Open Access Article
Open Access ArticleExploring substitution effects on the potential dominant conformations of NBF derivatives leading to functional conversion at the mu opioid receptor†
Ennian Lia,
Ahmed Redaa,
Hongguang Maa,
Samuel Woodardb,
James C. Gillespieb,
Dana E. Selleyb,
William L. Deweyb,
Piyusha P. Pagarea and
Yan Zhang *abcd
*abcd
aDepartment of Medicinal Chemistry, School of Pharmacy, Virginia Commonwealth University, 800 E Leigh Street, Richmond, Virginia 23298, USA. E-mail: yzhang2@vcu.edu
bDepartment of Pharmacology and Toxicology, School of Medicine, Virginia Commonwealth University, 410 North 12th Street, Richmond, Virginia 23298, USA
cCenter for Drug Discovery, Virginia Commonwealth University, 800 E Leigh Street, Richmond, Virginia 23298, USA
dInstitute for Drug and Alcohol Studies, Virginia Commonwealth University, 203 East Cary Street, Richmond, Virginia 23298, USA
First published on 5th May 2025
Abstract
We previously identified NBF (β-configuration at C6) and its 6α-counterpart as mu opioid receptor (MOR) antagonists. To explore the effect of C6 conformation of the epoxymorphinan ring on their MOR function, five pairs of NBF derivatives bearing both 6α and 6β configurations with substitutions on the 3′-position of the benzofuran ring were synthesized. In vitro and in vivo studies demonstrated that compounds carrying phenyl and 4-pyridine substituents retained their antagonistic properties independent of the C6 configuration. Halogen and methyl substituents with the 6α-configuration remained as MOR antagonists, while their 6β-counterparts switched to MOR agonists. Molecular modeling studies indicated that the C6 configuration and structural modification may collectively decide the orientation of the benzofuran ring, leading to conformation retention or a switch within the MOR binding pocket. These results together aid the understanding of the NBF structure–activity relationship (SAR) and provide insights for functional conversion at the MOR, supporting future endeavors to develop novel MOR ligands.
Introduction
It has been several decades since opioid binding sites were first proposed in the early 1950s,1 and several types of opioid receptors have been characterized by pharmacological studies.2–5 Among them, three main types of opioid receptors have been extensively studied, including the mu opioid receptor (MOR), the kappa opioid receptor (KOR), and the delta opioid receptor (DOR).6–9 These three opioid receptors share high sequence homology and belong to the superfamily of seven transmembrane-spanning G protein-coupled receptors (GPCRs).10 GPCRs play important roles in mediating the actions of many known neurotransmitters and hormones.11 Effects of opioids are mainly inhibitory, resulting in significant inhibition of nerve firing and reduction in neurotransmitter release. The rewarding and analgesic effects of opioids result mainly from activation of the MOR.12,13 KOR agonists have been shown to produce dysphoric and psychotomimetic effects,14 while DOR activation results in inhibition of anxiety and stress.15 As a result, a considerable number of opioid agonists and antagonists have been applied in the treatment of many diseases.16–18 Among them, MOR modulators with different functions and selectivities have gained significant attention for treating opioid use disorders (OUD).19,20As shown in Fig. 1, INTA was identified as a MOR agonist while NAN, with a structure similar to INTA, was identified as a MOR antagonist. To understand how introduction of an indole moiety with an α or β linkage at the C6 position of the epoxymorphinan skeleton would influence ligand function, our group previously conducted structural alteration on the “address” moiety of the MOR agonist INTA. By changing the substitution position on the indole ring or extending the length of the alkyl chain on the indole group of INTA, 6α/6β indolylacetamidonaltrexamine derivatives were identified as bitopic MOR modulators.21 This SAR study indicated that functional conversion between INTA (MOR agonist) and NAN (MOR antagonist) may be governed by substitution positions on the indole moiety and/or the α/β configurations, resulting in varying interactions with different domains of the MOR allosteric binding site. Furthermore, we applied the concept of “bivalent bioisostere” for structural modification of NAN and INTA, by replacing the –NH– moiety in the indole ring with –O– or –S–, and obtained two series of novel ligands bearing benzofuran or benzothiophene moieties. Interestingly, NBF bearing the 7′-benzofuryl moiety and a 6β-configuration was identified as a potent MOR partial agonist in vitro in MOR radioligand binding (Ki = 0.18 nM) and [35S]GTPγS functional assays (Emax = 27.31% of DAMGO) and in vivo (anti-analgesic potency, AD50 = 0.18 mg kg−1), while its α-configuration counterpart (compound 1) had a similar binding affinity (Ki = 0.28 nM) and with even lower efficacy (Emax = 12.89% of DAMGO) in vitro and showed only a marginal impact on the antinociceptive effect of morphine in vivo.22 Based on these observations we hypothesized that the α-configuration at the C6 of the epoxymorphinan may favor MOR antagonists, while the β-configuration seems to favor MOR agonists. However, as observed in the previous cases, such as 17-cyclopropylmethyl-3,14β-dihydroxy-4,5α-epoxy-6β-(4′-pyridylcarboxamido) morphinan (NAP), which bears a C6 β-configuration but acts as a MOR antagonist,23,24 the structure–activity relationship of the C6 stereochemistry may not be conclusive. Thus we decided to conduct structural modification of the 3′-position of the benzofuran ring of NBF with 6α/6β configurations, to further study the effect of substitutions on the distinct configurations and how they affect the MOR function. In order to study the substitution effects on the benzofuran moiety, we introduced various substitutions that affected the electronic and steric properties, including bromide, chloride and methyl groups, and the bulkier groups such as phenyl and pyridine. In vitro and in vivo pharmacological studies were conducted for the synthesized NBF derivatives to characterize their binding affinity and functional properties at the MOR, as well as their potency and efficacy compared to NBF and compound 1. Besides, computational studies provided a detailed depiction of the interaction of the synthesized ligands with the MOR binding site. These help comprehend how the substitutions with a C6 α/β-configuration complementarily contribute to a dominant conformation for the cyclohexane ring in the epoxymorphinan skeleton to achieve the functional conversion between MOR antagonists and agonists.
