Embedded bioprinting of dense cellular constructs in bone allograft-enhanced hydrogel matrices for bone tissue engineering†
Hang
Truong‡
 a,
Alperen
Abaci‡
a,
Hadis
Gharacheh
a and
Murat
Guvendiren
a,
Alperen
Abaci‡
a,
Hadis
Gharacheh
a and
Murat
Guvendiren
 *ab
*ab
aOtto H. York Department of Chemical and Materials Engineering, New Jersey Institute of Technology, Newark, NJ 07102, USA. E-mail: muratg@njit.edu
bDepartment of Biomedical Engineering, New Jersey Institute of Technology, Newark, NJ 07102, USA
First published on 25th February 2025
Abstract
Bone tissue engineering aims to address critical-sized defects by developing biomimetic scaffolds that promote repair and regeneration. This study introduces a material extrusion-based embedded bioprinting approach to fabricate dense cellular constructs within methacrylated hyaluronic acid (MeHA) hydrogels enhanced with bioactive microparticles. Composite matrices containing human bone allograft or tricalcium phosphate (TCP) particles were evaluated for their rheological, mechanical, and osteoinductive properties. High cell viability (>95%) and uniform strand dimensions were achieved across all bioprinting conditions, demonstrating the method's ability to preserve cellular integrity and structural fidelity. The inclusion of bone or TCP particles did not significantly alter the viscosity, crosslinking kinetics, or compressive modulus of the MeHA hydrogels, ensuring robust mechanical stability and shape retention. However, bone allograft particles significantly enhanced osteogenic differentiation of human mesenchymal stem cells (hMSCs), as evidenced by increased alkaline phosphatase (ALP) activity and calcium deposition. Notably, osteogenesis was observed even in basal media, with a dose-dependent response to bone particle concentration, highlighting the intrinsic bioactivity of allograft particles. This study demonstrates the potential of combining embedded bioprinting with bioactive matrices to create dense, osteoinductive cellular constructs. The ability to induce osteogenesis without external growth factors positions this platform as a scalable and clinically relevant solution for bone repair and regeneration.
Introduction
Advanced engineering technologies in biomaterials and biofabrication are revolutionizing tissue engineering, particularly in bone tissue regeneration, through the development of biomimetic scaffolds that address substantial bone defects caused by trauma or disease.1–4 The limitations of current bone grafting procedures, coupled with increasing prevalence of bone and joint disorders, have driven a shift towards scaffold-based tissue engineering strategies.5–7 Conventional fabrication techniques such as gas foaming,8–10 solvent casting combined with particle leaching,11–13 phase separation,14,15 freeze drying,16–18 and electrospinning19–21 are widely used for create porous scaffolds, but often fail to replicate the intricate 3D architecture of native bone accurately. In contrast, 3D bioprinting offers a promising alternative, enabling rapid and customizable fabrication of complex, cell-laden hydrogel scaffolds.22–27 These scaffolds can be tailored to individual patients anatomically, using medical images like X-rays or CT scans, and physiologically, by incorporating patent-derived stem cells and extracellular matrix materials.28–32The bioink formulations used in the bioprinting process are critical for fostering a microenvironment that mimics native bone, enabling the osteogenesis of stem cells, new tissue growth, and functional integration to the defect site. To replicate the highly mineralized nature of bone, bioceramics like hydroxyapatite (HA), tricalcium phosphate (TCP), and bioactive glass are commonly incorporated into bioinks, providing essential osteoinductive and osteoconductive properties.33–36 Naturally derived hydrogels complement these bioceramics by serving as carriers during printing, protecting cells and supporting attachment and growth. While bioceramic-based bioinks have significantly advanced bone tissue engineering, decellularized bone allograft bioinks—either as digested hydrogels28,37–41 or composite inks42–44—offer superior bioactive cues, including growth factors, for bone regeneration. Coupling decellularized bone-based bioinks with 3D bioprinting holds great potential for creating effective, personalized solutions, though achieving the high cell density required to match native tissues remains a challenge. Thus, there is a critical need to create dense cellular structures to better recapitulate the physiology of the native tissue.
With the development of embedded bioprinting techniques, soft materials like hydrogels or cell-laden hydrogels can be deposited into microgel support baths, enabling the fabrication of complex 3D structures that cannot be built in air.45–47 These temporary supports stabilize the printed shapes until they are removed, allowing the creation of intricate designs. Leveraging this approach, researchers have developed dense cellular structures by embedding cell aggregates or spheroids in support baths. For instance, Jeon et al. demonstrated that hMSCs could self-assemble into bone tissues within 28 days by embedding cell aggregates in an oxidized and methacrylated alginate bath and culturing them in osteogenic media with BMP-2, confirming osteogenesis through calcium deposition.48 Similarly, Ayan et al. fabricated large-scale dense structures using pre-differentiated osteogenic hMSC spheroids,49 while Banerjee et al. created an in vitro bone model for Gaucher disease using co-cultured spheroids of osteoblast and osteoclast lineages.50 Although these studies highlight significant progress in generating dense bone tissues, the support baths primarily serve as temporary mechanical aids and lack bone-mimetic cues, limiting their potential to further enhance bone tissue models.
