Open Access Article
Open Access ArticleLaccase-mediated degradation of emerging contaminants: unveiling a sustainable solution
Pooja
Thathola
 *ab,
Elda M.
Melchor-Martínez
cd,
Priyanka
Adhikari
e,
Saúl Antonio
Hernández Martínez
d,
Anita
Pandey
f and
Roberto
Parra-Saldívar
*ab,
Elda M.
Melchor-Martínez
cd,
Priyanka
Adhikari
e,
Saúl Antonio
Hernández Martínez
d,
Anita
Pandey
f and
Roberto
Parra-Saldívar
 cdg
cdg
aCenter for Land and Water Resource Management, Govind Ballabh Pant National Institute of Himalayan Environment, Kosi-Katarmal 263643, Almora, Uttarakhand, India. E-mail: poojathathola29@gmail.com
bAnalytical and Environmental Science Division and Centralized Instrument Facility, CSIR-Center Salt and Marine Chemicals Research Insititute, Bhavnagar 364002, Gujarat, India
cInstitute of Advanced Materials for Sustainable Manufacturing, Tecnologico de Monterrey, Monterrey 64849, Mexico
dSchool of Engineering and Sciences, Tecnologico de Monterrey, Monterrey 64849, Mexico
eCenter for GMP Extraction Facility (Dept. of Biotechnology), National Institute of Pharmaceutical Education and Research, Changsari 781101, Guwahati, Assam, India
fDepartment of Biotechnology, Graphic Era Deemed to be University, Dehradun 248002, Uttarakhand, India
gFacultad de Agronomía, Universidad Autónoma de Nuevo León, Francisco Villa, 66059 Colonia Ex hacienda El Canadá, General Escobedo, Nuevo León, Mexico
First published on 9th September 2024
Abstract
The excessive use of emerging contaminants (ECs) in various applications has led to a global health crisis. ECs are found in groundwater, surface water, soils, and wastewater treatment plants at concentrations ranging from ng L−1 to μg L−1. This review explores the sources of ECs and laccase's role in their degradation. ECs encompass diverse categories with potential implications for human health, animals, and the environment, and their adverse effects are examined. Laccase, a key mediator, can oxidize non-phenolic compounds, broadening its substrate range. The review discusses the intricacies of laccase-mediated degradation and highlights its potential to improve global water resource sustainability. Innovative strategies, like laccase immobilization, are explored for EC removal, benefiting environmental preservation. In summary, the review addresses the issue of excessive EC use, their presence in water sources, and their impact on health, wildlife, and the ecosystem. Laccase offers promise for EC degradation, emphasizing its mechanism and potential for sustainable water resource management. Advanced techniques, including laccase immobilization, further demonstrate the commitment to tackling EC-induced environmental challenges.
Environmental significanceThe review on Laccase-Mediated Degradation of Emerging Contaminants unveils a promising sustainable solution to the growing concern of environmental pollution caused by novel, unregulated compounds. Emerging contaminants, including pharmaceuticals, personal care products, and industrial chemicals, pose significant risks to ecosystems and human health due to their persistence and bioaccumulative potential. Traditional water treatment methods often fail to effectively remove these pollutants. Laccase, a naturally occurring enzyme, offers an eco-friendly and efficient approach to degrade these contaminants. By harnessing the catalytic power of laccase, this research demonstrates a viable pathway to mitigate the environmental impact of emerging contaminants, promoting cleaner water bodies and healthier ecosystems. The laccase-mediated process not only aligns with green chemistry principles but also supports the advancement of sustainable water treatment technologies, contributing to the broader efforts for environmental protection and public health safety. |
1. Introduction
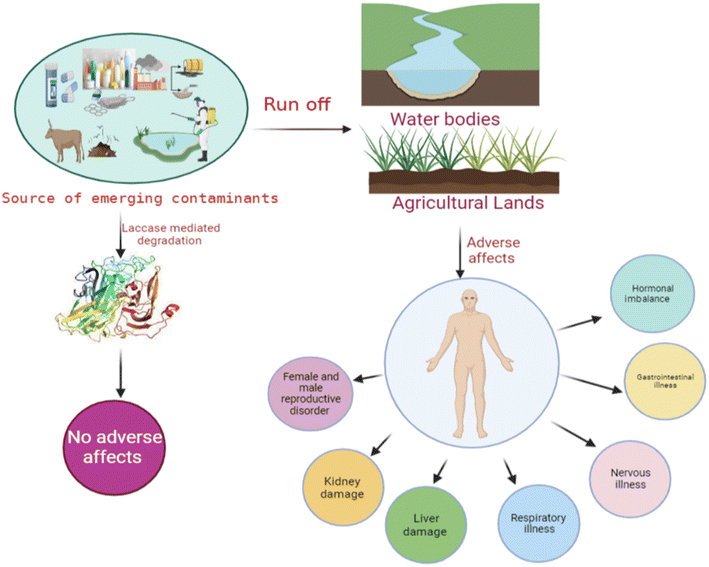
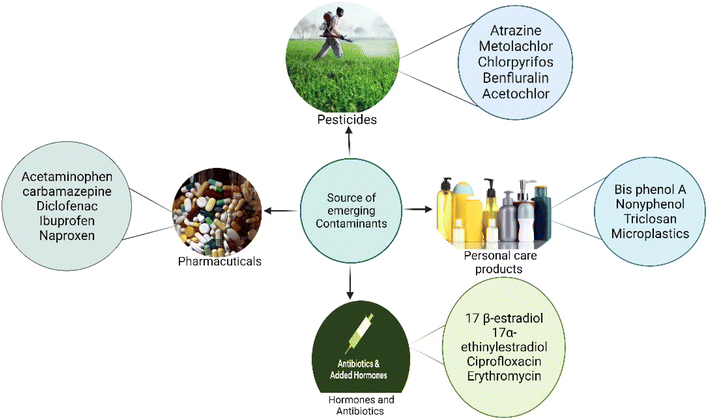
Occurrence of emerging contaminants (ECs) in the environment has become a significant concern due to their potential adverse effects on both humans and aquatic animals. The concentration of these ECs in soil and water bodies varies from a few nanograms to several micrograms per liter (μg L−1).1,2 These ECs (Table 1) are present in trace levels, and they are categorized into different types, i.e., pesticides, steroid hormones, pharmaceuticals, and chemicals from industries. Phytoestrogens are also one of the active ECs, which primarily constrain the sexual and reproductive development of wildlife.2,3 Therefore, the remediation of ECs has become the most significant concern in the current age. This portends a potential impact on broader ecology as well as human health (Fig. 1). Due to concerns about widespread human exposure and the potentially harmful health effects, or environmental impact of ECs, research on the treatment of these contaminants has recently gained increasing interest.4 Different chemical and physical methods, such as photodegradation,5 Fenton treatment,6 electrochemical treatment,7 ozonation,8 thermal degradation, and advanced oxidation processes,9 have been implemented for the treatment of ECs. However, several limitations, including high cost, generation of toxic by-products, and high energy consumption hinder their implementation.10 Thus, developing novel, efficient, and greener remediation processes has become an important objective for researchers, industrial chemists, and the scientific community. In this context, biodegradation appears to be a greener, sustainable, and cost-effective approach for treating ECs.11There are many innovations in the degradation of these pollutants, which mainly include filtration technology, advanced oxidation processes (AOPs), membrane bioreactors (MBRs), and adsorption and absorption processes. These methods are mainly based on their physical, chemical, and biological mechanisms. In the filtration process, ECs are effectively separated and studied in both pilot and full-scale systems.12,13 But the treatment of these ECs separated by filtration technology is also a main concern as it involves a high concentration of ECs. Adsorption and AOPs have also increased attention due to their efficiency in removing ECs.3 Nonetheless, these methods are normally used after primary or secondary treatments due to the higher content of organic carbon and turbidity. MBR technologies are an alternative to the conventional activated sludge processes for treatment. Many researchers have reported that MBRs effectively remove contaminants, but they are less effective in removing ECs.14,15 Recently, the enzymatic transformation and degradation of ECs using microorganisms have been determined as a promising and eco-friendly approach.16 Enzymes from microorganisms are probable biological agents for the degradation and transformation of these ECs.
