Assessment and application of GeneXpert rapid testing for respiratory viruses in school wastewater†
Jingjing
Wu
 a,
Katherine B.
Ensor
b,
Loren
Hopkins
bc and
Lauren B.
Stadler
a,
Katherine B.
Ensor
b,
Loren
Hopkins
bc and
Lauren B.
Stadler
 *a
*a
aDepartment of Civil and Environmental Engineering, Rice University, 6100 Main Street MS 519, Houston, TX 77005, USA. E-mail: lauren.stadler@rice.edu
bDepartment of Statistics, Rice University, Houston, 77005, USA
cHouston Health Department, 8000 N. Stadium Dr, Houston, TX 77054, USA
First published on 20th November 2024
Abstract
Schools are considered high-risk areas for the transmission of infectious respiratory diseases and thus could benefit from facility-level wastewater disease surveillance. This study assessed the quantitativeness, reproducibility, and feasibility of the rapid GeneXpert system for monitoring SARS-CoV-2, influenza A, influenza B, and RSV in school wastewater. We developed individual standard curves for each disease target using school wastewater spiked with known concentrations of target viruses to estimate viral concentrations in wastewater. Furthermore, we evaluated and compared the reproducibility of the GeneXpert system results against an established filtration-ddPCR workflow and compared the cost per sample of each method. Results show that the GeneXpert system can detect respiratory viruses in school wastewater. We used GeneXpert to perform daily wastewater monitoring for target respiratory viruses at four schools in Houston, TX, USA, over three months (from November 15, 2022, to February 15, 2023, n = 169). For SARS-CoV-2 and influenza A, we observed no significant differences between the positivity results (detection vs. not detected), and strong relationships between quantitative results from the two methods. The RSV detection using GeneXpert was less reproducible and sensitive, but overall consistent in terms of positive detections with the filtration-ddPCR workflow (90.5% consistent). The GeneXpert represents a cost-effective system for wastewater monitoring for respiratory viruses as compared to the filtration-ddPCR method, particularly when the number of samples is low. This study demonstrates the application of the GeneXpert system for facility-level wastewater surveillance and suggests that it represents an important technology for resource-constrained areas.
Water impactThis study assessed the feasibility of using a PCR rapid test system for monitoring SARS-CoV-2, influenza A, influenza B, and RSV in school wastewater. The findings demonstrate the potential of this rapid testing technology for efficient and cost-effective monitoring of respiratory viruses in facility-level wastewater, particularly for testing lower quantities of samples in resource-constrained areas. |
1. Introduction
Wastewater-based epidemiology (WBE) has been applied to monitor various infectious diseases. Numerous studies have shown a correlation between clinical data and the wastewater levels of different pathogens, including respiratory viruses such as SARS-CoV-2, influenza, and respiratory syncytial virus (RSV).1–10 While many wastewater monitoring systems collect and analyze samples from centralized wastewater treatment plants (WWTPs), upstream samples collected from lift (pump) stations and manholes have also been used to monitor smaller populations, such as those living in the same neighborhood, dormitory, congregate living facility, or attending the same school.11–19 Pathogen wastewater levels measured at facilities have also been shown to be associated with clinical cases.11,13,14,17,18,20,21 While wastewater monitoring at WWTPs can provide information on citywide disease dynamics, monitoring at upstream facilities can be used to identify outbreaks and inform targeted interventions to curb the spread of disease.22Schools are a targeted facility-level sampling site for wastewater surveillance. Schools are considered high-risk areas for the transmission of infectious respiratory diseases, as a large number of students and staff are in close proximity to one another.23–25 The detection of SARS-CoV-2 in school wastewater has been reported in multiple studies,13,17,19,26,27 and influenza and RSV to a lesser extent.19,28 Positive relationships were observed between wastewater monitoring results and the number of reported cases.13,17,19,26–28 School wastewater surveillance can help develop an early warning system to protect school-aged children from infectious diseases and identify appropriate interventions to mitigate impacts.13,23 Further, studies have shown that pathogen levels in school wastewater are representative of infections at the school, and also correlated to disease trends in the larger community and city.19,28 Thus, schools may also serve as valuable sentinel sites for certain diseases in the community.
Easy-to-use and rapid testing systems for wastewater monitoring could enable more frequent sampling and a fast turnaround time for results. Most wastewater surveillance systems involve collecting samples from sampling sites and transporting them to a centralized laboratory where they are analyzed. Proper storage conditions and minimized storage time are required during transportation and before the sample processing to reduce false-negative and false-positive results, as improper storage can lead to RNA degradation and cross-contamination.29 Analysis typically consists of a concentration step, followed by nucleic acid extraction and purification, followed by quantification of gene targets using (RT-)qPCR or (RT-)ddPCR. Depending on sample transport and analysis time and schedules, reporting of results typically takes 24 hours and sometimes a week or more. Recently, new technologies for wastewater disease target quantification that are field-deployable, rapid, and require fewer lab resources have been developed.
The Cepheid GeneXpert system is one technology that has been developed and applied for rapid wastewater testing (∼1 hour to analyze and generate results). Previous studies have applied the GeneXpert system for SARS-CoV-2, influenza, and RSV detection in wastewater samples.30,31 Daigle et al. (2022) compared the detection of SARS-CoV-2 using the GeneXpert with a laboratory-developed, solids-based extraction and RT-qPCR test and found that without a pre-concentration step, the results from the GeneXpert should be interpreted qualitatively.31 Asadi et al. (2023) used the GeneXpert system to detect SARS-CoV-2, influenza A, influenza B, and RSV in wastewater samples from WWTPs and compared the results with weekly clinical cases.30 Both studies indicate that the GeneXpert is a rapid test system that can be used for on-site wastewater surveillance of respiratory viruses and well-suited for resource-limited settings. However, no studies to date have used or evaluated the GeneXpert system for the detection of respiratory viruses in facility-level wastewater samples. Facility wastewater can differ in composition from downstream WWTP influent, as there is less dilution due to inflow and infiltration and industrial inputs. Further, viral concentrations are typically more variable in facility-level than WWTP samples.21 Research is needed to assess the sensitivity and application of the GeneXpert system for rapid, facility-level wastewater surveillance.
In this study, we apply the GeneXpert system to monitor school wastewater for quantification of SARS-CoV-2, influenza A, influenza B, and RSV. We test raw wastewater using the GeneXpert system and compare it against an established laboratory method that uses adsorption–extraction (AE) and RT-ddPCR quantification32,33 in terms of sensitivity, reproducibility, and quantitativeness. To estimate wastewater concentrations of each target, we developed individual standard curves for each disease target using school wastewater spiked with known concentrations of target viruses. We compared the detection of the GeneXpert system and the filtration-ddPCR workflow for SARS-CoV-2, influenza A, and RSV in daily wastewater samples collected over three months from four schools in Houston, TX, USA. Finally, we compared the startup and consumable costs per sample of each method.
