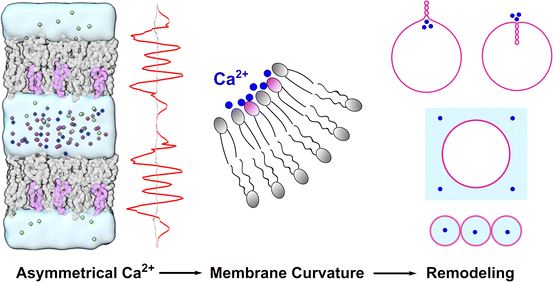
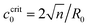Asymmetrical calcium ions induced stress and remodeling in lipid bilayer membranes†
Chang
Liu
 ab,
Qi
Zhong
c,
Kai
Kang
ab,
Qi
Zhong
c,
Kai
Kang
 ab,
Rui
Ma
ab,
Rui
Ma
 *c and
Chen
Song
*c and
Chen
Song
 *ab
*ab
aPeking-Tsinghua Center for Life Sciences, Academy for Advanced Interdisciplinary Studies, Peking University, Beijing 100871, China. E-mail: c.song@pku.edu.cn
bCenter for Quantitative Biology, Academy for Advanced Interdisciplinary Studies, Peking University, Beijing 100871, China
cCollege of Physical Science and Technology, Xiamen University, Xiamen 361005, China. E-mail: ruima@xmu.edu.cn
First published on 22nd November 2024
Abstract
Ca2+ ions play crucial roles in regulating many chemical and biological processes, but their impact on lipid bilayer membranes remains elusive, especially when the impacts on the two leaflets are asymmetrical. Using a recently developed multisite Ca2+ model, we performed molecular dynamics simulations to study the impact of Ca2+ on the properties of membranes composed of POPC and POPS and observed that both the structure and fluidity of the membranes were significantly affected. In particular, we examined the influence of asymmetrically distributed Ca2+ on asymmetric lipid bilayers and found that imbalanced stress in the two leaflets was generated, with the negatively charged leaflet on the Ca2+-rich side becoming more condensed, which in turn induced membrane curvature that bent the membrane away from the Ca2+-rich side. We employed continuum mechanics to study the large-scale deformations of a spherical vesicle and found that the vesicle can go through vesiculation to form a multi-spherical shape in which a number of spheres are connected with infinitesimal necks, depending on the specific Ca2+ distributions. These results provide new insights into the underlying mechanisms of many biological phenomena involving Ca2+-membrane interactions and may lead to new methods for manipulating the membrane curvature of vesicles in chemical, biological, and nanosystems.
Introduction
Biological membranes are essential for the integrity of cells, providing a barrier between the internal and external environments. They also accommodate a great diversity of proteins, which participate in a wide range of critical biological processes, such as energy or signal transduction, solute transport, cell–cell recognition, and trafficking.1 The physical properties of membranes are important for their barrier function, which can be regulated by the surrounding environmental factors, such as the presence of various molecules and ions. For example, previous studies have shown that the adsorption of Ca2+, one of the most important secondary messengers in cells, can influence many physical properties of membranes, such as the thicknesses,2 head group orientations of lipids,2–5 ordering of acyl chains,3,5–10 lipid dehydration,2,4–6,8 and lateral diffusion.10–16Phosphatidylcholine (PC) is the most abundant zwitterionic phospholipid in eukaryotic cell membranes and subcellular organelles.17 Phosphatidylserine (PS) is an anionic phospholipid present in most inner leaflets of cellular organelles, including the plasma membrane, where it accounts for as much as 28% of all lipids.18–21 Therefore, biological membranes are intrinsically asymmetric with different lipid compositions in the inner and outer leaflets, and a mixture of PC and PS is often used as a general model for membrane studies. Moreover, the concentration of calcium ions across membranes is often distributed asymmetrically. Intracellularly, the free calcium ion concentration typically ranges between 10 and 100 nM. However, within organelles such as the endoplasmic reticulum (ER) and sarcoplasmic reticulum (SR), the concentration can reach significantly higher levels, ranging from 60 to 500 μM, and in some cases, even up to 1 mM.22,23 In contrast, the extracellular free calcium ion concentration is approximately 1.8 mM.24 In addition, PS lipids were found to play additional roles by interacting with signaling proteins,25 regulating surface charges and protein localization,26 and inducing protein aggregation.27,28 Some proteins attract PS lipids by nonspecific electrostatic interactions, and the binding can be regulated by Ca2+.25 Therefore, the interaction between Ca2+ and the negatively charged PS is of particular interest.
It is well documented that the dehydrated head groups of PS and PIP2, induced by Ca2+ adsorption, could lead to separation of lipid components, aggregation of certain types of lipids, phase transitions of membranes, and even collapse of the membrane bilayer structure in experiments.13–16,29,30 Notably, there were two contradictory experimental results with respect to the Ca2+ induced membrane curvature, one showing that the presence of Ca2+ on one side of a membrane can induce a negative curvature31,32 such that the membrane bends away from Ca2+, and the other showing the opposite.33 To fully understand the experimental observations and the underlying mechanisms, the atomistic interactions between Ca2+ and membranes, particularly the impact of asymmetrically distributed Ca2+ on asymmetric membranes, need to be further investigated.
Molecular dynamics (MD) simulation is a powerful tool to reveal molecular interactions and has also been extensively utilized to study the interactions between Ca2+ and membranes.2–5,9–12,34,35 However, previous studies showed that the Ca2+ models used in classical nonpolarizable force fields were often inaccurate in describing the Ca2+-biomolecule interactions.36 Recently, we developed a multisite Ca2+ model, which significantly improved the calculation accuracy for the Ca2+–protein interaction.37 Considering that the dielectric constant of membranes is close to proteins, we expect that the new model would generate more reliable simulation results in the study of Ca2+-membrane interactions as well.
Another limitation of MD simulation in the membrane study is the use of periodic boundary conditions, which makes it difficult to observe large-scale deformations and remodeling of membranes. To solve this problem, a strategy of combining multiscale methods is helpful. Therefore, in this study, we combined MD simulations and continuum mechanics calculations to investigate the physical properties and lateral pressure profiles of POPC/POPS membranes in the presence of Ca2+, as well as the resulting curvature and steady shapes of membranes. Our results showed that the mechanical properties of lipid bilayers, especially PS-rich bilayers, can be significantly influenced by the presence of Ca2+, and asymmetrically distributed Ca2+ ions lead to asymmetrical lateral stress in the two leaflets of lipid bilayers. With this information adopted, continuum mechanics calculations showed that asymmetric stress can induce a negative curvature on membranes with respect to excessive Ca2+ ions, and the resulting steady shape of vesicle membranes, either locally or globally spherical or multi-spherical, depends on the specific Ca2+ distribution around the membrane. These results can improve our understanding of Ca2+-induced membrane remodeling, which may regulate many biological processes, such as endocytosis, exocytosis, membrane fusion and fission, and perhaps those activities mediated by mechanosensitive membrane proteins as well.18,38
Results
Adsorption and distribution of ions around lipid bilayer membranes
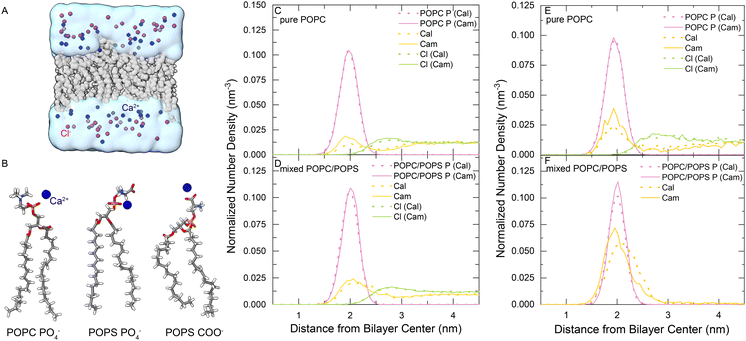
We performed MD simulations for membrane systems in a solution of CaCl2. The membranes were composed of either pure POPC or a mixture of POPC![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) POPS (3
POPS (3![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) (Fig. 1(A)), and both the recently developed multisite Ca2+ model37 and the default Ca2+ model of the CHARMM force field were used in the simulations for comparison, which were denoted Cam and Cal, respectively. The Ca2+ concentration was raised stepwise from 0 to 50, 100, 200, 500 and 1,000 mM. The majority of the membrane-bound Ca2+ ions bind to the phosphate groups of POPC and POPS, or the carboxylate groups of POPS lipids in simulations (Fig. 1(B)). Then, we analyzed the distribution of various groups along the membrane's normal direction in the equilibrated simulation systems using the SuAVE software.39,40 The density profiles were similar with various CaCl2 concentrations, so we only showed the results obtained for two representative cases: 1 M CaCl2 – a high-concentration regime (Fig. 1(C) and (D)) and 200 mM CaCl2 – a low-concentration regime (Fig. 1(E) and (F)). Simulations with the two Ca2+ models both showed that the density peak of phosphate groups, often viewed as the membrane boundary, was located approximately 2 nm away from the membrane center (Fig. 1(C)–(F) pink lines). The density peak of Ca2+ roughly overlapped with that of phosphate groups, indicating the adsorption and accumulation of Ca2+ under the membrane surface. Notably, more Ca2+ was observed around this region in the POPC/POPS system than in the pure POPC system (Fig. 1(C)–(F), yellow lines), which is reasonable since POPS is negatively charged and has a stronger binding affinity to Ca2+.
