Open Access Article
Open Access ArticleThree- and four-stranded nucleic acid structures and their ligands
Yoshiki
Hashimoto
,
Sumit
Shil
,
Mitsuki
Tsuruta
,
Keiko
Kawauchi
and
Daisuke
Miyoshi
 *
*
Frontiers of Innovative Research in Science and Technology, Konan University, 7-1-20 Minatojima-minamimachi, Chuo-ku, Kobe, Hyogo, 650-0047, Japan. E-mail: miyoshi@konan-u.ac.jp
First published on 19th February 2025
Abstract
Nucleic acids have the potential to form not only duplexes, but also various non-canonical secondary structures in living cells. Non-canonical structures play regulatory functions mainly in the central dogma. Therefore, nucleic acid targeting molecules are potential novel therapeutic drugs that can target ‘undruggable’ proteins in various diseases. One of the concerns of small molecules targeting nucleic acids is selectivity, because nucleic acids have only four different building blocks. Three- and four-stranded non-canonical structures, triplexes and quadruplexes, respectively, are promising targets of small molecules because their three-dimensional structures are significantly different from the canonical duplexes, which are the most abundant in cells. Here, we describe some basic properties of the triplexes and quadruplexes and small molecules targeting the triplexes and tetraplexes.
Introduction
Most small-molecule therapeutics in clinical use currently target proteins. Human cells express about 20![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 species of proteins, following the hypothesis of “one gene expresses one protein”. Within these proteins, up to 15%, corresponding to 3000 species of proteins, are disease related (Fig. 1).1–3 Within the disease-related proteins, many of the proteins are termed 'undruggable', which refers to proteins that are difficult to target pharmacologically. An undruggable protein often lacks a binding site for a small molecule. Intrinsically disordered proteins, lacking fixed or ordered structures, are also often undruggable. The most well-known examples of the former and the latter undruggable proteins are RAS4 and MYC,5 respectively, although these two proteins are important targets for cancer therapeutics. Moreover, recent interactome studies suggest that there are up to 650
000 species of proteins, following the hypothesis of “one gene expresses one protein”. Within these proteins, up to 15%, corresponding to 3000 species of proteins, are disease related (Fig. 1).1–3 Within the disease-related proteins, many of the proteins are termed 'undruggable', which refers to proteins that are difficult to target pharmacologically. An undruggable protein often lacks a binding site for a small molecule. Intrinsically disordered proteins, lacking fixed or ordered structures, are also often undruggable. The most well-known examples of the former and the latter undruggable proteins are RAS4 and MYC,5 respectively, although these two proteins are important targets for cancer therapeutics. Moreover, recent interactome studies suggest that there are up to 650![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 protein–protein interactions (PPIs) in humans.6 PPIs are also considered undruggable by small molecules because of the large, flat, and featureless surfaces of PPIs.7 In fact, only a few percent of these PPIs can be targeted with small drugs.8,9 Therefore, alternative strategies for the inactivation of these disease-related undruggable proteins are required.
000 protein–protein interactions (PPIs) in humans.6 PPIs are also considered undruggable by small molecules because of the large, flat, and featureless surfaces of PPIs.7 In fact, only a few percent of these PPIs can be targeted with small drugs.8,9 Therefore, alternative strategies for the inactivation of these disease-related undruggable proteins are required.
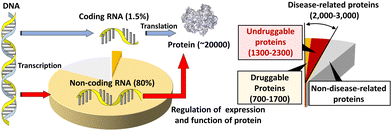 | ||
| Fig. 1 Schematic diagram depicting protein expression in the central dogma and the potential nucleic acid-targeted druggable genome. | ||
The 20![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 species of proteins in a human cell are encoded by only 1.5% of the human genome, which contains 3
000 species of proteins in a human cell are encoded by only 1.5% of the human genome, which contains 3![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000
000![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000
000![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 base pairs in total (Fig. 1).10,11 Among these proteins, fewer than 700 have been targeted by drugs. This corresponds to only 0.05% of the genetic information encoded in the human genome. However, around 80% of the human genome is transcribed to RNA. As described above, DNA coding amino acid information corresponds to 1.5% of the genome, so the remaining part of the 80% is non-coding RNA (ncRNA),12 which controls the central dogma at various steps, such as replication, transcription, processing and localization of mRNA, and translation. When compared with druggable proteins, there is far more opportunity to target RNAs to expand druggability and improve our understanding of biological processes governing the central dogma. If DNA and RNA can be targeted, protein products can be up- or down-regulated at the transcriptional and translational levels.
000 base pairs in total (Fig. 1).10,11 Among these proteins, fewer than 700 have been targeted by drugs. This corresponds to only 0.05% of the genetic information encoded in the human genome. However, around 80% of the human genome is transcribed to RNA. As described above, DNA coding amino acid information corresponds to 1.5% of the genome, so the remaining part of the 80% is non-coding RNA (ncRNA),12 which controls the central dogma at various steps, such as replication, transcription, processing and localization of mRNA, and translation. When compared with druggable proteins, there is far more opportunity to target RNAs to expand druggability and improve our understanding of biological processes governing the central dogma. If DNA and RNA can be targeted, protein products can be up- or down-regulated at the transcriptional and translational levels.
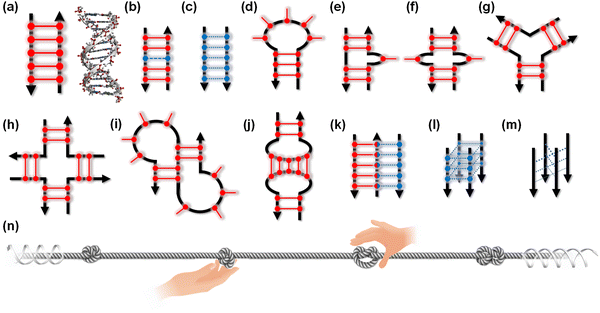
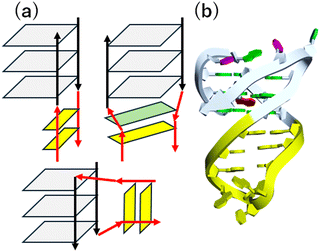
The canonical structures of DNA and RNA are B-form and A-form duplexes, respectively, with Watson–Crick base pairs. Canonical duplexes are suitable and highly adapted for their functions: storage, inheritance, and the transition of genetic information. However, both DNA and RNA can fold to form various non-canonical and transient structures (Fig. 2a–m), depending on the nucleotide sequence as well as the surrounding molecular environment. Similar to a knot providing a firm place to grip a rope (Fig. 2n), non-canonical structures can be recognition sites and controlling elements for a series of enzymes, including DNA and RNA polymerase, helicase, topoisomerase, nuclease, DNA and RNA modifiers, spliceosome, and ribosome, as well as transcriptional factors.13–15 In fact, roughly 1500 proteins are predicted to bind to RNA in a sequence- and structure-selective manner.16 Small molecules targeting DNA and RNA could regulate not only the expression of proteins but also the functions of the DNA- and RNA-binding proteins. Thus, small molecules targeting non-canonical structures have drawn attention as new therapeutic regimens.17–19 Risdiplam, sold under the brand name Evrysdi, represents a milestone given its status as the first small molecule drug that targets RNA.20
One of the concerns of small molecules targeting nucleic acids is selectivity, because nucleic acids have only four different building blocks, whereas proteins are composed of 20 amino acids. In addition, it is generally considered that nucleic acid structures are static and monotonous. In contrast, it has been discussed that there are up to 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 unique protein folds in nature.21–23 From this point of specificity, three- and four-stranded non-canonical structures, triplexes and quadruplexes, respectively, are promising because their three-dimensional structures are significantly different from the canonical duplexes. Noteworthy, recent progresses in bioinformatics, DNA and RNA sequencing, and bioimaging have demonstrated the existence of the multiplexes and their biological roles in living cells of various organisms from virus to human. In this review, we briefly describe the triplex and quadruplex structures of nucleic acids and their potential biological roles. We then describe recent progress in the development of small molecules that target these non-canonical nucleic acid structures.
000 unique protein folds in nature.21–23 From this point of specificity, three- and four-stranded non-canonical structures, triplexes and quadruplexes, respectively, are promising because their three-dimensional structures are significantly different from the canonical duplexes. Noteworthy, recent progresses in bioinformatics, DNA and RNA sequencing, and bioimaging have demonstrated the existence of the multiplexes and their biological roles in living cells of various organisms from virus to human. In this review, we briefly describe the triplex and quadruplex structures of nucleic acids and their potential biological roles. We then describe recent progress in the development of small molecules that target these non-canonical nucleic acid structures.
Triplexes
Structure and biological role of triplexes
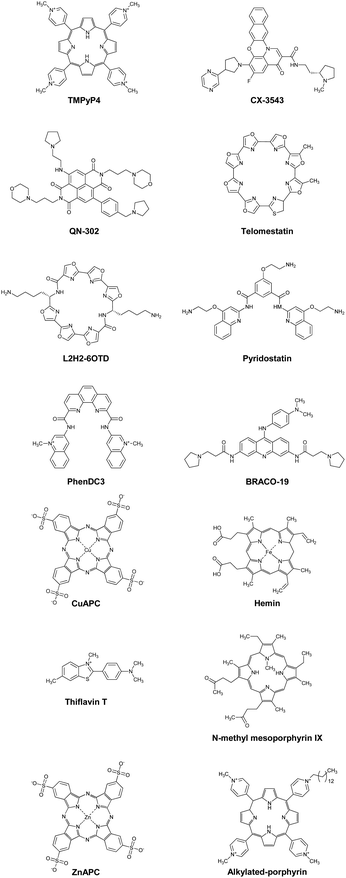
The canonical duplex has two grooves formed by the sugar-phosphate backbones of the two strands. The grooves play a vital role in interactions with other molecules, such as protein, small molecules, metal ion, and water. The grooves further play an important role in the sequence-specific recognition of the duplex by single-stranded nucleic acids, either DNA or RNA. These interactions are mediated by the formation of Hoogsteen-type hydrogen bonds between the bases of the third strand, often called a triplex-forming oligonucleotide (TFO), and the purine bases of the duplex formed by a polypurine-polypyrimidine sequence (Fig. 3a). The first example of a triplex was reported by Felsenfeld et al. in 1967. They found the formation of an intermolecular RNA triplex by two strands of polyribo(uridylic acid) and one strand of polyribo(adenylic acid).24 After the discovery of the triplex, many triplex structures under various conditions have been identified.25 There are two classes of triplex motifs, distinguished by the orientation of the TFO, which binds to the major groove of the target duplex. A polypyrimidine TFO binds to the polypurine strand of the duplex via Hoogsteen hydrogen bonds in a parallel fashion (in the same orientation as the purine strand of the duplex). In contrast, a polypurine TFO binds in an antiparallel orientation to the polypurine strand of the duplex via reverse-Hoogsteen hydrogen bonds.26,27 In an antiparallel triplex, the base pairs are G:G-C and A:A-T (“:” and “-“ indicate Hoogsteen and Watson–Crick base pairs, respectively). In a parallel triplex, the base pairs are C+:G-C and T:A-T (C+ represents a protonated cytosine). Thus, the ideal target of a TFO is a duplex of a homopurine sequence in one strand and a homopyrimidine sequence in the complementary strand. For both parallel and antiparallel motifs, contiguous homopurine-homopyrimidine runs of at least 10 base pairs are required for thermodynamically stable TFO binding.28 In addition, all TFOs adopt anti-conformations (Fig. 3b),29 although 8-oxoadenin in a TFO can adopt a syn-conformation.30 The anti-conformation in a TFO is the same as that of nucleotides in B- and A-form duplexes.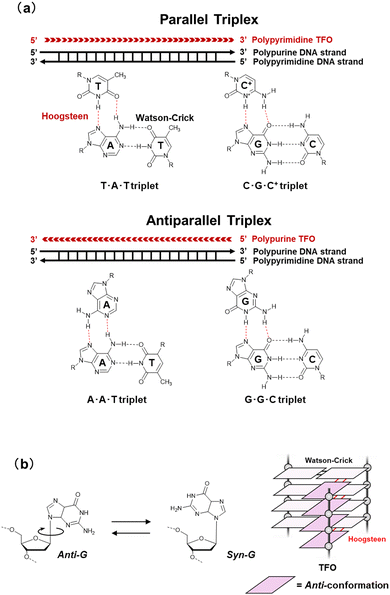 | ||
| Fig. 3 (a) Base pairs in parallel and antiparallel triplexes. (b) Schematic illustration of glycosidic conformation for nucleobases in TFO. | ||
Large numbers of potential triplex forming sites have been identified in the human genome.31 Most annotated genes in the human genome contain at least one triplex-forming site, and these sites are enriched in the promoter regions,32 indicating a potential role of triplex in controlling gene expression. These TFO-binding sites have attracted much attention because of their therapeutic potential to inhibit the expression of disease-related genes. Evidence for the existence of triplexes in cells comes from triplex-binding antibodies.33,34 However, small molecules probing the triplex are still required to observe the direct dynamic behavior of the triplex over time in living cells.
RNA triplexes
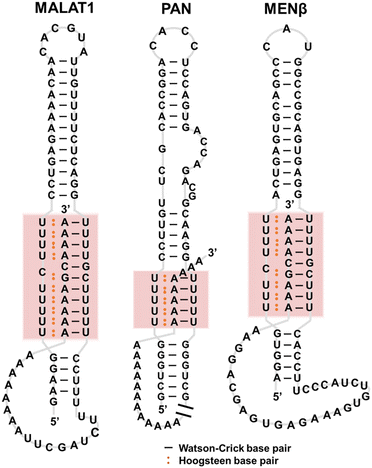
Triplexes have been observed in transcripts such as tRNAPhe, telomerase RNA, self-splicing group II introns, U2-U6 snRNAs, riboswitches, virus RNAs, and RNA stability elements.35 These RNA triplexes play roles in telomere synthesis, RNA splicing, binding to ligands and ions in metabolite-sensing riboswitches, and protecting RNA from degradation.Telomerase RNAs in various eukaryotes have a conserved pseudoknot in which a triplex exists.36,37 The triplex structure is essential for telomerase catalytic activity,38 demonstrating that the triplex structure rather than the nucleotide sequence is conserved and required for the ribonucleoprotein function. Triplexes observed in riboswitches are also important as binding sites of a series of ligands. For example, SAM-II riboswitch has triplex-forming sites that serve as structural platforms for ligand binding.39 RNA triplexes have also been proposed for various viruses, such as Kaposi's sarcoma-associated herpesvirus (KSHV).40 The triplexes protect the viral RNAs and inhibit decay by host nuclear RNA degradation to accumulate in KSHV-infected cells.41,42 Human metastasis-associated lung adenocarcinoma transcript 1 (MALAT1), a long noncoding RNA (lncRNA), folds to form a triplex (Fig. 4). MALAT1 is localized in nuclear speckles and is involved in the regulation of gene expression.43,44 MALAT1 is upregulated in multiple cancer types, depending on the triplex formation, making it a potential drug target.45,46 Unlike many lncRNAs that are rapidly degraded and thus expressed at near-undetectable levels, MALAT1, as well as MEN β and PAN, which are also known to form triplexes (Fig. 4), are stable transcripts with long half-lives.47 It has been proposed that the triplexes formed by these lncRNAs inhibit nuclear RNA decay.48 Although the biological roles of RNA stability elements remain unclear, the triplex supports the transport, stability, and translation of an RNA, allowing efficient repression by microRNAs.
Moreover, it was recently reported that lncRNAs bind target DNAs in a sequence-specific manner by forming RNA–DNA triplexes.49,50 This process allows lncRNAs to recruit proteins to specific genomic regions and to regulate gene expression. LncRNAs such as Fendrr,51 MEG3,52 KHPS1,53 PARTICLE,54,55 and HOTAIR56 bind to their target DNAs in a sequence-specific manner, utilizing the RNA–DNA triplexes. It has further been proposed by computational analysis that there are many triplex target sites of lncRNAs in the genome.57 Interestingly, such bioinformatic analysis has revealed a group of genomic regions that may have a very high propensity for triplex formation with a wide range of different RNAs.58 Excellent reviews have discussed the biological roles of RNA–DNA triplexes.59,60
Triplex-binding proteins
It has been reported that some proteins such as histones H1 and H2A, and topoisomerase II, bind to AT-rich sequences.61–63 A protein that binds preferentially to TAT DNA triplexes was reported for the first time in 1991, although its identification was not possible at that time.64 Proteins that specifically recognize certain DNA triplex motifs have been reported.65,66 Since the 2000s, many proteins have been identified as DNA triplex-binding proteins67–83 (Table 1). Although the biological roles of these DNA triplex-binding proteins are highly diverse, it is reasonable that some helicases are triplex-binding proteins. In addition, nuclear proteins are known to bind DNA/RNA triplexes, but their biological functions are unexplored.84| Protein | Function/target | Ref. |
|---|---|---|
| GAGA factor | Gene regulation and alteration of chromatin structure/pyrimidine motif | 67 |
| Stm1 | Involved in mitosis/purine motif | 68 |
| CDP1 | Chromosome segregation and histone displacement/purine motif | 69 |
| type III intermediate filaments | Gene regulation, DNA repair, and chromatin remodelling/pyrimidine motif | 70 |
| Loricrin | Keratinocyte cornification/purine motif | 71 |
| Orc4 | Origin organization during DNA replication/purine motif | 72 |
| hnRNPs | DNA and RNA binding/synthetic oligo DNAs | 73 |
| Tn7 | Transposon insertion/pyrimidine motif | 74,75 |
| XPC-hHR23B, XPA-RPA | DNA damage recognition/TFO-directed psoralen-interstrand crosslink | 76 |
| HMGB1 | Chromatin structure, transcriptional regulation/triplex-directed psoralen interstrand crosslinks | 77 |
| U2AF65, PSF, p54nrb | Splicing factors/purine motif | 78 |
| RecQ, BLM, WRN, | 3′ → 5′ helicase/pyrimidine motif with a free 3′ tail | 79 |
| FANCJ | 5′ → 3′ helicase/pyrimidine motif with a free 3′ tail | 80 |
| DHX9 | 3′ → 5′ helicase/purine motif with a free 3′ tail | 81 |
| SV40 large T-antigen | 3′ → 5′ helicase/pyrimidine motif with a free 3′ tail | 82 |
| p53 | Tumor suppressor/intra and intermolecular pyrimidine motif | 83 |
Small molecules that target triplexes
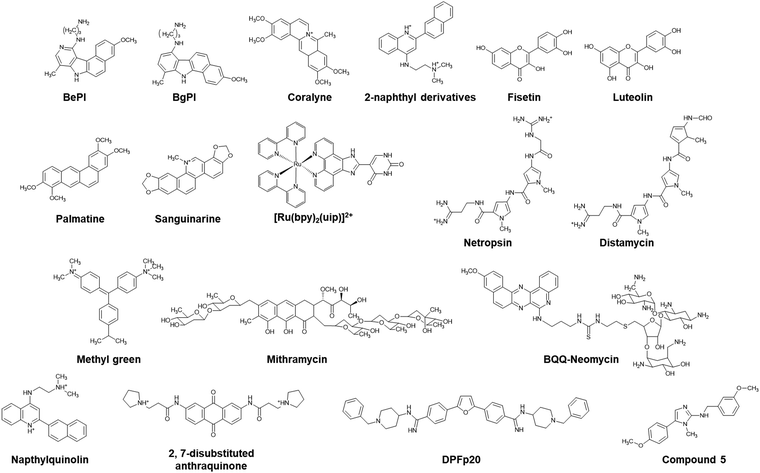
The Hoogsteen base pairs between the TFO and the target duplex are generally weaker than the Watson–Crick base pairs in the duplex.85 The instability of the triplex under physiological conditions limits its biological roles and applications. Of note, a parallel triplex is stable only at acidic pH, because the protonation of cytosines is required for Hoogsteen base pairing, although the stability of antiparallel triplexes does not depend on solution pH.86 Moreover, multivalent cations such as Mg2+ are required to neutralize the electrostatic repulsion between the three nucleic acid strands. The triplex but not the duplex is stabilized under molecular crowding conditions that mimic a cellular environment.87 To overcome the limited thermal stability, small molecules as well as chemical modifications of TFOs have been applied.Hélène and co-workers demonstrated the enhancement of triplex thermal stability by attaching small molecules such as acridine, ellipticine, and psoralen to the TFO.88–90 Small molecules that bind strongly and selectively to the triplex can also stabilize the triplex without covalent modification. Some triplex-binding small molecules are shown in Fig. 5. Ethidium bromide stabilizes a triplex containing only TAT base pairs,91 whereas it destabilizes a triplex containing both TAT and C+GC base pairs because of electrostatic repulsion with C+.92 Some of the earliest discovered small molecules that stabilize the TAT and C+GC of triplexes are benzopyroindole derivatives (BePI and BgPI) and coralyne.93–96 BePI stabilizes TAT-rich triplexes, whereas coralyne does not show sequence selectivity. BePI enhances the sequence-specific inhibition of transcription initiation of a specific gene by RNA polymerase by a TFO. Quinoxaline derivatives BfPQ and BQQ were further developed as analogues of BPI derivatives.97–99 These molecules have been shown to significantly stabilize triplexes under physiological conditions, whereas they bind poorly to duplexes. Notably, BQQ promotes a short TFO to bind to its target site in a plasmid and enhances the inhibition of restriction enzyme cleavage of the target site.
