Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Plasma protein corona on silica nanoparticles enhances exocytosis†
Laura
Dietz‡§
ab,
Julia
Simon‡§
ab,
Kai R.
Speth‡
ab,
Katharina
Landfester
 *a and
Volker
Mailänder
*a and
Volker
Mailänder
 *ab
*ab
aMax Planck Institute for Polymer Research, Ackermannweg 10, 55128 Mainz, Germany. E-mail: landfester@mpip-mainz.mpg.de; mailaender@mpip-mainz.mpg.de
bDepartment of Dermatology, University Medical Center Mainz, Langenbeckstraße 1, 55131 Mainz, Germany
First published on 5th May 2025
Abstract
While the influence of the protein corona on nanoparticle uptake in mammalian cells is well understood, little is known about the influence of the protein corona on nanoparticle exocytosis. However, the exocytosis of nanoparticles also contributes to the therapeutic efficacy as it influences the net delivery of nanoparticles to a cell. In this study we demonstrate that the exocytosis of silica nanoparticles from HCT 116 cells is enhanced by the pre-adsorption of a human plasma protein corona. This pre-adsorption effect also depends on the diameter of the nanoparticles. The exocytosis of small silica nanoparticles (10 nm) is less pronounced, while the exocytosis of larger silica nanoparticles (100 nm) is significantly increased in the presence of a protein corona. A proteomic analysis of the plasma protein corona of the different-sized silica nanoparticles (10 nm, 30 nm, 50 nm, and 100 nm) reveals different protein compositions. Apolipoproteins and coagulation proteins are enriched in a size-dependent manner with high amounts of apolipoproteins adsorbed to small silica nanoparticles. The findings underscore the significance of the nanoparticle protein corona for exocytosis and demonstrate the need to engineer nanocarriers that are not exocytosed rapidly to enhance the efficacy in drug delivery.
1. Introduction
Nanoparticles are promising for the targeted delivery of a range of therapeutically relevant cargo molecules including, but not limited to, nucleic acids, proteins, and small molecules for different treatment possibilities.1 In particular, the usage of lipid nanoparticles for vaccination against COVID-19 and other diseases demonstrated the clinical importance of this technology.2 To achieve that, extensive research has been conducted to understand the uptake of nanoparticles by target cells.3–5 However, the possibility that nanoparticles can be exocytosed by cells and the consequences thereof were rarely addressed. Nevertheless, the exocytosis of nanoparticles also contributes significantly to the overall efficacy of a drug-loaded nanoparticle as it determines the net delivery to a cell.6–8 A secondary problem is that exocytosed nanoparticles have adsorbed intracellular proteins or are covered with other biological components such as biomembranes.9,10 This has been shown to have significant impact on the reuptake behavior, which can result in undesirable side effects and toxicity.11 The formation of a protein corona has been identified as a crucial factor influencing not only the uptake of nanoparticles but also their subsequent exocytosis. The present study aims to investigate the influence of the nanoparticle plasma protein corona on exocytosis. The specific methodology of this study involves the investigation of the influence of a pre-formed protein corona and how this would not only influence uptake – a subject more widely studied in the field – but also exocytosis. We furthermore investigated whether the effect of a protein corona directing exocytosis could also be size-dependent.Nanomaterials in the size range between 10 nm and 100 nm show distinct interactions with biological cells. Particles with a diameter of 10 nm or less can sometimes even enter cells by passive penetration of the cell membrane, whereas particles with a diameter of 100 nm are likely taken up by clathrin-mediated endocytosis. However, nanoparticles in this size range can also be taken up by pinocytosis, caveolae-dependent endocytosis, and clathrin/caveolae-independent endocytosis.3 Depending on the uptake mechanism, nanoparticles can enter different intracellular trafficking pathways that determine their intracellular fate. After endocytosis, the nanoparticles are directed to their intracellular fate by the endosomal system ultimately resulting in cargo release, nanoparticle degradation, storage, or exocytosis.12,13 Nanoparticle exocytosis can occur via multiple pathways and often multiple pathways are involved simultaneously. The main routes of nanoparticle exocytosis are via the recycling endosome or lysosomal secretion, or are associated with exosomes.6,14,15 Nanoparticle exocytosis has been reported for many nanoparticle types. Mesoporous silica nanoparticles were rapidly endocytosed and exocytosis was reported as early as 40 min to a few hours after the removal of excess mesoporous silica nanoparticles.9,15 Exocytosis of gold nanoparticles with diameters between 2 nm and 100 nm was also reported.14,16–18 These studies demonstrated that nanoparticle size and surface modifications are critical factors for exocytosis.
The interaction of nanocarriers with cells is not only determined by the diameter or surface chemistry of the nanocarrier, but also crucially depends on the protein corona that forms on the nanoparticle surface. The protein corona is a layer of proteins that adsorbs to the nanocarrier upon contact with biological fluids such as blood or blood plasma.19,20 The process of protein corona formation is inevitable when administering a nanocarrier intravenously and is therefore highly important for drug delivery. The protein corona influences the biodistribution, cellular uptake, and intracellular trafficking of nanoparticles.21 This suggests that it may also influence exocytosis, but this was rarely investigated. A further layer of complexity is added as the composition of the protein corona is dependent on the nanoparticle size and hence nanoparticle uptake.22,23 The contributions of the individual factors, particle size and protein composition, for cellular uptake and exocytosis are difficult to separate as they are intertwined.
Here, we investigate the influence of a pre-formed human plasma protein corona on the exocytosis of SiNPs of different sizes. Therefore, we used flow cytometry analysis to determine the uptake and exocytosis of SiNPs with diameters of 10 nm, 30 nm, 50 nm, and 100 nm in the presence and absence of a pre-formed protein corona. In the presence of a protein corona, exocytosis increased in a size-dependent manner. SiNPs with a diameter of 100 nm were exocytosed more than SiNPs with a diameter of 10 nm. We analysed the composition of the protein corona and found a decrease in the amount of lipoproteins with increasing particle size.
2. Results
2.1. Uptake of SiNP depends on protein corona adsorption
In this study, we used fluorescent SiNPs to investigate how a pre-adsorbed protein corona affects the exocytosis of nanoparticles of different biologically relevant sizes from cells. First, we characterised the physicochemical properties of the four different SiNPs. TEM micrographs of the SiNPs showed intact particles with diameters of approximately 10 nm, 30 nm, 50 nm, and 100 nm (Fig. 1). We further analysed the hydrodynamic diameter of the nanoparticles (Dh) with multi-angle DLS and calculated the polydispersity index (PDI) from measurements at 90° (Table 1 and Fig. S1†). The Dh was in good agreement with the expected particle diameters. The PDI of the smaller SiNPs was higher, indicating a broader size distribution, whereas the size distribution of larger SiNPs was narrower. To assess the influence of nanoparticle diameter on exocytosis, other surface properties of the SiNPs must be similar despite the difference in size. Therefore, we analysed the ζ-potential of the SiNPs. The 10 nm and 30 nm SiNPs had a ζ-potential of approximately −25 mV and the 50 nm and 100 nm SiNPs had a ζ-potential of approximately −40 mV. The negative ζ-potential is caused by the free hydroxyl groups of the silica surface, indicating that the surface chemistry of all SiNPs is similar and no additional surfactant is used for stabilisation.| D h [nm] | PDI | ζ-potential [mV] ± SD | Fluorescence [AU] | |
|---|---|---|---|---|
| 10 nm | 10 | 0.340 | −23 ± 3.7 | 11![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 150 150 |
| 30 nm | 28 | 0.154 | −24 ± 2.9 | 9075 |
| 50 nm | 44 | 0.090 | −38 ± 0.3 | 8996 |
| 100 nm | 102 | 0.025 | −42 ± 3.2 | 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 784 784 |
Before investigating the influence of a pre-adsorbed protein corona on SiNP exocytosis, we tested the influence on the uptake of SiNPs.