2. Materials and methods
2.1 Wastewater sample collection
Wastewater samples were obtained Monday–Friday from four schools (Table S1† presents the type and enrollment of each school) across Houston, Texas, over a three-month period between November 15, 2022, to February 15, 2023 (n = 169). Samples were not collected during school holidays and occasionally samples were not available due to autosampler and/or staffing issues. Wastewater samples from schools across Houston were mixed together and used to test the GeneXpert rapid test reproducibility and develop the standard curves for converting the Ct values to target concentrations in wastewater. A refrigerated autosampler placed in or adjacent to a manhole that receives the wastewater from each school was used to collect a time-weighted composite sample by collecting an aliquot every 15 minutes between 6 am and 12 pm. On each sampling day, 250 mL of the composite wastewater sample was collected from each school, and they were transported on ice to Rice University and refrigerated at 4 °C in the laboratory for up to 24 hours before being aliquoted into two 50 mL centrifuge tubes (centrifuge tubes A and B) for further processing.2.2 Wastewater sample testing of SARS-CoV-2, influenza A, influenza B, and RSV using the GeneXpert system
The centrifuge tube A was used for the GeneXpert system test. The centrifuge tube was mixed by inverting the tube for 30 seconds before being placed on the bench for 2 minutes to let large solids settle. A disposable pipette in the GeneXpert kit was used to move 300 μL of the wastewater sample from the top 1/3 of the centrifuge tube into the Xpress-SARS-CoV-2/flu/RSV cartridge (Cepheid, Sunnyvale, CA) for nucleic acid extraction. The cartridge was analyzed using the GeneXpert® IV system following the manufacturer's instructions. The system uses RT-qPCR to provide a cycle threshold (Ct) value for SARS-CoV-2, influenza A, influenza B, and RSV in about 37 minutes. The GeneXpert Xpress-SARS-CoV-2/flu/RSV cartridge also includes an assay developed for avian influenza A detection, which was not included here because our study focuses on human strains. The target genes used for the detection were described previously.30,34 The RT-qPCR reactions were run for 45 cycles. The raw fluorescence value data after each cycle were exported from the software and used to calculate the Ct value and endpoint fluorescence using a converter provided by the manufacturer on Microsoft Excel for Microsoft 365 (version 2402). In our study, the sample was considered “positive” for a target if a Ct value was reported, and “weak positive” if an endpoint fluorescence value was reported and no Ct value was reported.312.3 Wastewater sample concentration, extraction, and quantification of SARS-CoV-2, influenza A, influenza B, and RSV using filtration-ddPCR workflow
The remaining wastewater samples in centrifuge tube A and tube B were used for the filtration-ddPCR workflow. Wastewater samples were concentrated using electronegative (HA) filtration followed by bead-beating, as described by Laturner et al. (2021) and Lou et al. (2022).32,33 The Chemagic™ Prime Viral DNA/RNA 300 Kit H96 (Chemagic, CMG-1433, PerkinElmer) was used for nucleic acid extraction as described by Lou et al. (2022).33The concentrations of SARS-CoV-2 N1 and N2 genes, influenza A, influenza B, and RSV, were quantified using 1-Step RT-ddPCR Advanced Kit for Probes (1864021, Bio-Rad). The automated droplet generator (1864101, Bio-Rad) was used to generate droplets before amplification using C1000 Touch™ Thermal Cycler (1851197, Bio-Rad). The QX600 AutoDG droplet digital PCR system (Bio-Rad) was used to read the droplet results. The results were analyzed using the QuantaSoft v1.7.4 software. Tables S2–S7† present the sequence information of the primers and probes, the RT-ddPCR reaction compositions, and the thermal cycler conditions. Negative control samples were included to assess contamination following the EMMI Guidelines.35 Details for quality control measures and the determination of limit of detection are described by Lou et al. (2022)33 and also briefly in the ESI.† We only quantified the concentrations of SARS-CoV-2 N1 and N2 genes, influenza A and RSV using the filtration-ddPCR method during the three-month school wastewater monitoring study, considering the overall low level of influenza B in the community.
2.4 Development of GeneXpert system standard curve to translate Ct values to viral concentration and the comparison of result reproducibility between GeneXpert system and filtration-ddPCR workflow
Heat-inactivated SARS-CoV-2, omicron variant (VR-3347HK, ATCC), influenza A virus (H1N1) (VR-1894, ATCC), influenza B virus (Victoria Lineage) (VR-1931, ATCC), and Zeptometrix respiratory syncytial virus type A stock (NATRSVA-STQ) were serially-diluted and spiked into 50 mL of school wastewater. The wastewater sample used tested negative for all the targets using the GeneXpert system, as described in section 2.2, prior to spiking. The stock concentrations were quantified by RT-ddPCR as described in section 2.3 after lysis at 95 °C for 10 minutes using C1000 Touch™ Thermal Cycler (1851197, Bio-Rad). For the standard curve development, the final concentrations of the targets ranged from 56 cp mL−1 to 1778 cp mL−1, and triplicates of each concentration were used. All school wastewater samples with spiked virus standards were tested using the Xpress-SARS-CoV-2/flu/RSV cartridge (Cepheid, Sunnyvale, CA) with the GeneXpert system (as described in section 2.2). To assess method reproducibility, a stock virus mixture was spiked at the same concentration into ten 50 mL replicate wastewater samples. Then, the viral concentration was determined using the GeneXpert system and filtration-ddPCR workflow, as described above. We calculated the recovery rate of the filtration-ddPCR workflow for each target by dividing the sample viral concentration determined using ddPCR by the spiked standard viral concentration (eqn (S4)†).2.5 Comparison of wastewater monitoring cost using GeneXpert system and filtration-ddPCR workflow
The cost analysis included an analysis of the costs associated with wastewater concentration, extraction, and quantification. The costs of wastewater monitoring using the GeneXpert system were compared to those of two laboratory-based wastewater monitoring workflows using RT-ddPCR and RT-qPCR, respectively. The cost analysis included the capital and operational costs associated with the analysis of SARS-CoV-2, influenza A, influenza B, and RSV in wastewater samples. Because the GeneXpert system uses one channel for SARS-CoV-2 quantification, for consistency, the cost analysis of laboratory-based filtration-ddPCR and filtration-qPCR workflows only included the quantification of SARS-CoV-2 targeting the N2 gene for SARS-CoV-2. Capital costs included instrument and equipment costs, and operational costs included instrument maintenance, consumables, and labor costs. All cost information was based on purchase history or list prices and was adjusted to 2023 U.S. dollars. The lifespan of the instruments and laboratory equipment was estimated as 12 years.36 Annual maintenance costs were assumed to be 14.1% of capital costs based on historical records in our laboratory. The labor costs were calculated using the estimated labor time required for wastewater sample processing for each method and assuming an hourly labor rate of $32.50, based on data from a public health department.2.6 Statistical analysis
Statistical analysis was conducted using RStudio (R version 4.2.0). We compared the consistency of results in terms of positivity between the GeneXpert system and ddPCR for the detection of SARS-CoV-2, influenza A, and RSV during the three-month school monitoring study. We used the McNemar chi-test to compare the detection of SARS-CoV-2, influenza A, and RSV using the GeneXpert system and filtration-ddPCR workflow. We developed the standard curve to translate the Ct values from the GeneXpert system to viral concentration using linear regression. The LOD of the GeneXpert system was determined using the standard curves with the Ct value set as 45. The Levene test was used to compare the variability of the viral concentration results generated by the GeneXpert system and filtration-ddPCR workflow from the reproducibility test.We estimated the correlation between the viral concentrations of school wastewater samples from the GeneXpert system and filtration-ddPCR workflow using Pearson's correlation tests. For the GeneXpert system, we imputed the wastewater measurements with no Ct value reported (weak positive and negative) as half the LOD. For ddPCR, we imputed the wastewater sample measurements with no positive droplets detected in any sample replicate as 110 copies per L of wastewater, half of the target concentration with one positive droplet out of 38![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 total droplets (average number of total droplets with replicate wells merged). Finally, we developed a linear regression between the target viral concentrations from ddPCR and the Ct values from the GeneXpert system when the target was deemed positive in the wastewater sample using both methods.
000 total droplets (average number of total droplets with replicate wells merged). Finally, we developed a linear regression between the target viral concentrations from ddPCR and the Ct values from the GeneXpert system when the target was deemed positive in the wastewater sample using both methods.
