Selective assembly and insertion of ubiquicidin antimicrobial peptide in lipid monolayers
Sonam
Raghav
a,
Prashant
Hitaishi†
a,
Rajendra P.
Giri‡
b,
Archana
Mukherjee
cd,
Veerendra K.
Sharma
 *de and
Sajal K.
Ghosh
*de and
Sajal K.
Ghosh
 *a
*a
aDepartment of Physics, School of Natural Sciences, Shiv Nadar Institution of Eminence, NH 91, Tehsil Dadri, G. B. Nagar, Uttar Pradesh 201314, India. E-mail: sajal.ghosh@snu.edu.in
bInstitut für Experimentelle und Angewandte Physik, Christian-Albrechts-Universität Zu Kiel, 24098 Kiel, Germany
cRadiopharmaceuticals Division, Bhabha Atomic Research Centre, Mumbai 400085, India
dHomi Bhabha National Institute, Mumbai, 400094, India. E-mail: sharmavk@barc.gov.in
eSolid State Physics Division, Bhabha Atomic Research Centre, Mumbai, 400085, India
First published on 16th October 2024
Abstract
Antimicrobial-resistant bacteria pose a significant threat to humans, prompting extensive research into developing new antimicrobial peptides (AMPs). The biomembrane is the first barrier of a biological cell, hence, comprehending the interaction and self-assembly of AMPs in and around such membranes is of great importance. In the present study, several biophysical techniques have been applied to explore the self-assembly of ubiquicidin (29–41), an archetypical AMP, in and around the phospholipid monolayers formed at air–water interface. Such a monolayer mimics one of the leaflets of a lipid bilayer. The surface pressure–area isotherm exhibits the strongest interaction with a negatively charged lipid, 1,2-dipalmitoyl-sn-glycero-3-phospho-(1′-rac-glycerol) (sodium salt) (DPPG). The weakest affinity was towards the zwitterionic lipid, 1,2-dipalmitoyl-sn-glycero-3-phosphocholine (DPPC). Another zwitterionic lipid, 1,2-dipalmitoyl-sn-glycero-3-phosphoethanolamine (DPPE), shows an intermediate affinity. This affinity was quantified by analyzing alterations in the effective mean molecular area of the lipid, the in-plane compressional modulus of the assembly, and the electrostatic potential induced by the presence of peptides. The precise organization of the peptide around the lipid monolayer at a sub-nanometre length scale was revealed using synchrotron-based X-ray reflectivity measurements from the air–water interface. Information about the selective interaction of the peptide with lipids and their varied orientation at the lipid–water interface could be useful in understanding the selectivity of AMP in developing new antibiotics.
1. Introduction
The breakthroughs of the ‘antibiotic era’ led people to conclude that infectious diseases were no longer a major scientific concern.1,2 However, the subsequent rise of antibiotic-resistant microbes has renewed the urgency for new antimicrobial agents. Consequently, the development of novel targets or more potent antibiotics are required to deal with resistant microbes, as infectious diseases continue to be a major worldwide health concern.3–5 In 2011, the World Health Organization (WHO) selected ‘antimicrobial resistance’ as a theme for World Health Day. This effort was made to highlight and bring attention to this public health threat.6Antimicrobial peptides (AMPs) have garnered attention for their pivotal role in host defence and innate immunity. They are identified across various species, including mammals, amphibians, and insects.7–13 Because of an ‘evolutionary arms race’ between hosts and infections, AMPs have become incredibly active against a variety of pathogens, including ones that are resistant to antibiotics.14–16 Over the past decade, there has been a significant increase in the study of AMPs due to their potential as a future therapeutic.17–20 Typically ranging from 10 to 150 amino acids in length and carrying a net charge, AMPs possess a diverse range of antimicrobial and antifungal properties.21,22 They modulate various components of the innate immune system, with short peptides (9–11 amino acids) showing particular promise as therapeutic agents due to their efficiency and advantages over longer peptides.2,23,24
Though, there are a few hypotheses which are specific to each peptide,25 a limitation is that, in most of the cases, the exact mechanism of action of AMPs at the molecular level is not well-understood. Importantly, the ability of AMPs to kill microorganisms without harming the host is a crucial requirement, making them particularly attractive for therapeutic development.26 Generally, the cellular membrane of a pathogen has been recognized as the target of a peptide molecule.27 Such a membrane is a fundamental structure in cells, primarily composed of lipids and proteins, and it can be conceptualized as two interconnected monomolecular lipid layers. Interestingly, the mammalian and bacterial membranes have different lipid composition in such layers. The outer leaflet of a mammalian cell membrane mainly consists of phosphatidylcholine (PC), sphingolipids, and various sterols, which are zwitterionic in nature.28–32 In contrast, bacterial cell membranes are rich in anionic phospholipids such as phosphatidylglycerol (PG), lysyl phosphatidylglycerol (LPG), and cardiolipin.33,34 This difference in lipid composition may form the basis for the selective interaction of AMPs with bacterial cell membranes. Therefore, selective interaction of AMPs with lipid membranes is still a focussed area of research.
Intracellular protein ubiquicidin (UBI) exhibits antibacterial properties against a range of infections causing microorganisms.35 It is a 59 amino acid protein that is rich in positively charged residues. From yeast to human cells, all eukaryotic cells share this protein. UBI isolated from human and mouse sources36–38 has demonstrated antibacterial action against a range of pathogens. The peptide fragments of UBI radiolabelled with diagnostic radionuclides have demonstrated promise as infection imaging probes in humans.39–41 UBI (29–41) is the amino acid fragment from positions 29–41 from the natural full length UBI of 59 amino acids.42 The electropositive groups, namely, 5 arginine and 1 lysine of UBI (29–41) contribute in selective binding to the electronegative groups on the surface of pathogenic bacteria. Recently, Bhatt et al.35 used circular dichroism (CD), isothermal titration calorimetry (ITC), and dynamic light scattering (DLS) techniques to investigate the interaction of UBI (29–41) and UBI (31–38) with model bacterial and mammalian cell membranes. The results show a stronger interaction of UBI (29–41) with a model bacterial membrane, which explains why preclinical research using radiolabelled UBI-derived peptides has shown encouraging results for in situ infection diagnosis.42 To comprehend these results, a quantitative analysis of the interaction of this peptide with specific lipids is necessary, which may pave the way to improving the efficacy of the molecule.
The main aim of this work is to investigate the assembly of UBI (29–41) near physiologically relevant lipid membranes and its consequence on the organization and physical property of lipids in the membrane. For that purpose, a lipid monolayer assembled at an air–water interface has been chosen to mimic one of the leaflets of a lipid bilayer. Due to their amphiphilic nature, the lipids self-assemble themselves at this interface attaching their heads to water and floating the chains in the air. Two zwitterionic lipids, 1,2-dipalmitoyl-sn-glycero-3-phosphocholine (DPPC) and 1,2-dipalmitoyl-sn-glycero-3-phosphoethanolamine (DPPE) and an anionic lipid 1,2-dipalmitoyl-sn-glycero-3-phospho-(1′-rac-glycerol) (sodium salt) (DPPG) are used to form the monolayer. Two complementary interfacial techniques, the surface pressure–area isotherm, and the surface potential, have been employed to decipher the details of the lipid monolayer in the absence and presence of the peptide. The synchrotron-based advanced X-ray reflectivity technique has addressed the structural details of the layer along with the assembly of the peptide in and around the layer.
2. Materials and methods
2.1. Materials
Zwitterionic lipids 1,2-dipalmitoyl-sn-glycero-3-phosphocholine (DPPC), 1,2-dipalmitoyl-sn-glycero-3-phosphoethanolamine (DPPE) and the negatively charged lipid 1,2-dipalmitoyl-sn-glycero-3-phospho-(1′-rac-glycerol) (sodium salt) (DPPG) were purchased from Avanti Polar Lipids (Alabaster, AL) in powder form and were used without further purification. The spectroscopy-grade solvents chloroform (HPLC), and methanol (HPLC) were purchased from Sigma Aldrich (USA). The peptide UBI (29–41) with sequence TGRAKRRMQYNRR was procured from ABI scientific (Sterling, US). The purity of the peptides was greater than 90%. The molecular structures of the lipids and the peptide used in the study are shown in Fig. 1.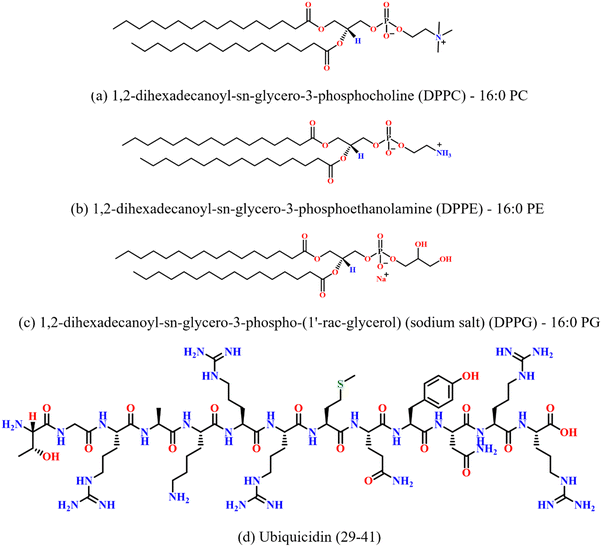 | ||
| Fig. 1 Chemical structures of (a) DPPC, (b) DPPE, (c) DPPG lipids, and (d) antimicrobial peptide ubiquicidin (29–41). | ||
2.2. Methods
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) methanol = 3
methanol = 3![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 (vol
1 (vol![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) vol). This helps the homogeneous mixing of the lipids and peptides. Such a solvent system is commonly employed in lipid–protein studies as it enables the effective mixing of amphiphilic lipids and peptides. The final mass concentration of the stock solution was 0.5 mg mL−1 with the respective molar concentrations of 0.68, 0.72 and 0.67 mM for DPPC, DPPE and DPPG. For monolayer experiments, this lipid–protein mixture in the solvent was spread on the water surface of the LB trough. Then, the solvent evaporates from the air–water interface leaving the lipids and peptides at the interface. This ensures the peptide activity and its interactions with the lipid membranes, preserving the integrity and functionality of the peptides.
vol). This helps the homogeneous mixing of the lipids and peptides. Such a solvent system is commonly employed in lipid–protein studies as it enables the effective mixing of amphiphilic lipids and peptides. The final mass concentration of the stock solution was 0.5 mg mL−1 with the respective molar concentrations of 0.68, 0.72 and 0.67 mM for DPPC, DPPE and DPPG. For monolayer experiments, this lipid–protein mixture in the solvent was spread on the water surface of the LB trough. Then, the solvent evaporates from the air–water interface leaving the lipids and peptides at the interface. This ensures the peptide activity and its interactions with the lipid membranes, preserving the integrity and functionality of the peptides.