1) (Fig. 1(A)), and both the recently developed multisite Ca2+ model37 and the default Ca2+ model of the CHARMM force field were used in the simulations for comparison, which were denoted Cam and Cal, respectively. The Ca2+ concentration was raised stepwise from 0 to 50, 100, 200, 500 and 1,000 mM. The majority of the membrane-bound Ca2+ ions bind to the phosphate groups of POPC and POPS, or the carboxylate groups of POPS lipids in simulations (Fig. 1(B)). Then, we analyzed the distribution of various groups along the membrane's normal direction in the equilibrated simulation systems using the SuAVE software.39,40 The density profiles were similar with various CaCl2 concentrations, so we only showed the results obtained for two representative cases: 1 M CaCl2 – a high-concentration regime (Fig. 1(C) and (D)) and 200 mM CaCl2 – a low-concentration regime (Fig. 1(E) and (F)). Simulations with the two Ca2+ models both showed that the density peak of phosphate groups, often viewed as the membrane boundary, was located approximately 2 nm away from the membrane center (Fig. 1(C)–(F) pink lines). The density peak of Ca2+ roughly overlapped with that of phosphate groups, indicating the adsorption and accumulation of Ca2+ under the membrane surface. Notably, more Ca2+ was observed around this region in the POPC/POPS system than in the pure POPC system (Fig. 1(C)–(F), yellow lines), which is reasonable since POPS is negatively charged and has a stronger binding affinity to Ca2+.
There were some differences in the distributions of Ca2+ around the membranes when using the two Ca2+ models. As shown in Fig. 1(C)–(F), more Cam Ca2+ was adsorbed into the POPC bilayer than Cal, although the difference in the mixed POPC/POPS was less significant. We analyzed the radial distribution function (RDF) of Ca2+ around the negatively charged groups of the lipid molecules (phosphate and carboxylate oxygen), which showed that the first density peak is within 3.2 Å (Fig. S1, ESI†). Therefore, we used 3.2 Å as the cutoff to determine the lipid-bound Ca2+ ions and the coordinated groups around them. We calculated the coordination numbers (Tabel S1, ESI†) by integrating the radial distribution function of its first peak. The RDFs of oxygen atoms of lipid molecules around the bound Ca2+ (Fig. S2, ESI†) showed that, compared to Cal, the Cam model greatly reduced the interaction with PS carboxylate groups, and increased the interaction with the phosphate groups in the mixed POPC/POPS bilayer systems. For example, for the Ca2+ ions bound to a mixed POPC/POPS bilayer in a 200-mM CaCl2 solution, there were approximately 0.99 phosphate (0.64 from POPC and 0.35 from POPS) and 0.28 carboxylate (from POPS) groups coordinated with Cam, while there were approximately 0.29 phosphate (0.19 from POPC and 0.10 from POPS) and 1.13 carboxylate (from POPS) groups coordinated with Cal (Table S1, ESI†). This might indicate an advantage of the multisite Cam model over the default Cal model in describing the Ca2+-membrane interactions, since previous experimental measurements showed that Ca2+ may have a stronger binding affinity to phosphate than to carboxylate groups in a POPS bilayer.2,41,42
Calcium ions alter the structure and fluidity of lipid bilayers
Area per lipid (APL) is a fundamental parameter describing the molecular packing of lipid bilayers and can reach stable values on a 100 nanosecond timescale in MD simulations. As shown in Fig. 2, APL gradually decreased in the process of adding more CaCl2 to the system (Fig. 2(A) and (B)), a phenomenon that is more significant in the mixed POPC/POPS membrane. When the Ca2+ concentration exceeded 200 mM, the change in APL became less pronounced. It appears that the effect of Ca2+ on the packing of membranes containing negatively charged PS is more significant. With saturated CaCl2 (200 mM), the APL was reduced by approximately 4.2% for the pure POPC bilayer (from ∼0.72 nm2 to ∼0.69 nm2) and 23.8% (from ∼0.88 nm2 to ∼0.67 nm2) for the mixed POPC/POPS bilayer. The reduction in APL was accompanied by an increase in bilayer thickness (Fig. 2(C) and (D)), which was also more significant in the mixed POPC/POPS bilayer. There was no significant difference in the APL and membrane thickness when using two different Ca2+ models, indicating that the observed impact of Ca2+ on the overall membrane size was rather robust and independent of the specific Ca2+ model used for simulations.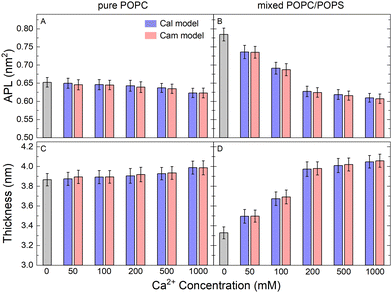 | ||
| Fig. 2 Structural properties of the simulated lipid bilayers. Area per lipid (APL) for pure POPC (A) and mixed POPC/POPS (B) lipid bilayers with increasing CaCl2 concentrations. The APL and bilayer thickness were calculated by the SuAVE software39,40(C) and (D). | ||
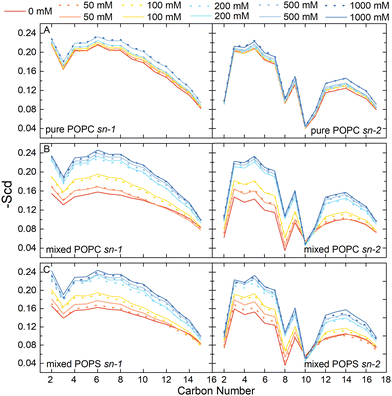
We also examined the structural properties of the lipid tails by analyzing the acyl chain C–H bond order parameters (eqn (1) and Fig. 3). Evidently, the lipid tails in the mixed POPC/POPS bilayer systems became more ordered with increasing concentrations of Ca2+ ions, which is consistent with previous findings,5,9,10 and the influence became less significant when the concentration of Ca2+ exceeded 200 mM. In contrast, the changes in the order parameters in the pure POPC bilayer systems were minor in all the simulations. The two Ca2+ models showed consistent effects on the acyl chain order parameters as well.
To further evaluate the performance of the Ca2+ models used in this study, we compared our simulation results with experimental data. The C–H bond order parameters obtained from NMR experiments serve as valuable metrics for validating the consistency of lipid structures in MD simulations with experimental data. These measures are particularly relevant as they are associated with key bilayer properties such as area per lipid, thickness, and the conformational variability of individual lipids.43 Previous MD studies used these metrics to evaluate the Ca2+ model with electronic continuum correction (ECC).34,35 Here, we also analyzed the changes of the headgroup C–H order parameters in our simulations with increasing CaCl2 concentrations and compared with experimental values.44,45 As can be seen, the change tendencies of the β, α, g3 order parameters were consistent with the experimental results when both the Cam and Cal models were used (Fig. S3 and S4, ESI†). However, it is also noticeable that most of the simulation results with the Cam model showed better consistency with the experimental data than the Cal model, except for the β order parameter in the pure POPC bilayer (Fig. S3B, ESI†). This indicates that our Cam model overall performed better than the default Cal model in simulating the lipid bilayer systems.