3. Results and discussion
3.1 Development of the standard curves to convert Ct values from the GeneXpert system to viral concentrations in school wastewater
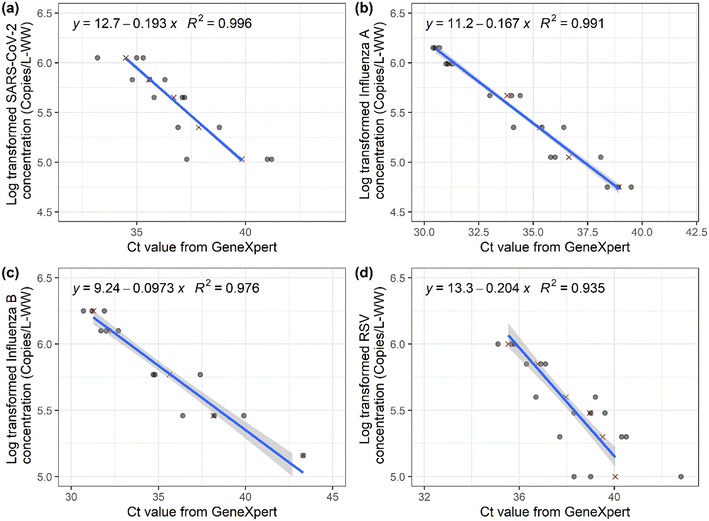
The output of the GeneXpert system is a Ct value and an endpoint fluorescence value for each of the targets. To convert the Ct values from the GeneXpert system into concentrations, we developed the standard curves for each of the targets by spiking the viruses into wastewater samples (Fig. 1). For all the targets, we observed a linear relationship between the Ct values and the amount of target virus spiked into the school wastewater samples, with the R2 values greater than 0.94. Previously, Becker et al. (2021)34 also developed a standard curve to convert Ct values from the GeneXpert system to SARS-CoV-2 concentrations for use with clinical specimens. Differences between the Becker et al. (2021)34 curve and ours are likely due to the media used for generating the standard curves: they used universal transport media (UTM) for target dilution, while we used wastewater, which is a complex mixture of substances and may contain various PCR inhibitors.29,37,38 These results illustrate the importance of generating a standard curve using media specific to the application or system to convert Ct values to concentrations. Our results also demonstrate that the GeneXpert system can provide quantitative results for SARS-CoV-2, influenza A, influenza B, and RSV monitoring in wastewater when the concentration is above 100![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 copies per L-wastewater. However, the LOD of the GeneXpert system is likely lower than 100
000 copies per L-wastewater. However, the LOD of the GeneXpert system is likely lower than 100![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 copies per L-wastewater. For example, a previous study reported the detection of 32
000 copies per L-wastewater. For example, a previous study reported the detection of 32![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 copies per L-wastewater of SARS-CoV-2 using the GeneXpert system.31 Here, we observed mean Ct values of 39.8, 40.0, and 36.6 for approximately 100
000 copies per L-wastewater of SARS-CoV-2 using the GeneXpert system.31 Here, we observed mean Ct values of 39.8, 40.0, and 36.6 for approximately 100![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 copies per L-wastewater of SARS-CoV-2, influenza A, and RSV. However, the maximum possible Ct value is 45, suggesting lower concentrations could be detected. Further evaluation is needed to establish the LOD of these viral targets in wastewater using the GeneXpert system.
000 copies per L-wastewater of SARS-CoV-2, influenza A, and RSV. However, the maximum possible Ct value is 45, suggesting lower concentrations could be detected. Further evaluation is needed to establish the LOD of these viral targets in wastewater using the GeneXpert system.
3.2 Reproducibility of GeneXpert and filtration-ddPCR results
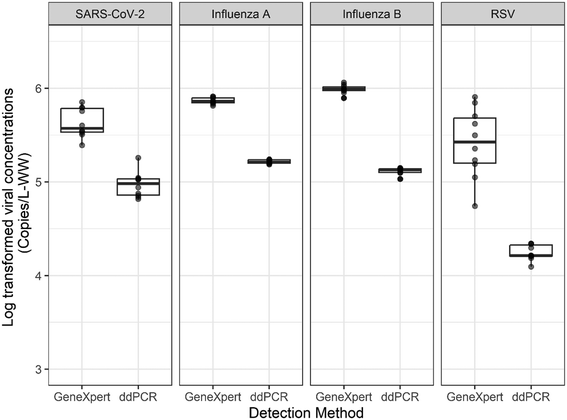
We also compared the reproducibility of the GeneXpert system and the filtration-ddPCR workflows by analyzing 10 sample replicates via both methods (Fig. 2). For all of the targets, we observed greater standard deviations of the viral concentrations from GeneXpert as compared to the filtration-ddPCR method. For RSV, the variance of the viral concentration determined by GeneXpert was significantly greater than that determined using ddPCR (p-value = 0.001). The variances of the viral concentrations using the GeneXpert and ddPCR methods were not significantly different for all other targets (p > 0.05). Our results indicate that the quantification results from the GeneXpert system are generally reproducible but have higher detection variances than the filtration-ddPCR workflow, especially for RSV. One possible reason for the overall increased detection variances of the GeneXpert system is that a total volume of 50 mL wastewater sample was used for virus concentration in the filtration-ddPCR method, while only 300 μL wastewater was processed in the GeneXpert system. We were not able to evaluate whether differences in assay design, such as primer and probe sequences, may have contributed to the observed variance because the primer and probe sequences used by the GeneXpert are proprietary. In correspondence with the manufacturer, Cepheid (Sunnyvale, CA), it was suggested that the RSV RT-PCR assay may be more susceptible to inhibitors in wastewater than the other target assays. Further studies on improving the wastewater processing steps before adding the sample to the cartridge, such as optimizing the mixing and settling duration, may help increase the reproducibility of the GeneXpert system.3.3 Comparison of respiratory viruses detection for daily monitoring of school wastewater using GeneXpert and filtration-ddPCR systems
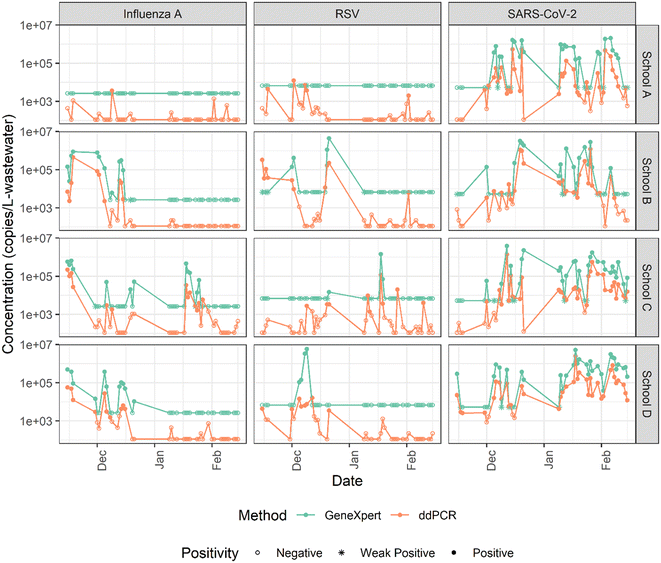
We detected SARS-CoV-2, influenza A, and RSV in school wastewater samples and compared the detection of each target in daily samples collected from four schools over three months (n = 169) using GeneXpert and filtration-ddPCR workflow (Table 1). No significant differences were observed between the positivity of samples (detected vs. not detected) determined using the GeneXpert system and the filtration-ddPCR workflow for SARS-CoV-2 and influenza A (p > 0.1). For SARS-CoV-2, we observed consistent positivity results for 147 of the 169 samples (87.0% consistent), where 119 samples were positive and 28 were negative by both methods. Influenza A and RSV were detected less frequently than SARS-CoV-2 in school wastewater samples. For influenza A, positivity results were consistent across methods for 159 of the 169 samples (94.1% consistent), where 31 samples were positive using both methods. Seven samples tested positive for influenza A using GeneXpert while negative using ddPCR, and 3 tested positive using ddPCR while negative using the GeneXpert system. This is consistent with a previous study that reported similar detection results for SARS-CoV-2 in WWTP and lift station samples using the GeneXpert and a laboratory-developed RT-qPCR workflow.31| GeneXpert system | |||||||
|---|---|---|---|---|---|---|---|
| SARS-CoV-2 | Influenza-A | RSV | |||||
| Positivity | Positive | Negative | Positive | Negative | Positive | Negative | |
| Filtration-ddPCR workflow | Positive | 119 | 11 | 31 | 3 | 19 | 13 |
| Negative | 11 | 28 | 7 | 128 | 3 | 134 | |
For RSV detection in wastewater, the filtration-ddPCR workflow was significantly more sensitive than GeneXpert based on McNemar's chi-test (p = 0.02). Positivity results were consistent for 153 samples (90.5% consistent), and both methods detected RSV in 19 school wastewater samples. Inconsistent positivity results were observed for 16 samples, where 3 samples tested positive for RSV using GeneXpert while negative using ddPCR, and 13 samples tested positive using ddPCR while negative using GeneXpert. The lower sensitivity of RSV detection in wastewater using GeneXpert as compared to the filtration-ddPCR workflow is also reflective of the reproducibility results as described in section 3.2, and may be due to the RSV RT-PCR assay being more sensitive to inhibitors in wastewater.
