Open Access Article
Open Access ArticleInherent antibacterial properties of mannose-containing polynorbornene glycomaterials†
Brady A.
Hall
ab,
Ophelia J.
Wadsworth
a,
Logan M.
Breiner
ac,
Jacob C.
Chappell
a,
Andrew S.
Brenner
a,
Jennifer P.
McCord
a,
Andrew N.
Lowell
 *ac and
Michael D.
Schulz
*ac and
Michael D.
Schulz
 *abc
*abc
aDepartment of Chemistry, Virginia Polytechnic Institute and State University, Blacksburg, VA 24061, USA. E-mail: alowell@vt.edu; mdschulz@vt.edu
bMacromolecules Innovation Institute, Virginia Polytechnic Institute and State University, Blacksburg, VA 24061, USA
cCenter for Emerging, Zoonotic, and Arthropod-borne Pathogens, Virginia Polytechnic Institute and State University, Blacksburg, VA 24061, USA
First published on 25th February 2025
Abstract
Monosaccharides are typically employed as targeting ligands in antimicrobial polymers, yet we discovered that certain glycopolymers prepared by ring-opening metathesis polymerization display inherent antibacterial activity, despite lacking conventional antimicrobial groups. Mannose-functionalized polymers proved potent against Escherichia coli, which could be rescued by adding excess mannose to the growth medium.
Bacterial infections are one of the leading causes of mortality worldwide.1 The emergence of antimicrobial-resistant (AMR) bacterial strains has diminished the effectiveness of antibiotics,2 necessitating development of new treatment methods. Beyond new small-molecule antibiotics, antimicrobial polymers hold promise for treating AMR pathogens because they often possess broad-spectrum antibacterial activity, sustained inhibition, improved biocompatibility, and less susceptibility to resistance development by bacteria.3,4
Carbohydrates, both in glycopolymers and as small molecules, often play a key role in antibacterial efficacy. For example, many small-molecule antibiotics, such as macrolides5 and aminoglycosides,6 use saccharides in their pharmacophore, while glycopeptides7 incorporate saccharides to enhance their affinity for bacterial cell membranes. More broadly, carbohydrates serve critical functions in many physiological pathways such as cell signaling, immunity, inflammation, and molecular recognition.8,9 For example, many pathogens initiate infection by docking to specific glycans on the surface of targeted cells,10 such as Escherichia coli, which adheres to oligomannosides.11–13 By mimicking these interactions, glycopolymers can target the bacterial surface. Consequently, these materials have attracted considerable attention in the context of antimicrobial countermeasures14–16 and other biomimetic interactions.17
Natural glycopolymers have been investigated for antimicrobial applications, but diverse sourcing and production processes often result in batch-to-batch variations.18,19 In contrast, synthetic glycopolymers reduce this variation,20 allowing reproducibility and control over a material's molecular structure and physical properties. This control enables the synthesis of materials that can leverage glycan-mediated recognition by incorporating specific mono-, di-, or oligosaccharides into the polymer.21,22 Moreover, advances in polymerization techniques have enabled the synthesis of well-defined glycopolymers with low dispersities.6,23 Ring-opening metathesis polymerization (ROMP) is an especially attractive choice for preparing synthetic glycopolymers due to its tolerance of a wide variety of functional groups.24–26 Typically, antimicrobial polymers use cationic groups27 or conjugated antibiotics28 as antimicrobial agents, with glycans serving as targeting ligands to enhance interactions with bacterial cell membranes. Here, we investigate the antibacterial properties of a norbornene-based glycopolymer series, revealing molecular weight dependent activity even in the absence of other antimicrobial moieties.
