Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Uncommon quadruple stacking topology in a honeycomb-sheet MOF compatible with through-space conduction†
Tappei Tanabe ab,
Liyuan Qu
ab,
Liyuan Qu b,
Kenta Uenoa,
Shinya Takaishi
b,
Kenta Uenoa,
Shinya Takaishi a,
Masahiro Yamashita
a,
Masahiro Yamashita ac,
Yuh Hijikata
ac,
Yuh Hijikata d,
Ryotaro Matsuda
d,
Ryotaro Matsuda b,
Ryota Sakamoto
b,
Ryota Sakamoto a and
Hiroaki Iguchi
a and
Hiroaki Iguchi *b
*b
aDepartment of Chemistry, Graduate School of Science, Tohoku University, 6-3 Aramaki-Aza-Aoba, Aoba-ku, Sendai, Miyagi 980-8578, Japan
bDepartment of Materials Chemistry, Graduate School of Engineering, Nagoya University, Chikusa-ku, Nagoya 464-8603, Japan. E-mail: hiroaki.iguchi@chembio.nagoya-u.ac.jp
cSchool of Chemical Science and Engineering, Tongji University, Shanghai 559 200092, P. R. China
dResearch Center for Net Zero Carbon Society, Institutes of Innovation for Future Society, Nagoya University, Chikusa-ku, Nagoya 464-8601, Japan
First published on 22nd May 2025
Abstract
Quadruple stacking topology with an unusual threefold stacking periodicity in four honeycomb sheets was realized in a flexible wavy honeycomb-sheet MOF (PMC-20). The infinite π-stacked arrays, formed exclusively at the π-conjugated core of the linker site, contribute to the highest electrical conductivity among the reported naphthalenediimide (NDI)-based crystals.
Honeycomb lattice, which is characterized by its 3-connected nodes and linkers, is commonly found in sheet materials such as graphene1,2 and metal oxides.3 It is one of the most extensively studied two-dimensional (2D) atomic scale structures due to its attractive electrical and magnetic properties. The research on 2D honeycomb lattices has recently extended to molecule-based framework.4–6 Notably, those with porosity such as metal–organic frameworks (MOFs) and covalent organic frameworks (COFs) have been of interest because of their bottom-up designability and various characteristic functions including ferromagnetism,7–9 carrier transport,5,10,11 photoluminescence,12,13 catalysis14–16 and gas separation.17,18 These functions originate from the molecular components, while they are also influenced by the stacking structure between honeycomb sheets.
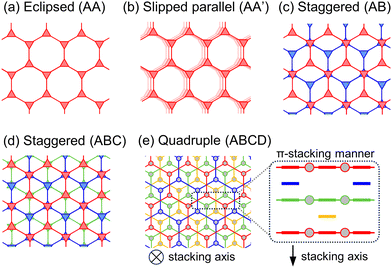
The π-stacking interaction is the typical interlayer interaction observed in honeycomb-sheet MOFs and COFs which consist of π-conjugated planar nodes and linkers. The interaction often extends infinitely to form columnar structures, enabling the conduction of carriers (electrons or holes) along the interlayer direction, so-called through-space conduction. The proper stacking of redox-active planar moieties19–21 and the extension of conjugation have been widely explored to enhance the electrical conductivity of MOFs, known as the through-space and through-bond approaches, respectively.11,22 Although intralayer (i.e. through-bond) conduction among π-conjugated nodes and linkers has been regarded as the major contributing factor to the electrical conductivity in these MOFs and COFs, the significant contribution of through-space conduction was recently reported in conductive honeycomb-sheet MOFs with lanthanide ions, which break through-bond π-conjugation.23,24 To the best of our knowledge, electrically conductive honeycomb-sheet MOFs and COFs exhibit one of the four stacking modes shown in Fig. 1a–d. In most cases, either an eclipsed (AA, Fig. 1a)25–27 or slipped parallel (AA′, Fig. 1b)28–31 stacking mode, which forms π-stacked columns at both node and linker sites, has been reported. The other minor case is known as a staggered stacking mode (AB stacking31,32 in Fig. 1c and ABC stacking27,31 in Fig. 1d), where π-stacking interaction works only at the node sites and the remaining linkers float in the nanospace. These studies indicate that planar π-conjugated nodes always contribute to the formation of π-stacked columnar structures in reported honeycomb-sheet MOFs and COFs. In contrast, to our knowledge, the formation of columnar structures only at the π-conjugated linker sites has never been reported. To overlap the π-conjugated planes of all linkers, quadruple stacking topology as shown in Fig. 1e, i.e., four honeycomb sheets with staggered stacking manner at linker sites, would be formed. In this case, however, each π-stacked linker site consists of only three of four honeycomb sheets, leaving an empty space between a trimer of π-stacked linkers. This geometric constraint would prevent the realization of the quadruple stacking topology in honeycomb-sheet MOFs and COFs.
