Open Access Article
Open Access ArticleSolid–liquid partitioning of dengue, West Nile, Zika, hepatitis A, influenza A, and SARS-CoV-2 viruses in wastewater from across the USA†
Laura
Roldan-Hernandez
 ,
Camila
Van Oost
,
Camila
Van Oost
 and
Alexandria B.
Boehm
and
Alexandria B.
Boehm
 *
*
Department of Civil & Environmental Engineering, School of Engineering and Doerr School of Sustainability, Stanford University, 473 Via Ortega, Stanford, CA 94305, USA. E-mail: aboehm@stanford.edu; Tel: +1 650 724 9128
First published on 7th May 2024
Abstract
Limited information is available on the fate of respiratory and arthropod-borne viruses in wastewater. Enteric viruses have been extensively studied in wastewater treatment plants, however partition coefficients have not been well documented. This information is essential for interpreting wastewater-based surveillance (WBS) data and optimizing sample collection and processing methods. In this study, we examined the solid–liquid partitioning behavior of dengue, West Nile, Zika, hepatitis A, influenza A, and SARS-CoV-2 viruses in wastewater. Samples were collected from the primary sludge line of eleven wastewater treatment plants across the United States and spiked with varying concentrations of each virus. Solid and liquid fractions were separated via centrifugation. Viral nucleic acids were extracted and quantified using reverse-transcription digital droplet PCR (RT-ddPCR). Partition coefficients (KF), determined using the Freundlich adsorption model, ranged from 4.0 × 102 mL g−1 to 3.9 × 106 mL g−1 (median = 1.1 × 104 mL g−1). We applied a multiple linear regression model to evaluate the effects of factors like viruses and wastewater treatment plants on virus partitioning. We found that the individual effects of those variables were not significant, however, their combined effect was significant. Specifically, significant differences were observed between KF for Zika and West Nile virus between wastewater treatment plants. Further research is needed to understand how wastewater characteristics might impact the partition of viral markers. The results from this experiment underscore the importance of considering wastewater solids for the early detection and monitoring of viral infectious diseases, particularly in regions with low prevalence of infections.
Water impactUnderstanding the fate of viruses in wastewater is essential for accurately interpreting WBS data. In this study, we found that viral markers can be highly enriched in solids, which underscores the importance of considering wastewater solids as a matrix for early detection and monitoring of viral infectious diseases, particularly in communities with low levels of infections. |
Introduction
Viral infectious diseases pose a significant global health threat due to their ease of transmission1 and potential for high mortality and morbidity rates.2 Timely detection and monitoring of new and re-emerging infectious diseases are crucial for effective public health responses and rapid implementation of mitigation strategies. However, traditional forms of public health surveillance, such as clinical testing, may be limited by test-seeking behaviors, test availability, and delays or biases in data collection.3,4 During the COVID-19 pandemic, wastewater-based surveillance (WBS), also referred to as wastewater-based epidemiology (WBE), emerged as a valuable complementary tool to monitor the spread of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2). It has also been effectively used to identify other acute respiratory illnesses (ARI) that might be silently circulating in the population such as influenza, respiratory syncytial virus, and human metapneumovirus.5–7 WBS continues to play a crucial role in public health surveillance post-pandemic by monitoring markers for SARS-CoV-2 variants and other common respiratory viruses.8A few recent studies have also assessed the feasibility of monitoring arboviral (arthropod-borne viral) diseases – such as dengue, Zika, and Chikungunya – using WBS.9–13 In 2023, the World Health Organization (WHO) reported over five million dengue cases and 5000 dengue-related deaths in over 80 countries. The magnitude and geographic spread of arboviral epidemics is expected to increase with climate change.14 In terms of viral shedding, dengue virus, West Nile virus, and Zika virus have been successfully detected in urine and saliva, while Zika virus has also been detected in genital secretions, sweat, tears, ammonitic fluids, placenta, and breast milk.15,16 However, a study by Lee et al.9 suggests that arboviral loads in wastewater might be lower compared to other viruses. Further research is needed to understand urinary and fecal viral shedding in asymptomatic and symptomatic people. Only a couple of studies have been able to detect arboviruses in wastewater. The first study monitored dengue (DENV) serotypes 1–4 at three wastewater treatment plants in Miami-Dade County, Florida; researchers were able to consistently detect DENV-3 when both travel-associated and locally acquired cases of Dengue 3 were identified in the county.17 The second study monitored dengue and Chikungunya at ten wastewater treatment plants across Portugal; dengue was consistently detected throughout the study whereas Chikungunya was rarely detected; associations between WBS and clinical surveillance data were not established given the lack of clinical data.18 Finally, a study in Singapore analyzed clinical, entomological, and wastewater-based surveillance data for Zika; wastewater samples were collected from manholes in the community and peak detections coincided with reported cases.12 Monitoring arboviral diseases through wastewater could help overcome some of the challenges and limitations faced by conventional forms of monitoring and testing mosquito-borne diseases.9,19
WBS has also been used to monitor enteric (intestinal) diseases caused by norovirus, adenovirus, astrovirus, hepatitis A and E virus, and enteroviruses.20–23 Enteric viruses are primarily spread through the fecal-oral route (by ingesting contaminated food or water) or direct person-to-person contact. They are often detected in raw wastewater because they are shed at high concentrations in the feces of symptomatic and asymptomatic individuals.24,25 Long-term fecal shedding has also been reported after weeks and months of infection.25,26 Multiple studies have reported a correlation between WBS and clinical surveillance data. For instance, a study in Ohio used WBS and metagenomics to examine the seasonal dynamics and circulation of enterovirus infections; researchers found similar trends in WBS data and reported clinical cases.27 Another study used WBS as a complementary tool to monitor hepatitis A viruses in centralized and decentralized sewage in Argentina and found similar results.28 WBS has also been used to monitor hepatitis A outbreaks in the United States; a study in Detroit showed that WBS data was significantly correlated with the number of cases reported after one week of wastewater sampling.29 Since 2016, over 45![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 cases and 400 deaths have been reported in the United States, according to the Centers for Disease Control and Prevention (CDC).
000 cases and 400 deaths have been reported in the United States, according to the Centers for Disease Control and Prevention (CDC).
Viruses and their genetic material can adsorbed onto the surface of wastewater solids as they travel through the sewer system; this process is likely driven by electrostatic and hydrophobic interactions between the virus and the wastewater solids.30 Even though wastewater surveillance has been widely applied in the last few years, only a few studies have examined the partitioning behavior of respiratory and enteric viruses as well as laboratory surrogates for pathogenic viruses in wastewater.20,31–34 To our knowledge, no studies have examined the partitioning behavior of arboviruses in wastewater. This information is crucial for optimizing wastewater sampling strategies and lab processing methods, particularly in sewersheds with a low prevalence of infections. It could also help improve the detection and monitoring of diseases with low viral shedding rates. The goal of this study is to determine the partition coefficient of dengue, West Nile, Zika, hepatitis A, influenza A, and SARS-CoV-2 viruses in wastewater. We achieved this by conducting a series of batch experiments, where we spiked different concentrations of each virus to wastewater samples from eleven wastewater treatment plants across the United States.
The results from this experiment will also help populate mechanistic models aiming to back-calculate the levels of community infections. For example, Wolfe et al.35 developed a mass balance model to compare SARS-CoV-2 RNA concentrations in wastewater solids to laboratory-confirmed COVID-19 cases using solid–liquid partitioning coefficients for SARS-CoV-2 among other factors; researchers found a positive and significant association between SARS-CoV-2 RNA and COVID-19 cases. However, they also emphasize the need for further data on partitioning coefficients to better understand differences in testing bias across regions and estimate true COVID-19 cases in a sewershed. Similarly, a study by Soller et al.36 derived three mechanistic WBS models to estimate levels of COVID-19 infections and found that solid–liquid partitioning coefficients of viruses can strongly influence model outputs.