Among all the reported enzymes, laccase is one of the lignin-modifying enzymes responsible for the effective degradation of these ECs. These enzymes are applied in many processes due to their oxidation capacity.17 Laccase enzymes in their crude, purified, and immobilized forms have been used for the degradation of several types of ECs in all categories of experiments and trials (i.e., continuous and batch experiments).18 These enzyme technologies are widespread and have promising applications in environmental biotechnology. Based on this, the present review aims to gather current trends and information on ECs, sources, harmful effects, and the role of laccase enzyme in the degradation process.
2. Aim and scope
This review will address emerging pollutants, their sources, and laccase-related studies with an emphasis on bioremediation. Laccases are responsible for breaking down these compounds without producing any harmful metabolites. Laccase enzyme-based bioremediation is highly effective and specific for treating pollutants because these enzymes have a higher efficiency of degradation as compared to the microorganisms as a whole. The quality of enzyme-based bioremediation, i.e., it requires mild conditions and low energy, makes it more suitable for the field of bioremediation.19,20 Laccase belongs to the diverse group of enzymes/biocatalysts within the oxidoreductase group which is produced by many microorganisms (e.g., fungi and bacteria). It has several biotechnological applications ranging from lignin degradation to organic pollutant biodegradation. Laccases are also involved in the transformation of various groups of ECs which mainly include polyaromatic hydrocarbons (PAHs), pharmaceutical and personal care products (PPCPs), phenols, and dyes.21,22 Although the applications of laccase degradation are well proven, most of the studies are limited to the lab scale. So further emphasis on pilot scale studies is necessary for economic feasibility. Therefore, this review expands upon bioremediation studies which will be further contextualized to address future studies. This review also focuses on immobilised laccase for bioremediation with different synthesis methods, hypothesized uptake mechanisms, and trends. Furthermore, the use of laccases as biocatalysts is discussed. Immediate gaps in the laccase bioremediation are highlighted to guide future studies, and potential mechanisms of laccase degradation are also discussed.3. Emerging contaminants and their sources
The annual production of ECs in both developed and developing countries has been steadily increasing over the past few decades, largely due to their persistent nature. They are commonly used in agriculture, pharmaceuticals, personal care products, and hormones.23 These compound residues are incessantly released into the water bodies via runoff and wash-off from hospitals and households to the surface, contaminating the agricultural land. Additionally, surface water contaminants enter groundwater bodies through leaching and filtration. Furthermore, the other routes for ECs to enter the diverse water bodies are unused medicine disposal, irrigation methods using wastewater, disposal of animal carcasses, and the treatment plants with wastewater and sewage water.16,23 The discharge of these ECs from wastewater plants is a major route for their entry into the aquatic environment.24 Poor management strategies and inefficient treatment technologies for EC pollution may lead to the release of these ECs into the environment (Fig. 1). Disposal of solid waste into the environment is the most common practice nowadays, which mainly increases the EC pollution in both the environment and wastewater treatment plants. However, the concentration is in the range of pg L−1.25 Several studies have indicated the presence of sweeteners. For instance, acesulfame, saccharin, and sucralose have been reported to be present in domestic wastewater via human excretion.26 On the other hand, food preservatives, i.e., parabens, have also been detected in groundwater.27 Surface water and groundwater are usually used for drinking in developing countries and some developed countries. These water bodies are a common source of ECs in drinking water. The presence of ECs in water bodies is a common source of contamination in drinking water. The concentration of herbicides detected in drinking water suggests that agricultural runoff is the primary pathway for transporting these contaminants from agricultural land to water bodies and finally to drinking water sources. These contaminants are detected in higher concentrations. The standard and frequently detected contaminants are atrazine, and terbuthylazine.28Non-sewered sanitation methods which mainly include urine-diverted dry toilets (UDDTs) are identified as a major source of EC pollution in developed countries. UDDTs, a means to separate human urine toilets, are built on a pilot scale in many developed countries. Still, there are few reports available on ECs from UDDTs. Ref. 29 described the presence of 12 ECs in source-separated urine. Pharmaceuticals are the major ECs detected, including hydrochlorothiazide, clarithromycin, darunavir, atenolol, atazanavir, atenolol acid, acetyl-sulfamethoxazole, diclofenac, emtricitabine, N4-ritonavir, and trimethoprim (TMP).30
The management strategies for these ECs are very poor, so these ECs are also detected in landfill leachate, which increases the possibility of these ECs contaminating freshwater sources. Consequently, the constant monitoring of these landfill leachates should be done, as it is the major factor in detecting these ECs in water bodies.31 Moreover, proper disposal of these ECs in the environment and the treatment strategies for these landfill leachates are growing concerns. Untreated disposal of this leachate into the sewer is a common practice (Environmental Affairs 2019), which may lead to loading higher EC concentrations onto municipal wastewater treatment plants (WWTPs). Many studies have reported the presence of ECs in landfill leachates. Daso et al. reported that landfill leachates in Cape Town are the main source of polybrominated diphenyl ether (PBDE) in waterbodies and the environment.32 Olukunle et al. described the presence of these ECs in the landfill at concentrations in the range of pg L−1.33
4. Adverse effects of ECs on the environment
ECs have an enormous destructive impact on the environment; when these environmental pollutants come in contact with the ecosystem, they tend to show diverse effects. These pollutants probably cause many acute and long-term effects (i.e., disruption of endocrine function, immunotoxicity, neurological disorders, cancers, etc.) on aquatic life, human health, and the environment, (Fig. 2). Increased release of these ECs in the environment provides many routes for their entry into the aquatic environment.34 ECs are incessantly discharged into the environment but they are efficiently removed only sometimes; subsequently many of these contaminants persist in the environment for a long time.35,36 Improper and incomplete degradation of these contaminants on waste treatment plants during the degradation process are one of the main reasons for contaminants to enter into aquatic environment.37 The risks associated with these ECs are chronic for living organisms, and therefore, they may cause lethal effects even at very low concentrations.38 Creatures or living bodies in the aquatic environment are continuously exposed to the mixtures of these ECs, and these contaminants may interact with aquatic organisms and produce synergistic or toxic effects.39 ECs are universally found in the environment primarily in soil and water bodies. These compounds, including organic compounds, brominated flame retardants, phthalates, plastics, microplastics, hormones, and polycyclic siloxanes, are responsible for causing deleterious effects on the health of humans and aquatic animals.40 These contaminants are considered one of the foremost reasons for endocrine disorders in humans, which mainly include the modifications of salivary glands, the thyroid, and various genetic disorders in the male body system. These ECs increase the risk of cancer in humans and act as antiandrogens in males, leading to feminizing effects.41 EC toxicity chiefly affects the endocrine cycle; many groups of these ECs may cause hormonal imbalance, DNA damage, carcinogenic effects, disruptions in thyroid function, impaired brain development, reduced fertility, and overall growth issues. Moreover, the genetic toxic effects of these ECs are abnormality in female implantation, breasts, and the liver, which may lead to increased hormones due to brominated flame retardants.42 Their excessive concentrations have been reported to cause abortion and maternity difficulties.435. Laccase enzyme