One possible reason for the similar sensitivity observed for SARS-CoV-2 and influenza A is that although the starting volume for GeneXpert and filtration-ddPCR methods were substantially different (300 μL versus 50 mL), the equivalent volume of wastewater analyzed in the ddPCR reaction was only 3 mL due to dilution during concentration, extraction, and quantification reaction preparation (50 mL of wastewater is filtered and the filter is added to 1 mL of lysis buffer; 300 μL of lysis buffer is used for nucleic acid extraction to generate a 50 μL extract; 10 μL of extract is used for quantification; calculation shows in eqn (S5)†). Furthermore, while the LOD of the filtration-ddPCR method was approximately 1320 copies per L-wastewater (see section S3†), this does not account for losses of RNA during the wastewater processing steps.29,37 We observed recovery rates of 21.9 ± 7.6% for SARS-CoV-2, and 5.9 ± 1.1% for RSV (Fig. S1†). Accounting for recovery, the LOD of the filtration-ddPCR workflow is actually much greater than 1320 copies per L-wastewater. Another reason that we may have observed similar sensitivity for the GeneXpert and filtration-ddPCR methods is because only a few school samples had viral concentrations between the LOD of these two methods. Compared to WWTP samples, viral concentrations in school wastewater tend to be more variable and more sensitive to active cases,21 and thus fewer samples have concentrations near the LOD.
Our results indicate that both methods were generally consistent with one another in terms of wastewater positivity for SARS-CoV-2 and influenza A detection in school wastewater and that both methods have a similar detection sensitivity. Our study also suggests that using GeneXpert for RSV detection in wastewater may result in a higher probability of false-negative results, especially when the viral concentrations are low, and indicates further investigations are needed to improve the sensitivity of RSV detection in wastewater using GeneXpert.
3.4 Comparison of respiratory viral concentration measurements for daily monitoring of school wastewater using GeneXpert and filtration-ddPCR systems
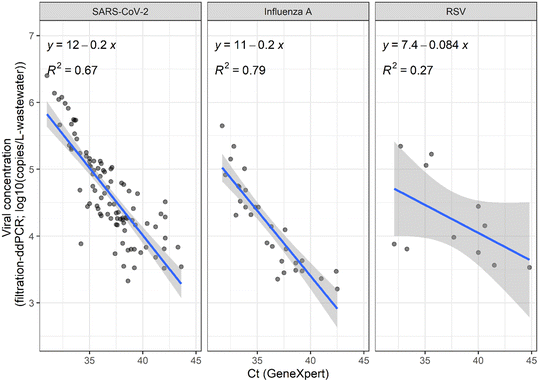
Next, we compared the quantitative viral concentrations determined using the GeneXpert system versus the filtration-ddPCR workflow. First, we converted the Ct values from GeneXpert to viral concentrations of SARS-CoV-2, influenza A, and RSV in school wastewater samples over three months using the standard curves described above. We then compared those concentrations to the viral concentrations determined using the filtration-ddPCR workflow (Fig. 3). Significant positive relationships were observed between the viral concentrations measured in school wastewater samples using GeneXpert and filtration-ddPCR for SARS-CoV-2 (Pearson's r = 0.90 p-values < 0.001) and influenza A (r = 0.79, p-values < 0.001); and a significant, moderate positive relationship was observed for RSV (r = 0.35, p-value < 0.001). In addition, we conducted Pearson's correlation test and linear regressions between the Ct values from GeneXpert and the viral concentrations from filtration-ddPCR for SARS-CoV-2 (n = 94), influenza A (n = 29), and RSV (n = 11) for the wastewater samples tested positive using both methods. We observed significant negative relationships between the GeneXpert Ct value and filtration-ddPCR viral concentrations for SARS-CoV-2 and influenza A (SARS-CoV-2: Pearson's r = −0.82, p-value < 0.001; influenza A: r = −0.89, p-value < 0.001), with the r-squared values for linear regression equals to 0.67 (SARS-CoV-2) and 0.79 (influenza A) (Fig. 4). However, a significant negative relationship was not observed between the GeneXpert Ct value and ddPCR RSV concentrations in school wastewater samples (p = 0.1). The poor correlation between the GeneXpert Ct value and filtration-ddPCR-based viral concentration may be due to several factors, including (1) quantitative RSV results (positive detections) only occurred in a limited number of school wastewater samples (n = 11) during the monitoring period, (2) the variance of the RSV concentration using the GeneXpert system was significantly higher than the variance based on filtration-ddPCR workflow (Fig. 2), and (3) the GeneXpert system was significantly less sensitive than the filtration-ddPCR workflow for RSV detection (Table 1).Another limitation of the GeneXpert system is that it does not include a fecal indicator target such as pepper mild mottle virus (PMMoV) that would enable normalization of viral concentrations in wastewater. A previous study showed that using PMMoV to normalize the SARS-CoV-2 RNA concentration in building-level wastewater samples can improve the strength of the correlation between the wastewater results and COVID-19 cases.17 Nevertheless, the same study also reported significant correlations between SARS-CoV-2 RNA concentration (unnormalized) and COVID-19 cases.17 Another study that performed wastewater surveillance for respiratory viruses in 51 preK-12 schools showed that SARS-CoV-2 RNA concentrations in school wastewater were strongly associated with the number of COVID-19 cases in school and community positivity rates.19 These results suggest that unnormalized viral concentrations in school wastewater can be used to estimate the trend of diseases in schools, and further work is needed to assess the value of normalization using an additional fecal indicator target.
Our results indicate that both the GeneXpert system and filtration-ddPCR workflow can be used to generate consistent quantitative concentrations of SARS-CoV-2 and influenza A, and consistent detections (detected versus not detected) of RSV in school wastewater samples. In addition, linear regression models can be developed to relate the Ct values from GeneXpert and ddPCR-based viral concentrations for SARS-CoV-2 and influenza A, such that the methods could be applied interchangeably and provide comparable information about trends in viral levels. This is important in scenarios where samples are analyzed by multiple labs using different methods and equipment. More RSV-positive wastewater samples should be analyzed to assess the quantitativeness of the GeneXpert system.