As embedded printing techniques advance,51–54 they have enabled the fabrication of complex 3D dense cellular structures, including cell aggregates or spheroids, for various tissue types, including bone. However, current support baths predominantly rely on shear-thinning or self-healing microgels, limiting the range of materials suitable for dense cell printing. To address this gap, we recently developed a new strategy involving the bioprinting of photocurable viscous hydrogel layers, within which cell aggregates can be deposited on demand.55,56 By adjusting the viscosity of the hydrogel ink, we enable needle motion and material extrusion without requiring shear-thinning and recovery properties, significantly expanding the available hydrogel inks that can serve as support media. Our previous studies demonstrated that this method facilitates the use of cell-instructive hydrogels as stable, long-term supports to modulate stem cell behavior, or as removable scaffolds to leave behind cell-only constructs.55 Furthermore, it allows the incorporation of multiple hydrogel inks to create localized matrix heterogeneity and multi-cellular bioprinted structures.
Building on this approach, in this study, we utilized an embedded bioprinting strategy to fabricate dense hMSC strands within methacrylated hyaluronic acid (MeHA) composite supports containing human bone allograft particles, with tricalcium phosphate (TCP) particles serving as a control. We investigated the influence of these particles on the rheological and mechanical properties of the support matrix and evaluated their effects on the osteogenic differentiation of bioprinted hMSC strands. This support matrix not only provides inherent bioactivity conducive to bone regeneration but also represents a significant advancement toward engineering dense microtissues for bone repair and regeneration.
Results and discussion
3D bioprinting of dense cellular structures within composite matrices
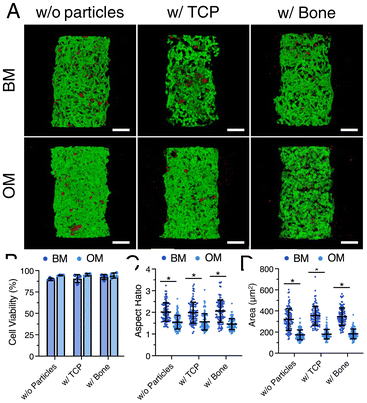
A material extrusion-based 3D bioprinting approach55,56 was employed to fabricate dense cellular constructs by depositing cell aggregates within a viscous, photocurable support matrix (Fig. 1A and ESI Movies S1 and S2†). In this method, alternating layers of a methacrylated hyaluronic acid (MeHA, ∼80% methacrylation) support ink (200–300 μm) and cell aggregates were deposited (ESI Movies S1 and S2†). MeHA, a linear polysaccharide derived from the extracellular matrix, was chosen for its biocompatibility and ability to form hydrogels via photopolymerization (λ = 405 nm, 5 s). This stepwise process produced a partially crosslinked, self-supporting matrix, enabling the creation of a 3D construct with embedded cell aggregates (Fig. 1B). Finally, the construct was exposed to light (75 s) to achieve complete crosslinking. Unlike conventional extrusion-based embedded bioprinting methods, which rely on shear-thinning properties, our approach utilizes a viscous photocurable hydrogel as a dynamic support matrix, enabling structural integrity while allowing the deposition of bioinks with a controlled viscosity range of 0.1 to 10 Pa s.55,56 This method provides fine control over construct geometry, with layer thickness typically ranging from 200–300 μm but adjustable to ∼80 μm for enhanced precision. Additionally, the tunable viscosity of the support hydrogel ensures compatibility with various bioinks, while the optimized crosslinking process balances mechanical stability and a cell-friendly environment.To investigate stem cell osteogenesis, composite support inks were prepared by incorporating human bone allograft particles (1 w/v%) or tricalcium phosphate (TCP) particles as controls. TCP particles are known to promote osteogenic differentiation in engineered microenvironments due to their calcium phosphate content and inherent bioactivity.1,57–59 Stem cell viability was assessed by bioprinting continuous strands of hMSCs within MeHA hydrogels (Fig. 1C), both with and without particles. Confocal microscopy revealed that the majority of cells stained green (live) compared to red (dead) across all conditions (Fig. 1D). Quantitative analysis showed consistent strand widths (∼350 μm) and high cell viability (99–95%), regardless of the matrix composition (Fig. 1E and F). These results demonstrate that our bioprinting approach maintains uniformity in cellular strand dimensions while preserving high cell viability, independent of the inclusion of bioactive particles. This highlights the potential of this method to create tailored 3D constructs with user-defined cellular and material properties for applications in bone tissue engineering.
Rheological and mechanical characterization of support inks
The rheological properties of support inks are critical for achieving optimal printability and shape fidelity. These properties influence not only the performance of the support ink itself but also the deposition quality of subsequent cell aggregate inks.60 Our previous findings demonstrated that the viscosity of the support matrix should range between 0.1–10 Pa s, corresponding to 5–10 wt% methacrylated hyaluronic acid (MeHA) solutions.55 This viscosity range facilitates needle motion within the support layer without requiring shear-thinning behavior, while preserving the structural integrity of printed constructs.55For this study, we used a 10 wt% MeHA solution with a viscosity of approximately 1 Pa s. Rheological measurements confirmed that the addition of particles did not significantly alter the ink viscosity (Fig. 2A). The gel point—defined as the time at which G′ = G′′—and the equilibrium crosslinking time, marked by the onset of the G′ plateau, were ∼5 s and ∼75 s, respectively, for MeHA (Fig. 2B). Importantly, these crosslinking parameters remained consistent upon the addition of particles. Support inks were printed using identical parameters: 125 kPa extrusion pressure, 10 mm s−1 print speed, and a 0.25 mm nozzle. The printed support layer was partially crosslinked for a duration equivalent to the gel point (∼5 s) to provide adequate mechanical support, followed by complete crosslinking over 75 s to ensure structural integrity.