The molar percentage of AMP was quantified using the expression, 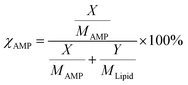 where χAMP is the molar percentage of AMP in a mixed system. Here, X and Y are the measured masses and MAMP and MLipid are the molecular masses of AMP and lipid, respectively.43,44 The LB trough and Platinum Wilhelmy plate were extensively cleaned with ethanol and DI water to remove any trace of organic contaminants. For a monolayer formation, 40 µL of the pristine lipid or mixed stock solution was homogenously spread at the air–water interface using a glass Hamilton micro-syringe. After that, the trough was left undisturbed for 45 min to allow the evaporation of the volatile solvent. The temperature of subphase (water) was maintained at 22 ± 1 °C using a water bath circulator (Equitron, India). The Delrin barriers were brought close to compress the monolayer symmetrically at a constant rate of 4 mm min−1. Finally, the surface pressure was recorded as a function of area per molecule to construct the surface pressure (π)–area (A) isotherm.
where χAMP is the molar percentage of AMP in a mixed system. Here, X and Y are the measured masses and MAMP and MLipid are the molecular masses of AMP and lipid, respectively.43,44 The LB trough and Platinum Wilhelmy plate were extensively cleaned with ethanol and DI water to remove any trace of organic contaminants. For a monolayer formation, 40 µL of the pristine lipid or mixed stock solution was homogenously spread at the air–water interface using a glass Hamilton micro-syringe. After that, the trough was left undisturbed for 45 min to allow the evaporation of the volatile solvent. The temperature of subphase (water) was maintained at 22 ± 1 °C using a water bath circulator (Equitron, India). The Delrin barriers were brought close to compress the monolayer symmetrically at a constant rate of 4 mm min−1. Finally, the surface pressure was recorded as a function of area per molecule to construct the surface pressure (π)–area (A) isotherm.
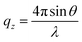 , where θ is the incident angle and λ is the wavelength of the X-rays.10 The enclosure of the trough was flushed with Helium gas to reduce the background scattering. The profile is fitted using Parratt's recursive technique using a MATLAB code, the details of which can be found elsewhere.48,50,51 In brief, a lipid monolayer at the interface is modelled with two boxes representing the head and tail region. An extra box for the peptide molecules was considered where required.52 Each of these boxes has a definite value of electron density and a thickness. Between two boxes, roughness has been considered for the smooth transition of electron density from one box to the other. The bulk subphase, an infinitely long water layer is considered for all samples with the same electron density of 0.334 e− Å−3 and a roughness of 3 Å. The electron density of air is taken to be zero.
, where θ is the incident angle and λ is the wavelength of the X-rays.10 The enclosure of the trough was flushed with Helium gas to reduce the background scattering. The profile is fitted using Parratt's recursive technique using a MATLAB code, the details of which can be found elsewhere.48,50,51 In brief, a lipid monolayer at the interface is modelled with two boxes representing the head and tail region. An extra box for the peptide molecules was considered where required.52 Each of these boxes has a definite value of electron density and a thickness. Between two boxes, roughness has been considered for the smooth transition of electron density from one box to the other. The bulk subphase, an infinitely long water layer is considered for all samples with the same electron density of 0.334 e− Å−3 and a roughness of 3 Å. The electron density of air is taken to be zero.
3. Results and discussion
Each of the lipids DPPC, DPPE, and DPPG provides various facets of membrane composition and function. Despite being a common lipid in eukaryotic membranes, DPPC is included here to provide a baseline for comparison because of its well-characterized nature with physical and chemical properties.53–55 As described by Stephani et al., DPPE is an accurate representative lipid of the inner membrane of bacteria, especially those that are Gram-negative.56 The abundant presence of this lipid is essential for maintaining the curvature and functionality of the membrane. DPPG is important for modelling bacterial outer membranes as it maintains the electrostatic nature of the membrane and controls its interaction with the external environment. The present work has shed light on the exact roles of these lipids in interacting with an antimicrobial peptide.3.1. Interaction of UBI (29–41) with the lipid monolayer
The surface pressure–area (π–A) isotherm measurement is an advanced, crucial, and useful technique to examine interactions between amphiphilic molecules and determine their self-assembly at the air–water interface.57–59 A lipid monolayer formed at this interface serves as a simplified system to mimic one of the leaflets of a biological cell membrane.60 An isotherm examines the phase behaviour of lipids in such a monolayer.61 One could also follow how such behaviour changes once any foreign molecules interact with the layer.In a typical isotherm, initially, the lipid molecules are far apart occupying a high mean molecular area (MMA) at the air–water interface. This phase is characterised as a gaseous (G) phase as there is negligible interaction between the molecules. On compression, the trough barriers bring the molecules closer introducing an interaction among these molecules, which leads to a rise in surface pressure. Here, a gaseous (G) to liquid expanded (LE) phase transition occurs. The area at which the molecules first show the non-zero surface pressure on the onset of the LE phase is termed as the lift-off area (ALO). On further compression, the monolayer finally reaches to a liquid condensed (LC) phase where the molecules are closely packed on the water surface. There is a highest critical pressure, termed as the ‘collapse pressure’ up to which this LC phase sustains. Beyond this pressure, the monolayer collapses, either forming multiple layers of lipids or losing the molecules into the bulk subphase. In the present study, all isotherms were measured at the water subphase temperature of 22 ± 1 °C. This temperature is below the gel to fluid phase transition temperature (Tm) of all three lipids DPPC (Tm ∼ 41 °C),62 DPPE (Tm ∼ 63 °C), and DPPG (Tm ∼ 41 °C).
An important physical property of a monolayer at the air–water interface is the compressional modulus (ECM), which can be calculated from the isotherm data using the equation,63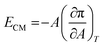 . As stated above, π is the surface pressure and A is the mean molecular area. The value of ECM is temperature (T) specific as the isotherm of a molecule itself depends on temperature. This ECM provides information about the compactness and packing of molecules in the monolayer.63
. As stated above, π is the surface pressure and A is the mean molecular area. The value of ECM is temperature (T) specific as the isotherm of a molecule itself depends on temperature. This ECM provides information about the compactness and packing of molecules in the monolayer.63
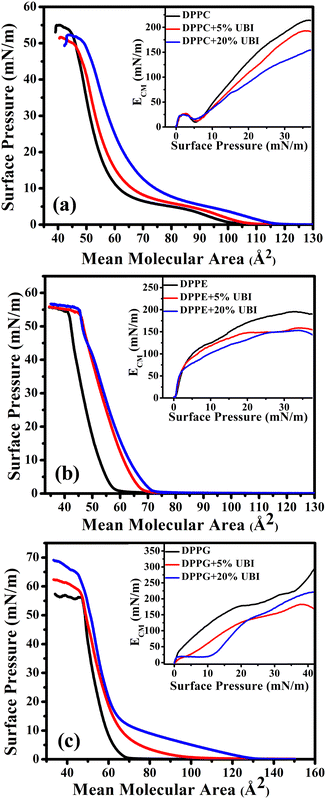
The isotherms of pristine lipids with added 5 and 20 mol% of UBI (29–41), henceforth termed as ‘UBI’, are shown in Fig. 2. The peptide concentrations of 5 and 20 mol% relative to the lipids were chosen based on previous studies demonstrating that these concentrations are relevant for investigating peptide–lipid interactions, particularly in mimicking biologically relevant systems.64–66 While the 5 mol% concentration is employed to investigate the initial step of interaction without causing drastic disruption in the membrane, 20 mol% is used to achieve a noticeable effect, which is easily quantifiable by the instruments. In the case of pure zwitterionic DPPC, the value of ALO is ∼105 Å2. The LE to LC phase transition occurs through a plateau region, which is consistent with the earlier studies.67,68 The monolayer collapses at a surface pressure of ∼55 mN m−1.69,70 The plateau region in the isotherm is due to the coexistence of two phases, which is a signature of a first-order phase transition.71 In the presence of UBI in the monolayer, the π–A isotherm shifts to the right exhibiting higher values of ALO. Note that for calculating the MMA for lipid/UBI mixed systems, only the number of lipid molecules in the system was considered. Therefore, the change in the isotherm is an indication of the affinity of the peptide to the lipid layer. In the case of the absence of any peptide in or around the lipid layer, the mixed system would have produced an isotherm identical to the pure lipid. The ALO value increases to ∼115 Å2 in the presence of 20 mol % of UBI, as shown in Fig. 2(a). Furthermore, the UBI causes a decrease in the value of ECM as evident in the inset of the figure. At 20 mN m−1, it has a value of 131 mN m−1 for the pure DPPC layer which drops to ∼90 mN m−1 in the presence of 20 mol% of UBI. At 30 mN m−1, the value drops from ∼189 to ∼132 mN m−1. The increasing nature of ECM with increasing surface pressure for a particular monolayer is readily understood which arises from the enhanced compactness in the arrangement of the lipids. The dip in the ECM at the lower surface pressure of around 5 mN m−1 is the region of coexistence of LE and LC phases.
