
.
- I. Tunn, G. Beaune, J. Tersteegen, T. Välisalmi, J. V. I. Timonen, F. Brochard-Wyart and M. B. Linder, Commun. Phys., 2024, 7, 157 CrossRef
.
- S. Alberti, A. Gladfelter and T. Mittag, Cell, 2019, 176, 419–434 CrossRef CAS PubMed
.
- D. Fedorov, F. Sammalisto, A. L. Harmat, M. Ahlberg, S. Koskela, M. P. Haataja, A. Scacchi, M. Sammalkorpi and M. B. Linder, Adv. Funct. Mater., 2024, 2410421 CrossRef
.
- P. Mohammadi, C. Jonkergouw, G. Beaune, P. Engelhardt, A. Kamada, J. V. I. Timonen, T. P. J. Knowles, M. Penttila and M. B. Linder, J. Colloid Interface Sci., 2020, 560, 149–160 CrossRef CAS PubMed
.
- J. Kirschbaum and D. Zwicker, J. R. Soc., Interface, 2021, 18, 20210255 CrossRef CAS PubMed
.
- A. Gangotra, M. Biviano, R. R. Dagastine, J. D. Berry and G. R. Willmott, Soft Matter, 2019, 15, 7286–7294 RSC
.
- D. T. McSwiggen, M. Mir, X. Darzacq and R. Tjian, Genes Dev., 2019, 33, 1619–1634 CrossRef CAS PubMed
.
- C. Roden and A. S. Gladfelter, Nat. Rev. Mol. Cell Biol., 2021, 22, 183–195 CrossRef CAS PubMed
.
- N. Chappidi, T. Quail, S. Doll, L. T. Vogel, R. Aleksandrov, S. Felekyan, R. Kühnemuth, S. Stoynov, C. A. M. Seidel, J. Brugués, M. Jahnel, T. M. Franzmann and S. Alberti, Cell, 2024, 187, 945–961 CrossRef CAS PubMed
.
- I. Alshareedah, M. M. Moosa, M. Raju, D. A. Potoyan and P. R. Banerjee, Proc. Natl. Acad. Sci. U. S. A., 2020, 117, 15650–15658 CrossRef CAS PubMed
.
- A. S. Rathore, S. E. Sobacke, T. J. Kocot, D. R. Morgan, R. L. Dufield and N. M. Mozier, J. Pharm. Biomed. Anal., 2003, 32, 1199–1211 CrossRef CAS PubMed
.
- K. Pilely, M. R. Johansen, R. R. Lund, T. Kofoed, T. K. Jørgensen, L. Skriver and E. Mørtz, Anal. Bioanal. Chem., 2022, 414, 747–758 CrossRef CAS PubMed
.
- K. Schacht and T. Scheibel, Biomacromolecules, 2011, 12, 2488–2495 CrossRef CAS PubMed
.
- L. Jia, Z. Ji, Y. Ji, C. Zhou, G. Xing and Y. Qiao, ChemSystemsChem, 2021, 3, e2000044 CrossRef CAS
.
- S. Yang, H. Yu, X. Xu, T. Yang, Y. Wei, R. Zan, X. Zhang, Q. Ma, H. C. Shum and Y. Song, ACS Nano, 2023, 17, 8195–8203 CrossRef CAS PubMed
.
- A. Sanz-Velasco, O. Amargós-Reyes, A. Kähäri, S. Lipinski, L. M. Cavinato, R. D. Costa, M. A. Kostiainen and E. Anaya-Plaza, Chem. Sci., 2024, 15, 2755–2762 RSC
.
- S. Biswas, A. L. Hecht, S. A. Noble, Q. Huang, R. E. Gillilan and A. Y. Xu, Biomacromolecules, 2023, 24, 4771–4782 CrossRef CAS PubMed
.
- M. Poudyal, K. Patel, L. Gadhe, A. S. Sawner, P. Kadu, D. Datta, S. Mukherjee, S. Ray, A. Navalkar, S. Maiti, D. Chatterjee, J. Devi, R. Bera, N. Gahlot, J. Joseph, R. Padinhateeri and S. K. Maji, Nat. Commun., 2023, 14, 6199 CrossRef CAS PubMed
.
- R. V. Pappu, S. R. Cohen, F. Dar, M. Farag and M. Kar, Chem. Rev., 2023, 123, 8945–8987 CrossRef CAS PubMed
.
- C. M. Fare, A. Villani, L. E. Drake and J. Shorter, Open Biol., 2021, 11, 210137 CrossRef CAS PubMed
.
- T. Kaur, M. Raju, I. Alshareedah, R. B. Davis, D. A. Potoyan and P. R. Banerjee, Nat. Commun., 2021, 12, 872 CrossRef CAS PubMed
.
- M. Feric, N. Vaidya, T. S. Harmon, D. M. Mitrea, L. Zhu, T. M. Richardson, R. W. Kriwacki, R. V. Pappu and C. P. Brangwynne, Cell, 2016, 165, 1686–1697 CrossRef CAS PubMed
.