The lateral diffusion coefficient of lipids is closely related to the structure and phase of membranes, which is one of the most important dynamic parameters of biomembranes. From our atomistic MD simulations, we calculated the lateral diffusion coefficients of the lipid molecules in our model membranes from the mean square displacement of lipid phosphorus atoms (Fig. S5, ESI†). The diffusion constant was calculated by fitting a straight line from 100 ns to 150 ns according to eqn (2). The values were approximately 0.4 × 10−7 cm2 s−1 to 1.4 × 10−7 cm2 s−1 depending on the Ca2+ concentration (Fig. 4). As a comparison, previous experimental measurements from fluorescence recovery after photobleaching,46 pulsed NMR,47 single particle tracking,48 and fluorescence correlation spectroscopy49 yielded the similar magnitude of the diffusion coefficients of approximately 0.5 × 10−7 cm2 s−1 to 1.0 × 10−7 cm2 s−1. Interestingly, the presence of Ca2+ significantly reduced the mobility of lipid molecules in the mixed POPC/POPS bilayers, but not to the same extent in the pure POPC bilayers (Fig. 4). Therefore, our results showed that membranes containing negatively charged lipids can become significantly less diffusive in the presence of Ca2+. Again, the two Ca2+ models did not show significant differences in affecting the lipid diffusion coefficient.
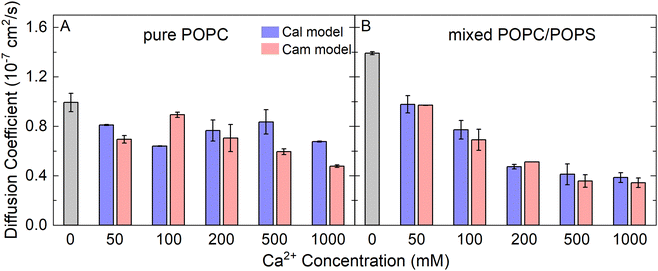 | ||
| Fig. 4 The lateral diffusion coefficients of lipid molecules in the pure POPC bilayers (A) and mixed POPC/POPS bilayers (B), with increasing CaCl2 concentrations. | ||
Asymmetrical calcium ions induced asymmetric pressure profiles across lipid bilayers
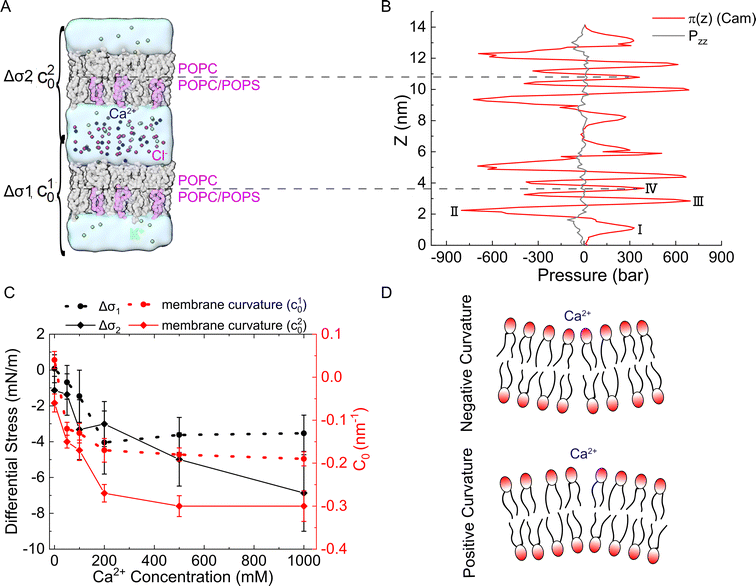
To further characterize the effect of Ca2+ ions on membrane properties, we computed the lateral pressure profiles (LPPs) across the lipid bilayers. Considering that PS lipids are mostly located in the inner leaflet of biological membranes, we set up a double-membrane and asymmetric simulation system as shown in Fig. 5(A), in which an asymmetric bilayer system was duplicated along the z direction. Please note that such a double-membrane system resulted in two isolated water compartments, as the top and bottom water regions were actually connected into one compartment due to the periodic boundary condition used in the simulations. K+ ions were used to neutralize the negative charges generated from PS and evenly distributed in the two water compartments, while different amounts of Ca2+ ions were added to the two water compartments to generate an asymmetrical Ca2+ distribution. Cl− ions were added to ensure that no charge imbalance was generated between the two water compartments. Thus, both the ion distribution and composition of lipid bilayers were asymmetrical, which can help to characterize the impact of asymmetrical Ca2+ distribution on biomembrane properties. For instance, the sarcoplasmic reticulum (SR) membrane faces a higher inner Ca2+ concentration than the outer, which resembles the upper lipid bilayer in Fig. 5(A), while the outer leaflet of plasma membranes may face a higher Ca2+ concentration than the inner leaflet that enriches POPS, which resembles the lower lipid bilayer in Fig. 5(A). Note that in our MD simulations, we used an NPT-ensemble such that the normal pressure pzz is kept almost constant along the z-axis (Fig. 5(B), black curve). This was achieved by adjusting the density difference of water across the membrane such that the osmotic pressure caused by Ca2+ density gradient is just balanced off by the opposite water density gradient. The same strategy was also used in the previous study.50The LPPs of the asymmetrical Ca2+ treated bilayer lipids are shown in Fig. 5(B). As can be seen, moving from the bulk solvent toward the bilayer center, LPPs displayed a moderate positive pressure peak (I) attributed to the repulsion of the lipid headgroups. A large negative peak (II) was observed in each leaflet, indicative of the lipid–water interfacial tension, followed by another positive peak (III) close to the center of the bilayer, which was demonstrated to align well with the location of the acyl chain double bonds and caused by the relative incompressibility of the double bonds in the unsaturated lipids.51–53 In addition, LPP had a positive peak at the bilayer center (IV), which was related to the geometries of the specific lipid types.54,55
We calculated the differential stress for each leaflet by integrating the lateral pressure π(z) along z from the bulk water to the bilayer center (Fig. 5(B)). The differential stress of the two leaflets was then obtained and denoted as Δσ1 and Δσ2 (Fig. 5(A)). We analyzed the lateral pressure profiles π(z) in simulation systems without Ca2+ to show the effect of asymmetric lipid compositions (Fig. S6, ESI†). The LPP was also aligned to the bilayer centers to give an easier visual comparison across bilayers (Fig. S6C and S7, ESI†). The Δσ1 and Δσ2 were 0.06 ± 0.68 mN m−1 and −1.05 ± 0.72 mN m−1 in this system, indicating that the lipid composition asymmetry in our simulations system did not lead to a significant differential stress between the two leaflets. Therefore, the differential stress shown in Fig. 5(C) was mostly caused by the addition Ca2+. As shown in Fig. 5(C), the addition of Ca2+ had a minor effect on Δσ1 (black dotted line), but the impact was significant for Δσ2 (black solid line), which was increased up to −6.37 mN m−1 while increasing Ca2+ concentration on the POPS-rich side. This indicates that the addition of Ca2+ to the negatively charged leaflet of membranes will lead to a significant change in the differential stress in this leaflet, and consequently, a stress imbalance will be generated in the two leaflets of the membrane.
Such a stress imbalance would cause a spontaneous curvature of the membrane. We calculated the curvature order parameter using the SuAVE software to explore the effects of Ca2+ on membrane shapes. The results showed no significant difference in the average order parameter across the simulation systems with various Ca2+ concentrations, and the values were consistent with those calculated for planar bilayers (Fig. S8, ESI†). This indicates that no significant curvature was generated by the presence of Ca2+ in the MD simulations, probably owing to the influence of the periodic boundary condition and the limited size of the simulation systems. Therefore, we had to use other methods to analyze the shape change of a continuous membrane.
