Open Access Article
Open Access ArticleDetailed mechanism of a DNA/RNA nucleobase substituting bridging ligand in diruthenium(II,III) and dirhodium(II,II) tetraacetato paddlewheel complexes: protonation of the leaving acetate is crucial
Iogann
Tolbatov
 ad,
Tiziano
Marzo
ad,
Tiziano
Marzo
 b,
Paolo
Umari
b,
Paolo
Umari
 a,
Diego
La Mendola
b and
Alessandro
Marrone
a,
Diego
La Mendola
b and
Alessandro
Marrone
 *c
*c
aDepartment of Physics and Astronomy, University of Padova, via F. Marzolo 8, 35131, Padova, Italy
bDepartment of Pharmacy, University of Pisa, Via Bonanno Pisano 6, 56126, Pisa, Italy
cDipartimento di Farmacia, Università degli Studi “G. D'Annunzio” Chieti-Pescara, Via dei Vestini 31, 66100 Chieti, Italy. E-mail: amarrone@unich.it
dDepartment of Chemical, Physical, Mathematical and Natural Sciences, University of Sassari, 07100 Sassari, Italy
First published on 12th November 2024
Abstract
Paddlewheel complexes of bimetallic scaffolds are emerging metallic agents in the bioinorganic chemistry landscape. In the most commonly employed construct, these complexes are decorated by the carboxylate moiety, prompting their possible deployment to target either protein or nucleic acid targets. In this study, density functional investigation was performed to assess viable mechanistic routes for the substitution of one acetate ligand with one chelating purine, i.e. adenine or guanine, in diruthenium and dirhodium tetraacetate paddlewheel complexes. This study evidenced the relevant stages of the process at an atomistic scale of resolution and provided for the encompassed rate-determining chemical events. Therefore, calculations indicated that acetate decomplexation as well as the concomitant nucleobase bridging proceeded gradually via a multistep process that included protonation of the leaving acetate. The present picture of the mechanism is envisioned to be relevant to the design and interpretation of experiments focused on the reaction of diruthenium and/or dirhodium tetracarboxylate complexes with nucleobases and eventuating in the formation of either nucleobase bridged-complexes or in the dismantling of the bimetallic construct.
Introduction
The genetic guidelines for the crucial mechanisms in all life forms and viruses are held by deoxyribonucleic (DNA) and ribonucleic (RNA) molecules. The cellular nucleus of eukaryotes contains DNA biopolymers with the instructions relevant on how the organism should correctly operate, develop, and replicate.1 The intricate network of metabolic directives within the cell is functionally held by RNAs, which not only serve as the prototypes for proteins,2 but are also capable of executing specialized biological tasks, e.g. non-coding RNA.3 The information both in DNA and RNA is stored in the form of ordered nucleobases, which include adenine (A), cytosine (C), guanine (G), and thymine (T) in DNA or its demethylated equivalent uracil (U) in RNA. By studying the coding sequences of nucleobases via either high-resolution experimental methods or via computational approaches, it is possible to comprehend their functionalities.4 The recent successes in the clustered regularly interspaced short palindromic repeats (CRISPR) methodology permit the modification of the genetic data, thus allowing the control of transcription, the alternation of epigenomes, and the conduction of genome-wide screens.5 Such a CRISPR-enabled genome editing produced tangible successes in targeted cancer and antimicrobial therapies, crop and livestock breeding, and regulating the disease-carrying insect proliferation.6–8Formulation and development of therapeutic agents which hamper the proper operation of DNA and RNA funnel into a promising research direction, which affects the cellular metabolism and results in apoptosis.9–11 Conversely, DNA and RNA could be undesired targets of drugs developed for other biomolecular targets, thus causing toxic side effects.12,13
The therapeutic agents targeting DNA are numerous;14–16 while there are only very few drugs that specifically target RNA,17,18 the development of such drugs has given rise to a novel trend in the field of medicinal chemistry since RNA plays the crucial role in cellular and viral metabolism.19,20 Development of drugs that selectively attack DNA or RNA is a challenging task; the composition and folding of their backbones are markedly different; both nucleic compounds bear essentially the same nucleobases, with the only exception of uracil and thymine found only in RNA and DNA, respectively. As said, the secondary and tertiary structures are very distinguishable with the very flexible RNA and the more rigidly composed DNA. Another possible route to design drugs that distinguish between these two biopolymers is the implementation of differing activation characteristics. Indeed, it is supposed that the drugs, activated in the nucleus, more favorably target DNA, whereas those activated in the cytoplasm attack RNA.21,22
The process for the development of drugs selectively targeting DNA or RNA includes the exploitation of the chemistry of nucleobases, the geometries of the whole molecules (steric and electrostatics effects of the secondary and tertiary structures), as well as the fine control of the chemical conditions in which the drug is activated.23,24 The drug that coordinates a nucleobase in vivo alters its chemical characteristics, thus hampering the correct functioning of the whole DNA/RNA chain.25 Another approach is the preliminary preparation of a nucleobase with an attached drug in vitro and its subsequent dispense into the cell. Thereupon, the drug-linked nucleobase occurs to be incorporated into the DNA/RNA chain, impeding its correct activity.26 This strategy, based on mimicking the nucleosides, showcased multiple successes.27–30
Some inorganic drugs are prominent examples of drugs specifically capable of coordinating nucleobases.31,32 For instance, cisplatin, the leading platinum-based anticancer drug with the formula [Pt(NH3)2Cl2], has been the first FDA approved anticancer metallodrug. Upon the activation that occurs in the cell by the release of chloride ligands, the prodrug converts into the corresponding active pharmacophore that, in turn, mainly binds at the N7 position of guanine residues.33–35 The latter event induces DNA distortions and alterations eventually leading to apoptosis. Many other transition metals are employed in metallodrugs, ruthenium being one of the most ubiquitous metal centers in use. Indeed, ruthenium-based metallodrugs NAMI-A and KP1019 reached the advanced phases of pharmacological testing.36