This approach ensured adequate mechanical stability to support bioprinted structures. Furthermore, the printed constructs exhibited negligible weight changes after swelling in PBS, indicating robust shape retention (Fig. 2C). Mechanical testing revealed that the compressive modulus of MeHA hydrogels remained largely unaffected by the inclusion of particles (Fig. 2D). The mean modulus values were ∼610 kPa for MeHA without particles, 596 kPa for MeHA with TCP particles, and 497 kPa for MeHA with bone particles. Overall, our findings demonstrate that the incorporation of particles does not significantly impact the viscosity, crosslinking kinetics, or stiffness of the support hydrogels.
Characterization of particle distribution within support hydrogels
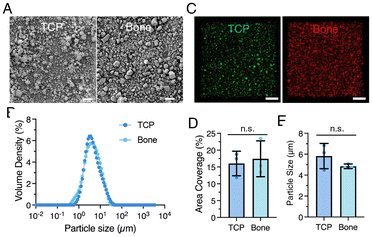
Bone allograft chips, provided by MTF Biologics, were pulverized via ball milling to produce particles of similar size to commercially obtained TCP particles. Both particle types exhibited comparable shapes and size distributions, with a mean particle size of approximately 4.5 μm (Fig. 3A and B). Confocal microscopy images (Fig. 3C) confirmed the uniform distribution of particles within the support hydrogels, with TCP particles labeled in red and bone particles in green. Qualitative analysis of the confocal images revealed consistent areal coverage of 16–17% for both particle types (Fig. 3D). The measured mean particle sizes were ∼4.8 μm for bone and ∼5.8 μm for TCP, with minimal evidence of particle aggregation (Fig. 3E). These findings demonstrate that both TCP and bone allograft microparticles possess similar size and uniform distribution within the MeHA matrix. Bone allograft microparticles (∼5 μm) were selected to match the size of TCP particles, ensuring comparability while avoiding potential degradation of native biological factors from prolonged milling. Although nano-sized bioceramics are often preferred in bone tissue engineering for their higher surface area and bioactivity, micro-sized bone allografts are commonly used due to processing limitations.43,61,62In vitro culture studies of bioprinted hMSC strands within composite supports
Following confirmation of high cell viability after printing, we investigated the effect of the composite support matrix on cell behavior over a 14-day in vitro period (Fig. 4A). For this purpose, hMSC aggregates were bioprinted as a continuous strand within a 10 wt% MeHA support matrix, either without particles or with 1 w/v% particles, and cultured in basal media (BM) or osteogenic induction media (OM).High cell viability (90–95%) was observed after 14 days of culture in both BM and OM, with no significant differences due to the inclusion of particles (Fig. 4B). Analysis of individual cells revealed that those cultured in BM exhibited significantly higher aspect ratios (AR ∼2) and larger cell areas (320–250 μm2) compared to cells in OM (AR ∼1.5, cell area ∼170–180 μm2) (Fig. 4C and D). The increased AR and cell area in BM are consistent with the expected reduction in functions such as proliferation and spreading during stem cell differentiation.63,64 While media conditions significantly influenced cell AR and area, the inclusion of particles did not have a notable impact on these parameters.
Osteogenic differentiation of hMSC strands within composite supports
To assess osteogenic differentiation, 3D bioprinted dense hMSC strands within MeHA hydrogels (w or w/o particles) were cultured in BM or OM for 14 days. Osteogenic differentiation was quantified by measuring alkaline phosphatase (ALP) activity and calcium deposition (using alizarin red staining, ARS). ARS images revealed minimal calcium deposition in MeHA hydrogels, while matrices with TCP or bone particles showed significant calcium deposition (Fig. 5A). Quantitative analysis indicated a ∼2-fold increase in calcium deposition in OM compared to BM, with bone particles yielding 1.2 times higher deposition than TCP particles in BM (Fig. 5B). Similarly, ALP activity was 2-fold higher in OM than in BM across all conditions, with bone particles showing a 3-fold increase in ALP activity compared to TCP, both in BM and OM (Fig. 5C). The bone allograft within the matrix on day 1 (Fig. 6A) and day 14 (Fig. 6B) exhibited a well-distributed microparticle arrangement across different z-sections, maintaining a consistent pattern over time. These results indicate that the microparticles remained evenly dispersed without sedimentation, which could otherwise compromise the scaffold's osteogenic potential.Overall, these results demonstrate that particle inclusion significantly enhances osteogenesis, with bone particles having the most pronounced effect. Notably, the enhanced osteogenesis in BM with bone particles suggests their potential to induce osteogenesis even without induction media, paving the way for applications in creating bone interfaces, such as osteochondral tissues.