Materials and methods
Overview
We conducted two batch experiments to determine the partition coefficient of dengue, West Nile, Zika, hepatitis A, influenza A, and SARS-CoV-2 viruses at eleven wastewater treatment plants. The family, genome type, structure, and shape of these viruses are shown in Table 1. The first partition experiment (batch 1) was conducted using a wastewater sample from the San José-Santa Clara Regional Wastewater Facility (plant A). The wastewater sample was aliquoted into seven 10 mL subsamples and spiked with viruses. Spiked subsamples were immediately stored at 4 °C, slowly mixed using a tube roller for approximately three hours, and then centrifuged to separate the liquid and solid fractions. The temperature was selected based on common storage practices for wastewater samples; our previous work suggests that temperature does not impact the partitioning behavior of viral markers in wastewater.31 Viral RNA was extracted from each fraction (aliquot) and the RNA specific to each virus was quantified using reverse-transcription-digital droplet PCR (RT-ddPCR). Partition coefficients were determined using the Linear and Freundlich models, which are well-established for the study of chemical adsorption. The second partition experiment (batch 2) was conducted using samples from ten wastewater treatment plants (plants B–K) located across the United States. Wastewater samples from each plant were aliquoted into three 10 mL subsamples, spiked with viruses, and processed using the previously described method. Methods are described in detail below. Reporting of methods follows the Minimum Information for Publication of Quantitative Digital PCR Experiments (dMIQE2020 guidelines37); see Fig. S1 and S2† for dMIQE2020 checklist and additional details.| Virus | Family/genus | Genome type | Structure | Shape | Primary route of transmission |
|---|---|---|---|---|---|
| Dengue | Flaviviridae | +ssRNA | Enveloped | Spherical | Arbovirus |
| West Nile virus | Flaviviridae | +ssRNA | Enveloped | Icosahedral | Arbovirus |
| Zika | Flaviviridae | +ssRNA | Enveloped | Spherical | Arbovirus |
| Influenza A | Orthomyxoviridae | −ssRNA | Enveloped | Spherical/icosahedral | Respiratory |
| Hepatitis A | Picornavirus | +ssRNA | Non-enveloped | Spherical | Enteric |
| SARS-CoV-2 | Coronaviridae | +ssRNA | Enveloped | Spherical | Respiratory |
Wastewater sample collection
For batch 1, a 500 mL wastewater sample was collected on 3 October 2023 from the primary sludge line of the San José-Santa Clara Regional Wastewater Facility (plant A). The plant adds FeCl3 and NaOCl upstream of the sample collection point for odor control. The plant serves approximately 1.4 million people and processes an average flow of 110 million gallons per day (MGD). The sample was transported on ice and stored at 4 °C for one day before spiking.Batch 2 was conducted in two parts (experiments) using 50 mL wastewater samples from ten wastewater treatment plants. The plants (B–K) are located in Michigan, Ohio, New Jersey, Kansas, North Carolina, Connecticut, and Idaho. Wastewater samples were collected from the primary sludge line of each plant. The plants do not add chemicals upstream of the sample collection point except for plants B and K. Plant B adds ferrous chloride and plant K adds aluminum sulfate, in both cases as a coagulant agent. Table S1† has details on the plant names, population served, sample collection date, and description of chemicals added upstream of the sample collection point for each wastewater treatment plant. The first experiment examined the partitioning of dengue, influenza A, hepatitis A, and SARS-CoV-2 viruses in wastewater; samples were collected between 23 and 27 October 2023 and stored at 4 °C for 4–8 days before spiking with viruses. The second experiment examined the partitioning of West Nile and Zika viruses; wastewater samples were collected on either 30 November or 1 December 2023 and stored at 4 °C for 11–12 days before spiking with viruses.
Purification of spiked viruses and preparation of virus cocktails
This study and its associated protocols were approved by the Stanford Biosafety Officer (Biosafety protocol 4260-AB0722). Dengue virus 1 (strain: Hawaii, catalog no. 0810088CF), influenza A virus (strain: California/07/09, H1N1 influenza virus strain, catalog no. 0810165CFHI), SARS-CoV-2 virus (strain: USA-WA1/202, catalog no. 0810587CFHI), West Nile virus (strain: NY 2001-6263, catalog no. 0810033CFHI), and Zika virus (strain: MR 766, catalog no. 0810092CF) were purchased from Zeptometrix (Buffalo, New York). Hepatitis A virus (strain: HM175/18f, catalog no. VR-1402) was purchased from the American Type Culture Collection (ATCC; Manassas, Virginia). Influenza A, SARS-CoV-2, and West Nile viruses were heat-inactivated by the vendor at 56–65 °C for 1–2 h, depending on the virus. The rest of the viruses were infectious. Virus stocks were purified using Amicon Ultra-0.5 mL centrifugal filters (100 kDa MWCO; Millipore UFC5100) following the methods described in Roldan-Hernandez and Boehm.31For batch 1, purified dengue, hepatitis A, influenza A, SARS-CoV-2, West Nile, and Zika viruses were diluted using autoclaved phosphate-buffered saline (PBS; Fisher BioReagents, Pittsburgh, Pennsylvania). These dilutions were then combined to achieve seven distinct virus cocktails, each one containing a mixture of the six viruses at different concentrations. Across virus cocktails, the minimum and maximum concentrations ranged from 1 × 104–2 × 108 cp ml−1 for dengue, 2 × 105–4 × 106 cp ml−1 for hepatitis A, 2 × 104–9 × 105 cp ml−1 for influenza A, 1 × 105–2 × 106 cp ml−1 for SARS-CoV-2, 6 × 103–3 × 108 cp ml−1 for West Nile virus, and 4 × 103–9 × 107 cp ml−1 for Zika. Table S2† provides the concentrations for each specific cocktail.
For batch 2, purified viruses were combined to achieve three distinct virus cocktails. For the first experiment, the minimum and maximum concentrations ranged from 2 × 105–2 × 107 cp ml−1 for dengue, 8 × 105–5 × 106 cp ml−1 for hepatitis A, 2 × 105–1 × 106 cp ml−1 for influenza A, and 1 × 106–2 × 106 cp ml−1 for SARS-CoV-2. For the second experiment, concentrations ranged from 2 × 106–2 × 107 cp ml−1 for West Nile virus and 3 × 105–5 × 106 cp ml−1 for Zika virus. Table S2† provides detailed concentrations for each virus cocktail.
Pre-analytical process
Wastewater samples were thoroughly mixed by inverting the sample 3–4 times. Then aliquoted into seven 10 mL subsamples. For batch 1, subsamples were spiked on 4 October 2023 with a mixture of dengue, hepatitis A, influenza A, SARS-CoV-2, West Nile virus, and Zika. Spiked subsamples (N = 7) were immediately stored at 4 °C and slowly mixed using a tube roller for approximately three hours to allow the system to equilibrate. The time needed to reach apparent equilibrium was determined based on the results of a preliminary experiment, see Fig. S3† for further details.For batch 2, the first set of subsamples was spiked with dengue, SARS-CoV-2, hepatitis A, and influenza A on 31 October 2023, The second set of subsamples was spiked with West Nile virus and Zika on 11 December 2023. The total number of spiked subsamples was 30 (3 subsamples × 10 plants) for the first experiment and 24 (3 subsamples × 8 plants) for the second experiment; fresh wastewater samples were not available for plants D and J and therefore samples from those plants were excluded from the second experiment.
After approximately three hours, spiked subsamples were centrifuged at 4 °C, 24![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 × g for 30 minutes. For each subsample, an aliquot (200 μl) was collected from the supernatant; this aliquot represents the liquid fraction of the wastewater sample. Liquid aliquots were spiked with 10 μl of bovine coronavirus vaccine (Zoetis; #CALF-GUARD) as an internal process control. The remaining supernatant was discarded. For solids, approximately 0.1 g of dewatered solids were aliquoted from the pellet and transferred to a 2 mL collection tube. Solids aliquots were resuspended in approximately 1.3 ml of BCoV spiked-in DNA/RNA shield (Zymo Research; cat. no. R1100-250) to achieve a final concentration of 75 mg of solids per ml of DNA/RNA shield. BCoV spiked-in DNA/RNA shield was prepared using 1.5 μL of BCoV vaccine per ml of DNA/RNA shield. Three grinding balls (OPS DIAGNOSTICS, GBSS 156-5000-01) were added to the 2 mL collection tubes and homogenized at 4 m s−1 for 1 min using the MP Bio Fastprep-24™ (MP Biomedicals, Santa Ana, CA). The solid aliquots were then centrifuged for 5 min at 5250 × g and 200 μL of the supernatant was transferred to a 2 mL microcentrifuge tube for RNA extraction. Liquid and solid aliquots were stored at 4 °C overnight and processed the next day.