 | ||
| Fig. 1 Chemical structures of MOR-targeted agonist INTA, antagonist NAN, partial agonist NBF and its 6α-counterpart (compound 1). | ||
Results and discussion
Chemistry
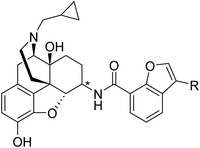
All newly designed compounds were synthesized according to previously reported procedures.25 Briefly, 6α- and 6β-naltrexamines (NTAs) were prepared by stereoselective reductive amination of naltrexone with benzylamine and dibenzylamine, followed by catalytic hydrogenation under acidic conditions, with yields of 75% and 50%, respectively.22 Various commercially available 3-substituted-benzofuran-7-carboxylic acids were then coupled with 6α- and 6β-naltrexamine using the EDCI/HOBt coupling method. 6-position monosubstituted free bases were then obtained in reasonable yields by treating them with K2CO3 in methanol (Scheme 1). These final compounds were obtained in moderate to high yields (38–83%), then converted to their hydrochloric acid salt forms, fully characterized, and applied for in vitro and in vivo pharmacological characterization.In vitro radioligand binding assay
The binding affinity of all synthesized ligands on the three opioid receptors was determined using competitive radioligand binding assays. Following previously reported protocols,21,26,27 opioid receptor-expressing Chinese hamster ovary (CHO) cell membranes were used, where the MOR was labeled with [3H]naloxone and the KOR and DOR with [3H]diprenorphine. The binding affinity (Ki) and selectivity data for the synthesized NBF derivatives are summarized in Table 1. Almost all compounds showed subnanomolar affinity for the MOR except compounds 5 and 6, which carry the α configuration at the C6 and are substituted with phenyl (Ph, MOR Ki = 1.38 nM) and 4-pyridine (4-Py, MOR Ki = 2.38 nM), respectively. Both 6α and 6β isomers retained high MOR binding affinity while showing improvement in KOR and DOR binding. 4-Pyridyl and phenyl groups, however, are the most promising substitutions for improving MOR binding affinity in the 6β configuration. 4-Pyridyl, in particular, appears to be the most selective and effective in increasing binding affinity at the MOR. For methyl, bromine, and chlorine substitutions, improving binding at the MOR seems to be more beneficial for KOR and DOR binding. In general, the 6β configuration seems to be beneficial across substitutions for enhancing MOR binding, with 4-pyrindyl and phenyl substitutions standing out for their more significant improvements in this regard.| Compound | α/β | R | Ki (nM) | Selectivity | MOR [35S]GTPγS | ||||
|---|---|---|---|---|---|---|---|---|---|
| MOR | KOR | DOR | K/M | D/M | EC50 (nM) | Emaxc (%) | |||
| a The values are the mean ± SEM of three independent experiments.b Data were reported in ref. 23.c Efficacy values (Emax) are expressed in percentage relative to maximal stimulation produced by DAMGO. | |||||||||
| Morphine28 | 34.4 ± 5.1 | 89 ± 17 | |||||||
| 1b | 6α | H | 0.28 ± 0.04 | 5.12 ± 0.96 | 29.8 ± 4.6 | 18 | 106 | 0.97 ± 0.19 | 12.9 ± 1.8 |
| 2 | Br | 0.57 ± 0.09 | 17.3 ± 1.2 | 87.8 ± 5.5 | 30 | 154 | 7.37 ± 2.32 | 16.0 ± 4.0 | |
| 3 | Cl | 0.50 ± 0.06 | 17.1 ± 1.3 | 138 ± 24 | 34 | 277 | 2.65 ± 0.47 | 21.6 ± 0.8 | |
| 4 | Me | 0.67 ± 0.08 | 15.7 ± 1.3 | 87.0 ± 5.3 | 23 | 130 | 4.52 ± 0.39 | 23.5 ± 1.6 | |
| 5 | Ph | 1.38 ± 0.24 | 33.8 ± 5.6 | 238 ± 28 | 24 | 173 | 27.4 ± 5.0 | 17.3 ± 1.0 | |
| 6 | 4-Py | 2.38 ± 0.16 | 8.7 ± 1.5 | 97.7 ± 16.3 | 3.7 | 41 | 12.0 ± 2.1 | 14.8 ± 1.6 | |
| 7, NBFb | 6β | H | 0.18 ± 0.03 | 0.81 ± 0.14 | 17.6 ± 3.2 | 4.5 | 98 | 0.95 ± 0.18 | 27.3 ± 3.3 |
| 8 | Br | 0.20 ± 0.03 | 0.21 ± 0.01 | 5.34 ± 0.99 | 1.0 | 27 | 0.49 ± 0.08 | 42.9 ± 3.9 | |
| 9 | Cl | 0.14 ± 0.02 | 0.16 ± 0.02 | 5.17 ± 1.13 | 1.1 | 37 | 0.35 ± 0.07 | 51.2 ± 3.4 | |
| 10 | Me | 0.23 ± 0.04 | 0.44 ± 0.08 | 13.1 ± 1.1 | 1.9 | 57 | 0.81 ± 0.14 | 32.7 ± 3.3 | |
| 11 | Ph | 0.18 ± 0.02 | 19.7 ± 2.2 | 22.1 ± 3.2 | 110 | 123 | 0.56 ± 0.07 | 8.89 ± 0.72 | |
| 12 | 4-Py | 0.21 ± 0.00 | 7.34 ± 0.95 | 45.0 ± 9.3 | 35 | 213 | 1.20 ± 0.38 | 11.4 ± 0.7 | |
In vitro MOR [35S]GTPγS functional assay
Subsequently, the [35S]GTPγS functional assay was carried out to determine the potency and efficacy of these derivatives at the MOR, and the results were interpreted as potency (EC50) and efficacy (%Emax relative to the full agonist DAMGO). The potency and efficacy results are summarized in Table 1. Not surprisingly, almost all of the 6β compounds showed subnanomolar potency except compound 12, which showed EC50 = 1.20 nM, which is consistent with the high MOR binding affinity, while 6α series compounds showed lower potency at the MOR. Furthermore, regarding MOR efficacy, 6α derivatives 2–6 showed a minimal variation compared to compound 1. Interestingly, 6β derivatives, however, compounds 8–10 with –Br, –Cl and –Me substitutions exhibited a higher efficacy (30–50% Emax of DAMGO), while the phenyl and 4-pyridine substituted derivatives (11 and 12) exhibited a lower efficacy of around 10% for the full MOR agonist DAMGO, which was even lower than that observed with the non-substituted compound 7 (NBF, 27.31%). This suggests that the bulkiness of the 3′-substituent groups may play a role in influencing the function of these MOR ligands. Overall, the in vitro functional studies suggested that the 6α derivatives resulted in lower MOR efficacy, while in the case of the 6β derivatives, their MOR functional efficacy varied depending on the substituents. In other words, the 6β derivatives showed a trend based on the bulkiness increment of 3′-substitutions, where the MOR agonism activity initially increased and then decreased accordingly, i.e. NBF with no substituion on the 3′ position exhibited 27.31% efficacy for DAMGO, and the NBF derivatives showed moderate efficacy with small substitution groups such as bromine (42.89%), chlorine (51.22%) and methyl (32.69%), finally dramatically returning back to lower efficacy with bulky group substituents such as phenyl (8.89%) and pyridine (11.36%).Warm-water tail immersion assay
To corroborate the findings of radioligand binding and MOR [35S]GTPγS functional assay, warm-water tail immersion assay was then conducted as described previously to evaluate the antinociceptive ability of NBF derivatives or their ability to antagonize the antinociception of morphine in vivo.24 The results were interpreted as the percentage of the maximum possible effect (% MPE). A higher % MPE indicates a stronger antinociception effect of the ligand.It was observed that compared to the vehicle (sterile-filtered distilled/deionized water), compounds 8, 9, and 10 showed significant antinociception at 10 mg kg−1, indicating that they may behave as agonists for opioid receptors (%MPE as 90.9%, 84.5% and 54.2% respectively), while the rest of the compounds showed no significant activities, consistent with their efficacy in vitro (Fig. 2). Here, the bulkiness effect elicited by the different sizes of substitution groups is once again worthy of attention.
To further study the pharmacological activity of the compounds that did not show antinociception effect, we tested their ability to reverse the antinociceptive effects of morphine at 10 mg kg−1. Collectively, the 6α series derivatives except compounds 1 and 5 antagonized the antinociceptive effects of morphine (Fig. 3). Taking the high in vitro MOR binding affinity and a low efficacy in the [35S]GTPγS functional assay into account, the observed lack of ability of compound 1 to inhibit morphine's antinociception effect could be due to compound 1 acting as an opioid partial agonist as in shown Fig. 2. Considering the relatively lower binding affinity of compound 5 (∼1.4 nM) compared to its counterpart compound 11 (0.18 nM), it seemed to be in line with its lower potency in vivo reflected by its significantly high AD50 (10.4 mg kg−1) compared to compound 11 (2.59 mg kg−1). Moreover, in 6β series, both compounds 11 and 12 showed considerable antagonism activity, which was stronger than that of the corresponding 6α ones. Overall, these studies suggested that both the C6 configuration and 3′-substitutions may play a vital role collectively in affecting the function of ligands at the MOR.