Laccases (EC 1.10.3.2) are monomeric glycoproteins that have great potential in the field of environmental microbiology.44–46 Over the last few years, the use of enzyme technology has attracted great interest in the field of green and sustainable technologies. Lacasse acts as a biocatalyst in conventional chemical-based processes and pharmaceutical industries.39,40 The use of laccases in the industrial and biotechnological sectors is a thriving area of research nowadays.47,48 The use of Laccases in industries, i.e., food, textile, paper, etc., as oxidative enzymes due to their ability to act on a wide range of substrates.49,50 Lacasse enzymes provide an eco-friendly substitute for conventional chemical reactions. Laccase was first discovered in the nineteenth century in the Japanese lacquer tree Rhus vernicifera.51 Although the presence of laccases has been reported in higher plants, microorganisms (fungi, bacteria), and insects, most researchers reported laccases from fungal sps., which mainly include those in genera ascomycetes, deuteromycetes, basidiomycetes, and cellulolytic fungi.52–56 Among these, basidiomycetes (white-rot fungi) laccases are most common and frequently reported and these are Phlebia radiata, Trametes (Coriolus) versicolor, Coriolopsis polyzona, T. hirsuta, T. ochracea, T. villosa, Pycnoporus cinnabarinus, T. gallica, Lentinus edodes, Pleurotus ostreatus, Coprinus cinereus, etc.52Monocillium indicum was the first characterized ascomycete enzyme.57,58 Laccases, isolated from fungi, are mainly responsible for fructification, detoxification, phytopathogenic, sporulation, and degradation of lignin (Table 2).| Source | Emerging contaminant | Biodegradation rate (%) | Reference |
|---|---|---|---|
| A. densiflorus | Rubin GFL (RGFL) dye | Rubin GFL (RGFL) dye (40 g L−1) up to 91% within 48 h | 59 |
| Tagetes patula, Aster amellus, Portulaca grandiflora and Gaillardia grandiflora | Textile effluent | Reduction in dyes 59, 50, 46 and 73% | 60 |
| Glandularia pulchella | Reactive orange HE2R, reactive yellow MEG4, reactive yellow GR, blue 2GL, remazol red, green HE4B, brown 3REL, blue 2RNL, patent blue, and malachite green | Decolorization of all the dyes | 61 |
| Brumea malcolmii | Brilliant blue R (BBR), malachite green, reactive red 2, direct red 5B and methyl orange | Compete degradation of dyes | 62 |
| Alternanthera philoxeroides | Sulfonated remazol red | Completely decolorize remazol red dye | 63 |
| Arabidopsis thaliana | Detoxification of herbicides atrazine (ATR) and isoproturon (IPU) | Detoxification or degradation of ATR or IPU | 64 |
| Pycnoporus sanguineus CCT-4518 | 17-Alpha-ethinylestradiol (EE2) | 80% of removal of EE2 after 24 h | 65 |
| Pycnoporus sanguineus | Estrogens | 96% of estrogens after 8 h of reaction | 66 |
| Trametes trogii | Amaranth, carmoisine, cochineal red, sunset yellow, patented blue, blue indigo and alizarin red S | All the dyes were decolorized up to 60% percent after 2 h | 67 |
| Myceliophthora thermophila | Anthraquinonic dyes acid blue 25 and acid green | RBBR and the diazo RB-5 were only decolorized with laccase/HBT, 31 and 60%, respectively, after 24 h | 68 |
| Trametes versicolor | Triphenylmethane dyes | Complete removal | 69 |
| Trametes trogii BAFC 463 | Indigo carmine, xylidine, malachite green, gentian violet, bromophenol blue | Decolorized 85% of all dyes | 70 |
| Trametes trogii | Azoic, indigoid, triarylmethane, and anthraquinonic with acetosyringone | 50–100% decolorization | 71 |
| Trametes versicolor | Bromocresol purple, safranin, malachite green, kristal violet, bromothymol blue, nigrosine and phenol red | 43%, 54%, 55%, 49%, 56%, 53% and 37% for bromocresol purple, safranin, malachite green, kristal violet, Bromothymol blue, nigrosine and phenol red, respectively | 72 |
| P. pastoris | Crystal violet | Decolorization rates 90.7% | 8 |
| Oudemansiella canarii | Congo red | 80% of 50 mg L−1 Congo red within 24 h | 73 |
| Ganoderma lucidum E47 strain | Decolorizing xanthene, azo and triarylmethane dyes | Decolorization | 74 |
| Pycnoporus sanguineus (CS43) | Bisphenol A | 100% degradation of bisphenol A (20 mg L−1) was achieved in less than 24 h | 16 |
| Trametes versicolor BAFC 2234 | Phenol | 84% phenol removal in 4 h | 75 |
| Trametes sanguineus | Phenanthrene and benzo[α]pyrene | Removed phenanthrene and benzo[α]pyrene (97 and 99% respectively | 69 |
| Pleurotus ostreatus | Phenol | 70% phenol removal | 76 |
| T. versicolor | Naphthalene, phenanthrene | 100% degradation | 77 |
| P. ostreatus | Atrazine | — | 78 |
| Pseudomonas putida | Chlorpyrifos | 90% of degradation after 8 days of incubation | 79 |
| P. pastoris | Atrazine, isoproturon | Complete degradation | 64 |
| T. versicolor | Amoxicillin, Ampicillin, cloxacillin, penicillin G, penicillin V, oxacillin | 54–100% after 24 h | 80 |
| T. versicolor | Sulfapyridine, sulfathiazole | 100% after 8 h | 81 |
| T. versicolor | Sulfadimethoxine | >90% after 72 h | 82 |
| Cerrena sp. HYB07 | Oxytetracycline, tetracycline | 80% after 12 h | 83 |
| A. oryzae | Ciprofloxacin | 51% after 5 h | 84 |
| Ganoderma lucidum | Triclosan (biocide) | 90% after 24 h | 85 |
Laccases isolated from bacteria have been reported to be the most stable laccases due to their resistance to high temperatures and wide pH ranges. While fungal laccases can be intra and extra-cellular, bacterial laccases are mostly intracellular, i.e., Marinomonas mediterranea, Azospirillum lipoferum, and Bacillus subtilis.59,60 Bacterial laccases normally show the best activity at a higher pH compared to laccases from fungal sources,61 with the optimum pH being acidic for the latter.62 Plant laccases have intracellular physiological properties in the neutral pH range.63,64 Plant laccases have a higher isoelectric point compared with fungal laccases. Most laccases have an optimum temperature between 50–70 °C.65 The thermal stability mainly depends on the type of microorganism; laccases from bacteria are more stable than those from fungi.66 Due to the higher glycosylated content in the plant enzymes, they have higher molecular weight when compared with bacterial and fungal laccases. Laccases are secondary metabolites formed under conditions of limited growth, mainly nitrogen, which negatively impacts the enzyme yield.67 The production of laccases is mainly achieved through the fermentation process (liquid and solid).68 Copper,69 ethanol,70 and aromatic compounds such as xylidine, anisidine, syringaldazine, gallic acid, veratrine alcohol, ferulic acid, and lignosulfonates71 are used to induce the production of laccases.