3.5 Cost analysis of school wastewater monitoring using the GeneXpert system and filtration-ddPCR workflow
To better understand the context and scenarios where one might choose the GeneXpert system over more standard laboratory workflows, we performed an economic analysis of the GeneXpert and filtration-ddPCR workflows. We compared the capital, maintenance, consumable, and labor costs of quantifying SARS-CoV-2, influenza A, influenza B, and RSV in wastewater samples using the GeneXpert system and the filtration-ddPCR workflow (Table 2) and assessed the impact of sample quantity on overall costs. At the time of this study, the capital cost of the GeneXpert system was $50![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000, 9.9% of the cost of the filtration-ddPCR workflow, which was approximately $503
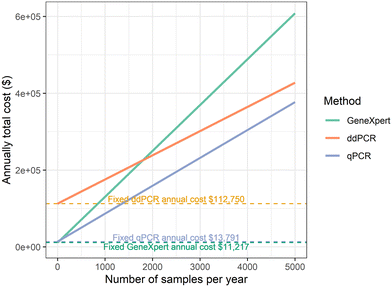
000, 9.9% of the cost of the filtration-ddPCR workflow, which was approximately $503![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000. The consumable and labor costs using the GeneXpert system were $119.43 per wastewater sample (including duplicate measurements and four respiratory targets), 1.9 times that of the filtration-ddPCR workflow, which was $62.96 per sample. Assuming a 12 year lifespan of all the instruments, we estimated the total annual cost based on the yearly testing load (Fig. 5). In general, wastewater monitoring using the GeneXpert system was more economical than the filtration-ddPCR workflow when the total number of wastewater samples was small. The total costs using the two methods were the same when 1799 wastewater samples were tested annually (about 35 samples per week). In addition, although not included in this study because of a lack of information, as a site-deployable wastewater monitoring system, using GeneXpert can also reduce the costs associated with sample transportation if the analysis is performed on site. Using the GeneXpert system may further reduce labor costs because it requires less skilled labor for processing wastewater sampling than the filtration-ddPCR workflow. In brief, our economic analysis shows that wastewater monitoring using the GeneXpert system requires less start-up cost but more processing costs compared to the filtration-ddPCR method. As a result, the GeneXpert is more cost-effective for testing lower quantities of samples.
000. The consumable and labor costs using the GeneXpert system were $119.43 per wastewater sample (including duplicate measurements and four respiratory targets), 1.9 times that of the filtration-ddPCR workflow, which was $62.96 per sample. Assuming a 12 year lifespan of all the instruments, we estimated the total annual cost based on the yearly testing load (Fig. 5). In general, wastewater monitoring using the GeneXpert system was more economical than the filtration-ddPCR workflow when the total number of wastewater samples was small. The total costs using the two methods were the same when 1799 wastewater samples were tested annually (about 35 samples per week). In addition, although not included in this study because of a lack of information, as a site-deployable wastewater monitoring system, using GeneXpert can also reduce the costs associated with sample transportation if the analysis is performed on site. Using the GeneXpert system may further reduce labor costs because it requires less skilled labor for processing wastewater sampling than the filtration-ddPCR workflow. In brief, our economic analysis shows that wastewater monitoring using the GeneXpert system requires less start-up cost but more processing costs compared to the filtration-ddPCR method. As a result, the GeneXpert is more cost-effective for testing lower quantities of samples.
| GeneXpert system | Filtration-ddPCR workflow | Filtration-qPCR workflow (manual extraction) | ||
|---|---|---|---|---|
| Capital cost ($) | Concentration | 50![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000.00 000.00 |
19![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 849.00 849.00 |
19![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 849.00 849.00 |
| Extraction and PCR preparation | 290![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 449.69 449.69 |
5165.50 | ||
| Quantification | 192![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 794.25 794.25 |
37![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 305.60 305.60 |
||
| Total |
50![[thin space (1/6-em)]](https://www.rsc.org/images/entities/b_char_2009.gif) 000.00 000.00
|
50![[thin space (1/6-em)]](https://www.rsc.org/images/entities/b_char_2009.gif) 3092.94 3092.94
|
62![[thin space (1/6-em)]](https://www.rsc.org/images/entities/b_char_2009.gif) 320.10 320.10
|
|
| Annual maintenance cost ($) | 7050.00 |
70![[thin space (1/6-em)]](https://www.rsc.org/images/entities/b_char_2009.gif) 826.25 826.25
|
8777.92 | |
| Consumable cost per sample for four targets ($) | Concentration | 110.76 | 6.02 | 6.02 |
| Extraction and PCR preparation | 10.90 | 26.98 | ||
| Quantification | 22.75 | 14.41 | ||
| Total | 110.76 | 39.67 | 47.41 | |
| Labor cost per sample for four targets ($) | Concentration | 8.67 | 9.75 | 9.75 |
| Extraction and PCR preparation | 10.83 | 11.92 | ||
| Quantification | 2.71 | 3.56 | ||
| Total | 8.67 | 23.29 | 25.23 |
There are multiple options for laboratory methods for quantifying disease targets in wastewater, and the cost per sample for each option depends on the equipment required, labor involved, and consumables. The choice of laboratory method also impacts the throughput of sample analysis and data quality. In this study, we used a Chemagic360 and Janus G3 liquid handling robot (CJM8002, PerkinElmer), which dominated the equipment costs, comprising 96.4% of the capital costs for extraction and PCR preparation. Cheaper alternatives for nucleic acid extraction are available but may not be as high throughput and thus will require more labor. Furthermore, other target quantification methods, such as qPCR, can be used instead of ddPCR, decreasing the quantification capital costs and detecting sensitivity and reproducibility.39 For example, we estimated the cost of the manual nucleic acid extraction QIAamp Viral RNA kits (Qiagen, Hilden, Germany), as an alternative for the Chemagic360 and manual PCR preparation, and qPCR (Table 2). The estimated total costs were lower than those of the GeneXpert system when more than 59 samples were tested annually (about 1 sample per week). To sum up, there are trade-offs in wastewater monitoring costs, testing capacity and throughput, and data quality, and the selection of wastewater sample processing methods requires careful consideration of available resources, including space, personnel, equipment, and budget. One limitation of the cost analysis is the assumption of a 12 year lifespan of all the included instruments, which may not reflect real-world conditions. Further studies are needed to assess the actual lifespans of different wastewater monitoring systems and evaluate their impacts on the overall costs based on the cost data presented in this study.
3.6 Potential applications and limitations of the GeneXpert system for wastewater monitoring
This study demonstrates that the performance of the GeneXpert system is similar to our filtration-ddPCR workflow in terms of sensitivity and quantitation for school wastewater monitoring of respiratory viruses, especially for SARS-CoV-2 and influenza A. The GeneXpert system represents a viable technology for on-site or mobile testing units for wastewater monitoring and could be used in community-level facilities, such as universities, nursing homes, hospitals, and airports. The GeneXpert system is also well-suited for enabling wastewater surveillance in resource-constrained areas, and/or areas where transporting samples is challenging or cumbersome. As a cartridge-based system, GeneXpert requires limited pretreatment and no additional equipment. In addition, while some consumables of PCR-based methods need to be shipped on dry ice and stored in a freezer, the GeneXpert cartridges can be shipped and stored at room temperature. The GeneXpert system is a viable option in areas that lack access to laboratory facilities. For example, in many low- and middle-income countries that may lack centralized WWTPs and/or facilities to support laboratory analysis, GeneXpert could be applied for community wastewater monitoring.40,41 The GeneXpert platform has been used for tuberculosis (TB), human immunodeficiency virus (HIV), and human papillomavirus (HPV) tests in multiple communities in Africa, where laboratory-based clinic testing is not widely available in their healthcare systems.42–45 Similarly, it is very likely that the GeneXpert system can also be used for wastewater surveillance in these areas to improve the data availability for public health response and early warning.Significant limitations associated with the GeneXpert system applications include the limited testing capacity and reliance on their cartridges. Compared to PCR-based workflow, where up to 96 samples can be processed in one plate, only 4 cartridges can be run simultaneously using a single GeneXpert system. The testing capacity of GeneXpert can be increased by using a 16-module machine or multiple instruments, but it will significantly increase the costs. The cartridge cost, nearly $110 per wastewater sample assuming replicates, may also limit the application of the GeneXpert system, especially in some developing countries. In addition, the GeneXpert CoV-2/flu/RSV cartridge used in this study can only be used to monitor these four viruses, and the assay is proprietary. Cartridges are currently only available for a limited number of targets, whereas assays can easily be changed using ddPCR workflows by using different primers and probes. Further, the nucleic acid extract is not recoverable from the GeneXpert cartridge, whereas a single nucleic acid extract in the ddPCR workflow can split and assayed for numerous targets. As a result, the filtration-ddPCR workflow may be more feasible and cost-effective if numerous targets are included in the monitoring system.
