Open Access Article
Open Access ArticleA sensitive fluorescent nanoprobe for sulfatase detection and imaging in living cells and in vivo†
Huijia Liu‡
,
Jiaqi Zhang‡,
Li Liu,
Wenqing Li,
Jing Yang* and
Peng Wang *
*
Department of Biomedical Engineering, School of Engineering, China Pharmaceutical University, Nanjing 210009, China. E-mail: wangpeng@cpu.edu.cn; yangjing@cpu.edu.cn
First published on 18th February 2025
Abstract
In this study, we developed a fluorescent probe TCF-SULF for sulfatase detection with satisfactory selectivity and sensitivity. After being encapsulated by the DSPE-PEG2000, the nanoprobe TCF-SULF NPs can be applied to imaging endogenous sulfatase in MCF-7 cells with low cytotoxicity. Importantly, TCF-SULF NPs were successfully used to monitor sulfatase in vivo.
Sulfatases are a class of proteases that are essential to biological organisms because they have the ability to hydrolyze sulfate esters. Currently, scientists have identified 17 genes associated with sulfatases, including ARSA, ARSB, and ARSC.1 The enzymes encoded by these genes play key roles in biological processes such as hormone synthesis, glycosaminoglycan, and glycolipid metabolism.2,3 Sulfatase dysregulation is intimately linked to the onset and progression of cancer.4,5 Steroid sulfatase overexpression, for instance, may promote the desulfation of estrone 3-sulfate, raising estrone levels and potentially causing breast cancer.6–8 Therefore, sulfatase as a biomarker for cancer detection has become a hot topic of interest in recent years.
The methods used to detect sulfatase mainly include immunoassays,9 fluorescent probes,10–12 gas chromatography-mass spectrometry (GC-MS),13 and analytical isotachophoresis.14 Among these, fluorescent probes have garnered a lot of interest due to their high sensitivity, low cost, and great selectivity.15 Currently, numerous fluorescent probe series have been created for sulfatase imaging both in vitro and in vivo.11,16–18 p-Nitrobenzene sulfate (p-NPS), and 4-methyl-umbelliferous sulfate (4-MUS) have been used for commercialization. Although these advanced probes exhibit excellent practical performance in vitro, they face challenges when monitoring sulfatase activity under live-cell conditions.8 The intricate environment found in living cells may have impacts on the probe's stability and reaction efficiency, leading to subpar monitoring outcomes. Thus, a pressing technical challenge in this area is how to modify probes to fit the complex environment inside living cells while preserving their sensitivity and specificity.
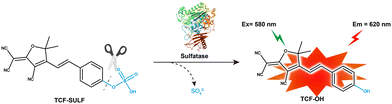
Here, we designed a fluorescent probe TCF-SULF, which uses the TCF fluorescent part and a sulfatase-cleavable unit. In the presence of sulfatase, the probe TCF-SULF exhibits a 778-fold increase in fluorescence intensity in vitro, and it has high specificity, enabling the specific detection of sulfatase activity. With a determined LOD of 2.12 × 10−3 U mL−1, the probe TCF-SULF demonstrates adequate sensitivity and selectivity towards sulfatase. This probe not only maintains stability in the live cell environment but also exhibits significant fluorescence signal changes based on the presence and activity levels of sulfatase. Therefore, this probe can be utilized for the activity screening of live cells. Importantly, the TCF-SULF probe was effectively utilized for non-invasive observation and documentation of the dynamic changes in sulfatase activity at the tumor site through intratumoral injection in the subcutaneous MCF-7 xenograft model.
TCF-OH is a donor–π–acceptor (D–π–A) structure with strong fluorescence. In the probe TCF-SULF we designed, the sulfate part acts as a recognition group and a protecting group to lock the phenolic hydroxyl of TCF-OH, causing the ICT effect to be interrupted and the photonic signal to be quenched. TCF-SULF can release TCF-OH under the catalysis of sulfatase, thereby achieving the purpose of detecting sulfatase.
The synthetic route is outlined in Scheme 1. According to previous reports, the starting materials 3-hydroxy-3-methyl-2-butanone and malononitrile could form the tricyanofuran intermediate in the presence of EtONa. Then, the tricyanofuran compound was reacted with 4-hydroxybenzaldehyde to give the fluorophore TCF-OH. Finally, the phenolic hydroxyl group of TCF-OH was sulfonated to afford the probe TCF-SULF. All synthesized compounds were characterized using 1H NMR, 13C NMR, and MS.