The π–A isotherm of another zwitterionic DPPE shows the ALO value of 60 Å2 (Fig. 2(b)), which is close to the value reported earlier.72 Note that this value is considerably smaller than the DPPC even though both the lipids have the same saturated chains and are in the gel phase. Due to the difference in the chemical structure, a PE head group is considerably smaller than that of a PC.73 Moreover, the DPPC lipid has a higher effective cross-sectional area due to a tilt in its tail. Such a tilt is absent in the DPPE molecule. The DPPE monolayer exhibits a steeper increase in pressure characterising a direct G to LC phase transition. In the presence of UBI, the monolayer shifts to a higher ALO value to 72 Å2 for the 20 mol % of UBI. The inset of Fig. 2(b) shows the plot of ECM as a function of surface pressure indicating qualitatively a similar behaviour of DPPC lipid except for the absence of any dip of coexistence of phases. However, the quantitative drop of the values of ECM is different when UBI is present in the lipid monolayer. At 30 mN m−1, the value of ECM decreases from ∼192 to ∼150 mN m−1 in the presence of 20 mol% of the peptide.
The negatively charged lipid DPPG shows a much steeper increase in surface pressure with a value of ALO ∼ 68 Å2, which aligns with previous studies.48,74 Even though this lipid is negatively charged, the electrostatic repulsion does not contribute to enhancing the inter molecular separation. However, when the molecules are closed enough, immediately after the ALO, the strong repulsion produces a steeper isotherm. Such a behaviour has already been observed for many other charged lipids with low values of ALO.75,76 Fundamentally, the steric repulsion depending upon the lipid chain conformation and the geometrical size of the head group is the deciding factor for providing the corresponding value of ALO. Surprisingly, addition of UBI causes a drastic increase in its value to ∼100 Å2 and ∼132 Å2 for the 5 and 20 mol% of UBI, respectively (Fig. 2(c)). In the inset of the figure, the presence of UBI in the monolayer causes a decrease in the compressional modulus. For example, at a surface pressure of 30 mN m−1, the value of ECM decreases from ∼205 to ∼171 mN m−1 in the presence of 20 mol% of UBI.
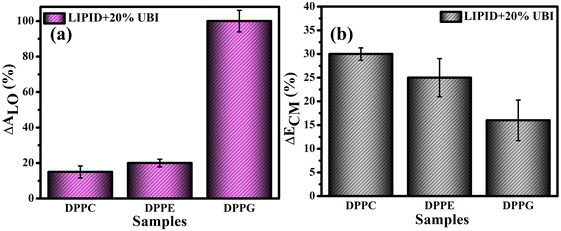
For direct comparison, the percentage increase in the values of lift-off area (ΔALO) for lipid monolayers in the presence of 20 mol% UBI with respect to the pure lipid monolayer are shown in Fig. 3(a). While the increase is 19% and 22% for zwitterionic DPPC and DPPE, respectively, it jumps to 95% for the anionic DPPG. Hence, it is evident that UBI affects the monolayer of the negatively charged lipid DPPG to the greatest extent.
Fig. 3(b) provides a quick visualization of the percentage change in the ΔECM of lipid layers at 30 mN m−1 in the presence of UBI with respect to the pure lipid layer. The physiological surface pressure of a cellular membrane is reported to be around 30 mN m−177,78 and hence this pressure is considered in this discussion. The decrease of the value of ECM, shown in the figure, indicates a flexible and loosely packed structure of the lipid/UBI composite film compared to the pristine lipid film.79–81 A peptide is a polymer composed of multiple amino acids. Such a macromolecule is inherently compressible and has a much lower compressional modulus compared to a pure lipid film. The trend in ΔECM is opposite to the ΔALO, which is counter intuitive. The difference in assembly of the peptide around the lipid may cause such a difference, which will be discussed in a later section.
Even though the mol% of added UBI is the same for all the used lipids, the strength of interaction between the lipid and the peptide decides the number and specific organization of the peptide in and around the lipid layer. The most important physical interactions are the electrostatic and the hydrophobic interactions.82,83 The UBI is predominantly hydrophilic, which means that it interacts well with water and other polar solvents. Therefore, this macromolecule was expected to stay dissolved in the bulk water subphase of the LB trough. However, the strong electrostatic interaction between the anionic head group of the DPPG and the cationic patches on the peptide due to the presence of arginine and lysine, aids the assembly of UBI in and around the DPPG layer. This interaction becomes weaker for the zwitterionic lipids which do not have any net charge but a dipole moment. Notably, within the two zwitterionic lipids, the PE head group is more interactive with the peptide. In all the cases, once the peptide is around the lipid layer, the short ranged hydrophobic interaction plays a role in the final assembly of the peptide. Matsuzaki et al. have already demonstrated this interaction to facilitate the insertion of peptide into the lipid layer leading to pore formation.84 Also, Huang elucidated that interaction between the non-polar portions of an AMP and the hydrophobic core of a lipid bilayer are critical to the insertion and assembly of a peptide.85
3.2. Modified electrostatics of the lipid monolayer
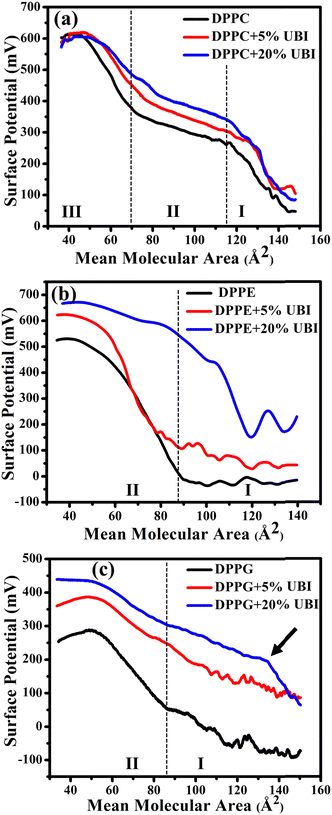
Surface potential (ΔV)–area measurements were conducted to follow the modified electrostatics of the surface of the lipid monolayer caused by the presence of UBI. This study is important as the charge state of the surface plays a vital role in protein or peptide adsorption on or insertion into cellular membranes.47,86 The overall electrostatics of the lipid monolayer at the air–water interface depends on the net dipole moment and the charge related to the two distinct interfaces, namely the water/lipid and the lipid/air.87,88 Furthermore, the net dipole moment of the interfaces depends on the head group, orientation of lipid molecules, reorientation of bound water to the head group and any other nearby foreign molecules. In the present study, the ΔV is the potential calculated by subtracting the potential of the fresh air–water interface from that of an organic molecular layer formed either by a pure lipid or by the composite layer of lipid and UBI.In Fig. 4(a), for the lipid DPPC, there are three distinct regions in the surface potential (ΔV)–area measurements, which is consistently aligned with the previous study.87,89 In region (I), up to a mean lipid area of ∼115 Å2, there is a notable rise in ΔV. On compression, as the molecules start coming closer, enhancing the number density of lipids on the surface and hence the dipole moments, the ΔV shows a rise. This is the characteristic region of the gas phase with a maximum potential of ∼250 mV. On the onset of the LE phase with a visible inflection, the potential curve enters region (II) and spans from ∼115 to ∼65 Å2. In this region, the rise in ΔV is much slower. The second inflection signifies the phase transition from LE to LC phase taking the ΔV curve into region III. In this region, the rise in ΔV is relatively faster compared to region II, but slower than region I. Since the molecules in region III are already closely packed, the contribution in ΔV is probably originated from the orientation of the lipid molecules reducing its tilt angle and the reorganization of water molecules at the interface. Finally, the curve enters the collapse region. Even though the phase boundaries do not strictly follow each other, qualitatively, this ΔV curve provides the details of the physical phases of the lipid monolayer, which was established by the π–A isotherm. In the presence of UBI, the overall curve shifts up while the phase boundary slightly moves to higher MMA. These observations coincide with the observation of π–A isotherms signifying the interaction of the peptide with the monolayer.
As shown in Fig. 4(b), there are only two distinct regions in the ΔV curve for pure DPPE lipid. In region (I), up to a mean molecular area of ∼90 Å2, there is no change in the surface potential, probably because of random orientation of the dipole of the lipids producing no net momentum. On the onset of region II with an inflection point, there is a rise of the potential as the monolayer directly enters into the LC phase. This region spans up to a mean molecular area of ∼40 Å2 with the highest value of ΔV ∼ 500 mV. Beyond this point, the curve enters the collapse phase. The feature of the curve is consistent with the study reported earlier.87 In the presence of UBI, the curve shifts upwards to a higher value of ΔV, which is qualitatively similar to the DPPC. However, in the presence of 20 mol% UBI, the G/LC phase boundary has a much higher shift to the right compared to that in DPPC. This indicates a better interaction of the UBI with the DPPE lipid than the DPPC. From the isotherm data, such a conclusion was drawn in the previous section.
Similar to earlier reports,90,91 the curve of ΔV of the negatively charged lipid DPPG shown in Fig. 4(c) exhibits two regions. In the beginning, there is a slow rise in the potential, which enhances after the inflection point. This point separates the two phases of G and the LC with a transition value of ∼50 mV at the mean molecular area of ∼85 Å2. The increase of the curve in region II is, as discussed for other lipids, due to the higher areal dipole density and their reorientation. The decrease in ΔV beyond ∼300 mV exhibits the collapse of the monolayer. The shift of this curve in the presence of UBI is qualitatively like the other lipids with a striking difference in moving the first inflection point to a very high value of mean molecular area. This has been demonstrated with an arrow in the figure Fig. 4(c). This is exactly the observation of very high increase in the lift-off area. Therefore, the two independent techniques unambiguously suggest the highest interaction of the peptide with the DPPG lipid. This interaction may alter the number density of the lipid at the interface along with changing the lipid orientation. Furthermore, the water molecules attached to the lipid head could themselves reorganise and reorient. Even the peptide itself contributes to the net dipole moment of the interface.