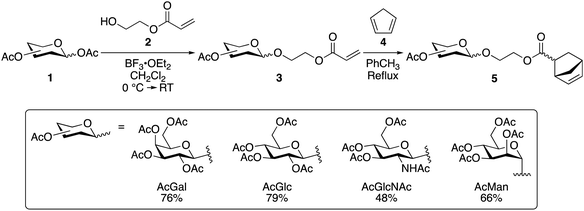
We synthesized monomers containing four different monosaccharides: β-D-galactose, β-D-glucose, β-D-N-acetyl glucose, or α-D-mannose. We initially attempted to polymerize acrylate-functionalized monosaccharides (3) directly, but high molecular weights and narrow dispersities proved challenging to obtain through controlled radical polymerization of these monomers, as per previous reports.29,30 Consequently, we investigated norbornene-based glycomonomers for ROMP.
Historically, norbornene-based glycomonomers are synthesized by installing an (oligo)saccharide onto an existing norbornene monomer; we instead developed an alternative synthetic strategy where the norbornene is formed directly from an acrylate-functionalized monosaccharide (Scheme 1). Coupling norbornene alcohols to a monosaccharide via a Koenigs–Knorr reaction proved problematic, presumably due to a competitive reaction between the norbornene alkene and BF3·OEt2. Instead, we directly modified each monosaccharide (1) with 2-hydroxyethyl acrylate (2) using the Koenigs–Knorr reaction, followed by a Diels–Alder reaction of the resulting acrylate glycomonomer (3) with cyclopentadiene (4) to generate the norbornene glycomonomers (5).
This protocol was convenient and could be completed without an intermediate purification step of 3, as 5 was easily chromatographed. The ethyl norbornene carboxylate glycomonomers (5) were isolated as clear oils with overall yields ranging from 48% for GlcNAc-5 to 79% for Glc-5, with a ratio of approximately 70/30 endo/exo norbornene. Hydroxyethyl norbornene carboxylate prepared in an analogous fashion gave a similar endo/exo ratio.
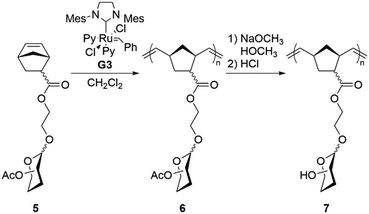
We polymerized these glycomonomers using ROMP to produce a series of glycopolymers (Scheme 2). Polymers with four degrees of polymerization (DP = 50, 100, 250, and 500) were targeted to evaluate polymer molecular weight effects on antibacterial activity. 1H NMR spectroscopy was used to monitor the conversion of 5 to 6, showing complete disappearance of 5 after one hour, and the molecular weights of 6 were analyzed using size-exclusion chromatography (Table 1). General agreement was observed between the theoretical and actual molecular weights. In some cases, however, the dispersity was higher than often seen in ROMP, possibly due to the nature of the glycomonomers themselves, which have several heteroatoms that may coordinate to the metal center and adversely affect the polymerization. As ruthenium is known to be cytotoxic, residual catalyst was removed by filtering each polymer through a pad of silica gel. Deacetylation of 6 was carried out using sodium methoxide to produce 7, as indicated by the complete disappearance of the acetyl CH3 signals in the NMR spectrum.
Disk diffusion assays showed deprotected glycopolymers (7) did not inhibit the growth of Staphylococcus aureus NorA knockout mutant (ΔNorA) but did produce zones of inhibition against E. coli TolC knockout mutant (ΔTolC) with the Man-7 glycopolymers (Fig. S33, ESI†). These strains were selected as weakened representatives of Gram-positive and Gram-negative bacteria. The selectivity towards E. coli is especially promising: Gram-negative bacteria are particularly difficult to kill due to their outer membrane, but this feature is presumably targeted by these glycomaterials based on the lack of activity against S. aureus.