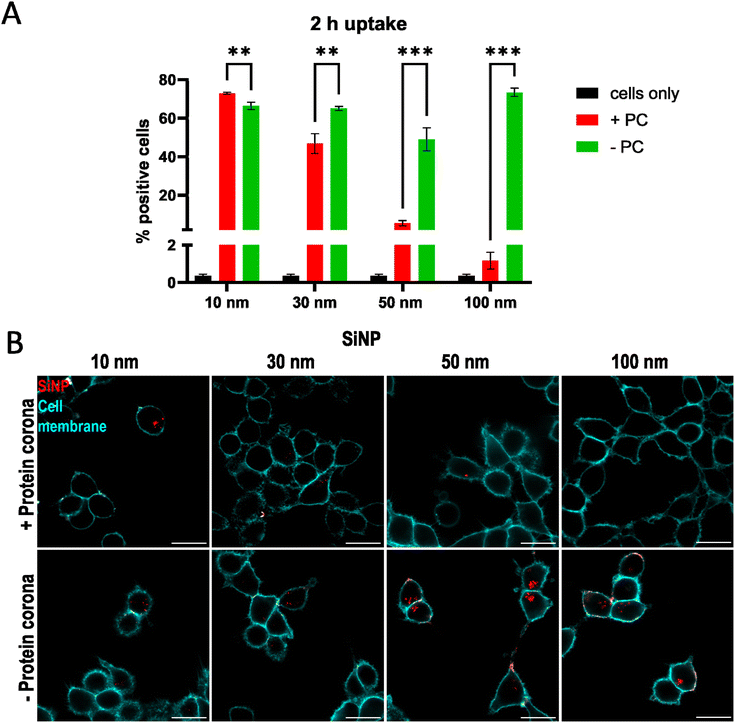
Fig. 2A shows the uptake of SiNPs in the presence and absence of a pre-adsorbed human plasma protein corona in HCT116 cells after 2 h incubation. In the absence of a protein corona, cells took up the different sized SiNPs to a similar extent. 50–70% of cells were positive for SiNPs and the uptake was not affected by particle size. In contrast, the adsorption of a protein corona affected the uptake of the different SiNPs differently. The uptake of 100 nm SiNPs was reduced to almost 0% positive cells upon the adsorption of a protein corona. The uptake of 50 nm SiNPs was also reduced to about 10%. The uptake of 30 nm SiNPs was reduced from 60% positive cells to approximately 45% positive cells. Interestingly, the uptake of the 10 nm SiNPs was not reduced by the adsorption of a protein corona. This trend was also reflected by the median fluorescence intensity (MFI) of the cells. Here, the MFI decreased with increasing size in the presence of a protein corona. The MFI of cells incubated with SiNPs without protein corona was similar between different SiNPs (Fig. 2A). The cell viability was not affected by incubation with SiNPs (Fig. S2B†). This experiment demonstrated that the influence of a protein corona on nanoparticle uptake is size-dependent. The uptake of small SiNPs was not affected by the adsorption of a protein corona, whereas the uptake of larger SiNPs was increasingly affected in the presence of a protein corona. The reduction of nanoparticle uptake in the presence of a protein corona that we observed here has been frequently observed in previous studies and is generally known as the stealth effect.24 In the absence of a protein corona, the nanoparticle surface interacts directly with the cell membrane. This can lead to the passive penetration of the cell membrane and therefore a closer contact between the nanoparticle and the cell surface. For very small nanoparticles direct entry into the cytosol has been observed.25
In addition to flow cytometry, fluorescence micrographs of SiNP uptake in the presence and absence of a protein corona after 2 h confirmed the dependence of the influence of the protein corona on particle size (Fig. 2B). The fluorescence micrographs confirmed that the SiNPs were located intracellularly after 2 h of uptake.
2.2. Exocytosis of SiNPs was increased by pre-adsorption of a protein corona
To investigate whether a pre-adsorbed protein corona has an effect on SiNP exocytosis, we incubated HCT 116 cells with different SiNPs for 2 h. Subsequently, the cell culture supernatant was discarded, and the cells were washed to remove any SiNPs attached to the cell surface. Particle-free medium was added to the cells and the intracellular particle content was assessed 2 h and 22 h later (Fig. 3 and Fig. S3, S4†). The particles added without pre-adsorption of a protein corona were not strongly exocytosed. Approximately 90% of the cells that were positive immediately after the uptake period remained particle-positive 22 h after removal of the SiNPs. In contrast, for particles with a pre-adsorbed protein corona a time- and size-dependent decrease of the intracellular particle signal was observed compared to the signal immediately after the uptake period. Exocytosis of SiNPs was rapid as we observed a decrease of intracellular signal 2 h after the initial uptake period. In our study, we observed the strongest exocytosis for 100 nm SiNPs which showed a decrease of particle-positive cells to approximately 30% of the initially positive cells 2 h after the medium change. The percentage of cells positive for 50 nm SiNPs was reduced to around 50% after 2 h of exocytosis and the signal for 30 nm SiNPs was only reduced to about 80%. The 10 nm SiNPs did not show a reduced intracellular signal after 2 h of exocytosis. The MFI of the cells incubated with the 10 nm and 30 nm SiNPs decreased during the exocytosis period as the cells were oversaturated with nanoparticles due to the high uptake in the absence of a protein corona (Fig. S3 and Fig. S4†). However, the percentage of positive cells is a more robust measure for particle uptake and exocytosis. Interestingly, the intracellular signals of 10 nm, 30 nm, and 50 nm SiNPs decreased even more after 22 h of exocytosis. For the 100 nm SiNPs, the intracellular signal increased again after 22 h of exocytosis, suggesting a re-uptake of exocytosed particles. We also measured the viability of cells subjected to different conditions (Fig. S5 and S6†). No severe toxicity was observed compared to that of untreated cells. Also, the SiNPs without protein corona did not have a cytotoxic effect at the concentration and incubation time used here. In conclusion, the exocytosis of SiNPs was greatly enhanced in the presence of a plasma-adsorbed protein corona, and smaller SiNPs were exocytosed less compared to larger SiNPs. Rapid exocytosis of 100 nm silica nanoparticles 6 h after uptake was also observed by Yanes et al.15 Furthermore, another study showed that exocytosis only occurred in the presence of serum, which could be explained by the involvement of a protein corona in the exocytosis of nanoparticles.26 In the absence of a protein corona, the nanoparticle surface interacts directly with the cell membrane and no active, receptor-mediated uptake process can take place. This can lead to the passive penetration of the cell membrane and direct entry into the cytosol, especially for very small nanoparticles.25 This uptake mechanism would not result in high levels of nanoparticle penetration into the endosomal system. As the endosomal system is the starting point for most exocytosis pathways this could suppress nanoparticle exocytosis.12,13 This mechanism is a possible explanation for the very low exocytosis of nanoparticles in the absence of a protein corona and a protein corona could be a prerequisite for exocytosis.2.3. The pre-adsorbed protein corona composition is particle size dependent