Among the most interesting and intensely studied types of metallodrugs are the paddlewheel complexes based on the diruthenium(II,III) and dirhodium(II,II) cores with a crown of the bridging acetate ligands. These complexes have a general formula [M2(O2CR)4]L2 (M = Rh, Ru; R = CH3−, CH3CH2−, etc., L = solvent molecule or anionic ligand). In the case of the Ru2 core, three uncoupled electrons on antibonding metal–metal bonds provide the mixed valence character(II,III) to the complex and +2.5 charge on each metal center.37,38 Encouraging results on the possible employment of Ru2 paddlewheel complexes in the targeting of RNA structures have been reported by Herrero et al.39 In contrast, the Rh2 core features a metal–metal bond of the first order, thus yielding the valence(II,II). This disparity is in the origin of the different selectivity of both paddlewheel complexes. The Rh2 construct has been found to effectively bind at the B-DNA structure via the coordination of one adenine nucleobase at the axial position of the paddlewheel scaffold.40 For the Ru complex with bridging acetates of the formula [Ru2(μ-O2CCH3)4Cl], a high selectivity toward the side chains of the protein residue aspartate has been reported,41 whereas its Rh-based equivalent shows a lower selectivity and binds to Asn, Asp, His, Lys, and the C-terminal.42–44
Depending on the ligand composition, both Ru2 and Rh2 paddlewheel complexes with an ascertained cytotoxicity were reported.45–48 The Rh complex was found to be effective against several cancers, such as Ehrlich-Lettre ascites carcinoma,49–51 L1210 tumors,52 and P388 leukemia.53 The most promising feature of the paddlewheel complexes relies on the chance to couple the medicinal properties of the dimetallic core together with the biological effects of the bound carboxylate-bearing ligands. Provided the latter are pharmacologically active entities, it is potentially possible to effectuate the simultaneous delivery of up to four equivalents of them. There have been multiple studies in which various active medicinal substances were tested as the bridging ligands, for instance, fatty acids or dipeptides.38 Nevertheless, the most auspicious results were produced with various non-steroidal anti-inflammatory drugs (NSAIDs), such as aspirin, ibuprofen, indomethacin, ketoprofen, and naproxen. Their integration allowed an advantageous combination of anti-inflammatory and cytotoxic properties.38,54
In a recent study,55 the thermodynamics of the first step of the coordination at the DNA and RNA nucleobases of the prototypical Ru- and Rh-based paddlewheel complexes [Ru2(μ-O2CCH3)4(H2O)Cl], [Ru2(μ-O2CCH3)4(OH)Cl], and [Rh2(μ-O2CCH3)4(H2O)2] was investigated. It was found that the Rh2 complexes tend to react more effectively with the nucleobases compared to the Ru2 complexes and that both bimetallic cores bind preferentially to purines, i.e. adenine and guanine.55 The computational analysis of the metal–acetate bonds witnessed their substantial weakening upon binding with a nucleobase at the axial position, and it was noted that this bond debilitation should favor the downstream dechelation of the bridging ligands. It is a very promising effect for the medicinal use of these complexes since the drugs replacing the bridging ligands should thus be released.
The present study focuses on a detailed mechanistic description of the process in which the purine nucleobase coordinates to the paddlewheel complexes [Ru2(μ-O2CCH3)4(H2O)Cl] and [Rh2(μ-O2CCH3)4(H2O)2]. The process starts with the η1-coordination of the nucleobase N7 at the axial position of the complex, by replacing one axial ligand, and eventuates into the substitution of one acetate ligand and the concomitant formation of a nucleobase-bridged complex. The studied reaction route consists of several steps which we described quantitatively as well as all the existing barriers, some of which were found to be rate-determining. Our investigation showed that the acetate decomplexation and nucleobase chelation are not simultaneous under the physiological conditions, but rather progress gradually and, more importantly, proceed via the protonation of the leaving acetate.
Computational details
The Gaussian 16 was employed for all DFT calculations.56 DFT allows for the accurate description of reactions of transition metal complexes,57–59 including Ru and Rh.60–62 All the optimizations were performed with the hybrid range-corrected functional ωB97X-D63 and the def2SVP basis set.64,65 Indeed, this functional is expressed by the sum of the ωB97X functional and the empirical term D that enhances the estimate of non-covalent interactions. The ωB97X-D has been indicated among the high performing functionals in the prediction of the CCSD geometry and the interaction energy in molecular dimers66 and showed overall good predictive performances in combination with the def2SVP basis set, as previously ascertained by us in the study of bioinorganic systems.67,68 That is why we regard the ωB97X-D/def2SVP methodology as adequate for the energy calculation focused on the disentanglement of the interactions of the studied metal scaffolds with the nucleobases. We performed frequency computations to verify the correct topology of the localized minimum and the transition state geometries. Also, these computations produced the Gibbs free energies (GFE) for each structure, the zero-point energies (ZPE) and vibrational corrections to the thermodynamic properties by means of the application of harmonic approximation. We used the Polarizable Continuum Model using the integral equation formalism variant (IEFPCM) technique69 to characterize the solvation free energy in water, as this approach yields accurate free energies both for neutral and charged complexes.70 Therefore, COSMO or PCM methods being endowed by very similar predictive and numerical performances when small to medium sized molecular systems are concerned,70 we opted for the IEFPCM method which is merely the PCM methodology in the g16 default set-up.The Gibbs free energy for the step III, i.e. the protonation of the partially decomplexed acetate, was computed by assuming the molar Gibbs free energy of the H+(aq) ion (298.15 K and 1.0 atm) of −265.9 kcal mol−1 reported by Kelly et al.71 The free energy correction of 1.98 kcal mol−1 was applied to each species to account for the different standard states in gas (unitary partial pressure) and in water (unitary molar concentration); it is worth noting that this correction cancels off in all but steps I and III, in which two moles of reactants yield one mole of product, i.e. the standard state correction is −1.98 kcal mol−1 in both cases.
The Natural Bond Orbital (NBO) analysis,72 performed in water at the level ωB97X-D/def2SVP, allowed the assessment of the strength of the hydrogen bond interactions in either free or metalated nucleobase pairs. In particular, the second order perturbation analysis (SOP)73 was used to estimate the energy gap in the donor-to-acceptor orbital interaction between the lone pair orbital on either N or O atoms and the virtual σ* orbital of either N–H or O–H bonds, thus providing for semi-quantitative estimation of the hydrogen bond strengths. All NBO calculations were performed by using the nbo (version 3) called and executed in the g16 script (i.e. population = nbo).