Next, we evaluated the in vivo potency of the three agonists i.e. compounds 8, 9, and 10. As shown in Table 2, the half-maximal effective dose (ED50) of compounds 8, 9 and 10 was determined as 0.94, 0.58 and 11.66 mg kg−1, respectively. Interesingly, the halide substituted compounds 8 and 9 in the 6β configuration showed over a ten-fold higher antinociceptive potency compared to the methyl substituted compound 10. It is to be noted that compounds 8 and 9 exhibited a higher MOR efficacy than the methyl substituted 10 (Table 1).
| Compound | α/β | R | Potency (mg kg−1, 95%CL) | |
|---|---|---|---|---|
| AD50 | ED50 | |||
| a The values are the mean ± SEM of three independent experiments.b Data were reported in ref. 23.NA = not applicable, ND = not determined. | ||||
| Morphine29 | NA | 2.34 (1.57–3.50) | ||
| 1b | 6α | H | ND | ND |
| 2 | Br | 2.23 (1.32–3.76) | NA | |
| 3 | Cl | 2.83 (1.03–7.78) | NA | |
| 4 | Me | 1.88 (1.45–2.42) | NA | |
| 5 | Ph | 10.36 (4.65–23.06) | NA | |
| 6 | 4-Py | 2.31 (1.24–4.32) | NA | |
| 7, NBFb | 6β | H | 0.18 (0.03–0.97) | NA |
| 8 | Br | NA | 0.94 (0.65–1.36) | |
| 9 | Cl | NA | 0.58 (0.23–1.47) | |
| 10 | Me | NA | 11.66 (9.09–14.95) | |
| 11 | Ph | 2.59 (1.88–3.56) | NA | |
| 12 | 4-Py | 3.04 (1.65–5.61) | NA | |
We then tested the antagonistic potency of the remaining compounds against 10 mg kg−1 morphine (Table 2). As shown in Table 2, AD50 means the half antagonistic effect dose against morphine's antinociception, while the EC50 means half maximal effective dose for antinociception. In the 6α series, except for compound 5 (AD50 = 10.36 mg kg−1), the remaining compounds showed single digit molar potency (AD50 of compounds 2, 3, 4 and 6 = 2.23, 2.83, 1.88, 2.31 mg kg−1). These results are also consistent with the trends observed in in vitro MOR binding and functional assay results. In addition, the 6β compounds 11 and 12 exhibited a similar degree of antagonism against morphine.
Molecular modelling studies
To further understand how structural modifications of these compounds influence functional activity at the MOR, molecular modeling studies were conducted. Initially, compounds 2–6, 11, and 12 were docked into the inactive MOR (PDB ID: 4DKL),30 while the agonists/partial agonists, compounds 8–10, were docked into the active MOR (PDB ID: 5C1M)31 (Fig. 4). The highest-ranking docking solutions, identified using the CHEM-PLP scoring function, were selected as the optimal binding conformations. | ||
| Fig. 4 Binding mode of (A) compounds 2, 3, and 4 with the inactive MOR receptor; (B) compounds 5 and 6 with the inactive MOR receptor; (C) compounds 8, 9, and 10 with the active MOR receptor; and (D) compounds 11 and 12 with the inactive MOR receptor (PDB; 4DKL). The MOR is shown as grey cartoons. NBF derivatives and key amino acid residues are shown on the sticks. Carbon atoms: 2, 8, (magenta); 3, 9, (cyan); 4, 10 (blue); 5, 11 (green); 6, 12 (orange); key amino acid residues (yellow); oxygen atoms (red); and nitrogen atoms (blue). | ||
The epoxymorphinan moiety of all compounds bound to the MOR orthosteric binding site, establishing direct interactions with key residues, including ASP147, TYR148, MET151, HIS297, TRP293, and TRP318, and TYR326, similar to other known opioid ligands. Specifically, the quaternary ammonium nitrogen of the epoxymorphinan formed an ionic interaction with ASP147, while the dihydrofuran oxygen formed hydrogen bond interactions with TYR148 (Fig. 4).8,30,31
The substituted benzofuran portions of the compounds extended towards the allosteric binding sites of the MOR driving the functional profile of the compounds (Fig. 4).30–33 The cyclohexyl ring of 6α compounds adopted a twisted chair conformation, enabling the binding of the benzofuran side chain with the inactive MOR allosteric site (Fig. S2A, ESI†). We have shown previously that the chair conformation of the NAN, an epoxymorphinan antagonist ligand, enables the amide side chain to interact with the inactive allosteric site of the MOR.21 Compounds 2–6 bound to the allosteric binding site in the inactive MOR and showed hydrophobic interactions with the key residues THR218, LEU219, PHE221, GLU229, LEU232, LYS233, VAL236, and VAL300 (Fig. 4A and B). These interactions have been previously shown to stabilize the inactive state of the MOR.21,30,32
To accommodate the 6β configuration of the benzofuran side chain, the cyclohexyl ring in compounds 8–12 adopted a twisted boat conformation (Fig. S2B, ESI†). This conformational arrangement enabled compounds with halogen or methyl substituents to form hydrophobic interactions with the active MOR allosteric site through residues GLN124, TRP133, and ILE144 in TM2, TM3, and ECL1 (Fig. 4C).21,31,34 Compounds 11 and 12 however displayed distinct binding characteristics, with their address portion shifting toward TM6 and TM7, forming hydrophobic interactions with TRP318 and potential hydrogen bonding interactions between the amide oxygen and LYS233 in the allosteric binding site of the inactive MOR.29,35 It was observed that the distance between the small substituents of benzofuran and amino residues at the active state binding site was relatively constrained, measuring 3.4 Å and 3.6 Å for ASN127 and TRP133 respectively (Fig. 4C). This tight spatial arrangement suggests limited accommodation for bulky substituents as seen in compounds 11 and 12, which could explain the orientation of the benzofuran ring towards TM6 and TM7, resulting in enhanced hydrophobic and hydrogen-bonding interactions (Fig. 4D). Consequently, the hydrophobic interactions with TM2 and TM3 were comparatively weaker than those with TM6 and TM7. Previous studies have also demonstrated that MOR antagonists such as NAP and NAQ exhibit similar interactions, with TRP318 mutation reducing MOR binding affinity.33 Additionally, the interaction between β-FNA, a known MOR antagonist, and LY233 in the allosteric binding site was shown to be critical in the MOR inactivation.30 These enhanced interactions could explain the 10-fold increase in the binding affinities, higher potency, and lower efficacy of compounds 11 and 12 compared to 5 and 6 respectively (Fig. 4D).
