 Open Access Article
Open Access ArticleIntracellular protein ligation and polyprotein synthesis using an asparaginyl endopeptidase core†
Renming Liu,
Jun Qiu,
Yifen Huang,
Ziyi Wang and
Peng Zheng *
*
State Key Laboratory of Coordination Chemistry, School of Chemistry and Chemical Engineering, Chemistry and Biomedicine Innovation Center (ChemBIC), Nanjing University, Nanjing, China. E-mail: pengz@nju.edu.cn
First published on 17th February 2025
Abstract
An intracellular protein ligation system using a truncated variant of OaAEP1 (OaAEP1 core) to ligate and polymerize Ig domains in E. coli is developed, enabling real-time polyprotein synthesis without enzyme activation and external ligation. This approach yields functional, mechanically stable polyproteins and expands AEP applications in synthetic biology and biomaterials.
Polyproteins, composed of multiple protein domains or repetitive units, play critical roles in biological systems.1,2 One prominent example is titin, a giant muscle protein with numerous immunoglobulin (Ig) domains essential for muscle elasticity and resilience.3,4 These multi-domain proteins perform vital functions and serve as templates for biomaterials and enzyme engineering.5,6 To replicate or surpass these natural systems, there is a growing need for modular approaches that enable the synthesis of polyproteins with enhanced functional and mechanical properties.7,8
Classic polyprotein construction relies on genetic engineering, where multiple domains are fused into a single open reading frame.9,10 While widely used, these methods are limited in their efficiency and applicability to more complex or toxic proteins, as they often result in incomplete expression, misfolding, or aggregation. Recently, chemical and enzymatic ligation methods have emerged as powerful alternatives.11–14 These post-translational approaches ligate purified protein monomers, enabling the assembly of polyproteins with greater control and precision.15–17 Enzymatic methods, such as Oldenlandia affinis asparaginyl endopeptidase 1 (OaAEP1),18 have demonstrated remarkable efficiency for peptide and protein ligation in vitro.19–22 Several studies, including our own, have utilized OaAEP1 to construct polyproteins by ligating purified monomeric domains.23,24 However, this in vitro approach requires multiple purification and activation steps, limiting scalability.
The success of in vitro polyprotein synthesis using OaAEP1 highlights its potential but also underscores the need for further innovation to streamline the process. This led us to explore whether AEPs could function efficiently in living cells for real-time polyprotein ligation. A truncated OaAEP1 variant was recently used for intracellular head-to-tail protein cyclization in E. coli,25 bypassing post-expression processing and enabling a simpler, greener method for stabilizing proteins. Inspired by this, we hypothesized that OaAEP1 could also facilitate intracellular polyprotein ligation and polymerization.
In this study, we developed an intracellular polyprotein ligation system using the OaAEP1 catalytic core domain, which enables the polymerization of immunoglobulin (Ig) domains in E. coli. By adapting the in vivo ligase activity of the OaAEP1 core, we have created a system that eliminates the need for complex post-expression modifications, allowing for real-time polyprotein synthesis directly within the bacterial cytoplasm.