5.1 Biochemical property characterization
Characterization parameters, like optimum temperature and pH, always play a significant role in the degradation process by any enzyme. Previously, many researchers have reported the role of characterization parameters in the laccase enzyme. Laccase (molecular weight (MW) 52 kDa) from Cyberlindnera fabianii has been reported for the degradation of 42.7% of Ca-alginate immobilized laccase (AIL) and 39.1% (Cu-AIL) of bisphenol A (BPA) (100 μM) after 24 h. The characterization of the native enzyme has shown an optimum pH and temperature of 5 and 40 °C, respectively.72 Likewise, laccase (MW 37 kDa) from Bacillus subtilis MTCC 2414 has been reported to degrade 81.72% of yellow GR dye after 120 h at 35 °C and 9 pH.73Aspergillus oryzae derived laccase (MM 57 kDa) with the potential of removing more than 80% of sulfamethoxazole, carbamazepine (CBZ), diclofenac, and bisphenol A at 30 °C and 7 pH,74 is also reported to biodegrade 75% of azo-dyes after consecutive cycles at pH 5 at room temperature.75Trametes versicolor derived laccase also works as a biosensor for detecting phenolic compound catechol. Additionally, laccase (MW 31 kDA) isolated from Bacillus atrophaeus has been stated to clarify 41–58% phenolic compounds by reducing the color turbidity in the juices at 35 °C and 5.5 pH.49Biochemically, the protein is extremely thermostable with a half-life of about 2 h at 80 °C.76 In contrast to fungal laccases, bacterial-derived laccases are biochemically more active at high pH values and highly stable at high temperatures. A high alkaline pH optimum of 8.5 to 9 was shown for several Streptomyces laccases for 2,2′-azino-bis(3-ethylbenzothiazoline-6-sulfonic acid) (ABTS),77,78 whereas an optimum temperature of 85 °C was reported for the multicopper oxidase protein (McoP) laccase derived from Pyrobaculumaerophilum.79 Furthermore, for T. thermophilus laccase, optimum temperature is 92 °C.80 Fascinatingly, the laccase retrieved from drained soil with industrial wastewater is biochemically stable in the optimum pH range of 3 to 10.6.81 Another equally important biochemical parameter is salt tolerance. Streptomyces ipomoea derived laccase holds 100% activity in 1 M sodium chloride (NaCl) at an optimum pH of 8.0.7 These astonishing biochemical features highlight laccases as a potential basis for robust compounds with valuable biotechnological applications.
5.2 Mechanism
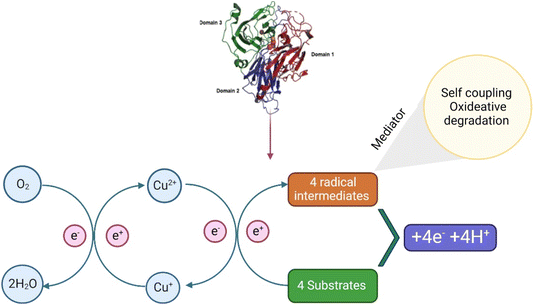
The biocatalytic processes of laccases have the potential application in the degradation of emerging compounds without any harmful effects; therefore, it increases the interest of the researchers to understand their mechanisms of degradation. Normally, laccases are reported for the oxidation of various ECs which mainly include phenols and aromatic amines; after degradation, they are directly transformed into free radicals (Fig. 3).82,83 These free radicals are commonly known to initiate domino reactions, primarily driving the complex transformations of ECs through biological processes, such as degradation by laccase and lignin synthesis.84 Moreover, the laccase reaction involves one electron (1e−), oxidations consecutive of four molecules of plummeting substrates, concurrently with doubling electron reduction (2 × 2e−) for oxygen atoms into H2O molecules of their respective mechanisms. This method is accompanied by a catalytic exchange of 4H+ between parallel molecules.85,86 The laccase reaction involves two half-reactions associated with an internal electron transfer (IET) step and is supported by catalytic copper ions located at the T1 copper (Cu) site and T2 Cu/T3 Cuα/T3 Cuβ trinuclear cluster (TNC) sites from the mechanistic, kinetical, and structural points of view.85,87–89 The complete conservation of the eleven residues (one Cys and ten His) forming the T1 copper and TNC laccase sites explains their essential role in the catalytic action. In many studies, comparisons related to sequences and mutagenic approaches have not been clearly demonstrated.83–85,90,91 Furthermore, highly conserved residues play important roles in various catalytic steps involved in laccase action.92 Despite these advances in the understanding of the action of laccases in terms of structure–function, a complete picture relating their molecular properties and mechanisms with their kinetic performance remains unclear. In brief, how these structural elements are automatically linked to catalytic copper center function and how the kinetic performance of these elements is influenced are listed below.In the first half semi-reaction, 1e− substrate oxidation at the T1 copper site takes place which is located in the substrate binding pocket at the bottom. In T. versicolor laccase, this substrate interaction region is delimited by several highly conserved hydrophobic residues, namely, phenol (Phe) 162, leucine (Leu) 164, Phe 265, Phe 332, and proline (Pro) 391, which is useful for the formation of a favourable environment for the docking of hydrophobic molecules and for the degradation of aromatic compounds substrates by laccase. Moreover, the residue of fully conserved Asp 206 (near His 458 of the T1 copper site), situated in the binding pocket of the substrate at the bottom, is useful for the substrate orientation and stabilization through interaction with O–H bonds at the catalytic copper site T1, by the participation of fully conserved His 458.93 Finally, the last residue should be exposed using a solvent in the interface for the binding cavity of the substrate. Furthermore, Asp 206 acts as a useful mechanistic component using electron subtraction promotion and the transfer of substrate donor molecules from the T1 copper ion (Cu2+ → Cu1+) with the His 458, direct interaction at T1 copper site. Moreover, the high E°′ detected on this T. versicolor is related directly to the Phe 463 residue at an axial position in the center.94
6. Immobilized laccase in biodegradation
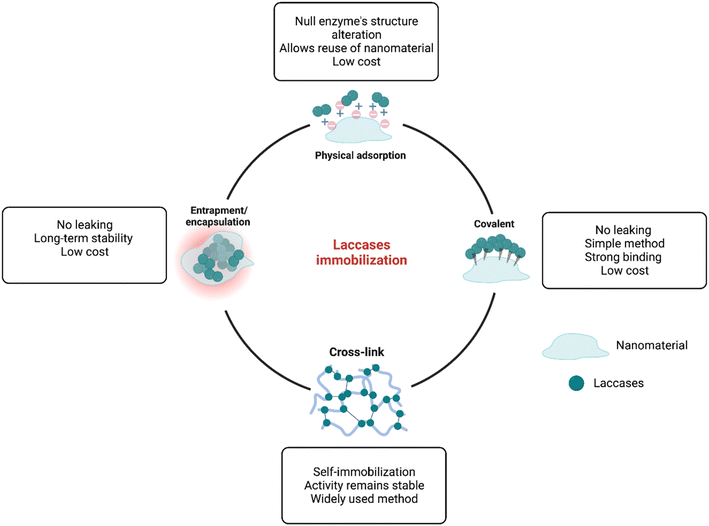
In general, biodegradation is the result of advances in molecular biotechnology, with important advantages over traditional degradation or catalytic processes, including higher catalytic behavior, higher specificity, higher reaction rate under mild conditions of pH and temperature, lower energy consumption, and most importantly, biodegradability.95 Basically, biodegradation of ECs can be achieved in three different ways: (1) whole-cell biodegradation, which involves using biological molecules as biocatalysts, (2) isolated enzyme biodegradation, which involves using isolated enzymes; and (3) isolated-immobilized enzyme biodegradation, which involves immobilizing enzymes into nanomaterials.96 The first two methods have been widely used for the biodegradation of ECs due to important properties, including high reaction rates and relatively low cost and operation time.97 For example,98 a high biodegradation rate (96%) for sertraline, clomipramine, mianserin, and paroxetine has been reported using Pleurotus ostreatus as a biocatalyst. However, both techniques might present problems, such as potential cross-reactivity, side reactions, and poor or null reusability.96,99 On the other hand, the immobilization of enzymes into suitable nanomaterials might provide an optimal environment to reach high biodegradation rates. Several important properties are enhanced during this process, i.e., higher thermal and pH stability, and improved storage stability, long-term operation, reusability, and higher enzymatic activity.100 Different enzymes have been reported as immobilized enzymes for the biodegradation of ECs, including peroxidases, oxidases, catalase, super oxidase dismutase, and laccases.101 Laccases have gained significant interest in developing novel immobilized materials for their application in the biodegradation of emerging pollutants since they can catalyze the biotransformation of a wide range of compounds, such as phenols, dyes, pharmaceuticals, and many others.102 Different nanomaterials, each one with different properties, have been applied for the immobilization of laccases and its further application in the biodegradation of ECs, including polymers,103 metals and metal oxides,96 and carbon-based materials.104 Thus, depending on the nanomaterial used and the procedure of immobilization, the characteristics and properties of the laccase immobilized system change.73 The immobilization of laccases into nanomaterials can be achieved by different methods (Fig. 4), which are briefly described below:• Physical adsorption immobilization is mainly characterized since it is the simplest immobilization method, which basically involves the interaction of the enzyme with the nanomaterial by the formation of weak ionic bonds that usually are formed by the dissolution of the enzyme at controlled pH and the subsequent addition of the host matrix96 Important advantages of adsorption immobilization are low operational costs and times, reusability, and increased catalytic stability. However, since the enzyme–nanomaterial interaction is governed by weak binding forces, changes in pH, temperature, or ionic strength, might produce a leaching effect.97,105