The effect of bone particle concentration on hMSC osteogenesis in basal media
Once we confirm the enhanced osteogenesis in the presence of bone particles even in BM, we investigated the effect of particle concentration on osteogenic differentiation of hMSC strands after 14 days of culture in BM. Here, we focused on lowering the bone particle concentration to enhance opacity of the support hydrogels, which could be important for qualitative characterization methods including staining and microscopy. We bioprinted dense hMSC strands within 10% MeHA with 0.25, 0.5 and 1% bone particles. We first confirmed that the composite ink viscosity (Fig. 7A), crosslinking kinetics (Fig. 7B and C), and composite support stiffness (Fig. 7D) were not affected with the particle concentration. SEM images revealed that MeHA's pore size remained ∼22 μm with 0.5% bone but increased to 29 μm at 1% bone content (ESI Fig. S2†). As compared to MeHA hydrogel without particles, calcium deposition increased 1.1-fold and 1.2-fold for 0.25% and 0.5% bone particles and significantly increased 1.6-folds for 1% bone particles (Fig. 7E). We observed a 2.8-fold to 3.6-fold and finally 4-fold increase in MeHA supports with 0.25%, 0.5% and 1% bone, respectively. ALP activity (Fig. 7F) increased gradually with increasing particle concentration such that a 2.8-fold, 3.7-fold and 4-fold increase observed for 0.25%, 0.5% and 1% bone particles, respectively. Our findings suggest that bone particles are effective in inducing osteogenic differentiation of stem cells in basal media, and increasing particle concentration up to 1% enhances osteogenesis. This could be explained by the fact that human allograft bone particles inherently contain native bioactive cues – minerals, growth factors and ECM components including collagen – driving stem cells into osteogenic lineage.Conclusions
This study demonstrates the potential of a material extrusion-based embedded bioprinting approach for creating dense cellular constructs within composite hydrogel matrices, advancing applications in bone tissue engineering. Using methacrylated hyaluronic acid (MeHA) hydrogels as the support matrix, with human bone allograft particles or tricalcium phosphate (TCP) as bioactive components, we achieved high cell viability, uniform hMSC strand dimensions, and stable mechanical properties. Notably, the incorporation of bone particles significantly enhanced hMSC osteogenesis, evidenced by increased alkaline phosphatase (ALP) activity and calcium deposition.Our key findings include:
1. Our bioprinting strategy successfully fabricated 3D constructs with embedded cell aggregates, maintaining high cell viability (>95%) and uniformity across conditions.
2. The addition of bioactive microparticles did not affect the viscosity, crosslinking kinetics, or compressive modulus of the MeHA support ink, ensuring robust shape fidelity and mechanical stability.
3. Bone particles outperformed TCP particles in promoting osteogenic differentiation, with a pronounced effect even in basal media, highlighting their potential as a source of bioactive cues. Increasing the bone particle concentration (up to 1%) further amplified osteogenic markers, underscoring the dose-dependent nature of their bioactivity.
These findings establish a versatile bioprinting platform that can integrate dense cellular structures with tailored bioactivity for bone repair and regeneration. The ability of bone particles to induce stem cell osteogenesis without specialized induction media broadens the scope for creating complex tissue interfaces, such as osteochondral constructs. Moreover, this approach eliminates reliance on external factors and making it more practical and scalable for clinical applications. Future work will focus on further characterizing the bioactive potential of allograft particles in diverse tissue engineering applications.
Materials and methods
Polymer synthesis
MeHA was synthesized following a previously described procedure.64,65 Briefly, 5 g of sodium hyaluronate (HA, 60 kDa, Lifecore) was dissolved in deionized water (1 w/v%) and reacted with 2 mL methacrylic anhydride (MA, Sigma) per gram of polymer (consumed within 1.5–2 h by dropwise addition) in an ice bath at 1–4 °C for 8 h, while pH was kept at 8–9 by simultaneous addition of 4 M sodium hydroxide (NaOH, Sigma) solution. After 8 h, the solution was kept in 4 °C overnight. The reaction was continued by dropwise addition of MA (1 ml MA per gram of HA, consumed within 1 h) at pH 8–9 in an ice bath for 4 h. The obtained solution was purified by dialysis in deionized water (Spectra/Por®1 dialysis membrane, molecular weight cutoff 6–8 kDa, Fisher) for 5 days, frozen in −80 °C and subsequently lyophilized. 1H NMR (Bruker) was used to confirm the percent modification (methacrylation) of the polymer (ESI Fig. S1†).Bone particle processing
Cancellous allograft bone was kindly provided by the Musculoskeletal Tissue Foundation (MTF) Biologics (Edison, NJ, USA). The crushed bone pieces were ball milled using a shaker miller fitted with zirconia beads (2 mm in diameter). The particle size distribution post-milling was analyzed using a Mastersizer 3000 particle size analyzer from Malvern Panalytical Inc. (Westborough, MA, USA).Support and cell-only bioink preparation
Neat hydrogel ink formulations were prepared by dissolving methacrylated hyaluronic acid (MeHA) in Dulbecco's phosphate-buffered saline (DPBS, Sigma) containing the photoinitiator lithium phenyl-2,4,6-trimethylbenzoylphosphinate (LAP, 405–410 nm, Sigma). Specifically, 10% (w/v) MeHA was combined with DPBS containing 0.05% (w/v) LAP in a 20 mL scintillation vial equipped with a magnetic stir bar. The vial was covered with aluminum foil to protect it from light and gently stirred overnight to ensure complete dissolution. For composite hydrogel ink formulations, tricalcium phosphate (TCP) particles (4 μm, Sigma-Aldrich) or bone particles were first dispersed into DPBS containing 0.05% (w/v) LAP in a 20 mL scintillation vial. The mixture was sonicated for 30 minutes to break up particle agglomerates. MeHA was then added to the vial, and the mixture was gently stirred overnight under light-protective conditions. Prior to printing, 1 mM RGD peptide (GRGDSPC, GenScript) was added to the ink formulations and incubated at room temperature for 30 min to enhance bioactivity.64,65Cell-only bioinks were prepared using aggregates of human mesenchymal stem cells (hMSCs). The hMSCs were expanded (passage 3) in growth media consisting of minimum essential medium α (MEM α, Gibco) supplemented with 10% fetal bovine serum (FBS, Gibco) and 1% penicillin–streptomycin (pen–strep, Gibco). The cells were cultured in 225 cm2 tissue culture flasks with media changes every four days. Upon reaching approximately 80% confluency, the cells were washed with DPBS and detached by incubating with TrypLE express (Gibco) at 37 °C for 10 min. The detached cells were collected into 50 mL centrifuge tubes containing fresh growth media and centrifuged. The resulting cell pellets were combined into a single 50 mL centrifuge tube and filtered through a 40 μm cell strainer to remove any large aggregates that could obstruct the needle during the printing process. Following a final centrifugation, the cell aggregates were drawn into a 3 mL syringe and used immediately as cell-only bioinks. A minimum of 50 million cells were prepared to ensure consistent and reliable bioprinting.