Asymmetrical stress induces membrane curvature and remodeling
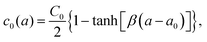
As a result of the asymmetrical stress within the lipid bilayer induced by the Ca2+ ions, the membrane tends to bend to release the stress. This tendency for the membrane to bend can be characterized by the spontaneous curvature c0 in the classical Helfrich model, which has been widely used to model the deformation of a membrane on a large scale.56–59 Herein, we studied the deformation of a spherical vesicle with a diameter of 100 nm with the Helfrich model, by employing the MD simulation results as the input physical parameters. Basically, we estimated the spontaneous curvature induced by the asymmetrically distributed Ca2+ ions viaeqn (6) based on the integral of the first moment of LPPs obtained from MD simulation analyses, which were then input to the Helfrich model to calculate the steady shape of the vesicle (see continuum mechanics calculation in the section of Materials and methods for a detailed description).Note that any asymmetries between the two leaflets could lead to spontaneous curvature of the membrane. Three types of asymmetries have been studied: (1) compositional asymmetry—one of the leaflets has a different lipid composition or different ratios of lipid compositions than the other; (2) number asymmetry—the two leaflets have different numbers of lipids; (3) environmental asymmetry —for example, the headgroups of the lipids in the two leaflets face different Ca2+ concentrations. The spontaneous curvature caused by differential stress as a result of number asymmetry (2) has been well characterized in ref. 50 and 60. In this paper, we studied both the compositional asymmetry (1) and environmental asymmetry (3), but neglected the number asymmetry by intentionally equalizing the number of lipid molecules in the two leaflets. As shown in Fig. 5(C), in the absence of Ca2+ concentration, compositional asymmetry alone could induce a spontaneous curvature c10 ≃ 0.04 nm−1 for the lower bilayer and c20 ≃ −0.06 nm−1 for the upper bilayer. Here we chose the sign convention that a negative spontaneous curvature tended to bend away from the middle water layer while a positive spontaneous curvatured tended to bend towards the middle water layer. The addition of Ca2+ ions significantly enhanced the upper bilayer's tendency to bend away from the middle water layer, and could reach up to c20 = − 0.3 nm−1 at high centration of Ca2+ ions. While for the lower bilayer, the presence of Ca2+ ions was able to reverse its bending direction, from towards the middle water layer to away from it. Combining the two results, we inferred that Ca2+ ions near a membrane tended to bend the membrane away from the Ca2+ ions. To verify that our observation was not caused by the specific double-membrane simulation setup, we calculated the curvature of another double membrane system, where the lower membrane is upside down, treated with 200 mM Ca2+ (Fig. S9, ESI†). We found that Ca2+ could still induce a negature curvature for the lower membrane (c10 ≃ −0.16 nm−1), which was qualitatively aligned with those of the original simulation system (Fig. 5(C)). Therefore, although the curvature values may vary depending on the specific simulation setup, the bending directions of the membranes are always consistent.
In order to calculate how a spherical vesicle of a diameter of 100 nm would deform in response to the change in spontaneous curvature c0, we needed to specify how the surface area A of the vesicle and the volume V enclosed by the vesicle varied when the spontaneous curvature c0 was changed. Here for simplicity, we considered two types of deformations: (i) in long timescale, the surface area A was assumed to be conserved, while the volume V was allowed to change to keep a fixed osmotic pressure p. (ii) In short timescale, the volume V was assumed to be conserved, while the surface area A was allowed to change to keep a fixed membrane tension σ. Our calculation results showed that the stable shapes under the two conditions are similar, therefore, we presented the results for type (i) deformation in the main text, while leaving the results for type (ii) deformation in the ESI† (Fig. S10 and S11).
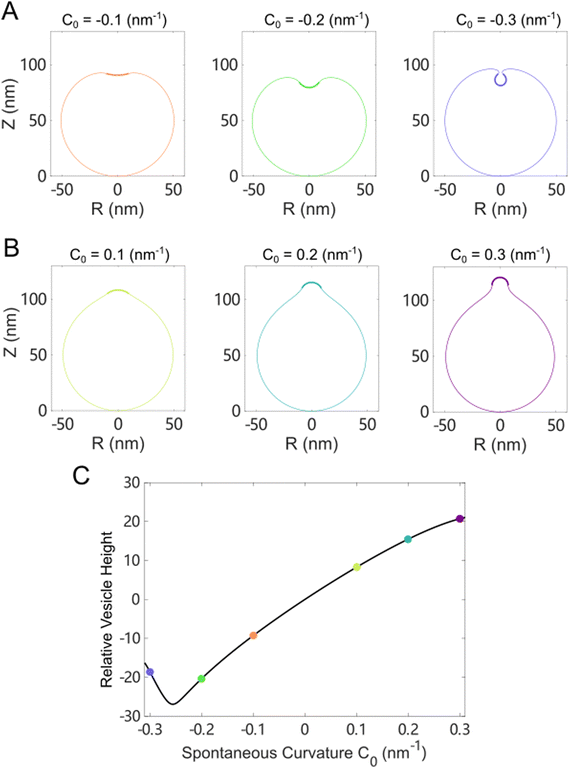
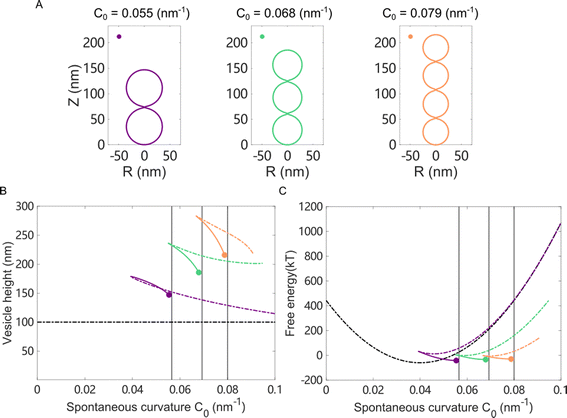
We first studied the case in which the area impacted by the Ca2+ ions was localized to a small patch (1% of the total surface area) of the vesicle near the north pole. For a vesicle of 100 nm in diameter, as the spontaneous curvature c0 was gradually increased from 0 to 0.3 nm−1, which matched the maximum spontaneous curvature estimated from eqn (6) in Fig. 5(C), the impacted membrane patch protruded further outward and developed into a hemispherical cap (Fig. 6). The positive spontaneous curvature could be induced by a local accumulation of Ca2+ in the inner part of the membrane. Similar trends were also observed for negative c0, in which the impacted membrane patch bent inward and developed into a small spherical vesicle with a diameter of ∼20 nm (Fig. 6(B)), as indicated by the relative height, which is the difference of the pole-to-pole distance between the deformed vesicle and the undeformed spherical vesicle (Fig. 6(C)). If the diameter of the undeformed spherical vesicle was increased to 200 nm, the impacted membrane patch (1% of the total surface area) would form a pearl-shaped structure in which multi-spherical vesicles are interconnected via narrow necks (Fig. S12, ESI†).
When the Ca2+ ions were uniformly distributed in the exterior or interior of the spherical vesicle, i.e., the spontaneous curvature of the entire vesicle was impacted by the asymmetric Ca2+ ions, the shape transformation showed dramatically different trends between negative and positive values of c0. For negative spontaneous curvature c0 < 0 (Ca2+ outside the vesicle), the spherical shape remained the most stable one (data not shown). However, for positive spontaneous curvature c0 > 0 (Ca2+ inside the vesicle), we found branches of solutions such that the vesicle exhibited a multi-spherical shape in which several smaller vesicles are connected with narrow necks (Fig. 7(A)). There existed critical values of 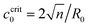 , where n is an integer, at which the neck is infinitesimally narrow (Fig. 7(B) and (C), indicated by the vertical lines) and the large vesicle would break into a number of n smaller vesicles, so-called vesiculation. The physics behind the vesiculation phenomenon was investigated in reference.61 For a spherical vesicle of R0, when the spontaneous curvature was increased to ccrit0, the vesicle would assume a shape of n connected spheres each with a radius of
, where n is an integer, at which the neck is infinitesimally narrow (Fig. 7(B) and (C), indicated by the vertical lines) and the large vesicle would break into a number of n smaller vesicles, so-called vesiculation. The physics behind the vesiculation phenomenon was investigated in reference.61 For a spherical vesicle of R0, when the spontaneous curvature was increased to ccrit0, the vesicle would assume a shape of n connected spheres each with a radius of 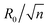 such that the total surface area of the n small vesicles equaled to the surface area of the original vesicle. The neck would assume a saddle shape whose mean curvature just matched ccrit0. In this configuration, the bending energy of the membrane was almost zero and, therefore was energetically more favorable.
such that the total surface area of the n small vesicles equaled to the surface area of the original vesicle. The neck would assume a saddle shape whose mean curvature just matched ccrit0. In this configuration, the bending energy of the membrane was almost zero and, therefore was energetically more favorable.
Note that our calculation of the membrane shape here assumes the cell is able to secret or expel other osmolytes to keep the osmotic pressure at a constant level via the osmoregulation process in the presence of Ca2+ gradient. Therefore, the major impact of the Ca2+ gradient on the membrane is to alter the spontaneous curvature of the membrane, while the osmotic pressure is kept constant. In fact, many organisms are able to keep the osmotic pressure at a relatively stable level.62 For instance, kidney cells play an important role in osmoregulation in humans by actively controlling the amount of hormones.