While the docking results provide some insights into the potential binding modes of ligands at the receptor, they offer a limited understanding of the dynamics factors involved in ligand–protein interactions and the stability of these interactions. Therefore, the docking poses of compounds 3 and 9 in their respective receptor constructs were chosen for molecular dynamics (MD) simulations to explore the underlying mechanism of the observed difference in their efficacy. The two systems achieved equilibrium after 90 ns MD simulation with RMSD values of protein backbones below 3 Å throughout the simulation period, indicating a stable receptor–ligand complex (Fig. S1, ESI†).36
The binding mode of the message part of compound 3 and 9 after MD simulations was similar to their binding modes from the molecular docking studies (Fig. 5), that is, the address moiety of compound 3 extended into TM4 and TM5 forming hydrophobic interactions with LEU232, LYS233, and VAL236. Moreover, the benzofuran side chain appeared to be shifted slightly towards TM7 forming hydrophobic interactions with TRP318 (Fig. 5A). On the other hand, the chloro substituent on the benzofuran ring of compound 9 slightly extended towards TM2 of the active MOR where it showed hydrophobic interactions with Ile144 of the allosteric binding site (Fig. 5B). This interaction might be responsible for the activation of MOR.34
In conclusion, molecular modeling studies demonstrated how the C6 configuration and substituent size may influence the MOR function of the NBF derivatives. Relatively smaller substituents (Cl, Br, and Me) produced opposite effects depending on the configuration: 6β derivatives promoted partial agonism through a twisted boat conformation, favoring receptor activation, while 6α ones retained antagonist activity via a chair conformation that stabilizes the inactive state. In contrast, bulky substituents (Ph and 4-Py) preserved antagonist activity regardless of the configuration by favoring inactive state binding modes. These structure–function relationships offer insight into developing MOR ligands with specific functional profiles.
Conclusion
In summary, a series of NBF derivatives were synthesized to investigate the effect of C6 configuration and 3′-benzofuran substitution on the MOR functional activity. This work resulted in novel ligands with high binding affinity and differentiated functional activity profiles at the MOR. In vitro and in vivo studies revealed that the 6α configuration preserved the MOR antagonist effect irrespective of the substitution at 3′-benzofuran. In the case of 6β configuration, compounds with small substituents like chloro, bromo or methyl at 3′-benzofuran demonstrated an agonistic effect at the MOR, while compounds with larger or bulky substituents like phenyl and pyridine resulted in a switch in their pharmacological activity demonstrating the antagonistic effect of the NBF. In vivo studies verified this observation that 3′-benzofuran substitution in the case of 6β conformation of NBF yields diversed pharmacological profiles, with compounds 8, 9 and 10 exhibiting the antinociceptive effect comparable with morphine, while their counterparts reversed the antinociceptive effect of morphine. Molecular modeling studies revealed how the C6 configuration and benzofuran modifications collectively influence ligand conformation at the MOR receptor, driving distinct pharmacological profiles. These structure–activity relationship insights help inform future MOR modulator design and development.Methods
Chemistry
General procedure A for the synthesis of 2–6 and 8–12
N-(3-Dimethylaminopropyl)-N′-ethylcarbo-diimide hydrochloride (2.5 equiv.), hydrobenzotriazole (2.5 equiv.), 4 Å molecular sieves, and trimethylamine (5 equiv.) were added to a solution of carboxylic acid (2.5 equiv.) in DMF on an ice-water bath under N2 protection. After 30 min, a solution of 6α/6β-naltrexamine (1 equiv.) in DMF was added dropwise. The mixture was kept stirring overnight at room temperature and filtered the next day. The filtrate was then concentrated under reduced pressure to remove the solvent. The residue was dissolved in MeOH, and potassium carbonate (2 equiv.) was added to the mixture. The reaction mixture was stirred overnight at room temperature. Next day, the mixture was concentrated, and the residue was purified with a silica gel column using methanol/dichloromethane (0.1% NH3·H2O) as the solvent system to obtain the target compounds.1H NMR (400 MHz, DMSO-d6) δ 9.21 (s, 1H), 8.90 (s, 1H), 8.38 (s, 1H), 8.01 (d, J = 8.1 Hz, 1H), 7.87 (d, J = 7.5 Hz, 1H), 7.75 (d, J = 7.8 Hz, 1H), 7.51 (t, J = 7.7 Hz, 1H), 6.72 (d, J = 8.1 Hz, 1H), 6.60 (d, J = 8.1 Hz, 1H), 6.39 (s, 1H), 4.84 (d, J = 4.1 Hz, 1H), 4.72–4.67 (m, 1H), 3.96 (d, J = 6.7 Hz, 1H), 3.32–3.23 (m, 3H), 3.10 (dd, J = 19.8, 7.2 Hz, 2H), 2.99–2.95 (m, 1H), 2.75–2.70 (m, 1H), 2.54–2.51 (m, 1H), 1.94 (dd, J = 16.3, 8.0 Hz, 1H), 1.68–1.64 (m, 2H), 1.48 (dd, J = 14.9, 9.1 Hz, 1H), 1.05–1.02 (m, 1H), 0.73–0.70 (m, 1H), 0.66–0.60 (m, 1H), 0.52–0.48 (m, 1H), 0.43–0.40 (m, 1H). 13C NMR (100 MHz, DMSO-d6) δ 162.97, 151.12, 146.16, 144.84, 139.47, 129.27, 127.88, 126.66, 124.45, 123.09, 122.54, 119.86, 118.81, 97.99, 87.57, 69.80, 61.52, 46.31, 45.76, 30.51, 29.60, 23.95, 20.53, 6.18, 5.64, 3.07. HRMS calcd for C29H30N2O5Br [M + H]+: 565.1333. Found: 565.1336. HPLC 97.10% at R.T. = 6.52 min.
1H NMR (400 MHz, DMSO-d6) δ 9.24 (s, 1H), 8.91 (s, 1H), 8.05 (d, J = 8.1 Hz, 1H), 7.87 (dd, J = 7.5, 0.9 Hz, 1H), 7.84 (dd, J = 7.8, 1.1 Hz, 1H), 7.52 (t, J = 7.7 Hz, 1H), 6.73 (d, J = 8.1 Hz, 1H), 6.61 (d, J = 8.1 Hz, 1H), 6.41 (s, 1H), 4.84 (d, J = 4.2 Hz, 1H), 4.72–4.64 (m, 1H), 3.96 (d, J = 6.7 Hz, 1H), 3.31–3.26 (m, 2H), 3.14–2.96 (m, 3H), 2.77–2.66 (m, 1H), 2.60–2.52 (m, 1H), 1.98–1.91 (m, 1H), 1.69–1.65 (m, 2H), 1.48 (dd, J = 14.6, 9.4 Hz, 1H), 1.09–0.98 (m, 2H), 0.72–0.68 (m, 1H), 0.66–0.63 (m, 1H), 0.52–0.48 (m, 1H), 0.43–0.40 (m, 1H). 13C NMR (100 MHz, DMSO-d6) δ 162.47, 150.45, 145.66, 142.52, 138.96, 128.76, 126.21, 125.95, 123.94, 122.05, 121.80, 119.54, 119.38, 118.28, 111.86, 87.06, 69.29, 64.88, 56.99, 45.81, 45.24, 30.01, 29.10, 23.44, 20.00, 5.68, 5.16, 2.56. HRMS calcd for C29H30N2O5Cl [M + H]+: 521.1838. Found: 521.1826. HPLC 95.65% at R.T. = 5.73 min.