Rheological and mechanical characterization of support inks
Kinexus Prime Ultra+ (NETZSCH) rheometer was used to study the rheology of the inks and mechanical behavior of printed constructs. Tests were performed using a flat upper plate geometry (20 mm diameter) at a constant gap size (0.50 mm) at room temperature. Shear viscosity was measured at shear rates from 1 to 1000 s−1. Time sweep tests were performed at 1 Hz frequency and 1% strain to investigate the change in elastic (G′) and viscous modulus (G′′) under light exposure. For this purpose, a UV light apparatus (NETZSCH) was connected to a UV light source (Omnicure S2000, 365 nm, 40 mW cm−2) and light intensity was adjusted to compensate for the difference in the wavelength of the printer light source (405 nm) according to the molar absorptivity spectrum of the photoinitiator (LAP).66–68 The ink was equilibrated for 2 min and exposed to UV light to investigate crosslinking kinetics.To measure compressive modulus (Young's modulus, E), three sets of disk-shaped hydrogel samples (1 mm thick and 25 mm in diameter) were equilibrated in DPBS overnight. The compression test was performed by applying a normal force to the construct using an 8 mm upper plate geometry. To ensure initial contact, an initial compressive force equal to 0.05 N was applied and the compressive force was increased continuously (0.1 mm s−1) up to 10 N. The compressive modulus was obtained by calculating the slope of stress–strain curve using the first linear range within 5% strain.
Bioprinting process
The BioX bioprinter (CellINK) was employed in this study. Inks were prepared in 3 mL syringes, with support hydrogels loaded into pneumatic printheads for efficient extrusion and cell-only bioinks loaded into syringe pump printheads to enable precise, controlled deposition. A glass slide was placed on the printer platform, and the tips of all needles were calibrated to the same position to ensure alignment. The printing process began with the deposition of a support hydrogel base layer, which was partially crosslinked for 5 s. A second hydrogel matrix layer was subsequently deposited, and cell-only bioink was carefully printed within this viscous layer. The second layer was then partially crosslinked for 5 s to stabilize the structure and prevent any disruption during the deposition of subsequent layers. Additional layers were built upon this foundation, with final crosslinking extended to 75 s to fully stabilize the construct. Following bioprinting, the constructs were transferred to 6-well plates and cultured in appropriate media. Media was refreshed every three days to maintain optimal conditions for further study.To determine the optimum printing parameters, hMSC aggregates were bioprinted as continuous struts (10 mm long) within support hydrogels (15 mm × 15 mm) made from neat or composite MeHA (10% w/v). Consistent strut sizes of approximately 350 μm were achieved at a flow rate of 0.1 μL s−1 and a print speed of 5 mm s−1. For each print, at least three individual lines were analyzed, with three random sections of each line imaged. Three width measurements per image were taken using ImageJ to assess the consistency of the printed strut dimensions.
Particle distribution within the support constructs was analyzed using confocal microscopy, leveraging the autofluorescence of TCP and bone particles for identification. The average particle size and coverage area were quantified using ImageJ (NIH). Furthermore, the distribution of bone particles within the support matrix was examined by generating profile plots in ImageJ from z-sections of confocal images of the bioprinted constructs at days 1 and 14 during differentiation studies described below.