000 × g for 30 minutes. For each subsample, an aliquot (200 μl) was collected from the supernatant; this aliquot represents the liquid fraction of the wastewater sample. Liquid aliquots were spiked with 10 μl of bovine coronavirus vaccine (Zoetis; #CALF-GUARD) as an internal process control. The remaining supernatant was discarded. For solids, approximately 0.1 g of dewatered solids were aliquoted from the pellet and transferred to a 2 mL collection tube. Solids aliquots were resuspended in approximately 1.3 ml of BCoV spiked-in DNA/RNA shield (Zymo Research; cat. no. R1100-250) to achieve a final concentration of 75 mg of solids per ml of DNA/RNA shield. BCoV spiked-in DNA/RNA shield was prepared using 1.5 μL of BCoV vaccine per ml of DNA/RNA shield. Three grinding balls (OPS DIAGNOSTICS, GBSS 156-5000-01) were added to the 2 mL collection tubes and homogenized at 4 m s−1 for 1 min using the MP Bio Fastprep-24™ (MP Biomedicals, Santa Ana, CA). The solid aliquots were then centrifuged for 5 min at 5250 × g and 200 μL of the supernatant was transferred to a 2 mL microcentrifuge tube for RNA extraction. Liquid and solid aliquots were stored at 4 °C overnight and processed the next day.
RNA extraction
Nucleic acids were extracted from the liquid and solid aliquots using the AllPrep PowerViral DNA/RNA kit (QIAGEN, catalog no. 28000-50). One extraction replicate was performed for each solid and liquid aliquot. RNase-Free water was used as a negative control for each batch of extraction. RNA extracts were eluted in 100 μl of RNase-Free water (provided in the extraction kit) and processed using the OneStep PCR Inhibitor Removal Kit (Zymo Research, catalog no. D6030) following the manufacturer's instructions. Purified RNA eluates were aliquoted into two separate PCR tubes (each one containing approximately 50 μl), immediately stored at −80 °C, and measured the next day using RT-ddPCR.RNA quantification
Viral nucleic acids were quantified using the One-Step RT-ddPCR Advanced Kit for Probes (Bio-Rad, catalog no. 1863021) following the methods described in Roldan-Hernandez and Boehm.31 See ESI† for additional details on reaction components and primer and probe concentrations. Primers and probes were obtained from previously published research papers and purchased from Integrated DNA Technologies (IDT; San Diego, CA). Table S3† contains the primer/probe sequences. An in silico analysis was performed in NCBI BLAST (Bethesda, MD) and Benchling (San Francisco, CA) to confirm the specificity of each primer and probe. Primers and probes were also tested in the lab using RNA extracts from virus stocks.Duplex assays were prepared for dengue, hepatitis A, influenza A, SARS-CoV-2, West Nile virus, and Zika; the assay mixes varied based on batch and experiment (see ESI†). BCoV was measured using a simplex assay and interpreted as a gross extraction and inhibition control. RNA extracts were used neat as templates and were processed in duplicate (two technical PCR replicates). RNase-Free water was used as a negative PCR control and RNA extracts from virus stocks were used as positive PCR controls. After preparing the PCR plates, droplets were generated using the AutoDG Automated Droplet Generator (Bio-Rad, Hercules, CA) and amplified using the C1000 Touch Thermal Cycler (Bio-Rad, Hercules, CA). The thermal cycling conditions of each assay are shown in Table S4.† After amplification, droplets were analyzed using the QX200 Droplet reader (BIORAD, catalog no. 1864003) and QuantaSoft Software (Bio-Rad, version 1.7). The QuantaSoft files were then exported to QuantaSoft™ Analysis Pro software (Bio-Rad, version 1.0.596.0525) for further analysis. All plates were manually thresholded in QuantaSoft™ Analysis Pro by setting a universal manual threshold for each plate. As a quality control, PCR wells with less than 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 droplets were discarded. Technical PCR replicates (wells) were merged before performing dimensional analysis. The estimated limit of detention is 3.0 copies per ml for liquids and 2000 copies per g for solids. Non-detects were replaced with half of the limit of detection to estimate partition coefficients.
000 droplets were discarded. Technical PCR replicates (wells) were merged before performing dimensional analysis. The estimated limit of detention is 3.0 copies per ml for liquids and 2000 copies per g for solids. Non-detects were replaced with half of the limit of detection to estimate partition coefficients.
Dimensional analysis, adsorption models, and statistical analysis
BCoV recovery was calculated by dividing the measured BCoV concentrations in liquid and solid aliquots by the stock concentration, which was subjected to the same extraction procedure described above. Viral RNA concentrations were converted from copies per reaction to copies per gram of dry weight (cp g−1) for the solids and copies per milliliter (cp ml−1) for liquids. See ESI† for details on the dimensional analysis.38 We used the linear (eqn (1)) and Freundlich (eqn (2)) adsorption models39 to determine the partition parameters K, KF, and n, respectively:| Cs = KCL | (1) |
log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) Cs = log Cs = log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) KF + n KF + n![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) log log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) CL CL | (2) |
Data from batch 1 and 2 were combined and modeled using a multiple linear regression42 (eqn (3)) to assess the main and interaction effects of log10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) CL (X), virus (V), and wastewater treatment plant (P) on log10
CL (X), virus (V), and wastewater treatment plant (P) on log10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) Cs (Y).
Cs (Y).
| Y = β0 + β1X + β2V + β3XV + β4P + β5XP + β6VP + β7XVP + ε | (3) |
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) KF) under reference conditions. β1, β2, and β4 represent the main effects of X (log10
KF) under reference conditions. β1, β2, and β4 represent the main effects of X (log10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) CL, continuous variable), V (virus, categorical dummy variable), and P (plant, categorical dummy variable), respectively. β3, β5, β6, β7 represent the interaction effects of XV, XP, VP, and XVP, respectively. ε is the residual standard error of the model. Coefficients with p-values < 0.05 were considered statistically different from 0. Post hoc comparisons were made using the estimated marginal means (EMMs) of the model adjusted for multiple comparisons using Tukey's method. All analyses were conducted in R (version 4.3.2) using the ln, emmeans, and pairs (adjusted by Tukey's method) functions.
CL, continuous variable), V (virus, categorical dummy variable), and P (plant, categorical dummy variable), respectively. β3, β5, β6, β7 represent the interaction effects of XV, XP, VP, and XVP, respectively. ε is the residual standard error of the model. Coefficients with p-values < 0.05 were considered statistically different from 0. Post hoc comparisons were made using the estimated marginal means (EMMs) of the model adjusted for multiple comparisons using Tukey's method. All analyses were conducted in R (version 4.3.2) using the ln, emmeans, and pairs (adjusted by Tukey's method) functions.
Results
RNA extraction and ddPCR controls
Positive and negative controls for extraction and PCR were positive and negative, respectively; indicating no cross-contamination when processing the wastewater samples. BCoV was spiked into solid and liquid aliquots and used as an internal process control to assess the validity of RNA extractions. The average BCoV recovery was 0.92 (interquartile range: 0.87–1.04) for liquid fractions and 1.40 (interquartile range: 1.19–1.67) for solid fractions. BCoV recoveries were not statistically different between liquid and solid fractions (Student's t-test, p > 0.05). Recoveries greater than one were likely associated with uncertainties in measuring spiked-in BCoV concentrations.Partition experiment: batch 1 (plant A)
Viral RNA concentrations were measured in the liquid and solid fractions of each subsample after they reached apparent equilibrium. For liquids, the minimum and maximum CL measured across subsamples ranged from 1.5–30![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 500 cp ml−1 for dengue, 39–3300 cp ml−1 for hepatitis A, 1.5–100 cp ml−1 for influenza A, 4–400 cp ml−1 for SARS-CoV-2, 1.5–8000 cp ml−1 for West Nile virus, and 1.5–8000 cp ml−1 for Zika. Dengue, influenza A, WNV, and Zika had CL values below the limit of detection; therefore the minimum range value reported in the previous sentence for those viruses is half the CL limit of the detection. For solids, Cs ranged from 5 × 103–6 × 107 cp g−1 for dengue, 4 × 104–3 × 106 cp g−1 for hepatitis A, 1 × 104–7 × 105 cp g−1 for influenza A, 2 × 104–2 × 106 cp g−1 for SARS-CoV-2, 1 × 103–9 × 107 cp g−1 for West Nile virus, and 1 × 103–2 × 107 cp g−1 for Zika. Zika had Cs values under the limit of detection; therefore the minimum range value reported in the previous sentence for Zika is half the Cs limit of the detection.