Wilson and co-workers proposed that the structural properties of a triplex intercalator include the following: (i) the compounds should be cationic because of the high negative charge density of the triplex. (ii) The compounds should have an aromatic surface to form stacking interactions with the three bases in the triplex. (iii) The aromatic compounds should have torsional flexibility, because the three bases in a triplex have torsional freedom.100 Based on these properties, they synthesized a series of quinoline derivatives and found that a 2-naphthyl derivative could bind to poly(dA)·2poly(dT) to a much greater extent than to the corresponding duplex.100 Other triplex intercalators, such as fisetin,101 luteolin,102 palmatine,103 sanguinarine,104 and ruthenium(II) complexes,105 are also shown in Fig. 5.
Groove binders have also been demonstrated to have affinity with DNA triplexes. It has been shown that netropsin, a drug that interacts with the minor groove of a DNA duplex, binds the minor groove of a triplex and destabilizes it, but stabilizes the duplex.106,107 Umemoto et al. showed by using distamycin 2, an analogue of netropsin, that the drug does not destabilize.108 Breslauer and coworkers showed that berenil, DAPI, ethidium, and netropsin induce the formation of a TAT DNA/RNA/DNA triplex.109 In addition, a series of small molecules such as methyl green and mitharamycin, which bind and destabilize triplexes, have been reported.110–112 Aminoglycosides, especially neomycin, stabilize a triplex, while they do not alter the stability of a duplex.113 Triplex stabilization by neomycin is salt- and pH-dependent, and increasing the concentrations of K+ and Mg2+ and the solution pH decreases the stabilization effects of neomycin.114,115 These aminoglycosides bind with not only DNA triplexes, but also RNA and DNA/RNA triplexes.115 A combination of intercalation and groove binding has also been reported. Intercalator–neomycin conjugates have been studied to enhance binding affinity with DNA triplexes. BQQ-neomycin enhances the binding affinity with a TAT DNA triplex up to almost 1000-fold more than that of neomycin through a dual recognition mode: groove binding and intercalation of neomycin and BQQ, respectively.116
As described above, helix-forming MALAT1 lncRNA is a potential anticancer target due to its overexpression in cancer cells, and its knockdown reduces tumor growth. Deletion and mutation studies in the triplex-forming region have shown that alterations in the stability of the triplex lead to significant changes in transcript levels.117,118 Several small molecules, such as diphenylfuran derivatives, have been reported as lncRNA triplex-targeting molecules.119 Small molecules targeting the MALAT1 triplex with high selectivity have also been reported, of which a representative example is compound 5 (Fig. 5).120 Compound 5 was obtained by small molecule microarray screening of a library comprising approximately 26![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 compounds. Compound 5 regulates MALAT expression. Since NEAT1 has a triplex structure, these results confirm that compound 5 selectively binds to the MALAT1 triplex in living cells. NMR experiments and MD simulations demonstrated that compound 5 binds within the deep and narrow major groove of the MALAT1 triplex through several van der Waals interactions with the phosphate and nucleobase groups of the Hoogsteen strand and the Watson–Crick strands. An RT-qPCR experiment showed that the MALAT1 expression level in mouse model mammary tumor organoids was suppressed by 51% following treatment with 1 μM of compound 5 whereas NEAT1 expression was unaffected,120 demonstrating the high sensitivity of compound 5. Molecules such as compound 5, which specifically target the MALAT1 triplex, are a promising new class of anticancer therapeutics and molecular probes for the treatment and investigation, respectively, of cancers driven by MALAT1.
000 compounds. Compound 5 regulates MALAT expression. Since NEAT1 has a triplex structure, these results confirm that compound 5 selectively binds to the MALAT1 triplex in living cells. NMR experiments and MD simulations demonstrated that compound 5 binds within the deep and narrow major groove of the MALAT1 triplex through several van der Waals interactions with the phosphate and nucleobase groups of the Hoogsteen strand and the Watson–Crick strands. An RT-qPCR experiment showed that the MALAT1 expression level in mouse model mammary tumor organoids was suppressed by 51% following treatment with 1 μM of compound 5 whereas NEAT1 expression was unaffected,120 demonstrating the high sensitivity of compound 5. Molecules such as compound 5, which specifically target the MALAT1 triplex, are a promising new class of anticancer therapeutics and molecular probes for the treatment and investigation, respectively, of cancers driven by MALAT1.
G-quadruplexes
A G-quadruplex (G4) structure of guanylic acid was reported for the first time by Gellert et al. in 1962.121 The G4 structure of a guanine-rich telomeric sequence was then revealed by Sen and Gilbert in 1988.122 These studies made us realize that guanine-rich sequences can form G4s (Fig. 6a). Putative G4-forming sequences are generally considered to have the following consensus: G2≦L1≦G2≦L1≦G2≦L1≦G2≦, in which guanine stretches composing more than two guanines are connected by linker loop sequences (L) including at least one nucleotide.123 These putative G4-forming motifs are observed ubiquitously in diverse organisms’ genomes. Among the various putative G4-forming sequences, the most well-studied G4 is that of telomere DNA. Human telomere DNA comprises repetitive sequences of the hexanucleotide d(TTAGGG)n repeat unit. In normal human somatic cells, telomere DNA ranges from 5–15 kb in length with an extended single-stranded 3′-overhang of a few hundred bases (Fig. 6b).124–126 In each cell division, the telomere DNA is shortened because of the end replication problem, which leads to apoptosis.127 In contrast, the telomere DNA length is maintained in cancer cells. In most cancers, telomerase, a ribonucleoprotein, extends the telomere 3′ overhang by its reverse transcription activity.128,129 Thus, telomerase inhibitors provide a new approach to cancer chemotherapy.130 The hybridization between the telomere DNA and its complementary cytosine-rich sequence in the telomerase RNA is essential for the reverse transcription activity.131 Formation and stabilization of the G4 in the telomere overhang by a small molecule can inhibit telomerase activity, and thus cancer cell proliferation.132,133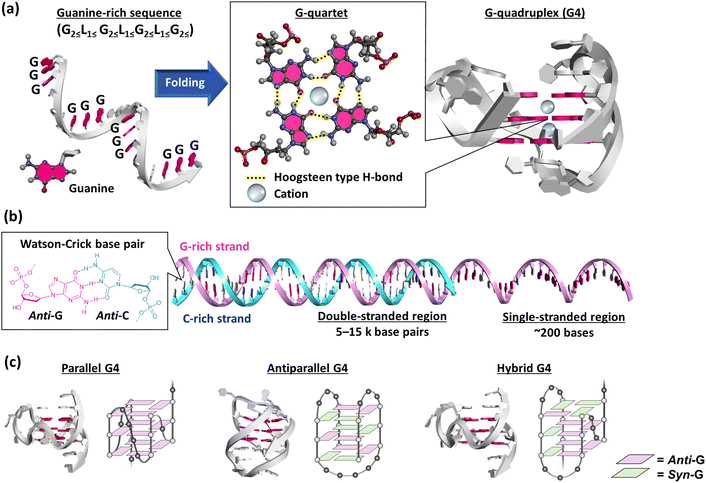 | ||
| Fig. 6 (a) Schematic illustration of the folding of a guanine-rich (G-rich) sequence. The G-rich sequence folds into a G-quadruplex (G4) via a G-quartet. The three-dimensional structure of G4 is adapted from PDB ID: 2HY9. Guanine bases are highlighted in red. (b) Schematic illustration of a telomeric end with a double-stranded G-rich strand (red) and cytosine-rich (C-rich) strand (blue) and with a single-stranded G-rich region. Guanine and cytosine bases are highlighted in red and blue, respectively. (c) Three-dimensional structure of parallel (PDB ID: 2M27, ref. 134), antiparallel (PDB ID: 6ZX7, ref. 135), and hybrid (PDB ID: 2HY9, ref. 136) G4s. | ||
A G4 exhibits various topologies, parallel, antiparallel, and hybrid, in an intermolecular and intramolecular manner (Fig. 6c).134–137 G4s show such highly structural polymorphisms depending on the nucleotide sequence as well as molecular environmental factors.138 Moreover, G4 guanines can adopt either an anti- or syn-conformation, resulting in different B-form duplexes. The resulting pronounced variations in the glycosidic torsion angles result in four grooves of different sizes, which could be important for the specificity of G4 ligands binding to target G4s. Parallel G4s comprised exclusively of guanines adopt the anti-glycosidic conformation (Fig. 6c)139 whereas in an antiparallel G4, guanines adopt both anti- and syn-conformations and at least one of the four strands must be oriented antiparallel to the other strands (Fig. 6c).139 In this chapter, factors affecting the structure and stability of G4 will be briefly explained.
Factors affecting the structure and stability of G4 originating from the nucleotide sequence
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 G4-duplex hybrids-forming sequences in the human genome. About 60% of these sequences were strand-specifically located in genic/promoter regions.172 These G4-duplex hybrids have been reported to play multiple roles in determination of topology, folding and unfolding kinetics, and cation binding of G4.173–176 Moreover, G4s involving the duplex showed more efficient transcriptional inhibition.170,177 Since the G4-duplex hybrid-forming DNA sequences were identified in the sense and antisense strands,172 corresponding RNA structure can exist in naturally occurring RNAs such as PIM1 mRNA (Fig. 8b)178 as well as synthetic RNA aptamer such as SC1179 and Spinach.180,181 The G4-duplex hybrid can be a promising target in diverse biological and pathological processes, because the junction between the G4 and the duplex gives distinct structure from the “canonical” G4, which has shorter loop length. Moreover, the duplex region could be useful for sequence selective binding for a target G4.
000 G4-duplex hybrids-forming sequences in the human genome. About 60% of these sequences were strand-specifically located in genic/promoter regions.172 These G4-duplex hybrids have been reported to play multiple roles in determination of topology, folding and unfolding kinetics, and cation binding of G4.173–176 Moreover, G4s involving the duplex showed more efficient transcriptional inhibition.170,177 Since the G4-duplex hybrid-forming DNA sequences were identified in the sense and antisense strands,172 corresponding RNA structure can exist in naturally occurring RNAs such as PIM1 mRNA (Fig. 8b)178 as well as synthetic RNA aptamer such as SC1179 and Spinach.180,181 The G4-duplex hybrid can be a promising target in diverse biological and pathological processes, because the junction between the G4 and the duplex gives distinct structure from the “canonical” G4, which has shorter loop length. Moreover, the duplex region could be useful for sequence selective binding for a target G4.
 | ||
| Fig. 8 (a) Schematic structure of the G4-duplex hybrids. G-quartet and Watson–Crick base pair are shown in gray and yellow. Green shows non-canonical base pairs. (grey), (b) three-dimensional G4-duplex hybrid structure in PIM1 gene (PDB: 7CV3). The duplex region is highlighted in yellow. | ||
Factors affecting the structure and stability of G4 originating from the surrounding environment
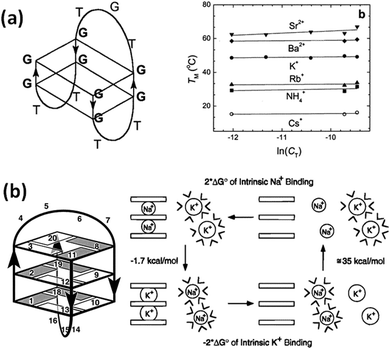 | ||
| Fig. 9 (a) Schematic structure of the thrombin binding aptamer (TBA) (left). Melting temperature (Tm) of the TBA in the presence of various cations (right). Adapted from ref. 186. Copyright © 2001 American Chemical Society. (b) Schematic structure of d(G3T4G3) (left) and the relationship between the dehydration energy of the cations and the thermodynamic stability of G4 (right). Adapted from ref. 192. Copyright © 1996 American Chemical Society. | ||
The ion radius of a cation is critical for binding with G4. It is generally considered that this is because there is an optimal size that fits into the cavity of the G-quartet(s). It is also evident that dehydration is required for cation coordination. Thus, the hydration energy is important to determine the structural stability of G4s.144,186 Hud et al. studied by NMR the competition between Na+ and K+ for coordination with G-quartets using the oligonucleotide d(G3T4G3), which forms an antiparallel G4 in the presence of Na+ and K+ (Fig. 9b).192 They found that the more favored binding of K+ than Na+ is driven by the greater dehydration energy of Na+ than K+. Hydration from cations is also important for the structure and stability of G4s under cell-mimicking molecular crowding conditions, as explain in the next section.
In many cases, molecular crowding conditions induced by the addition of neutral cosolute molecules increase the thermodynamic stability and folding kinetics of G4s.198–205 Stabilization effects of molecular crowding have been reported for other non-canonical structures such as triplexes, junctions, dangling ends, and loops.156,206,207 In contrast to the stabilization effects on non-canonical structures, molecular crowding destabilizes the canonical duplex.198,208 These opposite effects on non-canonical and canonical structures indicate that G4s and other non-canonical structures are thermodynamically more stable under molecularly crowded cellular conditions than has been considered based on test-tube experiments. Thus, G4s as well as other non-canonical structures of nucleic acids are promising candidates as bidding sites for proteins and small molecules. These molecular crowding effects are attributed to the osmotic pressure that stabilizes the less hydrated G4 and other non-canonical structures.200,201,209–212 In contrast, positive hydration changes through duplex formation led to destabilization under molecular crowding conditions, where the concentration and activity of water molecules decrease.213,214
In addition, structural polymorphisms of G4 are induced and regulated by molecular crowding. For example, molecular crowding induces a structural transition from a duplex to a G4 in a mixture of telomere G-rich and complementary C-rich strands.215 Molecular crowding further alters the strand orientation of G4s from antiparallel and mixed to parallel,202,216–219 which regulates the activity of enzymes such as telomerase and affects protein and ligand binding to G4s.220–222
| Protein | Function/Target | Ref. |
|---|---|---|
| Nucleolin | Regulation of transcript/c-MYC | 232 |
| DNA methyltransferase | Methylation of cytosine at C-5/c-MYC, CDKN1C and MEST | 233 |
| Nucleophosmin | Nucleolar phosphoprotein/c-MYC | 234 |
| ADAR | Convert adenosine to inosine/c-MYC | 235 |
| PARP1 | Gene regulation and DNA repair/BCL-2 | 236 |
| hnRNP1 | Gene regulation, DNA repair, and chromatin remodelling/KRAS | 237 |
| MAZ | DNA repair and chromatin remodelling/KRAS | 238,239 |
| FANCJ | 5′ → 3′ Helicase/Telomeric DNA | 240 |
| Pif1 | 5′ → 3′ Helicase/c-MYC | 241 |
| SMARCA4 | Chromatin remodelling/c-KIT | 230 |
| UHRF1 | Ubiquitin ligase/c-MYC and c-KIT | 230 |
| RBM22 | pre-mRNA-splicing factor/c-MYC and c-KIT | 230 |
| TTF2 | Transcription termination factor/c-MYC and c-KIT | 230 |
| DDX24 | Helicase/c-KIT | 230 |
| DDX1 | Helicase/c-MYC and c-KIT | 230 |
| CIRBP | cold-inducible RNA-binding protein/c-MYC | 228 |
| DHX36 | Helicase/c-MYC | 242 |
| DDX21 | Helicase/C9orf72 | 243 |
| DHX9 | 3′ → 5′ Helicase/TP-G4 | 244 |
| FUS | DNA repair and regulation of splicing/Telomeric DNA | 245 |
| TDP-43 | Regulation of transcription and splicing/Expanded C9orf72 gene | 246 |
| α-synuclein | DNA repair and rescuing neurons/Telomeric DNA | 247 |
Although many G4-binding proteins have been identified, there are still a very limited number of three-dimensional structures of G4–protein complexes. Three proteins, RHAU,258,259 RAP1,260 and a designed ankyrin protein utilize an α-helix to recognize a G-quartet (Fig. 10).261 A single α-helix of RHAU and RAP1 stacks on an exposed G-quartet. In contrast, ankyrin derivative uses a bundle of helices and loops to bind an exposed G-quartet. These binding modes lead to the specific recognition of the parallel G4. In these complexes, amino acids such as Arg, Ala, Tyr, Leu, and Met of the proteins form π–π and CH–π interactions with the bases and the sugar ring of the parallel G4. In addition, electrostatic and hydrogen bonding interactions between DNA and polar/basic residues contribute to stabilize the interactions.
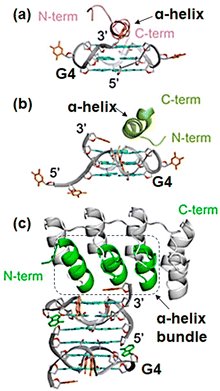 | ||
| Fig. 10 G4 recognition modes of (a) RHAU, (b) RAP1, and (c) ankyrin derivative. Adapted from ref. 261. Copyright © 2024 American Chemical Society. | ||
Because G4s in the genome inhibit replication and transcription, DNA G4s must be unfolded to proceed with these reactions. Thus, helicases are another category of G4-binding protein.262–264 A combination of G4 helicases and DNA replication proteins have therapeutic potential against cancer, because G4 helicases are upregulated in cancer cells.265 In fact, cancer cells deficient in a G4 helicase, FANCJ, have higher sensitivity to the G4 ligand telomestatin (Fig. 12).240 A series of 5′ to 3′ and 3′ to 5′ helicases proposed to bind G4s is listed in Table 2.236–238,245–247,266 Recently, Balasubramanian's group reported a co-binding-mediated protein profiling strategy with their own developed G4 ligand, pyridostatin (PDS, Fig. 12), to investigate the interactome of DNA G4s in native chromatin.230 The authors employed this approach in human cells and identified hundreds of putative G4-interacting proteins that comprised diverse functional classes, such as RNA binding, DNA binding, ATP binding, metal ion binding, ribonucleoprotein, hydrolase, repressor, helicase, activator, and chromatin regulator. Among them, SMARCA4, UHRF1, RBM22, TTF2, DDX24, and DDX1 were confirmed to bind to G4. Interestingly, the most frequently observed function of these proteins was RNA binding, even though the target was DNA G4 in these studies. However, this is consistent with the cold-inducible RNA-binding protein (CIRBP) being identified as a new G4 DNA-binding protein both in vitro and in cells.228 These results indicate that G4s participate in more diverse biological processes in cells than previously considered via interactions with various proteins.