Discussion
The electrostatic properties of the membrane interface are of paramount importance in numerous situations. Ca2+ is particularly interesting in this respect due to its divalent nature. Although sharing common properties with other divalent cations, Ca2+ is one of the most critical secondary messengers and functions by inducing fine-tuned electrostatic effects. For example, it was shown that blood clotting proteins associated with membrane surfaces become activated during the coagulation cascade, and Ca2+ enhances protein penetration into the lipid monolayer and induces flocculation of surface lipid aggregates.63 The high Ca2+ levels inside presynaptic vesicles can prevent even the most lipophilic transmitters, such as dopamine from adhering to the inner leaflet surface, thus rendering unhindered neurotransmitter release feasible.64 Ca2+ also regulates the interactions between annexins and membranes, which may influence cell membrane-related domain organization, endocytic, exocytic transport, and trafficking.65,66 When Ca2+ ions were pumped into cells by ion channels or transporters, they can generate large clusters by linking PIP2.29 In contrast, monovalent cations such as Na+ and K+ form smaller and less stable clusters. Moreover, a synergy exists between Ca2+ and K+, facilitating the growth of PIP2 clusters and the formation of more complex structures within the cell.30 Therefore, there have been extensive studies seeking to elucidate the effects of Ca2+ on membrane properties.As early as 2006, Pedersen et al. performed MD simulations to study the interactions between cations and negatively charged lipid bilayers,9 and found that Ca2+ ions exert a pronounced effect on bilayers by binding to anionic groups of lipid molecules. Later, further studies showed that the binding of Ca2+ made the membrane bilayers more condensed, with decreasing APL, increasing thickness, and dehydration of the membrane interface. Notably, the specific lipid composition and chemistry can also influence these physical–chemical properties of membranes.2,4,10,67,68 Overall, the binding affinity between Ca2+ ions and lipids was significantly stronger than that of Na+,4 K+ and Mg2+ ions, regardless of the composition of the lipid bilayers.5 Meanwhile, we speculate that divalent cations such as Zn2+ would produce similar results to Ca2+, while tri-valent ions may induce even stronger effects. Overall, our simulations obtained results consistent with the previous findings. The binding of Ca2+ to anionic groups of lipid molecules leads to a reduction in membrane surface area and an increase in membrane thickness. Notably, the effect is more significant for bilayers containing negatively charged lipid molecules. As a consequence, the acyl chains become more ordered, which may facilitate phase transitions.29 Our results also showed that these findings are rather robust, irrespective of the specific ion model and parameters adopted in the MD simulations.
There are still some details that are dependent on the ion model and parameters adopted in the MD simulations. With the new multisite ion model, Ca2+ ions were found to be stably bound to the anionic groups of lipid molecules, with similar affinity to the phosphate groups and carboxylate groups (Table S1, ESI†). This is different from the previous simulation results that revealed a strong binding preference of Ca2+ to carboxylate groups,2,4,9,11,12 as also observed in our simulations with the default Ca2+ model. Notably, previous infrared absorption spectroscopy experiments suggested that Ca2+ ions probably do not stably bind to the carboxylate groups in PS, but can bind to phosphate groups to induce dehydration.41,42 There were also experiments showing that Ca2+ can bind to both carboxylate and phosphate groups of PS.2,3 In this aspect, our new multisite Ca2+ model may have the advantage of reducing the binding bias to carboxylate groups. Nonetheless, although these local differences resulting from specific ion models are appreciable, they do not generate qualitative differences in the global properties of the bilayers discussed in this work.
The effect of Ca2+ ions on the dynamic properties of lipid bilayers has not been systematically studied before. Our analysis of the MD trajectories showed that the lipid molecules become less diffusive upon Ca2+ binding, for both the zwitterionic and negatively charged lipid bilayers. However, the effect is significant for bilayers containing negatively charged lipids, but only minor for zwitterionic lipid bilayers. We attribute this to the electrostatic “glue” effect of Ca2+ on the negatively charged lipids, which makes those lipids more sticky and less diffusive. It appears that when the concentration of Ca2+ reached a certain threshold (200 mM in our simulations), the effect became saturated, probably because the binding between Ca2+ and lipids was saturated above the threshold and therefore the bilayer did not adsorb more Ca2+ to become stickier.
It should be noted that the Ca2+ distribution can often be asymmetrical on the two sides of membranes. For example, the plasma membrane often faces a much higher Ca2+ concentration on the outer leaflet than on the inner leaflet, while the SR membrane usually experiences a higher Ca2+ concentration on the inner leaflet than on the outer leaflet. In fact, membranes themselves are often asymmetric. For example, the inner leaflet of plasma membranes contains much more negatively charged lipid molecules than the outer leaflet. Therefore, studying the effect of asymmetrical Ca2+ on asymmetric membranes is of particular interest. In line with previous findings, our atomistic simulations showed that the Ca2+-exposed leaflet of membranes becomes more packed. One would expect such a scenario to generate imbalanced stress in the two leaflets of membranes. Indeed, our pressure profiles analysis showed that the addition of Ca2+ to the negatively charged leaflet leads to a significant imbalance of the lateral pressure profiles across the membrane Fig. 5(C), while the effect is much less pronounced in the case of the Ca2+ ions added to the leaflet with only zwitterionic lipids. As a consequence, this imbalanced lateral pressure distribution can induce spontaneous curvature of the membrane, as indicated by our continuum mechanics calculation and other recent studies.31,32,69 There were two experimental results regarding the effect of Ca2+ on the membrane's bending: one suggests a negative curvature,32i.e., bending away from the Ca2+, while the other suggests the opposite.33 Our simulation results are in line with Graber et al.'s experimental observation that Ca2+ can induce a negative curvature of membranes.32 We suspect that the observation of the positive curvature was due to the different experimental setup,33 which may introduce other factors that were not accounted for in simulations.
Interestingly, our continuum mechanics calculations showed that, when the impact of Ca2+ is focused on a local surface area of a vesicle, a pearl chain extending toward the other side of the membrane can be generated (Fig. 6 and Fig. S10, S12, ESI†). This is similar to recent observations that vesicle chain-growth from the GUV membrane can be maintained as long as there is a supply of membrane material and Ca2+, analogous to native secretory vesicles.31,32 This kind of remodeling can only be generated by an asymmetric effect in the two leaflets of membranes, which may be related to membrane budding and fusion,70,71 morphology of ER exits,72 as well as the generation of interconnected vesicles.73 For example, dynamic membrane remodeling may be achieved by the interplay between lipids and Ca2+ to initiate the formation of tubules.74–76 In contrast, when the impact of Ca2+ is uniform on the membrane surface and Ca2+ ions are added to the outside, the vesicle tends to remain spherical. When low-concentration Ca2+ ions are uniformly added to the inside, vesicles tend to remain spherical as well. Intriguingly, the vesicle will become multi-spherical while more Ca2+ ions are added to the inside (larger spontaneous curvature). This suggests that a significant reduction in stress within the inner leaflet of membranes may facilitate vesicle fission, and could potentially aid cell division as well. A significant increase in stress within the outer leaflet of membranes should have a similar effect, and so does the combination of the two. It should be noted though, we did not explicitly consider the asymmetry of the lipid composition of membranes in our continuum mechanics calculations. Moreover, the actual membrane compositions and environments are much more complex than the ideal cases studied here, so the eventual shape may be the result of many factors, including the distribution of proteins.77 Nonetheless, the distribution of Ca2+ and negatively charged lipid molecules and their interplay may play important roles here.
Our MD simulations and continuum mechanics calculations provided additional atomistic details of Ca2+-membrane interactions, and further clarified the question about the controversial membrane curvature caused by Ca2+. Although the Ca2+ concentration used here was much greater than the average physiological Ca2+ concentration measured experimentally, the electrolytic environment around the biomembranes was by no means a static average distribution of ions. Significant localized concentration spikes of Ca2+ ions occur along Ca2+ signaling pathways, which are always challenging to measure experimentally and can span in excess of three orders of magnitude, and the Ca2+ concentration near membranes are known to be higher than that in bulk.78,79 Therefore, we believe that our results are not only quantitatively informative for chemical systems but also qualitatively meaningful for biological systems, which would be helpful for the understanding of many membrane stress/curvature-related processes.
A potential issue in our MD simulation was that we used the same number of lipids for the asymmetric bilayers to remove the effects of lipid numbers, which may lead to a somewhat frustrated state of the bilayer in the rectangular simulation box due to the size difference of the asymmetric lipids. To alleviate this concern, we implemented a correction by subtracting the lateral pressure of the system in the absence of calcium ions from the lateral pressure of each simulation scenario. This adjustment can effectively neutralize the effects of the potential frustration induced by the asymmetric lipid composition.