1H NMR (400 MHz, DMSO-d6) δ 9.30 (s, 1H), 8.90 (s, 1H), 8.03 (d, J = 8.2 Hz, 1H), 7.87 (d, J = 1.3 Hz, 1H), 7.84 (s, 1H), 7.82 (s, 1H), 7.41 (t, J = 7.6 Hz, 1H), 6.74 (d, J = 8.1 Hz, 1H), 6.62 (d, J = 8.1 Hz, 1H), 6.36 (s, 1H), 4.84 (d, J = 4.1 Hz, 1H), 4.74–4.66 (m, 1H), 3.95 (d, J = 6.4 Hz, 1H), 3.31–3.26 (m, 1H), 3.15–3.05 (m, 2H), 2.98–2.93 (m, 1H), 2.77–2.68 (m, 1H), 2.60–2.52 (m, 2H), 2.27 (d, J = 1.2 Hz, 3H), 1.99–1.90 (m, 1H), 1.70–1.66 (m, 2H), 1.52–1.47 (m, 1H), 1.09–0.96 (m, 2H), 0.73–0.67 (m, 1H), 0.65–0.63 (m, 1H), 0.52–0.49 (m, 1H), 0.43–0.40 (m, 1H). 13C NMR (100 MHz, DMSO-d6) δ 162.69, 151.31, 145.60, 142.42, 139.03, 129.56, 128.82, 124.97, 123.29, 122.68, 122.04, 119.45, 118.29, 117.95, 115.66, 87.22, 69.26, 64.89, 61.01, 57.00, 45.59, 45.25, 30.00, 29.19, 23.43, 20.27, 7.45, 5.66, 5.14, 2.55. HRMS calcd for C30H33N2O5 [M + H]+: 501.2384. Found: 501.2402. HPLC 98.08% at R.T. = 6.25 min.
1H NMR (400 MHz, DMSO-d6) δ 9.11 (s, 1H), 8.73 (s, 1H), 8.30 (s, 1H), 8.00–7.90 (m, 2H), 7.73 (d, J = 7.4 Hz, 1H), 7.63 (d, J = 7.1 Hz, 2H), 7.41 (t, J = 7.6 Hz, 2H), 7.37–7.27 (m, 2H), 6.59 (d, J = 8.1 Hz, 1H), 6.48 (d, J = 8.0 Hz, 1H), 6.19 (s, 1H), 4.72 (d, J = 3.7 Hz, 1H), 4.59–4.56 (m, 1H), 3.79 (d, J = 6.2 Hz, 1H), 3.15–3.10 (m, 2H), 2.99–2.95 (m, 2H), 2.88–2.77 (m, 2H), 2.64–2.61 (m, 1H), 2.43–2.40 (m, 1H), 1.84–1.75 (m, 1H), 1.56–1.49 (m, 2H), 1.41–1.33 (m, 1H), 0.94–0.91 (m, 1H), 0.56–0.52 (m, 1H), 0.51–0.48 (m, 1H), 0.37–0.34 (m, 1H), 0.0.29–0.26 (m, 1H). 13C NMR (100 MHz, DMSO-d6) δ 162.77, 151.94, 145.67, 142.90, 139.02, 130.72, 129.16, 128.81, 127.85, 127.30, 126.65, 125.34, 123.71, 123.53, 122.05, 121.41, 119.44, 118.96, 118.32, 87.19, 69.31, 64.87, 61.07, 57.05, 45.74, 45.29, 30.91, 30.65, 30.02, 29.16, 5.67, 5.14, 2.57. HRMS calcd for C35H35N2O5 [M + H]+: 563.2540. Found: 563.2535. HPLC 99.54% at R.T. = 7.23 min.
1H NMR (400 MHz, DMSO-d6) δ 9.25 (s, 1H), 9.05 (s, 1H), 8.94–8.88 (m, 3H), 8.35–8.30 (m, 3H), 8.13 (d, J = 7.6 Hz, 1H), 7.91 (d, J = 7.5 Hz, 1H), 7.59 (t, J = 7.5 Hz, 1H), 6.75 (d, J = 7.8 Hz, 1H), 6.62 (d, J = 7.8 Hz, 1H), 6.42 (s, 1H), 4.87 (d, J = 3.7 Hz, 1H), 4.75–4.70 (m, 1H), 3.98 (d, J = 5.4 Hz, 1H), 3.35–3.28 (m, 2H), 3.09–2.99 (m, 3H), 2.76–2.70 (m, 1H), 2.58–2.53 (m, 1H), 2.02–1.92 (m, 1H), 1.68 (d, J = 10.7 Hz, 2H), 1.50 (dd, J = 13.5, 10.7 Hz, 1H), 1.11–1.05 (m, 2H), 0.73–0.69 (m, 1H), 0.66–0.68 (m, 1H), 0.54–0.51 (m, 1H), 0.45–0.41 (m, 1H). 13C NMR (100 MHz, DMSO-d6) δ 163.20, 152.82, 148.72, 146.23, 139.49, 129.29, 126.50, 125.36, 124.84, 124.18, 123.63, 122.57, 120.33, 119.88, 118.82, 118.50, 87.61, 69.83, 61.51, 49.06, 46.37, 45.78, 30.56, 29.63, 23.98, 20.50, 6.19, 5.66, 3.07. HRMS calcd for C34H34N3O5 [M + H]+: 564.2493. Found: 564.2498. HPLC 95.62% at R.T. = 4.48 min.
1H NMR (400 MHz, DMSO-d6) δ 9.35 (s, 1H), 8.86 (s, 1H), 8.56 (d, J = 8.2 Hz, 1H), 8.45 (s, 1H), 7.79 (d, J = 6.9 Hz, 1H), 7.73 (d, J = 7.8 Hz, 1H), 7.49 (t, J = 7.7 Hz, 1H), 6.74 (d, J = 8.1 Hz, 1H), 6.67 (d, J = 8.1 Hz, 1H), 6.22 (s, 1H), 4.86 (d, J = 7.7 Hz, 1H), 3.87 (d, J = 5.1 Hz, 1H), 3.79–3.75 (m, 1H), 3.38–3.33 (m, 2H), 3.13–3.03 (m, 2H), 2.91–2.82 (m, 1H), 2.47–2.43 (m, 2H), 2.02–1.92 (m, 1H), 1.77 (d, J = 13.9 Hz, 1H), 1.69–1.62 (m, 1H), 1.49–1.40 (m, 2H), 1.07–1.04 (m, 1H), 0.72–0.66 (m, 1H), 0.62–0.58 (m, 1H), 0.54–0.50 (m, 1H), 0.44–0.41 (m, 1H). 13C NMR (100 MHz, DMSO-d6) δ 163.42, 151.10, 144.95, 142.66, 141.82, 130.12, 127.98, 126.30, 124.30, 122.79, 121.11, 120.50, 119.81, 118.44, 97.78, 90.39, 70.22, 57.18, 53.76, 51.76, 46.97, 46.16, 29.93, 27.83, 24.15, 23.50, 6.20, 5.59, 3.11. HRMS calcd for C29H30N2O5Br [M + H]+: 565.1333. Found: 565.1329. HPLC 96.56% at R.T. = 6.26 min.
1H NMR (400 MHz, DMSO-d6) δ 9.38 (s, 1H), 8.88 (s, 1H), 8.60 (d, J = 8.2 Hz, 1H), 8.49 (s, 1H), 7.84–7.78 (m, 2H), 7.50 (t, J = 7.7 Hz, 1H), 6.75 (d, J = 8.1 Hz, 1H), 6.68 (d, J = 8.1 Hz, 1H), 6.24 (s, 1H), 4.86 (d, J = 7.7 Hz, 1H), 3.88 (d, J = 5.1 Hz, 1H), 3.80–3.73 (m, 1H), 3.31–3.29 (m, 2H), 3.14–3.03 (m, 2H), 2.90–2.83 (m, 1H), 2.48–2.42 (m, 2H), 1.96 (q, J = 12.9 Hz, 1H), 1.78 (d, J = 13.7 Hz, 1H), 1.71–1.60 (m, 1H), 1.52–1.37 (m, 2H), 1.07–1.05 (m, 1H), 0.72–0.66 (m, 1H), 0.63–0.58 (m, 1H), 0.54–0.50 (m, 1H), 0.45–0.41 (m, 1H). 13C NMR (100 MHz, DMSO-d6) δ 162.90, 150.42, 142.61, 142.13, 141.31, 129.62, 126.04, 125.85, 123.79, 121.52, 120.62, 120.16, 119.32, 117.92, 111.65, 89.86, 69.70, 61.61, 56.65, 51.25, 46.46, 45.65, 29.41, 27.32, 23.65, 22.98, 5.70, 5.10, 2.59. HRMS calcd for C29H30N2O5Cl [M + H]+: 521.1838. Found: 521.1836. HPLC 95.95% at R.T. = 6.20 min.