Since the protein corona had a major effect on the uptake and exocytosis of SiNPs in a nanoparticle size-dependent manner, we wanted to further investigate the composition of the protein corona pre-adsorbed on the different sized particles. It is known that small particles have a high surface curvature and therefore, less protein adsorbs to their surface.27–29 We also observed this effect on 10 nm, 30 nm, and 50 nm SiNPs which adsorbed increasing amounts of protein per surface area (Fig. 4A). For the 100 nm SiNPs, the amount of protein adsorbed per surface area decreased slightly compared to the 50 nm SiNPs. The increasing amount of protein per surface area might explain the increasing influence that the protein corona had on cellular uptake and exocytosis. Interestingly, the composition of the protein corona was highly dependent on the particle size as well. In particular, the amount of lipoproteins and coagulation proteins crucially depended on particle size. The protein corona of 10 nm SiNPs contained high amounts of lipoproteins but comparably low amounts of coagulation proteins (Fig. 4B). This ratio was increasingly reversed with increasing particle size. The protein corona of 100 nm SiNPs contained high amounts of coagulation proteins but comparably small amounts of lipoproteins. In contrast, the abundance of other protein classes like acute phase proteins, immunoglobulins, and complement system proteins was unaffected by particle size. We also observed a strong dependence on particle size for individual proteins. The amount of apolipoproteins A-1, B-100, and E decreased with increasing particle size. In contrast, the abundance of coagulation factor XI, histidine-rich glycoprotein as well as plasminogen and α-2-macroglobulin (see SI_LC MS_protein list.xlsx†) which belong to or are associated with coagulation proteins, showed a tendency to increase with increasing particle size. Interestingly, the apolipoprotein C-III was adsorbed more to 100 nm SiNPs, although the total amount was lower than those of other apolipoproteins (see SI_LC MS_protein list.xlsx†).Very small SiNPs appeared to attract more apolipoproteins into the pre-adsorbed protein corona, which are proteins known to naturally decorate lipoprotein complexes. This differential adsorption of apolipoproteins to SiNPs with a diameter of around 10 nm could be caused by their similarity in size to high density lipoproteins (HDL). HDLs also exhibit diameters of around 9 to 15 nm.30 The 10 nm SiNPs were taken up equally well in the presence and absence of a protein corona. These particles, when carrying a protein corona, resembled HDLs in terms of size and apolipoprotein composition and thus may be able to use similar active uptake pathways. The mimicry of HDL by nanoparticles was also suggested by Barrán-Berdón et al., who observed apolipoprotein-dependent uptake of nanoparticles by scavenger receptor class B type 1 (SR-B1).31 which is present in many tissues including the endothelium and facilitates receptor-mediated endocytosis of lipoprotein particles in cells.32,33 However, in other experimental settings apolipoproteins have been found to mediate stealth properties,29 where apolipoprotein J (clusterin) was found to be mainly responsible for the stealth effect.29,34 Here, we also observed that the protein corona on 30 nm, 50 nm, and 100 nm SiNPs exhibited a stealth effect. However, apolipoprotein J was not found in high abundance in the protein corona of any of the SiNPs used here. Instead, the decrease in the amount of apolipoproteins present in the SiNP protein corona with increasing particle size may reduce the interaction with specific lipoprotein receptors. In addition, apolipoproteins adsorbed onto larger particles may have a different conformation from that on 10 nm SiNPs which may influence the recognition by lipoprotein receptors.35 Therefore, the protein corona of larger SiNPs exhibited stealth properties rather than mediating the active uptake of SiNPs.
In addition to providing insights into the relationship between the protein corona and SiNPs’ uptake, we observed an increase in exocytosis for all SiNPs. Similar to us, Panyam et al. observed an almost complete abolition of exocytosis in the absence of serum.36 They suggested that this was due to a lack of energy available in the cell culture medium and that exocytosis would be an energy-dependent process. Another possibility is that the presence of the protein corona alters the uptake route of the SiNPs and therefore their intracellular trafficking route. It is possible that the protein corona may also influence the exocytosis of SiNPs. It may be that the specific protein corona leads the SiNPs toward an intracellular trafficking route that is more likely to lead to exocytosis.
In order to investigate the role of the different most abundant proteins in the pre-adsorbed protein corona (Fig. 4C) in exocytosis, we incubated the particles additionally in human single protein solutions (100 μg mL−1) of kininogen I, histidine-rich glycoprotein (HRGP), apolipoprotein A-I and serum albumin and subsequently measured the exocytosis of SiNPs (here the only representative is 100 nm SiNPs) (Fig. S7†). Although we diluted human plasma to the same concentration as those of the single protein solutions (100 μg mL−1) we still could observe a reduced intracellular particle signal after 4 and 24 h of further incubation, both for % of positive cells (∼80% compared to the 2 h timepoint = 100%) and MFI, indicating exocytosis. In line with this, SiNPs pre-incubated in un-diluted human plasma (∼70 mg mL−1) showed a more pronounced exocytosis (Fig. 3). In addition, this time we detected a slightly higher intracellular particle signal for the pristine SiNPs in the exocytosis phases (4 h and 24 h) compared to the 2 h uptake condition. This appears to be counterintuitive, but one has to keep in mind that for the determination of the uptake after 2 h, cells are directly trypsinized, harvested and centrifuged to remove additional particles from the cell surface and remnants which could not be removed by the washing procedure from the extracellular environment. For the other samples (2 + 4 h and 2 + 24 h time points) these particles adhering to the cells on the surface and on the plastic are still in the cell culture. Exactly this amount of particles can still be taken up under the 4 h and 24 h exocytosis conditions, which eventually can cause a higher level of particle signal than the initial signal after 2 h. Similarly, for all single protein coating approaches the remaining particle signal was also higher than that after the corresponding signals after 2 h of uptake, excluding a prominent contribution of these proteins.
Hence, it appears that rather the ensemble of more or all pre-coated plasma proteins contributes to the observed exocytosis phenomena for the different SiNPs in this study. The cell viability was not affected when HCT 116 cells were treated with SiNPs pre-coated with one of the single proteins (Fig. S8†).
We also want to point out that the different protein corona compositions enable uptake by different uptake mechanisms – an aspect not investigated in this study.
Exocytosis of SiNPs increased with increasing particle size. This is consistent with the size-dependence of the protein corona composition observed here. Smaller SiNPs (10 nm) adsorbed high amounts of apolipoproteins and low amounts of coagulation proteins. This composition resulted in very low exocytosis. In contrast, large SiNPs (100 nm) adsorbed low amounts of apolipoproteins and high amounts of coagulation proteins.