500 cp ml−1 for dengue, 39–3300 cp ml−1 for hepatitis A, 1.5–100 cp ml−1 for influenza A, 4–400 cp ml−1 for SARS-CoV-2, 1.5–8000 cp ml−1 for West Nile virus, and 1.5–8000 cp ml−1 for Zika. Dengue, influenza A, WNV, and Zika had CL values below the limit of detection; therefore the minimum range value reported in the previous sentence for those viruses is half the CL limit of the detection. For solids, Cs ranged from 5 × 103–6 × 107 cp g−1 for dengue, 4 × 104–3 × 106 cp g−1 for hepatitis A, 1 × 104–7 × 105 cp g−1 for influenza A, 2 × 104–2 × 106 cp g−1 for SARS-CoV-2, 1 × 103–9 × 107 cp g−1 for West Nile virus, and 1 × 103–2 × 107 cp g−1 for Zika. Zika had Cs values under the limit of detection; therefore the minimum range value reported in the previous sentence for Zika is half the Cs limit of the detection.
Fig. 1, shows the Cs and CL concentrations for dengue, hepatitis A, influenza A, SARS-CoV-2, West Nile virus, and Zika from the seven spiked wastewater subsamples from plant A. For WNV and Zika, the results from only six spiked wastewater samples are displayed because one subsample (spiked with the virus cocktail containing the lowest concentrations of viruses) had Cs and CL both below the limits of detection.
Table 2 shows partition model parameters (K, KF, and n) for each virus obtained using the linear and Freundlich adsorption models. K ranged from 800–11![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 400 mL g−1 across viruses using the linear model. KF and n ranged from 500–7600 mL g−1 and 0.93–1.15, respectively, using the Freundlich model. The average relative error (ARE) ranged from 0.33–1.00 using the linear model and 0.12–0.56 using the Freundlich model. K and KF were similar across models, except in the cases of WNV and Zika, where the Freundlich model had a lower ARE (0.28 vs. 1.00 for WNV and 0.12 vs. 0.61 for Zika). For this reason, we decided to focus our analysis only on the Freundlich model. For batch 1 experiments, influenza A exhibited the greatest partition coefficients followed by SARS-CoV-2, WNV, dengue, hepatitis A, and Zika. KF values for SARS-CoV-2 and influenza A were significantly higher than Zika and hepatitis A virus, given that their 95% confidence intervals do not overlap. Partition coefficients for WNV were also significantly higher than Zika.
400 mL g−1 across viruses using the linear model. KF and n ranged from 500–7600 mL g−1 and 0.93–1.15, respectively, using the Freundlich model. The average relative error (ARE) ranged from 0.33–1.00 using the linear model and 0.12–0.56 using the Freundlich model. K and KF were similar across models, except in the cases of WNV and Zika, where the Freundlich model had a lower ARE (0.28 vs. 1.00 for WNV and 0.12 vs. 0.61 for Zika). For this reason, we decided to focus our analysis only on the Freundlich model. For batch 1 experiments, influenza A exhibited the greatest partition coefficients followed by SARS-CoV-2, WNV, dengue, hepatitis A, and Zika. KF values for SARS-CoV-2 and influenza A were significantly higher than Zika and hepatitis A virus, given that their 95% confidence intervals do not overlap. Partition coefficients for WNV were also significantly higher than Zika.
| Reference | Viral target | Linear model | Freundlich model | |||
|---|---|---|---|---|---|---|
| K (LE–UE) (mL g−1) | ARE (−) | K F (LE–UE) (mL g−1) | n (SE) (−) | ARE (−) | ||
| a SE, LE, and UE are the standard error, the lower SE bound, and the upper SE bound, respectively. ARE is the average relative error of the adsorption models. ARE and n are dimensionless. | ||||||
| This study (batch 1) | Dengue | 1800 (1800–1900) | 0.33 | 1900 (1000–3600) | 0.98 (0.05) | 0.31 |
| Hepatitis A | 800 (800–900) | 0.24 | 1100 (500–2200) | 0.94 (0.06) | 0.18 | |
| Influenza A | 6300 (5400–7100) | 0.53 | 7600 (2900–19![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 600) 600) |
0.93 (0.17) | 0.56 | |
| SARS-CoV-2 | 3900 (3500–4300) | 0.17 | 4800 (3200–7400) | 0.94 (0.05) | 0.18 | |
| West Nile virus | 11![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 400 (10 400 (10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 700–12 700–12![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 100) 100) |
1.00 | 3500 (2000–6100) | 1.14 (0.05) | 0.28 | |
| Zika | 1900 (1800–2000) | 0.61 | 500 (300–900) | 1.09 (0.05) | 0.12 | |
| Previous study (Roldan-Hernandez and Boehm)31 | SARS-CoV-2 | — | — | 18![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 (4100–41 000 (4100–41![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000) 000) |
0.81 (0.07) | 0.40 |
| RSV-A | — | — | 32![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 (2000–67 000 (2000–67![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000) 000) |
1.24 (0.02) | 0.25 | |
| RV-B | — | — | 13![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 (1500–28 000 (1500–28![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000) 000) |
0.84 (0.03) | 0.15 | |
Partition experiment: batch 2 (10 wastewater treatment plants across the United States)
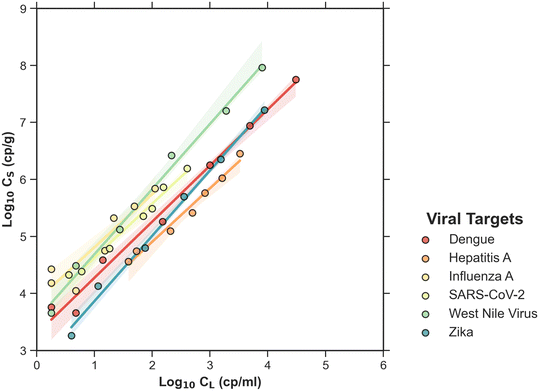
Across subsamples, the minimum and maximum measured CL ranged from 1.5–1700 cp ml−1 for dengue, 1.5–2000 cp ml−1 for hepatitis A, 1.5–20 cp ml−1 for influenza A, 1.5–400 cp ml−1 for SARS-CoV-2, 1.5–3000 cp ml−1 for West Nile virus, and 1.5–1700 cp ml−1 for Zika. All viruses had CL values under the limit of detection; therefore the minimum range value reported in the previous sentence for all viruses is half the CL limit of the detection. For solids, Cs ranged from 5 × 103–2 × 107 cp g−1 for dengue, 5 × 104–1 × 107 cp g−1 for hepatitis A, 7 × 103–2 × 106 cp g−1 for influenza A, 4 × 104–1 × 107 cp g−1 for SARS-CoV-2, 5 × 104–5 × 108 cp g−1 for West Nile virus, and 3 × 103–3 × 107 cp g−1 for Zika.Fig. 2, shows Cs and CL concentrations measured for dengue, hepatitis A, influenza A, SARS-CoV-2, West Nile virus, and Zika from 30 spiked wastewater subsamples from plants B–K (batch 2). For dengue, the results only from 29 spiked subsamples are displayed because one subsample had Cs and CL below the detection limit and the data are not plotted. For WNV and Zika, 24 spiked subsamples are displayed because fresh wastewater samples were not available for plants D and J on the date of the experiment. Fig. 2 also includes the results from batch 1 (plant A) to facilitate comparison between batch 1 and 2.
 | ||
Fig. 2 log10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) Cs and log10 Cs and log10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) CL for dengue, hepatitis A, influenza A, SARS-CoV-2, West Nile virus, and Zika spiked into wastewater samples from 11 plants (batch 1 and 2). CL for dengue, hepatitis A, influenza A, SARS-CoV-2, West Nile virus, and Zika spiked into wastewater samples from 11 plants (batch 1 and 2). | ||
For each virus and plant, partition parameters (KF and n) were obtained using the Freundlich adsorption model. Table 3 shows the total number of KF values calculated for each virus, the average, standard deviation, minimum, and maximum partition coefficients (KF) for dengue, hepatitis A, influenza A, SARS-CoV-2, West Nile virus, and Zika in spiked wastewater samples from 11 wastewater treatment plants (batch 1 and 2). KF and n were determined for all viruses and plants except: 1) influenza A and SARS-CoV-2 in plants C and G because CL values were all below the limit of detection and therefore a linear regression could not be performed, and 2) WNV and Zika for plant D and J because fresh wastewater samples were not available for the second experiment.