3.3. Structure of the lipid monolayer and assembly of the peptide
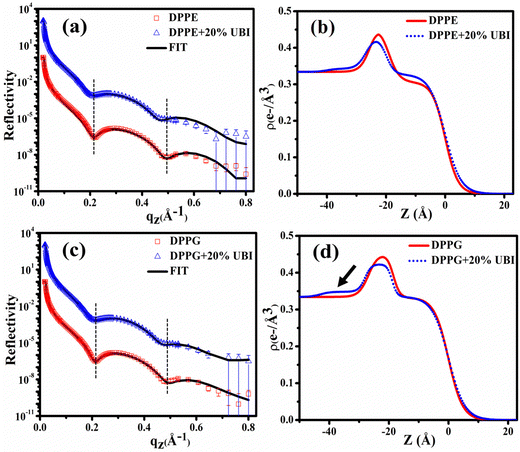
A synchrotron based X-ray source of high photon flux with a controllable beam size and energy is necessary to explore the structure of a single molecular layer at the air–water interface.48 In particular, XRR is suitable to follow the assembly of amphiphiles in such a layer. In this technique, an electron density (ED) profile is extracted by fitting the scattered intensity using Parratt's recursive method, the details of which can be found elsewhere.44,48,92 Even though the interaction of the UBI with the lipid layer with varying strength is already evident from the surface pressure–area isotherm and the surface potential measurements, the exact assembly of the peptide can be followed by this XRR study.The reflectivity curves for the pristine DPPC monolayer and the monolayer in the presence of 20 mol% of UBI are shown in Fig. 5(a). The surface pressure of all monolayers was maintained at 30 mN m−1 because of its physiological relevance.77,78 The reflectivity curve shows distinct Kiessig fringes, which is a signature of modulated ED of the air–water interface due to the presence of a lipid layer. This scenario is schematically described with the ED profile in Fig. 5(b). Without an assembly of these molecules at the smooth air–water interface, the reflectivity curve should have decayed monotonously following the Fresnel theory of reflectivity.51 The fitting parameters for this lipid are given in Table 1. As mentioned earlier, two slabs corresponding to the hydrophilic head attached to water and the hydrophobic tail floating in air were considered for fitting the XRR curve of the pristine DPPC (Fig. 5(b)). The estimated thickness of the tail and the head region of the monolayer are 16.99 and 6.08 Å, respectively. Their corresponding electron densities are 0.324 and 0.461 e− Å−3, aligning well with the previous reports.48,72 As shown in the schematic of Fig. 5(b), the peak in the ED profile corresponds to the lipid head group region as its chemical composition ensures the highest electron density. The schematic clearly depicts that the profile is along the normal of the plane of the lipid monolayer, which is achieved after the in-plane averaging of the electron density. As explained in earlier reports, the profile does not reach the discrete value of a slab due to the smearing effect of the interfacial roughness considered in the model analysis between any two slabs.48,92
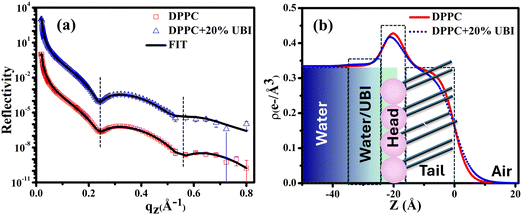 | ||
| Fig. 5 (a) X-ray reflectivity profiles of the monolayer at the air–water interface for pristine DPPC and in the presence of 20 mol% UBI at a surface pressure of 30 mN m−1. The reflectivity profile in (a) for the added UBI is vertically offset by a factor of 103 for distinct representation. (b) Corresponding electron density profiles of the lipid monolayer obtained by the best fit to the data in Fig. 5(a). The schematic of the lipid monolayer floating at the air–water interface is drawn to highlight various regions of the density profile. The dashed boxes represent the slabs to model different parts of the interface. | ||
| Sample | Tail | Head | UBI | χ 2 | ||||||
|---|---|---|---|---|---|---|---|---|---|---|
| d (Å) | ρ (e− Å−3) | σ (Å) | d (Å) | ρ (e− Å−3) | σ (Å) | d (Å) | ρ (e− Å−3) | σ (Å) | ||
| DPPC | 16.99 ± 0.20 | 0.324 ± 0.002 | 4.0 ± 0.1 | 6.08 ± 0.10 | 0.461 ± 0.004 | 3.23 ± 0.10 | No layer required | 0.80 | ||
| DPPC + 20%UBI | 17.2 ± 0.20 | 0.321 ± 0.001 | 5.19 ± 0.10 | 6.31 ± 0.10 | 0.445 ± 0.003 | 4.1 ± 0.10 | 12.77 ± 0.30 | 0.339 ± 0.003 | 2.12 ± 0.10 | 0.86 |
| DPPE | 19.39 ± 0.30 | 0.305 ± 0.009 | 3.9 ± 0.10 | 5.89 ± 0.10 | 0.480 ± 0.005 | 3.39 ± 0.10 | No layer required | 2.54 | ||
| DPPE + 20%UBI | 19.98 ± 0.40 | 0.327 ± 0.011 | 4.8 ± 0.10 | 6.77 ± 0.10 | 0.433 ± 0.007 | 2.52 ± 0.10 | 13.10 ± 0.40 | 0.342 ± 0.012 | 2.88 ± 0.40 | 2.19 |
| DPPG | 18.5 ± 0.20 | 0.323 ± 0.002 | 4.30 ± 0.10 | 8.37 ± 0.17 | 0.452 ± 0.003 | 2.0 ± 0.10 | No layer required | 1.25 | ||
| DPPG + 20%UBI | 18.91 ± 0.20 | 0.335 ± 0.003 | 4.92 ± 0.10 | 9.14 ± 0.20 | 0.423 ± 0.003 | 1.54 ± 0.10 | 14.95 ± 0.40 | 0.348 ± 0.003 | 1.97 ± 0.20 | 0.52 |
In the presence of 20 mol% of UBI, the reflectivity profile of the monolayer shows a slight shift of the fringe towards a lower value of qz, shown by a dotted line in Fig. 5(a), indicating an increase in overall film thickness. In the process of fitting of the profile, as shown in the schematic of Fig. 5(b), an additional layer below the lipid head group with an electron density value slightly more than that of water was necessary to achieve the best fit. The thickness of this layer is ∼12.76 Å. It indicates the presence of the peptide around the lipid layer. Even though there is no significant change in the size of the head and tail of the lipid, the electron density slightly decreases for both the layers. Such an observation is consistent with the enhanced area per molecule observed in the isotherm data in the presence of the peptide. The overall slight changes in the parameters suggest a weak interaction between the peptide and the lipid. The hydrophobic interactions, hydrogen bonding, and van der Waals forces could be the source of this weak interaction.
The XRR curves of the monolayer of pristine DPPE and DPPE in the presence of 20 mol% of UBI at 30 mN m−1 surface pressure are shown in Fig. 6(a) and the corresponding ED profiles in Fig. 6(b). As evident in the figure, the XRR curve shifts towards the lower qz, which is higher than that of the DPPC lipid in the presence of UBI. This signature can be visualized in the first and second dips in the XRR profile indicated by the vertical dotted lines. It should be noted that even though both the DPPC and DPPE have the same number of C-atoms in the hydrophobic chain, the best fit requires a higher slab size (19.39 Å) for DPPE compared to DPPC (16.99 Å) (Table 1). A DPPC molecule shows a tilt angle of about 30° with the surface normal producing a relatively thinner monolayer compared to a DPPE molecule, which does not have any tilt in its chain assembly.93 The fit parameters in Table 1 indicate that the presence of the UBI causes the overall lipid layer, which is the combination of the thickness of the head and chain, to increase from 25.28 to 26.75 Å. The increment is relatively weaker in DPPC lipid. The peptide also alters the electron density of both the chain and the head group of the lipid. These modified structural parameters of the lipid layer manifest an insertion of the peptide into the lipid monolayer. The third slab corresponding to the presence of the peptide below the lipid layer is found to have a size of 13.10 Å with an electron density of 0.342 e− Å−3. Both parameters are higher in their magnitude compared to the UBI layer below the DPPC monolayer. Therefore, although DPPC and DPPE are both zwitterionic lipids, the DPPE has higher capability to assemble the UBI near the lipid–water interface.
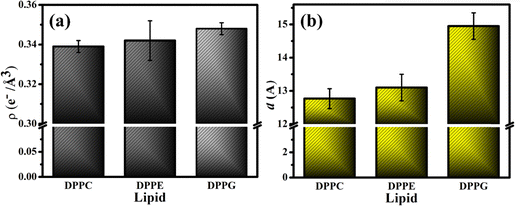
The fitting parameters to the XRR curves of the DPPG lipid are shown in Table 1 which are in good agreement with the earlier studies.19,48 As observed for DPPE, the dips in the curve also move to the left in the presence of UBI (Fig. 6(c)). The overall lipid layer increases from 26.87 to 28.05 Å, which is relatively higher than DPPC but lower than the DPPE. The most effective changes in the DPPG lipid sample are in the parameters related to the slab of the assembled peptide below the lipid layer. As shown by an arrow in the ED profile in Fig. 6(d), a compact layer of the highest density of 0.348 e− Å−3 and thickness of 14.95 Å is observed. These two parameters related to the peptide layer below the lipid head group are plotted in Fig. 7(a) and (b), respectively, to visualize the relative changes among the three lipids.