Results and discussion
Chelation is the most energetically effective coordination process and eventuates in the formation of generally highly stable metal complexes. The tetracarboxylate paddlewheel complexes of Ru2 and Rh2 are indeed highly stable, being basically structured by four O,O′-μ-coordinated equatorial ligands that confer a particularly high thermodynamic stability and low reactivity.38,74 On the other hand, the extraordinarily high chemical complexity of biological media may expose these complexes to the reaction with more effective chelating agents compared to acetate which are thus expected to dismantle their highly stable paddlewheel structure. Metal-coordinating residues settled on the surface of protein structures may give rise to outperforming chelating agents (Fig. 1). On the other hand, other biological scaffolds, i.e. either DNA or RNA nucleobases, may act as bidentate chelating ligands; for example, adenine and guanine may be considered as 1,4-chelating ligands via the involvement of the N7 and either N6 or O6 donor atoms, respectively (Fig. 1). In fact, by taking into account all tautomer forms, as well as deprotonation of either amino or six-membered NH groups, further 1,3 chelating sites can be retraced on both adenine and guanine scaffolds (Fig. 1). On the other hand, we think that the 1,3 chelating sites might be assumed as accessory or secondary if compared with the chelation involving the N7 position. Indeed, this pyridyl site has been recognized as the primary target of metal binding on the DNA because of its majorly exposed location into the duplex major groove.75Therefore, to better resemble the chemical environment of the A and G scaffolds encountered in biological systems, the N9 position was methylated, thus hampering any involvement of this site in the metal coordination. These considerations have prompted us to consider the purine nucleobases as viable ligands of the Ru2 and Rh2 paddlewheel complexes, capable to replace one of the μ-carboxylate moieties via either N7,N6 or N7,O6-μ-coordination.
We have recently shown that the coordination of purine nucleobase scaffolds on vacant axial coordination sites is not only a viable process, but it was also shown to be thermodynamically more favorable compared to the reaction of pyrimidines, and that the coordinating role of the N7 position is prominent.55 In the present study, the reaction of purine nucleobases with either ClRu2Ac4, 1a, or (H2O)Rh2Ac4, 1b (Ac = CH3COO−), i.e. the metal scaffolds with one vacant axial site obtained from [Ru2(μ-O2CCH3)4(H2O)Cl] and [Rh2(μ-O2CCH3)4(H2O)2], respectively, was investigated more comprehensively to assess both thermodynamics and kinetics of the adenine or guanine chelation at these bimetallic moieties. The coordination of water molecules at both axial positions is reputed to be the most probable initial configuration of the bimetallic complex. In this case, we chose to investigate the early coordination of the purine nucleobase on each paddlewheel complex in its native form, i.e. before hydrolysis or other ligand exchange processes are completed; hence, the second axial position was assumed to bear a chloro or an aquo ligand in the Ru2 or Rh2 complex, respectively. We assumed that the ligand on the second metal center would affect only marginally the calculated energy profiles. Furthermore, our choice allowed us to compare the reactivity of bimetallic Ru and Rh species bearing the same formal charge, thus enhancing the comparison of the calculated energy profiles. The above mentioned processes were investigated by means of density functional theory and by the use of pseudomolecular models; a general scheme of the overall processes is provided (Scheme 1). To model the early coordination of purines on the Ru2 and Rh2 complexes, we relied on both chemical intuition/knowledge and computational outcomes. The initial axial coordination of the N7 nucleobase was reputed as the most probable, and it was investigated by gradually approaching the nucleobase toward the metal center (and several orientations were explored). At this stage, we trusted the geometry optimization to show up the most stable coordination for the nucleobase. In all our attempts, the coordination of the N7 in the calculated minima was always obtained. Hence, we report that the experimental evidence of axial nucleobase chelation in X-ray structures does not necessary deny or conflict with our computational outcome. Indeed, the lack of water solvation and the influence of packing forces may explain the higher stability of nucleobase chelates detected in the X-ray structures. On the other hand, when the aqueous environment is concerned, the mono-N7 coordination of the purine allowed the favorable interactions of the other heteroatomic functional groups with the solvent, thus stabilizing the monodentate coordination. Both adenine and guanine bear a pyridyl nitrogen atom, N7, available to coordinate the vacant axial site and initiate the chelation process, thus, we hypothesized that the axial coordination of the N7 corresponds to the first step of the process (Scheme 2). As previously reported,55 the nucleobase coordination on the axial position of the Ru2 or Rh2 scaffold causes a weakening of the paddlewheel structure; hence, this chemical event can reasonably anticipate the upcoming acetate substitution. Despite this destabilizing effect, the axial coordination of the nucleobase cannot appreciably affect the “chelating effect” still operating on the metal–carboxylate coordination. For this reason, we hypothesized that the decomplexation of one acetate occurs gradually in a multistep pathway. The second step was thus sketched to operate the partial decomplexation of one acetate oxygen concertedly to the axial-to-equatorial position shift of the nucleobase (Scheme 2). The viability of our mechanistic hypothesis was assessed by the calculation of the kinetics and thermodynamics of the two earlier steps of the process, i.e. nucleobase coordination and partial acetate decomplexation (Scheme 3). As expected, the approach of either adenine or guanine at the vacant axial site of either 1a or 1b was always kinetically and thermodynamically highly favorable (Scheme 3). On the other hand, the axial-to-equatorial flip of the nucleobase ligand, accompanying the partial decomplexation of one acetate, was found to be