To determine whether the OaAEP1 core could catalyze efficient intracellular polyprotein construction, we first verify its ability to perform intracellular protein ligation by ligating two Ig domains in E. coli. OaAEP1 has been widely used for peptide cyclization, labeling and ligation. However, recombinant OaAEP1 is typically prepared as an inactive zymogen with a cap domain and the catalytic core domain (Fig. 1A).18 It is activated only after the cleavage of the cap domain under acidic solution, leaving only the core domain active. Recently, Prof. Mason's group developed a method that enables the co-expression of the OaAEP1 core and its substrate, murine dihydrofolate reductase (mDHFR), leading to the formation of a cyclic protein.25
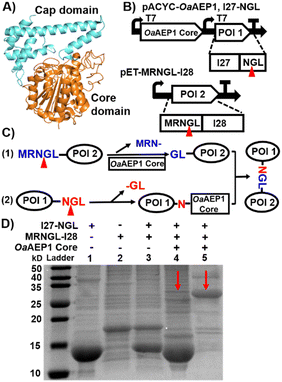 | ||
| Fig. 1 (A) Crystal structure of OaAEP1 (PDB: 5H01). Front view of the OaAEP1 structure represented as a cartoon with the catalytic core domain colored in orange and the cap domain in cyan. (B) Schematic co-expression system for ligating two proteins of interest (POI). The two protein precursors contain N- and C-terminal pro-peptides sequences (MRNGL and NGL, respectively), which are processed by the OaAEP1 core at the site indicated by the red triangles. (C) Scheme of the mechanism for substrate ligation catalyzed by the OaAEP1 core. (D) SDS-PAGE analysis of the purified two substrates, I27 and I28 (lanes 1 and 2), the reactant mixture without OaAEP1 core (control in lane 3), and the ligation product with both Histag and Streptag firstly purified by Ni-NTA affinity chromatography (red arrow in lane 4), and further purified by Strep-tag affinity chromatography (red arrow, in lane 5). Note: the unmodified SDS-PAGE gel image is presented in Fig. S3A (ESI†). | ||
Here, we hypothesize that the OaAEP1 core can also be used to ligate proteins and applied to build polyproteins in E. coli. To assess whether the OaAEP1 core could efficiently catalyze protein ligation intracellularly, we co-expressed the enzyme with a series of Ig domain constructs in E. coli. Our primary objective was to test the in vivo ligation activity of the OaAEP1 core by observing the polymerization of Ig domains directly within bacterial cells.
First, we inserted the genes encoding the OaAEP1 core only and its substrate I27-NGL (I27, the 27th immunoglobin domain of human titin) into the pACYCDuet-1 co-expression vector, creating plasmid pACYC-OaAEP1, I27-NGL for IPTG-inducible recombinant gene expression (both with Histag). The gene encoding the second substrate, MRNGL-I28, was incorporated into the pET30a(+) vector to produce plasmid pET-MRNGL-I28 with Streptag (Fig. 1B). Here, the specific OaAEP1 recognition sequences “GL” and “NGL” are appended to the N- and C-termini of the target protein, respectively.
The protease function of OaAEP1 was utilized to excise the N-terminal peptide sequence from the substrate MRNGL-I28, converting it into “GL-I28” and thereby exposing an N-terminal Gly-Leu sequence for subsequent ligation (Fig. 1C, step (1)). While an N-terminal MGL sequence could also be used, the rate of Met removal by endogenous methionine aminopeptidase is often slow, leading to insufficient cleavage to produce GL. To address this limitation, an “N-terminal MRNGL” sequence was designed, facilitating efficient exposure of the “GL” sequence through the hydrolytic activity of OaAEP1 on the “NGL” sequence, thereby promoting the ligation process.
Simultaneously, the C-terminal peptide “-NGL” of I27-NGL is cleaved by the OaAEP1 core (Fig. 1C, step (2)), yielding an enzyme-substrate intermediate that undergoes nucleophilic substitution by the “GL-I28” N-terminus, resulting in the formation of the product I27-NGL-I28 (Fig. 1C).
Then, we characterized the reaction. First, SDS-PAGE analysis of the purified protein extracts revealed a band at ∼25 kDa (Fig. 1D, lanes 4 and 5, red arrow), which matches the expected molecular weight (MW) of the I27-NGL-I28 ligated product (∼26.8 kDa), indicating the ligated Ig dimer formation. Moreover, MALDI-TOF-MS analysis revealed that the molecular weight of I27-NGL-I28 is 26![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 803 Da, in close concordance with the calculated theoretical molecular weight of 26
803 Da, in close concordance with the calculated theoretical molecular weight of 26![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 785 Da. The overall MW of the OaAEP1 core used in here is approximately 43.4 kDa. While it is not visible on the gel due to the absence of a Strep purification tag, its presence does not interfere with the detection of the product on the gel.25 The yield of I27-NGL-I28 was determined to be 11.5% through MALDI-TOF-MS and SDS-PAGE analysis (Fig. S1A, ESI† and Fig. 1D). This ligation efficiency is lower than in previous in vitro studies of OaAEP1, demonstrating that the activity was affected within the intracellular environment. In addition, size exclusion chromatography (SEC) also confirmed the presence of ligated Ig dimer (Fig. S1B, ESI†). These results confirmed the formation of protein dimer products in vivo.