• The covalent immobilization technique is widely used due to its important advantages, which include strong bindings between enzymes and supports, high stability across varying pH and temperature conditions, high uniformity, and good control over the immobilized enzyme amount.106 Covalent immobilization is a two-step process, in which first, the host carrier is activated by the addition of multifunctional reagents (e.g., glutaraldehyde and carbodiimide), which then promotes the formation of covalent bonds with functional groups of the enzyme, most commonly amino, carboxylic, phenolic, and hydroxyl groups.96 Even though covalent immobilization offers great advantages, due to the structural modification of the enzyme after the formation of the covalent bond, the active sites of the enzyme might be deactivated, thus affecting the catalytic properties.92
• Cross-linking immobilization involves the self-immobilization of enzymes by the formation of cross-linkages named cross-linked enzyme aggregates (CLEAs), which are formed by the addition of bifunctional reagents, such as glutaraldehyde, dialdehydes, diiminoesters, diisocyanates and diamines, and the subsequent formation of a three-dimensional complex structure through the interaction of amine groups of the enzyme and the activated bifunctional reagents.107 This method has the ability to form materials resistant to the effects of pH and temperature, but a major disadvantage is the loss of enzymatic activity after immobilization.106
• Entrapment and encapsulation are two similar methods involving a two-step process, in which the enzyme and a monomeric solution are mixed, and then polymerization of the monomer confines/entraps the enzyme within the polymeric network without chemical interactions.96 These two methods are characterized for providing significant protection from the external environmental conditions to the enzyme, enhancing the stability and decreasing denaturation.74 On the other hand, these methods have important mass transfer limitations, which means that substrates might have problems in reaching the active sites of the confined enzyme.70,97
7. Laccase as a biocatalyst for biodegradation
Immobilized laccases have been widely implemented as biocatalysts in the biodegradation of different ECs (Table 3). Even though the application of laccases as biocatalysts might be achieved either through free or immobilized enzymatic systems, it is well known that through the implementation of immobilized laccase biocatalysts, it is possible to overcome the major disadvantages of using free enzymes, which are discussed above.11 Different nanomaterials and immobilization methods have been applied to design laccase-based biocatalysts for the biodegradation of different ECs. For example, adsorption immobilization was reported to be effective for the biodegradation of CBZ, bisphenol A (BPA), and synthetic dyes, by the immobilization of laccases into polymers,108 carbon-based nanomaterials,109 and metal-based nanomaterials.110 Moreover, covalent, entrapment, and cross-link immobilization have been implemented to design novel biocatalysts by using polymers, metal oxide, and carbon-based nanomaterials.115| Laccase source | Support | Immobilization method | Emerging contaminant | Conditions | Biodegradation rate (%) | Reference |
|---|---|---|---|---|---|---|
| a Carbamazepine (CBZ), polyvinylidene fluoride (PVDF), multiwalled carbon nanotubes (MWCNTs), bisphenol A (BPA), sodium zeolite Y dealuminated form (DAY), graphene oxide (GO), 2,4-dichlorophenol (2,4-DCP), ultraporous alumina (UPA), rifampicin (RIF), isoniazid (INH), cross-linked. | ||||||
| Trametes versicolor | Polyimide aerogels | Covalent | CBZ | 20 ng mL−1 of CBZ, 200 rpm, 25 °C, 24 h | 74–76 | 108 |
| Trametes hirsuta | PVDF/MWCNTs | Adsorption | CBZ | 5 ppm of CBZ, 223 U mL−1 of catalysts, pH 5, 25 °C, 4 h | 95 | 109 |
| Trametes versicolor | CNTs | Adsorption | Dyes | 10 mg L−1 of dye, 0.08 g L−1 of catalyst, 35 °C, 3 h | 96 | 111 |
| Trametes versicolor | Fe3O4@C–Cu2+ | Adsorption | Dyes | 5–200 mg L−1 of dye, pH 4.5, 50 °C, 1 h | 75–99 | 112 |
| Trametes versicolor | DAY | Adsorption | BPA | 1 mL BPA 2 mM, pH 4.5, 5 mg of catalyst, 45 °C, 1 h | 86.7 | 110 |
| Aspergillus flavus | GO | Covalent | Azo dyes | 300 mg L−1 of catalyst, 10 ppm of dye, pH 5, 45 °C, 0.2 h | 48.7–88.7 | 76 |
| Trametes versicolor | Copper alginate | Entrapment | 2,4-DCP | 10 mg mL−1 of 2,4-DCP, pH 4.6, 40 °C, 10 mh | 96.4 | 113 |
| Trametes pubescens | Chitosan | Entrapment | Dyes | 50 mg mL−1 of dye, 10 g of catalyst, pH 5, 55 °C, 96 h | 37–79 | 114 |
| Trametes villosa | CLEAs | Cross-link | RIF, INH | 5 mg mL−1 of drug, 100 U L−1 of catalyst, pH 5, 40 °C, 24 h | 71–95 | 115 |
Moreover, Ma et al. (2018) also described the biodegradation of dyes, including CV, Congo red (CR), indigo blue (IB), and others by the entrapment of laccases from Trametes pubescens into a polymeric chitosan matrix. In this case, biodegradation rates reached about 79%. However, the biodegradation of dyes by implementing this biocatalyst was achieved after 96 h, which might be mainly attributed to the characteristic mass transfer limitations achieved after entrapment immobilization.74,114
The application of immobilized enzyme systems as biocatalysts for the biodegradation of ECs is an important approach to deal with the increasing environmental impact that ECs might have on human well-being and the environment.10 Thus, designing a novel and highly catalytic biocatalyst is a critical point as a prospect; a suitable biocatalyst might be designed in two main steps: (1) preparation of the host carrier material and (2) choosing a suitable immobilization method.96
8. Conclusion and future prospects
In summary, laccases play a crucial role in breaking down ECs. Leveraging modern protein engineering tools makes the use of laccases viable. However, optimizing various factors such as pH, temperature, suspended solids, and mechanical stress is essential to maximize laccase yield. Extensive optimization is pivotal for generating substantial laccase biomass, which holds the potential for addressing the pressing challenge of wastewater treatment and purification in the commercial sector. Presently, safeguarding natural water bodies from pollutants stemming from ECs is an urgent concern. These pollutants, encompassing plastics, herbicides, fertilizers, dyes, phthalates, pharmaceuticals, and personal care products, find their way into water bodies through direct runoff and disposal. The degradation of these substances is paramount. Laccases emerge as efficient biocatalytic agents, adept at converting these compounds into less harmful and inert derivatives. It's worth noting that complexities like the intricate composition of contaminated water (high salt concentrations and/or elevated pH levels) pose obstacles to efficient degradation. Nonetheless, contemporary techniques like laccase engineering offer promising solutions. Encouraging reports have highlighted successful EC degradation. Yet, transitioning these processes to pilot-scale bioprocesses hinges on economic feasibility. Immobilizing laccases presents a viable strategy for treatment procedures. Nevertheless, the time-intensive nature of laccase extraction and purification remains a challenge. In this regard, producing recombinant laccases with increased activity and stability holds immense promise. This advancement could streamline the enzyme purification process, significantly enhancing cost-effectiveness.Ethical approval
All data referenced in this review paper have been obtained from publicly available sources, with no use of confidential or sensitive information.Data availability
No primary research results, software or code have been included and no new data were generated or analysed as part of this review.Author contributions
Pooja Thathola: conceptualization, writing of the original draft, review & editing. Elda M. Melchor-Martínez: writing of the original draft, review & editing. Priyanka Adhikari: writing of the original draft, review & editing. Saúl Antonio Hernández Martínez: reviewing. Anita Pandey: conceptualization, reviewing. Roberto Parra-Saldívar: reviewing.Conflicts of interest
There are no conflicts to declare.Acknowledgements
The authors are grateful to their representative universities and institutes for providing literature facilities.References
- B. S. Rathi, P. S. Kumar and P. L. Show, J. Hazard. Mater., 2021, 409, 124413 CrossRef CAS.
