 Open Access Article
Open Access ArticleComparative analysis of sulfated L-idose and L-iduronic acid in neoproteoglycan cell surface engineering†
Rakesh
Raigawali‡
 a,
Saurabh
Anand‡
a,
Ankita
Chandra
a,
Virendrasinh
Mahida
a,
Preeti Ravindra
Bhoge
a,
Jancy Nixon
Abraham
a and
Raghavendra
Kikkeri
a,
Saurabh
Anand‡
a,
Ankita
Chandra
a,
Virendrasinh
Mahida
a,
Preeti Ravindra
Bhoge
a,
Jancy Nixon
Abraham
a and
Raghavendra
Kikkeri
 *ab
*ab
aIndian Institute of Science Education and Research, Pune 411008, India
bDepartment of CPAS, Jackson State University, Jackson 39217, USA. E-mail: rkikkeri@iiserpune.ac.in; Fax: +91-20-25908207; Tel: +91-20-25908207
First published on 5th May 2025
Abstract
Herein, we report the synthesis of heparan sulfate (HS) proteoglycan mimetics bearing iduronic acid (IdoA) and sulfated L-idose (Ido) hexasaccharides to assess how these isostructural sugars with similar charge density influence neoproteoglycan display on the cell membrane. PG@I2, carrying sulfated L-idose, showed rapid internalization in both cancerous and normal cells, whereas PG@I1, containing native IdoA expressed on the cell membrane and slowly internalized, underscoring the role of IdoA in HSPG cell surface engineering.
Heparan sulfate (HS) is a polysulfated glycosaminoglycan (GAG) widely present on cell surfaces and within the extracellular matrix. HS engages in interactions with a multitude of proteins, including growth factors, chemokines, and enzymes, playing a pivotal role in signaling pathways critical to cellular growth, differentiation, immune responses, and angiogenesis.1 Structurally, HS consists of repeating disaccharide units composed of D-glucuronic acid (GlcA) or its epimer L-iduronic acid (IdoA), linked to glucosamine (GlcN) residues.2 These units exhibit diverse sulfation patterns and acetylation. The conformational flexibility of IdoA, particularly its ability to adopt multiple forms such as the chair (1C4), skew-boat (2S0), and boat conformations, enables specific and dynamic interactions with proteins.3 This structural diversity endows HS with exceptional functional versatility and specificity in biological recognition processes. Consequently, there is considerable research interest in understanding the specific roles of each component of HS in detail. Among these, L-iduronic acid is particularly intriguing due to its synthetic challenges and conformational flexibility.4 Previously, L-iduronic acid in the anticoagulant drug fondaparinux and idraparinux was substituted with isostructural sugars, including D-glucuronic acid, D-xylose and 6-deoxy-L-talose moiety respectively. These substitutions resulted in the loss of anticoagulant activity, emphasizing the critical importance of L-iduronic acid in the anticoagulation activity of the drugs.5,6 To elucidate the importance of the specific conformation of L-iduronic acid in anticoagulation activity, 1C4 and 2S0-conformation of L-iduronic acid were locked in idraparinux and their anticoagulation activities were compared.7 These studies underscore the crucial importance of the 2S0 conformational of L-iduronic acid in idraparinux for its anticoagulant function. Recently, we reported the synthesis of sulfated homo-oligo L-iduronic acid and elucidated the significance of the trisaccharide as the minimal binding motif required for FGF2 recognition.4 Another significant characteristic of HS is its anionic sulfate-carboxylate composition, which plays a key role in biological recognition. However, the rationale behind nature's selection of such anionic combination remains unclear, as does the potential impact of fully sulfated HS. To explore this question, herein, we synthesized two hexasaccharides: one incorporating the native L-iduronic acid ligand and the other containing sulfated L-idose as the uronic acid residue and conjugated onto a neoproteoglycan backbone to investigate their role in cell surface engineering.8 Despite both molecules possessing 12 anionic groups, they are expected to exhibit distinct cell surface arrangement and internalization behaviors. These findings highlight the critical importance of the L-iduronic acid of HSPGs cell surface engineering (Fig. 1).