1H NMR (400 MHz, DMSO-d6) δ 9.37 (s, 1H), 8.87 (s, 1H), 8.41 (d, J = 8.2 Hz, 1H), 7.93 (d, J = 1.2 Hz, 1H), 7.80 (dd, J = 7.7, 1.1 Hz, 1H), 7.74–7.71 (m, 1H), 7.38 (t, J = 7.7 Hz, 1H), 6.74 (d, J = 8.1 Hz, 1H), 6.67 (d, J = 8.1 Hz, 1H), 6.23 (s, 1H), 4.90 (d, J = 7.8 Hz, 1H), 3.87 (d, J = 4.7 Hz, 1H), 3.81–3.76 (m, 1H), 3.30–3.27 (m, 2H), 3.13–3.03 (m, 2H), 2.89–2.81 (m, 1H), 2.49–2.41 (m, 2H), 2.27 (d, J = 1.1 Hz, 3H), 2.02–1.92 (m, 1H), 1.76 (d, J = 13.9 Hz, 1H), 1.70–1.62 (m, 1H), 1.50–1.40 (m, 2H), 1.08–1.06 (m, 1H), 0.72–0.69 (m, 1H), 0.63–0.60 (m, 1H), 0.54–0.50 (m, 1H), 0.45–0.40 (m, 1H). 13C NMR (100 MHz, DMSO-d6) δ 163.39, 151.29, 142.52, 142.17, 141.30, 129.66, 124.60, 123.17, 122.82, 122.42, 121.94, 120.61, 119.30, 118.83, 117.92, 115.42, 89.99, 69.74, 61.62, 56.66, 51.17, 46.49, 45.69, 29.49, 27.32, 22.97, 7.50, 5.70, 5.08, 2.59. HRMS calcd for C30H33N2O5 [M + H]+: 501.2384. Found: 501.2389. HPLC 96.09% at R.T. = 5.92 min.
1H NMR (400 MHz, DMSO-d6) δ 9.24 (s, 1H), 8.73 (s, 1H), 8.43 (d, J = 8.2 Hz, 1H), 8.38 (s, 1H), 7.96 (d, J = 7.9 Hz, 1H), 7.68–7.62 (m, 3H), 7.42 (t, J = 7.6 Hz, 2H), 7.32 (dt, J = 11.6, 7.6 Hz, 2H), 6.62 (d, J = 8.1 Hz, 1H), 6.55 (d, J = 8.1 Hz, 1H), 6.08 (s, 1H), 4.77 (d, J = 7.8 Hz, 1H), 3.75 (d, J = 4.7 Hz, 1H), 3.70–3.65 (m, 1H), 3.28–3.24 (m, 2H), 3.02–2.90 (m, 2H), 2.74–2.71 (m, 1H), 2.34–2.31 (m, 2H), 1.87 (dd, J = 14.8, 9.8 Hz, 1H), 1.65 (d, J = 13.8 Hz, 1H), 1.56–1.53 (m, 1H), 1.38–1.33 (m, 2H), 0.95–0.93 (m, 1H), 0.60–0.53 (m, 1H), 0.52–0.44 (m, 1H), 0.43–0.36 (m, 1H), 0.34–0.27 (m, 1H). 13C NMR (100 MHz, DMSO-d6) δ 163.36, 151.94, 143.08, 142.21, 141.33, 130.90, 129.67, 129.14, 127.76, 127.25, 126.70, 124.94, 123.33, 121.27, 120.62, 119.78, 119.30, 117.96, 89.98, 69.75, 64.87, 61.67, 51.27, 46.50, 45.70, 27.33, 23.70, 23.01, 5.71, 5.10, 2.62. HRMS calcd for C35H35N2O5 [M + H]+: 563.2540. Found: 563.2534. HPLC 100% at R.T. = 7.09 min.
1H NMR (400 MHz, DMSO-d6) δ 9.37 (s, 1H), 9.09 (s, 1H), 8.93–8.90 (m, 3H), 8.68 (d, J = 7.9 Hz, 1H), 8.33–8.31 (m, 2H), 7.85 (d, J = 7.4 Hz, 1H), 7.57 (t, J = 7.7 Hz, 1H), 6.76 (d, J = 8.0 Hz, 1H), 6.68 (d, J = 8.0 Hz, 1H), 6.30 (s, 1H), 4.91 (d, J = 7.4 Hz, 1H), 3.91 (d, J = 4.7 Hz, 1H), 3.82–3.80 (m, 1H), 3.34–3.29 (m, 2H), 3.14–3.01 (m, 3H), 2.91–2.88 (m, 1H), 2.45–2.42 (m, 1H), 2.01 (d, J = 13.1 Hz, 1H), 1.81 (d, J = 12.9 Hz, 1H), 1.72–1.68 (m, 1H), 1.47–1.42 (m, 2H), 1.12–1.14 (m, 1H), 0.72–0.69 (m, 1H), 0.63–0.61 (m, 1H), 0.54–0.51 (m, 1H), 0.46–0.42 (m, 1H). 13C NMR (100 MHz, DMSO-d6) δ 163.60, 152.82, 148.72, 145.50, 145.30, 145.22, 142.67, 141.83, 130.16, 126.11, 125.48, 124.69, 123.94, 123.88, 123.47, 121.14, 120.91, 119.81, 118.45, 117.80, 90.40, 70.23, 62.13, 57.19, 51.82, 46.99, 46.18, 29.94, 27.85, 24.20, 23.51, 6.22, 5.61, 3.12. HRMS calcd for C34H34N3O5 [M + H]+: 564.2493. Found: 564.2473. HPLC 95.15% at R.T. = 4.53 min.