less favorable with an endergodicity of 7.8–13.7 kcal mol−1 and the activation free energy in the range of 18.8–27.3 kcal mol−1 (Scheme 3) and, more importantly, evidencing a mechanistic differentiation between Ru2 and Rh2 scaffolds. Indeed, while the activation free energies for the formation of the adenine and guanine complexes of the Ru2 scaffold 3a and 3a′ of 19 and 23 kcal mol−1, respectively, are consistent with a viable process, a barrier of almost 27 kcal mol−1, calculated for the formation of 3b and 3b′, is consistent with kinetically hampered processes. Some technical considerations on these data must be expounded. The activation free energies for the axial-to-equatorial flip of the nucleobases were calculated as the difference between the free energy of the corresponding transition state, TS2, and the free energy of species 2; this means that the higher the stability of species 2 is, the higher the activation free energy is for the formation of species 3. However, the stages of the reaction process, obtained as minimum energy structures by DFT calculations, are representative of the transformation of the chemical system at 0 K, whereas, at higher temperature, higher energy structures are sampled. Based on this consideration, we envisioned that the overall activation free energy for the combination of step I and II could be better estimated with the free energy of TS2, which, conversely, evidences how the formation of 3a, 3a′, 3b, and 3b′ is kinetically viable (Scheme 3). The species 3 were characterized by a partially decomplexated acetate ligand and two possible mechanistic continuations of the process: on one hand, the monocoordinated nucleobase on species 3 may coordinate at the second metal center upon the release of the acetate ligand; on the other hand, the dangling and solvent-exposed acetate oxygen may be protonated by the bulk, i.e. hydrolysis, and the formed carboxylic ligand may be replaced by a nucleobase in a subsequent step. Both mechanistic alternatives were investigated, and the corresponding results are shown in Scheme 3. The direct replacement of the dangling acetate to afford the nucleobase-bridged complex was found to be affected by a huge activation free energy, thus making it substantially unviable (Scheme 3). On the other hand, the protonation of the partially decomplexated acetate in species 3 yielded an unexpected outcome: the carboxylic ligand, formed in species 4, is first released in the bulk, and the nucleobase chelation is accomplished subsequently. The carboxylic ligand release yielding species 5, i.e. the complexes with the tricarboxylate bimetallic moiety monocoordinated by the nucleobase, was found to be rate-determining and affected by an activation free energy barrier of 15–20 kcal mol−1, while the further step leading to the final species 6 was very fast and highly exergonic (Scheme 3).
 | ||
| Scheme 2 Rendition of the five-step mechanistic hypothesis investigated in the present theoretical study. | ||
The comparison of the calculated free energy profiles spotlighted several interesting differences in the reactivity of Ru2 and Rh2 tetraacetato with adenine and guanine.
The rate-determining step for the reaction of the Rh2 scaffold with nucleobases was represented by the axial-to-equatorial flip of the monocoordinated nucleobase (step II), whereas the downstream steps, III–V, are all kinetically and thermodynamically much favorable. On the other hand, the step IV, i.e. the release of the carboxylic ligand, was found to be rate-determining in the reaction of Ru2 with nucleobases. We envision that the presence in the reaction medium of a high concentration of nucleobase might be more effective to prompt the reaction of Rh2 compared to the Ru2 complex, which would increase the concentration of species 2. On the other hand, the low pH condition should majorly prompt the nucleobase chelation of Ru2 compared to Rh2 complexes due to an enhanced formation of species 4. Although the presented computational results and their interpretation are still limited to the stage of speculation, we hope they provided new stimuli and addresses to the investigation of the binding of purine nucleobase at bimetallic paddlewheel complexes.
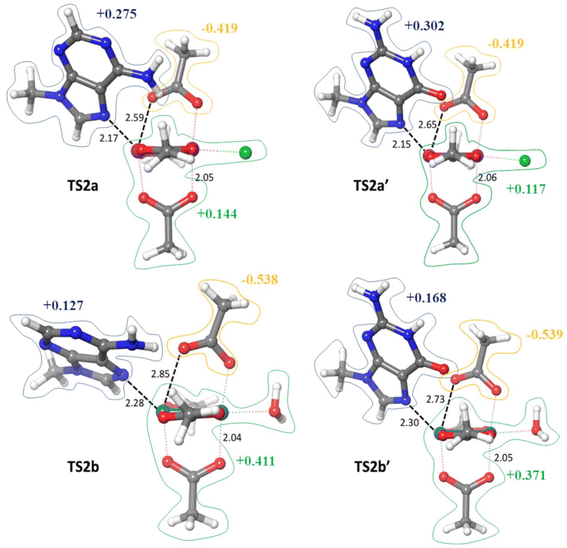
Transition states TS2 describe the breaking of the M–O(Ac) bond with a concomitant axial → equatorial shift of the coordinated nucleobase (Fig. 2). The M–M–N angle, where N is the nitrogen of the nucleobase, is around 45° in all instances. The M–N bond ranges from 2.15 to 2.30 Å, whereas the M–O(Ac) breaking bond is within the range 2.59–2.85 Å, i.e. corresponding to the 125–138% of the non-elongated bond which is around 2.05 Å. TS2 is rate-determining in the case of the Rh2 scaffold.
Transition states TS3 describe the complete detachment of acetate with a concomitant formation of the second bond between the nucleobase and the second metal center, i.e. the formation of the nucleobase-bridged complex (Fig. 3). These states are described by the geometry where there already exists a coordination of the nucleobase at one of the metal centers (via M–N bonds of 2.06–2.10 Å). The protonated acetate moves out, being substituted by the adenine's N6 or guanine's O6 atom. The transition state is characterized by the presence of multiple hydrogen bonds in each case. In the instance of the Ru2 scaffold, the leaving acetic acid interacts with chloride via only one hydrogen bond (2.32–2.42 Å). Moreover, the detachment of acetate from the adenine-bound complex is facilitated by a strong hydrogen bond with a length of 2.00 Å (Fig. 3). In the case of the Rh2 scaffold, the breaking of the M–O(Ac) bond is eased by the presence of the water molecule in the axial position, which forms a H-bond with the O of acetate (around 1.90–2.00 Å). On the other hand, this release is further facilitated by the formation of a hydrogen bond between an oxygen of one of the remaining bridging acetates with the hydrogen of protonated acetate (1.72–1.83 Å). The presence of these two hydrogen bonds in the case of the Rh2 scaffold was envisioned to stabilize appreciably the TS3, thus resulting in the axial → equatorial shift of the coordinated nucleobase as the only rate-determining step. In contrast, in the case of the Ru2 scaffold, there is only one hydrogen bond that forms between the leaving (protonated) acetate and chloride.