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 loci where G4s form within the transcriptome in humans have been identified.271 RNA G4s are enriched in functionally important regions, including 5′- and 3′-UTRs271–273 and introns, although some G4s are observed in exons.
000 loci where G4s form within the transcriptome in humans have been identified.271 RNA G4s are enriched in functionally important regions, including 5′- and 3′-UTRs271–273 and introns, although some G4s are observed in exons.
As shown for DNA G4 helicases, several helicases, including DHX36, DDX21, and DHX9, bind to and unwind RNA G4s,242–244 because RNAs must be at least partly unfolded for their messenger function. Moreover, RNA G4-binding proteins observed in RNA modification reactions, viral pathogenesis, and mitochondria have been reviewed.274 Other classes of RNA G4-binding proteins are ribosomal proteins, the AFF family, and hnRNPs.257 Ribosomal RNAs (rRNAs) have many G4-forming sequences with three and two G-quartets,275,276 which exist as rRNA tentacles extending for hundreds of Å from ribosomal surfaces.277 Thus, it is reasonable that ribosomal proteins bind to these RNA G4s. The AFF family of genes includes four members: AFF1/AF4, AFF2/FMR2, AFF3/LAF4, and AFF4/AF5q31. hnRNP proteins play critical roles in the packaging, transport, and splicing of mRNAs, and some hnRNP proteins, such as A1 and A2, have been reported to bind to RNA G4s.278,279
G4 in liquid–liquid phase separation
Interestingly, many of the RNA G4 binding proteins discussed above are involved in liquid–liquid phase separation (LLPS) to form membrane-less organelles (biomolecular condensates and liquid droplets) in cells, which play critical roles in cellular processes and diseases. LLPS is a hot topic in many research fields from cell biology to soft matter physics.280,281 It is generally considered that LLPS is induced by intrinsically disordered proteins (IDPs) or that include intrinsic disorder regions (IDRs).282 Nucleic acids (DNA and RNA) also undergo LLPS with binding to proteins to regulate transcription and translation at various stages coupled with other molecular regulatory mechanisms.283 Multimolecular bindings among proteins and nucleic acids via electrostatic, hydrogen bonding, π–π stacking, cation–π, and CH–π interactions are essentials for LLPS.284 Notably, it has been proposed that DNA and RNA G4s are critical for LLPS,239,285–287 despite the fact that IDP and IDR are essential for LLPS. Fig. 11 shows the LLPS model system with a G4-forming RNA oligonucleotide, derived from repeat expansion sequence in FMR1 mRNA, and Arg- and Gly-rich model peptide, derived from RGG motif of FMRP. It was demonstrated that the RNA–peptide mixture undergoes LLPS, depending on the formation of RNA G4. Moreover, aberrant LLPS may trigger protein aggregation in neurodegenerative diseases such as frontotemporal dementia associated with FUS, amyotrophic lateral sclerosis associated with TDP-43, Parkinson's disease associated with α-synuclein, and Alzheimer's disease associated with tau.288 In some of these neurodegenerative diseases, the expansion of G-rich repeat sequences, which may form G4s at the genomic and mRNA levels, participate in the onset mechanism.289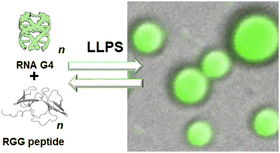 | ||
| Fig. 11 LLPS model system, consisting of RNA G4 and Arg- and Gly-rich peptide. Adapted from ref. 289. Copyright © 2022 Royal Society of Chemistry. | ||
G-quadruplex-targeting molecules
The fact that stabilized DNA or RNA G4s inhibit series of enzymes, involving DNA and RNA polymerases, ribosome, and reverse transcriptase, has spurred a huge interest in the structure-based design and synthesis of ligands that bind and stabilize G4s (G4 ligands). As of November 2024, the G4 ligand database (G4LDB) opened.290 G4LDB includes over 4800 ligands targeting quadruplexes (G4s and i-motifs of cytosine-rich sequences) entries. Several G4 ligands are in clinical trials, as we discuss later.291,292 The structures of some of these ligands are shown in Fig. 12 and are introduced here. Moreover, most G4 ligands possess cationic functional groups that form electrostatic interactions with anionic phosphates of nucleic acids and that make them water soluble. TMPyP4 is a typical and well-known G4 ligand. TMPyP4 shows high affinity for G4s due to its wide π-plane as well as its positive charges (Fig. 12).293 TMPyP4 has been shown to inhibit human telomere elongation and gene expression by inhibiting telomerase and RNA polymerase, respectively.294 However, the inhibitory effect on enzymatic activity is not structure selective. In telomerase reverse transcription assays in the presence of DNA duplexes, the inhibition effect on telomerase activity is significantly decreased.221,295 This non-structure-selective binding of G4 ligands may decrease their inhibitory effects on enzymes of interest. At least partly due to their low selectivity, no G4 ligand has advanced beyond phase II trials yet. Quarfloxin (CX-3543, Fig. 12) is the ligand that has reached phase II (NCT00780663).296 CX-3543 binds selectively to G4s formed in rDNA and disrupts rRNA interactions with nucleolin.297 Another G4 ligand, QN-302 (Fig. 12), has most recently entered clinical phase I trials (NCT06086522).291 We briefly discuss the development of QN-302 later in this manuscript.Thus, structure-selectivity has also attracted attention in G4 ligand design. Telomestatin, which is isolated from Streptomyces anulatus 3533-SV4, selectively binds to and stabilizes telomeric DNA G4s, resulting in highly efficient telomerase inhibition (IC50 = 5 nM).299 Nagasawa and co-workers reported that L2H2-6OTD (Fig. 12), a macrocyclic hexa-oxazole, strongly stabilizes human telomere G4 and shows potent inhibition activity against telomerase.300 Recently, a L2H2-6OTD derivate has also been used as a G4 imaging tool in living cells.301 PDS was designed and synthesized by Balasubramanian in 2008 and has been used in various biological assays.230,302–305 PDS was rationally designed with an expanded planar surface and hydrogen bond sites to efficiently bind to G4 structures. PDS binds and stabilizes telomeric G4 while having no effect on DNA duplexes, indicating high structure-selective binding.302 Because of this property, a modified PDS analogue has also been used in intracellular G4 imaging applications. For example, Balasubramanian and co-workers reported that SiR-pyPDS, a fluorophore-modified PDS,305 exhibits remarkable fluorescence turn-on properties upon binding to G4. PhenDC3, a bisquinolinium family compound, strongly selectively binds to and stabilizes G4s over DNA duplexes.295,306,307 BRACO-19 is an aromatic molecule containing protonatable sites that was designed based on crystal structure and simulation studies to bind to three G4 grooves. BRACO-19 exhibits 31-fold higher affinity for G4s than duplexes.
Another strategy for obtaining structure-selective G4 ligands is the development of small molecules with anionic charges. Cationic ligands bind nucleic acids with non-structure-selectivity due to the negative charges on their backbones. Therefore, it is useful to develop an anionic ligand that causes electrostatic repulsion with the phosphate groups of nucleic acids. Copper(II) phthalocyanine 3,4′,4′′,4′′′-tetrasulfonic acid (CuAPC, Fig. 12), an anionic ligand, has an IC50 of 1.6 μM against telomerase activity in the presence of excess and competitive DNA duplexes.221 Hemin is also a typical negatively charged G4 ligand (Fig. 12).308 Hemin suppresses xlr3b gene expression through its binding to a G4 formed at the CpG island in ATR-X model mice, leading to the recovery of cognitive deficits associated with ATR-X syndrome.309 Although many developed G4 ligands aim to inhibit transcription or telomere elongation, ligands involved in other biological processes have been reported. Tetrandrine binds to G4 selectively and stabilizes it strongly.310 Recently, it has been shown that stabilization of RNA G4 formed by an miR-23b/27b/24-1 cluster by using tetrandrine prevents the binding of Drosha to the miR cluster, resulting in the recovery of cardiac function.311 Thioflavin T (ThT, Fig. 12) is also a selective fluorescent probe for G4, although it has a positive charge.312N-Methyl mesoporphyrin IX (NMM, Fig. 12) is another G4-selective ligand. Bolton et al. found that NMM binds to DNA G4 but not to duplexes by fluorescence spectroscopy.313 NMM enhances fluorescence intensity upon binding to G4s, which is useful for their detection. Later, it was reported that NMM does not bind to other nucleic acid conformations314 and is selective for the parallel topology. The Kd value of NMM for the parallel topology is nearly 10 times less than for the mixed topology and 100 times less than for the antiparallel topology.315
Breaking down target RNA G4s is another therapeutic drug strategy. Our group reported that ZnAPC (Fig. 12) selectively binds to RNA G4 formed by the NRAS mRNA 5′-UTR. ZnAPC breaks down the target RNA G4 via reactive oxygen species upon photoirradiation, resulting in NRAS suppression.316 Xodo and co-workers also reported that an alkylated porphyrin (Fig. 12) bind to KRAS mRNA G4 and cleaves it upon photoirradiation.317 These results suggest that photodynamic therapy targeting mRNA G4s is promising as a new modality for cancer treatment. A remarkable recent progress in this field is ribonuclease targeting chimera (RIBOTAC), which can recruit the endogenous ribonuclease RNase L to a RNA target, resulting in the target RNA cleavage in living cells.318 This strategy has been applied to G4-binding proteins and RNA G4s.319–321 However, selectivity of G4 ligand is the main issue to apply this technology for a practical and clinical use.322
Systematic comparisons of the three-dimensional structures of G4 ligand/G4 complexes are important for improving the affinity and selectivity of G4 ligands. Yang and colleagues reported the solution structures of c-MYC G4 with phenyl–ethenyl–quinoline (PEQ),298 a specific c-MYC G4 binder, and of three other G4 ligands (Fig. 13).323 This c-MYC G4 sequence contains G23-to-T and G14-to-T mutations in the 3′-flanking and second loop, respectively. This sequence was used as a model parallel G4 structure. Yang and colleagues found that the flexible flanking regions are recruited in a conserved and sequence-specific way, and that these regions have further potential for selective ligand–G4 hydrogen-bond interactions. For example, a combination of π–π stacking interactions with a G-quartet core and hydrogen bonds with other regions such as the flanking and loop regions have been observed in other G4 ligands, including PDS and its derivatives.324 Other systematic and detailed examples of G4 ligand/G4 complexes have been reported in the literature,325–327 involving contributions of other binding modes, such as cation–π and hydrophobic interactions. A quantitative study of nonselective interactions of G4 ligands with long DNA duplexes using a single molecule stretching technique was recently published.328 Those results showed that five of eight G4 ligands commonly used in diverse studies bind the DNA duplex with a binding affinity similar to that of ethidium bromide, a typical DNA duplex binder. This structure-selectivity of G4 ligands therefore holds promise for practical applications in living cells, since there are many more DNA and RNA bases involved in duplex structures than in G4 structures.
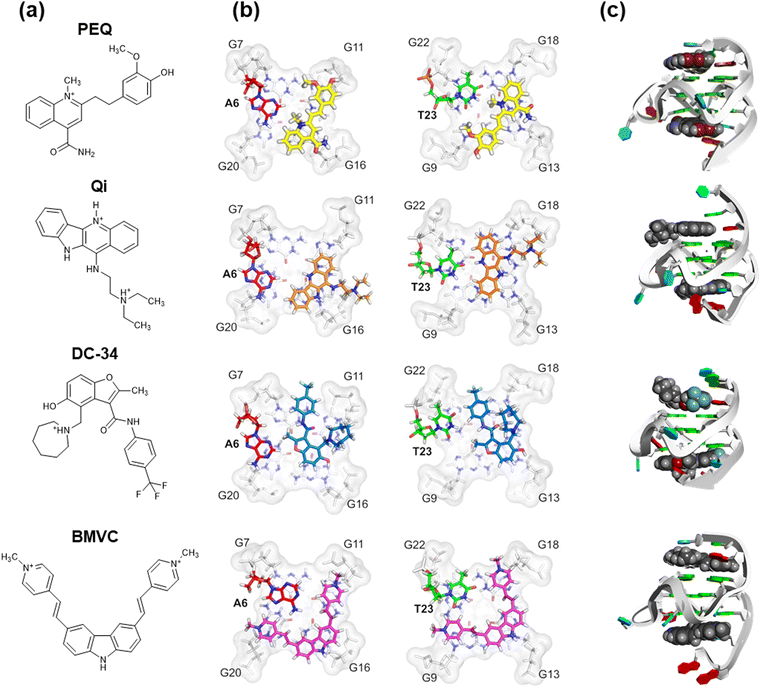 | ||
| Fig. 13 (a) Chemical structures of phenyl–ethenyl–quinoline (PEQ), Quindoline-I (Qi), DC-34, and BMVC. (b) Comparisons of the high-resolution structure of c-MYC G4 with PEQ (PDB ID: 7KCX), Qi (PDB OD: 2L7V), DC-34 (PDB ID: 5W77), and BMVC (PDB ID: 6O2L) binding to the 5′-site and the 3′-site. (c) Three-dimensional structures of the PEQ, Qi, DC-34, and BMVC complexes with c-MYC G4 (PDB ID: 7KBX). Adapted from ref. 298. Copyright © 2021 Oxford University Press. | ||
i-Motif
Structure, sequence, and biological role of the i-motif
The i-motif, another tetraplex, is formed by cytosine-rich (C-rich) sequences via cytosine·cytosine+ base pairs (Fig. 14a). The first DNA i-motif was reported in 1993 for d(TCCCCC) (Fig. 14b and c).329 Since protonation of cytosine is required, this structure is particularly stable in acidic conditions. Although the pKa of free cytidine is 4.2, the pKa is shifted in RNA and DNA.330,331 The value is further shifted to be around 6.5 in the i-motif.332 Moreover, the pKa value increases as the number of cytosines per tract increases from two to seven.333 Loop regions connecting cytosine stretches and capping nucleotides next to the stretches also affect the thermodynamics of i-motifs.334–336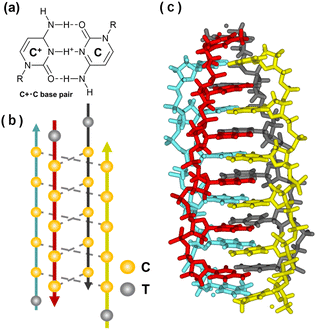 | ||
| Fig. 14 Characteristics of the i-motif structure. (a) Chemical structure of a C:C+ base pair. (b) Illustration of the i-motif structure folded by a C-rich sequence, 5′-TCCCC-3′. (c) Crystal structure of the tetrameric i-motif (PDB ID: 2N89). | ||
The molecular environment also affects the structural stability of i-motifs. In contrast to G4, cation species do not affect i-motifs, but the cation concentration does. It has been reported that a higher cation concentration destabilizes i-motifs.337,338 A lower stability with higher cation concentrations is observed only for the i-motif, but not for other structures of DNA. In contrast to the opposing effects of the cation concentration, molecular crowding stabilizes i-motifs, as with G4s. It has been found that C-rich sequences adopt the i-motif structure at neutral pH under molecular crowding conditions.339,340 Given the stabilization effects of molecular crowding on G4s, an intramolecular structure of an i-motif with a C-rich sequence + a G4 of a complementary G-rich sequence could be more stable than an intermolecular duplex of G- and C-rich sequences, which is generally thermodynamically more stable than the sum of the intramolecular structures (G4 and i-motif) in a physiological buffer.341 In fact, it has been reported that molecular crowding induces a structural transition of telomeric G-rich and C-rich sequences from an intramolecular G4 and i-motif to an intermolecular duplex.215 Another factor that affects thermal stability of i-motifs is negative superhelicity, which unwinds the duplex.342 Various biological processes such as transcription, replication, recombination, and DNA damage repair produce negative supercoiling.343 Thus, during these processes, the formation of intramolecular structures, including i-motifs, could be accelerated. The C-rich sequence of the human telomere can form an i-motif in the S-phase of replicating human cells.344 In addition, the stabilization of a G4 of a G-rich sequence further aids the complementary C-rich sequence to form an i-motif by reducing structural competition with the intermolecular duplex. Thus, it is possible that the i-motif exists in living cells. In fact, evidence for the existence of i-motifs in living cells has been provided by in-cell NMR and an i-motif antibody.345,346
As a complementary sequence to putative quadruplex-forming sequences, putative i-motif-forming sequences (PIS) have been identified in various genes. The PIS in the c-MYC gene folds into several different i-motif structures with the stability being remarkable at pH 7.347 There are more than 60 structures of i-motifs, with sequences including methylated and other modified cytosines, that are registered in the protein data bank (September 2021). As there are many PIS, there are also proteins that bind to i-motifs and PIS oligonucleotides. Poly-C binding protein is a ubiquitous oligonucleotide-binding protein in eukaryotic cells that regulates gene expression.348 This family consists of the hnRNP K (heterogeneous nuclear ribonucleoprotein K) and isoforms of αCP1, including αCP1-4 and αCP-KL. hnRNP K is known as a transcription factor that binds to CT elements in the promoter region of c-MYC.349 αCP1 also recognizes the C-rich sequence of human telomere DNA with high affinity.350 Lacroix and coworkers studied the interaction of hnRNP K and ASF/SF2 with telomeric C-rich sequences under various pH conditions. Their results suggested that the protein binds to the unfolded sequence.351 A later study indicated that hnRNA interacts with single-stranded DNAs.352 In addition, hnRNP K and hnRNP LL, which are transcriptional factors, have been reported to bind to C-rich sequences of VEGF and BCL-2, respectively.353,354 As the protein unfolds the i-motif structure and forms a stable single-stranded DNA–protein complex, the i-motif may play an important role in controlling gene expression. Moreover, i-motifs have been proposed to have various biological functions through their structure and protein binding. These biological roles include the inhibition of telomerase activity, a structural element that provides long-range interactions between laterally associated centromeric nucleosomes, transcriptional regulation, and stalling DNA polymerase and thus impeding DNA replication and repair. These biological roles of i-motifs have received thorough recent reviews.355–357
i-Motif targeting molecules
The first discovered i-motif ligand was TMPyP4, which was also known as a G4 ligand.358,359 Since it is highly cationic, TMPyP4 binds to duplexes in a non-specific manner, and the binding to G4 was overestimated.295 This original study stimulated many researchers to develop i-motif ligands.As with the G4 ligands discussed above, many of i-motif ligands have a planar structure (Fig. 15). Such ligands include derivatives of phenanthroline,360 acridine,361 macrocyclic poly-oxazole,362 and metal complexes.363,364 NMR studies have demonstrated that i-ligands such as L2H2-4OTD and IMC-48 form complexes through stacking interactions with C–C+ base pairs and interactions with loops.362,365 Although the detailed binding modes (in addition to the stacking interactions) were not identified, these findings nonetheless suggest that interactions with the C–C+ base pairs and the loops are critical for stabilizing the i-motif. Phenanthroline derivatives show preferential binding to telomeric i-motifs over duplexes and G4. Acridine derivatives are based on BRACO-19, which is a well-known G4 ligand. This compound has been demonstrated to suppress the proliferation of cancer cells.366 One poly-oxazole derivative has been reported to interact with a telomeric i-motif, but not G4.362 Interestingly, carboxyl-modified single-walled carbon nanotubes (SWNTs) induce and stabilize human telomeric i-motifs. K562 and HeLa cells were treated with SWNTs, then the inhibition of telomere elongation was confirmed using a telomeric repeat amplification protocol assay. Colocalization of SWNTs and a telomere restriction fragment, a telomere marker, suggested that SWNTs were located in the telomere region and stabilized telomeric i-motif DNA, causing folding of telomeric G4 and inhibiting telomere elongation.367,368
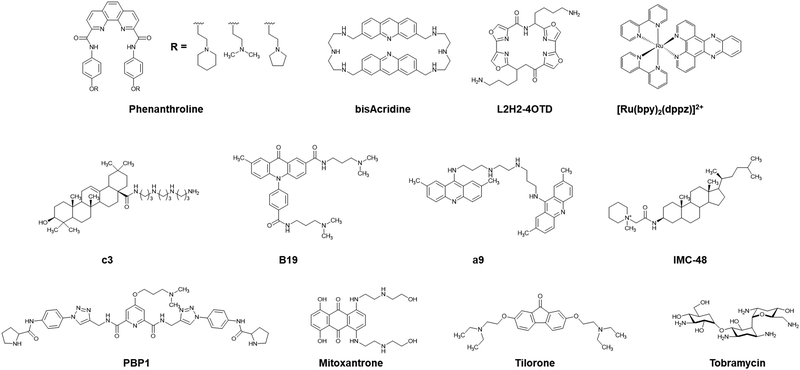 | ||
| Fig. 15 Chemical structures of i-motif-targeting molecules phenanthroline, bisacridine, L2H2-4OTD, [Ru(bpy)2(dppz)]2+, c3, B19, a9, IMC-48, PBP1, mitoxantrone, tilorone, and tobramycin. | ||
It has been shown that small molecules targeting i-motifs regulate the transcription of many genes. B19 and a9 bind to the c-MYC i-motif and downregulate transcription.369,370 Oleanolic acid derivatives interact with the VEGF i-motif and also downregulate transcription.371,372 In contrast, IMC48 and PBP1 bind to the BCL2 i-motif and upregulate transcription.370,373 Although these results suggest that i-motifs are important as cis-acting gene regulators, it is still unclear how genes are down- and up-regulated by i-motif formation and ligand binding. The fluorescent intercalator displacement assay is a powerful method to screen ligands that target i-motifs as well as other structures of nucleic acids. Waller and co-workers have discovered i-motif ligands, such as the antitumor agent mitoxantrone and antiviral agents tilorone and tobramycin, by fluorescent intercalator displacement assays utilizing thiazole orange.374,375 However, thiazole orange emits fluorescence not only with i-motifs but also other structures, including G4s and duplexes. Thus, an i-motif fluorescent probe will be useful to develop a more simplified and high-throughput screening system for i-motif-specific ligands.