4. Comments and further discussions
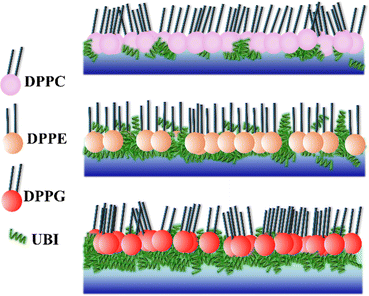
From the three independent measurements described in the above sections, a comparative analysis of the assembly of the peptide and its consequences on the lipid layer can be drawn. The drop in the head group electron density for all the lipids is justified due to enhanced mean molecular area observed in the surface pressure area isotherms. X-ray reflectivity studies on Langmuir monolayers of lipids have reported a decrease in the head group electron density, accompanied by an increase in mean molecular area, after the addition of peptides.94,95 However, the chains of the lipids do not follow this trend. While it remains almost unchanged for DPPC, there is an increase for the lipids DPPE and DPPG. Given the chemical composition and effective molecular volume, the electron density of UBI is lower than the head group of all the lipids but higher than their chains. The XRR data analysis also has established this fact (Table 1). Therefore, the insertion of the peptide well into the DPPE and DPPG lipid layer ensures an increase of average electron density of the chain region. The effect of enhanced mean molecular area is overshadowed by the presence of the peptides into the lipid. A compelling observation is that the rise in chain electron density and the drop in the head group electron density is higher in DPPE than DPPG, hinting at more insertion of the peptide into the PE lipid compared to the PG one. Therefore, in the case of insertion capacity of the peptide, the lipids can be arranged as DPPE > DPPG > DPPC.A remarkable observation is that the assembly of the peptide just below the lipid layer does not follow the exact story of insertion into the lipid layer. As quantified in Fig. 7, the sequence of the strength of compactness and thickness of the peptide layer with respect to the lipids is DPPG > DPPE > DPPC. In the case of DPPG, the positively charged sites of the peptide are strongly attached to the negatively charged head group ensuring many of them to assemble at and around the head group. The hydrophobic interaction may not be capable of overpowering this electrostatic force to bring all the peptides into the chain region. Of course, some peptides would insert into the chain due to abundant availability near the lipid layer. This sequence of lipids to assemble the peptide layer is amiably supported by the quantified surface potential. At the surface pressure of 30 mN m−1 at which the XRR measurements are done, all the lipids are in the liquid condensed phase. In this phase, at the surface area of 60 Å2, the increase in potential is found to be 67% for DPPG (241.6 to 403.7 mV) with the number 51% and 16% for DPPE (420.6 to 635.98 mV) and DPPC (479.0 to 560 mV), respectively. Following these understandings, the lipid monolayers, and the assembly of the peptide in and around the layer is schematically depicted in Fig. 8.
The presence of the UBI peptide causes the in-plane compressional modulus to drop for all the lipids.79–81,96 However, the drop is the least for the DPPG lipid compared to the other two. As depicted in Fig. 8, the most compact peptide layer below the DPPG layer and the inserted peptides into this lipid layer probably make it more rigid compared to the other two lipids when peptides are around them. There are more peptides inserted into the DPPE compared to the DPPC and hence the drop in the modulus is less in DPPE. Even though there are peptides below the head group of DPPE, they are not as compact as the DPPG. Hence, the compressional modulus plotted in Fig. 3(b) can be realized in the phenomenon of UBI–lipid interaction established from the XRR data.
Researchers have utilized a variety of experimental methods to explore the interactions between an AMP and a membrane. While isothermal titration calorimetry (ITC) provides the binding of the peptide with the membrane,97–101 dynamic light scattering (DLS) investigates how AMPs affect the size of a liposome.101,102 Other techniques, such as fluorescence correlation spectroscopy, quasi-elastic neutron scattering (QENS),103,104 and nuclear magnetic resonance (NMR),105,106 have been used to investigate how AMPs affect membrane dynamics. In the present study, a further step has been taken to shed more lights on the assembly and insertion of the UBI peptide, which is selective to specific lipids.
While the major component of outer leaflet of the mammalian cell membrane is the zwitterionic PC, it is the negatively charged PG that primarily composes the bacterial cell membrane.107 It has been established earlier35,108 and quantified in the present study that the UBI peptide can differentiate between these two membranes effectively. As stated above, the bacterial membrane may attract the positively charged UBI by the long range electrostatic interaction organizing the peptide near the membrane.109,110 Once the peptides are available near the membrane, the PE lipid, which is another major component in the bacterial membrane, probably pulls the peptides into the hydrophobic core of the membrane to generate various conformational assemblies of the peptides. These combined effects of the PG and PE on UBI are likely the reason that the peptide is toxic to a bacterium but not to humans.
The results reveal that UBI selectively interacts with simplified, single-component membranes. While this model is valuable for understanding the specific role of individual lipids in interacting with an AMP, it does not fully mimic a real bacterial membrane. Actual membranes are chemically complex comprising various lipids, transmembrane proteins and other macromolecules. Studying such a complex system would provide more physiologically relevant insights and could be explored in future research.
Other factors, including the peptide length, could control the peptide–membrane interaction and corresponding assembly of peptides. Therefore, future experiments need to be designed with varying peptide lengths, as this determines the exact conformation, such as coil and helix, in a membrane environment.35 Further understanding could be achieved by monitoring membrane permeability using a dye leakage assay and observing pore formation using a bio-AFM. High resolution bio-AFM could clearly show how a peptide perturbs the membrane organization in various ways, such as formation of lipid–peptide domains inducing nanoholes.111 Furthermore, small angle X-ray scattering (SAXS) from unilamellar and multilamellar vesicles could provide detailed membrane structure and the exact location of the peptide in a lipid bilayer. Diffuse X-ray scattering could quantify the bulk and bending moduli of the membrane. The physical parameters of all these planned investigations will be the subjects of future communications.
5. Conclusions
In this study, multiple biophysics techniques have been utilized to investigate the interaction and assembly of a fraction of a ubiquicidin derived peptide, UBI (29–41). The surface pressure–area isotherms of the lipid monolayer formed at the air–water interface show that the mean molecular area for zwitterionic and anionic lipids is enhanced in the presence of this peptide. Consequently, the in-plane compressional modulus of these lipids decreases. The peptide causes the lipid monolayer to enhance the surface potential. This study also quantified the exact effect of the peptide on the self-assembly of lipids in the monolayer and the corresponding assembly of the peptide in and around the lipid layer. When it comes to peptide insertion, the PE is found to be most effective, while PC shows the least effect. However, the negatively charged PG has the highest propensity to form an assembly of the peptide near the lipid layer. The long-range attraction of PG lipids to the cationic peptides assembles the peptides near a membrane, and then, successively the PE lipids insert them into the membrane to form various self-assembled structures of the peptides that exhibit adverse effects on bacteria.Data availability
The data supporting this article have been included in the main text of the article.Conflicts of interest
There are no conflicts to declare.Acknowledgements
The authors are thankful to Board of Research in Nuclear Sciences (BRNS), Department of Atomic Energy (DAE), Govt. of India for the financial support to conduct the research through a project with sanction number 58/14/13/2022-BRNS/37059. Financial support by the Department of Science & Technology (Government of India) provided within the framework of the India@DESY collaboration to perform the X-ray scattering experiment at PETRA III is greatly acknowledged. We also thank the beamline staff at P08 of PETRA III for their support during the beamtime. Special thanks to Dr Bridget M. Murphy, Dr Jyotsna Bhatt Mitra, Dr Anupam Bandyopadhyay and Mr Nicolas Hayen for useful scientific discussions.References
- J. Lederberg, Infectious History, Science, 2000, 288(5464), 287–293, DOI:10.1126/science.288.5464.287.
- E. N. Frigini, R. D. Porasso, T. Beke-Somfai, J. J. López Cascales, R. D. Enriz and S. Pantano, The Mechanism of Antimicrobial Small-Cationic Peptides from Coarse-Grained Simulations, J. Chem. Inf. Model., 2023, 63(21), 6877–6889, DOI:10.1021/acs.jcim.3c01348.
- U. Theuretzbacher, Accelerating Resistance, Inadequate Antibacterial Drug Pipelines and International Responses, Int. J. Antimicrob. Agents, 2012, 39(4), 295–299 CrossRef CAS PubMed.
- D. J. Payne, M. N. Gwynn, D. J. Holmes and D. L. Pompliano, Drugs for Bad Bugs: Confronting the Challenges of Antibacterial Discovery, Nat. Rev. Drug Discovery, 2007, 6(1), 29–40 CrossRef CAS.
- L. L. Silver, Challenges of Antibacterial Discovery, Clin. Microbiol. Rev., 2011, 24(1), 71–109, DOI:10.1128/CMR.00030-10.
- E. Leung, D. E. Weil, M. Raviglione and H. Nakatani, The WHO Policy Package to Combat Antimicrobial Resistance, Bull. W. H. O., 2011, 89(5), 390–392, DOI:10.2471/BLT.11.088435.
- C. L. Bevins and M. Zasloff, PEPTIDES FROM FROG SKIN, Annu. Rev. Biochem., 1990, 59(1), 395–414, DOI:10.1146/annurev.bi.59.070190.002143.
- H. G. Boman, Peptide Antibiotics and Their Role in Innate Immunity, Annu. Rev. Immunol., 1995, 13(1), 61–92, DOI:10.1146/annurev.iy.13.040195.000425.
- H. G. Boman, Antibacterial Peptides: Key Components Needed in Immunity, Cell, 1991, 65(2), 205–207 CrossRef CAS.
- Z. Oren and Y. Shai, Mode of Action of Linear Amphipathic Alpha-Helical Antimicrobial Peptides, Biopolymers, 1998, 47(6), 451–463, DOI:10.1002/(sici)1097-0282(1998)47:6<451::aid-bip4>3.0.co;2-f.