Competitive radioligand binding and [35S]GTPγS functional studies
In the competition binding assay, 30 μg of membrane protein was incubated with the corresponding radioligand in the presence of different concentrations of test compounds in TME buffer (50 mM Tris, 3 mM MgCl2, and 0.2 mM EGTA, pH = 7.4). The bound radioligand was separated by filtration using the Brandel harvester. Specific (i.e., opioid receptor-related) binding at the MOR, KOR, and DOR was determined as the difference in binding obtained in the absence and presence of 5 μM naltrexone, U50,488, and SNC80, respectively. The IC50 values were determined and converted to Ki values using the Cheng–Prusoff equation. In the [35S]GTPγS functional assay, 10 μg of MOR-CHO membrane protein was incubated with 10 μM GDP, 0.1 nM [35S]GTPγS, assay buffer (TME + 100 mM NaCl) and varying concentrations of the compounds under investigation for 90 min in a 30 °C water bath. Nonspecific binding was determined using 20 μM unlabeled GTPγS. 3 μM DAMGO was included in the assay as maximally effective concentration of a full agonist for the MOR. All assays were determined in duplicate and repeated at least 4 times. Percent DAMGO-stimulated [35S]GTPγS binding was defined as (net-stimulated binding by ligand/net-stimulated binding by 3 μM DAMGO) × 100.Data analysis of [35S]GTPγS binding assays
All samples were assayed in duplicate and the assay was repeated at least four times for a total of ≥4 independent determinations. Results were reported as mean values ± SEM. Concentration-effect curves were fit by nonlinear regression to a one-site binding model, using GraphPad Prism software, to determine EC50 and Emax values. IC50 values were obtained from Hill plots and analyzed by linear regression using GraphPad Prism software. Binding Ki values were determined from IC50 values using the Cheng–Prusoff equation: Ki = IC50/[1 + ([L]/KD)], where [L] is the concentration of competitor and KD is the KD of the radioligand.Warm water tail immersion test
Swiss Webster mice (5 male mice for each group, 7–8 weeks, ENVIGO) were used for this experiment. All test compounds including morphine were dissolved in sterile-filtered distilled/deionized water as the vehicle. Water bath temperature was maintained at 56 ± 0.1 °C. The baseline latency (control) was determined before the test compound was injected subcutaneously (s.c.) into the mice. The average baseline latency obtained for this experiment was 3.0 ± 0.1 s, and only mice with a baseline latency of 2–4 s were used. For the agonism study, tail immersion was conducted 20 min (time that morphine's anti-nociceptive effect starts to peak) after the test compound was injected. To prevent tissue damage, a 10 s maximum cutoff time was imposed. Antinociception response was calculated as the percentage maximum possible effect (%MPE), where %MPE = [(test − control)/(10 − control)] × 100. For the antagonism study, the test compound was given 5 min before morphine. Tail immersion was then conducted 20 min after giving morphine. %MPE was calculated for each mouse using at least five mice per drug. AD50 values were calculated using the least-squares linear regression analysis followed by calculation of 95% confidence interval by the Bliss method.Statistical analysis
One-way ANOVA followed by the posthoc Dunnett test was performed to assess significance using Prism 6.0 software (GraphPad Software, San Diego, CA).Molecular modeling study
The crystal structures of the antagonist-bound MOR (PDB ID: 4DKL)29 and agonist-bound MOR (PDB ID: 5C1M)31 were downloaded from the Protein Data Bank (https://www.rcsb.org) and prepared for docking by adding hydrogen atoms and removing water molecules as well as the bound ligands and modeling the missing residues in ICL-3 of 4DKL. Compounds were sketched in Sybyl-X 2.0. Each compound was energy minimized (10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 iterations) to a gradient of 0.05 with Gasteiger–Huckel charges assigned under the Tripos force field (TFF). The genetic algorithm docking program GOLD 2020 was used to conduct the docking studies.37 The atoms within 10 Å of the γ-carbon atom of ASP147 in both crystal structures were used to define the binding site. The distance constraint of 4 Å between the piperidine quaternary ammonium nitrogen atom of the compounds’ epoxymorphinan nucleus and ASP147 and the hydrogen bond between the compounds’ dihydrofuran oxygen atom and the phenolic oxygen atom of TYR148 were set up during the automated docking processes. The highest-scored solutions were selected and merged into the respective analyzed protein conformation using the PyMOL Molecular Graphics System. Molecular dynamics (MD) simulations were performed using the Amber 2020 package.38 Initially, the membrane system was constructed using the CHARMGUI web service, incorporating POPC lipids, a TIP3P water box, and 0.15 M sodium and chloride ions, which form a complex with the protein and ligand.39,40 A 90 ns MD simulation was then carried out on the system under the NPT ensemble (P = 1 atm and T = 310 K) with periodic boundary conditions. Temperature was maintained using the Langevin thermostat, and long-range electrostatic interactions were calculated using the particle mesh ewald (PME) method.41 Non-bonded van der Waals interactions were truncated at 10 Å. Following the simulations, the MD trajectories were further analyzed using the visual molecular dynamic (VMD) software.42
000 iterations) to a gradient of 0.05 with Gasteiger–Huckel charges assigned under the Tripos force field (TFF). The genetic algorithm docking program GOLD 2020 was used to conduct the docking studies.37 The atoms within 10 Å of the γ-carbon atom of ASP147 in both crystal structures were used to define the binding site. The distance constraint of 4 Å between the piperidine quaternary ammonium nitrogen atom of the compounds’ epoxymorphinan nucleus and ASP147 and the hydrogen bond between the compounds’ dihydrofuran oxygen atom and the phenolic oxygen atom of TYR148 were set up during the automated docking processes. The highest-scored solutions were selected and merged into the respective analyzed protein conformation using the PyMOL Molecular Graphics System. Molecular dynamics (MD) simulations were performed using the Amber 2020 package.38 Initially, the membrane system was constructed using the CHARMGUI web service, incorporating POPC lipids, a TIP3P water box, and 0.15 M sodium and chloride ions, which form a complex with the protein and ligand.39,40 A 90 ns MD simulation was then carried out on the system under the NPT ensemble (P = 1 atm and T = 310 K) with periodic boundary conditions. Temperature was maintained using the Langevin thermostat, and long-range electrostatic interactions were calculated using the particle mesh ewald (PME) method.41 Non-bonded van der Waals interactions were truncated at 10 Å. Following the simulations, the MD trajectories were further analyzed using the visual molecular dynamic (VMD) software.42
Author contributions
EL and HM performed the chemical synthesis supervised by YZ. SW and JCG performed the cell assays supervised by DES and WLD. PPP performed the animal studies. AR performed the computational studies. The manuscript was prepared by EL, AR and PPP and reviewed by all authors.Data availability
The data supporting this article have been included as part of the ESI.†Conflicts of interest
There are no conflicts to declare.Acknowledgements
This work is partially supported by NIH/NIDA grants UG3DA054785 and UG3/UH3DA050311. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institute on Drug Abuse or the National Institutes of Health.References
- A. H. Beckett and A. F. Casy, J. Pharm. Pharmacol., 1954, 6, 986–1001 CrossRef CAS PubMed.
- P. S. Portoghese, J. Med. Chem., 1965, 8, 609–616 CrossRef CAS PubMed.
- L. Terenius, Acta Pharmacol. Toxicol., 1973, 32, 317–320 CrossRef CAS PubMed.
- E. J. Simon, J. M. Hiller and I. Edelman, Proc. Natl. Acad. Sci. U. S. A., 1973, 70, 1947–1949 CrossRef CAS PubMed.
- C. B. Pert and S. H. Snyder, Science, 1973, 179, 1011–1014 CrossRef CAS PubMed.
- S. J. Paterson, L. E. Robson and H. W. Kosterlitz, Br. Med. Bull., 1983, 39, 31–36 CrossRef CAS PubMed.
- G. W. Pasternak, Neuropharmacology, 2014, 76, 198–203 CrossRef CAS PubMed.
- Y. Shang and M. Filizola, Eur. J. Pharmacol., 2015, 763, 206–213 CrossRef CAS PubMed.
- Y. Xia and G. G. Haddad, Brain Res., 1991, 549, 181–193 CrossRef CAS PubMed.
- G. Wittert, P. Hope and D. Pyle, Biochem. Biophys. Res. Commun., 1996, 218, 877–881 CrossRef CAS PubMed.
- D. M. Rosenbaum, S. G. F. Rasmussen and B. K. Kobilka, Nature, 2009, 459, 356–363 CrossRef CAS PubMed.
- H. W. D. Matthes, R. Maldonade, F. Simonin, O. Valverde, S. Slowe, I. Kitchen, K. Befort, A. Dierich, M. L. Meur, P. Dolle, E. Tzavara, J. Hanoune, B. P. Roques and B. L. Kieffer, Nature, 1996, 383, 819–823 CrossRef CAS PubMed.
- A. H. Li and H. Wang, J. Neurochem., 2001, 77, 435–444 CrossRef CAS PubMed.