Targeting three- and four-stranded nucleic acids molecules toward clinical trials
Nucleic acid-targeting small molecule as approved drug
It is essential to consider clinical applications even at the earliest stages of drug development. For example, risdiplam (Evrysdi®) is an approved small molecule therapeutic that targets nucleic acids to treat spinal muscular atrophy, and it is the first small molecule to be identified as a splicing modifier.376 Risdiplam was identified by phenotypic screening rather than by structure-based screening.377 Screening was performed using a human embryonic kidney cell line harboring an SMN2 minigene to increase the inclusion of exon 7 during SMN2 pre-mRNA splicing. Various functional groups were introduced to enhance oral availability.378 For example, the insertion of a pyridopyrimidinone moiety reduced the phototoxicity of the compound and a cyclic N-alkyl group prevented nonspecific protein binding. These design steps and modifications allow risdiplam to strongly promote SMN protein expression with low cytotoxicity.379–381 The biological activity of the compounds was confirmed using animal models of SMA. Pharmacokinetic studies showed that the compounds penetrated the brains of adult mice. Finally, the efficiency and safety of risdiplam was confirmed in a clinical development program with SMA patients.G4 ligands for clinical applications
Similar problems need to be solved by chemical modifications for small molecules targeting non-canonical structures of nucleic acids. After screening or structure-based rational design the lead compounds must be optimized, as shown by the example of G4 ligands being developed for clinical use. BRACO-19 (Fig. 12), developed by Neidle and coworkers, has been modified, with one derivative exhibiting high selectivity against duplexes and strong inhibition of telomerase activity.366 This compound persistently arrested the growth of a breast carcinoma cell line366 and is the first G4 ligand showing an anticancer effect and proven anticancer activity in human tumor xenograft models. Neidle and colleagues also developed and optimized compounds with a naphthalene diimide (NDI) core using structure-based design, medicinal chemistry and pharmacology. QN-302, an NDI derivative with a benzyl-pyrrolidine group, significantly inhibits the growth of pancreatic ductal adenocarcinoma cells in animal models, with GI50 values of 1–2 nM and superior potency compared to derivatives with a methoxy group replacing the benzyl-pyrrolidine.382 QN-302 binds tightly to various parallel G4s and is highly specific for G4 compared to duplexes.382 QN-302 began clinical trials in 2023.291,378The detailed binding mode of QN-302 with G4 has been studied.383–385 Molecular modelling studies suggest that QN-302 binds at the duplex-G4 junction through the NDI core via stacking interactions with the G4 terminal G-quartet. The benzyl-pyrrolidine substituent of QN-302 protrudes significantly into the groove. The phenyl ring stacks efficiently onto the adjacent guanine of the lower G-quartet. The benzyl-pyrrolidine group allows enhanced G4 binding affinity and selectivity, enhanced cellular and in vivo potency, and greater selectivity in the pattern of downregulated gene expression. Furthermore, the pharmacokinetic properties of QN-302, such as half-life in vivo and bioavailability, are not apparently responsible for its superior activity in vivo. These results further demonstrate the importance of the binding affinity and selectivity of G4 ligands for clinical applications.
G4 ligands with higher selectivity
As the above examples show, the selectivity of the compound for its target is of utmost importance. High selectivity is critical for improving drug efficacy and reducing side effects, as it allows the highly efficient inhibition of G4-targeted enzymes and prevents off-target enzyme inhibition. High selectivity against the duplex is also important to reduce the genotoxicity of G4 ligands and small compounds targeting other three- and four-stranded nucleic acid structures. High selectivity for G4 ligands requires the involvement of directional interactions in addition to stacking interactions. The energy of electrostatic interactions is highest within non-covalent interactions, but it depends only on the distance between the charges, making it difficult to confer selectivity. This can be particularly detrimental to discriminating between G4 and canonical duplex structures. The energy of hydrogen bonding varies with direction, so many G4 ligands have been modified to introduce hydrogen-bonding donors and acceptors to allow hydrogen bonds with the target G4, especially with the loops, grooves, and flanking regions (Fig. 7). The importance of these three regions for specificity is discussed in this subsection.![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 844 possible sequences with three loops in intramolecular G4s with 1–7 nucleotides of which 20
844 possible sequences with three loops in intramolecular G4s with 1–7 nucleotides of which 20![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 492 are found at least once in the human genome.139 The loop region was recently reported to significantly impact the topology and thermal stability of G4, and the kinetics of G4 formation.123,386,387 This suggests that the binding of G4 ligands to the loop regions can significantly alter the overall G4 structure and thus modulate protein binding and enzymatic activity. In addition, a G4 structure can adapt other nucleic acid structural motifs, such as a hairpin loop. A duplex can be attached to a G4.167,168 The duplex formed within the loops stabilizes the overall G4 structure compared to an unstructured loop and accelerates G4 folding kinetics.169,175–177
492 are found at least once in the human genome.139 The loop region was recently reported to significantly impact the topology and thermal stability of G4, and the kinetics of G4 formation.123,386,387 This suggests that the binding of G4 ligands to the loop regions can significantly alter the overall G4 structure and thus modulate protein binding and enzymatic activity. In addition, a G4 structure can adapt other nucleic acid structural motifs, such as a hairpin loop. A duplex can be attached to a G4.167,168 The duplex formed within the loops stabilizes the overall G4 structure compared to an unstructured loop and accelerates G4 folding kinetics.169,175–177
Flanking: The 5′ and 3′ flanking regions adjacent to the G4 core region are also targets for hydrogen bonding interactions. The stacking and hydrogen bonding interactions formed between the flanking nucleotide and another flanking nucleotide, the adjacent G-quartet, and the neighboring loop, all contribute to the overall structure and thermodynamics of G4. The flanking nucleotides reportedly influence the binding affinity of the G4 ligand. As discussed above, the importance of detailed structural information on the loop, groove and flanking regions will increase as the design of G4 ligands progresses.
The three-dimensional structure of the loop regions, and the depth and width of the grooves, depend on the nucleotide sequence. The nucleotide sequences for the 5′ and 3′-flanking regions determine the target. These differences are important not only for structural selectivity against the canonical duplex, but also for sequence selectivity towards the many G4s present in cells, although only a few examples of G4 ligands have been reported to show sequence selectivity. Selectivity for DNA G4 and RNA G4 may also be important for precise targeting. More three-dimensional G4 structures are needed to predict the overall topology of G4 from the sequence and also the fine structure of the loops, grooves, and flanking regions.
Small molecules that highly selectively target triplex and i-motifs have been reported, as shown for the triplex of MALAT1 discussed above; however, only a few studies show the effects of these molecules on cellular and organoid functions.120,390 As mentioned above, even small molecules with high selectivity require toxicity testing, improved oral dosing, animal models, and confirmation of compound efficacy and pharmacokinetics. More systematic and quantitative studies of the relationship between structure, selectivity, and function in vitro and in vivo are required.
Conclusion and perspective
DNA and RNA sequences encode not only genetic information but also higher order information, such as three-dimensional structures and stabilizing factors, and they respond to environmental factors, all of which are critical for gene regulation. Moreover, it has been found that DNA and RNA undergo LLPS in living cells to regulate the central dogma at many steps involving the replication of genomic DNA and viral RNA; the transcription of mRNA, rRNA, and non-coding RNA; the processing and localization of RNAs; and translation from RNA to protein.391–395 One fundamental property of LLPS is responsiveness to cellular stresses, such as high temperatures, nutrient starvation, toxins, and reactive oxygen species. The structure and stability of nucleic acids depend on the molecular environment as we described in this review. Interestingly, recent studies suggest that LLPS of nucleic acids is dependent of their secondary structures.239,285 Thus, DNA and RNA sequences may further encode phase behavior. Despite our knowledge of the biology, chemistry, and physics of non-canonical nucleic acid structures, many aspects are still opening for further investigation. In this regard, researches on the non-canonical structures of DNA and RNA utilizing small ligands in vitro and in vivo are essential for elucidating their roles in different biological processes. In spite of recent findings of ligands that target non-canonical structures, the number of structure-specific ligands is very limited. Moreover, non-canonical structural ligands that can target a specific sequence are further needed. To develop structure and sequence-selective ligands, at least the following are required.(1) Three-dimensional structures of nucleic acid-ligand complexes: Structural information is essential for the rational design of ligands. Even though biochemical and biophysical information for G4 ligand complexes is relatively abundant, there are only a limited number of RNA and DNA G4 ligand complexes with structures that have been solved. Moreover, the structure of an i-motif and ligand complex has not been reported yet.
(2) Dynamic properties of non-canonical structures: Non-canonical structures are not static. Their formation and thermal stability largely depend on molecular environmental factors and protein binding. In the case of DNA, non-canonical structures always compete against the canonical duplex. In the case of RNA, structural switches among the most stable and metastable structures may be important for their biological roles as in the case of riboswitches. Thus, the dynamic and kinetic properties of non-canonical structures should be studied under cell-mimicking conditions and in living cells.
(3) High-throughput screening systems: since not only high affinity but also high selectivity of ligands are required for cellular applications, screening systems that can evaluate biorthogonality among canonical and non-canonical structures are needed. For example, many i-motif and G4 ligands also bind G4s and i-motifs, respectively. Moreover, it is difficult to eliminate non-specific binding of ligands to the duplex. In addition to structure-specific ligands, sequence-specific ligands are required, because as shown for triplexes and quadruplexes, there are many putative sequences that form certain non-canonical structures.
A wide variety of G4 ligand screening systems, such as fluorescent intercalator displacement (FID), are used to identify ligands,396–398 but these methods are low-throughput. Obtaining structure- or sequence-selective ligands using such methods requires multiple steps. Our group established new G4 ligand screening systems that are optimized to provide structure- or sequence-selective G4 ligands,399,400 although a molecular probe exhibiting target selectivity is required. Furthermore, cell-based FID assays can provide ligands that bind intracellular G4s efficiently.401 These reports demonstrate that G4 ligands exhibiting structure selectivity can efficiently target G4s within cells. Further structural information on target-selective ligands can enrich target-selective ligand databases such as G4 LDB290 and NALDB.402 A systematic quantitative structure–activity relationship (QSAR) workflow in 2022 was reported to broadly discover RNA ligands using HIV-1 transactivation response RNA as a model system.403 This method can evaluate binding parameters, including affinity and binding kinetics, bypassing the need for structural information. Therefore, the discovery of target-selective ligands not only aids understanding of the scaffolds, but also the rational design of ligands for target-selective binding.
(4) Widely useable databases: bioinformatic and omics studies have provided a massive amount of data sets to researchers, but it is almost impossible to make effective use of them by examining individual studies. Thus, to unify and utilize a large amount of data, databases and machine learning systems for wide and open use are desired.
In conclusion, structure- and sequence-selective ligands could be the key to decoding hidden information in nucleic acids and to develop small molecular drugs targeting nucleic acids. Nucleic acids -DNA, RNA, as well as artificial ones- are now coming of age as drug targets and targeting biomolecules.
Data availability
No primary research results, software or code have been included, and no new data were generated or analyzed as part of this review.Conflicts of interest
There are no conflicts to declare.Acknowledgements
This work was supported by JSPS KAKENHI, grant numbers 24K21801, 21H02062, and 20K21259, a Research Grant of the Asahi Glass Foundation, Japan, the Hirao Taro Foundation and the Tateno Junzo Foundation of Konan Gakuen for Academic Research, Japan, and JST SPRING, grant number JPMJSP2117.Notes and references
- A. L. Hopkins and C. R. Groom, Nat. Rev. Drug Discovery, 2002, 1, 727–730 CrossRef PubMed.
- J. P. Overington, B. Al-Lazikani and A. L. Hopkins, Nat. Rev. Drug Discovery, 2006, 5, 993–996 CrossRef PubMed.
- S. J. Dixon and B. R. Stockwell, Curr. Opin. Chem. Biol., 2009, 13, 549–555 CrossRef CAS PubMed.
- A. D. Cox, S. W. Fesik, A. C. Kimmelman, J. Luo and C. J. Der, Nat. Rev. Drug Discovery, 2014, 13, 828–851 CrossRef CAS PubMed.
- C. V. Dang, Cell, 2012, 149, 22–35 CrossRef CAS PubMed.
- L. Mabonga and A. P. Kappo, Biophys. Rev., 2019, 11, 559–581 CrossRef CAS PubMed.
- A. Sawyer, Biotechniques, 2020, 69, 239–241 CrossRef CAS PubMed.
- R. Kahan, D. J. Worm, G. V. Castro, S. Ng and A. Barnard, RSC Chem. Biol., 2021, 2, 387–409 RSC.
- K. S. McPherson and D. M. Korzhnev, RSC Chem. Biol., 2021, 2, 1167–1195 RSC.
- M. Clamp, B. Fry, M. Kamal, X. Xie, J. Cuff, M. F. Lin, M. Kellis, K. Lindblad-Toh and E. S. Lander, Proc. Natl. Acad. Sci. U. S. A., 2007, 104, 19428–19433 CrossRef CAS PubMed.
- I. Ezkurdia, D. Juan, J. M. Rodriguez, A. Frankish, M. Diekhans, J. Harrow, J. Vazquez, A. Valencia and M. L. Tress, Hum. Mol. Genet., 2014, 23, 5866–5878 CrossRef CAS PubMed.
- M. N. Cabili, C. Trapnell, L. Goff, M. Koziol, B. Tazon-Vega, A. Regev and J. L. Rinn, Genes Dev., 2011, 25, 1915–1927 CrossRef CAS PubMed.
- J. Zell, S. F. Rota, S. Britton and D. Monchaud, RSC Chem. Biol., 2020, 2, 47–76 RSC.
- G. D. Stormo, Quant. Biol., 2013, 1, 115–130 CrossRef CAS PubMed.
- W. W. Wasserman and A. Sandelin, Nat. Rev. Genet., 2004, 5, 276–287 CrossRef CAS PubMed.
- E. L. Van Nostrand, P. Freese, G. A. Pratt, X. Wang, X. Wei, R. Xiao, S. M. Blue, J. Y. Chen, N. A. L. Cody, D. Dominguez, S. Olson, B. Sundararaman, L. Zhan, C. Bazile, L. P. B. Bouvrette, J. Bergalet, M. O. Duff, K. E. Garcia, C. Gelboin-Burkhart, M. Hochman, N. J. Lambert, H. Li, M. P. McGurk, T. B. Nguyen, T. Palden, I. Rabano, S. Sathe, R. Stanton, A. Su, R. Wang, B. A. Yee, B. Zhou, A. L. Louie, S. Aigner, X. D. Fu, E. Lécuyer, C. B. Burge, B. R. Graveley and G. W. Yeo, Nature, 2020, 583, 711–719 CrossRef CAS PubMed.
- H. Tateishi-Karimata and N. Sugimoto, Chem. Commun., 2020, 56, 2379–2390 RSC.
- H. Tateishi-Karimata and N. Sugimoto, Nucleic Acids Res., 2021, 49, 7839–7855 CrossRef CAS PubMed.
- J. P. Falese, A. Donlic and A. E. Hargrove, Chem. Soc. Rev., 2021, 50, 2224–2243 RSC.
- C. Sheridan, Nat. Biotechnol., 2021, 39, 6–8 CrossRef CAS PubMed.
- C. A. Orengo, D. T. Jones and J. M. Thornton, Nature, 1994, 372, 631–634 CrossRef CAS PubMed.
- S. Govindarajan, R. Recabarren and R. A. Goldstein, Proteins, 1999, 35, 408–414 CrossRef CAS.
- R. D. Schaeffer and V. Daggett, Protein Eng., Des. Sel., 2011, 24, 11–19 CrossRef CAS PubMed.
- G. Felsenfeld, D. R. Davies and A. Rich, J. Am. Chem. Soc., 1957, 79, 2023–2024 CrossRef CAS.
- A. Jain, G. Wang and K. M. Vasquez, Biochimie, 2008, 90, 1117–1130 CrossRef CAS PubMed.
- P. A. Beal and P. B. Dervan, Science, 1991, 251, 1360–1363 CrossRef CAS PubMed.
- A. G. Letai, M. A. Palladino, E. Fromm, V. Rizzo and J. R. Fresco, Biochemistry, 1988, 27, 9108–9112 CrossRef CAS PubMed.
- D. M. Gowers and K. R. Fox, Nucleic Acids Res., 1997, 25, 3787–3794 CrossRef CAS PubMed.
- D. S. Pilch, C. Levenson and R. H. Shafer, Biochemistry, 1991, 30, 6081–6088 CrossRef CAS PubMed.