- V. Uddameri, A. Ghaseminejad and E. A. Hernandez, Agric. Water Manag., 2020, 238, 106226 CrossRef.
- D. Saidulu, B. Gupta, A. K. Gupta and P. S. Ghosal, J. Environ. Chem. Eng., 2021, 9, 105282 CrossRef CAS.
- Y. H. Pang, Y. Y. Huang, L. Wang, X. F. Shen and Y. Y. Wang, Environ. Pollut., 2020, 263, 114616 CrossRef CAS.
- S. Kashefi, S. M. Borghei and N. M. Mahmoodi, J. Mol. Liq., 2019, 276, 153–162 CrossRef CAS.
- P. Wu, C. Zhou, Y. Li, M. Zhang, P. Tao, Q. Liu and W. Cui, Appl. Surf. Sci., 2021, 540, 148362 CrossRef CAS.
- V. Cristino, L. Pasti, N. Marchetti, S. Berardi, C. A. Bignozzi, A. Molinari, F. Passabi, S. Caramori, L. Amidani, M. Orlandi, N. Bazzanella, A. Piccioni, J. Kopula Kesavan, F. Boscherini and L. Pasquini, Photochem. Photobiol. Sci., 2019, 18, 2150–2163 CrossRef CAS.
- J. Wang and H. Chen, Sci. Total Environ., 2020, 704, 135249 CrossRef CAS.
- R. Bai, W. Yan, Y. Xiao, S. Wang, X. Tian, J. Li, X. Xiao, X. Lu and F. Zhao, Chem. Eng. J., 2020, 397, 125501 CrossRef CAS.
- R. Morsi, K. A. Al-Maqdi, M. Bilal, H. M. N. Iqbal, A. Khaleel, I. Shah and S. S. Ashraf, Biomolecules, 2021, 11(6), 904 CrossRef CAS.
- R. A. Sheldon and J. M. Woodley, Chem. Rev., 2018, 118, 801–838 CrossRef CAS.
- D. Wan, H. Wang, I. P. Pozdnyakov, C. Wang, J. Su, Y. Zhang, Y. Zuo, D. D. Dionysiou and Y. Chen, Water Res., 2020, 184, 116002 CrossRef CAS.
- J. García, M. J. García-Galán, J. W. Day, R. Boopathy, J. R. White, S. Wallace and R. G. Hunter, Bioresour. Technol., 2020, 307, 123228 CrossRef PubMed.
- N. H. Tran and K. Y. H. Gin, Sci. Total Environ., 2017, 599–600, 1503–1516 CrossRef CAS PubMed.
- M. Patel, R. Kumar, K. Kishor, T. Mlsna, C. U. Pittman and D. Mohan, Chem. Rev., 2019, 119, 3510–3673 CrossRef CAS PubMed.
- C. Barrios-Estrada, M. de J. Rostro-Alanis, A. L. Parra, M. P. Belleville, J. Sanchez-Marcano, H. M. N. Iqbal and R. Parra-Saldívar, Int. J. Biol. Macromol., 2018, 108, 837–844 CrossRef CAS.
- C. A. Lutterbeck, G. S. Colares, N. Dell'Osbel, F. P. da Silva, L. T. Kist and Ê. L. Machado, J. Cleaner Prod., 2020, 273, 122851 CrossRef CAS.
- Y. Chen, J. Vymazal, T. Březinová, M. Koželuh, L. Kule, J. Huang and Z. Chen, Sci. Total Environ., 2016, 566–567, 1660–1669 CrossRef CAS PubMed.
- S. M. Mousavi, S. A. Hashemi, S. M. Iman Moezzi, N. Ravan, A. Gholami, C. W. Lai, W. H. Chiang, N. Omidifar, K. Yousefi and G. Behbudi, Biochem. Res. Int., 2021, 5599204 CAS.
- T. D. H. Bugg, J. J. Williamson and G. M. M. Rashid, Curr. Opin. Chem. Biol., 2020, 55, 26–33 CrossRef CAS.
- M. Bilal, M. Adeel, T. Rasheed, Y. Zhao and H. M. N. Iqbal, Environ. Int., 2019, 124, 336–353 CrossRef CAS.
- L. Arregui, M. Ayala, X. Gómez-Gil, G. Gutiérrez-Soto, C. E. Hernández-Luna, M. Herrera De Los Santos, L. Levin, A. Rojo-Domínguez, D. Romero-Martínez, M. C. N. Saparrat, M. A. Trujillo-Roldán and N. A. Valdez-Cruz, Microb. Cell Factories, 2019, 18(1), 1–33 CrossRef.
- K. Balakrishna, A. Rath, Y. Praveenkumarreddy, K. S. Guruge and B. Subedi, Ecotoxicol. Environ. Saf., 2017, 137, 113–120 CrossRef CAS PubMed.
- J. M. Brausch and G. M. Rand, Chemosphere, 2011, 82, 1518–1532 CrossRef CAS PubMed.
- H. Wang, H. Xi, L. Xu, M. Jin, W. Zhao and H. Liu, Sci. Total Environ., 2021, 788, 147819 CrossRef CAS PubMed.
- F. Freeling, N. A. Alygizakis, P. C. von der Ohe, J. Slobodnik, P. Oswald, R. Aalizadeh, L. Cirka, N. S. Thomaidis and M. Scheurer, Sci. Total Environ., 2019, 681, 475–487 CrossRef CAS PubMed.
- M. E. Scheurer, M. L. Bondy, K. D. Aldape, T. Albrecht and R. El-Zein, Acta Neuropathol., 2008, 116, 79 CrossRef CAS PubMed.
- J. M. Dabrowski, S. Afr. J. Sci., 2015, 111, 1–7 CrossRef PubMed.
- H. N. Bischel, B. D. Özel Duygan, L. Strande, C. S. McArdell, K. M. Udert and T. Kohn, Water Res., 2015, 85, 57–65 CrossRef CAS PubMed.
- S. A. Madhi, C. Cutland, K. Ismail, C. O'Reilly, A. Mancha and K. P. Klugman, Clin. Infect. Dis., 2002, 35, 1120–1126 CrossRef CAS PubMed.
- K. Narasimhulu, Innovations in Microbiology and Biotechnology, 2022, vol. 5, pp. 70–81 Search PubMed.
- A. P. Daso, O. S. Fatoki, J. P. Odendaal and O. O. Olujimi, Environ. Monit. Assess., 2013, 185, 431–439 CrossRef CAS PubMed.
- O. I. Olukunle, I. v. Sibiya, O. J. Okonkwo and A. O. Odusanya, Environ. Sci. Pollut. Res. Int., 2015, 22, 2145–2154 CrossRef CAS PubMed.