Cell viability measurements
Cell viability was evaluated post-printing using live/dead staining (Invitrogen). Printed constructs were incubated in a staining solution containing calcein AM (live, 0.5 μL mL−1 DPBS) and ethidium homodimer-1 (dead, 2 μL mL−1 DPBS) for 30 minutes at room temperature. Following staining, constructs were washed three times with DPBS and immediately imaged using a confocal laser scanning microscope (Leica) to obtain 3D scans of the printed structures. Cell viability was quantified using ImageJ (NIH) by calculating the percentage of viable cells, expressed as the ratio of green (live) cells to the total number of cells (green + red). When precise cell counting became challenging due to the high cellular density, the area of green and red regions was used.Aspect ratio and area measurements of individual hMSCs
Live/dead staining images were also used to measure aspect ratio and area of individual hMSCs, since calcein AM shows the morphology of individual cells clearly. Aspect ratio of the individual cells was calculated from width/length measurements of individual cells on ImageJ. Area of the individual cells were measured using ImageJ. 100 cells were used in total for measurements (25 cells from four individual lines).Differentiation studies
For differentiation studies, 3D bioprinted constructs (10 mm long hMSC struds embedded within 15 mm × 15 mm × 0.5 mm MeHA hydrogels) were cultured in osteogenic induction media (OM) or basal media (BM) for 14 days. To characterize differentiation of the samples, ALP activity and calcium deposition were analyzed. ALP activity was measured using QuantiChrom™ alkaline phosphatase assay kit (BioAssay Systems). Constructs were pulverized in 400 μL lysis buffer (0.25% Triton-X in DI water) and incubated overnight in 1.5 mL microcentrifuge tubes. Each vial was centrifuged, and the supernatant (50 μL) was reacted with p-nitrophenyl phosphate working solution in 96-well plates. Absorbance values were measured at 405 nm using a plate reader (Infinite M200 Pro, Tecan). To normalize ALP activity values by DNA content, the same supernatants were used to quantify DNA content through PicoGreen assay (Quant-iT™ PicoGreen™ dsDNA assay kit, Invitrogen). Fluorescence intensity values were measured using a plate reader (Infinite M200 Pro, Tecan) at 480 nm excitation and 520 nm emission wavelengths. ALP readings at corresponding culture time points were normalized by the DNA contents. Calcium deposition was evaluated through Alizarin red staining solution (Sigma). Constructs were first rinsed with DPBS (2×) and then fixed in ice-cold 75% ethanol for 2 h. Then, the constructs were rinsed with DI water (2×), followed by staining in Alizarin red solution (3 mL in 6-well plates) overnight. After overnight staining, the constructs were washed extensively with DI water until the samples stopped releasing Alizarin red into the freshly added DI water. When the samples were ready for analysis, optical microscope images were taken and the images were converted to 8-bit, inverted, and mean staining intensity was measured using ImageJ from three random areas using three independent samples.Statistical analysis
Data were analyzed using Minitab® statistical software (Minitab, LLC). Unless otherwise specified, data were reported as mean ± SD for n ≥ 3. One-way or two-way ANOVA with Tukey's post hoc test was used to make comparisons. Significance determined by p < 0.05.Author contributions
Formal analysis, methodology, and visualization: H. T. and A. A., conceptualization: H. T., A. A. and M. G., investigation, validation: H. T., A. A. and H. G., writing – original draft: A. A. and H. T., funding accusation, project administration, supervision, writing – review and editing: M. G.Data availability
Data (csv and tiff/jpeg) will be available publicly through google drive share. https://drive.google.com/drive/folders/1jFC5d_nhVDno_NQn2uzlJ-J5mUC_ZYM_?usp=share_link.Conflicts of interest
There are no conflicts to declare.Acknowledgements
This work was partially supported by the NSF/DMR CAREER award (2044479) and a research grant from the New Jersey Health Foundation (NJHF). We thank MTF Biologics for providing the human bone allografts. We also appreciate Prof. Edward Dreizin and his research group for providing access to ball milling equipment.References
- S. Bose, S. Vahabzadeh and A. Bandyopadhyay, Mater. Today, 2013, 16, 496–504 CrossRef CAS.
- A. Oryan, S. Alidadi, A. Moshiri and N. Maffulli, J. Orthop. Surg. Res., 2014, 9, 18 CrossRef PubMed.
- G. L. Koons, M. Diba and A. G. Mikos, Nat. Rev. Mater., 2020, 5, 584–603 CrossRef CAS.
- F. R. Maia, A. R. Bastos, J. M. Oliveira, V. M. Correlo and R. L. Reis, Bone, 2022, 154, 116256 CrossRef CAS PubMed.
- R. Dimitriou, E. Jones, D. McGonagle and P. V. Giannoudis, BMC Med., 2011, 9, 66 CrossRef PubMed.
- G. Battafarano, M. Rossi, V. De Martino, F. Marampon, L. Borro, A. Secinaro and A. Del Fattore, Int. J. Mol. Sci., 2021, 22, 1128 CrossRef CAS PubMed.
- J. A. Shibli, B. E. Nagay, L. J. Suárez, C. Urdániga Hung, M. Bertolini, V. A. R. Barão and J. G. S. Souza, Tissue Eng., Part C, 2022, 28, 179–192 CrossRef CAS PubMed.
- H. J. Kim, I. K. Park, J. H. Kim, C. S. Cho and M. S. Kim, Tissue Eng. Regener. Med., 2012, 9, 63–68 CrossRef CAS.
- I. Manavitehrani, T. Y. L. Le, S. Daly, Y. Wang, P. K. Maitz, A. Schindeler and F. Dehghani, Mater. Sci. Eng., C, 2019, 96, 824–830 CrossRef CAS PubMed.
- P. Kazimierczak, A. Benko, K. Palka, C. Canal, D. Kolodynska and A. Przekora, J. Mater. Sci. Technol., 2020, 43, 52–63 CrossRef CAS.
- N. Thadavirul, P. Pavasant and P. Supaphol, J. Biomed. Mater. Res., Part A, 2014, 102, 3379–3392 CrossRef PubMed.