Nevertheless, we herein report the first 2D honeycomb network assembly that forms infinite π-stacked arrays exclusively at the π-conjugated core of the linkers. This network consists of Cd2+ ions as the non-planar 3-connected nodes, where π-stacking interaction is not available, and N,N′-di(4-pyridyl)-1,4,5,8-naphthalenetetracarboxdiimide (NDI-py) as the linkers. The flexibility of the honeycomb coordination networks plays a crucial role in forming 3-fold stacking periodicity in four different honeycomb sheets, realizing an infinite π-stacked array with high electrical conductivity. Because this 2D network contains both pores and partially reduced conductive π-stacked arrays, it is also regarded as the first porous molecular conductor (PMC)33–35 with a 2D framework.
The naphthalenediimide (NDI) core has widely been used as the ligand in MOFs,36 whereas the NDI-based conductive MOFs are still limited.20,33,35,37 To induce the formation of a π-stacked array by enhancing aromatic interaction with π-radicals, we applied preliminary reduction of the NDI-py ligand using cobaltocene as a reducing agent.33 The rod-shaped black crystals were synthesized by liquid–liquid diffusion of a N,N-dimethylacetamide (DMA) solution of CdBr2 to N-methylpyrrolidone (NMP) solution of NDI-py with cobaltocene under an inert atmosphere. This compound is hereafter abbreviated as PMC-20. The single-crystal X-ray diffraction (SXRD) analysis reveals that a Cd2+ ion is coordinated by three pyridyl groups of NDI-py ligands and some disordered atoms (Fig. 2a). As shown in the coordination geometry around the Cd2+ ion (Fig. S1a–d in ESI†), an H2O molecule (O5 or O6) and bromide ion in the axial position and the oxygen atom (O4 or O4A) of the solvent molecules in the equatorial position are disordered. As a result, the formula of the coordination network of PMC-20 is determined to be [Cd(NDI-py)1.5Br0.56(OH2)1.44(solvent)], which is consistent with the elemental analysis.
The equatorial coordination geometry is distorted from square planar geometry. The N–Cd–N bond angles of the cis and trans positions are 98.51(15)° and 163.0(3)°, respectively (Fig. S1e, ESI†), making a Cd2+ ion as the distorted triangle-like 3-connected node of the honeycomb sheet as shown in Fig. 2b and Fig. S1f (ESI†). The crystal structure of PMC-20 projected along the a axis (Fig. 2c) indicates that four honeycomb sheets (shown in different colours) are stacked at the NDI core of the linker with staggered stacking manner. Each π-stacked array consists of only three NDI cores of four honeycomb sheets. Although a vacancy between the trimer of π-stacked linkers is expected in the case of flat honeycomb sheets as shown in Fig. 1e, the actual sheets form a wavy structure to fill the vacancy and achieve infinite stacking as shown in Fig. 2c. The flexibility of the Cd2+ coordination geometry enables the formation of the wavy honeycomb sheet. The π-stacked columnar structure of NDI-py linkers is highlighted in Fig. 2d. The NDI cores are stacked in 3-fold periodicity with the stacking distances of 3.23, 3.22, and 3.22 Å. Their transfer integral values are calculated to be 274, 261, and 261 meV, respectively, which are slightly smaller than the values of previously reported PMCs33–35 but still larger than those of neutral NDI-based organic semiconductors (typically < 100 meV).38,39 These values indicate strong aromatic interactions enhanced by the presence of π-radicals, which would also contribute significantly to the formation of wavy honeycomb sheets and infinite π-stacked arrays. The sharp electron spin resonance (ESR) signal (g = 2.0038) confirms the presence of NDI radical species (Fig. S2, ESI†). Ignoring the solvent molecules both in the pores and coordinating to Cd2+ ions, two types of pores are formed in PMC-20 as shown in Fig. 2e.