- K. R. Fox, Curr. Med. Chem., 2000, 7, 17–37 CrossRef CAS PubMed.
- F. A. Buske, D. C. Bauer, J. S. Mattick and T. L. Bailey, Genome Res., 2012, 22, 1372–1381 CrossRef CAS PubMed.
- S. S. Gaddis, Q. Wu, H. D. Thames, J. DiGiovanni, E. F. Walborg, M. C. MacLeod and K. M. Vasquez, Oligonucleotides, 2006, 16, 196–201 CrossRef CAS PubMed.
- Y. M. Agazie, J. S. Lee and G. D. Burkholder, J. Biol. Chem., 1994, 269, 7019–7023 CrossRef CAS PubMed.
- Y. M. Agazie, G. D. Burkholder and J. S. Lee, Biochem. J., 1996, 316(Pt 2), 461–466 CrossRef CAS PubMed.
- J. A. Brown, Wiley Interdiscip. Rev.: RNA, 2020, 11, e1598 CrossRef CAS PubMed.
- D. Sacks, B. Baxter, B. C. V. Campbell, J. S. Carpenter, C. Cognard, D. Dippel, M. Eesa, U. Fischer, K. Hausegger, J. A. Hirsch, M. Shazam Hussain, O. Jansen, M. V. Jayaraman, A. A. Khalessi, B. W. Kluck, S. Lavine, P. M. Meyers, S. Ramee, D. A. Rüfenacht, C. M. Schirmer and D. Vorwerk, Int. J. Stroke, 2018, 13, 612–632 Search PubMed.
- C. A. Theimer, C. A. Blois and J. Feigon, Mol. Cell, 2005, 17, 671–682 CrossRef CAS PubMed.
- K. Shefer, Y. Brown, V. Gorkovoy, T. Nussbaum, N. B. Ulyanov and Y. Tzfati, Mol. Cell. Biol., 2007, 27, 2130–2143 CrossRef CAS PubMed.
- S. D. Gilbert, R. P. Rambo, D. Van Tyne and R. T. Batey, Nat. Struct. Mol. Biol., 2008, 15, 177–182 CrossRef CAS PubMed.
- K. T. Tycowski, M. D. Shu, S. Borah, M. Shi and J. A. Steitz, Cell Rep., 2012, 2, 26–32 CrossRef CAS PubMed.
- C. Cianciolo Cosentino, N. I. Skrypnyk, L. L. Brilli, T. Chiba, T. Novitskaya, C. Woods, J. West, V. N. Korotchenko, L. McDermott, B. W. Day, A. J. Davidson, R. C. Harris, M. P. de Caestecker and N. A. Hukriede, J. Am. Soc. Nephrol., 2013, 24, 943–953 CrossRef PubMed.
- N. K. Conrad, Virus Res., 2016, 212, 53–63 CrossRef CAS PubMed.
- J. N. Hutchinson, A. W. Ensminger, C. M. Clemson, C. R. Lynch, J. B. Lawrence and A. Chess, BMC Genomics, 2007, 8, 39 CrossRef PubMed.
- R. Miyagawa, K. Tano, R. Mizuno, Y. Nakamura, K. Ijiri, R. Rakwal, J. Shibato, Y. Masuo, A. Mayeda, T. Hirose and N. Akimitsu, RNA, 2012, 18, 738–751 CrossRef CAS PubMed.
- T. Gutschner, M. Hämmerle and S. Diederichs, J. Mol. Med., 2013, 91, 791–801 CrossRef CAS PubMed.
- F. A. Abulwerdi, W. Xu, A. A. Ageeli, M. J. Yonkunas, G. Arun, H. Nam, J. S. Schneekloth, Jr., T. K. Dayie, D. Spector, N. Baird and S. F. J. Le Grice, ACS Chem. Biol., 2019, 14, 223–235 CrossRef CAS PubMed.
- J. A. Brown, M. L. Valenstein, T. A. Yario, K. T. Tycowski and J. A. Steitz, Proc. Natl. Acad. Sci. U. S. A., 2012, 109, 19202–19207 CrossRef CAS PubMed.
- J. E. Wilusz, C. K. JnBaptiste, L. Y. Lu, C. D. Kuhn, L. Joshua-Tor and P. A. Sharp, Genes Dev., 2012, 26, 2392–2407 CrossRef CAS PubMed.
- K. C. Wang and H. Y. Chang, Mol. Cell, 2011, 43, 904–914 CrossRef CAS PubMed.
- T. Li, X. Mo, L. Fu, B. Xiao and J. Guo, Oncotarget, 2016, 7, 8601–8612 CrossRef PubMed.
- P. Grote, L. Wittler, D. Hendrix, F. Koch, S. Währisch, A. Beisaw, K. Macura, G. Bläss, M. Kellis, M. Werber and B. G. Herrmann, Dev. Cell, 2013, 24, 206–214 CrossRef CAS PubMed.
- T. Mondal, S. Subhash, R. Vaid, S. Enroth, S. Uday, B. Reinius, S. Mitra, A. Mohammed, A. R. James, E. Hoberg, A. Moustakas, U. Gyllensten, S. J. Jones, C. M. Gustafsson, A. H. Sims, F. Westerlund, E. Gorab and C. Kanduri, Nat. Commun., 2015, 6, 7743 CrossRef CAS PubMed.
- A. Postepska-Igielska, A. Giwojna, L. Gasri-Plotnitsky, N. Schmitt, A. Dold, D. Ginsberg and I. Grummt, Mol. Cell, 2015, 60, 626–636 CrossRef CAS PubMed.
- V. B. O'Leary, S. V. Ovsepian, L. G. Carrascosa, F. A. Buske, V. Radulovic, M. Niyazi, S. Moertl, M. Trau, M. J. Atkinson and N. Anastasov, Cell Rep., 2015, 11, 474–485 CrossRef PubMed.
- V. B. O'Leary, J. Smida, F. A. Buske, L. G. Carrascosa, O. Azimzadeh, D. Maugg, S. Hain, S. Tapio, W. Heidenreich, J. Kerr, M. Trau, S. V. Ovsepian and M. J. Atkinson, Sci. Rep., 2017, 7, 7163 CrossRef PubMed.
- M. Kalwa, S. Hänzelmann, S. Otto, C. C. Kuo, J. Franzen, S. Joussen, E. Fernandez-Rebollo, B. Rath, C. Koch, A. Hofmann, S. H. Lee, A. E. Teschendorff, B. Denecke, Q. Lin, M. Widschwendter, E. Weinhold, I. G. Costa and W. Wagner, Nucleic Acids Res., 2016, 44, 10631–10643 CrossRef CAS PubMed.
- C. C. Kuo, S. Hänzelmann, N. Sentürk Cetin, S. Frank, B. Zajzon, J. P. Derks, V. S. Akhade, G. Ahuja, C. Kanduri, I. Grummt, L. Kurian and I. G. Costa, Nucleic Acids Res., 2019, 47, e32 CrossRef PubMed.
- I. Antonov and Y. A. Medvedeva, F1000Res, 2018, 7, 76 Search PubMed.
- A. Bacolla, G. Wang and K. M. Vasquez, PLoS Genet., 2015, 11, e1005696 CrossRef PubMed.
- Y. Li, J. Syed and H. Sugiyama, Cell Chem. Biol., 2016, 23, 1325–1333 CrossRef CAS PubMed.
- E. Käs, E. Izaurralde and U. K. Laemmli, J. Mol. Biol., 1989, 210, 587–599 CrossRef PubMed.
- E. Winter and A. Varshavsky, EMBO J., 1989, 8, 1867–1877 CrossRef CAS PubMed.
- Y. Adachi, E. Käs and U. K. Laemmli, EMBO J., 1989, 8, 3997–4006 CrossRef CAS PubMed.
- R. Kiyama and R. D. Camerini-Otero, Proc. Natl. Acad. Sci. U. S. A., 1991, 88, 10450–10454 CrossRef CAS PubMed.
- M. Musso, L. D. Nelson and M. W. Van Dyke, Biochemistry, 1998, 37, 3086–3095 CrossRef CAS PubMed.
- A. L. Guieysse, D. Praseuth and C. Hélène, J. Mol. Biol., 1997, 267, 289–298 CrossRef CAS PubMed.
- E. Jiménez-García, A. Vaquero, M. L. Espinás, R. Soliva, M. Orozco, J. Bernués and F. Azorín, J. Biol. Chem., 1998, 273, 24640–24648 CrossRef PubMed.
- L. D. Nelson, M. Musso and M. W. Van Dyke, J. Biol. Chem., 2000, 275, 5573–5581 CrossRef CAS PubMed.
- M. Musso, G. Bianchi-Scarrà and M. W. Van Dyke, Nucleic Acids Res., 2000, 28, 4090–4096 CrossRef CAS PubMed.
- G. Li, G. V. Tolstonog and P. Traub, DNA Cell Biol, 2002, 21, 163–188 CrossRef CAS PubMed.
- P. Ciotti, M. W. Van Dyke, G. Bianchi-Scarrà and M. Musso, Eur. J. Biochem., 2001, 268, 225–234 CrossRef CAS PubMed.
- J. Kusic, B. Tomic, A. Divac and S. Kojic, Mol. Biol. Rep., 2010, 37, 2317–2322 CrossRef CAS PubMed.
- F. Guillonneau, A. L. Guieysse, J. P. Le Caer, J. Rossier and D. Praseuth, Nucleic Acids Res., 2001, 29, 2427–2436 CrossRef CAS PubMed.
- J. E. Rao and N. L. Craig, J. Mol. Biol., 2001, 307, 1161–1170 CrossRef CAS PubMed.
- J. E. Rao, P. S. Miller and N. L. Craig, Proc. Natl. Acad. Sci. U. S. A., 2000, 97, 3936–3941 CrossRef CAS PubMed.
- B. S. Thoma, M. Wakasugi, J. Christensen, M. C. Reddy and K. M. Vasquez, Nucleic Acids Res., 2005, 33, 2993–3001 CrossRef CAS PubMed.
- S. S. Lange, M. C. Reddy and K. M. Vasquez, DNA Repair, 2009, 8, 865–872 CrossRef CAS PubMed.
- L. D. Nelson, C. Bender, H. Mannsperger, D. Buergy, P. Kambakamba, G. Mudduluru, U. Korf, D. Hughes, M. W. Van Dyke and H. Allgayer, Mol. Cancer, 2012, 11, 38 CrossRef CAS PubMed.
- R. M. Brosh, Jr., A. Majumdar, S. Desai, I. D. Hickson, V. A. Bohr and M. M. Seidman, J. Biol. Chem., 2001, 276, 3024–3030 CrossRef PubMed.
- J. A. Sommers, N. Rawtani, R. Gupta, D. V. Bugreev, A. V. Mazin, S. B. Cantor and R. M. Brosh, Jr., J. Biol. Chem., 2009, 284, 7505–7517 CrossRef CAS PubMed.
- A. Jain, A. Bacolla, P. Chakraborty, F. Grosse and K. M. Vasquez, Biochemistry, 2010, 49, 6992–6999 CrossRef CAS PubMed.
- V. Kopel, A. Pozner, N. Baran and H. Manor, Nucleic Acids Res., 1996, 24, 330–335 CrossRef CAS PubMed.
- M. Brázdová, V. Tichý, R. Helma, P. Bažantová, A. Polášková, A. Krejčí, M. Petr, L. Navrátilová, O. Tichá, K. Nejedlý, M. L. Bennink, V. Subramaniam, Z. Bábková, T. Martínek, M. Lexa and M. Adámik, PLoS One, 2016, 11, e0167439 CrossRef PubMed.
- C. Pasquier, S. Agnel and A. Robichon, G3, 2017, 7, 2295–2304 CrossRef CAS PubMed.
- G. E. Plum, Y. W. Park, S. F. Singleton, P. B. Dervan and K. J. Breslauer, Proc. Natl. Acad. Sci. U. S. A., 1990, 87, 9436–9440 CrossRef CAS PubMed.
- J. S. Lee, D. A. Johnson and A. R. Morgan, Nucleic Acids Res., 1979, 6, 3073–3091 CrossRef CAS PubMed.
- S. Nakano, D. Miyoshi and N. Sugimoto, Chem. Rev., 2014, 114, 2733–2758 CrossRef CAS PubMed.
- J. S. Sun, J. C. François, T. Montenay-Garestier, T. Saison-Behmoaras, V. Roig, N. T. Thuong and C. Hélène, Proc. Natl. Acad. Sci. U. S. A., 1989, 86, 9198–9202 CrossRef CAS PubMed.
- L. Perrouault, U. Asseline, C. Rivalle, N. T. Thuong, E. Bisagni, C. Giovannangeli, T. Le Doan and C. Hélène, Nature, 1990, 344, 358–360 CrossRef CAS PubMed.
- M. Takasugi, A. Guendouz, M. Chassignol, J. L. Decout, J. Lhomme, N. T. Thuong and C. Hélène, Proc. Natl. Acad. Sci. U. S. A., 1991, 88, 5602–5606 CrossRef CAS PubMed.
- P. V. Scaria and R. H. Shafer, J. Biol. Chem., 1991, 266, 5417–5423 CrossRef CAS PubMed.
- J. L. Mergny, D. Collier, M. Rougée, T. Montenay-Garestier and C. Hélène, Nucleic Acids Res., 1991, 19, 1521–1526 CrossRef CAS PubMed.
- J. L. Mergny, G. Duval-Valentin, C. H. Nguyen, L. Perrouault, B. Faucon, M. Rougée, T. Montenay-Garestier, E. Bisagni and C. Hélène, Science, 1992, 256, 1681–1684 CrossRef CAS PubMed.
- D. S. Pilch, M. T. Martin, C. H. Nguyen, J. S. Sun, E. Bisagni, T. Garestier and C. Helene, J. Am. Chem. Soc., 1993, 115, 9941–9951 Search PubMed.
- C. Escude, C. H. Nguyen, J. L. Mergny, J. S. Sun, E. Bisagni, T. Garestier and C. Helene, J. Am. Chem. Soc., 1995, 117, 10212–10219 CrossRef CAS.
- T. Biver, A. Boggioni, B. García, J. M. Leal, R. Ruiz, F. Secco and M. Venturini, Nucleic Acids Res., 2010, 38, 1697–1710 CrossRef CAS PubMed.
- C. Escudé, C. H. Nguyen, S. Kukreti, Y. Janin, J. S. Sun, E. Bisagni, T. Garestier and C. Hélène, Proc. Natl. Acad. Sci. U. S. A., 1998, 95, 3591–3596 CrossRef PubMed.
- C. H. Nguyen, E. Fan, J. F. Riou, M. C. Bissery, P. Vrignaud, F. Lavelle and E. Bisagni, Anticancer Drug Des., 1995, 10, 277–297 CAS.
- C. H. Nguyen, C. Marchand, S. Delage, D. Sun, T. Garestier, C. Hélène and E. Bisagni, J. Am. Chem. Soc., 1998, 120, 2501–2507 CrossRef CAS.
- W. D. Wilson, F. A. Tanious, S. Mizan, S. Yao, A. S. Kiselyov, G. Zon and L. Strekowski, Biochemistry, 1993, 32, 10614–10621 CrossRef CAS PubMed.
- S. Bhuiya, L. Haque, R. Goswami and S. Das, J. Phys. Chem. B, 2017, 121, 11037–11052 CrossRef CAS PubMed.
- R. Tiwari, L. Haque, S. Bhuiya and S. Das, Int. J. Biol. Macromol., 2017, 103, 692–700 CrossRef CAS PubMed.
- R. Sinha and G. S. Kumar, J. Phys. Chem. B, 2009, 113, 13410–13420 CrossRef CAS PubMed.
- S. Das, G. S. Kumar, A. Ray and M. Maiti, J. Biomol. Struct. Dyn., 2003, 20, 703–714 CrossRef CAS PubMed.
- X. J. He and L. F. Tan, Inorg. Chem., 2014, 53, 11152–11159 CrossRef CAS PubMed.
- Y. W. Park and K. J. Breslauer, Proc. Natl. Acad. Sci. U. S. A., 1992, 89, 6653–6657 CrossRef CAS PubMed.
- M. Durand, N. T. Thuong and J. C. Maurizot, J. Biol. Chem., 1992, 267, 24394–24399 CrossRef CAS PubMed.
- K. Umemoto, M. H. Sarma, G. Gupta, J. Luo and R. H. Sarma, J. Am. Chem. Soc., 1990, 112, 4539–4545 CrossRef CAS.
- D. S. Pilch and K. J. Breslauer, Proc. Natl. Acad. Sci. U. S. A., 1994, 91, 9332–9336 CrossRef CAS PubMed.
- R. Wadhwa, S. C. Kaul, Y. Komatsu, Y. Ikawa, A. Sarai and Y. Sugimoto, FEBS Lett., 1993, 315, 193–196 CrossRef CAS PubMed.
- N. Vigneswaran, C. A. Mayfield, B. Rodu, R. James, H. G. Kim and D. M. Miller, Biochemistry, 1996, 35, 1106–1114 CrossRef CAS PubMed.
- A. K. Jain and S. Bhattacharya, Bioconjugate Chem., 2010, 21, 1389–1403 CrossRef CAS PubMed.
- D. P. Arya, Acc. Chem. Res., 2011, 44, 134–146 CrossRef CAS PubMed.
- D. P. Arya and R. L. Coffee, Jr., Bioorg. Med. Chem. Lett., 2000, 10, 1897–1899 CrossRef CAS PubMed.
- D. P. Arya, R. L. Coffee, Jr., B. Willis and A. I. Abramovitch, J. Am. Chem. Soc., 2001, 123, 5385–5395 CrossRef CAS PubMed.
- L. Xue, H. Xi, S. Kumar, D. Gray, E. Davis, P. Hamilton, M. Skriba and D. P. Arya, Biochemistry, 2010, 49, 5540–5552 CrossRef CAS PubMed.
- P. Ji, S. Diederichs, W. Wang, S. Böing, R. Metzger, P. M. Schneider, N. Tidow, B. Brandt, H. Buerger, E. Bulk, M. Thomas, W. E. Berdel, H. Serve and C. Müller-Tidow, Oncogene, 2003, 22, 8031–8041 CrossRef PubMed.
- T. Gutschner, M. Hämmerle, M. Eissmann, J. Hsu, Y. Kim, G. Hung, A. Revenko, G. Arun, M. Stentrup, M. Gross, M. Zörnig, A. R. MacLeod, D. L. Spector and S. Diederichs, Cancer Res., 2013, 73, 1180–1189 CrossRef CAS PubMed.
- A. Donlic, M. Zafferani, G. Padroni, M. Puri and A. E. Hargrove, Nucleic Acids Res., 2020, 48, 7653–7664 CrossRef CAS PubMed.
- F. A. Abulwerdi, W. Xu, A. A. Ageeli, M. J. Yonkunas, G. Arun, H. Nam, J. S. Schneekloth, Jr., T. K. Dayie, D. Spector, N. Baird and S. F. J. Le Grice, ACS Chem. Biol., 2019, 14, 223–235 CrossRef CAS PubMed.
- M. Gellert, M. N. Lipsett and D. R. Davies, Proc. Natl. Acad. Sci. U. S. A., 1962, 48, 2013–2018 CrossRef CAS PubMed.
- D. Sen and W. Gilbert, Nature, 1988, 334, 364–366 CrossRef CAS PubMed.
- M. Cheng, Y. Cheng, J. Hao, G. Jia, J. Zhou, J. L. Mergny and C. Li, Nucleic Acids Res., 2018, 46, 9264–9275 CrossRef CAS PubMed.