Conclusions
This study evaluated the use of the GeneXpert system for SARS-CoV-2, influenza A, influenza B, and RSV surveillance in school wastewater and compared the results to data generated using a filtration-ddPCR workflow. Our results show that the GeneXpert system can efficiently and cost-effectively be used to monitor the dynamics of respiratory viruses in school wastewater with limited testing volume, providing both qualitative results on positivity and quantitative viral concentrations. When we compared the methods on daily wastewater samples collected from four schools over three months (n = 169), we observed no significant difference between the GeneXpert and filtration-ddPCR positivity results and significant positive relationships between the quantitative results using the two methods for SARS-CoV-2 and influenza A detection in wastewater and recommended further investigation for influenza B and RSV detection using GeneXpert. This research further demonstrates the feasibility of rapid and on-site wastewater surveillance in schools and suggests the potential broader applicability of the GeneXpert system in more facilities for virus surveillance through wastewater monitoring. In particular, it may be advantageous for communities that lack access to laboratory resources and analyze a limited number of samples.Data availability
The data underlying this study are available in the published article and its ESI.†Conflicts of interest
There are no conflicts of interest to declare.Acknowledgements
This work was supported by the Houston Health Department and Centers for Disease Control and Prevention (Contract No/Grant No: NU50CK000557-01). We thank Muhammad Tanveer Farhad, MS, Douglas Jennings BS, Francesca Ruby German from Houston Health Department for collecting wastewater samples. We also thank Kaavya Domakonda, Rebecca Schneider, and Anthony Mulenga from the Houston Health Department for their contributions to the Houston Wastewater Epidemiology system. We thank Lauren Bauhs, Kyle Palmer, Madeline Wolken, Russell Carlson Stadler, and Whitney Rich in the Stadler group at Rice University for their assistance with wastewater sample processing and analysis for the filtration-ddPCR workflow. Lastly, we thank James Harbridge, Scott Tucker, and Aaron Anson from Hach Company for providing the instrument and the kits for the GeneXpert.References
- A. B. Boehm, M. K. Wolfe, B. J. White, B. Hughes, D. Duong and A. Bidwell, More than a Tripledemic: Influenza A Virus, Respiratory Syncytial Virus, SARS-CoV-2, and Human Metapneumovirus in Wastewater during Winter 2022–2023, Environ. Sci. Technol. Lett., 2023, 10, 622–627 CrossRef CAS PubMed.
- T. de Melo, G. Islam, D. B. D. Simmons, J.-P. Desaulniers and A. E. Kirkwood, An alternative method for monitoring and interpreting influenza A in communities using wastewater surveillance, Front. Public Health, 2023, 11, 1141136 CrossRef PubMed.
- R. Dumke, M. Geissler, A. Skupin, B. Helm, R. Mayer, S. Schubert, R. Oertel, B. Renner and A. H. Dalpke, Simultaneous Detection of SARS-CoV-2 and Influenza Virus in Wastewater of Two Cities in Southeastern Germany, January to May 2022, Int. J. Environ. Res. Public Health, 2022, 19, 13374 CrossRef CAS PubMed.
- E. K. Hayes, M. T. Gouthro, J. J. LeBlanc and G. A. Gagnon, Simultaneous detection of SARS-CoV-2, influenza A, respiratory syncytial virus, and measles in wastewater by multiplex RT-qPCR, Sci. Total Environ., 2023, 889, 164261 CrossRef CAS PubMed.
- B. Hughes, D. Duong, B. J. White, K. R. Wigginton, E. M. G. Chan, M. K. Wolfe and A. B. Boehm, Respiratory Syncytial Virus (RSV) RNA in Wastewater Settled Solids Reflects RSV Clinical Positivity Rates, Environ. Sci. Technol. Lett., 2022, 9, 173–178 CrossRef CAS.
- M. Koureas, K. Mellou, A. Vontas, M. Kyritsi, I. Panagoulias, A. Koutsolioutsou, V. A. Mouchtouri, M. Speletas, D. Paraskevis and C. Hadjichristodoulou, Wastewater Levels of Respiratory Syncytial Virus Associated with Influenza-like Illness Rates in Children—A Case Study in Larissa, Greece (October 2022–January 2023), Int. J. Environ. Res. Public Health, 2023, 20, 5219 CrossRef CAS.
- A. J. Lee, S. Carson, M. I. Reyne, A. Marshall, D. Moody, D. M. Allen, P. Allingham, A. Levickas, A. Fitzgerald, S. H. Bell, J. Lock, J. D. Coey, C. McSparron, B. F. Nejad, D. G. Courtney, G. G. Einarsson, J. P. McKenna, D. J. Fairley, T. Curran, J. M. McKinley, D. F. Gilpin, K. Lemon, J. W. McGrath and C. G. G. Bamford, medRxiv, 2023, preprint, DOI:10.1101/2023.08.08.23293833.
- E. Mercier, P. M. D'Aoust, O. Thakali, N. Hegazy, J.-J. Jia, Z. Zhang, W. Eid, J. Plaza-Diaz, M. P. Kabir, W. Fang, A. Cowan, S. E. Stephenson, L. Pisharody, A. E. MacKenzie, T. E. Graber, S. Wan and R. Delatolla, Municipal and neighbourhood level wastewater surveillance and subtyping of an influenza virus outbreak, Sci. Rep., 2022, 12, 15777 CrossRef CAS PubMed.
- M. K. Wolfe, D. Duong, K. M. Bakker, M. Ammerman, L. Mortenson, B. Hughes, P. Arts, A. S. Lauring, W. J. Fitzsimmons, E. Bendall, C. E. Hwang, E. T. Martin, B. J. White, A. B. Boehm and K. R. Wigginton, Wastewater-Based Detection of Two Influenza Outbreaks, Environ. Sci. Technol. Lett., 2022, 9(8), 687–692 CrossRef CAS.
- A. Zafeiriadou, L. Kaltsis, M. Kostakis, V. Kapes, N. S. Thomaidis and A. Markou, Wastewater surveillance of the most common circulating respiratory viruses in Athens: The impact of COVID-19 on their seasonality, Sci. Total Environ., 2023, 900, 166136 CrossRef CAS PubMed.
- V. Castro-Gutierrez, F. Hassard, M. Vu, R. Leitao, B. Burczynska, D. Wildeboer, I. Stanton, S. Rahimzadeh, G. Baio, H. Garelick, J. Hofman, B. Kasprzyk-Hordern, R. Kwiatkowska, A. Majeed, S. Priest, J. Grimsley, L. Lundy, A. C. Singer and M. Di Cesare, Monitoring occurrence of SARS-CoV-2 in school populations: A wastewater-based approach, PLoS One, 2022, 17, e0270168 CrossRef CAS PubMed.
- L. M. Colosi, K. E. Barry, S. M. Kotay, M. D. Porter, M. D. Poulter, C. Ratliff, W. Simmons, L. I. Steinberg, D. D. Wilson, R. Morse, P. Zmick and A. J. Mathers, Development of wastewater pooled surveillance of SARS-CoV-2 from congregate living settings, medRxiv, 2020, preprint, DOI:10.1101/2020.10.10.20210484.