Considering the intrinsically asymmetric nature of plasma membranes, with mostly zwitterionic lipids on the outer leaflet and abundant negatively charged lipids on the inner leaflet, it may be reasonable to think that evolution has made the outer surface of cells more inert to external cation distribution in order to maintain a stable shape, while the inner surface of cells is more sensitive to Ca2+ for responding to internal signaling. In addition, by influencing the mechanical properties of membranes, Ca2+ may also regulate membrane-bound proteins, especially mechanosensitive proteins. For example, mechanosensitive ion channels are involved in many physiological processes, such as touch, hearing, balance, and the sensation of pain,80,81 which can be activated by forces exerted on membranes and the resulting surface tension, imbalanced stress, and membrane curvature. Therefore, the imbalanced stress across membranes and the resulting membrane curvature induced by Ca2+ might provide an additional regulatory mechanism for the activation of mechanosensitive channels. The addition of Ca2+ ions to vesicles containing negatively charged lipids may provide a straightforward way to manipulate membrane curvature, which can be useful for regulating many membrane-involved biological, chemical, and nanoscale processes, such as membrane fusion, drug delivery, and even capturing the activated structures of mechanosensitive ion channels. In addition, Ca2+ may also have indirect effects on membrane structures, for example through calcium-activated scramblases. When those proteins are activated by Ca2+, PS lipids would flip from the inner leaflet to the outer leaflet, which might also change the membrane structures.52
Conclusion
In this work, we combined MD simulations and continuum mechanics to study how Ca2+ ions influence the microscopic properties and macroscopic deformation of lipid membranes composed of POPC and POPS. We found that both the structure and fluidity of the lipid membranes were significantly affected by the presence of Ca2+ ions. A gradient of Ca2+ across the lipid bilayer could create imbalanced stress in the two leaflets, with the negatively charged leaflet on the Ca2+-rich side becoming more condensed, thus inducing membrane curvature that bent the membrane away from the Ca2+-rich side. For a spherical vesicle, the asymmetric distribution of Ca2+ on the two sides of the membrane could deform the vesicle into a locally or a globally multi-spherical shape in which a number of spheres are connected with infinitesimal necks. These results are summarized in Fig. 8, which can help clarify the question about the controversial membrane curvature caused by Ca2+ and may lead to new methods for manipulating the membrane curvature of vesicles in chemical, biological, and nanosystems.Materials and nethods
Simulation systems
The molecular dynamics simulation systems were composed of pure POPC or mixed POPC/POPS in an aqueous CaCl2 solution. Each single-membrane system was composed of 128 lipids and approximately 4368 water molecules, with varying concentrations of CaCl2 (Table 1). The bilayers, composed of either POPC or POPC/POPS, were symmetric in these single-membrane systems (Fig. 1(A)).| No. of bilayers | Bilayer composition | Lipid amounts | [Ca2+] (mM) | Duration (ns) | Repeats |
|---|---|---|---|---|---|
| Single | POPC | 128 | [0, 50, 100, 200, 500, 1000] | 400 | 1 |
| POPC/POPS | 96/32 | 400 | 1 | ||
| Double | POPC![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) POPC/POPS POPC/POPS |
128![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 96/32 96/32 |
500 | 5 |
To study the effect of asymmetrical Ca2+ on the lateral stress of asymmetric membranes, double-membrane systems were generated by duplicating a single-membrane system along the z direction (Fig. 5(A)). The outer leaflet and inner leaflet of each asymmetric lipid bilayer consisted of POPC (64) and POPC![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) POPS (48
POPS (48![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 16), respectively. In these double-membrane systems, K+ ions were added to neutralize the negative charges of PS lipids. Ca2+ ions were added to only one water compartment. Cl− ions were added to neutralize the systems and eliminate the transmembrane potential caused by ion distributions. The details of all the simulation systems are listed in Table 1.
16), respectively. In these double-membrane systems, K+ ions were added to neutralize the negative charges of PS lipids. Ca2+ ions were added to only one water compartment. Cl− ions were added to neutralize the systems and eliminate the transmembrane potential caused by ion distributions. The details of all the simulation systems are listed in Table 1.
Molecular dynamics simulations
All MD simulations were performed using GROMACS software 2018.6.82 The CHARMM36 force field and the CHARMM implementation of the TIP3 water model were used for the simulations.83–85 In addition to the default ion models (Ca2+, K+, Cl−) of the CHARMM force field, we utilized the recently developed multisite Ca2+ model that was optimized to achieve more accurate binding affinity with proteins.37 The temperature was kept constant at 310 K using the Nose–Hoover thermostat.86,87 The pressure was maintained at 1 bar using the semi-isotropic Parinello–Rahman barostat,88,89 with a time constant of 1 ps and compressibility of 4.5 × 10−5 bar−1. A cutoff of 1.2 nm was used for the Lennard-Jones interaction, with a smooth shift function from 1.0 to 1.2 nm. The particle mesh Ewald (PME) method was used for the long-range electrostatics.90 After energy minimization, we ran 200-ps NVT and 500-ps NPT simulations to preliminarily equilibrate the systems, and then we carried out 400-ns production simulations for the single-membrane systems and 500-ns production simulations for the double-membrane systems (Table 1).Analysis of MD trajectories
If not otherwise stated, the last 300 ns trajectories were used for the analysis of the single-membrane systems; while for the double-membrane systems, the last 200 ns trajectories were used for analysis.The order parameters of lipid tails were analyzed using the following definition:
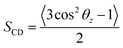 | (1) |
The lateral diffusion coefficient of lipids was calculated with the following equation:
| 〈r2(t)〉 = 〈(r(t) −r(0))2〉 = 4Dt | (2) |
The lateral pressure profiles (LPPs) were calculated using the GROMACS-LS tool.51 The calculation of the LPPs was based on the last 200 ns of the simulation data (Table 1), with the coordinates and velocities saved every 10 ps. An electrostatic cutoff of 2.0 nm was used for the LPP calculation.51 The profiles were calculated with about 140 slabs, corresponding to an approximate slab width of 1 Å. The outputs of GROMACS-LS were the components of the stress tensor, σ, as a function of the z coordinate (bilayer normal). LPP was calculated using the formula
| π(z) = PL(z) − PN | (3) |
Continuum mechanics calculations
We first explained how to estimate the spontaneous curvature caused by asymmetric Ca2+ concentration from the MD simulations. When two lipid monolayers were combined to form a bilayer, we used the mid-plane between the two monolayers to represent the shape of the bilayer, which was parameterized as X. This position was denoted as z = 0 along the normal direction of the bilayer. Assume the thickness of the two monolayers were d+ and d−, the differential stress Δσ is defined as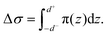 | (4) |
We calculated the spontaneous curvature of each monolayer via the integral50,60
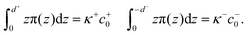 | (5) |
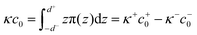 | (6) |
For both the bilayer and the monolayer, the first moment of the LLP π(z) gave the product of the bending rigidity and the spontaneous curvature. In order to obtain the spontaneous curvature, the values for the bending rigidity κ and κ± were needed which in principle could be obtained by a buckling simulation.91 Here for simplicity we assumed κ± = 12.5 kBT and κ = κ+ + κ− = 25 kBT, which were the typical values for the plasma membrane made of POPC.92–94 Here we neglected the effect that the bilayer bending rigidity κ could be different for the two monolayers due to compositional asymmetry. In fact, the bending rigidity can be influenced by the lipid composition, as well as ions in the external environment. If the membrane is made of a mixture of POPC and POPS (70![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 30), the surface charge might stiffen the membrane and increase the bending rigidity from 25 kB to 30 kBT.93 However, the presence of ions can counteract the effect of surface charge and soften the membrane.95 As a result of the complicated nature of the bending rigidity, the approximation κ± = 12.5 kBT may affect the quantitative accuracy of the calculated spontaneous curvature, but not the sign of it (bending direction of membranes).
30), the surface charge might stiffen the membrane and increase the bending rigidity from 25 kB to 30 kBT.93 However, the presence of ions can counteract the effect of surface charge and soften the membrane.95 As a result of the complicated nature of the bending rigidity, the approximation κ± = 12.5 kBT may affect the quantitative accuracy of the calculated spontaneous curvature, but not the sign of it (bending direction of membranes).