| Viral target | Total number of KF values | Median KF (mL g−1) | IQR KF (mL g−1) | Minimum (mL g−1) | Maximum (mL g−1) |
|---|---|---|---|---|---|
| Dengue | 11 | 6000 | 2000–10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 000 |
2000 | 16![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 000 |
| Hepatitis A | 11 | 13![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 000 |
7000–42![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 000 |
1000 | 103![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 000 |
| Influenza A | 9 | 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 000 |
6000–16![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 000 |
5000 | 40![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 000 |
| SARS-CoV-2 | 9 | 25![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 000 |
7000–53![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 000 |
2000 | 155![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 000 |
| West Nile virus | 9 | 24![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 000 |
19![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000–1 000–1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 375 375![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 000 |
3000 | 3![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 912 912![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 000 |
| Zika | 9 | 6000 | 2000–14![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 000 |
400 | 19![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 000 |
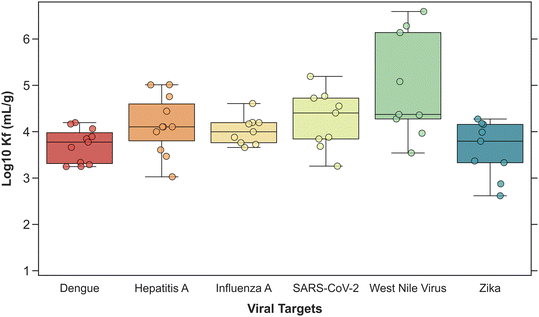
Across wastewater treatment plants (batch 1 and 2), KF values in increasing rank order of medians (IQR) were: 5.9 × 103 mL g−1 (2.0 × 103–9.7 × 103) for dengue, 6.2 × 103 mL g−1 (2.1 × 103–1.4 × 104) for Zika, 9.9 × 103 mL g−1 (5.8 × 103–1.6 × 104) for influenza A, 1.3 × 104 mL g−1 (7.0 × 103–4.2 × 104) for hepatitis A, 2.5 × 104 mL g−1 (7.0 × 103–5.3 × 104) for SARS-CoV-2, and 2.4 × 104 mL g−1 (1.9 × 104–1.4 × 106) for West Nile virus. Partition coefficients (KF) for dengue, hepatitis A, influenza A, SARS-CoV-2, West Nile virus, and Zika are shown in Fig. 3. Tables S5 and S6† show the partition coefficients (KF) and intensity of adsorption (n) for each virus and wastewater treatment plant.
Results from the multiple linear regression model indicated that the coefficients for the variables V (virus) and P (plant) were not statistically significant, while coefficients for their interaction term (VP) were significant. Therefore, a post hoc Tukey contrast test was conducted to identify differences for specific viruses between plants. For Zika, KF was smaller at plant A than plant B, C, E, G, I, and K. For WNV, KF was smaller at plant A than plant B, C, G, H, and I. In addition, WNV KF was higher at plant C than plant H. KF was not significantly different between plants for other viruses.
Discussion
Viral RNA concentrations were orders of magnitude higher in solids than in the liquid fraction of wastewater samples, for all viruses. When combining the results from the 11 wastewater treatment plants, minimum and maximum partition coefficients (KF) ranged from 4.0 × 102 mL g−1 to 3.9 × 106 mL g−1 across viruses. These results are similar to our previous study measuring the partitioning of SARS-CoV-2, rhinovirus type B (RV-B), and respiratory syncytial virus type A (RSV-A) RNA in wastewater, as shown in Table 2.31 In this study, KF values were not significantly different between viruses. However, significant differences were observed between certain pairs of wastewater treatment plants for Zika and West Nile virus. Overall, our results are consistent with previous studies examining the solid–liquid partitioning behavior of viruses and viral genetic markers in wastewater. While several studies have examined the partition of SARS-CoV-2 RNA38,43–45 in wastewater, only two studies have examined the partition of influenza A RNA.7,46 The fate and removal of enteric viruses, such as hepatitis A, has also been extensively studied in wastewater treatment plants, however, partitioning coefficients are not often reported. To our knowledge, this is the first study to examine the partitioning of viral markers for dengue, Zika, and WNV in wastewater.Previous studies suggest that some respiratory viruses, naturally present in wastewater – “endogenous”, may partition more favorably to wastewater solids. Viral RNA concentrations of endogenous SARS-CoV-2 and influenza A have been reported several orders of magnitude higher in solids of raw wastewater influent and primary sludge samples compared to the liquid fraction of wastewater. For example, a study by Mercier et al.46 assessed different enrichment and concentration methods for measuring viral markers for endogenous influenza A in wastewater matrices; settled solids from influent and primary sludge samples were separated via centrifugation and the supernatant was then further processed using polyethylene glycol (PEG) precipitation or a 0.45 μm filter, depending on the type of sample. Researchers found that over 85% of the viral RNA signal was detected in settled solids of both wastewater matrices.
Higher concentrations of endogenous SARS-CoV-2 and influenza A RNA have also been reported in solids of primary sludge samples than in paired wastewater influent samples.7,43,47,48 The results from these studies underscore the importance of considering wastewater solids for early detection and monitoring of acute respiratory diseases through WBS, particularly in regions with low prevalence of infections.
Enteric viruses are readily adsorbed onto wastewater solids.49 However, the partition (or distribution) coefficient of these viruses has not been well documented. Before the COVID-19 pandemic, WBS was also used to monitor outbreaks and seasonal dynamics of enteric viruses such as hepatitis A and E, norovirus GI and GII, adenovirus, enteroviruses, and rotavirus.20–22,28,29,50 Most of these studies focused on analyzing the liquid fraction of raw wastewater influent samples. However, a few recent studies have also started to monitor enteric viruses using solids from raw wastewater influent and primary sludge samples. Results from both wastewater matrices have shown strong correlations with the number of reported cases.35,38,43,51–53 Similar to respiratory viruses, higher viral concentrations of enteric viruses have been reported in solids of primary sludge samples than in influent samples, with distribution coefficients ranging from 650–26![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 mL g−1 for enterovirus, norovirus GI and GII, adenovirus, and rotavirus.20 The partition coefficients measured for hepatitis A in our study align with these previously reported values. For enteric viruses, both liquids and solids might be a sensitive and representative approach for monitoring acute gastrointestinal diseases. However, wastewater solids might require less sample volume to achieve similar sensitivities.
000 mL g−1 for enterovirus, norovirus GI and GII, adenovirus, and rotavirus.20 The partition coefficients measured for hepatitis A in our study align with these previously reported values. For enteric viruses, both liquids and solids might be a sensitive and representative approach for monitoring acute gastrointestinal diseases. However, wastewater solids might require less sample volume to achieve similar sensitivities.
A study by Lee et al.9 suggests that arboviral loads in wastewater might be lower than other viruses. It remains unclear whether arboviral diseases can be effectively monitored through wastewater; only three studies have successfully detected viral markers for arboviruses in wastewater.12,17,18 The first study, conducted by Wolfe et al.,17 monitored dengue (serotypes 1–4) using wastewater solids from 50 ml raw influent samples; researchers were able to detect dengue serotype 3 in a population with an estimated weekly incidence rate of 0.77–4.23 cases/1 million people, using a solid-optimized enrichment method. The second study, by Monteiro et al.,18 monitored dengue (non-specific serotype assay) by processing 1 liter raw wastewater samples using hollow fiber filtration and PEG precipitation. The apparent case detection limit was not reported given the lack of clinical case data. The third study, by Wong et al.,12 monitored viral markers for Zika virus in mosquito pools, individual mosquitoes captured, and wastewater samples and compared those results to clinical cases reported in a community in Singapore. Wastewater samples were collected from manholes and processed using ultrafiltration. The sample volume was not specified in the study, but peak detections in wastewater and mosquitoes coincided with reported cases in the area. A study by Chandra et al.54 evaluated different clarification and viral concentration methods for optimizing the detection of arboviruses in wastewater. However, all the methods were primarily focused on the liquid fraction of wastewater samples. Our results indicate that viral markers for dengue, Zika, and West Nile virus may partition several orders of magnitudes higher in solids than in liquids. Thus, wastewater solids from raw influent and primary sludge samples may be a more advantageous medium for detecting and monitoring viral markers for arboviral diseases.