- R. Fielding-Miller, S. Karthikeyan, T. Gaines, R. S. Garfein, R. A. Salido, V. J. Cantu, L. Kohn, N. K. Martin, A. Wynn, C. Wijaya, M. Flores, V. Omaleki, A. Majnoonian, P. Gonzalez-Zuniga, M. Nguyen, A. V. Vo, T. Le, D. Duong, A. Hassani, S. Tweeten, K. Jepsen, B. Henson, A. Hakim, A. Birmingham, P. D. Hoff, A. M. Mark, C. A. Nasamran, S. B. Rosenthal, N. Moshiri, K. M. Fisch, G. Humphrey, S. Farmer, H. M. Tubb, T. Valles, J. Morris, J. Kang, B. Khaleghi, C. Young, A. D. Akel, S. Eilert, J. Eno, K. Curewitz, L. C. Laurent, T. Rosing, R. Knight, N. A. Baer, T. Barber, A. Castro-Martinez, M. Chacón, W. Cheung, E. S. Crescini, E. R. Eisner, L. F. Vargas, A. Hakim, C. Hobbs, A. L. Lastrella, E. S. Lawrence, N. L. Matteson, K. Gangavarapu, T. T. Ngo, P. Seaver, E. W. Smoot, R. Tsai, B. Xia, S. Aigner, C. Anderson, P. Belda-Ferre, S. Sathe, M. Zeller, K. G. Andersen, G. W. Yeo and E. Kurzban, Safer at school early alert: an observational study of wastewater and surface monitoring to detect COVID-19 in elementary schools, Lancet Reg. Health Am., 2023, 19, 100449 Search PubMed.
- C. Gibas, K. Lambirth, N. Mittal, M. A. I. Juel, V. B. Barua, L. Roppolo Brazell, K. Hinton, J. Lontai, N. Stark, I. Young, C. Quach, M. Russ, J. Kauer, B. Nicolosi, D. Chen, S. Akella, W. Tang, J. Schlueter and M. Munir, Implementing building-level SARS-CoV-2 wastewater surveillance on a university campus, Sci. Total Environ., 2021, 782, 146749 CrossRef CAS.
- W. Johnson, K. Reeves, J. Liebig, A. Feula, C. Butler, M. Alkire, S. Singh, S. Litton, K. O'Conor, K. Jones, N. Ortega, T. Shimek, J. Witteman, Sampling Team Collaborators, K. K. Bjorkman and C. Mansfeldt, Effectiveness of building-level sewage surveillance during both community-spread and sporadic-infection phases of SARS-CoV-2 in a university campus population, FEMS Microbes, 2022, 3, xtac024 CrossRef.
- A. Pico-Tomàs, C. Mejías-Molina, I. Zammit, M. Rusiñol, S. Bofill-Mas, C. M. Borrego and L. Corominas, Surveillance of SARS-CoV-2 in sewage from buildings housing residents with different vulnerability levels, Sci. Total Environ., 2023, 162116 CrossRef PubMed.
- L. C. Scott, A. Aubee, L. Babahaji, K. Vigil, S. Tims and T. G. Aw, Targeted wastewater surveillance of SARS-CoV-2 on a university campus for COVID-19 outbreak detection and mitigation, Environ. Res., 2021, 200, 111374 CrossRef CAS PubMed.
- R. R. Spurbeck, A. Minard-Smith and L. Catlin, Feasibility of neighborhood and building scale wastewater-based genomic epidemiology for pathogen surveillance, Sci. Total Environ., 2021, 789, 147829 CrossRef CAS PubMed.
- M. Wolken, T. Sun, C. McCall, R. Schneider, K. Caton, C. Hundley, L. Hopkins, K. Ensor, K. Domakonda, P. Kalvapalle, D. Persse, S. Williams and L. B. Stadler, Wastewater surveillance of SARS-CoV-2 and influenza in preK-12 schools shows school, community, and citywide infections, Water Res., 2023, 231, 119648 CrossRef CAS.
- L. Haak, B. Delic, L. Li, T. Guarin, L. Mazurowski, N. G. Dastjerdi, A. Dewan and K. Pagilla, Spatial and temporal variability and data bias in wastewater surveillance of SARS-CoV-2 in a sewer system, Sci. Total Environ., 2022, 805, 150390 CrossRef CAS PubMed.
- R. H. Holm, A. Mukherjee, J. P. Rai, R. A. Yeager, D. Talley, S. N. Rai, A. Bhatnagar and T. Smith, SARS-CoV-2 RNA abundance in wastewater as a function of distinct urban sewershed size, Environ. Sci.: Water Res. Technol., 2022, 8, 807–819 RSC.
- Y. Wang, P. Liu, J. VanTassell, S. P. Hilton, L. Guo, O. Sablon, M. Wolfe, L. Freeman, W. Rose, C. Holt, M. Browning, M. Bryan, L. Waller, P. F. M. Teunis and C. L. Moe, When case reporting becomes untenable: Can sewer networks tell us where COVID-19 transmission occurs?, Water Res., 2023, 229, 119516 CrossRef CAS.
- F. Hassard, S. Singh, F. Coulon and Z. Yang, Can wastewater monitoring protect public health in schools?, Lancet Reg. Health Am., 2023, 20, 100475 Search PubMed.
- C. Jackson, P. Mangtani, J. Hawker, B. Olowokure and E. Vynnycky, The Effects of School Closures on Influenza Outbreaks and Pandemics: Systematic Review of Simulation Studies, PLoS One, 2014, 9, e97297 CrossRef.
- A. F. Murray, A. Emanuels, C. Wolf, N. Franko, L. Starita, J. A. Englund and H. Y. Chu, School-Based Surveillance of Respiratory Pathogens on “High-Touch” Surfaces, Front. Pediatr., 2021, 9, 686386 CrossRef.
- J. Crowe, A. T. Schnaubelt, S. SchmidtBonne, K. Angell, J. Bai, T. Eske, M. Nicklin, C. Pratt, B. White, B. Crotts-Hannibal, N. Staffend, V. Herrera, J. Cobb, J. Conner, J. Carstens, J. Tempero, L. Bouda, M. Ray, J. V. Lawler, W. S. Campbell, J.-M. Lowe, J. Santarpia, S. Bartelt-Hunt, M. Wiley, D. Brett-Major, C. Logan and M. J. Broadhurst, Assessment of a Program for SARS-CoV-2 Screening and Environmental Monitoring in an Urban Public School District, JAMA Netw. Open, 2021, 4, e2126447 CrossRef PubMed.
- B. A. Petros, J. S. Paull, C. H. Tomkins-Tinch, B. C. Loftness, K. C. DeRuff, P. Nair, G. L. Gionet, A. Benz, T. Brock-Fisher, M. Hughes, L. Yurkovetskiy, S. Mulaudzi, E. Leenerman, T. Nyalile, G. K. Moreno, I. Specht, K. Sani, G. Adams, S. V. Babet, E. Baron, J. T. Blank, C. Boehm, Y. Botti-Lodovico, J. Brown, A. R. Buisker, T. Burcham, L. Chylek, P. Cronan, A. Dauphin, V. Desreumaux, M. Doss, B. Flynn, A. Gladden-Young, O. Glennon, H. D. Harmon, T. V. Hook, A. Kary, C. King, C. Loreth, L. Marrs, K. J. McQuade, T. T. Milton, J. M. Mulford, K. Oba, L. Pearlman, M. Schifferli, M. J. Schmidt, G. M. Tandus, A. Tyler, M. E. Vodzak, K. K. Bevill, A. Colubri, B. L. MacInnis, A. Z. Ozsoy, E. Parrie, K. Sholtes, K. J. Siddle, B. Fry, J. Luban, D. J. Park, J. Marshall, A. Bronson, S. F. Schaffner and P. C. Sabeti, Multimodal surveillance of SARS-CoV-2 at a university enables development of a robust outbreak response framework, Med, 2022, 3, 883–900.e13 CrossRef.
- V. Vo, A. Harrington, C.-L. Chang, H. Baker, M. A. Moshi, N. Ghani, J. Y. Itorralba, R. L. Tillett, E. Dahlmann, N. Basazinew, R. Gu, T. D. Familara, S. Boss, F. Vanderford, M. Ghani, A. J. Tang, A. Matthews, K. Papp, E. Khan, C. Koutras, H.-Y. Kan, C. Lockett, D. Gerrity and E. C. Oh, Identification and genome sequencing of an influenza H3N2 variant in wastewater from elementary schools during a surge of influenza A cases in Las Vegas, Nevada, Sci. Total Environ., 2023, 872, 162058 CrossRef CAS PubMed.