To quantify the activity, broth dilution assays using the four series of glycopolymers were tested with E. coli ΔTolC along with a low-molecular-weight polyethyleneimine (PEI, 600 Da) as a positive control. Glycomaterials functionalized with mannose (Man-7) were most effective (Table 2). The activity of the Man-7 polymers increased with DP, indicating that the molecular weight likely plays a role in the mode of action. Indeed, the deacetylated D-mannose glycomonomer had MIC values of 5 mg mL−1 or greater, indicating that the polymeric structure enhances activity. We performed dynamic light scattering (DLS) measurements on the mannose-functionalized materials and observed aggregates of ∼100–200 nm at all molecular weights, with small populations of micron-scale aggregates occurring with higher molecular weight materials. Consequently, these polymers are likely interacting with the bacteria in an aggregated form, at least initially. The fact that the aggregates are of similar sizes for all molecular weights, however, suggests that the observed molecular weight-dependent inhibition may be driven by non-aggregated polymer–bacteria interactions.
| MIC (mg mL−1) | ||||
|---|---|---|---|---|
| Gal-7 | Glc-7 | GlcNAc-7 | Man-7 | |
| Monomer | >5 | >5 | 5 | 5 |
| DP = 50 | >5 | >5 | 2.5 | 2.5 |
| DP = 100 | 5 | 5 | 5 | 2.5 |
| DP = 250 | 5 | 5 | 5 | 1.25 |
| DP = 500 | 5 | 5 | 5 | 1.25 |
While the MIC values for the larger mannose glycomaterials are relatively high, the fact that these glycomaterials show any antimicrobial activity is surprising. Previous studies treated sugars only as targeting agents to potentiate the activity of other moieties, rather than exploring the possibility of their inherent antimicrobial effects. Man-7 polymers were bacteriostatic at these concentrations with bactericidal concentrations being 4-fold greater for the most active polymers (Table S3, ESI†).
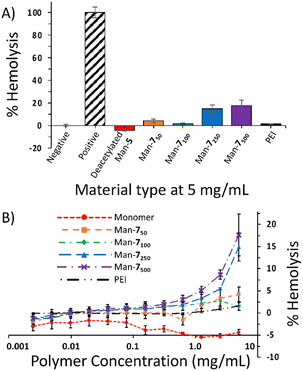
As a preliminary evaluation of toxicity against non-bacterial cells, we conducted a hemolysis assay using the Man-7 polymer series. The hemolysis assay demonstrated that Man-7 minimally lysed red blood cells (RBCs) at lower molecular weights (Fig. 1A) with Man-750 and Man-7100 causing 4.2% and 1.5% lysis, respectively, at the highest concentration tested (5 mg mL−1) (Fig. 1B). However, the Man-7250 and Man-7500 (the best inhibitors of E. coli with MICs of 1.25 mg mL−1) caused 15% and 18% lysis, respectively, at 5 mg mL−1, indicating that membrane interactions and lysis by these mannose-containing glycomaterials are not restricted to bacteria. The PEI control did not show significant lysis of RBCs at the concentrations and incubation time used.
As the disk diffusion assay results showed no activity against Gram-positive S. aureus, the mechanism of action is likely some form of interaction with the outer membrane that is present exclusively in Gram-negative bacteria. In a previous study by Liu et al.,31 modifying PEI with mannose increased the activity of PEI 22-fold against E. coli through enhanced interaction with the cell membrane. PEI kills bacteria by rupturing membranes, a mode of action driven by the positive charges on the polymer. Mannose-functionalized PEI is hypothesized to bind to FimH proteins,32 resulting in more effective delivery of PEI to the E. coli, thereby enhancing activity. While attraction to fimbriae or another component of the outer membrane is likely with our mannose-functionalized polymers, the lack of a positive charge on the polynorbornene backbone suggests the activity of Man-7 may function through a different mechanism.
Cationic amphiphilic polymers are attracted to the negative charge of the bacterial cell membranes.14,33 After binding, their hydrophobic groups create small pores, enabling fluid entry that leads to lysis. Man-7 may cause lysis in the same manner, but its lack of positive charge suggests that the attractive force is derived from the mannose. Consequently, we hypothesize that the pendent mannoses are recognized by the cell surfaces, causing an attractive interaction. Once the mannose glycopolymer and cell surface come into contact, the hydrophobic backbone may interact with the lipid bilayer, causing disruption, with higher molecular weight polymers having an amplified interaction. Indeed, hydrophobicity has been shown to enhance antibacterial properties and produce greater hemolytic activity due to cell membrane disruption.34 However, this explanation is undermined by the lack of activity of the lower molecular weight materials, which have the same ratio of hydrophobic to hydrophilic moieties. Molecular weight is clearly playing a central role, as is mannose functionalization, since the other glycopolymers did not display similar activity despite having the same backbone.