3. Conclusion
Here, we show that not only endocytosis but also exocytosis of SiNPs was strongly influenced by the presence of a protein corona. The SiNPs acquire a protein corona that enhances exocytosis in a nanoparticle size-dependent manner. Furthermore, the depletion of apolipoproteins E, A1, and B-100 was associated with enhanced exocytosis. Small SiNPs were less prone to exocytosis even in the presence of a protein corona. In contrast, the formation of a protein corona enhanced the exocytosis of larger SiNPs. In the context of nanoparticle drug delivery, these findings have important implications. To achieve a high intracellular drug delivery, it is important to prolong the residence time of the nanoparticles inside the cell. Therefore, rapid exocytosis of nanoparticles would be unfavorable and could reduce the efficacy of drug-loaded nanoparticles. In our study, we show that the presence of a human serum protein corona increases nanoparticle exocytosis in vitro. However, the formation of a protein corona upon injection of nanoparticles into the bloodstream is inevitable and is likely to enhance the exocytosis of nanoparticles in an in vivo setting. This raises the question whether the development of nanocarriers with a reduced exocytosis capacity is necessary to improve the net delivery of nanocarriers. Our study showed that it would be beneficial to use smaller SiNPs that acquire a protein corona that prevents exocytosis. On the other hand, there are attempts to use nanoparticle exocytosis as a mechanism for deeper tumor penetration or further systemic spreading of nanoparticles in cellular vesicles.37 For this approach it would be interesting to employ larger SiNPs. This requires collection of exocytosed nanoparticles of much higher quantity and therefore is not a focus of this study. In conclusion, the protein corona is an important factor in nanoparticle drug delivery that is not only able to drastically reduce the uptake of nanoparticles into cells but can also enhance their exocytosis. Therefore, the protein corona influences the net delivery of nanoparticles which ultimately determines the efficacy of a drug delivery system.4. Experimental section/methods
4.1. Silica nanoparticles
Mesoporous silica nanoparticles (SiNPs) with sizes 10 nm, 30 nm, 50 nm, and 100 nm from the sicastar®-redF series were purchased from Micromod Partikeltechnologie GmbH (Rostock, Germany). According to the manufacturer, particles were produced via the Stöber process38 and contained covalently bound rhodamine b for fluorescent labeling (excitation: 569 nm, emission: 585 nm).4.2. Cell culture
HCT 116 human colorectal cancer cells were cultivated in high glucose Dulbecco's Modified Eagle's Medium (DMEM) containing fetal bovine serum (FBS) (10%), penicillin (100 U mL−1) and streptomycin (100 mg mL−1). Cells were grown under 37 °C, CO2 (5%), and 95% humidity conditions. For passaging, the cells were incubated for 2 min with trypsin-EDTA (0.25%) at 37 °C under CO2 (5%), and then centrifuged (22 °C and 300g) for 5 min (all reagents were from Thermo Fisher Scientific, Waltham, USA).4.3. Protein corona adsorption
The protein corona was adsorbed as described previously by our group.39–41 In brief, human citrate plasma (0.5 mL) and SiNP (1 mg mL−1) were incubated for 1 h at 37 °C and 300 rpm. Plasma was removed by centrifugation at 20![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000g and 4 °C for 1 h. Subsequently, the pellet was resuspended in phosphate buffered saline (PBS) and centrifuged again. For cellular uptake experiments, the pellet was washed twice and resuspended in DMEM without FBS after the last run. For fluorescence calibration, both particles with and without protein corona were measured with an M1000 plate reader (Tecan, Männedorf, Switzerland) with 50 flashes for each sample. By using triplicate measurements the standard deviation was <7.5% for fluorescence measurements with all experimental errors included. For liquid chromatography-mass spectrometry (LC-MS) measurement, the pellet was washed three times with PBS. The final pellet was resuspended in desorption buffer (SDS (2% (w/v)) + Tris*HCL (62.5 mM) in H2O (3 mL)) and incubated at 95 °C for 5 min to desorb the proteins of the hard protein corona. Finally the suspension was centrifuged a last time to separate the remaining particles. The supernatant containing the desorbed proteins was subjected to digestion for LC-MS sample preparation.
000g and 4 °C for 1 h. Subsequently, the pellet was resuspended in phosphate buffered saline (PBS) and centrifuged again. For cellular uptake experiments, the pellet was washed twice and resuspended in DMEM without FBS after the last run. For fluorescence calibration, both particles with and without protein corona were measured with an M1000 plate reader (Tecan, Männedorf, Switzerland) with 50 flashes for each sample. By using triplicate measurements the standard deviation was <7.5% for fluorescence measurements with all experimental errors included. For liquid chromatography-mass spectrometry (LC-MS) measurement, the pellet was washed three times with PBS. The final pellet was resuspended in desorption buffer (SDS (2% (w/v)) + Tris*HCL (62.5 mM) in H2O (3 mL)) and incubated at 95 °C for 5 min to desorb the proteins of the hard protein corona. Finally the suspension was centrifuged a last time to separate the remaining particles. The supernatant containing the desorbed proteins was subjected to digestion for LC-MS sample preparation.
4.4. Single protein coatings
We based our experimental procedure on the experience of the group in the single protein coating experiments.42,43 Hence, the already described procedure for the protein corona coating was applied in a similar fashion. We incubated 500 μg of 100 nm SiNP, accounting for a particle surface area of ∼0.015 m2 with 30 μg of single protein solutions in a total volume of 300 μl (ensuring a protein concentration of 100 μg mL−1) for 1 h at 37 °C at 300 rpm. As single protein solutions we have chosen the most abundant proteins as revealed by our LC-MS analysis for the composition of the pre-coated protein corona on SiNPs, namely: kininogen I (Sigma Aldrich), histidine rich glycoprotein (MedChemExpress), apolipoprotein A-I (MyBioSource) and human serum albumin (Sigma Aldrich). Instead of applying an excess amount of the complex physiological human plasma control, we diluted it to the same final protein concentration of 100 μg mL−1 (named ‘PC diluted’ accordingly) to ensure comparability to the single protein solutions as also done elsewhere.44 Further preparation of different coated SiNPs was done as described above in the section Protein corona adsorption for cellular uptake experiments.4.5. Protein quantification per particle surface area
As written in the previous section, 500 μg of SiNPs were incubated with 0.5 mL of human plasma. Using the values of the density of the particles (2.0 g cm−3), the solid content and the diameter (all values as provided by the manufacturer), the amount of adsorbed proteins per surface area (in mg protein per m2) can be calculated. Using the diameter and assuming a sphere (as also suggested by the manufacturer and as is evident from our TEM images) one can obtain the area and the volume of a single particle. Using the density and multiplying it with the volume of a single particle one obtains the mass of a single particle. By dividing the solid content (given as mass concentration in g mL−1) with the mass of a single particle one obtains the number of particles per mL (which was also provided by the manufacturer as a control). By multiplying the area of a single particle with the number of particles per mL one obtains the area of all particles per mL. For the SiNPs with the sizes 10, 30 and 50 nm with a concentration of 25 mg mL−1 we used a volume of 40 μL and for the SiNPs with 100 nm with a concentration of 50 mg mL−1, we used a volume of 20 μL to account for 500 μg of the particles accordingly. Based on these results, one can calculate back the area of particles provided in these aliquots as 0.3 m2 (for 10 nm), 0.1 m2 (for 30 nm), 0.06 m2 (for 50 nm) and 0.03 m2 (for 100 nm). The desorbed proteins were collected in 100 μl of desorption buffer and subsequently the protein concentration was determined via Pierce assay. The protein amount was finally calculated back to obtain 1 m2.The concentration of the desorbed corona proteins was determined by using a Pierce™ 660 nm Protein Assay Reagent (Thermo Scientific, Germany) following the manufacturer's instructions. Due to the presence of SDS in the desorption buffer the assay reagent was supplemented with the Ionic Detergent Compatibility Reagent (Thermo Scientific, Germany). A standard calibration curve was recorded with bovine serum albumin (Sigma-Aldrich, Germany). The absorption was measured with an Infinite M1000 plate reader (Tecan, Switzerland) at 660 nm.