- H. A. Pickett, J. D. Henson, A. Y. Au, A. A. Neumann and R. R. Reddel, Hum. Mol. Genet., 2011, 20, 4684–4692 CrossRef CAS PubMed.
- V. F. S. Kahl, J. A. M. Allen, C. B. Nelson, A. P. Sobinoff, M. Lee, T. Kilo, R. S. Vasireddy and H. A. Pickett, Front. Cell Dev. Biol., 2020, 8, 493 CrossRef PubMed.
- W. E. Wright, V. M. Tesmer, K. E. Huffman, S. D. Levene and J. W. Shay, Genes Dev., 1997, 11, 2801–2809 CrossRef CAS PubMed.
- M. Z. Levy, R. C. Allsopp, A. B. Futcher, C. W. Greider and C. B. Harley, J. Mol. Biol., 1992, 225, 951–960 CrossRef CAS PubMed.
- N. W. Kim, M. A. Piatyszek, K. R. Prowse, C. B. Harley, M. D. West, P. L. Ho, G. M. Coviello, W. E. Wright, S. L. Weinrich and J. W. Shay, Science, 1994, 266, 2011–2015 CrossRef CAS PubMed.
- G. B. Morin, Cell, 1989, 59, 521–529 CrossRef CAS PubMed.
- A. N. Elayadi, D. A. Braasch and D. R. Corey, Biochemistry, 2002, 41, 9973–9981 CrossRef CAS PubMed.
- J. Feng, W. D. Funk, S. S. Wang, S. L. Weinrich, A. A. Avilion, C. P. Chiu, R. R. Adams, E. Chang, R. C. Allsopp, J. Yu, S. Le, D. W. West, C. B. Harley, W. H. Andrews, C. W. Greider and B. Villeponteau, Science, 1995, 269, 1236–1241 CrossRef CAS PubMed.
- A. M. Zahler, J. R. Williamson, T. R. Cech and D. M. Prescott, Nature, 1991, 350, 718–720 CrossRef CAS PubMed.
- S. Neidle, Febs J., 2010, 277, 1118–1125 CrossRef CAS PubMed.
- P. Agrawal, E. Hatzakis, K. Guo, M. Carver and D. Yang, Nucleic Acids Res., 2013, 41, 10584–10592 CrossRef CAS PubMed.
- S. Bielskutė, J. Plavec and P. Podbevšek, Nucleic Acids Res., 2021, 49, 2346–2356 CrossRef PubMed.
- J. Dai, C. Punchihewa, A. Ambrus, D. Chen, R. A. Jones and D. Yang, Nucleic Acids Res., 2007, 35, 2440–2450 CrossRef CAS PubMed.
- H. Yaku, T. Fujimoto, T. Murashima, D. Miyoshi and N. Sugimoto, Chem. Commun., 2012, 48, 6203–6216 RSC.
- H. Yaku, T. Murashima, D. Miyoshi and N. Sugimoto, Chem. Commun., 2010, 46, 5740–5742 RSC.
- S. Burge, G. N. Parkinson, P. Hazel, A. K. Todd and S. Neidle, Nucleic Acids Res., 2006, 34, 5402–5415 CrossRef CAS PubMed.
- J. Gros, F. Rosu, S. Amrane, A. De Cian, V. Gabelica, L. Lacroix and J. L. Mergny, Nucleic Acids Res., 2007, 35, 3064–3075 CrossRef CAS PubMed.
- C. J. Lech, B. Heddi and A. T. Phan, Nucleic Acids Res., 2013, 41, 2034–2046 CrossRef CAS PubMed.
- P. Hobza and J. Sponer, J. Am. Chem. Soc., 2002, 124, 11802–11808 CrossRef CAS PubMed.
- J. Gu and J. Leszczynski, J. Phys. Chem. A, 2000, 104, 6308–6313 CrossRef CAS.
- C. Roxo, W. Kotkowiak and A. Pasternak, Int. J. Mol. Sci., 2021, 22, 2940–2958 CrossRef PubMed.
- N. Q. Do and A. T. Phan, Chemistry, 2012, 18, 14752–14759 CrossRef CAS PubMed.
- N. Q. Do, K. W. Lim, M. H. Teo, B. Heddi and A. T. Phan, Nucleic Acids Res., 2011, 39, 9448–9457 CrossRef CAS PubMed.
- M. Adrian, D. J. Ang, C. J. Lech, B. Heddi, A. Nicolas and A. T. Phan, J. Am. Chem. Soc., 2014, 136, 6297–6305 CrossRef CAS PubMed.
- T. C. Marsh, J. Vesenka and E. Henderson, Nucleic Acids Res., 1995, 23, 696–700 CrossRef CAS PubMed.
- D. Miyoshi, H. Karimata and N. Sugimoto, Angew. Chem., Int. Ed., 2005, 44, 3740–3744 CrossRef CAS PubMed.
- D. Miyoshi, H. Karimata, Z. M. Wang, K. Koumoto and N. Sugimoto, J. Am. Chem. Soc., 2007, 129, 5919–5925 CrossRef CAS PubMed.
- D. Miyoshi and N. Sugimoto, Methods Mol. Biol., 2011, 749, 93–104 CrossRef CAS PubMed.
- K. Bose, C. J. Lech, B. Heddi and A. T. Phan, Nat. Commun., 2018, 9, 1959 CrossRef PubMed.
- A. M. Varizhuk, A. D. Protopopova, V. B. Tsvetkov, N. A. Barinov, V. V. Podgorsky, M. V. Tankevich, M. A. Vlasenok, V. V. Severov, I. P. Smirnov, E. V. Dubrovin, D. V. Klinov and G. E. Pozmogova, Nucleic Acids Res., 2018, 46, 8978–8992 CrossRef CAS PubMed.
- C. Saintomé, S. Amrane, J. L. Mergny and P. Alberti, Nucleic Acids Res., 2016, 44, 2926–2935 CrossRef PubMed.
- H. Q. Yu, D. Miyoshi and N. Sugimoto, J. Am. Chem. Soc., 2006, 128, 15461–15468 CrossRef CAS PubMed.
- H. Yu, X. Gu, S. Nakano, D. Miyoshi and N. Sugimoto, J. Am. Chem. Soc., 2012, 134, 20060–20069 CrossRef CAS PubMed.
- A. Risitano and K. R. Fox, Nucleic Acids Res., 2004, 32, 2598–2606 CrossRef CAS PubMed.
- P. Hazel, J. Huppert, S. Balasubramanian and S. Neidle, J. Am. Chem. Soc., 2004, 126, 16405–16415 CrossRef CAS PubMed.
- N. Kumar and S. Maiti, Nucleic Acids Res., 2008, 36, 5610–5622 CrossRef CAS PubMed.
- C. Ghimire, S. Park, K. Iida, P. Yangyuoru, H. Otomo, Z. Yu, K. Nagasawa, H. Sugiyama and H. Mao, J. Am. Chem. Soc., 2014, 136, 15537–15544 CrossRef CAS PubMed.
- A. Guédin, J. Gros, P. Alberti and J. L. Mergny, Nucleic Acids Res., 2010, 38, 7858–7868 CrossRef PubMed.
- E. Hatzakis, K. Okamoto and D. Yang, Biochemistry, 2010, 49, 9152–9160 CrossRef CAS PubMed.
- A. A. Bizyaeva, D. A. Bunin, V. L. Moiseenko, A. S. Gambaryan, S. Balk, V. N. Tashlitsky, A. M. Arutyunyan, A. M. Kopylov and E. G. Zavyalova, Int. J. Mol. Sci., 2021, 22, 2409–2422 CrossRef CAS PubMed.
- K. W. Lim, S. Amrane, S. Bouaziz, W. Xu, Y. Mu, D. J. Patel, K. N. Luu and A. T. Phan, J. Am. Chem. Soc., 2009, 131, 4301–4309 CrossRef CAS PubMed.
- Z. Zhang, J. Dai, E. Veliath, R. A. Jones and D. Yang, Nucleic Acids Res., 2010, 38, 1009–1021 CrossRef CAS PubMed.
- Y. Chen and D. Yang, Current Protocols in Nucleic Acid Chemistry, 2012, ch. 17, Unit17.15 Search PubMed.
- K. W. Lim and A. T. Phan, Angew. Chem., Int. Ed., 2013, 52, 8566–8569 CrossRef CAS PubMed.
- K. W. Lim, T. Q. Nguyen and A. T. Phan, J. Am. Chem. Soc., 2014, 136, 17969–17973 CrossRef CAS PubMed.
- K. W. Lim, Z. J. Khong and A. T. Phan, Biochemistry, 2014, 53, 247–257 CrossRef CAS PubMed.
- S. Ravichandran, M. Razzaq, N. Parveen, A. Ghosh and K. K. Kim, Nucleic Acids Res., 2021, 49, 10689–10706 CrossRef CAS PubMed.
- H. Feng and C. K. Kwok, RSC Chem. Biol., 2022, 3, 431–435 RSC.
- K. W. Lim, P. Jenjaroenpun, Z. J. Low, Z. J. Khong, Y. S. Ng, V. A. Kuznetsov and A. T. Phan, Nucleic Acids Res., 2015, 43, 5630–5646 CrossRef CAS PubMed.
- B. Karg, S. Mohr and K. Weisz, Angew. Chem., Int. Ed., 2019, 58, 11068–11071 CrossRef CAS PubMed.
- Y. Cheng, Q. Tang, Y. Li, Y. Zhang, C. Zhao, J. Yan and H. You, J. Biol. Chem., 2019, 294, 5890–5895 CrossRef CAS PubMed.
- T. Q. N. Nguyen, K. W. Lim and A. T. Phan, J. Phys. Chem. B, 2020, 124, 5122–5130 CrossRef CAS PubMed.
- M. Nakata, N. Kosaka, K. Kawauchi and D. Miyoshi, ACS Omega, 2024, 9, 35028–35036 CrossRef CAS PubMed.
- M. Nakata, N. Kosaka, K. Kawauchi and D. Miyoshi, Biochemistry, 2024, 64, 609–619 CrossRef PubMed.
- D. J. Y. Tan, F. R. Winnerdy, K. W. Lim and A. T. Phan, Nucleic Acids Res., 2020, 48, 11162–11171 CrossRef CAS PubMed.
- A. T. Phan, V. Kuryavyi, J. C. Darnell, A. Serganov, A. Majumdar, S. Ilin, T. Raslin, A. Polonskaia, C. Chen, D. Clain, R. B. Darnell and D. J. Patel, Nat. Struct. Mol. Biol., 2011, 18, 796–804 CrossRef CAS PubMed.
- K. D. Warner, M. C. Chen, W. Song, R. L. Strack, A. Thorn, S. R. Jaffrey and A. R. Ferré-D'Amaré, Nat. Struct. Mol. Biol., 2014, 21, 658–663 CrossRef CAS PubMed.
- H. Huang, N. B. Suslov, N. S. Li, S. A. Shelke, M. E. Evans, Y. Koldobskaya, P. A. Rice and J. A. Piccirilli, Nat. Chem. Biol., 2014, 10, 686–691 CrossRef CAS PubMed.
- D. Bhattacharyya, G. Mirihana Arachchilage and S. Basu, Front. Chem., 2016, 4, 38 Search PubMed.
- J. S. Lee, Nucleic Acids Res., 1990, 18, 6057–6060 CrossRef CAS PubMed.
- C. C. Hardin, T. Watson, M. Corregan and C. Bailey, Biochemistry, 1992, 31, 833–841 CrossRef CAS PubMed.
- E. A. Venczel and D. Sen, Biochemistry, 1993, 32, 6220–6228 CrossRef CAS PubMed.
- B. I. Kankia and L. A. Marky, J. Am. Chem. Soc., 2001, 123, 10799–10804 CrossRef CAS PubMed.
- A. Włodarczyk, P. Grzybowski, A. Patkowski and A. Dobek, J. Phys. Chem. B, 2005, 109, 3594–3605 CrossRef PubMed.
- F. M. Chen, Biochemistry, 1992, 31, 3769–3776 CrossRef CAS PubMed.
- S. W. Blume, V. Guarcello, W. Zacharias and D. M. Miller, Nucleic Acids Res., 1997, 25, 617–625 CrossRef CAS PubMed.
- C. C. Hardin, A. G. Perry and K. White, Biopolymers, 2000, 56, 147–194 CrossRef CAS PubMed.
- D. Miyoshi, A. Nakao, T. Toda and N. Sugimoto, FEBS Lett., 2001, 496, 128–133 CrossRef CAS PubMed.
- N. V. Hud, F. W. Smith, F. A. Anet and J. Feigon, Biochemistry, 1996, 35, 15383–15390 CrossRef CAS PubMed.
- M. Beck, V. Lucić, F. Förster, W. Baumeister and O. Medalia, Nature, 2007, 449, 611–615 CrossRef CAS PubMed.
- R. J. Ellis, Curr. Opin. Struct. Biol., 2001, 11, 114–119 CrossRef CAS PubMed.
- D. S. Goodsell, Trends Biochem. Sci., 1991, 16, 203–206 CrossRef CAS PubMed.
- S. B. Zimmerman and S. O. Trach, J. Mol. Biol., 1991, 222, 599–620 CrossRef CAS PubMed.
- K. Luby-Phelps, Int. Rev. Cytol., 2000, 192, 189–221 CAS.
- D. Miyoshi, H. Karimata and N. Sugimoto, J. Am. Chem. Soc., 2006, 128, 7957–7963 CrossRef CAS PubMed.
- T. Fujimoto, S. Nakano, N. Sugimoto and D. Miyoshi, J. Phys. Chem. B, 2013, 117, 963–972 CrossRef CAS PubMed.
- D. Miyoshi, K. Nakamura, H. Tateishi-Karimata, T. Ohmichi and N. Sugimoto, J. Am. Chem. Soc., 2009, 131, 3522–3531 CrossRef CAS PubMed.
- Y. M. Ueda, Y. K. Zouzumi, A. Maruyama, S. I. Nakano, N. Sugimoto and D. Miyoshi, Sci. Technol. Adv. Mater., 2016, 17, 753–759 CrossRef CAS PubMed.
- Y. Xue, Z. Y. Kan, Q. Wang, Y. Yao, J. Liu, Y. H. Hao and Z. Tan, J. Am. Chem. Soc., 2007, 129, 11185–11191 CrossRef CAS PubMed.
- K. W. Zheng, Z. Chen, Y. H. Hao and Z. Tan, Nucleic Acids Res., 2010, 38, 327–338 CrossRef CAS PubMed.
- Z. Y. Kan, Y. Yao, P. Wang, X. H. Li, Y. H. Hao and Z. Tan, Angew. Chem., Int. Ed., 2006, 45, 1629–1632 CrossRef CAS PubMed.
- A. Marchand, R. Ferreira, H. Tateishi-Karimata, D. Miyoshi, N. Sugimoto and V. Gabelica, J. Phys. Chem. B, 2013, 117, 12391–12401 CrossRef CAS PubMed.
- S. Muhuri, K. Mimura, D. Miyoshi and N. Sugimoto, J. Am. Chem. Soc., 2009, 131, 9268–9280 CrossRef CAS PubMed.
- H. Tateishi-Karimata, S. Pramanik, S. Nakano, D. Miyoshi and N. Sugimoto, ChemMedChem, 2014, 9, 2150–2155 CrossRef CAS PubMed.
- S. Nakano, H. Karimata, T. Ohmichi, J. Kawakami and N. Sugimoto, J. Am. Chem. Soc., 2004, 126, 14330–14331 CrossRef CAS PubMed.
- H. Tateishi-Karimata, D. Banerjee, T. Ohyama, S. Matsumoto, D. Miyoshi, S. I. Nakano and N. Sugimoto, Biochem. Biophys. Res. Commun., 2020, 525, 177–183 CrossRef CAS PubMed.
- D. Miyoshi, T. Fujimoto and N. Sugimoto, Top. Curr. Chem., 2013, 330, 87–110 CrossRef CAS PubMed.
- M. C. Miller, R. Buscaglia, J. B. Chaires, A. N. Lane and J. O. Trent, J. Am. Chem. Soc., 2010, 132, 17105–17107 CrossRef CAS PubMed.
- L. Petraccone, B. Pagano and C. Giancola, Methods, 2012, 57, 76–83 CrossRef CAS PubMed.
- D. Miyoshi and N. Sugimoto, Biochimie, 2008, 90, 1040–1051 CrossRef CAS PubMed.
- M. Tsuruta, Y. Sugitani, N. Sugimoto and D. Miyoshi, Int. J. Mol. Sci., 2021, 22, 947–962 CrossRef CAS PubMed.
- D. Miyoshi, S. Matsumura, S. Nakano and N. Sugimoto, J. Am. Chem. Soc., 2004, 126, 165–169 CrossRef CAS PubMed.
- D. Miyoshi, A. Nakao and N. Sugimoto, Biochemistry, 2002, 41, 15017–15024 CrossRef CAS PubMed.
- D. Miyoshi, H. Karimata and N. Sugimoto, Angew. Chem., Int. Ed., 2005, 44, 3740–3744 CrossRef CAS PubMed.
- B. Heddi and A. T. Phan, J. Am. Chem. Soc., 2011, 133, 9824–9833 CrossRef CAS PubMed.
- R. Buscaglia, M. C. Miller, W. L. Dean, R. D. Gray, A. N. Lane, J. O. Trent and J. B. Chaires, Nucleic Acids Res., 2013, 41, 7934–7946 CrossRef CAS PubMed.
- H. Q. Yu, D. H. Zhang, X. B. Gu, D. Miyoshi and N. Sugimoto, Angew. Chem., Int. Ed., 2008, 47, 9034–9038 CrossRef CAS PubMed.
- H. Yaku, T. Murashima, D. Miyoshi and N. Sugimoto, J. Phys. Chem. B, 2014, 118, 2605–2614 CrossRef CAS PubMed.
- H. Yaku, T. Murashima, H. Tateishi-Karimata, S. Nakano, D. Miyoshi and N. Sugimoto, Methods, 2013, 64, 19–27 CrossRef CAS PubMed.
- S. Cogoi, M. Paramasivam, B. Spolaore and L. E. Xodo, Nucleic Acids Res., 2008, 36, 3765–3780 CrossRef CAS PubMed.
- B. Pagano, L. Margarucci, P. Zizza, J. Amato, N. Iaccarino, C. Cassiano, E. Salvati, E. Novellino, A. Biroccio, A. Casapullo and A. Randazzo, Chem. Commun., 2015, 51, 2964–2967 RSC.
- V. González, K. Guo, L. Hurley and D. Sun, J. Biol. Chem., 2009, 284, 23622–23635 CrossRef PubMed.
- L. T. Gray, A. C. Vallur, J. Eddy and N. Maizels, Nat. Chem. Biol., 2014, 10, 313–318 CrossRef CAS PubMed.
- V. Brázda, J. Červeň, M. Bartas, N. Mikysková, J. Coufal and P. Pečinka, Molecules, 2018, 23, 2341–2356 CrossRef PubMed.
- Z. L. Huang, J. Dai, W. H. Luo, X. G. Wang, J. H. Tan, S. B. Chen and Z. S. Huang, J. Am. Chem. Soc., 2018, 140, 17945–17955 CrossRef CAS PubMed.
- M. M. Makowski, C. Gräwe, B. M. Foster, N. V. Nguyen, T. Bartke and M. Vermeulen, Nat. Commun., 2018, 9, 1653 CrossRef PubMed.