In our study, similar partitioning behaviors were observed across all viruses. However, further research is needed to understand how virus characteristics, such as envelope structure, capsid proteins, and particle size, might influence the fate and transport of viruses and viral genetic markers in wastewater. The results from our study also suggest that partition coefficients might be similar between wastewater treatment plants, however, further research is needed to determine how wastewater characteristics might impact the partition of viruses. For example, a study by Guo et al.55 indicates that ferric chloride might enhance the adsorption of viruses to wastewater solid particles. Other studies also suggest that the pH levels and the presence of organic matter might also impact viral adsorption.39,49,56 In our study, we did not observe a clear difference in KF values in plants that added chemicals upstream of the sample collection point compared to those that did not. Overall, the results from our experiments could help optimize the enrichment and concentration methods for the recovery/quantification of viral markers in wastewater and primary sludge samples.
This study has several limitations. First, we used exogenous viruses, some of which were heat-inactivated (i.e., influenza A, SARS-CoV-2, and West Nile). In our study, partition coefficients of heat-inactivated viruses were similar to those that were infectious, suggesting that heat inactivation might not significantly impact the solid–liquid partitioning behavior of viruses in wastewater. However, exogenous viruses may be in different physiological states than endogenous viruses in wastewater. For instance, endogenous viruses may or may not have an intact lipid membrane (if enveloped) or intact capsid. Additionally, viral nucleic acids in wastewater may not be protected by a capsid. Limited work has investigated the physiological state of viruses in wastewater. Robinson et al.57 concluded, using detergents, that SARS-CoV-2 RNA in wastewater was present in a lipid membrane, while Wurtzer et al.58 indicated that viruses in wastewater may be present with damaged as well as intact capsids. While conducting partitioning experiments with endogenous viruses may be ideal as it would best represent the conditions of the virus in wastewater, in practice it is difficult because high titers of virus are needed to measure their partitioning in wastewater. In our previous study, we found that partition coefficients (KF) of exogenous viruses were similar to the distribution coefficients (Kd) of the endogenous viruses in wastewater. This finding suggests that spiking viruses into wastewater gives a valid assessment of how endogenous viruses partition in wastewater.31
Conflicts of interest
There are no conflicts to declare.Acknowledgements
We thank Sergey Brin Family Foundation and the Knight-Hennessy Program for financially supporting this work. Graphical abstract was prepared with BioRender.References
- J. Barker, D. Stevens and S. F. Bloomfield, Spread and Prevention of Some Common Viral Infections in Community Facilities and Domestic Homes, J. Appl. Microbiol., 2001, 91(1), 7–21, DOI:10.1046/j.1365-2672.2001.01364.x.
- Z. A. Bhutta, J. Sommerfeld, Z. S. Lassi, R. A. Salam and J. K. Das, Global Burden, Distribution, and Interventions for Infectious Diseases of Poverty, Infect. Dis. Poverty, 2014, 3(1), 21, DOI:10.1186/2049-9957-3-21.
- P. A. McElfish, R. Purvis, L. P. James, D. E. Willis and J. A. Andersen, Perceived Barriers to COVID-19 Testing, Int. J. Environ. Res. Public Health, 2021, 18(5), 2278, DOI:10.3390/ijerph18052278.
- C. L. Gibbons, M.-J. J. Mangen, D. Plass, A. H. Havelaar, R. J. Brooke, P. Kramarz, K. L. Peterson, A. L. Stuurman, A. Cassini, E. M. Fèvre and M. E. Kretzschmar, Measuring Underreporting and Under-Ascertainment in Infectious Disease Datasets: A Comparison of Methods, BMC Public Health, 2014, 14(1), 147, DOI:10.1186/1471-2458-14-147.
- A. B. Boehm, M. K. Wolfe, B. J. White, B. Hughes, D. Duong and A. Bidwell, More than a Tripledemic: Influenza A Virus, Respiratory Syncytial Virus, SARS-CoV-2, and Human Metapneumovirus in Wastewater during Winter 2022–2023, Environ. Sci. Technol. Lett., 2023, 10(8), 622–627, DOI:10.1021/acs.estlett.3c00385.
- R. Dumke, M. Geissler, A. Skupin, B. Helm, R. Mayer, S. Schubert, R. Oertel, B. Renner and A. H. Dalpke, Simultaneous Detection of SARS-CoV-2 and Influenza Virus in Wastewater of Two Cities in Southeastern Germany, January to May 2022, Int. J. Environ. Res. Public Health, 2022, 19(20), 13374, DOI:10.3390/ijerph192013374.
- M. K. Wolfe, D. Duong, K. M. Bakker, M. Ammerman, L. Mortenson, B. Hughes, P. Arts, A. S. Lauring, W. J. Fitzsimmons, E. Bendall, C. E. Hwang, E. T. Martin, B. J. White, A. B. Boehm and K. R. Wigginton, Wastewater-Based Detection of Two Influenza Outbreaks, Environ. Sci. Technol. Lett., 2022, 9(8), 687–692, DOI:10.1021/acs.estlett.2c00350.
- D. Valencia, A. T. Yu, A. Wheeler, L. Hopkins, I. Pray, L. Horter, D. J. Vugia, S. Matzinger, L. Stadler, N. Kloczko, M. Welton, S. Bertsch-Merbach, K. Domakonda, D. Antkiewicz, H. Turner, C. Crain, A. Mulenga, M. Shafer, J. Owiti, R. Schneider, K. H. Janssen, M. K. Wolfe, S. L. McClellan, A. B. Boehm, A. Roguet, B. White, M. K. Schussman, M. S. Rane, J. Hemming, C. Collins, A. Abram, E. Burnor, R. Westergaard, J. N. Ricaldi, J. Person and N. Fehrenbach, Notes from the Field: The National Wastewater Surveillance System's Centers of Excellence Contributions to Public Health Action During the Respiratory Virus Season — Four U.S. Jurisdictions, 2022–23, Morb. Mortal. Wkly. Rep., 2023, 72(48), 1309–1312, DOI:10.15585/mmwr.mm7248a4.
- W. L. Lee, X. Gu, F. Armas, M. Leifels, F. Wu, F. Chandra, F. J. D. Chua, A. Syenina, H. Chen, D. Cheng, E. E. Ooi, S. Wuertz, E. J. Alm and J. Thompson, Monitoring Human Arboviral Diseases through Wastewater Surveillance: Challenges, Progress and Future Opportunities, Water Res., 2022, 223, 118904, DOI:10.1016/j.watres.2022.118904.
- F. Chandra, W. L. Lee, F. Armas, M. Leifels, X. Gu, H. Chen, S. Wuertz, E. J. Alm and J. Thompson, Persistence of Dengue (Serotypes 2 and 3), Zika, Yellow Fever, and Murine Hepatitis Virus RNA in Untreated Wastewater, Environ. Sci. Technol. Lett., 2021, 8(9), 785–791, DOI:10.1021/acs.estlett.1c00517.
- O. Thakali, S. Raya, B. Malla, S. Tandukar, A. Tiwari, S. P. Sherchan, J. B. Sherchand and E. Haramoto, Pilot Study on Wastewater Surveillance of Dengue Virus RNA: Lessons, Challenges, and Implications for Future Research, Environ. Challenges, 2022, 9, 100614, DOI:10.1016/j.envc.2022.100614.
- J. C. C. Wong, M. Tay, H. C. Hapuarachchi, B. Lee, G. Yeo, D. Maliki, W. Lee, N.-A. Mohamed Suhaimi, K. Chio, W. C. H. Tan and L. C. Ng, Case report: Zika surveillance complemented with wastewater and mosquito testing, EBioMedicine, 2024, 101, 105020 CrossRef PubMed.
- K. Zhu, C. Hill, A. Muirhead, M. Basu, J. Brown, M. A. Brinton, M. J. Hayat, C. Venegas-Vargas, M. G. Reis, A. Casanovas-Massana, J. S. Meschke, A. I. Ko, F. Costa and C. E. Stauber, Zika Virus RNA Persistence and Recovery in Water and Wastewater: An Approach for Zika Virus Surveillance in Resource-Constrained Settings, Water Res., 2023, 241, 120116, DOI:10.1016/j.watres.2023.120116.
- M. Girard, C. B. Nelson, V. Picot and D. J. Gubler, Arboviruses: A Global Public Health Threat, Vaccine, 2020, 38(24), 3989–3994, DOI:10.1016/j.vaccine.2020.04.011.
- M. Niedrig, P. Patel, A. A. El Wahed, R. Schädler and S. Yactayo, Find the Right Sample: A Study on the Versatility of Saliva and Urine Samples for the Diagnosis of Emerging Viruses, BMC Infect. Dis., 2018, 18(1), 707, DOI:10.1186/s12879-018-3611-x.