4.6. In-solution digestion and LC-MS measurement
The in-solution digestion and LC-MS measurements were performed as previously described by our group.45–47 In short, SDS was removed by using Pierce Detergent Removal Spin Columns (Thermo Fisher Scientific, Waltham, USA) followed by protein precipitation using a ProteoExtract protein precipitation kit (Merck Millipore, Darmstadt, Germany). After isolation, proteins were resuspended in RapiGest SF (Waters, Milford, USA), reduced with dithiothreitol (Sigma-Aldrich, St. Louis, USA), and alkylated with iodoacetamide (Sigma-Aldrich, St. Louis, USA). Tryptic digestion of the protein was performed at a protein![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) trypsin ratio of 50
trypsin ratio of 50![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 for 18 h at 37 °C. After stopping the digestion by adding HCl (2 μL) (Sigma-Aldrich, St. Louis, USA) and removing the degradation products of RapiGest by centrifugation, the peptides were subjected to LC-MS measurements. Therefore, the samples were diluted with formic acid (0.1%) and spiked with Hi3 E. coli (50 fmol μL−1) (Waters, Milford, USA). Measurements were performed using a nanoACQUITY UPLC system coupled to a Synapt G2-SI mass spectrometer (Waters, Milford, USA). Electrospray ionization was performed in positive mode with a NanoLockSpray source. As a reference, Glu-fibrinopeptide (150 fmol μL−1) at a flow rate of 0.5 μL min−1 was injected and a sample flow rate of 0.3 μL min−1 was set. The mass spectrometer was operated in a resolution mode performing data-independent acquisition (MSE). Data were processed using MassLynx 4.1 and proteins were identified using Progenesis QI 2.0. A reviewed human database downloaded from UniProt was used for protein identification. Noise reduction thresholds were set for low energy, high energy, and peptide intensity at 120, 25, and 750 counts, respectively. A maximum protein mass of 600 kDa, one missed cleavage, fixed carbamidomethyl modification for cysteine, variable oxidation of methionine, and a false discovery rate of 4% for proteins were set for protein and peptide identification. For peptide identification at least three assigned fragments and for protein identification at least two assigned peptides and five assigned fragments are required. The TOP3/HI3 approach was used for the quantification of each protein in fmol.48
1 for 18 h at 37 °C. After stopping the digestion by adding HCl (2 μL) (Sigma-Aldrich, St. Louis, USA) and removing the degradation products of RapiGest by centrifugation, the peptides were subjected to LC-MS measurements. Therefore, the samples were diluted with formic acid (0.1%) and spiked with Hi3 E. coli (50 fmol μL−1) (Waters, Milford, USA). Measurements were performed using a nanoACQUITY UPLC system coupled to a Synapt G2-SI mass spectrometer (Waters, Milford, USA). Electrospray ionization was performed in positive mode with a NanoLockSpray source. As a reference, Glu-fibrinopeptide (150 fmol μL−1) at a flow rate of 0.5 μL min−1 was injected and a sample flow rate of 0.3 μL min−1 was set. The mass spectrometer was operated in a resolution mode performing data-independent acquisition (MSE). Data were processed using MassLynx 4.1 and proteins were identified using Progenesis QI 2.0. A reviewed human database downloaded from UniProt was used for protein identification. Noise reduction thresholds were set for low energy, high energy, and peptide intensity at 120, 25, and 750 counts, respectively. A maximum protein mass of 600 kDa, one missed cleavage, fixed carbamidomethyl modification for cysteine, variable oxidation of methionine, and a false discovery rate of 4% for proteins were set for protein and peptide identification. For peptide identification at least three assigned fragments and for protein identification at least two assigned peptides and five assigned fragments are required. The TOP3/HI3 approach was used for the quantification of each protein in fmol.48
4.7. Flow cytometry
HCT 116 cells were seeded into 24 well plates (Greiner Bio-one, Frickenhausen, Germany). 150,000 cells were seeded per well in FBS-containing DMEM. The next day, 1 h prior to uptake with SiNPs the culture medium was removed and replaced with FBS-free medium. Next, the cells were incubated with SiNPs (5 μg mL−1) in the presence or absence of a pre-formed protein corona in serum-free medium for 2 h, 4 h, or 24 h. For measuring the exocytosis of SiNPs, the cell culture medium was exchanged after 2 h, the cells were washed thoroughly 3x times with Dulbecco's Phosphate-Buffered Saline (DPBS) to remove the remaining particles and subsequently were incubated for another 2 h or 22 h in FBS-free DMEM. As a control for dead cells, cells were incubated for 2 h, 4 h, or 24 h with dimethyl sulfoxide (DMSO) (20%) (Sigma-Aldrich, St. Louis, USA). The cells were harvested by trypsin detachment and subsequently centrifuged at 300 g and 22 °C for 5 min. Two of the three pellets of one condition were stained with a LIVE/DEAD™ Fixable Green dead cell staining kit (Thermo Fisher Scientific, Waltham, USA) according to the manufacturer‘s instructions. The pellets were resuspended in 1 mL PBS and at least 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 cells (excluding debris) were measured with an Attune NxT flow cytometer (Thermo Fisher Scientific, Waltham, USA). For the excitation of the particles, a yellow laser (561 nm) and an emission filter in the range of 530/30 nm were used. For the detection of LVE/DEADTM Fixable Green, a blue laser (488 nm) and an emission filter in the range of 585/16 nm were used. For data analysis, the Attune NxT software was used. Forward scatter/side scatter (FSC/SSC) plots were used to discriminate the cell population and events were depicted as percentages of gated cells and MFI. Afterward, the fluorescence intensity of the SiNP dispersion was used to adjust the difference in fluorescence for particles with and without protein corona. The results were depicted using GraphPad Prism 9 (Dotmatics, Boston, USA).
000 cells (excluding debris) were measured with an Attune NxT flow cytometer (Thermo Fisher Scientific, Waltham, USA). For the excitation of the particles, a yellow laser (561 nm) and an emission filter in the range of 530/30 nm were used. For the detection of LVE/DEADTM Fixable Green, a blue laser (488 nm) and an emission filter in the range of 585/16 nm were used. For data analysis, the Attune NxT software was used. Forward scatter/side scatter (FSC/SSC) plots were used to discriminate the cell population and events were depicted as percentages of gated cells and MFI. Afterward, the fluorescence intensity of the SiNP dispersion was used to adjust the difference in fluorescence for particles with and without protein corona. The results were depicted using GraphPad Prism 9 (Dotmatics, Boston, USA).