- D. Andreu and L. Rivas, Animal Antimicrobial Peptides: An Overview, Biopolymers, 1998, 47(6), 415–433, DOI:10.1002/(SICI)1097-0282(1998)47:6<415::AID-BIP2>3.0.CO;2-D.
- R. E. Hancock, Peptide Antibiotics, Lancet, 1997, 349(9049), 418–422 CrossRef CAS PubMed.
- K. T. Miyasaki and R. I. Lehrer, β-Sheet Antibiotic Peptides as Potential Dental Therapeutics, Int. J. Antimicrob. Agents, 1998, 9(4), 269–280 CrossRef CAS PubMed.
- J. B. Mitra, V. K. Sharma, A. Dash, A. Mukherjee and M. Kumar, Deciphering Interaction of UBI (23-33) with Model Bacterial and Mammalian Membranes, AIP Conf. Proc., 2020, 2265(1), 030026 CrossRef CAS.
- R. I. Lehrer and T. Ganz, Antimicrobial Peptides in Mammalian and Insect Host Defence, Curr. Opin. Immunol., 1999, 11(1), 23–27 CrossRef CAS PubMed.
- R. E. W. Hancock and D. S. Chapple, Peptide Antibiotics, Antimicrob. Agents Chemother., 1999, 43(6), 1317–1323, DOI:10.1128/AAC.43.6.1317.
- B. Rivas-Santiago, C. J. Serrano and J. A. Enciso-Moreno, Susceptibility to Infectious Diseases Based on Antimicrobial Peptide Production, Infect. Immun., 2009, 77(11), 4690–4695, DOI:10.1128/IAI.01515-08.
- O. Barak, J. R. Treat and W. D. James, Antimicrobial Peptides: Effectors of Innate Immunity in the Skin, Adv. Dermatol., 2005, 21, 357–374 CrossRef PubMed.
- F. Neville, Y. Ishitsuka, C. S. Hodges, O. Konovalov, A. J. Waring, R. Lehrer, K. Y. C. Lee and D. Gidalevitz, Protegrin Interaction with Lipid Monolayers: Grazing Incidence X-Ray Diffraction and X-Ray Reflectivity Study, Soft Matter, 2008, 4(8), 1665–1674 RSC.
- H. Steiner, D. Hultmark, Å. Engström, H. Bennich and H. G. Boman, Sequence and Specificity of Two Antibacterial Proteins Involved in Insect Immunity, Nature, 1981, 292(5820), 246–248 CrossRef CAS PubMed.
- D. A. Devine and R. E. Hancock, Mammalian Host Defense Peptides, Cambridge University Press, 2004, vol. 6 Search PubMed.
- G. Wang, Human Antimicrobial Peptides and Proteins, Pharmaceuticals, 2014, 7(5), 545–594 CrossRef CAS.
- C. Montis, E. Marelli, F. Valle, F. B. Bombelli and C. Pigliacelli, Engineering the Interaction of Short Antimicrobial Peptides with Bacterial Barriers, Mol. Syst. Des. Eng., 2024, 9(6), 541–560 RSC.
- S. Chou, H. Guo, F. G. Zingl, S. Zhang, J. Toska, B. Xu, Y. Chen, P. Chen, M. K. Waldor and W. Zhao, Synthetic Peptides That Form Nanostructured Micelles Have Potent Antibiotic and Antibiofilm Activity against Polymicrobial Infections, Proc. Natl. Acad. Sci. U. S. A., 2023, 120(4), e2219679120 CrossRef CAS.
- M. Akbarian, A. Khani, S. Eghbalpour and V. N. Uversky, Bioactive Peptides: Synthesis, Sources, Applications, and Proposed Mechanisms of Action, Int. J. Mol. Sci., 2022, 23(3), 1445 CrossRef CAS PubMed.
- G. Idiong, A. Won, A. Ruscito, B. O. Leung, A. P. Hitchcock and A. Ianoul, Investigating the Effect of a Single Glycine to Alanine Substitution on Interactions of Antimicrobial Peptide Latarcin 2a with a Lipid Membrane, Eur. Biophys. J., 2011, 40, 1087–1100 CrossRef CAS.
- J. Lei, L. Sun, S. Huang, C. Zhu, P. Li, J. He, V. Mackey, D. H. Coy and Q. He, The Antimicrobial Peptides and Their Potential Clinical Applications, Am. J. Transl. Res., 2019, 11(7), 3919 CAS.
- K. Lohner and E. J. Prenner, Differential Scanning Calorimetry and X-Ray Diffraction Studies of the Specificity of the Interaction of Antimicrobial Peptides with Membrane-Mimetic Systems, Biochim. Biophys. Acta, Biomembr., 1999, 1462(1–2), 141–156 CrossRef CAS PubMed.
- A. Rothnie, D. Theron, L. Soceneantu, C. Martin, M. Traikia, G. Berridge, C. F. Higgins, P. F. Devaux and R. Callaghan, The Importance of Cholesterol in Maintenance of P-Glycoprotein Activity and Its Membrane Perturbing Influence, Eur. Biophys. J., 2001, 30, 430–442 CrossRef CAS.
- B. P. Navas, K. Lohner, G. Deutsch, E. Sevcsik, K. A. Riske, R. Dimova, P. Garidel and G. Pabst, Composition Dependence of Vesicle Morphology and Mixing Properties in a Bacterial Model Membrane System, Biochim. Biophys. Acta, Biomembr., 2005, 1716(1), 40–48 CrossRef.
- S. L. Keller, W. H. Pitcher, W. H. Huestis and H. M. McConnell, Red Blood Cell Lipids Form Immiscible Liquids, Phys. Rev. Lett., 1998, 81(22), 5019–5022, DOI:10.1103/PhysRevLett.81.5019.
- G. van Meer and A. I. de Kroon, Lipid Map of the Mammalian Cell, J. Cell Sci., 2011, 124(1), 5–8 CrossRef CAS PubMed.
- D. C. White and F. E. Frerman, Extraction, Characterization, and Cellular Localization of the Lipids of Staphylococcus Aureus, J. Bacteriol., 1967, 94(6), 1854–1867, DOI:10.1128/jb.94.6.1854-1867.1967.
- C. Sohlenkamp and O. Geiger, Bacterial Membrane Lipids: Diversity in Structures and Pathways, FEMS Microbiol. Rev., 2016, 40(1), 133–159 CrossRef CAS PubMed.
- J. Bhatt Mitra, V. K. Sharma, A. Mukherjee, V. Garcia Sakai, A. Dash and M. Kumar, Ubiquicidin Derived Peptides Selectively Interact with the Anionic Phospholipid Membrane, Langmuir, 2019, 36, 397–408, DOI:10.1021/acs.langmuir.9b03243.
- P. S. Hiemstra, M. T. van den Barselaar, M. Roest, P. H. Nibbering and R. van Furth, Ubiquicidin, a Novel Murine Microbicidal Protein Present in the Cytosolic Fraction of Macrophages, J. Leukocyte Biol., 1999, 66(3), 423–428 CrossRef CAS.
- M. Tollin, P. Bergman, T. Svenberg, H. Jörnvall, G. H. Gudmundsson and B. Agerberth, Antimicrobial Peptides in the First Line Defence of Human Colon Mucosa, Peptides, 2003, 24(4), 523–530 CrossRef CAS.
- C. P. Brouwer, S. J. Bogaards, M. Wulferink, M. P. Velders and M. M. Welling, Synthetic Peptides Derived from Human Antimicrobial Peptide Ubiquicidin Accumulate at Sites of Infections and Eradicate (Multi-Drug Resistant) Staphylococcus Aureus in Mice, Peptides, 2006, 27(11), 2585–2591 CrossRef CAS.
- S. M. Ferreira, H. C. Carneiro, R. B. Alves, A. C. S. Batista, E. N. da Silva Junior, G. G. Dias, J. M. Resende, D. A. Santos, D. L. Oliveira and M. L. Rodrigues, A UBI 31-38 Peptide-Coumarin Conjugate: Photophysical Features, Imaging Tracking and Synergism with Amphotericin B Against Cryptococcus, Curr. Top. Med. Chem., 2018, 18(2), 157–163 CrossRef CAS PubMed.
- M. M. Welling, A. Lupetti, H. S. Balter, S. Lanzzeri, B. Souto, A. M. Rey, E. O. Savio, A. Paulusma-Annema, E. K. Pauwels and P. H. Nibbering, 99mTc-Labeled Antimicrobial Peptides for Detection of Bacterial and Candida Albicans Infections, J. Nucl. Med., 2001, 42(5), 788–794 CAS.
- M. S. Akhtar, A. Qaisar, J. Irfanullah, J. Iqbal, B. Khan, M. Jehangir, M. A. Nadeem, M. A. Khan, M. S. Afzal and M. B. Imran, Antimicrobial Peptide 99mTc-Ubiquicidin 29–41 as Human Infection-Imaging Agent: Clinical Trial, J. Nucl. Med., 2005, 46(4), 567–573 CAS.
- D. Beiki, G. Yousefi, B. Fallahi, M. Tahmasebi, A. Gholamrezanezhad, A. Fard-Esfahani, M. Erfani and M. Eftekhari, Tc-Ubiquicidin [29–41], a Promising Radiopharmaceutical to Differentiate Orthopedic Implant Infections from Sterile Inflammation, Iran. J. Pharm. Res. IJPR, 2013, 12, 347–353 CAS.
- P. Hitaishi, P. Mandal and S. Ghosh, Partitioning of a Hybrid Lipid in Domains of Saturated and Unsaturated Lipids in a Model Cellular Membrane, ACS Omega, 2021, 6(50), 34546–34554 CrossRef CAS PubMed.