785 Da. The overall MW of the OaAEP1 core used in here is approximately 43.4 kDa. While it is not visible on the gel due to the absence of a Strep purification tag, its presence does not interfere with the detection of the product on the gel.25 The yield of I27-NGL-I28 was determined to be 11.5% through MALDI-TOF-MS and SDS-PAGE analysis (Fig. S1A, ESI† and Fig. 1D). This ligation efficiency is lower than in previous in vitro studies of OaAEP1, demonstrating that the activity was affected within the intracellular environment. In addition, size exclusion chromatography (SEC) also confirmed the presence of ligated Ig dimer (Fig. S1B, ESI†). These results confirmed the formation of protein dimer products in vivo.
Building on the successful intracellular protein–protein ligation, we next explored whether the OaAEP1 core could catalyze the polymerization of Ig domain constructs in vivo. We subcloned a new gene encoding MRNGL-I27-NGL with the OaAEP1 core domain into plasmid pACYCDuet-1 (Fig. 2A and Fig. S2, ESI†). After induction by additional IPTG, the protease activity of the OaAEP1 core was employed to excise the N-terminal peptide sequence “MRN-” from the precursor substrate, leading to GL-I27-NGL, exposing an N-terminal “GL-” sequence poised for oligomerization (Fig. 2B, step (1)). Subsequently, the C-terminal peptide “-NGL” was cleaved by the OaAEP1 core (Fig. 2B, step (2)), forming an enzyme-I27 complex that underwent nucleophilic substitution by another substrate exposed N-terminal peptide “GL-” (Fig. 2B, step 3). Ultimately, through automatic cycles of cleavage and nucleophilic substitution, polyproteins were synthesized naturally in vivo (Fig. 2B).
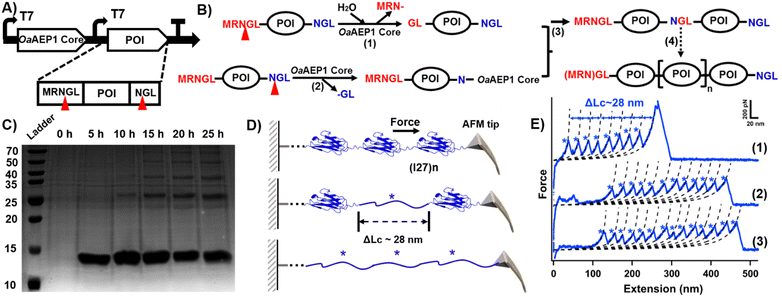 | ||
| Fig. 2 (A) Schematic co-expression system for generating POI oligomer. The protein contains N- and C-terminal peptide sequences, which are cleaved by the OaAEP1 core. (B) Scheme of the mechanism for substrate oligomerization. (C) SDS-PAGE analysis of time-dependent I27 polymerization. (D) The schematic displays the AFM unfolding process of (I27)n, which results in a peak with ΔLc of ∼28 nm from the I27 domain. (E) Representative force-extension curves showing the unfolding events of (I27)n (marked by blue star). The curves confirm the successful polymerization of the I27 domain, with curves 1–3 corresponding to polymers containing at least 8, 11, and 13 domains, respectively. Note: the unmodified SDS-PAGE gel image is presented in Fig. S3B (ESI†). | ||
We next monitored the results in a time-dependent fashion. In the first five hours after induction, only one band of I27 monomer was observed. Five hours later, another band with MW of ∼25 kDa can be observed, indicating the formation of I27 dimer. When the induction time reached 15 h, trimers, tetramers and even pentamers can be observed (Fig. 2C). The presentation of I27 oligomers increased at 20 h and 25 h, with the monomer and dimer species being the most abundant (Fig. 2C and Fig. S4, ESI†). These series of bands corresponded to ligated products of increasing molecular weight, indicating successful intracellular polymerization of the Ig domains. The yields of the respective oligomer are presented in Table S1 (ESI†).