- W. Ahmed, S. L. Simpson, P. M. Bertsch, K. Bibby, A. Bivins, L. L. Blackall, S. Bofill-Mas, A. Bosch, J. Brandão, P. M. Choi, M. Ciesielski, E. Donner, N. D'Souza, A. H. Farnleitner, D. Gerrity, R. Gonzalez, J. F. Griffith, P. Gyawali, C. N. Haas, K. A. Hamilton, H. C. Hapuarachchi, V. J. Harwood, R. Haque, G. Jackson, S. J. Khan, W. Khan, M. Kitajima, A. Korajkic, G. La Rosa, B. A. Layton, E. Lipp, S. L. McLellan, B. McMinn, G. Medema, S. Metcalfe, W. G. Meijer, J. F. Mueller, H. Murphy, C. C. Naughton, R. T. Noble, S. Payyappat, S. Petterson, T. Pitkänen, V. B. Rajal, B. Reyneke, F. A. Roman, J. B. Rose, M. Rusiñol, M. J. Sadowsky, L. Sala-Comorera, Y. X. Setoh, S. P. Sherchan, K. Sirikanchana, W. Smith, J. A. Steele, R. Sabburg, E. M. Symonds, P. Thai, K. V. Thomas, J. Tynan, S. Toze, J. Thompson, A. S. Whiteley, J. C. C. Wong, D. Sano, S. Wuertz, I. Xagoraraki, Q. Zhang, A. G. Zimmer-Faust and O. C. Shanks, Minimizing errors in RT-PCR detection and quantification of SARS-CoV-2 RNA for wastewater surveillance, Sci. Total Environ., 2022, 805, 149877 CrossRef CAS PubMed.
- M. Asadi, D. Hamilton, C. Shomachuk, F. F. Oloye, C. De Lange, X. Pu, C. A. Osunla, J. Cantin, S. El-Baroudy, E. M. Mejia, B. Gregorchuk, M. G. Becker, C. Mangat, M. Brinkmann, P. D. Jones, J. P. Giesy and K. N. McPhedran, Assessment of rapid wastewater surveillance for determination of communicable disease spread in municipalities, Sci. Total Environ., 2023, 901, 166541 CrossRef CAS PubMed.
- J. Daigle, K. Racher, J. Hazenberg, A. Yeoman, H. Hannah, D. Duong, U. Mohammed, D. Spreitzer, B. S. J. Gregorchuk, B. M. Head, A. F. A. Meyers, P. A. Sandstrom, A. Nichani, J. I. Brooks, M. R. Mulvey, C. S. Mangat and M. G. Becker, A Sensitive and Rapid Wastewater Test for SARS-COV-2 and Its Use for the Early Detection of a Cluster of Cases in a Remote Community, Appl. Environ. Microbiol., 2022, 88, e01740-21 CrossRef PubMed.
- Z. W. Laturner, D. M. Zong, P. Kalvapalle, K. R. Gamas, A. Terwilliger, T. Crosby, P. Ali, V. Avadhanula, H. H. Santos, K. Weesner, L. Hopkins, P. A. Piedra, A. W. Maresso and L. B. Stadler, Evaluating recovery, cost, and throughput of different concentration methods for SARS-CoV-2 wastewater-based epidemiology, Water Res., 2021, 197, 117043 CrossRef CAS.
- E. G. Lou, N. Sapoval, C. McCall, L. Bauhs, R. Carlson-Stadler, P. Kalvapalle, Y. Lai, K. Palmer, R. Penn, W. Rich, M. Wolken, P. Brown, K. B. Ensor, L. Hopkins, T. J. Treangen and L. B. Stadler, Direct comparison of RT-ddPCR and targeted amplicon sequencing for SARS-CoV-2 mutation monitoring in wastewater, Sci. Total Environ., 2022, 833, 155059 CrossRef CAS.
- M. G. Becker, T. Taylor, S. Kiazyk, D. R. Cabiles, A. F. A. Meyers and P. A. Sandstrom, Recommendations for sample pooling on the Cepheid GeneXpert® system using the Cepheid Xpert® Xpress SARS-CoV-2 assay, PLoS One, 2020, 15(11), e0241959 CrossRef CAS.
- M. A. Borchardt, A. B. Boehm, M. Salit, S. K. Spencer, K. R. Wigginton and R. T. Noble, The Environmental Microbiology Minimum Information (EMMI) Guidelines: qPCR and dPCR Quality and Reporting for Environmental Microbiology, Environ. Sci. Technol., 2021, 55, 10210–10223 CrossRef CAS PubMed.
- Department of the Treasury, Depreciation of Scientific Instruments, Department of the Treasury, 1990 Search PubMed.
- Y. Cao, J. F. Griffith, S. Dorevitch and S. B. Weisberg, Effectiveness of qPCR permutations, internal controls and dilution as means for minimizing the impact of inhibition while measuring Enterococcus in environmental waters, J. Appl. Microbiol., 2012, 113, 66–75 CrossRef CAS.
- C. Schrader, A. Schielke, L. Ellerbroek and R. Johne, PCR inhibitors – occurrence, properties and removal, J. Appl. Microbiol., 2012, 113, 1014–1026 CrossRef CAS PubMed.
- M. Ciesielski, D. Blackwood, T. Clerkin, R. Gonzalez, H. Thompson, A. Larson and R. Noble, Assessing sensitivity and reproducibility of RT-ddPCR and RT-qPCR for the quantification of SARS-CoV-2 in wastewater, J. Virol. Methods, 2021, 297, 114230 CrossRef CAS.
- S. Adhikari and R. U. Halden, Opportunities and limits of wastewater-based epidemiology for tracking global health and attainment of UN sustainable development goals, Environ. Int., 2022, 163, 107217 CrossRef CAS PubMed.
- P. Gahlot, K. D. Alley, S. Arora, S. Das, A. Nag and V. K. Tyagi, Wastewater surveillance could serve as a pandemic early warning system for COVID-19 and beyond, Wiley Interdiscip. Rev.: Water, 2023, 10, e1650 CrossRef.
- I. V. Bassett, L. S. Forman, S. Govere, H. Thulare, S. C. Frank, B. Mhlongo and E. Losina, Test and Treat TB: a pilot trial of GeneXpert MTB/RIF screening on a mobile HIV testing unit in South Africa, BMC Infect. Dis., 2019, 19, 110 CrossRef PubMed.
- H. A. Cubie, D. Morton, E. Kawonga, M. Mautanga, I. Mwenitete, N. Teakle, B. Ngwira, H. Walker, G. Walker, S. Kafwafwa, B. Kabota, R. Ter Haar and C. Campbell, HPV prevalence in women attending cervical screening in rural Malawi using the cartridge-based Xpert® HPV assay, J. Clin. Virol., 2017, 87, 1–4 CrossRef PubMed.
- M. B. Fitzpatrick, R. S. Dube Mandishora, D. A. Katzenstein, K. McCarty, J. Weber, M. K. Sahoo, J. Manasa, Z. M. Chirenje and B. A. Pinsky, hrHPV prevalence and type distribution in rural Zimbabwe: A community-based self-collection study using near-point-of-care GeneXpert HPV testing, Int. J. Infect. Dis., 2019, 82, 21–29 CrossRef PubMed.
- V. Mwenda, J.-P. Bor, M. Nyangasi, J. Njeru, S. Olwande, P. Njiri, M. Arbyn, S. Weyers, P. Tummers and M. Temmerman, Integrating human papillomavirus testing as a point-of care service using GeneXpert platforms: Findings and lessons from a Kenyan pilot study (2019–2020), PLoS One, 2023, 18, e0286202 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d4ew00526k |
| This journal is © The Royal Society of Chemistry 2025 |