- I. Peran and T. Mittag, Curr. Opin. Struct. Biol., 2020, 60, 17–26 CrossRef CAS PubMed
.
- J. Dinic, A. B. Marciel and M. V. Tirrell, Curr. Opin. Colloid Interface Sci., 2021, 54, 101457 CrossRef CAS
.
- T. Välisalmi and M. B. Linder, Protein Sci., 2024, 33, e4907 CrossRef PubMed
.
- A. Leppert, J. Feng, V. Railaite, T. Bohn Pessatti, C. P. Cerrato, C. Mörman, H. Osterholz, D. P. Lane, F. R. N. C. Maia, M. B. Linder, A. Rising and M. Landreh, J. Am. Chem. Soc., 2024, 146, 19555–19565 CrossRef CAS PubMed
.
- A. K. Varanko, J. C. Su and A. Chilkoti, Annu. Rev. Biomed. Eng., 2020, 22, 343–369 CrossRef CAS PubMed
.
- H. J. Kim, B. H. Hwang, S. Lim, B.-H. Choi, S. H. Kang and H. J. Cha, Biomaterials, 2015, 72, 104–111 CrossRef CAS PubMed
.
- E. Y. Jeon, S. Um, J. Park, Y. Jung, C. Cheon, H. Jeon and J. J. Chung, Small, 2022, 18, 2200416 CrossRef CAS PubMed
.
- M. J. Harrington, R. Mezzenga and A. Miserez, Nat. Rev. Bioeng., 2023, 2, 260–278 CrossRef
.
- C. Fredriksson, M. Hedhammar, R. Feinstein, K. Nordling, G. Kratz, J. Johansson, F. Huss and A. Rising, Materials, 2009, 2, 1908–1922 CrossRef CAS
.
- M. Widhe, N. D. Shalaly and M. Hedhammar, Biomaterials, 2016, 74, 256–266 CrossRef CAS PubMed
.
- L. Baoyong, Z. Jian, C. Denglong and L. Min, Burns, 2010, 36, 891–896 CrossRef PubMed
.
- T. S. Harmon, A. S. Holehouse and R. V. Pappu, New J. Phys., 2018, 20, 045002 CrossRef
.
- H.-X. Zhou, V. Nguemaha, K. Mazarakos and S. Qin, Trends Biochem. Sci., 2018, 43, 499–516 CrossRef CAS PubMed
.
- Y. Shin and C. P. Brangwynne, Science, 2017, 357, eaaf4382 CrossRef PubMed
.
- Z. Feng, X. Chen, X. Wu and M. Zhang, J. Biol. Chem., 2019, 294, 14823–14835 CrossRef CAS PubMed
.
- M. Heidenreich, J. M. Georgeson, E. Locatelli, L. Rovigatti, S. K. Nandi, A. Steinberg, Y. Nadav, E. Shimoni, S. A. Safran, J. P. K. Doye and E. D. Levy, Nat. Chem. Biol., 2020, 16, 939–945 CrossRef CAS PubMed
.
- R. Mammen Regy and J. Mittal, Nat. Chem. Biol., 2020, 16, 934–935 CrossRef CAS PubMed
.
- J. A. Villegas, M. Heidenreich and E. D. Levy, Nat. Chem. Biol., 2022, 18, 1319–1329 CrossRef CAS PubMed
.
- D. Huemmerich, C. W. Helsen, S. Quedzuweit, J. Oschmann, R. Rudolph and T. Scheibel, Biochemistry, 2004, 43, 13604–13612 CrossRef CAS PubMed
.
- I. Tunn, G. Beaune, J. Tersteegen, T. Välisalmi, J. V. I. Timonen, F. Brochard-Wyart and M. B. Linder, Commun. Phys., 2024, 7(1), 157 CrossRef
.
- D. Fedorov, N. Roas-Escalona, D. Tolmachev, A. L. Harmat, A. Scacchi, M. Sammalkorpi, A. S. Aranko and M. B. Linder, Small, 2024, 20, 2306817 CrossRef CAS PubMed
.
Footnote |
| † Electronic supplementary information (ESI) available: Fig. S1: protein sequence for CBM-AQ12-CBM; Fig. S2: SDS-PAGE of CBM-AQ12-CBM and BG lysate; Fig. S3: fluorescence microscopy images with free eGFP; Fig. S4: AIE structures; Fig. S5: inverse capillary velocity (ICV) analysis; Fig. S6: FTIR analysis; Fig. S7: light microscopy images with ssDNA from salmon testes; Table S1: overview of the values for the micropipette aspiration; Video S1: micropipette aspiration of IMAC silk; Video S2: HT silk coalescence; Video S3: IMAC silk + BG lysate coalescence; Video S4: IMAC silk coalescence. See DOI: https://doi.org/10.1039/d4tb01422g |
| This journal is © The Royal Society of Chemistry 2024 |