In addition, we characterized the I27 polyprotein at the single-molecule level by using atomic force microscopy-based single-molecule force spectroscopy (AFM-SMFS), which is widely used to study single molecules.26–29 AEP also plays a significant role for polyprotein construction and immobilization.30–34 The characteristic sawtooth-like force-extension curves obtained from AFM-SMFS provided direct evidence of successful polyprotein synthesis (Fig. 2D).35 Each peak in the force-extension curve corresponds to the mechanical unfolding of a single Ig domain, with a contour length increment (ΔLc) of approximately 28 nm (Fig. 2E and Fig. S5, ESI†), consistent with previously reported values for Ig domains.36,37 In addition, the number of peaks indicates the polymerization degree of the polyprotein.15,23,38 For example, curves (1)–(3) represent the polymerization degree of the I27 polyproteins, indicating that at least 8, 11, and 13 monomers were unfolded by AFM-SMFS, respectively. Each peak corresponds to the unfolding of a single I27 domain (Fig. 2E), demonstrating the high ligation efficiency. Hence, both SDS-PAGE and AFM measurements corroborated the in vivo synthesis of I27 oligomers by the OaAEP1 core.
Previously, the gene of the OaAEP1 core and the target gene are subcloned into the same plasmid. If we wanted to build another polyprotein, a completely new plasmid is needed. To simplify the process, we attempted to use the same competent strain BLR (DE3), which harbors the pACYC-OaAEP1 core, but introduced an independent plasmid with the target protein gene for polymerization. Upon successful execution, only a new plasmid is needed for transformation into the same competent cell with the OaAEP1 core gene. Moreover, we selected a larger Ig tetramer, I27–I28–I29–I30 (I27–I30), as the “monomer” to assess the scalability of the ligation process. pET-MRNGL-(I27–I30)-NGL was constructed (Fig. S2B, ESI†). The co-expression system is illustrated in Fig. 3A. And the single plasmid co-expression system was also tested for comparison (Fig. S2C, ESI†).
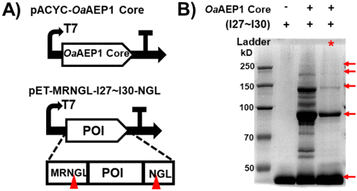 | ||
| Fig. 3 (A) Schematic co-expression system for generating oligo Ig domain. (B) SDS-PAGE result of (I27–I30)n produced by the single (lane 2) or dual (lane 3) vector co-expression system (indicated by red star). The result without the OaAEP1 core is shown in lane 1 with a MW of ∼43 kDa. Note: the unmodified SDS-PAGE gel image is presented in Fig. S3C (ESI†). | ||
The molecular weight of I27–I30 is about 43 kDa (Fig. 3B, lane 1). The SDS-PAGE analysis further indicated that the (I27–I30)n oligomer was successfully produced for both single and dual plasmid methods (Fig. 3B, red arrow, lanes 2 and 3). Consistent with our expectations, the oligomerization of the Ig domains was found to extend up to a pentamer at least, thereby demonstrating the capacity of the dual-plasmid co-expression system within the cytoplasm to synthesize oligomers. The yields of (I27–I30)n oligomers produced by the single or dual vector co-expression system are presented in Tables S2 and S3 (ESI†). Quantitative comparison revealed that the yield of the dual-plasmid expression system is similar to the single-plasmid expression system for dimer formation (∼30%), yet with a higher purity (Fig. 3B). Consequently, we have developed a versatile “plug and play” platform to produce polyproteins or oligomers in E. coli, utilizing the pACYC-OaAEP1 core plasmid as a key component.
In this study, we developed an intracellular ligation system using a truncated variant of Oldenlandia affinis asparaginyl endopeptidase, OaAEP1 core, to achieve polyprotein construction within E. coli. Our work extends two significant lines of research: in vitro polyprotein construction and intracellular protein cyclization, both utilizing AEP. While previous studies have demonstrated the power of AEPs for in vitro ligation of proteins and the potential for intracellular AEP-mediated protein cyclization, this study demonstrates the enzyme's capacity for intracellular ligation and protein polymerization, marking a step forward in protein engineering.
This intracellular ligation system offers several advantages over the previously established in vitro polyprotein construction approach. Firstly, by means of enabling in vivo ligation, we bypass the need for enzyme activation, purification steps and stepwise ligation, making the process much simpler and faster. The intracellular environment provides natural folding conditions that may help preserve protein integrity, further reducing the risk of misfolding or aggregation often encountered during in vitro processes.