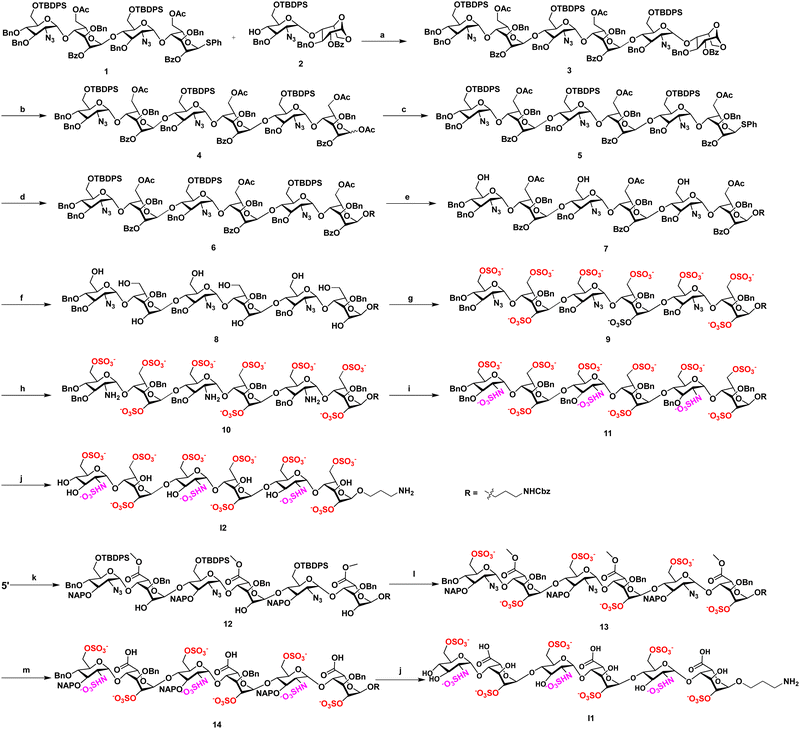
We synthesized sulfated L-idose and L-iduronic acid HS hexasaccharides using a [4+2] glycosylation strategy involving donor 1 and acceptor 2, both prepared according to previously reported procedures.9 The glycosylation of 1 and 2, in the presence of N-iodosuccinimide (NIS) and trimethylsilyl trifluoromethanesulfonate (TMSOTf) as a promoter, resulted in hexasaccharide 3. This intermediate underwent acetolysis using acetic anhydride and copper(II) trifluoromethanesulfonate as a catalyst. Subsequent treatment with phenyl trimethylsilyl sulfide and ZnI2 generated the corresponding thioglycoside donor 5. Further, linker glycosylation was performed, followed by selective deprotection of TBDPS groups using a hydrogen fluoride: pyridine complex in pyridine. Deacetylation and debenzoylation using a lithium hydroxide solution, along with O-sulfation in the presence of SO3·Et3N in DMF, yielded the sulfated L-idose hexasaccharide precursor 9 (Scheme 1). The C-2 azide group of 9 was then reduced to an amine using trimethylphosphine, resulting in compound 10. N-Sulfation of the amine was achieved using the SO3·pyridine complex. Finally, global deprotection through hydrogenolysis afforded the desired I2. Similarly, deacetylation of intermediate 5′, followed by TEMPO-mediated oxidation and subsequent methyl esterification, yielded compound 12. Compound 12 was further subjected to selective TBDPS deprotection, O-sulfation, reduction, N-sulfation, and global deprotection following the previously described procedures, yielded I1.9 Both hexasaccharide were conjugated to DBCO linker and the crude DBCO conjugates were functionalized with a fluorescent amphiphilic peptide PG@N3, to obtain desired neoproteoglycans (neoPGs: PG@I1 and PG@I2). NeoPGs were purified by HPLC using MeOH/H2O as an eluent. The product purity and conjugation were confirmed from IR, HPLC and mass spectra of the complex.