4.8. Confocal laser scanning microscopy (cLSM)
HCT 116 cells were seeded into an IBIDI 8 well chamber (ibidi, Gräfelfing, Germany). Cells (100![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000) were seeded per well. The next day, the cells were incubated with SiNPs in the presence and absence of a pre-formed protein corona (10 μg mL−1) in serum-free DMEM for 2 h. The plasma membrane was stained with CellMask™ Deep Red plasma membrane stain (Thermo Fisher Scientific, Waltham, USA) and lysosomes were stained with Invitrogen™ LysoTracker™ Green DND-26 (Thermo Fisher Scientific, Waltham, USA) according to the manufacturer's instructions. The cells were measured live. The images were acquired with an LSM SP5 STED Leica Laser Scanning Confocal Microscope (Leica, Wetzlar, Germany) and a Leica TCS SP8 (Leica, Wetzlar, Germany), equipped with a multi-laser combination and five detectors (range of 400–800 nm). SiNPs were excited at 561 nm and detected at 575–640 nm. CellMask™ Deep Red plasma membrane stain was excited at 633 nm and detected at 644–800 nm. LysoTracker™ Green DND-26 was excited at 469 nm and detected at 503–541 nm. Images were taken by sequential scanning using the LAS X software. For processing and analysis, Fiji was used. Brightness and contrast were adjusted to reflect differences in fluorescence intensity of the different-sized SiNPs.
000) were seeded per well. The next day, the cells were incubated with SiNPs in the presence and absence of a pre-formed protein corona (10 μg mL−1) in serum-free DMEM for 2 h. The plasma membrane was stained with CellMask™ Deep Red plasma membrane stain (Thermo Fisher Scientific, Waltham, USA) and lysosomes were stained with Invitrogen™ LysoTracker™ Green DND-26 (Thermo Fisher Scientific, Waltham, USA) according to the manufacturer's instructions. The cells were measured live. The images were acquired with an LSM SP5 STED Leica Laser Scanning Confocal Microscope (Leica, Wetzlar, Germany) and a Leica TCS SP8 (Leica, Wetzlar, Germany), equipped with a multi-laser combination and five detectors (range of 400–800 nm). SiNPs were excited at 561 nm and detected at 575–640 nm. CellMask™ Deep Red plasma membrane stain was excited at 633 nm and detected at 644–800 nm. LysoTracker™ Green DND-26 was excited at 469 nm and detected at 503–541 nm. Images were taken by sequential scanning using the LAS X software. For processing and analysis, Fiji was used. Brightness and contrast were adjusted to reflect differences in fluorescence intensity of the different-sized SiNPs.
4.9. ζ-potential measurement
A Zetasizer Nano Z (Malvern Panalytical GmbH, Germany) was used for zeta potential determination of SiNPs. For the measurement, the SiNP stock solution (5 μL) was diluted in potassium chloride solution (KCl) (1 mL, 1 mM). The measurement was performed at 20 °C after an equilibration time of 2 min. Each measurement was performed in triplicate and mean values as well as standard deviations were calculated. The ζ-potential was approximated according to the model of Smoluchowski (see also ref. 42).4.10. Multi-angle dynamic light scattering (DLS)
An ALV spectrometer equipped with a goniometer and an ALV-5004 multiple-tau full-digital correlator (320 channels) was used. The light source was a He–Ne laser at a wavelength of 632.8 nm. To control the temperature a thermostat (Julabo) was used. The water used for dilutions was filtered with a GS filter (0.2 μm) (Merck Millipore, Billerica, USA) and the particle stock solution was added unfiltered. The stock solution of the 10 nm SiNPs was filtered with an LCR filter (0.45 μm) (Merck Millipore, Billerica, USA) before dilution. The measurements were performed in quartz cuvettes with an inner radius of 9 mm (Hellma, Mühlheim, Germany).49 The data were analyzed using the CONTIN algorithm as previously reported.504.11. Transmission electron microscopy (TEM)
The samples were dropcast on grids (Plano, Wetzlar, Germany) with a 10 nm carbon film. Images were acquired with a JEOL JEM1400 transmission electron microscope (JEOL, Freising, Germany).4.12. Statistical analysis
Data visualization and statistical analysis was carried out with GraphPad Prism 9 and 10.2.2 (GraphPad Software, USA). The presented flow cytometry data and the protein corona quantification are described by means of descriptive statistics (mean and standard deviation). For statistical analysis of the flow cytometry data multiple unpaired t tests were performed with the Holm–Šídák method for multiple comparisons using a P value threshold of alpha = 0.05. Alternatively, a two-way ANOVA test was performed with the Dunnett‘s multiple comparison test using a P value threshold of alpha = 0.05.Declaration of compliance with regulations
All experiments were performed in compliance with relevant laws or guidelines. All experiments followed institutional guidelines. Human citrate blood plasma was taken from healthy donors at the Department of Transfusion Medicine in Mainz after a physical examination and after obtaining written informed consent in accordance with the Declaration of Helsinki. The blood plasma of 10 healthy donors was pooled and stored at −20 °C. The study was approved by the local ethics committee “Landesärztekammer Rheinland-Pfalz” (Bearbeitungsnummer: 837.439.12 (8540-F)).Data availability
The data supporting this article have been included as part of the ESI.†Data are available on request from the authors.
Conflicts of interest
There are no conflicts to declare.Acknowledgements
This work was supported by DFG-SFB1066 (“Nanodimensional polymer therapeutics for tumor therapy”) and the Max Planck Graduate Center with the Johannes Gutenberg-Universität Mainz (MPGC). The authors acknowledge the support from the Max Planck Society. The authors thank Christoph Sieber for TEM imaging of the nanoparticles and Christine Rosenauer for multi-angle DLS measurements. None of the authors has a conflict of interest in publishing this manuscript. Open access funding is provided by the Max Planck Society.References
- M. J. Mitchell, M. M. Billingsley, R. M. Haley, M. E. Wechsler, N. A. Peppas and R. Langer, Engineering precision nanoparticles for drug delivery, Nat. Rev. Drug Discovery, 2021, 20(2), 101–124 Search PubMed.
- F. P. Polack, S. J. Thomas, N. Kitchin, J. Absalon, A. Gurtman, S. Lockhart, J. L. Perez, G. P. Marc, E. D. Moreira, C. Zerbini, R. Bailey, K. A. Swanson, S. Roychoudhury, K. Koury, P. Li, W. V. Kalina, D. Cooper, R. W. Frenck, L. L. Hammitt, Ö. Türeci, H. Nell, A. Schaefer, S. Ünal, D. B. Tresnan, S. Mather, P. R. Dormitzer, U. Şahin, K. U. Jansen and W. C. Gruber, Safety and Efficacy of the BNT162b2 mRNA Covid-19 Vaccine, N. Engl. J. Med., 2020, 383(27), 2603–2615 Search PubMed.
- M. Sousa de Almeida, E. Susnik, B. Drasler, P. Taladriz-Blanco, A. Petri-Fink and B. Rothen-Rutishauser, Understanding nanoparticle endocytosis to improve targeting strategies in nanomedicine, Chem. Soc. Rev., 2021, 50(9), 5397–5434 Search PubMed.
- S. Subramaniam, P. Joyce, C. E. Conn and C. A. Prestidge, Cellular uptake and in vitro antibacterial activity of lipid-based nanoantibiotics are influenced by protein corona, Biomater. Sci., 2024, 12(13), 3411–3422 Search PubMed.