Alternatively, a combination of mannose-driven attraction and backbone rigidity may drive activity. The stiffness of the rings and double bonds of the polymer backbone may transfer to the cell membrane, producing greater rigidity and reducing fluctuations, thus preventing growth. The observed bacteriostatic inhibition (Table S3, ESI†) supports an interaction of this type. This effect would be expected to increase with higher molecular weights and is in accord with the observed results (Table 2).35 Furthermore, we probed the ability of free mannose to rescue E. coli from the inhibitory polymer by adding 1× and 10× mass equivalencies of mannose along with the polymer to the bacterial culture (Table S4, ESI†). No significant change in inhibition was observed when an equal mass of mannose was present in the growth medium, but at 10-fold mannose no inhibition was observed. This finding suggests that sufficiently high concentrations of free mannose can outcompete binding of the Man-7 polymer to the bacterial membrane. These results underscore the role of mannose recognition in the polymer's antibacterial activity.
While these data support interaction with the cell surface, another possibility is that the higher DP polymers may simply better permeate the membrane and inhibit bacterial growth via interaction with an intracellular target. Regardless of the mechanism, the higher molecular weight Man-7 polymers showed greater antibiotic activity, demonstrating that mannose-functionalized polymers are themselves inherently antimicrobial, even without traditionally active functional groups.
In conclusion, glycopolymers with pendant monosaccharides were synthesized to evaluate how polymer composition and molecular weight impact antibacterial properties. We developed a facile synthesis of norbornene glycomonomers (5) that required only a single purification and could be polymerized using ROMP. Higher molecular weight polymers (DP = 250 and 500) were effective at inhibiting E. coli, and MIC assays revealed that the mannose-functionalized polymers showed bacteriostatic inhibition at concentrations as low as 1.25 mg mL−1, similar to the known antimicrobial polymer PEI. These results demonstrate that saccharides cannot be treated merely as targeting agents on antibacterial polymers but can contribute antimicrobial activity themselves.
This work was supported by GlycoMIP, an NSF Materials Innovation Platform funded through Cooperative Agreement DMR-1933525, and Virginia Tech's Department of Chemistry.
Data availability
The data supporting this article have been included as part of the ESI.†Conflicts of interest
There are no conflicts to declare.Notes and references
- E. J. Toone, Bacterial Infections Remains a Leading Cause of Death in Both Western and Developing World, John Wiley & Sons, Hoboken, New Jersey, 2011 Search PubMed.
- R. Laxminarayan, et al. , Lancet Infect. Dis., 2013, 13(12), 1057 CrossRef PubMed.
- A. Arora and A. Mishra, Mater. Today Proc., 2018, 5, 17156 CrossRef CAS.
- L. Timofeeva and N. Kleshcheva, Appl. Microbiol. Biotechnol., 2011, 89(3), 475 Search PubMed.
- A. G. Myers and R. B. Clark, Acc. Chem. Res., 2021, 54, 1635 CrossRef CAS PubMed.
- T. Pelras and K. Loos, Prog. Polym. Sci., 2021, 117, 101393 CrossRef CAS.
- E. van Groesen, P. Innocenti and N. I. Martin, ACS Infect. Dis., 2022, 8, 1381 CrossRef CAS PubMed.
- P. M. Rudd, T. Elliott, P. Cresswell, I. A. Wilson and R. A. Dwek, Science, 2001, 291(5512), 2370 CrossRef CAS PubMed.