We next explained how to calculate the deformation of the membrane as a result of the spontaneous curvature c0 induced by a local asymmetric Ca+ concentration between the inside and outside of the membrane. Assuming that only a small patch of the membrane of area a0 near the north pole changed its spontaneous curvature to a value of C0, while the other part of the membrane has a zero spontaneous curvature, c0 is modeled as a function of the surface area a calculated from the north pole
 | (7) |
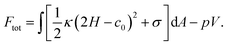 | (8) |
| X(u,ϕ) = [r(u)cos(ϕ),r(u)sin(ϕ),z(u)] | (9) |
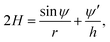 | (10) |
r′ = h![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) cos cos![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) ψ, z′ = −h ψ, z′ = −h![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) sin sin![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) ψ. ψ. | (11) |
Author contributions
C. S. and R. M. designed and supervised the research. C. L. performed molecular dynamics simulations and analysis. Q. Z. conducted continuum mechanics calculation. K. K. parallelized the GROMACS-LS code for the stress analysis. All the authors contributed to the manuscript writing.Data availability
The multisite calcium ion model used in this work can be downloaded from https://github.com/ComputBiophys/Multisite_Calcium. The simulation data are available upon reasonable request.Conflicts of interest
There are no conflicts to declare.Acknowledgements
This work was supported by grants from the National Natural Science Foundation of China (32071251 and 21873006 to C. S., 12474199 to R. M.), and Fundamental Research Funds for Central Universities (20720240144 to R. M.). The MD simulations were performed on the computing platform of the Center for Life Sciences at Peking University and the National Supercomputer Center in Tianjin.References
- M. Edidin, Nat. Rev. Mol. Cell Biol., 2003, 4, 414–418 CrossRef CAS.
- A. Melcrová, S. Pokorna, S. Pullanchery, M. Kohagen, P. Jurkiewicz, M. Hof, P. Jungwirth, P. S. Cremer and L. Cwiklik, Sci. Rep., 2016, 6, 1–12 CrossRef.
- J. M. Boettcher, R. L. Davis-Harrison, M. C. Clay, A. J. Nieuwkoop, Y. Z. Ohkubo, E. Tajkhorshid, J. H. Morrissey and C. M. Rienstra, Biochemistry, 2011, 50, 2264–2273 CrossRef CAS PubMed.
- P. T. Vernier, M. J. Ziegler and R. Dimova, Langmuir, 2009, 25, 1020–1027 CrossRef CAS.
- H.-H. G. Tsai, W.-X. Lai, H.-D. Lin, J.-B. Lee, W.-F. Juang and W.-H. Tseng, Biochim. Biophys. Acta, Biomembr., 2012, 1818, 2742–2755 CrossRef CAS.
- P. Garidel, A. Blume and W. Hübner, Biochim. Biophys. Acta, Biomembr., 2000, 1466, 245–259 CrossRef CAS PubMed.
- H. Akutsu and J. Seelig, Biochemistry, 1981, 20, 7366–7373 CrossRef CAS.
- H. Binder and O. Zschörnig, Chem. Phys. Lipids, 2002, 115, 39–61 CrossRef CAS.
- U. R. Pedersen, C. Leidy, P. Westh and G. H. Peters, Biochim. Biophys. Acta, Biomembr., 2006, 1758, 573–582 CrossRef CAS PubMed.
- A. Melcrová, S. Pokorna, M. Vošahlíková, J. Sýkora, P. Svoboda, M. Hof, L. Cwiklik and P. Jurkiewicz, Langmuir, 2019, 35, 11358–11368 CrossRef.
- M. L. Valentine, A. E. Cardenas, R. Elber and C. R. Baiz, Biophys. J., 2018, 115, 1541–1551 CrossRef CAS PubMed.
- A. Martín-Molina, C. Rodríguez-Beas and J. Faraudo, Biophys. J., 2012, 102, 2095–2103 CrossRef.
- S. Nir, J. Wilschut, J. Bentz, C. Newton, A. Portis and D. Papahadjopoulos, et al. , J. Membr. Biol., 1981, 59, 115–125 CrossRef PubMed.
- G. Feigenson, Biochemistry, 1986, 25, 5819–5825 CrossRef CAS PubMed.
- D. Leckband, C. Helm and J. Israelachvili, Biochemistry, 1993, 32, 1127–1140 CrossRef CAS PubMed.
- T. Ito and S.-I. Ohnishi, Biochim. Biophys. Acta, Biomembr., 1974, 352, 29–37 CrossRef CAS.
- T. A. Lagace and N. D. Ridgway, Biochim. Biophys. Acta, Mol. Cell Res., 2013, 1833, 2499–2510 CrossRef CAS PubMed.
- L. Li, X. Shi, X. Guo, H. Li and C. Xu, Trends Biochem. Sci., 2014, 39, 130–140 CrossRef CAS PubMed.
- S. E. Gordesky and G. Marinetti, Biochem. Biophys. Res. Commun., 1973, 50, 1027–1031 CrossRef CAS PubMed.
- G. D. Fairn, N. L. Schieber, N. Ariotti, S. Murphy, L. Kuerschner, R. I. Webb, S. Grinstein and R. G. Parton, J. Cell Biol., 2011, 194, 257–275 CrossRef CAS.
- J. Lorent, K. Levental, L. Ganesan, G. Rivera-Longsworth, E. Sezgin, M. Doktorova, E. Lyman and I. Levental, Nat. Chem. Biol., 2020, 16, 644–652 CrossRef CAS.
- M. J. Phillips and G. K. Voeltz, Nat. Rev. Mol. Cell Biol., 2016, 17, 69–82 CrossRef CAS PubMed.
- A. P. Ziman, C. W. Ward, G. G. Rodney, W. J. Lederer and R. J. Bloch, Biophys. J., 2010, 99, 2705–2714 CrossRef CAS PubMed.
- A. Bogdanova, A. Makhro, J. Wang, P. Lipp and L. Kaestner, Int. J. Mol. Sci., 2013, 14, 9848–9872 CrossRef PubMed.
- P. A. Leventis and S. Grinstein, Annu. Rev. Biophys., 2010, 39, 407–427 CrossRef CAS.
- T. Yeung, G. E. Gilbert, J. Shi, J. Silvius, A. Kapus and S. Grinstein, Science, 2008, 319, 210–213 CrossRef CAS PubMed.
- H. Zhao, E. K. Tuominen and P. K. Kinnunen, Biochemistry, 2004, 43, 10302–10307 CrossRef CAS.
- G. P. Gorbenko and P. K. Kinnunen, Chem. Phys. Lipids, 2006, 141, 72–82 CrossRef CAS.
- E. Bilkova, R. Pleskot, S. Rissanen, S. Sun, A. Czogalla, L. Cwiklik, T. Rog, I. Vattulainen, P. S. Cremer and P. Jungwirth, et al. , J. Am. Chem. Soc., 2017, 139, 4019–4024 CrossRef CAS PubMed.
- K. Han, S. H. Kim, R. M. Venable and R. W. Pastor, Proc. Natl. Acad. Sci. U. S. A., 2022, 119, e2202647119 CrossRef CAS.
- O. Tarun, H. I. Okur, P. Rangamani and S. Roke, Commun. Chem., 2020, 3, 1–8 CrossRef PubMed.
- Z. Graber, Z. Shi and T. Baumgart, Phys. Chem. Chem. Phys., 2017, 19, 15285–15295 RSC.
- M. Simunovic, K. Y. C. Lee and P. Bassereau, Soft Matter, 2015, 11, 5030–5036 RSC.
- J. Melcr, H. Martinez-Seara, R. Nencini, J. Kolafa, P. Jungwirth and O. S. Ollila, J. Phys. Chem. B, 2018, 122, 4546–4557 CrossRef CAS PubMed.
- J. Melcr, T. M. Ferreira, P. Jungwirth and O. S. Ollila, J. Chem. Theory Comput., 2019, 16, 738–748 CrossRef PubMed.
- H. Li, V. Ngo, M. C. Da Silva, D. R. Salahub, K. Callahan, B. Roux and S. Y. Noskov, J. Phys. Chem. B, 2015, 119, 9401–9416 CrossRef CAS.
- A. Zhang, H. Yu, C. Liu and C. Song, Nat. Commun., 2020, 11, 1–10 CrossRef.
- M. J. Berridge, Biochim. Biophys. Acta, Mol. Cell Res., 2009, 1793, 933–940 CrossRef CAS PubMed.
- D. E. Santos, F. J. Pontes, R. D. Lins, K. Coutinho and T. A. Soares, J. Chem. Inf. Model., 2019, 60, 473–484 CrossRef.
- D. E. Santos, K. Coutinho and T. A. Soares, J. Chem. Inf. Model., 2022, 62, 4690–4701 CrossRef CAS.
- R. Dluhy, D. G. Cameron, H. H. Mantsch and R. Mendelsohn, Biochemistry, 1983, 22, 6318–6325 CrossRef CAS.
- W. Hübner and A. Blume, Chem. Phys. Lipids, 1998, 96, 99–123 CrossRef.