- S. Subramaniam, P. Joyce, L. Donnellan, C. Young, A. Wignall, P. Hoffmann and C. A. Prestidge, Protein adsorption determines pulmonary cell uptake of lipid-based nanoparticles, J. Colloid Interface Sci., 2023, 641, 36–47 Search PubMed.
- R. Sakhtianchi, R. F. Minchin, K.-B. Lee, A. M. Alkilany, V. Serpooshan and M. Mahmoudi, Exocytosis of nanoparticles from cells: Role in cellular retention and toxicity, Adv. Colloid Interface Sci., 2013, 201–202, 18–29 CrossRef CAS PubMed.
- I. I. Slowing, J. L. Vivero-Escoto, Y. Zhao, K. Kandel, C. Peeraphatdit, B. G. Trewyn and V. S. Lin, Exocytosis of mesoporous silica nanoparticles from mammalian cells: from asymmetric cell-to-cell transfer to protein harvesting, Small, 2011, 7(11), 1526–1532 CrossRef CAS PubMed.
- I. Stayton, J. Winiarz, K. Shannon and Y. Ma, Study of uptake and loss of silica nanoparticles in living human lung epithelial cells at single cell level, Anal. Bioanal. Chem., 2009, 394(6), 1595–1608 CrossRef CAS PubMed.
- I. I. Slowing, J. L. Vivero-Escoto, Y. Zhao, K. Kandel, C. Peeraphatdit, B. G. Trewyn and V. S.-Y. Lin, Exocytosis of Mesoporous Silica Nanoparticles from Mammalian Cells: From Asymmetric Cell-to-Cell Transfer to Protein Harvesting, Small, 2011, 7(11), 1526–1532 CrossRef CAS PubMed.
- T. Yong, X. Zhang, N. Bie, H. Zhang, X. Zhang, F. Li, A. Hakeem, J. Hu, L. Gan, H. A. Santos and X. Yang, Tumor exosome-based nanoparticles are efficient drug carriers for chemotherapy, Nat. Commun., 2019, 10(1), 3838 CrossRef PubMed.
- S. Krol, R. Macrez, F. Docagne, G. Defer, S. Laurent, M. Rahman, M. J. Hajipour, P. G. Kehoe and M. Mahmoudi, Therapeutic Benefits from Nanoparticles: The Potential Significance of Nanoscience in Diseases with Compromise to the Blood Brain Barrier, Chem. Rev., 2013, 113(3), 1877–1903 CrossRef CAS PubMed.
- Y.-B. Hu, E. B. Dammer, R.-J. Ren and G. Wang, The endosomal-lysosomal system: from acidification and cargo sorting to neurodegeneration, Transl. Neurodegener., 2015, 4(1), 18 CrossRef PubMed.
- M. Jovic, M. Sharma, J. Rahajeng and S. Caplan, The early endosome: a busy sorting station for proteins at the crossroads, Histol. Histopathol., 2010, 25(1), 99–112 CAS.
- L. W. C. Ho, C. K. W. Chan, R. Han, Y. F. Y. Lau, H. Li, Y.-P. Ho, X. Zhuang and C. H. J. Choi, Mammalian Cells Exocytose Alkylated Gold Nanoparticles via Extracellular Vesicles, ACS Nano, 2022, 16(2), 2032–2045 CrossRef CAS PubMed.
- R. E. Yanes, D. Tarn, A. A. Hwang, D. P. Ferris, S. P. Sherman, C. R. Thomas, J. Lu, A. D. Pyle, J. I. Zink and F. Tamanoi, Involvement of lysosomal exocytosis in the excretion of mesoporous silica nanoparticles and enhancement of the drug delivery effect by exocytosis inhibition, Small, 2013, 9(5), 697–704 CrossRef CAS PubMed.
- C. S. Kim, N. D. B. Le, Y. Xing, B. Yan, G. Y. Tonga, C. Kim, R. W. Vachet and V. M. Rotello, The Role of Surface Functionality in Nanoparticle Exocytosis, Adv. Healthcare Mater., 2014, 3(8), 1200–1202 CrossRef CAS PubMed.
- N. Oh and J.-H. Park, Surface Chemistry of Gold Nanoparticles Mediates Their Exocytosis in Macrophages, ACS Nano, 2014, 8(6), 6232–6241 CrossRef CAS PubMed.
- B. D. Chithrani and W. C. W. Chan, Elucidating the Mechanism of Cellular Uptake and Removal of Protein-Coated Gold Nanoparticles of Different Sizes and Shapes, Nano Lett., 2007, 7(6), 1542–1550 CrossRef CAS PubMed.
- M. Bruckner, J. Simon, S. Jiang, K. Landfester and V. Mailander, Preparation of the protein corona: How washing shapes the proteome and influences cellular uptake of nanocarriers, Acta Biomater., 2020, 114, 333–342 CrossRef PubMed.
- J. Simon, G. Kuhn, M. Fichter, S. Gehring, K. Landfester and V. Mailander, Unraveling the In Vivo Protein Corona, Cells, 2021, 10(1), 132 CrossRef CAS PubMed.
- R. da Costa Marques, N. Hüppe, K. R. Speth, J. Oberländer, I. Lieberwirth, K. Landfester and V. Mailänder, Proteomics reveals time-dependent protein corona changes in the intracellular pathway, Acta Biomater., 2023, 172, 355–368 CrossRef CAS PubMed.
- M. Lundqvist, J. Stigler, G. Elia, I. Lynch, T. Cedervall and K. A. Dawson, Nanoparticle size and surface properties determine the protein corona with possible implications for biological impacts, Proc. Natl. Acad. Sci. U. S. A., 2008, 105(38), 14265–14270 CrossRef CAS PubMed.
- S. Tenzer, D. Docter, S. Rosfa, A. Wlodarski, J. Kuharev, A. Rekik, S. K. Knauer, C. Bantz, T. Nawroth, C. Bier, J. Sirirattanapan, W. Mann, L. Treuel, R. Zellner, M. Maskos, H. Schild and R. H. Stauber, Nanoparticle size is a critical physicochemical determinant of the human blood plasma corona: a comprehensive quantitative proteomic analysis, ACS Nano, 2011, 5(9), 7155–7167 CrossRef CAS PubMed.
- S. Schöttler, K. Landfester and V. Mailänder, Controlling the Stealth Effect of Nanocarriers through Understanding the Protein Corona, Angew. Chem., Int. Ed., 2016, 55(31), 8806–8815 CrossRef PubMed.
- A. Lesniak, F. Fenaroli, M. P. Monopoli, C. Åberg, K. A. Dawson and A. Salvati, Effects of the presence or absence of a protein corona on silica nanoparticle uptake and impact on cells, ACS Nano, 2012, 6(7), 5845–5857 CrossRef CAS PubMed.
- J. Panyam and V. Labhasetwar, Dynamics of endocytosis and exocytosis of poly(D,L-lactide-co-glycolide) nanoparticles in vascular smooth muscle cells, Pharm. Res., 2003, 20(2), 212–220 CrossRef CAS PubMed.
- G. Berrecoso, J. Crecente-Campo and M. J. Alonso, Unveiling the pitfalls of the protein corona of polymeric drug nanocarriers, Drug Delivery Transl. Res., 2020, 10(3), 730–750 CrossRef CAS PubMed.