The 1H NMR spectrum of PMC-20 dissolved in DMSO-d6 suggests that NMP and DMA molecules are present in a 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2 ratio (Fig. S3, ESI†). On the basis of the 1H NMR spectrum and elemental analysis, we concluded that the unit formula of PMC-20 contains one NMP, two DMA and 3.44 H2O molecules including the coordinating solvent. The thermogravimetric analysis (TGA) showed the weight loss with two unclear steps, which roughly correspond to the liberation of 3.44 H2O molecules (up to 50 °C) and that of one NMP and two DMA molecules (from 50 to 200 °C) (Fig. S4, ESI†). The variable-temperature powder X-ray diffraction (PXRD) measurement (Fig. S5, ESI†) indicates that the PXRD patterns are maintained up to 60 °C, demonstrating structural stability under the liberation of H2O molecules. However, the patterns gradually broadened and weakened above 60 °C, reflecting a loss of crystallinity due to the liberation of NMP and DMA molecules from the pores. We next tried to exchange NMP and DMA molecules to other solvent molecules such as acetone and methanol by soaking the crystals into each solvent for a week. 1H NMR spectra of the dissolved crystals after soaking show no peaks attributable to NMP and DMA molecules, confirming the completion of the solvent exchange reaction (Fig. S6, ESI†). The PXRD patterns of the soaked crystals (Fig. S7, ESI†) indicate that the crystal structure was maintained under the exchange with methanol and acetone. This phenomenon contrasts with the previously reported PMC-1, whose 1D coordination framework was rearranged into a non-porous structure upon soaking,35 reflecting the improved robustness of the 2D framework in PMC-20.
2 ratio (Fig. S3, ESI†). On the basis of the 1H NMR spectrum and elemental analysis, we concluded that the unit formula of PMC-20 contains one NMP, two DMA and 3.44 H2O molecules including the coordinating solvent. The thermogravimetric analysis (TGA) showed the weight loss with two unclear steps, which roughly correspond to the liberation of 3.44 H2O molecules (up to 50 °C) and that of one NMP and two DMA molecules (from 50 to 200 °C) (Fig. S4, ESI†). The variable-temperature powder X-ray diffraction (PXRD) measurement (Fig. S5, ESI†) indicates that the PXRD patterns are maintained up to 60 °C, demonstrating structural stability under the liberation of H2O molecules. However, the patterns gradually broadened and weakened above 60 °C, reflecting a loss of crystallinity due to the liberation of NMP and DMA molecules from the pores. We next tried to exchange NMP and DMA molecules to other solvent molecules such as acetone and methanol by soaking the crystals into each solvent for a week. 1H NMR spectra of the dissolved crystals after soaking show no peaks attributable to NMP and DMA molecules, confirming the completion of the solvent exchange reaction (Fig. S6, ESI†). The PXRD patterns of the soaked crystals (Fig. S7, ESI†) indicate that the crystal structure was maintained under the exchange with methanol and acetone. This phenomenon contrasts with the previously reported PMC-1, whose 1D coordination framework was rearranged into a non-porous structure upon soaking,35 reflecting the improved robustness of the 2D framework in PMC-20.
The solid-state absorption spectrum of PMC-20 contains various absorption bands as shown in Fig. 3. On the basis of the previous report,35,40–42 the broad absorption band attributed to the charge transfer between the NDI radical and neutral species was observed near 0.4 eV, as well as the relatively weak absorption band attributable to the charge transfer between two NDI radical species observed around 1 eV. The optical band gap of PMC-20 is estimated from the former band (0.4 eV). In addition, the absorption band around 2.7 eV and 3.5 eV can be attributed to the intramolecular π–π* transition of NDI radical and neutral species, respectively. The intense band of the radical species (2.7 eV) compared with the neutral one (3.5 eV) is consistent with the large mean charge of the NDI core (−0.96) calculated from the chemical formula of PMC-20.