- T. R. Poloni, A. S. Oliveira, H. L. Alfonso, L. R. Galvão, A. A. Amarilla, D. F. Poloni, L. T. Figueiredo and V. H. Aquino, Detection of Dengue Virus in Saliva and Urine by Real Time RT-PCR, Virol. J., 2010, 7(1), 22, DOI:10.1186/1743-422X-7-22.
- M. K. Wolfe, A. H. Paulos, A. Zulli, D. Duong, B. Shelden, B. J. White and A. B. Boehm, Wastewater Detection of Emerging Arbovirus Infections: Case Study of Dengue in the United States, Environ. Sci. Technol. Lett., 2024, 11(1), 9–15, DOI:10.1021/acs.estlett.3c00769.
- S. Monteiro, R. Pimenta, F. Nunes, M. V. Cunha and R. Santos, Wastewater-Based Surveillance for Tracing the Circulation of Dengue and Chikungunya Viruses, SSRN, 2023, preprint, DOI:10.2139/ssrn.4618839.
- W. Gu, T. R. Unnasch, C. R. Katholi, R. Lampman and R. J. Novak, Fundamental Issues in Mosquito Surveillance for Arboviral Transmission, Trans. R. Soc. Trop. Med. Hyg., 2008, 102(8), 817–822, DOI:10.1016/j.trstmh.2008.03.019.
- A. B. Boehm, B. Shelden, D. Duong, N. Banaei, B. J. White and M. K. Wolfe, A retrospective longitudinal study of adenovirus group F, norovirus GI and GII, rotavirus, and enterovirus nucleic acids in wastewater solids at two wastewater treatment plants: solid-liquid partitioning and relation to clinical testing data, mSphere, 2024, 9, e00736-23 CrossRef PubMed.
- M. Bisseux, J. Colombet, A. Mirand, A.-M. Roque-Afonso, F. Abravanel, J. Izopet, C. Archimbaud, H. Peigue-Lafeuille, D. Debroas, J.-L. Bailly and C. Henquell, Monitoring Human Enteric Viruses in Wastewater and Relevance to Infections Encountered in the Clinical Setting: A One-Year Experiment in Central France, 2014 to 2015, Eurosurveillance, 2018, 23(7), 16–26, DOI:10.2807/1560-7917.ES.2018.23.7.17-00237.
- L. Chacón, E. Morales, C. Valiente, L. Reyes and K. Barrantes, Wastewater-Based Epidemiology of Enteric Viruses and Surveillance of Acute Gastrointestinal Illness Outbreaks in a Resource-Limited Region, Am. J. Trop. Med. Hyg., 2021, 105(4), 1004–1012, DOI:10.4269/ajtmh.21-0050.
- C. McCall, H. Wu, B. Miyani and I. Xagoraraki, Identification of Multiple Potential Viral Diseases in a Large Urban Center Using Wastewater Surveillance, Water Res., 2020, 184, 116160, DOI:10.1016/j.watres.2020.116160.
- N. S. Upfold, G. A. Luke and C. Knox, Occurrence of Human Enteric Viruses in Water Sources and Shellfish: A Focus on Africa, Food Environ. Virol., 2021, 13(1), 1–31, DOI:10.1007/s12560-020-09456-8.
- P. F. M. Teunis, F. H. A. Sukhrie, H. Vennema, J. Bogerman, M. F. C. Beersma and M. P. G. Koopmans, Shedding of Norovirus in Symptomatic and Asymptomatic Infections, Epidemiol. Infect., 2015, 143(8), 1710–1717, DOI:10.1017/S095026881400274X.
- B. Kapusinszky, P. Minor and E. Delwart, Nearly Constant Shedding of Diverse Enteric Viruses by Two Healthy Infants, J. Clin. Microbiol., 2012, 50(11), 3427–3434, DOI:10.1128/JCM.01589-12.
- N. E. Brinkman, G. S. Fout and S. P. Keely, Retrospective Surveillance of Wastewater To Examine Seasonal Dynamics of Enterovirus Infections, mSphere, 2017, 2(3) DOI:10.1128/mSphere.00099-17.
- A. Fantilli, G. D. Cola, G. Castro, P. Sicilia, A. M. Cachi, M. De Los Ángeles Marinzalda, G. Ibarra, L. López, C. Valduvino, G. Barbás, S. Nates, G. Masachessi, M. B. Pisano and V. Ré, Hepatitis A Virus Monitoring in Wastewater: A Complementary Tool to Clinical Surveillance, Water Res., 2023, 241, 120102, DOI:10.1016/j.watres.2023.120102.
- C. McCall, H. Wu, E. O'Brien and I. Xagoraraki, Assessment of Enteric Viruses during a Hepatitis Outbreak in Detroit MI Using Wastewater Surveillance and Metagenomic Analysis, J. Appl. Microbiol., 2021, 131(3), 1539–1554, DOI:10.1111/jam.15027.
- A. Armanious, M. Aeppli, R. Jacak, D. Refardt, T. Sigstam, T. Kohn and M. Sander, Viruses at Solid–Water Interfaces: A Systematic Assessment of Interactions Driving Adsorption, Environ. Sci. Technol., 2016, 50(2), 732–743, DOI:10.1021/acs.est.5b04644.
- L. Roldan-Hernandez and A. B. Boehm, Adsorption of Respiratory Syncytial Virus, Rhinovirus, SARS-CoV-2, and F+ Bacteriophage MS2 RNA onto Wastewater Solids from Raw Wastewater, Environ. Sci. Technol., 2023, 57(36), 13346–13355, DOI:10.1021/acs.est.3c03376.
- Z. Yin, T. C. Voice, V. V. Tarabara and I. Xagoraraki, Sorption of Human Adenovirus to Wastewater Solids, J. Environ. Eng., 2018, 144(11), 06018008, DOI:10.1061/(ASCE)EE.1943-7870.0001463.
- P. R. Breadner, H. A. Dhiyebi, A. Fattahi, N. Srikanthan, S. Hayat, M. G. Aucoin, S. J. Boegel, L. M. Bragg, P. M. Craig, Y. Xie, J. P. Giesy and M. R. Servos, A Comparative Analysis of the Partitioning Behaviour of SARS-CoV-2 RNA in Liquid and Solid Fractions of Wastewater, Sci. Total Environ., 2023, 895, 165095, DOI:10.1016/j.scitotenv.2023.165095.
- Y. Ye, R. M. Ellenberg, K. E. Graham and K. R. Wigginton, Survivability, Partitioning, and Recovery of Enveloped Viruses in Untreated Municipal Wastewater, Environ. Sci. Technol., 2016, 50(10), 5077–5085, DOI:10.1021/acs.est.6b00876.
- M. K. Wolfe, A. Archana, D. Catoe, M. M. Coffman, S. Dorevich, K. E. Graham, S. Kim, L. M. Grijalva, L. Roldan-Hernandez, A. I. Silverman, N. Sinnott-Armstrong, D. J. Vugia, A. T. Yu, W. Zambrana, K. R. Wigginton and A. B. Boehm, Scaling of SARS-CoV-2 RNA in Settled Solids from Multiple Wastewater Treatment Plants to Compare Incidence Rates of Laboratory-Confirmed COVID-19 in Their Sewersheds, Environ. Sci. Technol. Lett., 2021, 8(5), 398–404, DOI:10.1021/acs.estlett.1c00184.
- J. Soller, W. Jennings, M. Schoen, A. Boehm, K. Wigginton, R. Gonzalez, K. E. Graham, G. McBride, A. Kirby and M. Mattioli, Modeling Infection from SARS-CoV-2 Wastewater Concentrations: Promise, Limitations, and Future Directions, J. Water Health, 2022, 20(8), 1197–1211, DOI:10.2166/wh.2022.094.
- The dMIQE Group, A. S. Whale, W. De Spiegelaere, W. Trypsteen, A. A. Nour, Y.-K. Bae, V. Benes, D. Burke, M. Cleveland, P. Corbisier, A. S. Devonshire, L. Dong, D. Drandi, C. A. Foy, J. A. Garson, H.-J. He, J. Hellemans, M. Kubista, A. Lievens, M. G. Makrigiorgos, M. Milavec, R. D. Mueller, T. Nolan, D. M. O'Sullivan, M. W. Pfaffl, S. Rödiger, E. L. Romsos, G. L. Shipley, V. Taly, A. Untergasser, C. T. Wittwer, S. A. Bustin, J. Vandesompele and J. F. Huggett, The Digital MIQE Guidelines Update: Minimum Information for Publication of Quantitative Digital PCR Experiments for 2020, Clin. Chem., 2020, 66(8), 1012–1029, DOI:10.1093/clinchem/hvaa125.