- B. L. Roth, K. Baner, R. Westkaemper, D. Siebert, K. C. Rice, S. Steinberg, P. Ernsberger and R. B. Rothman, Proc. Natl. Acad. Sci. U. S. A., 2002, 99, 11934–11939 CrossRef CAS PubMed.
- A. Saitoh, Y. Yoshikawa, K. Onodera and J. Kamei, Psychopharmacology, 2005, 182, 327–334 CrossRef CAS PubMed.
- D. M. Zimmerman and J. D. Leander, J. Med. Chem., 1990, 33, 895–902 CrossRef CAS PubMed.
- M. Eguchi, Med. Res. Rev., 2004, 24, 182–212 CrossRef CAS PubMed.
- Y. Feng, X. He, Y. Yang, D. Chao, L. H. Lazarus and Y. Xia, Curr. Drug Targets, 2012, 13, 230–246 CrossRef CAS PubMed.
- J. C. Veilleux, P. J. Colvin, J. Anderson, C. York and A. J. Heinz, Clin. Psychol. Rev., 2010, 30, 155–166 CrossRef PubMed.
- G. Bart, J. Addict. Dis., 2012, 31, 207–225 CrossRef PubMed.
- S. Obeng, H. Wang, A. Jali, D. L. Stevens, H. I. Akbarali, W. L. Dewey, D. E. Selley and Y. Zhang, ACS Chem. Neurosci., 2019, 10, 1075–1090 CrossRef CAS PubMed.
- H. Ma, S. Obeng, H. Wang, Y. Zheng, M. Li, A. M. Jali, D. L. Stevens, W. L. Dewey, D. E. Selley and Y. Zhang, J. Med. Chem., 2019, 62, 11399–11415 CrossRef CAS PubMed.
- P. P. Pagare, M. Li, Y. Zheng, A. S. Kulkarni, S. Obeng, B. Huang, C. Ruiz, J. C. Gillespie, R. E. Mendez, D. L. Stevens, J. L. Poklis, M. S. Halquist, W. L. Dewey, D. E. Selley and Y. Zhang, J. Med. Chem., 2022, 65, 5095–5112 CrossRef CAS PubMed.
- H. Ma, H. Wang, J. C. Gillespie, R. E. Mendez, D. E. Selley and Y. Zhang, Bioorg. Med. Chem. Lett., 2021, 41, 127953 CrossRef CAS PubMed.
- G. Li, L. C. Aschenbach, J. Chen, M. P. Cassidy, D. L. Stevens, B. H. Gabra, D. E. Selley, W. L. Dewey, R. B. Westkaemper and Y. Zhang, J. Med. Chem., 2009, 52, 1416–1427 CrossRef CAS PubMed.
- G. Li, L. C. K. Aschenbach, H. He, D. E. Selley and Y. Zhang, Bioorg. Med. Chem. Lett., 2009, 19, 1825–1829 CrossRef CAS PubMed.
- Y. Yuan, S. A. Zaidi, O. Elbegdorj, L. C. K. Aschenbach, G. Li, D. L. Stevens, K. L. Scoggins, W. L. Dewey, D. E. Selley and Y. Zhang, J. Med. Chem., 2013, 56, 9156–9169 CrossRef CAS PubMed.
- T. B. Haddou, S. Beni, S. Hosztafi, D. Malfacini, G. Calo, H. Schmidhammer and M. Spetea, PLoS One, 2014, 9, e99231 CrossRef PubMed.
- K. Luttfy, P. Doan, M. Nugyen and E. Weber, Pharmacol. Res., 1998, 38, 453–460 CrossRef PubMed.
- A. Manglik, A. C. Kruse, T. S. Kobilka, F. S. Thian, J. M. Mathiesen, R. K. Sunahara, L. Pardo, W. I. Weis, B. K. Kobilka and S. Granier, Nature, 2012, 485, 321–326 CrossRef CAS PubMed.
- W. Huang, A. Manglik, A. J. Venkatakrishnan, T. Laeremans, E. N. Feinberg, A. L. Sanborn, H. E. Kato, K. E. Livingston, T. S. Thorsen, R. C. Kling, S. Granier, P. Gmeiner, S. M. Husbands, J. R. Traynor, W. I. Weis, J. Steyaert, R. O. Dror and B. K. Kobilka, Nature, 2015, 524, 315–321 CrossRef CAS PubMed.
- S. A. Zaidi, C. K. Arnatt, H. He, D. E. Selley, P. D. Mosier, G. E. Kellogg and Y. Zhang, Bioorg. Med. Chem., 2013, 21, 6405–6413 CrossRef CAS PubMed.
- H. Wang, S. A. Zaidi and Y. Zhang, Bioorg. Med. Chem., 2017, 25, 2463–2471 CrossRef CAS PubMed.
- Y. Zhuang, Y. Wang, B. He, X. He, X. E. Zhou, S. Guo, Q. Rao, J. Yang, J. Liu, Q. Zhou, X. Wang, M. Liu, W. Liu, X. Jiang, D. Yang, H. Jiang, J. Shen, K. Melcher, H. Chen, Y. Jiang, X. Cheng, M.-W. Wang, X. Xie and H. E. Xu, Cell, 2022, 185, 4361–4375.e19 CrossRef CAS PubMed.
- Z. Feng, G. Hu, S. Ma and X.-Q. Xie, AAPS J., 2015, 17, 1080–1095 CrossRef CAS PubMed.
- M. De Vivo, M. Masetti, G. Bottegoni and A. Cavalli, J. Med. Chem., 2016, 59, 4035–4061 CrossRef CAS PubMed.
- G. Jones, P. Willett, R. C. Glen, A. R. Leach and R. Taylor, J. Mol. Biol., 1997, 267, 727–748 CrossRef CAS PubMed.
- D. A. Case, K. Belfon, I. Y. Ben-Shalom, S. R. Brozell, D. S. Cerutti, T. E. Cheatham, III, V. W. D. Cruzeiro, T. A. Darden, R. E. Duke, G. Giambasu, M. K. Gilson, H. Gohlke, A. W. Goetz, R. Harris, S. Izadi, S. A. Iz-mailov, K. Kasavajhala, A. Kovalenko, R. Krasny, T. Kurtzman, T. S. Lee, S. LeGrand, P. Li, C. Lin, J. Liu, T. Luchko, R. Luo, V. Man, K. M. Merz, Y. Miao, O. Mikhailovskii, G. Monard, H. Nguyen, A. Onufriev, F. Pan, S. Pantano, R. Qi, D. R. Roe, A. Roitberg, C. Sagui, S. Schott-Verdugo, J. Shen, C. L. Simmerling, N. R. Skrynnikov, J. Smith, J. Swails, R. C. Walker, J. Wang, L. Wilson, R. M. Wolf, X. Wu, Y. Xiong, Y. Xue, D. M. York and P. A. Kollman, AMBER 2020, University of California, San Francisco, 2020 Search PubMed.
- S. Jo, T. Kim, V. G. Iyer and W. Im, J. Comput. Chem., 2008, 29, 1859–1865 CrossRef CAS PubMed.
- S. Jo, T. Kim and W. Im, PLoS One, 2007, 2, e880 CrossRef PubMed.
- T. Darden, D. York and L. Pedersen, J. Chem. Phys., 1993, 98, 10089–10092 CrossRef CAS.
- W. Humphrey, A. Dalke and K. Schulten, J. Mol. Graphics, 1996, 14, 33–38 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d5cb00036j |
| This journal is © The Royal Society of Chemistry 2025 |