- M. H. Wu, C. C. Lee, A. S. Hsiao, S. M. Yu, A. H. J. Wang and T. H. D. Ho, FEBS Open Bio, 2018, 8, 1230–1246 CrossRef CAS PubMed.
- A. M. Deegan, B. Shaik, K. Nolan, K. Urell, M. Oelgemöller, J. Tobin and A. Morrissey, Int. J. Environ. Sci. Technol., 2011, 8(3), 649–666 CrossRef CAS.
- S. Mohapatra, C. H. Huang, S. Mukherji and L. P. Padhye, Chemosphere, 2016, 159, 526–535 CrossRef CAS PubMed.
- A. Agüera, M. J. Martínez Bueno and A. R. Fernández-Alba, Environ. Sci. Pollut. Res., 2013, 20(6), 3496–3515 CrossRef.
- M. C. V. M. Starling, C. C. Amorim and M. M. D. Leão, J. Hazard. Mater., 2019, 372, 17–36 CrossRef CAS PubMed.
- T. K. Kasonga, M. A. A. Coetzee, I. Kamika, V. M. Ngole-Jeme and M. N. Benteke Momba, J. Environ. Manage., 2021, 277, 111485 CrossRef CAS.
- P. Chaturvedi, P. Shukla, B. S. Giri, P. Chowdhary, R. Chandra, P. Gupta and A. Pandey, Environ. Res., 2021, 194, 110664 CrossRef CAS.
- Y. Atih, M. ALBashtawy, A. Alkhawaldeh, B. Albashtawy, S. Albashtawy, A. Ibnian, Z. Albashtawy, A. Kazaleh, A. Ayed, M. Suliman, K. al Dameery, E. Rn and F. Jordanian, J. Hospital, 2017, 1, 7 Search PubMed.
- H. Ramírez-Malule, D. H. Quiñones-Murillo and D. Manotas-Duque, Emerging Contam., 2020, 6, 179–193 CrossRef.
- S. Zahmatkesh, A. Bokhari, M. Karimian, M. M. A. Zahra, M. Sillanpää, H. Panchal, A. J. Alrubaie and Y. Rezakhani, Environ. Monit. Assess., 2022, 194(12), 1–15 CrossRef.
- R. R. Nair, P. Demarche and S. N. Agathos, New Biotechnol., 2013, 30, 814–823 CrossRef CAS.
- K. Agrawal, V. Chaturvedi and P. Verma, Bioresour Bioprocess, 2018, 5, 1–12 CrossRef.
- J. Zdarta, K. Antecka, R. Frankowski, A. Zgoła-Grześkowiak, H. Ehrlich and T. Jesionowski, Sci. Total Environ., 2018, 615, 784–795 CrossRef CAS.
- M. Bilal, T. Rasheed, F. Nabeel, H. M. N. Iqbal and Y. Zhao, J. Environ. Manage., 2019, 234, 253–264 CrossRef CAS.
- D. M. Mate and M. Alcalde, Microb. Biotechnol., 2017, 10, 1457–1467 CrossRef CAS.
- L. K. Narnoliya, N. Agarwal, S. N. Patel and S. P. Singh, J. Microbiol., 2019, 57(10), 900–909 CrossRef CAS.
- F. Lassouane, H. Aït-Amar, S. Amrani and S. Rodriguez-Couto, Bioresour. Technol., 2019, 271, 360–367 CrossRef CAS.
- H. Yoshida, J. Chem. Soc. Trans., 1883,(43), 472–486 RSC.
- P. Baldrian, FEMS Microbiol. Rev., 2006, 30, 215–242 CrossRef CAS.
- P. Schneider, M. B. Caspersen, K. Mondorf, T. Halkier, L. K. Skov, P. R. Østergaard, K. M. Brown, S. H. Brown and F. Xu, Enzyme Microb. Technol., 1999, 25, 502–508 CrossRef CAS.
- P. Sharma, R. Goel and N. Capalash, World J. Microbiol. Biotechnol., 2006, 23(6), 823–832 CrossRef.
- A. Pandey, G. Szakacs, C. R. Soccol, J. A. Rodriguez-Leon and V. T. Soccol, Bioresour. Technol., 2001, 77, 203–214 CrossRef CAS.
- A. Hatakka, FEMS Microbiol. Rev., 1994, 13, 125–135 CrossRef CAS.
- H. Patel, V. K. Yadav, K. K. Yadav, N. Choudhary, H. Kalasariya, M. M. Alam, A. Gacem, M. Amanullah, H. A. Ibrahium, J. W. Park, S. Park and B. H. Jeon, Water, 2022, 14(19), 3163 CrossRef CAS.
- J. M. Khaled, S. A. Alyahya, R. Govindan, C. K. Chelliah, M. Maruthupandy, N. S. Alharbi, S. Kadaikunnan, R. Issac, S. Murugan and W. J. Li, Environ. Res., 2022, 207, 112211 CrossRef CAS.
- O. v. Morozova, G. P. Shumakovich, S. V. Shleev and Y. I. Yaropolov, Appl. Biochem. Microbiol., 2007, 43(5), 523–535 CrossRef CAS.
- F. Rosconi, L. F. Fraguas, G. Martínez-Drets and S. Castro-Sowinski, Enzyme Microb. Technol., 2005, 36, 800–807 CrossRef CAS.
- J. Margot, C. Bennati-Granier, J. Maillard, P. Blánquez, D. A. Barry and C. Holliger, AMB Express, 2013, 3, 1–14 CrossRef.
- K. Dhakar and A. Pandey, Enzym. Res., 2013, 869062 Search PubMed.
- C. Pezzella, V. Lettera, A. Piscitelli, P. Giardina and G. Sannia, Appl. Microbiol. Biotechnol., 2012, 97(2), 705–717 CrossRef.
- U. N. Dwivedi, P. Singh, V. P. Pandey and A. Kumar, J. Mol. Catal. B Enzym., 2011, 68, 117–128 CrossRef CAS.
- B. Chefetz, Y. Chen and Y. Hadar, Appl. Environ. Microbiol., 1998, 64, 3175–3179 CrossRef CAS.
- K. Hildén, T. K. Hakala and T. Lundell, Biotechnol. Lett., 2009, 31(8), 1117–1128 CrossRef.
- L. Gianfreda, F. Xu and J. M. Bollag, Bioremediat. J., 2010, 3, 1–26 Search PubMed.
- S. Mazumder, S. K. Basu and M. Mukherjee, Eng. Life Sci., 2009, 9, 45–52 CrossRef CAS.
- G. Palmieri, P. Giardina, C. Bianco, B. Fontanella and G. Sannia, Appl. Environ. Microbiol., 2000, 66, 920–924 CrossRef CAS.
- B. Bertrand, F. Martínez-Morales, R. Tinoco, S. Rojas-Trejo, L. Serrano-Carreón and M. R. Trejo-Hernández, World J. Microbiol. Biotechnol., 2013, 30(1), 135–142 CrossRef.
- J. A. Saraiva, A. P. M. Tavares and A. M. R. B. Xavier, Appl. Biochem. Biotechnol., 2012, 167(4), 685–693 CrossRef CAS.
- F. M. Olajuyigbe, O. Y. Adetuyi and C. O. Fatokun, Int. J. Biol. Macromol., 2019, 125, 856–864 CrossRef CAS.
- L. N. Nguyen, F. I. Hai, A. Dosseto, C. Richardson, W. E. Price and L. D. Nghiem, Bioresour. Technol., 2016, 210, 108–116 CrossRef CAS.
- H. H. Nguyen and M. Kim, Appl. Sci. Converg. Technol., 2017, 26, 157–163 CrossRef.
- S. Kashefi, S. M. Borghei and N. M. Mahmoodi, J. Mol. Liq., 2019, 276, 153–162 CrossRef CAS.