- C.-J. Liao, C.-F. Chen, J.-H. Chen, S.-F. Chiang, Y.-J. Lin and K.-Y. Chang, J. Biomed. Mater. Res., 2002, 59, 676–681 CrossRef CAS PubMed.
- S. Taherkhani and F. Moztarzadeh, J. Appl. Polym. Sci., 2016, 133, 43523 CrossRef.
- S. Chen, X. Zhao and C. Du, Eur. Polym. J., 2018, 109, 303–316 CrossRef CAS.
- M. S. Aydin, M. Sahin, Z. Dogan and G. Kiziltas, ACS Omega, 2023, 8, 47595–47605 CrossRef CAS PubMed.
- L. Nie, D. Chen, J. Suo, P. Zou, S. Feng, Q. Yang, S. Yang and S. Ye, Colloids Surf., B, 2012, 100, 169–176 CrossRef CAS PubMed.
- Z. Shahbazarab, A. Teimouri, A. N. Chermahini and M. Azadi, Int. J. Biol. Macromol., 2018, 108, 1017–1027 CrossRef CAS PubMed.
- M. Haghbin, J. Esmaeilzadeh and S. Kahrobaee, Macromol. Res., 2020, 28, 1232–1240 CrossRef CAS.
- C. Li, C. Vepari, H.-J. Jin, H. J. Kim and D. L. Kaplan, Biomaterials, 2006, 27, 3115–3124 CrossRef CAS PubMed.
- T. Xu, J. M. Miszuk, Y. Zhao, H. Sun and H. Fong, Adv. Healthcare Mater., 2015, 4, 2238–2246 CrossRef CAS PubMed.
- M. P. Prabhakaran, J. Venugopal and S. Ramakrishna, Acta Biomater., 2009, 5, 2884–2893 CrossRef CAS PubMed.
- C.-Y. Liaw and M. Guvendiren, Biofabrication, 2017, 9, 024102 CrossRef PubMed.
- Z. Yazdanpanah, J. D. Johnston, D. M. L. Cooper and X. Chen, Front. Bioeng. Biotechnol., 2022, 10, 824156 CrossRef PubMed.
- S. Adepu, N. Dhiman, A. Laha, C. S. Sharma, S. Ramakrishna and M. Khandelwal, Curr. Opi. Biomed. Eng., 2017, 2, 22–28 CrossRef.
- S. Ji and M. Guvendiren, APL Bioeng., 2021, 5, 011508 CrossRef PubMed.
- H. Gharacheh and M. Guvendiren, MRS Bull., 2023, 48, 668–675 CrossRef CAS.
- M. Guvendiren, J. Molde, R. M. D. Soares and J. Kohn, ACS Biomater. Sci. Eng., 2016, 2, 1679–1693 CrossRef CAS PubMed.
- A. Abaci and M. Guvendiren, Adv. Healthcare Mater., 2020, 9, 2000734 CrossRef CAS PubMed.
- S. V. Murphy and A. Atala, Nat. Biotechnol., 2014, 32, 773–785 CrossRef CAS PubMed.
- H. W. Kang, S. J. Lee, I. K. Ko, C. Kengla, J. J. Yoo and A. Atala, Nat. Biotechnol., 2016, 34, 312–319 CrossRef CAS PubMed.
- T. Gonzalez-Fernandez, A. J. Tenorio, K. T. Campbell, E. A. Silva and J. K. Leach, Tissue Eng., Part A, 2021, 27, 1168–1181 CrossRef CAS PubMed.
- A. Abaci, G. Camci-Unal, M. Guvendiren and E. Guest, MRS Bull., 2023, 48, 624–631 CrossRef CAS.
- P. M. Sivakumar, A. A. Yetisgin, E. Demir, S. B. Sahin and S. Cetinel, Int. J. Biol. Macromol., 2023, 250, 126237 CrossRef CAS PubMed.
- T. Nonoyama, Polym. J., 2020, 52, 709–716 CrossRef CAS.
- S. Kuttappan, D. Mathew and M. B. Nair, Int. J. Biol. Macromol., 2016, 93, 1390–1401 CrossRef CAS PubMed.
- G. Turnbull, J. Clarke, F. Picard, P. Riches, L. Jia, F. Han, B. Li and W. Shu, Bioact. Mater., 2018, 3, 278–314 Search PubMed.
- S. Das and J. Jang, J. 3D Print. Med., 2018, 2, 69–87 CrossRef CAS.
- F. Pati, J. Jang, D.-H. Ha, S. Won Kim, J.-W. Rhie, J.-H. Shim, D.-H. Kim and D.-W. Cho, Nat. Commun., 2014, 5, 3935 CrossRef CAS PubMed.
- M. J. Sawkins, W. Bowen, P. Dhadda, H. Markides, L. E. Sidney, A. J. Taylor, F. R. A. J. Rose, S. F. Badylak, K. M. Shakesheff and L. J. White, Acta Biomater., 2013, 9, 7865–7873 CrossRef CAS PubMed.
- S. Chae, Y. Sun, Y.-J. Choi, D.-H. Ha, I. Jeon and D.-W. Cho, Biofabrication, 2021, 13, 035005 CrossRef CAS PubMed.
- J. Lee, J. Hong, W. Kim and G. H. Kim, Carbohydr. Polym., 2020, 250, 116914 CrossRef CAS PubMed.
- G. Ratheesh, C. Vaquette and Y. Xiao, Adv. Healthcare Mater., 2020, 9, 2001323 CrossRef CAS PubMed.