- X. Zhang, J. Spiegel, S. Martínez Cuesta, S. Adhikari and S. Balasubramanian, Nat. Chem., 2021, 13, 626–633 CrossRef CAS PubMed.
- S. K. Mishra, A. Tawani, A. Mishra and A. Kumar, Sci. Rep., 2016, 6, 38144 CrossRef CAS PubMed.
- V. González and L. H. Hurley, Biochemistry, 2010, 49, 9706–9714 CrossRef PubMed.
- S. L. Cree, R. Fredericks, A. Miller, F. G. Pearce, V. Filichev, C. Fee and M. A. Kennedy, FEBS Lett., 2016, 590, 2870–2883 CrossRef CAS PubMed.
- A. Gallo, C. Lo Sterzo, M. Mori, A. Di Matteo, I. Bertini, L. Banci, M. Brunori and L. Federici, J. Biol. Chem., 2012, 287, 26539–26548 CrossRef CAS PubMed.
- H. J. Kang, T. V. Le, K. Kim, J. Hur, K. K. Kim and H. J. Park, J. Mol. Biol., 2014, 426, 2594–2604 CrossRef CAS PubMed.
- V. A. Soldatenkov, A. A. Vetcher, T. Duka and S. Ladame, ACS Chem. Biol., 2008, 3, 214–219 CrossRef CAS PubMed.
- M. Paramasivam, A. Membrino, S. Cogoi, H. Fukuda, H. Nakagama and L. E. Xodo, Nucleic Acids Res., 2009, 37, 2841–2853 CrossRef CAS PubMed.
- S. Cogoi, M. Paramasivam, A. Membrino, K. K. Yokoyama and L. E. Xodo, J. Biol. Chem., 2010, 285, 22003–22016 CrossRef CAS PubMed.
- M. Mimura, S. Tomita, Y. Shinkai, T. Hosokai, H. Kumeta, T. Saio, K. Shiraki and R. Kurita, J. Am. Chem. Soc., 2021, 143, 9849–9857 CrossRef CAS PubMed.
- Y. Wu, K. Shin-ya and R. M. Brosh, Jr., Mol. Cell. Biol., 2008, 28, 4116–4128 CrossRef CAS PubMed.
- A. K. Byrd and K. D. Raney, J. Biol. Chem., 2015, 290, 6482–6494 CrossRef CAS PubMed.
- B. Giri, P. J. Smaldino, R. G. Thys, S. D. Creacy, E. D. Routh, R. R. Hantgan, S. Lattmann, Y. Nagamine, S. A. Akman and J. P. Vaughn, Nucleic Acids Res., 2011, 39, 7161–7178 CrossRef CAS PubMed.
- E. K. S. McRae, E. P. Booy, A. Moya-Torres, P. Ezzati, J. Stetefeld and S. A. McKenna, Nucleic Acids Res., 2017, 45, 6656–6668 CrossRef CAS PubMed.
- P. Chakraborty and F. Grosse, DNA Repair, 2011, 10, 654–665 CrossRef CAS PubMed.
- K. Takahama, A. Takada, S. Tada, M. Shimizu, K. Sayama, R. Kurokawa and T. Oyoshi, Chem. Biol., 2013, 20, 341–350 CrossRef CAS PubMed.
- A. Ishiguro, N. Kimura, Y. Watanabe, S. Watanabe and A. Ishihama, Genes Cells, 2016, 21, 466–481 CrossRef CAS PubMed.
- J. M. Knop, S. K. Mukherjee, R. Oliva, S. Möbitz and R. Winter, J. Am. Chem. Soc., 2020, 142, 18299–18303 CrossRef CAS PubMed.
- T. D. McKnight and D. E. Shippen, Plant Cell, 2004, 16, 794–803 CrossRef CAS PubMed.
- M. A. Islam, S. D. Thomas, V. V. Murty, K. J. Sedoris and D. M. Miller, J. Biol. Chem., 2014, 289, 8521–8531 CrossRef CAS PubMed.
- S. K. Madden, A. D. de Araujo, M. Gerhardt, D. P. Fairlie and J. M. Mason, Mol. Cancer, 2021, 20, 3 CrossRef PubMed.
- R. Hänsel-Hertsch, D. Beraldi, S. V. Lensing, G. Marsico, K. Zyner, A. Parry, M. Di Antonio, J. Pike, H. Kimura, M. Narita, D. Tannahill and S. Balasubramanian, Nat. Genet., 2016, 48, 1267–1272 CrossRef PubMed.
- F. Kouzine, D. Wojtowicz, L. Baranello, A. Yamane, S. Nelson, W. Resch, K. R. Kieffer-Kwon, C. J. Benham, R. Casellas, T. M. Przytycka and D. Levens, Cell Syst., 2017, 4, 344–356.e347 CrossRef CAS PubMed.
- J. Spiegel, S. M. Cuesta, S. Adhikari, R. Hänsel-Hertsch, D. Tannahill and S. Balasubramanian, Genome Biol., 2021, 22, 117 CrossRef CAS PubMed.
- S. Roychoudhury, S. Pramanik, H. L. Harris, M. Tarpley, A. Sarkar, G. Spagnol, P. L. Sorgen, D. Chowdhury, V. Band, D. Klinkebiel and K. K. Bhakat, Proc. Natl. Acad. Sci. U. S. A., 2020, 117, 11409–11420 CrossRef CAS PubMed.
- S. C. Ha, J. Choi, H. Y. Hwang, A. Rich, Y. G. Kim and K. K. Kim, Nucleic Acids Res., 2009, 37, 629–637 CrossRef CAS PubMed.
- S. Eddy, L. Maddukuri, A. Ketkar, M. K. Zafar, E. E. Henninger, Z. F. Pursell and R. L. Eoff, Biochemistry, 2015, 54, 3218–3230 CrossRef CAS PubMed.
- V. Brázda, L. Hároníková, J. C. Liao and M. Fojta, Int. J. Mol. Sci., 2014, 15, 17493–17517 CrossRef PubMed.
- B. Heddi, V. V. Cheong, H. Martadinata and A. T. Phan, Proc. Natl. Acad. Sci. U. S. A., 2015, 112, 9608–9613 CrossRef CAS PubMed.
- M. C. Chen, R. Tippana, N. A. Demeshkina, P. Murat, S. Balasubramanian, S. Myong and A. R. Ferré-D'Amaré, Nature, 2018, 558, 465–469 CrossRef CAS PubMed.
- A. Traczyk, C. W. Liew, D. J. Gill and D. Rhodes, Nucleic Acids Res., 2020, 48, 4562–4571 CrossRef CAS PubMed.
- K. H. Ngo, C. W. Liew, B. Heddi and A. T. Phan, J. Am. Chem. Soc., 2024, 146, 13709–13713 CrossRef CAS PubMed.
- M. Sauer and K. Paeschke, Biochem. Soc. Trans., 2017, 45, 1173–1182 CrossRef CAS PubMed.
- O. Mendoza, A. Bourdoncle, J. B. Boulé, R. M. Brosh, Jr. and J. L. Mergny, Nucleic Acids Res., 2016, 44, 1989–2006 CrossRef CAS PubMed.
- K. N. Estep, T. J. Butler, J. Ding and R. M. Brosh, Curr. Med. Chem., 2019, 26, 2881–2897 CrossRef CAS PubMed.
- R. M. Brosh, Jr., Nat. Rev. Cancer, 2013, 13, 542–558 CrossRef PubMed.
- S. Cogoi, S. Zorzet, V. Rapozzi, I. Géci, E. B. Pedersen and L. E. Xodo, Nucleic Acids Res., 2013, 41, 4049–4064 CrossRef CAS PubMed.
- A. Bugaut and S. Balasubramanian, Nucleic Acids Res., 2012, 40, 4727–4741 CrossRef CAS PubMed.
- S. Kumari, A. Bugaut, J. L. Huppert and S. Balasubramanian, Nat. Chem. Biol., 2007, 3, 218–221 CrossRef CAS PubMed.
- G. Biffi, M. Di Antonio, D. Tannahill and S. Balasubramanian, Nat. Chem., 2014, 6, 75–80 CrossRef CAS PubMed.
- E. G. Conlon, L. Lu, A. Sharma, T. Yamazaki, T. Tang, N. A. Shneider and J. L. Manley, Elife, 2016, 5, 17820–17847 CrossRef PubMed.
- C. K. Kwok, G. Marsico, A. B. Sahakyan, V. S. Chambers and S. Balasubramanian, Nat. Methods, 2016, 13, 841–844 CrossRef CAS PubMed.
- J. Eddy and N. Maizels, Nucleic Acids Res., 2008, 36, 1321–1333 CrossRef CAS PubMed.
- J. L. Huppert, A. Bugaut, S. Kumari and S. Balasubramanian, Nucleic Acids Res., 2008, 36, 6260–6268 CrossRef CAS PubMed.
- L. Dumas, P. Herviou, E. Dassi, A. Cammas and S. Millevoi, Trends Biochem. Sci., 2021, 46, 270–283 CrossRef CAS PubMed.
- S. Mestre-Fos, P. I. Penev, S. Suttapitugsakul, M. Hu, C. Ito, A. S. Petrov, R. M. Wartell, R. Wu and L. D. Williams, J. Mol. Biol., 2019, 431, 1940–1955 CrossRef CAS PubMed.
- S. Mestre-Fos, P. I. Penev, J. C. Richards, W. L. Dean, R. D. Gray, J. B. Chaires and L. D. Williams, PLoS One, 2019, 14, e0226177 CrossRef CAS PubMed.
- J. C. Bowman, A. S. Petrov, M. Frenkel-Pinter, P. I. Penev and L. D. Williams, Chem. Rev., 2020, 120, 4848–4878 CrossRef CAS PubMed.
- S. Khateb, P. Weisman-Shomer, I. Hershco, L. A. Loeb and M. Fry, Nucleic Acids Res., 2004, 32, 4145–4154 CrossRef CAS PubMed.
- A. von Hacht, O. Seifert, M. Menger, T. Schütze, A. Arora, Z. Konthur, P. Neubauer, A. Wagner, C. Weise and J. Kurreck, Nucleic Acids Res., 2014, 42, 6630–6644 CrossRef CAS PubMed.
- S. Boeynaems, S. Alberti, N. L. Fawzi, T. Mittag, M. Polymenidou, F. Rousseau, J. Schymkowitz, J. Shorter, B. Wolozin, L. Van Den Bosch, P. Tompa and M. Fuxreiter, Trends Cell Biol., 2018, 28, 420–435 CrossRef CAS PubMed.
- S. Alberti and D. Dormann, Annu. Rev. Genet., 2019, 53, 171–194 CrossRef CAS PubMed.
- Q. Li, X. Peng, Y. Li, W. Tang, J. Zhu, J. Huang, Y. Qi and Z. Zhang, Nucleic Acids Res., 2020, 48, D320–d327 CrossRef CAS PubMed.
- K. Kohata and D. Miyoshi, Biophys. Rev., 2020, 12, 669–676 CrossRef CAS PubMed.
- Y. R. Kamimura and M. Kanai, Bull. Chem. Soc. Jpn., 2021, 94, 1045–1058 CrossRef CAS.
- Y. Zhang, M. Yang, S. Duncan, X. Yang, M. A. S. Abdelhamid, L. Huang, H. Zhang, P. N. Benfey, Z. A. E. Waller and Y. Ding, Nucleic Acids Res., 2019, 47, 11746–11754 CAS.
- M. Tsuruta, T. Torii, K. Kohata, K. Kawauchi, H. Tateishi-Karimata, N. Sugimoto and D. Miyoshi, Chem. Commun., 2022, 58, 12931–12934 RSC.
- S. Shil, M. Tsuruta, K. Kawauchi and D. Miyoshi, ChemMedChem, 2024, e202400460, DOI:10.1002/cmdc.202400460.
- A. Zbinden, M. Pérez-Berlanga, P. De Rossi and M. Polymenidou, Dev. Cell, 2020, 55, 45–68 CrossRef CAS PubMed.
- E. Wang, R. Thombre, Y. Shah, R. Latanich and J. Wang, Nucleic Acids Res., 2021, 49, 4816–4830 CrossRef CAS PubMed.
- Q. F. Yang, X. R. Wang, Y. H. Wang, X. H. Wu, R. Y. Shi, Y. X. Wang, H. N. Zhu, S. Yang, Y. L. Tang and F. Li, Nucleic Acids Res., 2024, 91–98 CAS.
- S. Neidle, Molecules, 2024, 29, 3653–3674 CrossRef CAS PubMed.
- J. Hilton, K. Gelmon, P. L. Bedard, D. Tu, H. Xu, A. V. Tinker, R. Goodwin, S. A. Laurie, D. Jonker, A. R. Hansen, Z. W. Veitch, D. J. Renouf, L. Hagerman, H. Lui, B. Chen, D. Kellar, I. Li, S. E. Lee, T. Kono, B. Y. C. Cheng, D. Yap, D. Lai, S. Beatty, J. Soong, K. I. Pritchard, I. Soria-Bretones, E. Chen, H. Feilotter, M. Rushton, L. Seymour, S. Aparicio and D. W. Cescon, Nat. Commun., 2022, 13, 3607 CrossRef CAS PubMed.
- F. X. Han, R. T. Wheelhouse and L. H. Hurley, J. Am. Chem. Soc., 1999, 121, 3561–3570 CrossRef CAS.
- E. Izbicka, R. T. Wheelhouse, E. Raymond, K. K. Davidson, R. A. Lawrence, D. Sun, B. E. Windle, L. H. Hurley and D. D. Von Hoff, Cancer Res., 1999, 59, 639–644 CAS.
- A. De Cian, G. Cristofari, P. Reichenbach, E. De Lemos, D. Monchaud, M. P. Teulade-Fichou, K. Shin-Ya, L. Lacroix, J. Lingner and J. L. Mergny, Proc. Natl. Acad. Sci. U. S. A., 2007, 104, 17347–17352 CrossRef CAS PubMed.
- Y. X. Yao, B. H. Xu and Y. Zhang, Biomed. Res. Int., 2018, 2018, 9701957 Search PubMed.
- D. Drygin, A. Siddiqui-Jain, S. O'Brien, M. Schwaebe, A. Lin, J. Bliesath, C. B. Ho, C. Proffitt, K. Trent, J. P. Whitten, J. K. Lim, D. Von Hoff, K. Anderes and W. G. Rice, Cancer Res., 2009, 69, 7653–7661 CrossRef CAS PubMed.
- J. Dai, M. Carver, L. H. Hurley and D. Yang, J. Am. Chem. Soc., 2011, 133, 17673–17680 CrossRef CAS PubMed.
- K. Shin-ya, K. Wierzba, K. Matsuo, T. Ohtani, Y. Yamada, K. Furihata, Y. Hayakawa and H. Seto, J. Am. Chem. Soc., 2001, 123, 1262–1263 CrossRef CAS PubMed.
- G. A. Blankson, D. S. Pilch, A. A. Liu, L. F. Liu, J. E. Rice and E. J. LaVoie, Bioorg. Med. Chem., 2013, 21, 4511–4520 CrossRef CAS PubMed.
- M. Yasuda, Y. Ma, S. Okabe, Y. Wakabayashi, D. Su, Y. T. Chang, H. Seimiya, M. Tera and K. Nagasawa, Chem. Commun., 2020, 56, 12905–12908 RSC.
- R. Rodriguez, S. Müller, J. A. Yeoman, C. Trentesaux, J. F. Riou and S. Balasubramanian, J. Am. Chem. Soc., 2008, 130, 15758–15759 CrossRef CAS PubMed.
- D. Koirala, S. Dhakal, B. Ashbridge, Y. Sannohe, R. Rodriguez, H. Sugiyama, S. Balasubramanian and H. Mao, Nat. Chem., 2011, 3, 782–787 CrossRef CAS PubMed.
- R. Rodriguez, K. M. Miller, J. V. Forment, C. R. Bradshaw, M. Nikan, S. Britton, T. Oelschlaegel, B. Xhemalce, S. Balasubramanian and S. P. Jackson, Nat. Chem. Biol., 2012, 8, 301–310 CrossRef CAS PubMed.
- M. Di Antonio, A. Ponjavic, A. Radzevičius, R. T. Ranasinghe, M. Catalano, X. Zhang, J. Shen, L. M. Needham, S. F. Lee, D. Klenerman and S. Balasubramanian, Nat. Chem., 2020, 12, 832–837 CrossRef CAS PubMed.
- D. Monchaud, C. Allain, H. Bertrand, N. Smargiasso, F. Rosu, V. Gabelica, A. De Cian, J. L. Mergny and M. P. Teulade-Fichou, Biochimie, 2008, 90, 1207–1223 CrossRef CAS PubMed.
- A. De Cian, P. Grellier, E. Mouray, D. Depoix, H. Bertrand, D. Monchaud, M. P. Teulade-Fichou, J. L. Mergny and P. Alberti, Chembiochem, 2008, 9, 2730–2739 CrossRef CAS PubMed.
- X. J. Luo, Q. P. Qin, K. B. Huang, Y. C. Liu and H. Liang, Asian J. Chem., 2013, 25, 6396–6400 CrossRef CAS.
- N. Shioda, Y. Yabuki, K. Yamaguchi, M. Onozato, Y. Li, K. Kurosawa, H. Tanabe, N. Okamoto, T. Era, H. Sugiyama, T. Wada and K. Fukunaga, Nat. Med., 2018, 24, 802–813 CrossRef CAS PubMed.
- Y. Qi, H. Chen, W. Tan, Y. Li, G. Yuan and M. Xu, Rapid Commun. Mass Spectrom., 2015, 29, 1611–1616 CrossRef CAS PubMed.
- M. Zhu, J. Gao, X. J. Lin, Y. Y. Gong, Y. C. Qi, Y. L. Ma, Y. X. Song, W. Tan, F. Y. Li, M. Ye, J. Gong, Q. H. Cui, Z. H. Huang, Y. Y. Zhang, X. J. Wang, F. Lan, S. Q. Wang, G. Yuan, Y. Feng and M. Xu, Nucleic Acids Res., 2021, 49, 2522–2536 CrossRef CAS PubMed.
- V. Gabelica, R. Maeda, T. Fujimoto, H. Yaku, T. Murashima, N. Sugimoto and D. Miyoshi, Biochemistry, 2013, 52, 5620–5628 CrossRef CAS PubMed.
- H. Arthanari, S. Basu, T. L. Kawano and P. H. Bolton, Nucleic Acids Res., 1998, 26, 3724–3728 CrossRef CAS PubMed.
- J. Ren and J. B. Chaires, Biochemistry, 1999, 38, 16067–16075 CrossRef CAS PubMed.
- M. Perenon, H. Bonnet, T. Lavergne, J. Dejeu and E. Defrancq, Phys. Chem. Chem. Phys., 2020, 22, 4158–4164 RSC.
- K. Kawauchi, W. Sugimoto, T. Yasui, K. Murata, K. Itoh, K. Takagi, T. Tsuruoka, K. Akamatsu, H. Tateishi-Karimata, N. Sugimoto and D. Miyoshi, Nat. Commun., 2018, 9, 2271 CrossRef PubMed.
- A. Ferino, G. Nicoletto, F. D'Este, S. Zorzet, S. Lago, S. N. Richter, A. Tikhomirov, A. Shchekotikhin and L. E. Xodo, J. Med. Chem., 2020, 63, 1245–1260 CrossRef CAS PubMed.
- M. G. Costales, Y. Matsumoto, S. P. Velagapudi and M. D. Disney, J. Am. Chem. Soc., 2018, 140, 6741–6744 CrossRef CAS PubMed.