Direct current (DC) electrical conductivity measurements of single crystals were performed by the two-probe method. The temperature dependence of electrical conductivity indicates semiconducting behaviour with a conductivity of 7.62 × 10−3 S cm−1 at room temperature. Notably, it is higher than the record of electrical conductivity of NDI-based and TTF-based MOFs (Table S2, ESI†). The activation energy (Ea) is calculated to be 87.7 meV by fitting the data with the Arrhenius equation σ = σ0![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) exp(Ea/kT), where σ0 is the prefactor, k is the Boltzmann constant and T is the temperature. Although the smaller transfer integrals suggest lower carrier mobility, the larger mean charge on the NDI core (–0.96), i.e., the higher carrier concentration, is likely to play a more important role in the higher conductivity of PMC-20 compared with that of reported PMCs.
exp(Ea/kT), where σ0 is the prefactor, k is the Boltzmann constant and T is the temperature. Although the smaller transfer integrals suggest lower carrier mobility, the larger mean charge on the NDI core (–0.96), i.e., the higher carrier concentration, is likely to play a more important role in the higher conductivity of PMC-20 compared with that of reported PMCs.
In conclusion, we synthesized PMC-20 as the first porous molecular conductor (PMC) constructed from a 2D coordination network. The conductive infinite π-stacked arrays were constructed at the π-conjugated core of the naphthalenediimide (NDI) ligands in PMC-20, resulting in an uncommon honeycomb-sheet framework that forms a columnar structure only at the linker sites. This 3-fold stacking periodicity in four honeycomb sheets has not been realized in flat honeycomb sheets due to the geometric constraint. The flexible nature of the 3-connected node with the Cd2+ ion plays a key role in forming the wavy honeycomb sheet, which in turn enables the achievement of the uncommon quadruple stacking structure. The relatively robust 2D framework of PMC-20 enables solvent exchange while maintaining the structure. Moreover, it exhibited the highest electrical conductivity and lowest activation energy among previously reported single-crystalline PMCs and NDI derivatives. This work opens new avenues for the synthesis of conductive honeycomb-sheet materials with unusual stacking topology by enhancing the flexibility of the framework.
H. I. conceived and designed the project. T. T., L. Q. and K. U. carried out the synthesis and crystal structure analysis. T. T. also performed characterization and the measurement of physical properties. T. T., L. Q. and Y. H. carried out calculation of transfer integrals. S. T., M. Y., R. M., R. S. and H. I. supervised this work and carried out discussions. T. T. and H. I. wrote the paper.
T. T. thanks the Transformative Research Area (A) “Condensed Conjugation” for the fellowship for junior scientists. This work was supported by a Grant-in-Aid of Tohoku University, Division for Interdisciplinary Advanced Research and Education (T. T.), by JST FOREST Program (Grant Number JPMJFR236B, Japan) (H. I.), by JSPS KAKENHI Grant Numbers JP23K13718 (L. Q.), JP21H01901 (H. I.), JP21K18971 (H. I.), JP23H04024 (H. I.), and JP24K01453 (H. I.), by the Toyota Riken Scholar Program (H. I.), by Iketani Science and Technology Foundation 0331101-A (H. I.), by The Murata Science Foundation (H. I.), and by TOBE MAKI Scholarship Foundation 22-JA-005 (H. I.). Crystal structure analysis of PMC-20 was performed under the approval of the Photon Factory Program Advisory Committee (Proposal No. 2021U002 and 2024G583, beamline 5A and 17A).
Data availability
The data supporting this article have been included as part of the ESI.† Crystallographic data for PMC-20 has been deposited at the CCDC under 2413297.Conflicts of interest
There are no conflicts to declare.Notes and references
- K. S. Novoselov, A. K. Geim, S. V. Morozov, D. Jiang, Y. Zhang, S. V. Dubonos, I. V. Grigorieva and A. A. Firsov, Science, 2004, 306, 666–669 CrossRef CAS PubMed.
- Y.-W. Son, M. L. Cohen and S. G. Louie, Nature, 2006, 444, 347–349 CrossRef CAS PubMed.
- G. M. Kanyolo, T. Masese, N. Matsubara, C.-Y. Chen, J. Rizell, Z.-D. Huang, Y. Sassa, M. Månsson, H. Senoh and H. Matsumoto, Chem. Soc. Rev., 2021, 50, 3990–4030 RSC.
- L. Liu, R. Yu, L. Yin, N. Zhang and G. Zhu, Chem. Sci., 2024, 15, 1924–1937 RSC.