- O. S. Ollila and G. Pabst, Biochim. Biophys. Acta, Biomembr., 2016, 1858, 2512–2528 CrossRef CAS PubMed.
- C. Altenbach and J. Seelig, Biochemistry, 1984, 23, 3913–3920 CrossRef CAS.
- M. Roux and M. Bloom, Biochemistry, 1990, 29, 7077–7089 CrossRef CAS.
- L. Song, E. Hennink, I. Young and H. Tanke, Biophys. J., 1995, 68, 2588–2600 CrossRef CAS.
- A. L. Kuo and C. G. Wade, Biochemistry, 1979, 18, 2300 CrossRef CAS PubMed.
- T. Fujiwara, K. Ritchie, H. Murakoshi and J. A. Kusumi, J. Cell Biol., 2002, 157, 1071–1081 CrossRef CAS PubMed.
- P. Schwille, J. Korlach and W. W. Webb, Cytometry, 2015, 36, 176–182 CrossRef.
- B. Różycki and R. Lipowsky, J. Chem. Phys., 2015, 142, 02B601_1 CrossRef PubMed.
- J. M. Vanegas, A. Torres-Sánchez and M. Arroyo, J. Chem. Theory Comput., 2014, 10, 691–702 CrossRef CAS.
- M. Doktorova, J. L. Symons and I. Levental, Nat. Chem. Biol., 2020, 16, 1321–1330 CrossRef CAS.
- M. Doktorova, M. V. LeVine, G. Khelashvili and H. Weinstein, Biophys. J., 2019, 116, 487–502 CrossRef CAS PubMed.
- K. J. Boyd, N. N. Alder and E. R. May, Biophys. J., 2018, 114, 2116–2127 CrossRef CAS.
- C.-P. Chng, Y. Sadovsky, K. J. Hsia and C. Huang, Extreme Mech. Lett., 2021, 43, 101174 CrossRef PubMed.
- W. Helfrich, Z. Naturforsch., C: J. Biosci., 1973, 28, 693–703 CrossRef CAS.
- U. Seifert, K. Berndl and R. Lipowsky, Phys. Rev. A: At., Mol., Opt. Phys., 1991, 44, 1182 CrossRef CAS.
- U. Seifert, Adv. Phys., 1997, 46, 13–137 CrossRef CAS.
- A. Agrawal and D. J. Steigmann, Biomech. Model. Mechanobiol., 2009, 8, 371–379 CrossRef.
- A. Hossein and M. Deserno, Biophys. J., 2020, 118, 624–642 CrossRef CAS PubMed.
- L. Miao, B. Fourcade, M. Rao, M. Wortis and R. K. Zia, Phys. Rev. A: At., Mol., Opt. Phys., 1991, 43, 6843 CrossRef CAS PubMed.
- E. Solomon, L. Berg and D. Martin, Biology, Brooks/Cole Publishing, Belmont, CA, 6th edn, 2002 Search PubMed.
- J. Craft, K. Epps, S. Brancato and A. Serfis, J. Colloid Interface Sci., 2003, 268, 181–187 CrossRef CAS.
- S. Mokkila, P. A. Postila, S. Rissanen, H. Juhola, I. Vattulainen and T. Rog, ACS Chem. Neurosci., 2017, 8, 1242–1250 CrossRef CAS PubMed.
- V. Gerke, C. E. Creutz and S. E. Moss, Nat. Rev. Mol. Cell Biol., 2005, 6, 449–461 CrossRef CAS.
- Y.-H. Ma, B. Li, J. Yang, X. Han, Z. Chen and X. Lu, Chem. Commun., 2020, 56, 1653–1656 RSC.
- D. E. Santos, A. De Nicola, V. F. Dos Santos, G. Milano and T. A. Soares, J. Phys. Chem. B, 2023, 127, 6694–6702 CrossRef CAS.
- A. Nascimento, F. J. Pontes, R. D. Lins and T. A. Soares, Chem. Commun., 2014, 50, 231–233 RSC.
- C. Allolio and D. Harries, ACS Nano, 2021, 15, 12880–12887 CrossRef CAS PubMed.
- C. G. Sinn, M. Antonietti and R. Dimova, Colloids Surf., A, 2006, 282, 410–419 CrossRef.
- P. Drücker, M. Pejic, H.-J. Galla and V. Gerke, J. Biol. Chem., 2013, 288, 24764–24776 CrossRef.
- A. V. Weigel, C.-L. Chang, G. Shtengel, C. S. Xu, D. P. Hoffman, M. Freeman, N. Iyer, J. Aaron, S. Khuon and J. Bogovic, et al. , Cell, 2021, 184, 2412–2429 CrossRef CAS PubMed.
- B. Ali Doosti, D. Fjällborg, K. Kustanovich, A. Jesorka, A.-S. Cans and T. Lobovkina, Sci. Rep., 2020, 10, 1–7 CrossRef.
- V. Kirejev, B. Ali Doosti, M. Shaali, G. D. Jeffries and T. Lobovkina, Small, 2018, 14, 1703541 CrossRef.
- S. Jones, A. Huynh, Y. Gao and Y. Yu, Mater. Chem. Front., 2018, 2, 603–608 RSC.
- A. Doosti, W. Pezeshkian, D. S. Bruhn, J. H. Ipsen, H. Khandelia, G. D. Jeffries and T. Lobovkina, Langmuir, 2017, 33, 11010–11017 CrossRef PubMed.
- C. Kluge, M. Pöhnl and R. A. Böckmann, Biophys. J., 2022, 121, 671–683 CrossRef CAS PubMed.
- N. Xu, M. Francis, D. L. Cioffi and T. Stevens, Studies on the resolution of subcellular free calcium concentrations: a technological advance. Focus on “Detection of differentially regulated subsarcolemmal calcium signals activated by vasoactive agonists in rat pulmonary artery smooth muscle cells”, 2014.
- M. J. Berridge and R. F. Irvine, Nature, 1984, 312, 315–321 CrossRef CAS PubMed.
- S. S. Ranade, R. Syeda and A. Patapoutian, Neuron, 2015, 87, 1162–1179 CrossRef CAS.
- S. Sukharev and F. Sachs, J. Cell Sci., 2012, 125, 3075–3083 CAS.
- M. J. Abraham, T. Murtola, R. Schulz, S. Páll, J. C. Smith, B. Hess and E. Lindahl, SoftwareX, 2015, 1, 19–25 CrossRef.
- J. B. Klauda, R. M. Venable, J. A. Freites, J. W. OConnor, D. J. Tobias, C. Mondragon-Ramirez, I. Vorobyov, A. D. MacKerell Jr and R. W. Pastor, J. Phys. Chem. B, 2010, 114, 7830–7843 CrossRef CAS.
- A. D. MacKerell Jr, D. Bashford, M. Bellott, R. L. Dunbrack Jr, J. D. Evanseck, M. J. Field, S. Fischer, J. Gao, H. Guo and S. Ha, et al. , J. Phys. Chem. B, 1998, 102, 3586–3616 CrossRef PubMed.
- S. R. Durell, B. R. Brooks and A. Ben-Naim, J. Phys. Chem., 1994, 98, 2198–2202 CrossRef CAS.
- S. Nosé, Mol. Phys., 1984, 52, 255–268 CrossRef.
- W. G. Hoover, Phys. Rev. A: At., Mol., Opt. Phys., 1985, 31, 1695 CrossRef.
- M. Parrinello and A. Rahman, J. Appl. Phys., 1981, 52, 7182–7190 CrossRef CAS.
- S. Nosé and M. Klein, Mol. Phys., 1983, 50, 1055–1076 CrossRef.
- T. Darden, D. York and L. Pedersen, J. Chem. Phys., 1993, 98, 10089–10092 CrossRef CAS.
- M. Hu, P. Diggins and M. Deserno, J. Chem. Phys., 2013, 138, 214110 CrossRef.
- M. Deserno, Chem. Phys. Lipids, 2015, 185, 11–45 CrossRef CAS PubMed.
- M. Doktorova, D. Harries and G. Khelashvili, Phys. Chem. Chem. Phys., 2017, 19, 16806–16818 RSC.
- M. A. S. Karal, M. M. Billah, M. Ahmed and M. K. Ahamed, Soft Matter, 2023, 19, 8285–8304 RSC.
- H. A. Faizi, S. L. Frey, J. Steinkühler, R. Dimova and P. M. Vlahovska, Soft Matter, 2019, 15, 6006–6013 RSC.
- R. Ma and J. Berro, Biophys. J., 2021, 120, 1625–1640 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d4cp01715c |
| This journal is © the Owner Societies 2025 |