- M. F. Hullo, I. Moszer, A. Danchin and I. Martin-Verstraete, J. Bacteriol., 2001, 183, 5426 CrossRef CAS.
- M. C. Machczynski, E. Vijgenboom, B. Samyn and G. W. Canters, Protein Sci., 2004, 13, 2388–2397 CrossRef CAS.
- K. N. Niladevi, P. S. Sheejadevi and P. Prema, Appl. Biochem. Biotechnol., 2008, 151, 9–19 CrossRef CAS.
- A. T. Fernandes, J. M. Damas, S. Todorovic, R. Huber, M. C. Baratto, R. Pogni, C. M. Soares and L. O. Martins, FEBS J., 2010, 277, 3176–3189 CrossRef CAS.
- K. Miyazaki, Extremophiles, 2005, 9, 415–425 CrossRef CAS.
- J. Bains, N. Capalash and P. Sharma, Biotechnol. Lett., 2003, 25, 1155–1159 CrossRef CAS.
- T. Kupper, C. Plagellat, R. C. Brändli, L. F. de Alencastro, D. Grandjean and J. Tarradellas, Water Res., 2006, 40, 2603–2612 CrossRef CAS.
- A. Wick, O. Marincas, Z. Moldovan and T. A. Ternes, Water Res., 2011, 45, 3638–3652 CrossRef CAS.
- S. Comtois-Marotte, T. Chappuis, S. Vo Duy, N. Gilbert, A. Lajeunesse, S. Taktek, M. Desrosiers, É. Veilleux and S. Sauvé, Chemosphere, 2017, 166, 400–411 CrossRef CAS.
- A. Palardy, J. P. Gagné and L. Tremblay, J Xenobiot, 2016, 5, 37–39 Search PubMed.
- S. N. Grove and C. E. Bracker, J. Bacteriol., 1970, 104, 989–1009 CrossRef CAS.
- H. Kojima, F. Sata, S. Takeuchi, T. Sueyoshi and T. Nagai, Toxicology, 2011, 280, 77–87 CrossRef CAS.
- S. Foteinis, A. G. L. Borthwick, Z. Frontistis, D. Mantzavinos and E. Chatzisymeon, J. Cleaner Prod., 2018, 182, 8–15 CrossRef CAS.
- J. A. Rivera-Jaimes, C. Postigo, R. M. Melgoza-Alemán, J. Aceña, D. Barceló and M. López de Alda, Sci. Total Environ., 2018, 613–614, 1263–1274 CrossRef CAS.
- L. You, V. T. Nguyen, A. Pal, H. Chen, Y. He, M. Reinhard and K. Y. H. Gin, Sci. Total Environ., 2015, 536, 955–963 CrossRef CAS.
- L. You, V. T. Nguyen, A. Pal, H. Chen, Y. He, M. Reinhard and K. Y. H. Gin, Sci. Total Environ., 2015, 536, 955–963 CrossRef CAS.
- S. Zhou, C. Di Paolo, X. Wu, Y. Shao, T. B. Seiler and H. Hollert, Environ. Int., 2019, 128, 1–10 CrossRef CAS.
- T. Tsuda, T. Igawa, K. Tanaka and D. Hirota, Bull. Environ. Contam. Toxicol., 2011, 87(3), 307–311 CrossRef CAS.
- A. S. Mansano, R. A. Moreira, M. Pierozzi, T. M. A. Oliveira, E. M. Vieira, O. Rocha and M. H. Regali-Seleghim, Environ. Pollut., 2016, 213, 160–172 CrossRef CAS.
- E. L. Bell, W. Finnigan, S. P. France, A. P. Green, M. A. Hayes, L. J. Hepworth, S. L. Lovelock, H. Niikura, S. Osuna, E. Romero, K. S. Ryan, N. J. Turner and S. L. Flitsch, Nat. Rev. Methods Primers, 2021, 1(1), 1–21 CrossRef.
- S. A. H. Martínez, E. M. Melchor-Martínez, J. A. R. Hernández, R. Parra-Saldívar and H. M. N. Iqbal, Fuel, 2022, 312, 122927 CrossRef.
- D. M. Liu, J. Chen and Y. P. Shi, TrAC, Trends Anal. Chem., 2018, 102, 332–342 CrossRef CAS.
- Y. Chen, J. Vymazal, T. Březinová, M. Koželuh, L. Kule, J. Huang and Z. Chen, Sci. Total Environ., 2016, 566–567, 1660–1669 CrossRef CAS.
- M. Bilal and H. M. N. Iqbal, Coord. Chem. Rev., 2019, 388, 1–23 CrossRef CAS.
- F. Darvishi, M. Moradi, C. Jolivalt and C. Madzak, Ecotoxicol. Environ. Saf., 2018, 165, 278–283 CrossRef CAS.
- D. O. Lopez-Cantu, R. B. González-González, E. M. Melchor-Martínez, S. A. H. Martínez, R. G. Araújo, L. Parra-Arroyo, J. E. Sosa-Hernández, R. Parra-Saldívar and H. M. N. Iqbal, Int. J. Biol. Macromol., 2022, 194, 676–687 CrossRef CAS.
- L. Alvarado-Ramírez, M. Rostro-Alanis, J. Rodríguez-Rodríguez, C. Castillo-Zacarías, J. E. Sosa-Hernández, D. Barceló, H. M. N. Iqbal and R. Parra-Saldívar, Int. J. Biol. Macromol., 2021, 181, 683–696 CrossRef.
- S. Aslam, M. Asgher, N. A. Khan and M. Bilal, J. Water Proc. Eng., 2021, 40, 101971 CrossRef.
- H. M. Xu, X. F. Sun, S. Y. Wang, C. Song and S. G. Wang, Sep. Purif. Technol., 2018, 204, 255–260 CrossRef CAS.
- W. Zhou, W. Zhang and Y. Cai, Chem. Eng. J., 2021, 403, 126272 CrossRef CAS.
- X. Lyu, R. Gonzalez, A. Horton and T. Li, Catalysts, 2021, 11, 1211 CrossRef CAS.
- D. M. Liu, J. Chen and Y. P. Shi, Trends Anal. Chem., 2018, 102, 332–342 CrossRef CAS.
- C. Simón-Herrero, M. Naghdi, M. Taheran, S. Kaur Brar, A. Romero, J. L. Valverde, A. Avalos Ramirez and L. Sánchez-Silva, J. Hazard. Mater., 2019, 376, 83–90 CrossRef.
- M. Masjoudi, M. Golgoli, Z. Ghobadi Nejad, S. Sadeghzadeh and S. M. Borghei, Chemosphere, 2021, 263, 128043 CrossRef CAS.
- T. Taghizadeh, A. Talebian-Kiakalaieh, H. Jahandar, M. Amin, S. Tarighi and M. A. Faramarzi, J. Hazard. Mater., 2020, 386, 121950 CrossRef CAS.
- S. Zhang, Z. Wu, G. Chen and Z. Wang, Catalysts, 2018, 8, 286 CrossRef.
- H. F. Ma, G. Meng, B. K. Cui, J. Si and Y. C. Dai, Chem. Eng. Res. Des., 2018, 132, 664–676 CrossRef CAS.
- S. Zhang, Z. Wu, G. Chen and Z. Wang, Catalysts, 2018, 8(7), 286 CrossRef.
- Z. Li, Z. Chen, Q. Zhu, J. Song, S. Li and X. Liu, J. Hazard. Mater., 2020, 399, 123088 CrossRef CAS.
- H. de P. Riedi, M. V. de Liz, D. M. Braga, A. B. Ianoski, T. de Freitas Pereira, T. Brugnari, C. W. I. Haminiuk and G. M. Maciel, Int. J. Environ. Res., 2022, 16(3), 1–12 Search PubMed.
| This journal is © The Royal Society of Chemistry 2024 |