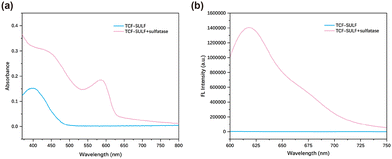
We evaluated the spectral properties of the probe TCF-SULF in PBS solution (1% DMSO, 10 mM, pH = 7.4). In the absence of sulfatase, TCF-SULF exhibited an absorption peak at 400 nm. However, after adding sulfatase, the absorption peak red-shifted, and a new absorption peak appeared at 580 nm (Fig. 1a). At the same time, under 580 nm excitation, the fluorescence emission at 620 nm was significantly enhanced (Fig. 1b). Moreover, the fluorescence quantum yield increased to 4.82% after the probe TCF-SULF responded to sulfatase. Therefore, the probe TCF-SULF can be used as a tool to detect sulfatase.
An important parameter to consider for enzyme-activated fluorescent probes is their time response. Under the conditions of 37 °C, the response time of sulfatase and the probe in PBS buffer solution was studied (Fig. S1, ESI†). When sulfatase (20 U mL−1) was present, a significant enhancement in the fluorescence signal could be detected within 2 minutes, indicating that the probe TCF-SULF had a rapid response capability to sulfatase (under 5 minutes). During the 0–30 min period, the fluorescence intensity continuously increased over time, reaching equilibrium at 30 minutes. This indicated the rapid response of the probe TCF-SULF to sulfatase.
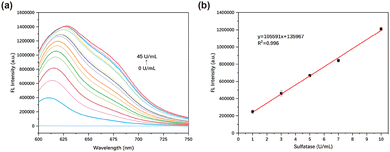
We investigated the fluorescence spectrum of TCF-SULF in the presence of different concentrations of sulfatase. When the concentration of sulfatase (0–45 U mL−1) increased, the fluorescence of TCF-SULF at 620 nm was significantly enhanced (Fig. 2a). The fluorescence intensity at 620 nm and the sulfatase concentration (0–10 U mL−1) exhibited an excellent linear connection in the probe–sulfatase reaction, with R2 = 0.996 (Fig. 2b). This indicated that the probe TCF-SULF had a satisfactory sensitivity for sulfatase detection. Furthermore, using the formula LOD = 3σ/k, it was found that the detection limit of the probe could be as low as 2.12 × 10−3 U mL−1, where σ represents the standard deviation of the blank measurement, and k represents the slope of the fluorescence intensity versus sulfatase concentration.
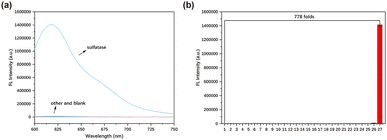
To validate the specificity of the probe, various interfering substances were introduced into the PBS solution containing TCF-SULF (1% DMSO, 10 mM, pH = 7.4) to assess the alterations in the solution's fluorescence. After adding other interfering substances, the fluorescence did not significantly increase. After adding sulfatase, the fluorescence increased by approximately 778 times compared to the blank group (Fig. 3). The research results indicated that, compared to other biomolecules and enzymes, the probe TCF-SULF had a higher selectivity for sulfatase.
When TCF-SULF interacted with sulfatase in a series of buffer solutions with pH values ranging from 3.0 to 10.0, a fluorescence enhancement phenomenon was observed for the probe at pH values of 3.0–8.0 (Fig. S2, ESI†). This phenomenon indicated that even under conditions where physiological parameters changed significantly (such as from acidic to alkaline ranges), TCF-SULF can still react with sulfatase to release fluorophores, thereby exhibiting a fluorescent signal. Therefore, this probe had the potential to be used as a biosensor for real-time monitoring of sulfatase activity changes in vivo or in vitro.
To deeply explore the activation mechanism of the probe TCF-SULF with sulfatase, we specifically conducted a docking study with sulfatase (PDB: 1AUK). The docking results revealed that TCF-SULF can easily embed itself into the hydrophobic cavity of the sulfatase (Fig. 4). It is worth noting that the anionic sulfate group of TCF-SULF was cleverly encapsulated within a catalytic pocket formed by two positively charged residues (Lys123 and Lys302). This structural adjustment significantly shortened the distance between them, thereby greatly enhancing the efficiency of the hydrolysis reaction. Furthermore, the sulfate group of the probe formed stable hydrogen bond interactions with other key residues including FGL69, Lys123, and Lys302, which was consistent with previous research reports.19,20 This indicated that the sulfate ester group played a crucial role in the recognition and binding process with sulfatases, primarily through hydrogen bond interactions.