However, the complex environment within E. coli may also influence the intracellular ligation efficiency. For instance, after a certain cultivation time, no higher polymerization occurs, possibly due to limited bacterial capacity for large proteins. In addition, the acidic reaction environment for OaAEP1 is optimal, and the physiological cytoplasmic pH may inhibit its ligation efficiency, leaving unreacted monomers in the cytoplasm. Future optimization of the ligation environment and OaAEP1 core expression level (Fig. S6, ESI†) may further improve its efficiency.
In comparison to the intracellular cyclization approach, which focuses on producing head-to-tail cyclic proteins, our work reveals that OaAEP1 can catalyze not only cyclization but also ligation and polymerization of multiple proteins. The ability to ligate domains in real-time within cells offers a valuable tool for synthetic biology and protein design. Additionally, we established a versatile “plug and play” platform to produce polyproteins or oligomers in E. coli with higher purity, utilizing the pACYC-OaAEP1 core plasmid as an essential component. And by introducing distinct secondary plasmids, the system can be easily modified to accommodate a range of target proteins.
This work is supported by the National Natural Science Foundation of China (22222703, 22477058) and the Fundamental Research Funds for the Central Universities (020514380335).
Data availability
All data are available in the main text or the ESI.†Conflicts of interest
The authors declare no competing interests.References
- J. Yang, X. Xie, N. Xiang, Z.-X. Tian, R. Dixon and Y.-P. Wang, Proc. Natl. Acad. Sci. U. S. A., 2018, 115, E8509–E8517 CAS.
- K. A. Scott, A. Steward, S. B. Fowler and J. Clarke, J. Mol. Biol., 2002, 315, 819–829 CrossRef CAS PubMed.
- M. Rief, M. Gautel, F. Oesterhelt, J. M. Fernandez and H. E. Gaub, Science, 1997, 276, 1109–1112 CrossRef CAS PubMed.
- E. C. Eckels, R. Tapia-Rojo, J. A. Rivas-Pardo and J. M. Fernández, Annu. Rev. Physiol., 2018, 80, 327–351 CrossRef CAS PubMed.
- E. M. Pelegri-O’Day and H. D. Maynard, Acc. Chem. Res., 2016, 49, 1777–1785 CrossRef PubMed.
- Y. J. Yang, A. L. Holmberg and B. D. Olsen, Annu. Rev. Chem. Biomol. Eng., 2017, 8, 549–575 CrossRef CAS PubMed.
- Z.-H. Cui, H. Zhang, F.-H. Zheng, J.-H. Xue, Q.-H. Yin, X.-L. Xie, Y.-X. Wang, T. Wang, L. Zhou and G.-M. Fang, Org. Biomol. Chem., 2025, 23, 188–196 RSC.
- H. Li, Adv. NanoBiomed Res., 2021, 1, 2100028 CrossRef CAS.
- M. Carrion-Vazquez, A. F. Oberhauser, S. B. Fowler, P. E. Marszalek, S. E. Broedel, J. Clarke and J. M. Fernandez, Proc. Natl. Acad. Sci. U. S. A., 1999, 96, 3694–3699 CrossRef CAS PubMed.
- T. Hoffmann, K. M. Tych, T. Crosskey, B. Schiffrin, D. J. Brockwell and L. Dougan, ACS Nano, 2015, 9, 8811–8821 CrossRef CAS PubMed.
- J. L. Zimmermann, T. Nicolaus, G. Neuert and K. Blank, Nat. Protoc., 2010, 5, 975–985 CrossRef CAS PubMed.
- H. Dietz, F. Berkemeier, M. Bertz and M. Rief, Proc. Natl. Acad. Sci. U. S. A., 2006, 103, 12724–12728 CrossRef CAS PubMed.
- P. Zheng, Y. Cao and H. Li, Langmuir, 2011, 27, 5713–5718 CrossRef CAS PubMed.
- N. H. Fischer, M. T. Oliveira and F. Diness, Biomater. Sci., 2023, 11, 719–748 RSC.
- H. Liu, D. T. Ta and M. A. Nash, Small Methods, 2018, 2, 1800039 CrossRef.
- S. Garg, G. S. Singaraju, S. Yengkhom and S. Rakshit, Bioconjugate Chem., 2018, 29, 1714–1719 CrossRef CAS PubMed.