- R. Sakamoto, N. Fukui, H. Maeda, R. Toyoda, S. Takaishi, T. Tanabe, J. Komeda, P. Amo-Ochoa, F. Zamora and H. Nishihara, Coord. Chem. Rev., 2022, 472, 214787 CrossRef CAS.
- Y. Shuku, A. Mizuno, R. Ushiroguchi, C. S. Hyun, Y. J. Ryu, B.-K. An, J. E. Kwon, S. Y. Park, M. Tsuchiizu and K. Awaga, Chem. Commun., 2018, 54, 3815–3818 RSC.
- G. M. Espallargas and E. Coronado, Chem. Soc. Rev., 2018, 47, 533–557 RSC.
- A. E. Thorarinsdottir and T. D. Harris, Chem. Rev., 2020, 120, 8716–8789 CrossRef CAS PubMed.
- H. Miyasaka, Bull. Chem. Soc. Jpn., 2021, 94, 2929–2955 CrossRef CAS.
- M. Ko, L. Mendecki and K. A. Mirica, Chem. Commun., 2018, 54, 7873–7891 RSC.
- L. S. Xie, G. Skorupskii and M. Dincă, Chem. Rev., 2020, 120, 8536–8580 CrossRef CAS PubMed.
- Q.-R. Fang, G.-S. Zhu, Z. Jin, Y.-Y. Ji, J.-W. Ye, M. Xue, H. Yang, Y. Wang and S.-L. Qiu, Angew. Chem., Int. Ed., 2007, 46, 6638–6642 CrossRef CAS PubMed.
- S. Kimura, M. Uejima, W. Ota, T. Sato, S. Kusaka, R. Matsuda, H. Nishihara and T. Kusamoto, J. Am. Chem. Soc., 2021, 143, 4329–4338 CrossRef CAS PubMed.
- X. Sun, K.-H. Wu, R. Sakamoto, T. Kusamoto, H. Maeda, X. Ni, W. Jiang, F. Liu, S. Sasaki, H. Masunaga and H. Nishihara, Chem. Sci., 2017, 8, 8078–8085 RSC.
- R. Dong, M. Pfeffermann, H. Liang, Z. Zheng, X. Zhu, J. Zhang and X. Feng, Angew. Chem., Int. Ed., 2015, 54, 12058–12063 CrossRef CAS PubMed.
- A. J. Clough, J. W. Yoo, M. H. Mecklenburg and S. C. Marinescu, J. Am. Chem. Soc., 2015, 137, 118–121 CrossRef CAS PubMed.
- J. Zhang, Y.-Y. Xue, P. Zhang, H.-P. Li, Y. Wang, J. Xu, S.-N. Li and Q.-G. Zhai, Cryst. Growth Des., 2022, 22, 469–477 CrossRef CAS.
- L. Zhang, L.-N. Ma, G.-D. Wang, L. Hou, Z. Zhu and Y.-Y. Wang, J. Mater. Chem. A, 2023, 11, 2343–2348 RSC.
- D. Chen, H. Xing, Z. Su and C. Wang, Chem. Commun., 2016, 52, 2019–2022 RSC.
- X. Kuang, S. Chen, L. Meng, J. Chen, X. Wu, G. Zhang, G. Zhong, T. Hu, Y. Li and C. Lu, Chem. Commun., 2019, 55, 1643–1646 RSC.
- G. Valente, M. Esteve-Rochina, A. Paracana, A. Rodríguez-Diéguez, D. Choquesillo-Lazarte, E. Ortí, J. Calbo, M. Ilkaeva, L. Mafra, M. A. Hernández-Rodríguez, J. Rocha, H. Alves and M. Souto, Mol. Syst. Des. Eng., 2022, 7, 1065–1072 RSC.
- B. Ding, M. B. Solomon, C. F. Leong and D. M. D’Alessandro, Coord. Chem. Rev., 2021, 439, 213891 CrossRef CAS.
- G. Skorupskii, B. A. Trump, T. W. Kasel, C. M. Brown, C. H. Hendon and M. Dinca, Nat. Chem., 2020, 12, 131–136 CrossRef CAS PubMed.
- G. Skorupskii, K. N. Le, D. L. M. Cordovahttps, L. Yang, T. Chen, C. H. Hendon, M. Q. Arguilla and M. Dincă, Proc. Natl. Acad. Sci. U. S. A., 2022, 119, e2205127119 CrossRef CAS PubMed.
- J. A. DeGayner, I.-R. Jeon, L. Sun, M. Dincă and T. D. Harris, J. Am. Chem. Soc., 2017, 139, 4175–4184 CrossRef CAS PubMed.
- L. M. Salonen, D. D. Medina, E. Carbó-Argibay, M. G. Goesten, L. Mafra, N. Guldris, J. M. Rotter, D. G. Stroppa and C. Rodríguez-Abreua, Chem. Commun., 2016, 52, 7986–7989 RSC.
- Z. Wang, Y. Zhang, T. Wang, E. Lin, T. Wang, Y. Chen, P. Cheng and Z. Zhang, Small, 2023, 19, 2303684 CrossRef CAS PubMed.
- A. M. Pütz, M. W. Terban, S. Bette, F. Haase, R. E. Dinnebier and B. V. Lotsch, Chem. Sci., 2020, 11, 12647–12654 RSC.
- D. Sheberla, L. Sun, M. A. Blood-Forsythe, S. Er, C. R. Wade, C. K. Brozek, A. Aspuru-Guzik and M. Dincă, J. Am. Chem. Soc., 2014, 136, 8859–8862 CrossRef CAS PubMed.
- N. Huang, L. Zhai, D. E. Coupry, M. A. Addicoat, K. Okushita, K. Nishimura, T. Heine and D. Jiang, Nat. Commun., 2016, 7, 12325–12336 CrossRef PubMed.
- S. Ghosh, A. Nakada, M. A. Springer, T. Kawaguchi, K. Suzuki, H. Kaji, I. Baburin, A. Kuc, T. Heine, H. Suzuki, R. Abe and S. Seki, J. Am. Chem. Soc., 2020, 142, 9752–9762 CAS.
- T. Kambe, R. Sakamoto, K. Hoshiko, K. Takada, M. Miyachi, J.-H. Ryu, S. Sasaki, J. Kim, K. Nakazato, M. Takata and H. Nishihara, J. Am. Chem. Soc., 2013, 135, 2462–2465 CrossRef CAS PubMed.
- S. Koyama, T. Tanabe, S. Takaishi, M. Yamashita and H. Iguchi, Chem. Commun., 2020, 56, 13109–13112 RSC.
- M. Cui, R. Murase, Y. Shen, T. Sato, S. Koyama, K. Uchida, T. Tanabe, S. Takaishi, M. Yamashita and H. Iguchi, Chem. Sci., 2022, 13, 4902–4908 RSC.
- L. Qu, H. Iguchi, S. Takaishi, F. Habib, C. F. Leong, D. M. D'Alessandro, T. Yoshida, H. Abe, E. Nishibori and M. Yamashita, J. Am. Chem. Soc., 2019, 141, 6802–6806 CrossRef CAS PubMed.
- Y. Zhou and L. Han, Coord. Chem. Rev., 2021, 430, 213665 CrossRef CAS.
- J. Li, A. Kumar, B. A. Johnson and S. Ott, Nat. Commun., 2023, 14, 4388 CrossRef CAS PubMed.
- T. Kakinuma, H. Kojima, M. Ashizawa, H. Matsumoto and T. Mori, J. Mater. Chem. C, 2013, 1, 5395–5401 RSC.
- H. Abe, A. Kawasaki, T. Takeda, N. Hoshino, W. Matsuda, S. Seki and T. Akutagawa, J. Am. Chem. Soc., 2021, 143, 1046–1060 CrossRef CAS PubMed.
- C. F. Leong, B. Chan, T. B. Faust and D. M. D'Alessandro, Chem. Sci., 2014, 5, 4724–4728 RSC.
- P. M. Usov, C. Fabian and D. M. D’Alessandro, Chem. Commun., 2012, 48, 3945–3947 RSC.
- A. Takai, T. Yasuda, T. Ishizuka, T. Kojima and M. Takeuchi, Angew. Chem., Int. Ed., 2013, 52, 9167–9171 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available: Experimental details, crystallographic details, thermogravimetric analysis, PXRD patterns, and NMR and ESR spectroscopy. CCDC 2413297. For ESI and crystallographic data in CIF or other electronic format see DOI: https://doi.org/10.1039/d5cc00084j |
| This journal is © The Royal Society of Chemistry 2025 |