The calculated GFE profiles provided not only a clear energy picture of the purine nucleobase chelation at either Ru2 or Rh2 paddlewheel scaffolds, but also allowed us to analyze in detail the structural features of the relevant transition states. The atomic charges calculated via the natural bond orbitals (NBO) analysis on both TS2 and TS3 structures confirmed that protonation is determinant to induce the release of one acetate ligand. Indeed, the axial-to-equatorial flip of the nucleobase yields the η1-coordination of one acetate with the exposure of the dangling oxygen atom to the aqueous bulk, acquisition of a partial negative charge (Fig. 2), and propensity to hydrolyze. On the other hand, the NBO atomic charges calculated on TS3 structures showed that the leaving carboxylic moiety is featured by a small positive charge (Fig. 3), conferring this group a slightly higher acidity compared to the free acetic acid. On the other hand, the purine nucleobase moieties retrieved in the TS3 structures were also found to acquire a positive charge (Fig. 3). This evidence anticipates that the leaving carboxylic ligand may disclose a hydrogen bond donor character, with the OH group being able to form strong hydrogen bond interactions. Indeed, we noted that the hydrogen bond interactions play a crucial role in the stabilization of the structures TS3. In order to estimate the strength of the hydrogen bonds in these structures, we performed the second order perturbation analysis (SOP) in the framework of NBO calculations (Table 1). In the structure TS3a, two hydrogen bond interactions were detected, involving (i) the adenine N6–H bond with metal-bound oxygen of the leaving carboxylic ligand, and (ii) the carboxylic bond of the leaving acetate and chloride ligand. SOP analysis quantified the strength of the tiled two interactions with 7.5 and 11.8 kcal mol−1, respectively. Since these interactions are accentuated by the decomplexation of the carboxylic moiety, i.e. the carbonyl part becoming more basic and carboxyl more acidic, they are expected to stabilize the TS3a species, thus favoring the carboxylic release process.
| Structure | H-bond | Length, Å | Angle, ° | E 2, kcal mol−1 |
|---|---|---|---|---|
| TS3a | Adenine–N6H2⋯{O![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C(OH)CH3} C(OH)CH3} |
2.00 | 142.2 | 7.5 |
{CH3C(![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O)–OH}⋯Cl O)–OH}⋯Cl |
2.32 | 168.4 | 11.8 | |
| TS3a′ | {CH3C(![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O)–OH}⋯[O–C( O)–OH}⋯[O–C(![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O)CH3] O)CH3] |
2.00 | 140.3 | 6.8 |
| TS3b | Adenine–N6H2⋯[O–C(![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O)CH3] O)CH3] |
2.19 | 161.9 | 1.7 |
{CH3C(![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O)–OH}⋯[O–C( O)–OH}⋯[O–C(![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O)CH3] O)CH3] |
1.83 | 157.2 | 14.9 | |
OH2⋯{O![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C(OH)CH3} C(OH)CH3} |
1.96 | 139.1 | 4.0 | |
| TS3b′ | {CH3C(![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O)–OH}⋯[O–C( O)–OH}⋯[O–C(![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O)CH3] O)CH3] |
1.72 | 162.9 | 22.2 |
OH2⋯{O![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C(OH)CH3} C(OH)CH3} |
1.89 | 143.3 | 8.5 |
Interestingly, the same effect was not detected in TS3a′, in which there is only a very weak H-bond interaction between the leaving carboxylic ligand and one μ-coordinated acetate with the value of 6.8 kcal mol−1. We presume that the reason is that the geometry in which the OH bond is not well directed toward the oxygen of bridging acetate, and the interatomic distance of 2.0 Å turns out to be poorly effective. Moreover, NBO detected no significant interaction (<1 kcal mol−1) between the axial Cl and leaving carboxylic group in this species.
The H-bonding in the TS3b species resembles slightly the case of TS3a. Indeed, there is present a weak interaction between the NH2 group of adenine (1.7 kcal mol−1) and the other two interactions: between the leaving and bridging acetates (14.9 kcal mol−1) and between water and leaving acetate (4.0 kcal mol−1). These data demonstrate that the contribution which substantially stabilizes this structure is the hydrogen bond between leaving and bridging acetates.
In the case of the structure TS3b′, we have a strong H-bond (22.2 kcal mol−1) between the leaving and bridging carboxylic groups. Moreover, there is also present a weak (8.5 kcal mol−1) interaction between the axial water ligand OH bond and the leaving acetate oxygen. Both interactions are expected to stabilize the TS3b′ species and intensify along the reaction coordinate (when the carboxylic group is completely detached from the metal fragment).
The SOP analysis provided the important picture in which the trend in the strengths of hydrogen bonds fairly correlates with the order of barrier heights for TS3. Indeed, the summed strengths of all present H-bonds are 19.3, 6.8, 20.6, and 30.7 for TS3a, TS3a′, TS3b, and TS3b′, respectively, whereas the height of these barriers are 20.3, 19.1, 18.7, and 15.9 kcal mol−1, the TS3a′ data representing the only outlier within the tiled correlation.
The analysis of NBO charges is consistent with the charge distributions expected for the processes intercepting TS2 and TS3, i.e. the axial-to-equatorial shift of the nucleobase and release of the carboxylic ligand, respectively. The analysis of the charge distribution displayed almost no significant differences when either A/G nucleobases or Ru/Rh scaffolds are compared (Fig. 2 and 3).
Conclusions
Diruthenium and dirhodium paddlewheel complexes are metal complexes with great medical potential due to the ease with which their novel derivative complexes may be synthesized by varying the composition of the bridging ligands. Indeed, these complexes have been demonstrated as optimal for the design of dual acting prodrugs via integration of various pharmaceutically-active bidentate ligands, such as NSAIDs.38,54 Additionally, these structures may easily load up to four releasable, medicinally active ligands whose properties could be enhanced owing to the synergistic combination with the metal properties.38In the present computational study, we investigated the possible mechanisms for the substitution of one bridging acetate with one chelating purine, i.e. adenine or guanine, in both diruthenium and dirhodium tetraacetato paddlewheel complexes and identified several stages in which the acetate decomplexation and the nucleobase chelation concomitantly took place (Scheme 4).