- K. E. Graham, S. K. Loeb, M. K. Wolfe, D. Catoe, N. Sinnott-Armstrong, S. Kim, K. M. Yamahara, L. M. Sassoubre, L. M. Mendoza Grijalva, L. Roldan-Hernandez, K. Langenfeld, K. R. Wigginton and A. B. Boehm, SARS-CoV-2 RNA in Wastewater Settled Solids Is Associated with COVID-19 Cases in a Large Urban Sewershed, Environ. Sci. Technol., 2021, 55(1), 488–498, DOI:10.1021/acs.est.0c06191.
- Y. Jin and M. Flury, Fate and Transport of Viruses in Porous Media, in Advances in Agronomy, Elsevier, 2002, vol. 77, pp. 39–102, DOI:10.1016/S0065-2113(02)77013-2.
- E. K. Hayes, C. L. Sweeney, M. Fuller, G. B. Erjavec, A. K. Stoddart and G. A. Gagnon, Operational Constraints of Detecting SARS-CoV-2 on Passive Samplers Using Electronegative Filters: A Kinetic and Equilibrium Analysis, ACS ES&T Water, 2022, 2(11), 1910–1920, DOI:10.1021/acsestwater.1c00441.
- N. Ayawei, A. N. Ebelegi and D. Wankasi, Modelling and Interpretation of Adsorption Isotherms, J. Chem., 2017, 2017, 1–11, DOI:10.1155/2017/3039817.
- N. H. Bingham and J. M. Fry, Regression, Springer, London, New York, 2010 Search PubMed.
- S. Kim, L. C. Kennedy, M. K. Wolfe, C. S. Criddle, D. H. Duong, A. Topol, B. J. White, R. S. Kantor, K. L. Nelson, J. A. Steele, K. Langlois, J. F. Griffith, A. G. Zimmer-Faust, S. L. McLellan, M. K. Schussman, M. Ammerman, K. R. Wigginton, K. M. Bakker and A. B. Boehm, SARS-CoV-2 RNA is enriched by orders of magnitude in primary settled solids relative to liquid wastewater at publicly owned treatment works, Environ. Sci.: Water Res. Technol., 2022, 8, 757–770 RSC.
- S. Kim and A. B. Boehm, Wastewater Monitoring of SARS-CoV-2 RNA at K-12 Schools: Comparison to Pooled Clinical Testing Data, PeerJ, 2023, 11, e15079, DOI:10.7717/peerj.15079.
- B. Li, D. Y. W. Di, P. Saingam, M. K. Jeon and T. Yan, Fine-Scale Temporal Dynamics of SARS-CoV-2 RNA Abundance in Wastewater during A COVID-19 Lockdown, Water Res., 2021, 197, 117093, DOI:10.1016/j.watres.2021.117093.
- E. Mercier, P. M. D'Aoust, O. Thakali, N. Hegazy, J.-J. Jia, Z. Zhang, W. Eid, J. Plaza-Diaz, M. P. Kabir, W. Fang, A. Cowan, S. E. Stephenson, L. Pisharody, A. E. MacKenzie, T. E. Graber, S. Wan and R. Delatolla, Municipal and Neighbourhood Level Wastewater Surveillance and Subtyping of an Influenza Virus Outbreak, Sci. Rep., 2022, 12(1), 15777, DOI:10.1038/s41598-022-20076-z.
- B. Bhattarai, S. Q. Sahulka, A. Podder, S. Hong, H. Li, E. Gilcrease, A. Beams, R. Steed and R. Goel, Prevalence of SARS-CoV-2 Genes in Water Reclamation Facilities: From Influent to Anaerobic Digester, Sci. Total Environ., 2021, 796, 148905, DOI:10.1016/j.scitotenv.2021.148905.
- S. Balboa, M. Mauricio-Iglesias, S. Rodriguez, L. Martínez-Lamas, F. J. Vasallo, B. Regueiro and J. M. Lema, The Fate of SARS-COV-2 in WWTPS Points out the Sludge Line as a Suitable Spot for Detection of COVID-19, Sci. Total Environ., 2021, 772, 145268, DOI:10.1016/j.scitotenv.2021.145268.
- C. J. Hurst and C. P. Gerba, Fate of Viruses during Wastewater Sludge Treatment Processes, Crit. Rev. Environ. Control, 1989, 18(4), 317–343, DOI:10.1080/10643388909388352.
- M. Hellmér, N. Paxéus, L. Magnius, L. Enache, B. Arnholm, A. Johansson, T. Bergström and H. Norder, Detection of Pathogenic Viruses in Sewage Provided Early Warnings of Hepatitis A Virus and Norovirus Outbreaks, Appl. Environ. Microbiol., 2014, 80(21), 6771–6781, DOI:10.1128/AEM.01981-14.
- J. Peccia, A. Zulli, D. E. Brackney, N. D. Grubaugh, E. H. Kaplan, A. Casanovas-Massana, A. I. Ko, A. A. Malik, D. Wang, M. Wang, J. L. Warren, D. M. Weinberger, W. Arnold and S. B. Omer, Measurement of SARS-CoV-2 RNA in Wastewater Tracks Community Infection Dynamics, Nat. Biotechnol., 2020, 38(10), 1164–1167, DOI:10.1038/s41587-020-0684-z.
- G. Medema, L. Heijnen, G. Elsinga, R. Italiaander and A. Brouwer, Presence of SARS-Coronavirus-2 RNA in Sewage and Correlation with Reported COVID-19 Prevalence in the Early Stage of the Epidemic in The Netherlands, Environ. Sci. Technol. Lett., 2020, 7(7), 511–516, DOI:10.1021/acs.estlett.0c00357.
- P. M. D'Aoust, É. Mercier, D. Montpetit, J.-J. Jia, I. Alexandrov, N. Neault, A. T. Baig, J. Mayne, X. Zhang, T. Alain, M.-A. Langlois, M. R. Servos, M. MacKenzie, D. Figeys, A. E. MacKenzie, T. E. Graber and R. Delatolla, Quantitative Analysis of SARS-CoV-2 RNA from Wastewater Solids in Communities with Low COVID-19 Incidence and Prevalence, Water Res., 2020, 116560, DOI:10.1016/j.watres.2020.116560.
- F. Chandra, F. Armas, G. Kwok, H. Chen, F. J. Desmond Chua, M. Leifels, N. Amir-Hamzah, X. Gu, S. Wuertz, W. L. Lee, E. J. Alm and J. Thompson, Comparing Recovery Methods for Wastewater Surveillance of Arthropod-Borne and Enveloped Viruses, ACS ES&T Water, 2023, 3(4), 974–983, DOI:10.1021/acsestwater.2c00460.
- Y. Guo, J. Shi, E. Sharma, S. Gao, X. Zhou, Y. Liu, M. Sivakumar and G. Jiang, Fate of Coronaviruses during the Wastewater Coagulation with Ferric Chloride, ACS ES&T Water, 2023, 3(10), 3206–3214, DOI:10.1021/acsestwater.3c00112.
- W. Yang, C. Cai and X. Dai, Interactions between Virus Surrogates and Sewage Sludge Vary by Viral Analyte: Recovery, Persistence, and Sorption, Water Res., 2022, 210, 117995, DOI:10.1016/j.watres.2021.117995.
- C. A. Robinson, H.-Y. Hsieh, S.-Y. Hsu, Y. Wang, B. T. Salcedo, A. Belenchia, J. Klutts, S. Zemmer, M. Reynolds, E. Semkiw, T. Foley, X. Wan, C. G. Wieberg, J. Wenzel, C.-H. Lin and M. C. Johnson, Defining Biological and Biophysical Properties of SARS-CoV-2 Genetic Material in Wastewater, Sci. Total Environ., 2022, 807, 150786, DOI:10.1016/j.scitotenv.2021.150786.
- S. Wurtzer, P. Waldman, A. Ferrier-Rembert, G. Frenois-Veyrat, J. M. Mouchel, M. Boni, Y. Maday, V. Marechal and L. Moulin, Several Forms of SARS-CoV-2 RNA Can Be Detected in Wastewaters: Implication for Wastewater-Based Epidemiology and Risk Assessment, Water Res., 2021, 198, 117183, DOI:10.1016/j.watres.2021.117183.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d4ew00225c |
| This journal is © The Royal Society of Chemistry 2025 |