- R. Gupta, S. Mitra, S. Chowdhury, G. Das, R. Priyadarshini, M. K. Mukhopadhyay and S. K. Ghosh, Discerning Perturbed Assembly of Lipids in a Model Membrane in Presence of Violacein, Biochim. Biophys. Acta, Biomembr., 2021, 1863(9), 183647, DOI:10.1016/j.bbamem.2021.183647.
- L. B. Dreier, C. Bernhard, G. Gonella, E. H. G. Backus and M. Bonn, Surface Potential of a Planar Charged Lipid–Water Interface. What Do Vibrating Plate Methods, Second Harmonic and Sum Frequency Measure?, J. Phys. Chem. Lett., 2018, 9(19), 5685–5691, DOI:10.1021/acs.jpclett.8b02093.
- E. E. Ambroggio, F. Separovic, J. Bowie and G. D. Fidelio, Surface Behaviour and Peptide–Lipid Interactions of the Antibiotic Peptides, Maculatin and Citropin, Biochim. Biophys. Acta, 2004, 1664(1), 31–37, DOI:10.1016/j.bbamem.2004.03.013.
- H. Brockman, Dipole Potential of Lipid Membranes, Chem. Phys. Lipids, 1994, 73(1–2), 57–79 CrossRef CAS.
- P. Mandal, R. Giri, B. Murphy and S. Ghosh, Self-Assembly of Graphene Oxide Nanoflakes in a Lipid Monolayer at the Air–Water Interface, ACS Appl. Mater. Interfaces, 2021, 13, DOI:10.1021/acsami.1c19004.
- B. Murphy, M. Greve, B. Runge, C. Koops, A. Elsen, J. Stettner, O. Seeck and O. Magnussen, A Novel X-Ray Diffractometer for Studies of Liquid-Liquid Interfaces, J. Synchrotron Radiat., 2014, 21, 45–56, DOI:10.1107/S1600577513026192.
- L. G. Parratt, Surface Studies of Solids by Total Reflection of X-Rays, Phys. Rev., 1954, 95(2), 359–369, DOI:10.1103/PhysRev.95.359.
- J. Als-Nielsen and D. McMorrow, Elements of Modern X-Ray Physics, John Wiley & Sons, 2011 Search PubMed.
- J. Als-Nielsen, Elements of Modern X-Ray Physics, 2002, vol. 55 DOI:10.1002/9781119998365.
- R. Gupta, V. K. Sharma, J. Gupta and S. K. Ghosh, 1, 3 Dialkylated Imidazolium Ionic Liquid Causes Interdigitated Domains in a Phospholipid Membrane, Langmuir, 2022, 38(11), 3412–3421 CrossRef CAS PubMed.
- P. Mandal, G. Bhattacharya, A. Bhattacharyya, S. S. Roy and S. K. Ghosh, Unravelling the Structural Changes of Phospholipid Membranes in Presence of Graphene Oxide, Appl. Surf. Sci., 2021, 539, 148252 CrossRef CAS.
- E. Hermans and J. Vermant, Interfacial Shear Rheology of DPPC under Physiologically Relevant Conditions, Soft Matter, 2014, 10(1), 175–186 RSC.
- J. C. Stephani, L. Gerhards, B. Khairalla, I. A. Solov’yov and I. Brand, How Do Antimicrobial Peptides Interact with the Outer Membrane of Gram-negative Bacteria? Role of Lipopolysaccharides in Peptide Binding, Anchoring, and Penetration, ACS Infect. Dis., 2024, 10(2), 763–778 CrossRef CAS PubMed.
- M. D. Phan, K. Y. Lee, J. Lee, S. K. Satija and K. Shin, The Effect of Cholesterol on Membrane Binding and Self-Assembly of Collagen Fibrils, Langmuir, 2020, 36(26), 7259–7267, DOI:10.1021/acs.langmuir.0c00574.
- G. Sharma, A. Seth, R. P. Giri, N. Hayen, B. M. Murphy and S. K. Ghosh, Ionic Liquid-Induced Assembly of DNA at Air–Water Interface, Langmuir, 2023, 39(45), 16079–16089, DOI:10.1021/acs.langmuir.3c02212.
- M.-J. Hwang and K. Kim, Poly(Ethylenimine) as a Subphase Stabilizer of Stearic Acid Monolayers at the Air/Water Interface: Surface Pressure–Area Isotherm and Infrared Spectroscopy Study, Langmuir, 1999, 15(10), 3563–3569, DOI:10.1021/la9804029.
- G. Bhattacharya, S. Mitra, P. Mandal, S. Dutta, R. Giri and S. Ghosh, Thermodynamics of Interaction of Ionic Liquids with Lipid Monolayer, Biophys. Rev., 2018, 10, DOI:10.1007/s12551-017-0390-3.
- R. P. Giri, A. Chakrabarti and M. K. Mukhopadhyay, Cholesterol-Induced Structural Changes in Saturated Phospholipid Model Membranes Revealed through X-Ray Scattering Technique, J. Phys. Chem. B, 2017, 121(16), 4081–4090, DOI:10.1021/acs.jpcb.6b12587.
- J. R. Silvius, Thermotropic Phase Transitions of Pure Lipids in Model Membranes and Their Modifications by Membrane Proteins, Lipid-Protein Interact., 1982, 2, 239–281 CAS.
- V. P. Geraldo, F. J. Pavinatto, T. M. Nobre, L. Caseli and O. N. Oliveira Jr, Langmuir Films Containing Ibuprofen and Phospholipids, Chem. Phys. Lett., 2013, 559, 99–106 CrossRef CAS.
- R. E. Hancock and H.-G. Sahl, Antimicrobial and Host-Defense Peptides as New Anti-Infective Therapeutic Strategies, Nat. Biotechnol., 2006, 24(12), 1551–1557 CrossRef CAS PubMed.
- L. Thøgersen, B. Schiøtt, T. Vosegaard, N. C. Nielsen and E. Tajkhorshid, Peptide Aggregation and Pore Formation in a Lipid Bilayer: A Combined Coarse-Grained and All Atom Molecular Dynamics Study, Biophys. J., 2008, 95(9), 4337–4347 CrossRef.
- D. Argudo, N. Bethel, F. Marcoline and M. Grabe, New Approaches Bridg. Comput. Exp. Membr. Proteins, Biochim. Biophys. Acta, Biomembr., 1858, 1619–1634 Search PubMed.
- E. Guzmán, L. Liggieri, E. Santini, M. Ferrari and F. Ravera, Mixed DPPC–Cholesterol Langmuir Monolayers in Presence of Hydrophilic Silica Nanoparticles, Colloids Surf., B, 2013, 105, 284–293, DOI:10.1016/j.colsurfb.2013.01.020.
- A. Benedetto, Ionic Liquids Meet Lipid Bilayers: A State-of-the-Art Review, Biophys. Rev., 2024, 15, DOI:10.1007/s12551-023-01173-3.
- E. Guzmán, L. Liggieri, E. Santini, M. Ferrari and F. Ravera, DPPC–DOPC Langmuir Monolayers Modified by Hydrophilic Silica Nanoparticles: Phase Behaviour, Structure and Rheology, Colloids Surf., A, 2012, 413, 174–183, DOI:10.1016/j.colsurfa.2011.12.059.
- A. Aroti, E. Leontidis, E. Maltseva and G. Brezesinski, Effects of Hofmeister Anions on DPPC Langmuir Monolayers at the Air–Water Interface, J. Phys. Chem. B, 2004, 108(39), 15238–15245, DOI:10.1021/jp0481512.
- T. Kamilya, P. Pal and G. B. Talapatra, Interaction of Ovalbumin with Phospholipids Langmuir–Blodgett Film, J. Phys. Chem. B, 2007, 111(5), 1199–1205, DOI:10.1021/jp063377l.
- C. E. Miller, D. D. Busath, B. Strongin and J. Majewski, Integration of Ganglioside GT1b Receptor into DPPE and DPPC Phospholipid Monolayers: An X-Ray Reflectivity and Grazing-Incidence Diffraction Study, Biophys. J., 2008, 95(7), 3278–3286 CrossRef CAS PubMed.
- E. Madrid and S. L. Horswell, Effect of Headgroup on the Physicochemical Properties of Phospholipid Bilayers in Electric Fields: Size Matters, Langmuir, 2013, 29(5), 1695–1708, DOI:10.1021/la304455d.
- D. Vollhardt, V. B. Fainerman and S. Siegel, Thermodynamic and Textural Characterization of DPPG Phospholipid Monolayers, J. Phys. Chem. B, 2000, 104(17), 4115–4121, DOI:10.1021/jp992529s.
- J. Li; Q. He and X. Yan, Molecular Assembly of Biomimetic Systems, John Wiley & Sons, 2010 Search PubMed.
- W. M. Gelbart; A. Ben-Shaul and D. Roux, Micelles, Membranes, Microemulsions, and Monolayers, Springer Science & Business Media, 2012 Search PubMed.
- W. Liu, Z. Wang, L. Fu, R. M. Leblanc and E. C. Y. Yan, Lipid Compositions Modulate Fluidity and Stability of Bilayers: Characterization by Surface Pressure and Sum Frequency Generation Spectroscopy, Langmuir, 2013, 29(48), 15022–15031, DOI:10.1021/la4036453.
- R. C. MacDonald and R. I. MacDonald, Membrane Surface Pressure Can Account for Differential Activities of Membrane-Penetrating Molecules, J. Biol. Chem., 1988, 263(21), 10052–10055 CrossRef CAS PubMed.
- R. Krivanek, P. Rybar, E. J. Prenner, R. N. McElhaney and T. Hianik, Interaction of the Antimicrobial Peptide Gramicidin S with Dimyristoyl–Phosphatidylcholine Bilayer Membranes: A Densitometry and Sound Velocimetry Study, Biochim. Biophys. Acta, Biomembr., 2001, 1510(1–2), 452–463 CrossRef CAS PubMed.