The involvement in the metal binding of the heteroatomic group on the position 6 of the purine nucleobase, expected to be not accessible due to the Watson–Crick base pairing, must be better circumstantiated. The first consideration concerns the presence of the A and G scaffolds not only in polynucleotides, but also in soluble cellular cofactors such as NADH, ATP, and GMP, in which all heteroatomic functional groups (with the exception of N9) can be assumed available to the metal coordination. Therefore, even in the polynucleotide structures, the hydrogen bond complementarity may interrupt in certain conditions, for instance, during the biochemical processes inducing a partial detachment of the complementary monostrands (duplication, transcription) and exposure of the involved nucleobases. Another consideration may be drawn concerning the possible employment of the reaction between paddlewheel complexes and nucleobases. Indeed, we think that, alternatively to the use of paddlewheel complexes as metallodrugs targeting the purine scaffolds in the cell, preformed bimetallic complexes with nucleobases (or modified nucleobase scaffolds) could be prepared and tested as prodrugs (or dual acting metallodrugs when biologically-active modified nucleobases are concerned). The first important conclusion that we made was that the protonation of the leaving acetate is crucial since it lowers the barrier for the acetate release by >20 kcal mol−1, making this step accessible under physiological conditions.
Another important observation is that the two paddlewheel complexes exhibit differences in their reaction profiles. While the rate-determining step for the reaction of the Rh2 scaffold with nucleobases is the axial-to-equatorial flip of the monocoordinated nucleobase, the rate-determining step in the case of the Ru2 scaffold is the release of the carboxylic ligand. We conclude that the substitution of the bridging acetate by the nucleobase can be effectively prompted at a high concentration of nucleobase in the case of the dirhodium paddlewheel complex, while a low pH should facilitate the nucleobase chelation of diruthenium compared to dirhodium complexes.
The overall insight into the energy landscape where the reaction of purines with Ru2 and Rh2 paddlewheel complexes takes place, provided by the calculated reaction free energy profiles (Scheme 3), inferred the feasibility of the examined processes. Indeed, calculations showed that the bridging coordination of either A or G on the Rh2 paddlewheel scaffold is endowed by the highest exergodicity, i.e. an essentially right-shifted process. On the other hand, the coordination of A and G on the Ru2 paddlewheel complex is much less exergonic, consistent with an equilibrium. Thus, while Rh2 complexes are expected to be the most effective in the targeting of purine scaffolds in the cell or in other biological contexts, the Ru2 scaffold might be taken into account to prepare ClRu2(acetate)3(purine) paddlewheel complexes in which acetate and purine are endowed by a similar lability. We envision that dual acting metallodrugs based on the paddlewheel paradigm could then be designed by incorporating a modified purine featured by an intrinsic biological or therapeutic activity.
This study sheds light upon the reactivity of diruthenium and/or dirhodium tetracarboxylate complexes with nucleobases and the correlation between the chelation of nucleobases and the destabilization of these bimetallic constructs. We anticipate that these computational outcomes will support and induct the forthcoming experimental and theoretical investigations on the interaction of bimetallic paddlewheel complexes towards DNA and RNA.
Author contributions
IT and AM contributed to conceptualization, investigation, data duration, writing – first draft, and writing – review & editing; PU, TM and DLM contributed to conceptualization and writing – review & editing.Data availability
The authors confirm that the data supporting the findings of this investigation are available within the article. Moreover, computational data and information to be used for reproducing the findings of this study are available from the corresponding author, upon reasonable request.Conflicts of interest
There are no conflicts to declare.Acknowledgements
This work has been funded by the European Union – Next-Generation EU (“PNRR M4C2-Investimento 1.4- CN00000041”). We acknowledge the CINECA award under the ISCRA initiative, for the availability of high performance computing resources and support. IT gratefully acknowledges the usage of HPC resources from Direction du Numérique – Centre de Calcul de l'Université de Bourgogne (DNUM CCUB). TM acknowledges the financial support from Ministero Italiano dell’ Universita e della Ricerca (MUR) under the program PRIN 2022-Progetti di Rilevante Interesse Nazionale, project code: 2022ALJRPL “Biocompatible nanostructures for the chemotherapy treatment of prostate cancer”.References
- Y. Hu and B. Stillman, Mol. Cell, 2023, 83, 352–372 CrossRef CAS PubMed.
- G. Faure, A. Y. Ogurtsov, S. A. Shabalina and E. V. Koonin, Nucleic Acids Res., 2016, 44, 10898–10911 CrossRef CAS.
- A. F. Palazzo and E. S. Lee, Front. Genet., 2015, 6, 2 Search PubMed.
- J. Li, J. Luo, L. Liu, H. Fu and L. Tang, Medicine, 2018, 97, e12884 CrossRef CAS PubMed.
- R. Barrangou and J. A. Doudna, Nat. Biotechnol., 2016, 34, 933–941 CrossRef CAS.
- T. Zhan, N. Rindtorff, J. Betge, M. P. Ebert and M. Boutros, Semin. Cancer Biol., 2019, 55, 106–119 CrossRef CAS PubMed.
- D. Jaganathan, K. Ramasamy, G. Sellamuthu, S. Jayabalan and G. Venkataraman, Front. Plant Sci., 2018, 9, 985 CrossRef PubMed.
- C. N. T. Taning, B. Van Eynde, N. Yu, S. Ma and G. Smagghe, J. Insect Physiol., 2017, 98, 245–257 CrossRef CAS.
- T. Hermann, Angew. Chem., Int. Ed., 2000, 39(11), 1890–1904 CrossRef.
- Z. Topcu, J. Clin. Pharm. Ther., 2001, 26, 405–416 CrossRef CAS PubMed.
- I. Tolbatov, P. Umari, T. Marzo, L. Chiaverini, D. La Mendola and A. Marrone, Chem. Phys. Lett., 2024, 842, 141197 CrossRef CAS.
- G. Bischoff and S. Hoffmann, DNA-binding of drugs used in medicinal therapies, Curr. Med. Chem., 2002, 9, 321–348 CrossRef CAS.