- A. Kara, T. Distler, C. Polley, D. Schneidereit, H. Seitz, O. Friedrich, F. Tihminlioglu and A. R. Boccaccini, Mater. Today Bio, 2022, 15, 100309 CrossRef CAS PubMed.
- H. Gharacheh and M. Guvendiren, Polymers, 2022, 14, 3788 CrossRef CAS PubMed.
- T. Bhattacharjee, S. M. Zehnder, K. G. Rowe, S. Jain, R. M. Nixon, W. G. Sawyer and T. E. Angelini, Sci. Adv., 2015, 1, e1500655 CrossRef PubMed.
- T. J. Hinton, Q. Jallerat, R. N. Palchesko, J. H. Park, M. S. Grodzicki, H.-J. Shue, M. H. Ramadan, A. R. Hudson and A. W. Feinberg, Sci. Adv., 2015, 1, e1500758 CrossRef PubMed.
- C. B. Highley, C. B. Rodell and J. A. Burdick, Adv. Mater., 2015, 27, 5075–5079 CrossRef CAS PubMed.
- O. Jeon, Y. B. Lee, H. Jeong, S. J. Lee, D. Wells and E. Alsberg, Mater. Horiz., 2019, 6, 1625–1631 RSC.
- B. Ayan, N. Celik, Z. Zhang, K. Zhou, M. H. Kim, D. Banerjee, Y. Wu, F. Costanzo and I. T. Ozbolat, Commun. Phys., 2020, 3, 183 CrossRef CAS PubMed.
- D. Banerjee, M. M. Ivanova, N. Celik, M. H. Kim, I. D. Derman, R. P. Limgala, I. T. Ozbolat and O. Goker-Alpan, Biofabrication, 2023, 15, 045023 CrossRef CAS PubMed.
- J. G. Roth, L. G. Brunel, M. S. Huang, Y. Liu, B. Cai, S. Sinha, F. Yang, S. P. Paşca, S. Shin and S. C. Heilshorn, Nat. Commun., 2023, 14, 4346 CrossRef CAS PubMed.
- J. A. Brassard, M. Nikolaev, T. Hübscher, M. Hofer and M. P. Lutolf, Nat. Mater., 2021, 20, 22–29 CrossRef CAS PubMed.
- M. H. Kim, Y. P. Singh, N. Celik, M. Yeo, E. Rizk, D. J. Hayes and I. T. Ozbolat, Nat. Commun., 2024, 15, 10083 CrossRef CAS PubMed.
- A. C. Daly, M. D. Davidson and J. A. Burdick, Nat. Commun., 2021, 12, 753 CrossRef CAS PubMed.
- A. Abaci and M. Guvendiren, Biofabrication, 2024, 16, 035027 CrossRef CAS PubMed.
- M. Guvendiren, A. Abaci and S. Ji, US Pat, 11806444B2, 2023 Search PubMed.
- J. Lu, H. Yu and C. Chen, RSC Adv., 2018, 8, 2015–2033 RSC.
- S. Samavedi, A. R. Whittington and A. S. Goldstein, Acta Biomater., 2013, 9, 8037–8045 CrossRef CAS PubMed.
- Y.-R. V. Shih, Y. Hwang, A. Phadke, H. Kang, N. S. Hwang, E. J. Caro, S. Nguyen, M. Siu, E. A. Theodorakis, N. C. Gianneschi, K. S. Vecchio, S. Chien, O. K. Lee and S. Varghese, Proc. Natl. Acad. Sci. U. S. A., 2014, 111, 990–995 CrossRef CAS PubMed.
- M. E. Cooke and D. H. Rosenzweig, APL Bioeng., 2021, 5, 011502 CrossRef PubMed.
- H. Chen, J. Zhang, X. Li, L. Liu, X. Zhang, D. Ren, C. Ma, L. Zhang, Z. Fei and T. Xu, Biofabrication, 2019, 11, 045007 CrossRef CAS PubMed.
- E. Nyberg, A. Rindone, A. Dorafshar and W. L. Grayson, Tissue Eng., Part A, 2016, 23, 503–514 CrossRef PubMed.
- N. Huebsch, P. R. Arany, A. S. Mao, D. Shvartsman, O. A. Ali, S. A. Bencherif, J. Rivera-Feliciano and D. J. Mooney, Nat. Mater., 2010, 9, 518–526 CrossRef CAS PubMed.
- S. Khetan, M. Guvendiren, W. R. Legant, D. M. Cohen, C. S. Chen and J. A. Burdick, Nat. Mater., 2013, 12, 458–465 CrossRef CAS PubMed.
- M. Guvendiren and J. A. Burdick, Nat. Commun., 2012, 3, 792 CrossRef PubMed.
- B. D. Fairbanks, M. P. Schwartz, C. N. Bowman and K. S. Anseth, Biomaterials, 2009, 30, 6702–6707 CrossRef CAS PubMed.
- S. Ji, A. Abaci, T. Morrison, W. M. Gramlich and M. Guvendiren, Bioprinting, 2020, 18, e00083 CrossRef.
- S. Ji, E. Almeida and M. Guvendiren, Acta Biomater., 2019, 95, 214–224 CrossRef CAS PubMed.
Footnotes |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d4bm01616e |
| ‡ Contributed equally. |
| This journal is © The Royal Society of Chemistry 2025 |