- V. Kaur, S. Garg and S. Rakshit, Chem. Commun., 2023, 59, 6946–6955 RSC.
- R. Yang, Y. H. Wong, G. K. T. Nguyen, J. P. Tam, J. Lescar and B. Wu, J. Am. Chem. Soc., 2017, 139, 5351–5358 CrossRef CAS PubMed.
- T. M. S. Tang, D. Cardella, A. J. Lander, X. Li, J. S. Escudero, Y.-H. Tsai and L. Y. P. Luk, Chem. Sci., 2020, 11, 5881–5888 RSC.
- T. J. Harmand, N. Pishesha, F. B. H. Rehm, W. Ma, W. B. Pinney, Y. J. Xie and H. L. Ploegh, ACS Chem. Biol., 2021, 16, 1201–1207 CrossRef CAS PubMed.
- Z. Lu, Y. Liu, Y. Deng, B. Jia, X. Ding, P. Zheng and Z. Li, Chem. Commun., 2022, 58, 8448–8451 RSC.
- W. Ott, E. Durner and H. E. Gaub, Angew. Chem., Int. Ed., 2018, 57, 12666–12669 CrossRef CAS PubMed.
- Y. Deng, T. Wu, M. Wang, S. Shi, G. Yuan, X. Li, H. Chong, B. Wu and P. Zheng, Nat. Commun., 2019, 10, 2775–2785 CrossRef PubMed.
- F. B. H. Rehm, T. J. Harmand, K. Yap, T. Durek, D. J. Craik and H. L. Ploegh, J. Am. Chem. Soc., 2019, 141, 17388–17393 CrossRef CAS PubMed.
- T. M. S. Tang and J. M. Mason, JACS Au, 2023, 3, 3290–3296 CrossRef CAS PubMed.
- P. Zhao, C.-Q. Xu, C. Sun, J. Xia, L. Sun, J. Li and H. Xu, Polym. Chem., 2020, 11, 7087–7093 RSC.
- Y. Xue, X. Li, H. Li and W. Zhang, Nat. Commun., 2014, 5, 4348–4356 CrossRef CAS PubMed.
- Y. Shi, W. Shi, S. Zhang and X. Wang, CCS Chem., 2023, 5, 2956–2965 CrossRef CAS.
- S. Lu, W. Cai, N. Cao, H.-J. Qian, Z.-Y. Lu and S. Cui, ACS Mater. Lett., 2022, 4, 329–335 CrossRef CAS.
- Y. Liu, D. Song, S. Li, Z. Guo and P. Zheng, J. Am. Chem. Soc., 2024, 146, 13126–13132 CrossRef CAS PubMed.
- S. Shi, Z. Wang, Y. Deng, F. Tian, Q. Wu and P. Zheng, CCS Chem., 2022, 4, 598–604 CrossRef CAS.
- Z. Wang, Z. Zhao, Z. Yang, G. Li and P. Zheng, J. Phys. Chem. B, 2023, 127, 2934–2940 CrossRef CAS PubMed.
- Y. Xiao, B. Zheng, X. Ding and P. Zheng, Chem. Commun., 2023, 59, 11268–11271 RSC.
- B. Zheng, Y. Xiao, B. Tong, Y. Mao, R. Ge, F. Tian, X. Dong and P. Zheng, JACS Au, 2023, 3, 1902–1910 CrossRef CAS PubMed.
- H. Wang and H. Li, Chem. Sci., 2020, 11, 12512–12521 RSC.
- M. Muddassir, B. Manna, P. Singh, S. Singh, R. Kumar, A. Ghosh and D. Sharma, Chem. Commun., 2018, 54, 9635–9638 RSC.
- J. Oroz, M. Bruix, D. V. Laurents, A. Galera-Prat, J. Schönfelder, F. J. Cañada and M. Carrión-Vázquez, Structure, 2016, 24, 606–616 CrossRef CAS PubMed.
- Z. Y. Wang, M. D. Wang, Z. X. Zhao and P. Zheng, Protein Sci., 2022, 32, e4583 CrossRef PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d4cc06617k |
| This journal is © The Royal Society of Chemistry 2025 |