- J. M. Sasso, B. J. B. Ambrose, R. Tenchov, R. S. Datta, M. T. Basel, R. K. DeLong and Q. A. Zhou, J. Med. Chem., 2022, 65, 6975–7015 CrossRef CAS.
- V. Brabec, O. Hrabina and J. Kasparkova, Coord. Chem. Rev., 2017, 351, 2–31 CrossRef CAS.
- B. A. Carneiro and W. S. El-Deiry, Nat. Rev. Clin. Oncol., 2020, 17, 395–417 CrossRef PubMed.
- A. R. Timerbaev, C. G. Hartinger, S. S. Aleksenko and B. K. Keppler, Chem. Rev., 2006, 106, 2224–2248 CrossRef CAS PubMed.
- L. Chen, G. A. Calin and S. Zhang, J. Chem. Inf. Model., 2012, 52, 2741–2753 CrossRef CAS.
- A. E. Hargrove, Chem. Commun., 2020, 56, 14744–14756 RSC.
- L. I. Messer, J. G. Levin and S. K. Chattopadhyay, J. Virol., 1981, 40, 683–690 CrossRef CAS.
- M. G. Katze and R. M. Krug, Mol. Cell. Biol., 1984, 4, 2198–2206 CAS.
- D. J. Huggins, W. Sherman and B. Tidor, J. Med. Chem., 2012, 55, 1424–1444 CrossRef CAS PubMed.
- K. D. Warner, C. E. Hajdin and K. M. Weeks, Nat. Rev. Drug Discovery, 2018, 17, 547–558 CrossRef CAS.
- M. J. O'Connor, Mol. Cell, 2015, 60, 547–560 CrossRef.
- S. T. Crooke, J. L. Witztum, C. F. Bennett and B. F. Baker, Cell Metab., 2018, 27, 714–739 CrossRef CAS PubMed.
- A.-M. Yu, Y. H. Choi and M.-J. Tu, Pharmacol. Rev., 2020, 72, 862–898 CrossRef CAS PubMed.
- K. Paunovska, D. Loughrey and J. E. Dahlman, Nat. Rev. Genet., 2022, 23, 265–280 CrossRef CAS.
- R. Taylor, P. Kotian, T. Warren, R. Panchal, S. Bavari, J. Julander, S. Dobo, A. Rose, Y. El-Kattan, B. Taubenheim, Y. Babu and W. P. Sheridan, J. Infect. Public Health, 2016, 9, 220–226 CrossRef PubMed.
- R. J. Geraghty, M. T. Aliota and L. F. Bonnac, Viruses, 2021, 13, 667 CrossRef CAS.
- J. M. Thomson and I. L. Lamont, Front. Microbiol., 2019, 10, 952 CrossRef PubMed.
- M. Guinan, C. Benckendorff, M. Smith and G. J. Miller, Molecules, 2020, 25, 2050 CrossRef CAS.
- E. J. Anthony, E. M. Bolitho, H. E. Bridgewater, O. W. L. Carter, J. M. Donnelly, C. Imberti, E. C. Lant, F. Lermyte, R. J. Needham, M. Palau, P. J. Sadler, H. Shi, F.-X. Wang, W.-Y. Zhang and Z. Zhang, Chem. Sci., 2020, 11, 12888–12917 RSC.
- D. Cirri, T. Marzo, I. Tolbatov, A. Marrone, F. Saladini, I. Vicenti, F. Dragoni, A. Boccuto and L. Messori, Biomolecules, 2021, 11, 1858 CrossRef CAS.
- B. Chiavarino, M. E. Crestoni, S. Fornarini, D. Scuderi and J.-Y. Salpin, J. Am. Chem. Soc., 2013, 135, 1445–1455 CrossRef CAS PubMed.
- L. A. S. Costa, T. W. Hambley, W. R. Rocha, W. B. De Almeida and H. F. Dos Santos, Int. J. Quantum Chem., 2006, 106, 2129–2144 CrossRef CAS.
- R. Paciotti, I. Tolbatov, V. Graziani, A. Marrone, N. Re and C. Coletti, AIP Conf. Proc., 2018, 2040, 20019 CrossRef.
- E. Alessio and L. Messori, Molecules, 2019, 24, 1995 CrossRef CAS PubMed.
- M. A. S. Aquino, Coord. Chem. Rev., 2004, 248, 1025–1045 CrossRef CAS.
- I. Tolbatov, E. Barresi, S. Taliani, D. La Mendola, T. Marzo and A. Marrone, Inorg. Chem. Front., 2023, 10, 2226–2238 RSC.
- G. Lozano, R. Jimenez-Aparicio, S. Herrero and E. Martinez-Salas, RNA, 2016, 22, 330–338 CrossRef CAS PubMed.
- G. Tito, R. Troisi, G. Ferraro, A. Geri, L. Massai, L. Messori, F. Sica and A. Merlino, Dalton Trans., 2023, 52, 6992–6996 RSC.
- L. Messori, T. Marzo, R. N. F. Sanches, H. Ur Rehman, D. de Oliveira Silva and A. Merlino, Angew. Chem., Int. Ed., 2014, 53, 6172–6175 CrossRef CAS.
- G. Ferraro, A. Pratesi, L. Messori and A. Merlino, Dalton Trans., 2020, 49, 2412–2416 RSC.
- D. Loreto, G. Ferraro and A. Merlino, Int. J. Mol. Sci., 2021, 22, 1496 CrossRef CAS PubMed.
- G. Ribeiro, M. Benadiba, A. Colquhoun and D. de Oliveira Silva, Polyhedron, 2008, 27, 1131–1137 CrossRef CAS.
- A. Terán, G. Ferraro, P. Imbimbo, A. E. Sánchez-Peláez, D. M. Monti, S. Herrero and A. Merlino, Int. J. Biol. Macromol., 2023, 253, 126666 CrossRef PubMed.
- A. M. Angeles-Boza, H. T. Chifotides, J. D. Aguirre, A. Chouai, P. K.-L. Fu, K. R. Dunbar and C. Turro, J. Med. Chem., 2006, 49, 6841–6847 CrossRef CAS PubMed.
- R. Hrdina, Eur. J. Inorg. Chem., 2021, 2021, 501–528 CrossRef CAS.