- K. M. Patil, D. Chin, H. L. Seah, Q. Shi, K. W. Lim and A. T. Phan, Chem. Commun., 2021, 57, 12816–12819 RSC.
- S. Mikutis, M. Rebelo, E. Yankova, M. Gu, C. Tang, A. R. Coelho, M. Yang, M. E. Hazemi, M. Pires de Miranda, M. Eleftheriou, M. Robertson, G. S. Vassiliou, D. J. Adams, J. P. Simas, F. Corzana, J. S. Schneekloth, Jr., K. Tzelepis and G. J. L. Bernardes, ACS Cent. Sci., 2023, 9, 892–904 CrossRef CAS PubMed.
- Y. Zhang, L. Wang, F. Wang, X. Chu and J. H. Jiang, J. Am. Chem. Soc., 2024, 146, 15815–15824 CrossRef CAS PubMed.
- M. Xiao, J. Zhao, Q. Wang, J. Liu and L. Ma, Biomolecules, 2022, 12, 1257–1271 CrossRef CAS PubMed.
- J. Dickerhoff, J. Dai and D. Yang, Nucleic Acids Res., 2021, 49, 5905–5915 CrossRef CAS PubMed.
- L. Y. Liu, T. Z. Ma, Y. L. Zeng, W. Liu and Z. W. Mao, J. Am. Chem. Soc., 2022, 144, 11878–11887 CrossRef CAS PubMed.
- F. S. Di Leva, E. Novellino, A. Cavalli, M. Parrinello and V. Limongelli, Nucleic Acids Res., 2014, 42, 5447–5455 CrossRef CAS PubMed.
- H. J. Sullivan, C. Readmond, C. Radicella, V. Persad, T. J. Fasano and C. Wu, ACS Omega, 2018, 3, 14788–14806 CrossRef CAS PubMed.
- T. Santos, G. F. Salgado, E. J. Cabrita and C. Cruz, Pharmaceuticals, 2021, 14, 769–808 CrossRef CAS PubMed.
- T. Liu, Y. Wu, L. Qin, Q. Luo, W. Li, Y. Cheng, Y. Tu and H. You, J. Phys. Chem. B, 2023, 127, 5859–5868 CrossRef CAS PubMed.
- K. Gehring, J. L. Leroy and M. Guéron, Nature, 1993, 363, 561–565 CrossRef CAS PubMed.
- P. Rajagopal and J. Feigon, Nature, 1989, 339, 637–640 CrossRef CAS PubMed.
- A. Lupták, A. R. Ferré-D'Amaré, K. Zhou, K. W. Zilm and J. A. Doudna, J. Am. Chem. Soc., 2001, 123, 8447–8452 CrossRef PubMed.
- J. L. Leroy, Nucleic Acids Res., 2009, 37, 4127–4134 CrossRef CAS PubMed.
- E. P. Wright, J. L. Huppert and Z. A. E. Waller, Nucleic Acids Res., 2017, 45, 2951–2959 CrossRef CAS PubMed.
- T. Fujii and N. Sugimoto, Phys. Chem. Chem. Phys., 2015, 17, 16719–16722 RSC.
- A. M. Fleming, K. M. Stewart, G. M. Eyring, T. E. Ball and C. J. Burrows, Org. Biomol. Chem., 2018, 16, 4537–4546 RSC.
- A. L. Lieblein, B. Fürtig and H. Schwalbe, Chembiochem, 2013, 14, 1226–1230 CrossRef CAS PubMed.
- J. L. Mergny, L. Lacroix, X. G. Han, J. L. Leroy and C. Helene, J. Am. Chem. Soc., 1995, 117, 8887–8898 CrossRef CAS.
- H. A. Day, P. Pavlou and Z. A. Waller, Bioorg. Med. Chem., 2014, 22, 4407–4418 CrossRef CAS PubMed.
- A. Rajendran, S. Nakano and N. Sugimoto, Chem. Commun., 2010, 46, 1299–1301 RSC.
- J. Cui, P. Waltman, V. H. Le and E. A. Lewis, Molecules, 2013, 18, 12751–12767 CrossRef CAS PubMed.
- L. Ying, J. J. Green, H. Li, D. Klenerman and S. Balasubramanian, Proc. Natl. Acad. Sci. U. S. A., 2003, 100, 14629–14634 CrossRef CAS PubMed.
- D. Sun and L. H. Hurley, J. Med. Chem., 2009, 52, 2863–2874 CrossRef CAS PubMed.
- N. Gilbert and J. Allan, Curr. Opin. Genet. Dev., 2014, 25, 15–21 CrossRef CAS PubMed.
- G. Cimino-Reale, E. Pascale, E. Alvino, G. Starace and E. D'Ambrosio, J. Biol. Chem., 2003, 278, 2136–2140 CrossRef CAS PubMed.
- S. Dzatko, M. Krafcikova, R. Hänsel-Hertsch, T. Fessl, R. Fiala, T. Loja, D. Krafcik, J. L. Mergny, S. Foldynova-Trantirkova and L. Trantirek, Angew. Chem., Int. Ed., 2018, 57, 2165–2169 CrossRef CAS PubMed.
- M. Zeraati, D. B. Langley, P. Schofield, A. L. Moye, R. Rouet, W. E. Hughes, T. M. Bryan, M. E. Dinger and D. Christ, Nat. Chem., 2018, 10, 631–637 CrossRef CAS PubMed.
- T. Simonsson, M. Pribylova and M. Vorlickova, Biochem. Biophys. Res. Commun., 2000, 278, 158–166 CrossRef CAS PubMed.
- A. V. Makeyev and S. A. Liebhaber, RNA, 2002, 8, 265–278 CrossRef CAS PubMed.
- E. F. Michelotti, G. A. Michelotti, A. I. Aronsohn and D. Levens, Mol. Cell. Biol., 1996, 16, 2350–2360 CrossRef CAS PubMed.
- Z. Du, J. Yu, Y. Chen, R. Andino and T. L. James, J. Biol. Chem., 2004, 279, 48126–48134 CrossRef CAS PubMed.
- L. Lacroix, H. Liénard, E. Labourier, M. Djavaheri-Mergny, J. Lacoste, H. Leffers, J. Tazi, C. Hélène and J. L. Mergny, Nucleic Acids Res., 2000, 28, 1564–1575 CrossRef CAS PubMed.
- A. Bandiera, G. Tell, E. Marsich, A. Scaloni, G. Pocsfalvi, A. Akintunde Akindahunsi, L. Cesaratto and G. Manzini, Arch. Biochem. Biophys., 2003, 409, 305–314 CrossRef CAS PubMed.
- D. J. Uribe, K. Guo, Y. J. Shin and D. Sun, Biochemistry, 2011, 50, 3796–3806 CrossRef CAS PubMed.
- H. J. Kang, S. Kendrick, S. M. Hecht and L. H. Hurley, J. Am. Chem. Soc., 2014, 136, 4172–4185 CrossRef CAS PubMed.
- S. L. Brown and S. Kendrick, Pharmaceuticals, 2021, 14, 96–119 CrossRef CAS PubMed.
- S. Sedghi Masoud and K. Nagasawa, Chem. Pharm. Bull., 2018, 66, 1091–1103 CrossRef PubMed.
- H. Abou Assi, M. Garavís, C. González and M. J. Damha, Nucleic Acids Res., 2018, 46, 8038–8056 CrossRef PubMed.
- O. Y. Fedoroff, A. Rangan, V. V. Chemeris and L. H. Hurley, Biochemistry, 2000, 39, 15083–15090 CrossRef CAS PubMed.
- A. Siddiqui-Jain, C. L. Grand, D. J. Bearss and L. H. Hurley, Proc. Natl. Acad. Sci. U. S. A., 2002, 99, 11593–11598 CrossRef CAS PubMed.
- L. Wang, Y. Wu, T. Chen and C. Wei, Int. J. Biol. Macromol., 2013, 52, 1–8 CrossRef CAS PubMed.
- P. Alberti, J. Ren, M. P. Teulade-Fichou, L. Guittat, J. F. Riou, J. Chaires, C. Hélène, J. P. Vigneron, J. M. Lehn and J. L. Mergny, J. Biomol. Struct. Dyn., 2001, 19, 505–513 CrossRef CAS PubMed.
- S. Sedghi Masoud, Y. Yamaoki, Y. Ma, A. Marchand, F. R. Winnerdy, V. Gabelica, A. T. Phan, M. Katahira and K. Nagasawa, Chembiochem, 2018, 19, 2268–2272 CrossRef CAS PubMed.
- H. Xu, H. Zhang and X. Qu, J. Inorg. Biochem., 2006, 100, 1646–1652 CrossRef CAS PubMed.
- S. Shi, X. Geng, J. Zhao, T. Yao, C. Wang, D. Yang, L. Zheng and L. Ji, Biochimie, 2010, 92, 370–377 CrossRef CAS PubMed.
- S. Kendrick, H. J. Kang, M. P. Alam, M. M. Madathil, P. Agrawal, V. Gokhale, D. Yang, S. M. Hecht and L. H. Hurley, J. Am. Chem. Soc., 2014, 136, 4161–4171 CrossRef CAS PubMed.
- A. M. Burger, F. Dai, C. M. Schultes, A. P. Reszka, M. J. Moore, J. A. Double and S. Neidle, Cancer Res., 2005, 65, 1489–1496 CrossRef CAS PubMed.
- X. Li, Y. Peng, J. Ren and X. Qu, Proc. Natl. Acad. Sci. U. S. A., 2006, 103, 19658–19663 CrossRef CAS PubMed.
- Y. Chen, K. Qu, C. Zhao, L. Wu, J. Ren, J. Wang and X. Qu, Nat. Commun., 2012, 3, 1074 CrossRef PubMed.
- B. Shu, J. Cao, G. Kuang, J. Qiu, M. Zhang, Y. Zhang, M. Wang, X. Li, S. Kang, T. M. Ou, J. H. Tan, Z. S. Huang and D. Li, Chem. Commun., 2018, 54, 2036–2039 RSC.
- G. Kuang, M. Zhang, S. Kang, D. Hu, X. Li, Z. Wei, X. Gong, L. K. An, Z. S. Huang, B. Shu and D. Li, J. Med. Chem., 2020, 63, 9136–9153 CrossRef CAS PubMed.
- L. Meng, Y. Su, F. Yang, S. Xiao, Z. Yin, J. Liu, J. Zhong, D. Zhou and F. Yu, Bioorg. Med. Chem., 2019, 27, 115147 CrossRef CAS PubMed.
- H. Zeng, S. Kang, Y. Zhang, K. Liu, Q. Yu, D. Li and L. K. An, Int. J. Mol. Sci., 2021, 22, 1711–1727 CrossRef CAS PubMed.
- S. Kendrick, H. J. Kang, M. P. Alam, M. M. Madathil, P. Agrawal, V. Gokhale, D. Yang, S. M. Hecht and L. H. Hurley, J. Am. Chem. Soc., 2016, 138, 11408 CrossRef CAS PubMed.
- E. P. Wright, H. A. Day, A. M. Ibrahim, J. Kumar, L. J. Boswell, C. Huguin, C. E. Stevenson, K. Pors and Z. A. Waller, Sci. Rep., 2016, 6, 39456 CrossRef CAS PubMed.
- Q. Sheng, J. C. Neaverson, T. Mahmoud, C. E. M. Stevenson, S. E. Matthews and Z. A. E. Waller, Org. Biomol. Chem., 2017, 15, 5669–5673 RSC.
- S. Dhillon, Drugs, 2020, 80, 1853–1858 CrossRef CAS PubMed.
- N. A. Naryshkin, M. Weetall, A. Dakka, J. Narasimhan, X. Zhao, Z. Feng, K. K. Ling, G. M. Karp, H. Qi, M. G. Woll, G. Chen, N. Zhang, V. Gabbeta, P. Vazirani, A. Bhattacharyya, B. Furia, N. Risher, J. Sheedy, R. Kong, J. Ma, A. Turpoff, C. S. Lee, X. Zhang, Y. C. Moon, P. Trifillis, E. M. Welch, J. M. Colacino, J. Babiak, N. G. Almstead, S. W. Peltz, L. A. Eng, K. S. Chen, J. L. Mull, M. S. Lynes, L. L. Rubin, P. Fontoura, L. Santarelli, D. Haehnke, K. D. McCarthy, R. Schmucki, M. Ebeling, M. Sivaramakrishnan, C. P. Ko, S. V. Paushkin, H. Ratni, I. Gerlach, A. Ghosh and F. Metzger, Science, 2014, 345, 688–693 CrossRef CAS PubMed.
- H. Ratni, G. M. Karp, M. Weetall, N. A. Naryshkin, S. V. Paushkin, K. S. Chen, K. D. McCarthy, H. Qi, A. Turpoff, M. G. Woll, X. Zhang, N. Zhang, T. Yang, A. Dakka, P. Vazirani, X. Zhao, E. Pinard, L. Green, P. David-Pierson, D. Tuerck, A. Poirier, W. Muster, S. Kirchner, L. Mueller, I. Gerlach and F. Metzger, J. Med. Chem., 2016, 59, 6086–6100 CrossRef CAS PubMed.
- S. Campagne, S. Boigner, S. Rüdisser, A. Moursy, L. Gillioz, A. Knörlein, J. Hall, H. Ratni, A. Cléry and F. H. Allain, Nat. Chem. Biol., 2019, 15, 1191–1198 CrossRef CAS PubMed.
- H. Ratni, L. Mueller and M. Ebeling, Prog. Med. Chem., 2019, 58, 119–156 Search PubMed.
- M. Sivaramakrishnan, K. D. McCarthy, S. Campagne, S. Huber, S. Meier, A. Augustin, T. Heckel, H. Meistermann, M. N. Hug, P. Birrer, A. Moursy, S. Khawaja, R. Schmucki, N. Berntenis, N. Giroud, S. Golling, M. Tzouros, B. Banfai, G. Duran-Pacheco, J. Lamerz, Y. Hsiu Liu, T. Luebbers, H. Ratni, M. Ebeling, A. Cléry, S. Paushkin, A. R. Krainer, F. H. Allain and F. Metzger, Nat. Commun., 2017, 8, 1476 CrossRef PubMed.
- A. A. Ahmed, R. Angell, S. Oxenford, J. Worthington, N. Williams, N. Barton, T. G. Fowler, D. E. O'Flynn, M. Sunose, M. McConville, T. Vo, W. D. Wilson, S. A. Karim, J. P. Morton and S. Neidle, ACS Med. Chem. Lett., 2020, 11, 1634–1644 CrossRef CAS PubMed.
- A. A. Ahmed, S. Chen, M. Roman-Escorza, R. Angell, S. Oxenford, M. McConville, N. Barton, M. Sunose, D. Neidle, S. Haider, T. Arshad and S. Neidle, Sci. Rep., 2024, 14, 3447 CrossRef CAS PubMed.
- C. Marchetti, K. G. Zyner, S. A. Ohnmacht, M. Robson, S. M. Haider, J. P. Morton, G. Marsico, T. Vo, S. Laughlin-Toth, A. A. Ahmed, G. Di Vita, I. Pazitna, M. Gunaratnam, R. J. Besser, A. C. G. Andrade, S. Diocou, J. A. Pike, D. Tannahill, R. B. Pedley, T. R. J. Evans, W. D. Wilson, S. Balasubramanian and S. Neidle, J. Med. Chem., 2018, 61, 2500–2517 CrossRef CAS PubMed.
- A. A. Ahmed, W. Greenhalf, D. H. Palmer, N. Williams, J. Worthington, T. Arshad, S. Haider, E. Alexandrou, D. Guneri, Z. A. E. Waller and S. Neidle, Molecules, 2023, 28, 2452–2468 CrossRef CAS PubMed.
- J. Jana, Y. M. Vianney, N. Schröder and K. Weisz, Nucleic Acids Res., 2022, 50, 7161–7175 CrossRef CAS PubMed.
- A. T. Phan, K. N. Luu and D. J. Patel, Nucleic Acids Res., 2006, 34, 5715–5719 CrossRef CAS PubMed.
- M. Webba da Silva, M. Trajkovski, Y. Sannohe, N. Ma'ani Hessari, H. Sugiyama and J. Plavec, Angew. Chem., Int. Ed., 2009, 48, 9167–9170 CrossRef CAS PubMed.
- K. Li, L. Yatsunyk and S. Neidle, Nucleic Acids Res., 2021, 49, 519–528 CrossRef CAS PubMed.
- M. Zafferani and A. E. Hargrove, Cell Chem. Biol., 2021, 28, 594–609 CrossRef CAS PubMed.
- S. F. Banani, H. O. Lee, A. A. Hyman and M. K. Rosen, Nat. Rev. Mol. Cell Biol., 2017, 18, 285–298 CrossRef CAS PubMed.
- Y. Shin and C. P. Brangwynne, Science, 2017, 357, 1253–1263 CrossRef CAS PubMed.
- A. R. Strom, A. V. Emelyanov, M. Mir, D. V. Fyodorov, X. Darzacq and G. H. Karpen, Nature, 2017, 547, 241–245 CrossRef CAS PubMed.
- Y. Lin, D. S. Protter, M. K. Rosen and R. Parker, Mol. Cell, 2015, 60, 208–219 CrossRef CAS PubMed.
- B. R. Sabari, A. Dall’Agnese, A. Boija, I. A. Klein, E. L. Coffey, K. Shrinivas, B. J. Abraham, N. M. Hannett, A. V. Zamudio, J. C. Manteiga, C. H. Li, Y. E. Guo, D. S. Day, J. Schuijers, E. Vasile, S. Malik, D. Hnisz, T. I. Lee, I. I. Cisse, R. G. Roeder, P. A. Sharp, A. K. Chakraborty and R. A. Young, Science, 2018, 361, 379–389 CrossRef CAS PubMed.
- D. Monchaud, C. Allain and M. P. Teulade-Fichou, Bioorg. Med. Chem. Lett., 2006, 16, 4842–4845 CrossRef CAS PubMed.
- A. De Cian, L. Guittat, M. Kaiser, B. Saccà, S. Amrane, A. Bourdoncle, P. Alberti, M. P. Teulade-Fichou, L. Lacroix and J. L. Mergny, Methods, 2007, 42, 183–195 CrossRef CAS PubMed.
- C. Platella, D. Musumeci, A. Arciello, F. Doria, M. Freccero, A. Randazzo, J. Amato, B. Pagano and D. Montesarchio, Anal. Chim. Acta, 2018, 1030, 133–141 CrossRef CAS PubMed.
- Y. Hashimoto, Y. Imagawa, K. Nagano, R. Maeda, N. Nagahama, T. Torii, N. Kinoshita, N. Takamiya, K. Kawauchi, H. Tatesishi-Karimata, N. Sugimoto and D. Miyoshi, Chem. Commun., 2023, 59, 4891–4894 RSC.
- Y. Hashimoto, H. Kubo, K. Kawauchi and D. Miyoshi, Chem. Commun., 2024, 60, 13179–13182 RSC.
- S. Zhang, H. Sun, D. Yang, Y. Liu, X. Zhang, H. Chen, Q. Li, A. Guan and Y. Tang, Anal. Chim. Acta: X, 2019, 2, 100017 CAS.
- S. Kumar Mishra and A. Kumar, Database, 2016, 2016, baw002 CrossRef PubMed.
- Z. Cai, M. Zafferani, O. M. Akande and A. E. Hargrove, J. Med. Chem., 2022, 65, 7262–7277 CrossRef CAS PubMed.
| This journal is © The Royal Society of Chemistry 2025 |