- H. E. Murrey and L. C. Hsieh-Wilson, Chem. Rev., 2008, 108(5), 1708 Search PubMed.
- A. Imberty and A. Varrot, Curr. Opin. Struct. Biol., 2008, 18(5), 567 Search PubMed.
- J. Bouckaert, et al. , Mol. Microbiol., 2005, 55(2), 441 CrossRef CAS PubMed.
- M. Cauwel, et al. , Chem. Commun., 2019, 55(68), 10158 RSC.
- F. Lupo, M. A. Ingersoll and M. A. Pineda, Immunology, 2021, 164, 3 CrossRef CAS PubMed.
- A. C. Englar, N. Wiradharma, Z. Y. Ong, D. J. Coady, J. L. Hedrick and Y.-Y. Yang, Nano Today, 2012, 7, 201 CrossRef.
- R. H. Bianculli, J. D. Mase and M. D. Schulz, Macromolecules, 2020, 53, 9158 CrossRef CAS.
- U. I. M. Gerling-Driessen, M. Hoffman, S. Schmidt, N. L. Snyder and L. Hartmann, Chem. Soc. Rev., 2023, 52, 2617–2642 RSC.
- A. G. Kruger, et al. , ACS Cent. Sci., 2021, 7(4), 624 CrossRef CAS PubMed.
- J. J. Lundquist and E. J. Toone, Chem. Rev., 2002, 102(2), 555 Search PubMed.
- J. Li and S. Zhuang, Eur. Polym. J., 2020, 138, 109984 CrossRef CAS.
- Y. Miura, Y. Hoshino and H. Seto, Chem. Rev., 2016, 116(4), 1673 CrossRef CAS PubMed.
- N. Jayaraman, Chem. Soc. Rev., 2009, 38(12), 3463 RSC.
- J. L. J. Blanco, C. O. Mellet and J. M. G. Fernández, Chem. Soc. Rev., 2013, 42(11), 4518 Search PubMed.
- I. Pramudya and H. Chung, Biomat. Sci., 2019, 7(12), 4848 RSC.
- K. H. Mortell, M. Gingras and L. L. Kiessling, J. Am. Chem. Soc., 1994, 116(26), 12053 CrossRef CAS.
- S. Meier, H. Reisinger, R. Haag, S. Mecking, R. Mülhaupt and F. Stelzer, Chem. Commun., 2001, 855 RSC.
- Z. Liu, Y. Zhu, W. Ye, T. Wu, D. Miao, W. Deng and M. Liu, Polym. Chem., 2019, 10(29), 4006 RSC.
- D. Pranantyo, L. Q. Xu, Z. Hou, E.-T. Kang and M. B. Chan-Park, Polym. Chem., 2017, 8(21), 3364 RSC.
- J. Chen, et al. , Biomaterials, 2019, 195, 38 Search PubMed.
- P. Escalé, S. R. S. Ting, A. Khoukh, L. Rubatat, M. Save, M. H. Stenzel and L. Billon, Macromolecules, 2011, 44(15), 5911 CrossRef.
- A. B. Lowe, B. S. Sumerlin and C. L. McCormick, Polymer, 2003, 44(22), 6761 CrossRef CAS.
- M. Liu, J. Li and B. Li, Langmuir, 2018, 34(4), 1574 CrossRef CAS PubMed.
- P. Klemm and M. A. Schembri, Int. J. Med. Microbiol., 2000, 290, 27 CrossRef CAS PubMed.
- I. Sovadinova, E. F. Palermo, R. Huang, L. M. Thoma and K. Kuroda, Biomacromolecules, 2011, 12, 260–268 CrossRef CAS PubMed.
- M. F. Ilker, K. Nüsslein, G. N. Tew and E. B. Coughlin, J. Am. Chem. Soc., 2004, 126, 15870–15875 CrossRef CAS PubMed.
- S. Salinas-Almaguer, et al. , Sci. Rep., 2022, 12, 933 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d5cc00590f |
| This journal is © The Royal Society of Chemistry 2025 |