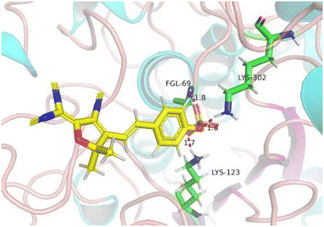 | ||
| Fig. 4 The binding site between TCF-SULF and sulfatase (PDB: 1AUK). The hydrogen bonds between the three residues (FGL69, Lys123, and Lys302) are represented in red. | ||
As shown in Scheme 2, the sensing mechanism of TCF-SULF for sulfatase. When TCF-SULF and sulfatase came into contact, their fluorescence and UV absorption spectra coincided, indicating that the reaction of TCF-SULF with sulfatase resulted in the release of the fluorophore TCF-OH (Fig. S3, ESI†). The identical peak as the TCF-OH molecular weight was seen at m/z 302.28 when the product was described using mass spectrometry (Fig. S4, ESI†). Thus, providing more evidence for the expected response mechanism.
To enhance the cell membrane permeability, probe TCF-SULF was encapsulated with DSPE-PEG2000 to afford the nanoprobe TCF-SULF NPs. Using a Malvern particle size analyzer and transmission electron microscopy techniques, a detailed characterization study of the nanoprobes was conducted (Fig. S5, ESI†). The hydrated average particle size of the TCF-SULF NPs reached 159 nm, while the PDI value was 0.227, indicating that the nanoprobes belong to a system with moderate dispersion and maintain an ideal distribution state. Further observation of the transmission electron microscope results revealed that the nanoparticles exhibited a perfect spherical shape and uniform size, approximately 50 nm.
Before bioimaging TCF-SULF NPs in living cells, the cytotoxicity of TCF-SULF NPs on MCF-7 cells was evaluated using the MTT assay (Fig. S6, ESI†). Over 80% of MCF-7 cells were still alive after a 24-hour incubation period, suggesting that TCF-SULF NPs exhibited low cytotoxicity and were suitable for application in vivo and in living cells.
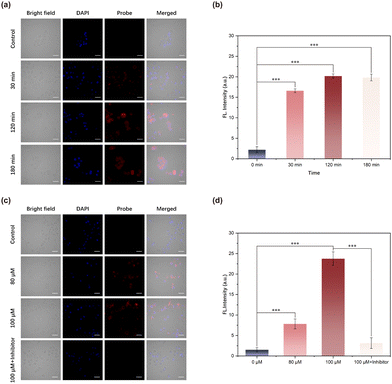
After co-incubating TCF-SULF NPs with cells for 0.5 hours, red fluorescence can be observed within the cells. The fluorescence signal increased with time and stabilized after approximately 2 hours of incubation (Fig. 5a and b). It indicated the potential of the probe TCF-SULF NPs to enable long-term detection of sulfatase. MCF-7 cells were incubated with different concentrations of TCF-SULF NPs (0, 80, and 100 μM) for 2 hours, rinsed with PBS buffer solution after incubation, and then the nuclei were counterstained with DAPI and imaged under an inverted fluorescence microscope (Fig. 5c and d). As the probe concentration increased, the intracellular fluorescence signal gradually intensified, while cells pre-incubated with the esterase inhibitor showed a significant reduction in intracellular fluorescence signal. This indicated the ability of the probe TCF-SULF NPs to specifically detect sulfatase.
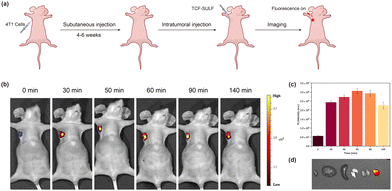
To further explore the imaging potential of TCF-SULF NPs on sulfatase overexpression in mouse tumors in vivo, a mouse xenograft tumor model was created by transplanting MCF-7 cells (1 × 106 cells) overexpressing sulfatase into the left axilla of female nude mice. After tumor formation, anesthesia was applied to the mice. After anesthesia, TCF-SULF NPs (400 μM, 50 μL) were injected into the tumor, and in vivo imaging of the nude mice was performed using a small animal imaging system (Fig. 6a). Due to the rapid cleavage of sulfate ester bonds by sulfatase, strong fluorescence was observed at the tumor site after 30 minutes, reaching its peak at 60 minutes, accompanied by a high tumor-to-normal tissue (T/N) ratio (Fig. 6b and c). The organs were imaged in vitro after the mice were put to sleep. Using fluorescence imaging, tumor tissue with a T/N ratio of 6 could be easily identified from other organs (Fig. 6d). Therefore, the nano TCF-SULF NP probe could serve as an auxiliary tool for resection of the tumor as a contrast agent.
In summary, we have designed and synthesized a TCF-based NIR fluorescent probe TCF-SULF, which uses sulfate esters as recognition sites and exhibits high selectivity and sensitivity towards sulfatase in PBS buffer. Compared to the buffer without sulfatase, the fluorescence intensity increased by 778 times, with an LOD of 2.1 × 10−3 U mL−1. Moreover, the probe has low cytotoxicity and can effectively image the overexpressed sulfatase in MCF-7. Importantly, in the mouse xenograft model, the probe TCF-SULF is capable of imaging tumors. Given its excellent properties, TCF-SULF is expected to become a promising tool for monitoring sulfatases.
We acknowledge funding support from the National Natural Science Foundation of China (NSFC 82071979 and 82202237).
Data availability
The data supporting this article have been included as part of the ESI.†Conflicts of interest
There are no conflicts to declare.Notes and references
- D. Ghosh, Cell. Mol. Life Sci., 2007, 64, 2013–2022 CrossRef CAS PubMed.
- R. Hobkirk, Trends Endocrinol. Metab., 1993, 4, 69–74 CrossRef CAS PubMed.
- M. Morimoto-Tomita, K. Uchimura, Z. Werb, S. Hemmerich and S. D. Rosen, J. Biol. Chem., 2002, 277, 49175–49185 CrossRef CAS PubMed.
- R. Shah, J. Singh, D. Singh, A. S. Jaggi and N. Singh, Eur. J. Med. Chem., 2016, 114, 170–190 CrossRef CAS PubMed.
- M. Sinreih, T. Knific, M. Anko, N. Hevir, K. Vouk, A. Jerin, S. Frković Grazio and T. L. Rižner, Front. Pharmacol., 2017, 8, 368 CrossRef PubMed.
- D. J. Ye, Y. J. Kwon, S. Shin, H. S. Baek, D. W. Shin and Y. J. Chun, Biomol. Ther., 2017, 25, 321–328 CrossRef CAS PubMed.
- P. Nussbaumer and A. Billich, Med. Res. Rev., 2004, 24, 529–576 CrossRef CAS PubMed.
- H. Y. Yoon, J. H. Lee, S. B. Park, S.-H. Choi, J.-S. Lee and J.-I. Hong, Dyes Pigm., 2022, 205, 110517 CrossRef CAS.
- M. P. Cosma, S. Pepe, I. Annunziata, R. F. Newbold, M. Grompe, G. Parenti and A. Ballabio, Cell, 2003, 113, 445–456 CrossRef CAS PubMed.
- H.-J. Park, H.-W. Rhee and J.-I. Hong, Bioorg. Med. Chem. Lett., 2012, 22, 4939–4941 CrossRef CAS PubMed.
- J. Chen, Z. Peng, M. Ji and P. Wang, Bioorg. Chem., 2023, 138, 106655 CrossRef CAS PubMed.
- R. Maltais, A. Ngueta Djiemeny, J. Roy, X. Barbeau, J.-P. Lambert and D. Poirier, Bioorg. Med. Chem., 2020, 28, 115368 CrossRef CAS PubMed.
- T. H. V. Vu, H.-H. Lim and H.-S. Shin, Bull. Korean Chem. Soc., 2020, 41, 424–432 CrossRef CAS.
- S. Pajarola, C. Weißenberg, F. Baysal, G. Bruchelt, I. Krägeloh-Mann and J. Böhringer, J. Chromatogr. B: Biomed. Sci. Appl., 2019, 1124, 109–113 CrossRef CAS PubMed.
- L. Xu, Y. Deng, H. Gao, Y. Yao, X. Liu, W. Zhan, G. Liang and X. Sun, Nanoscale, 2024, 16, 11538–11541 RSC.
- W. Li, S. Yin, X. Gong, W. Xu, R. Yang, Y. Wan, L. Yuan and X. Zhang, Chem. Commun., 2020, 56, 1349–1352 RSC.
- H.-H. Deng, H.-P. Peng, K.-Y. Huang, S.-B. He, Q.-F. Yuan, Z. Lin, R.-T. Chen, X.-H. Xia and W. Chen, ACS Sens., 2019, 4, 344–352 CrossRef CAS PubMed.
- Y. Chen, W. Shi, Y. Xu and P. Wang, Chem. Commun., 2023, 59, 9754–9757 RSC.
- S. R. Hanson, M. D. Best and C.-H. Wong, Angew. Chem., Int. Ed., 2004, 43, 5736–5763 CrossRef CAS PubMed.
- Y. Xu, M. Cui, W. Zhang, T. Liu, X. Ren, Y. Gu, C. Ran, J. Yang and P. Wang, Chem. Eng. J., 2022, 428, 132514 CrossRef CAS.
Footnotes |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d5cc00196j |
| ‡ These authors contributed equally. |
| This journal is © The Royal Society of Chemistry 2025 |