- T. Cedervall, I. Lynch, S. Lindman, T. Berggård, E. Thulin, H. Nilsson, K. A. Dawson and S. Linse, Understanding the nanoparticle–protein corona using methods to quantify exchange rates and affinities of proteins for nanoparticles, Proc. Natl. Acad. Sci. U. S. A., 2007, 104(7), 2050–2055 CrossRef CAS PubMed.
- M. P. Monopoli, D. Walczyk, A. Campbell, G. Elia, I. Lynch, F. B. Bombelli and K. A. Dawson, Physical−Chemical Aspects of Protein Corona: Relevance to in Vitro and in Vivo Biological Impacts of Nanoparticles, J. Am. Chem. Soc., 2011, 133(8), 2525–2534 CrossRef CAS PubMed.
- M. Zhang, R. Charles, H. Tong, L. Zhang, M. Patel, F. Wang, M. J. Rames, A. Ren, K.-A. Rye, X. Qiu, D. G. Johns, M. A. Charles and G. Ren, HDL surface lipids mediate CETP binding as revealed by electron microscopy and molecular dynamics simulation, Sci. Rep., 2015, 5(1), 8741 CrossRef CAS PubMed.
- A. L. Barrán-Berdón, D. Pozzi, G. Caracciolo, A. L. Capriotti, G. Caruso, C. Cavaliere, A. Riccioli, S. Palchetti and A. Laganà, Time evolution of nanoparticle-protein corona in human plasma: relevance for targeted drug delivery, Langmuir, 2013, 29(21), 6485–6494 CrossRef PubMed.
- B. Sun, E. R. Eckhardt, S. Shetty, D. R. van der Westhuyzen and N. R. Webb, Quantitative analysis of SR-BI-dependent HDL retroendocytosis in hepatocytes and fibroblasts, J. Lipid Res., 2006, 47(8), 1700–1713 CrossRef CAS PubMed.
- D. Wüstner, M. Mondal, A. Huang and F. R. Maxfield, Different transport routes for high density lipoprotein and its associated free sterol in polarized hepatic cells, J. Lipid Res., 2004, 45(3), 427–437 CrossRef PubMed.
- S. Schöttler, G. Becker, S. Winzen, T. Steinbach, K. Mohr, K. Landfester, V. Mailänder and F. R. Wurm, Protein adsorption is required for stealth effect of poly(ethylene glycol)- and poly(phosphoester)-coated nanocarriers, Nat. Nanotechnol., 2016, 11(4), 372–377 CrossRef PubMed.
- J. B. German, J. T. Smilowitz and A. M. Zivkovic, Lipoproteins: When size really matters, Curr. Opin. Colloid Interface Sci., 2006, 11(2–3), 171–183 CrossRef CAS PubMed.
- J. Panyam and V. Labhasetwar, Dynamics of Endocytosis and Exocytosis of Poly(D,L-Lactide-co-Glycolide) Nanoparticles in Vascular Smooth Muscle Cells, Pharm. Res., 2003, 20(2), 212–220 CrossRef CAS PubMed.
- R. E. Serda, A. Mack, A. L. van de Ven, S. Ferrati, K. Dunner Jr, B. Godin, C. Chiappini, M. Landry, L. Brousseau, X. Liu, A. J. Bean and M. Ferrari, Logic-Embedded Vectors for Intracellular Partitioning, Endosomal Escape, and Exocytosis of Nanoparticles, Small, 2010, 6(23), 2691–2700 CrossRef CAS PubMed.
- W. Stöber, A. Fink and E. Bohn, Controlled growth of monodisperse silica spheres in the micron size range, J. Colloid Interface Sci., 1968, 26(1), 62–69 CrossRef.
- M. Kokkinopoulou, J. Simon, K. Landfester, V. Mailander and I. Lieberwirth, Visualization of the protein corona: towards a biomolecular understanding of nanoparticle-cell-interactions, Nanoscale, 2017, 9(25), 8858–8870 RSC.
- J. Simon, L. K. Muller, M. Kokkinopoulou, I. Lieberwirth, S. Morsbach, K. Landfester and V. Mailander, Exploiting the biomolecular corona: pre-coating of nanoparticles enables controlled cellular interactions, Nanoscale, 2018, 10(22), 10731–10739 RSC.
- J. Simon, T. Wolf, K. Klein, K. Landfester, F. R. Wurm and V. Mailänder, Hydrophilicity Regulates the Stealth Properties of Polyphosphoester-Coated Nanocarriers, Angew. Chem., Int. Ed., 2018, 57(19), 5548–5553 CrossRef CAS PubMed.
- S. Ritz, S. Schottler, N. Kotman, G. Baier, A. Musyanovych, J. Kuharev, K. Landfester, H. Schild, O. Jahn, S. Tenzer and V. Mailander, Protein corona of nanoparticles: distinct proteins regulate the cellular uptake, Biomacromolecules, 2015, 16(4), 1311–1321 CrossRef CAS PubMed.
- J. Simon, J. Muller, A. Ghazaryan, S. Morsbach, V. Mailander and K. Landfester, Protein denaturation caused by heat inactivation detrimentally affects biomolecular corona formation and cellular uptake, Nanoscale, 2018, 10(45), 21096–21105 RSC.
- A. Aliyandi, C. Reker-Smit, R. Bron, I. S. Zuhorn and A. Salvati, Correlating Corona Composition and Cell Uptake to Identify Proteins Affecting Nanoparticle Entry into Endothelial Cells, ACS Biomater. Sci. Eng., 2021, 7(12), 5573–5584 CrossRef CAS PubMed.
- M. Kokkinopoulou, J. Simon, K. Landfester, V. Mailänder and I. Lieberwirth, Visualization of the protein corona: towards a biomolecular understanding of nanoparticle-cell-interactions, Nanoscale, 2017, 9(25), 8858–8870 RSC.
- J. Simon, L. K. Müller, M. Kokkinopoulou, I. Lieberwirth, S. Morsbach, K. Landfester and V. Mailänder, Exploiting the biomolecular corona: pre-coating of nanoparticles enables controlled cellular interactions, Nanoscale, 2018, 10(22), 10731–10739 RSC.
- J. Simon, T. Wolf, K. Klein, K. Landfester, F. R. Wurm and V. Mailander, Hydrophilicity Regulates the Stealth Properties of Polyphosphoester-Coated Nanocarriers, Angew. Chem., Int. Ed., 2018, 57(19), 5548–5553 CrossRef CAS PubMed.
- J. C. Silva, M. V. Gorenstein, G. Z. Li, J. P. Vissers and S. J. Geromanos, Absolute quantification of proteins by LCMSE: a virtue of parallel MS acquisition, Mol. Cell. Proteomics, 2006, 5(1), 144–156 CrossRef CAS PubMed.
- C. D. Thurmond, Control of dust in solution for turbidimetry, J. Polym. Sci., 1952, 8(6), 607–609 CrossRef CAS.
- K. Rausch, A. Reuter, K. Fischer and M. Schmidt, Evaluation of nanoparticle aggregation in human blood serum, Biomacromolecules, 2010, 11(11), 2836–2839 CrossRef CAS PubMed.
Footnotes |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d4bm01189a |
| ‡ Shared first authors. |
| § These authors contributed equally to this work. |
| This journal is © The Royal Society of Chemistry 2025 |