- R. Maget-Dana, The Monolayer Technique: A Potent Tool for Studying the Interfacial Properties of Antimicrobial and Membrane-Lytic Peptides and Their Interactions with Lipid Membranes, Biochim. Biophys. Acta, Biomembr., 1999, 1462(1–2), 109–140 CrossRef CAS PubMed.
- Z. Leonenko, E. Finot, H. Ma, T. Dahms and D. Cramb, Investigation of Temperature-Induced Phase Transitions in DOPC and DPPC Phospholipid Bilayers Using Temperature-Controlled Scanning Force Microscopy, Biophys. J., 2004, 86(6), 3783–3793 CrossRef CAS PubMed.
- E. N. Frigini, R. D. Porasso, T. Beke-Somfai, J. J. López Cascales, R. D. Enriz and S. Pantano, The Mechanism of Antimicrobial Small-Cationic Peptides from Coarse-Grained Simulations, J. Chem. Inf. Model., 2023, 63(21), 6877–6889, DOI:10.1021/acs.jcim.3c01348.
- M. R. Yeaman and N. Y. Yount, Mechanisms of Antimicrobial Peptide Action and Resistance, Pharmacol. Rev., 2003, 55(1), 27–55 CrossRef CAS.
- K. Matsuzaki, O. Murase, N. Fujii and K. Miyajima, An Antimicrobial Peptide, Magainin 2, Induced Rapid Flip-Flop of Phospholipids Coupled with Pore Formation and Peptide Translocation, Biochemistry, 1996, 35(35), 11361–11368 CrossRef CAS.
- H. W. Huang, Action of Antimicrobial Peptides: Two-State Model, Biochemistry, 2000, 39(29), 8347–8352 CrossRef CAS PubMed.
- S. McLaughlin, The Electrostatic Properties of Membranes, Annu. Rev. Biophys. Biophys. Chem., 1989, 18(1), 113–136, DOI:10.1146/annurev.bb.18.060189.000553.
- V. Vogel and D. Möbius, Local Surface Potentials and Electric Dipole Moments of Lipid Monolayers: Contributions of the Water/Lipid and the Lipid/Air Interfaces, J. Colloid Interface Sci., 1988, 126(2), 408–420 CrossRef CAS.
- D. M. Taylor, O. N. De Oliveira Jr and H. Morgan, Models for Interpreting Surface Potential Measurements and Their Application to Phospholipid Monolayers, J. Colloid Interface Sci., 1990, 139(2), 508–518 CrossRef CAS.
- A. A. Hidalgo, W. Caetano, M. Tabak and O. N. Oliveira Jr, Interaction of Two Phenothiazine Derivatives with Phospholipid Monolayers, Biophys. Chem., 2004, 109(1), 85–104 CrossRef CAS PubMed.
- H. M. Sauro and B. Ingalls, Conservation Analysis in Biochemical Networks: Computational Issues for Software Writers, Biophys. Chem., 2004, 109(1), 1–15 CrossRef CAS PubMed.
- G. P. Borissevitch, M. Tabak and O. N. Oliveira, Interaction of Dipyridamole with Lipids in Mixed Langmuir Monolayers, Biochim. Biophys. Acta, Biomembr., 1996, 1278(1), 12–18 CrossRef.
- S. Mitra, V. K. Sharma, J. B. Mitra, S. Chowdhury, M. K. Mukhopadhyay, R. Mukhopadhyay and S. K. Ghosh, Thermodynamics and Structure of Model Bio-Membrane of Liver Lipids in Presence of Imidazolium-Based Ionic Liquids, Biochim. Biophys. Acta, Biomembr., 2021, 1863(6), 183589 CrossRef CAS PubMed.
- C. Stefaniu and G. Brezesinski, Grazing Incidence X-Ray Diffraction Studies of Condensed Double-Chain Phospholipid Monolayers Formed at the Soft Air/Water Interface, Adv. Colloid Interface Sci., 2014, 207, 265–279 CrossRef CAS PubMed.
- D. Gidalevitz, Y. Ishitsuka, A. S. Muresan, O. Konovalov, A. J. Waring, R. I. Lehrer and K. Y. C. Lee, Interaction of Antimicrobial Peptide Protegrin with Biomembranes, Proc. Natl. Acad. Sci. U. S. A., 2003, 100(11), 6302–6307 CrossRef CAS PubMed.
- Š. Málková, F. Long, R. V. Stahelin, S. V. Pingali, D. Murray, W. Cho and M. L. Schlossman, X-Ray Reflectivity Studies of cPLA2α-C2 Domains Adsorbed onto Langmuir Monolayers of SOPC, Biophys. J., 2005, 89(3), 1861–1873 CrossRef PubMed.
- A. B. López-Oyama, P. Taboada, M. G. Burboa, E. Rodríguez, V. Mosquera and M. A. Valdez, Interaction of the Cationic Peptide Bactenecin with Mixed Phospholipid Monolayers at the Air–Water Interface, J. Colloid Interface Sci., 2011, 359(1), 279–288 CrossRef.
- T. Wieprecht and J. Seelig, Isothermal Titration Calorimetry for Studying Interactions between Peptides and Lipid Membranes, Curr. Top. Membr., 2002, 52, 31–56 CAS.
- T. Guide ITC Data Analysis in Origin®.
- L. Lombardi, M. I. Stellato, R. Oliva, A. Falanga, M. Galdiero, L. Petraccone, G. D’Errico, A. De Santis, S. Galdiero and P. Del Vecchio, Antimicrobial Peptides at Work: Interaction of Myxinidin and Its Mutant WMR with Lipid Bilayers Mimicking the P. Aeruginosa and E. Coli Membranes, Sci. Rep., 2017, 7(1), 44425 CrossRef CAS PubMed.
- R. Oliva, A. Emendato, G. Vitiello, A. De Santis, M. Grimaldi, A. M. D’Ursi, E. Busi, P. Del Vecchio, L. Petraccone and G. D’Errico, On the Microscopic and Mesoscopic Perturbations of Lipid Bilayers upon Interaction with the MPER Domain of the HIV Glycoprotein Gp41, Biochim. Biophys. Acta, Biomembr., 2016, 1858(8), 1904–1913 CrossRef CAS PubMed.
- T. M. Domingues, B. Mattei, J. Seelig, K. R. Perez, A. Miranda and K. A. Riske, Interaction of the Antimicrobial Peptide Gomesin with Model Membranes: A Calorimetric Study, Langmuir, 2013, 29(27), 8609–8618, DOI:10.1021/la401596s.
- P. Wadhwani, J. Reichert, J. Bürck and A. S. Ulrich, Antimicrobial and Cell-Penetrating Peptides Induce Lipid Vesicle Fusion by Folding and Aggregation, Eur. Biophys. J., 2012, 41(2), 177–187, DOI:10.1007/s00249-011-0771-7.
- V. K. Sharma, E. Mamontov, M. Tyagi and V. S. Urban, Effect of α-Tocopherol on the Microscopic Dynamics of Dimyristoylphosphatidylcholine Membrane, J. Phys. Chem. B, 2016, 120(1), 154–163, DOI:10.1021/acs.jpcb.5b10417.
- W. T. Heller, A. J. Waring, R. I. Lehrer and H. W. Huang, Multiple States of β-Sheet Peptide Protegrin in Lipid Bilayers, Biochemistry, 1998, 37(49), 17331–17338, DOI:10.1021/bi981314q.
- V. K. Sharma, E. Mamontov, D. Anunciado, H. O’Neill and V. Urban, Effect of Antimicrobial Peptide on the Dynamics of Phosphocholine Membrane: Role of Cholesterol and Physical State of Bilayer, Soft Matter, 2015, 11, 10.1039/C5SM01562F.
- V. K. Sharma, H. Srinivasan, S. Mitra, V. Garcia-Sakai and R. Mukhopadhyay, Effects of Hydrotropic Salt on the Nanoscopic Dynamics of DTAB Micelles, J. Phys. Chem. B, 2017, 121(22), 5562–5572, DOI:10.1021/acs.jpcb.7b02976.
- H. Strahl and J. Errington, Bacterial Membranes: Structure, Domains, and Function, Annu. Rev. Microbiol., 2017, 71(1), 519–538, DOI:10.1146/annurev-micro-102215-095630.
- O. Parravicini, C. Somlai, S. A. Andujar, A. D. Garro, B. Lima, A. Tapia, G. Feresin, A. Perczel, G. Tóth, J. L. Cascales, A. M. Rodríguez and R. D. Enriz, Small Peptides Derived from Penetratin as Antibacterial Agents, Arch. Pharm., 2016, 349(4), 242–251, DOI:10.1002/ardp.201500419.
- Y. Jiang and J. Zhang, Current Status of and Perspectives on Radiolabelled Ubiquicidin 29–41 Derivatives for Bacterial Infection Imaging, Mini-Rev. Med. Chem., 2023, 23, 1500–1506, DOI:10.2174/1389557523666230131100654.
- M. M. Welling, A. Paulusma-Annema, H. S. Balter, E. K. Pauwels and P. H. Nibbering, Technetium-99m Labelled Antimicrobial Peptides Discriminate between Bacterial Infections and Sterile Inflammations, Eur. J. Nucl. Med., 2000, 27, 292–301 CrossRef CAS PubMed.
- R. Brasseur, M. Deleu, M. Mingeot-Leclercq, G. Francius and Y. F. Dufrêne, Probing Peptide–Membrane Interactions Using AFM, Surf. Interface Anal., 2008, 40(3–4), 151–156, DOI:10.1002/sia.2682.
Footnotes |
| † Present address: Institut für Experimentelle und Angewandte Physik, Christian-Albrechts-Universität Zu Kiel, 24098 Kiel, Germany. |
| ‡ Present address: Department of Physics, Indian Institute of Technology (ISM) Dhanbad, Jharkhand 826004, India. |
| This journal is © The Royal Society of Chemistry 2024 |