To decode the role of L-idose and L-iduronic acid in the synthesized compounds PG@I1 and PG@I2 in cell surface engineering, MDA-MB-468 (aggressive breast cancer cell line), MCF-7 (mild breast cancer cells) and NIH-3T3 (normal fibroblast cell line) were used. A solution of neoPGs (2 μM) was incubated with the cells for 30 minutes, followed by washing to remove the excess neoPGs from the medium. For understanding the internalization of neoPGs in the cell line, time dependent confocal imaging was performed. Confocal images revealed significant differences in the plasma membrane (PM) expression and internalization of the neoPGs. In MDA-MB-468 cells, PG@I2 demonstrated internalization within 30 minutes (Fig. 2(ii)), whereas PG@I1 exhibited intense fluorescence on the PM and continued to localize there even after 4 hours. The flow cytometric measurement after 1.5 h showed similar amount of neoPGs internalization, indicating that the structural heterogeneity on neoPGs modulates the cell surface engineering process (Fig. 2(v)) This noteworthy difference in the colocalization process suggests that PG@I1 is more effective in decorating the cell membrane compared to PG@I2. To confirm the decoration of the cell membrane and internalization of PG@I1 and PG@I2, co-staining was performed using a green fluorescent anti-cadherin antibody (Fig. 2(iii)). Surprisingly even at 4 h of incubation of neoPGs there was a clear difference between PG@I1 (Pearson coefficient: p ∼ 0.5) and PG@I2 (p ∼ 0.29). A distinct coalescing of both green and red fluorescence on the cell membrane was observed for PG@I1, revealing a better ability of glycocalyx engineering. Interestingly, PG@I1 exhibited slightly faster internalisation in MCF-7 and NIH-3T3 cells compared to MDA-MB-468. In contrast, PG@I2 showed no evident cell surface decoration. These observations align with our previous findings,8a highlighting that variations in glycocalyx composition and surface receptors between normal, mild and triple negative breast cancer cells influence neoproteoglycan cell surface presentation.
This differential behaviour can be attributed to two key structural factors. Firstly, PG@I2 exhibits a significantly higher negative charge than PG@I1 at physiological pH, which enhances its interaction with positively charged domains on cell surface receptors. This stronger electrostatic attraction facilitates more efficient endocytosis. Given that NIH-3T3, MCF-7, and MDA-MB-468 cells possess distinct surface zeta potentials,10 molecules with higher negative charge densities are internalized more rapidly than their less charged counterparts. Secondly, the native I1 is likely to interact with a broader spectrum of cell surface receptors and exhibits greater stabilisation on the cell membrane compared to I2. Overall, native heparan sulphate (HS) ligands appear to be crucial for the expression of neoPGs on the cell surface, whereas the highly sulphated PG@I2 primarily promotes endocytosis and may serve as a promising platform for cargo delivery applications.
In summary, we successfully synthesized HS hexasaccharides of L-iduronic acid (I1) and 6-O-sulfated L-idose residue (I2) using a [4+2] glycosylation strategy. These HS ligands were functionalized on amphiphilic glycopeptides through a copper-free click reaction. Cell surface engineering experiments revealed marked differences between the two molecules. PG@I1 remained associated with the cell membrane for an extended period, while PG@I2 was internalized within minutes. These findings highlight the critical role of the L-iduronic acid in the cell surface decoration of proteoglycans, whereas complete sulfation of HS ligands promotes endocytosis.8a We are currently exploring the potential cargo delivery applications of PG@I2 and the glycocalyx remodeling capabilities of PG@I1.
This work is supported by IISER, Pune, DST (Grant No. SR/NM/NS-1113/2016), DBT (Grant No. BT/PR21934/NNT/28/1242/2017), P. R. B. acknowledges DST WOS-A grant SR/WOS-A/CS-72/2019 for financial support. All cells lines are procured from NCCS cell repository, Pune. India.
Data availability
The data supporting this article is provided in the ESI.†Conflicts of interest
There are no conflicts to declare.Notes and references
- (a) I. Capila and R. J. Linhardt, Angew. Chem., Int. Ed., 2002, 41, 391–412 CrossRef; (b) J. R. Bishop, M. Schuksz and J. D. Esko, Nature, 2007, 446, 1030–1037 CrossRef CAS PubMed; (c) D. Xu and J. D. Esko, Annu. Rev. Biochem., 2014, 83, 129–157 CrossRef CAS PubMed.
- (a) C. Noti and P. H. Seeberger, Chem. Biol., 2005, 12, 731–756 CrossRef CAS PubMed; (b) J. T. Gallagher and J. E. Turnbull, Glycobiology, 1992, 2, 523–528 CrossRef CAS PubMed; (c) D. Spillmann and U. Lindahl, Curr. Opin. Struct. Biol., 1994, 4, 677–682 CrossRef CAS; (d) K. S. Rostand and J. D. Esko, Infect. Immun., 1997, 65, 1–8 CrossRef CAS PubMed.