- H. Ur Rehman, T. E. Freitas, R. N. Gomes, A. Colquhoun and D. de Oliveira Silva, J. Inorg. Biochem., 2016, 165, 181–191 CrossRef.
- M. S. Nothenberg, S. B. Zyngicr, A. M. Giesbrecht, M. T. P. Gambardella, H. A. Santos, E. Kimura and R. Najjar, J. Braz. Chem. Soc., 1994, 5, 23 CrossRef CAS.
- S. Zyngier, E. Kimura and R. Najjar, Braz. J. Med. Biol. Res., 1989, 22, 397–401 CAS.
- A. Erck, L. Rainen, J. Whileyman, I.-M. Chang, A. P. Kimball and J. Bear, Proc. Soc. Exp. Biol. Med., 1974, 145, 1278–1283 CrossRef CAS PubMed.
- R. A. Howard, A. P. Kimball and J. L. Bear, Cancer Res., 1979, 39, 2568–2573 CAS.
- T. R. Serio, A. G. Cashikar, A. S. Kowal, G. J. Sawicki, J. J. Moslehi, L. Serpell, M. F. Arnsdorf and S. L. Lindquist, Science, 2000, 289, 1317–1321 CrossRef CAS PubMed.
- D. O. de Silva, Anticancer Agents Med. Chem., 2010, 10, 312–323 CrossRef.
- I. Tolbatov, P. Umari and A. Marrone, J. Mol. Graphics Modell., 2024, 131, 108806 CrossRef CAS PubMed.
- M. J. Frisch, G. W. Trucks, H. B. Schlegel, G. E. Scuseria, M. A. Robb, J. R. Cheeseman, G. Scalmani, V. Barone, G. A. Petersson, H. Nakatsuji, X. Li, M. Caricato, A. V. Marenich, J. Bloino, B. G. Janesko, R. Gomperts, B. Mennucci, H. P. Hratchian, J. V. Ortiz, A. F. Izmaylov, J. L. Sonnenberg, D. Williams-Young, F. Ding, F. Lipparini, F. Egidi, J. Goings, B. Peng, A. Petrone, T. Henderson, D. Ranasinghe, V. G. Zakrzewski, J. Gao, N. Rega, G. Zheng, W. Liang, M. Hada, M. Ehara, K. Toyota, R. Fukuda, J. Hasegawa, M. Ishida, T. Nakajima, Y. Honda, O. Kitao, H. Nakai, T. Vreven, K. Throssell, J. A. Montgomery Jr., J. E. Peralta, F. Ogliaro, M. J. Bearpark, J. J. Heyd, E. N. Brothers, K. N. Kudin, V. N. Staroverov, T. A. Keith, R. Kobayashi, J. Normand, K. Raghavachari, A. P. Rendell, J. C. Burant, S. S. Iyengar, J. Tomasi, M. Cossi, J. M. Millam, M. Klene, C. Adamo, R. Cammi, J. W. Ochterski, R. L. Martin, K. Morokuma, O. Farkas, J. B. Foresman and D. J. Fox, Gaussian, Inc., Wallingford CT, 2016.
- L. Chiaverini, A. Pratesi, D. Cirri, A. Nardinocchi, I. Tolbatov, A. Marrone, M. Di Luca, T. Marzo and D. La Mendola, Molecules, 2022, 27, 2578 CrossRef CAS.
- I. Tolbatov and A. Marrone, Pharmaceutics, 2023, 15, 985 CrossRef CAS.
- I. Tolbatov, D. Cirri, M. Tarchi, T. Marzo, C. Coletti, A. Marrone, L. Messori, N. Re and L. Massai, Inorg. Chem., 2022, 61, 3240–3248 CrossRef CAS PubMed.
- P. Piazzetta, T. Marino, N. Russo and D. R. Salahub, ACS Catal., 2015, 5, 5397–5409 CrossRef CAS.
- J.-E. Sánchez-Aparicio, G. Sciortino, E. Mates-Torres, A. Lledós and J.-D. Maréchal, Faraday Discuss., 2022, 234, 349–366 RSC.
- J. A. Platts, B. M. Kariuki and P. D. Newman, Molecules, 2024, 29, 1150 CrossRef CAS PubMed.
- J.-D. Chai and M. Head-Gordon, Phys. Chem. Chem. Phys., 2008, 10, 6615–6620 RSC.
- D. Andrae, U. Häußermann, M. Dolg, H. Stoll and H. Preuß, Theor. Chim. Acta, 1990, 77, 123–141 CrossRef CAS.
- F. Weigend and R. Ahlrichs, Phys. Chem. Chem. Phys., 2005, 7, 3297–3305 RSC.
- K. Remya and C. H. Suresh, J. Comput. Chem., 2013, 34, 1341–1353 CrossRef CAS PubMed.
- I. Tolbatov, L. Storchi and A. Marrone, Inorg. Chem., 2022, 61, 15664–15677 CrossRef CAS PubMed.
- I. Tolbatov and A. Marrone, Inorg. Chem., 2022, 61, 746–754 CrossRef CAS.
- J. Tomasi, B. Mennucci and E. Cancès, J. Mol. Struct.: THEOCHEM, 1999, 464, 211–226 CrossRef CAS.
- A. Klamt, C. Moya and J. Palomar, J. Chem. Theory Comput., 2015, 11, 4220–4225 CrossRef CAS PubMed.
- C. P. Kelly, C. J. Cramer and D. G. Truhlar, J. Phys. Chem. B, 2006, 110, 16066–16081 CrossRef CAS.
- J. P. Foster and F. Weinhold, J. Am. Chem. Soc., 1980, 102, 7211–7218 CrossRef CAS.
- A. E. Reed, L. A. Curtiss and F. Weinhold, Chem. Rev., 1988, 88, 899–926 CrossRef CAS.
- I. Tolbatov and A. Marrone, Inorg. Chem., 2022, 61, 16421–16429 CrossRef CAS.
- E. R. Jamieson and S. J. Lippard, Chem. Rev., 1999, 99, 2467–2498 CrossRef CAS.
| This journal is © The Royal Society of Chemistry 2025 |