- (a) J. Kreuger, D. Spillmann, J. P. Li and U. Lindahl, J. Cell Biol., 2006, 174, 323–327 CrossRef CAS PubMed; (b) C. Noti and P. H. Seeberger, Chem. Biol., 2005, 12, 731–756 CrossRef CAS PubMed; (c) J. C. Muñoz-García, J. López-Prados, J. Angulo, I. Díaz-Contreras, N. Reichardt, J. L. de Paz, M. Martín-Lomas and P. M. Nieto, Chem. – Eur. J., 2012, 18, 16319–16331 CrossRef PubMed.
- C. D. Shanthamurthy, A. Gimeno, S. Leviatan Ben-Arye, N. V. Kumar, P. Jain, V. Padler-Karavani, J. Jiménez-Barbero and R. Kikkeri, ACS Chem. Biol., 2021, 16, 2481–2489 CrossRef CAS PubMed.
- C. A. A. Van Boeckel and M. Petitou, Angew. Chem., Int. Ed. Engl., 1993, 32, 1671–1690 CrossRef.
- (a) F. Demeter, T. Demeter, Z. Bereczky, K. E. Kövér, M. Herczeg and A. Borbás, Sci. Rep., 2018, 8, 13736 CrossRef PubMed; (b) M. Herczeg, F. Demeter, E. Lisztes, M. Racskó, B. I. Tóth, I. Timári, Z. Bereczky, K. E. Kövér and A. Borbás, J. Org. Chem., 2022, 87, 15830–15836 CrossRef CAS PubMed.
- S. K. Das, J. M. Mallet, J. Esnault, P. A. Driguez, P. Duchaussoy, P. Sizun, J. P. Herault, J. Herbert, M. Petitou and P. Sinaÿ, Chem. –Eur. J., 2001, 7, 4821–4834 CrossRef CAS PubMed.
- (a) S. Anand, P. R. Bhoge, R. Raigwali, S. V. Saladi and R. Kikkeri, Chem. Sci., 2024, 15, 19962–19969 RSC; (b) S. Anand, S. Mardhekar, P. R. Bhoge, S. K. Mishra and R. Kikkeri, Chem. Commun., 2024, 60, 4495–4498 RSC.
- (a) P. Jain, C. D. Shanthamurthy, P. M. Chaudhary and R. Kikkeri, Chem. Sci., 2021, 12, 4021–4027 RSC; (b) S. Mardhekar, B. Subramani, P. Samudra, P. Srikanth, V. Mahida, P. Ravindra Bhoge, S. Toraskar, N. M. Abraham and R. Kikkeri, Chem. – Eur. J., 2023, 29(7), e202202622 CrossRef CAS PubMed; (c) S. Spijkers-Shaw, R. Devlin, N. J. Shields, X. Feng, T. Peck, G. Lenihan-Geels, C. Davis, S. L. Young, A. C. La Flamme and O. V. Zubkova, Angew. Chem., Int. Ed., 2024, 63, e20231691 CrossRef PubMed; (d) P. Chopra, T. Yadavalli, F. Palmieri, S. A. K. Jongkees, L. Unione, D. Shukla and G. J. Boons, Angew. Chem., Int. Ed., 2023, 62, e202309838 CrossRef CAS PubMed; (e) Y. P. Hu, Y. Q. Zhong, Z. G. Chen, C. Y. Chen, Z. Shi, M. M. Zulueta, C. C. Ku, P. Y. Lee, C. C. Wang and S. C. Hung, J. Am. Chem. Soc., 2012, 134, 20722–20727 CrossRef CAS PubMed; (f) K. N. Baryal, S. Ramadan, G. Su, C. Huo, Y. Zhao, J. Liu, L. C. Hsieh-Wilson and X. Huang, Angew. Chem., Int. Ed., 2023, 62, e202211985 CrossRef CAS PubMed.
- (a) Y. Zhang, M. Yang, Z. H. Park, J. Singelyn, H. Ma, M. J. Sailor, E. Ruoslahti, M. Ozkan and C. Ozkan, Small, 2009, 5(17), 1990–1996 CrossRef CAS PubMed; (b) R. B. Selvi, S. Chatterjee, D. Jagadeesan, P. Chaturbedy, B. S. Sumar, M. Sswaramoorthy and T. K. Kundu, J. Nanobiotechnol., 2012, 10, 35 CrossRef CAS PubMed.
Footnotes |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d5cc00527b |
| ‡ Equal contribution. |
| This journal is © The Royal Society of Chemistry 2025 |


![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
: