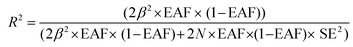Open Access Article
Open Access ArticleComment on “Circulating fatty acids and risk of severe non-alcoholic fatty liver disease in the UK biobank: a prospective cohort of 116![[thin space (1/6-em)]](https://www.rsc.org/images/entities/h2_char_2009.gif) 223 individuals” by P. Zhuang, Y. Ao, X. Liu, H. Ye, H. Li, X. Wan, Y. Zhang and J. Jiao, Food & Function, 2024, 15, 10527†
223 individuals” by P. Zhuang, Y. Ao, X. Liu, H. Ye, H. Li, X. Wan, Y. Zhang and J. Jiao, Food & Function, 2024, 15, 10527†
Zhitao
Chen
,
Yangjun
Gu
,
Shusen
Zheng
* and
Qiyong
Li
 *
*
Department of Hepatobiliary Surgery, Shulan (Hangzhou) Hospital Affiliated to Zhejiang Shuren University Shulan International Medical College, 848# Dongxin Road, Hangzhou, 310003, China. E-mail: shusenzheng@zju.edu.cn; liqiyong@zju.edu.cn
First published on 22nd April 2025
We read with great interest the recent article by Zhuang et al.,1 titled “Circulating fatty acids and risk of severe non-alcoholic fatty liver disease in the UK biobank: a prospective cohort of 116![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 223 individuals”. The authors conducted an impressive prospective study involving over 116
223 individuals”. The authors conducted an impressive prospective study involving over 116![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 participants to explore the relationship between circulating fatty acid (FA) profiles and the risk of severe non-alcoholic fatty liver disease (NAFLD). Although they adjusted for key confounders such as age, sex, ethnicity, and education, the observational nature of the study limits the ability to draw causal conclusions from the findings, as the authors themselves noted.
000 participants to explore the relationship between circulating fatty acid (FA) profiles and the risk of severe non-alcoholic fatty liver disease (NAFLD). Although they adjusted for key confounders such as age, sex, ethnicity, and education, the observational nature of the study limits the ability to draw causal conclusions from the findings, as the authors themselves noted.
Mendelian randomization (MR) analysis, using genetic variants as instrumental variables (IVs) for specific risk factors, is widely employed to clarify causal links between exposures and outcomes, including various human diseases.2 For our MR study on FAs and NAFLD, we sourced datasets from OpenGWAS (https://gwas.mrcieu.ac.uk/). The exposure dataset on FAs included 115![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 006 samples and 11
006 samples and 11![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 590
590![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 339 single nucleotide polymorphisms (SNPs), while the outcome dataset for NAFLD (ebi-a-GCST90091033) comprised 778
339 single nucleotide polymorphisms (SNPs), while the outcome dataset for NAFLD (ebi-a-GCST90091033) comprised 778![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 614 samples and 6
614 samples and 6![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 784
784![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 388 SNPs. The data sources used in the present MR study are summarized in Table S1.† Using the summary data from the aforementioned genome-wide association study (GWAS), we followed a rigorous protocol to identify eligible SNPs as IVs. First, SNPs had to show a strong association with the exposure, meeting the genome-wide significance threshold of p < 5 × 10−8. Second, linkage disequilibrium (LD) analysis was performed to select independent SNPs, defined by LD R2 < 0.001 and an LD distance of 10
388 SNPs. The data sources used in the present MR study are summarized in Table S1.† Using the summary data from the aforementioned genome-wide association study (GWAS), we followed a rigorous protocol to identify eligible SNPs as IVs. First, SNPs had to show a strong association with the exposure, meeting the genome-wide significance threshold of p < 5 × 10−8. Second, linkage disequilibrium (LD) analysis was performed to select independent SNPs, defined by LD R2 < 0.001 and an LD distance of 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 kb. Third, the strength of the IVs was evaluated using the F statistic, with a threshold of F > 10 deemed sufficient to minimize weak instrument bias. The strength of each IV was calculated using the following formulas:
000 kb. Third, the strength of the IVs was evaluated using the F statistic, with a threshold of F > 10 deemed sufficient to minimize weak instrument bias. The strength of each IV was calculated using the following formulas:
Here, EAF represents the effect allele frequency, β and SE denote the effect size and standard error of the SNP–antibody association, respectively, and N indicates the sample size. A total of 778 SNPs were selected as IVs for 15 distinct FA types and their composition ratios, following the established IV selection criteria (Table S2†). The inverse variance weighted (IVW) method, commonly used with summary statistics data, was the primary approach for assessing the potential causal effect of FAs on NAFLD risk, allowing for the calculation of causal effect sizes without the need for individual-level data.
In our recent MR study, we examined the causal relationship between FAs and NAFLD, with a focus on different fatty acid subtypes. Our results indicated that monounsaturated fatty acids (MUFAs) significantly increase the risk of developing NAFLD (OR = 1.232, 95% CI: 1.040 to 1.461, P = 0.016). In contrast, omega-6/total FA (OR = 0.776, 95% CI: 0.640 to 0.941, P = 0.009), polyunsaturated fatty acids (PUFAs)/total FA (OR = 0.722, 95% CI: 0.620 to 0.839, P < 0.001), and PUFA/MUFA ratios (OR = 0.795, 95% CI: 0.708 to 0.894, P < 0.001) demonstrated a protective effect against NAFLD (Fig. 1). Meanwhile, pleiotropy analysis indicated that our findings are not influenced by pleiotropy, further strengthening the reliability of the results (Table S3†). These findings align with and complement Zhuang et al.'s work, adding to the growing evidence of the complex role fatty acid subtypes play in liver disease development.
The difference between the present MR study, which found no significant link between circulating saturated fatty acids (SFAs) and NAFLD, and Zhuang et al.'s findings, which identified SFAs as a risk factor, can be explained by several factors: (1) Zhuang et al.'s observational cohort study is susceptible to confounding, reverse causation, and residual confounding, even with adjustments for various factors. In contrast, MR analysis uses genetic variants as IVs, reducing these biases and providing more reliable insights into causal relationships. (2) Zhuang et al. focused on severe NAFLD cases, identified through hospitalization or death records, while our study may have included a broader range of NAFLD cases, including milder forms. The association between SFAs and NAFLD could be stronger in more advanced cases, possibly explaining the differing results.3 (3) Zhuang et al. measured baseline plasma SFAs, which reflect short-term dietary intake or metabolic status at a single time point and can fluctuate due to changes in diet, lifestyle, or metabolism. In contrast, our MR study used genetic variants related to lifelong FA metabolism, offering a more consistent long-term exposure measure, which may account for the absence of a significant association with NAFLD in our results.
In conclusion, we applaud Zhuang et al. for their valuable contribution to understanding the role of circulating fatty acids in NAFLD. We believe that integrating prospective cohort data with genetic approaches like Mendelian randomization can provide deeper insights into the prevention and management of NAFLD. Therefore, we encourage future studies to incorporate genetic data, particularly focusing on fatty acid metabolism genes such as FADS1/2, to further explore these associations.
Author contributions
All authors made substantial contributions to the conception and design, acquisition of data, and analysis and interpretation of data; took part in drafting the article and revising it critically for important intellectual content; agreed to submit the manuscript to the current journal; gave final approval of the version to be published; and agreed to be accountable for all aspects of the work.Abbreviations
| MR | Mendelian randomization |
| FAs | Fatty acids |
| NAFLD | Non-alcoholic fatty liver disease |
| IVs | Instrumental variables (IVs) |
| IVW | Inverse variance weighted |
| MUFAs | Monounsaturated fatty acids |
| PUFAs | Polyunsaturated fatty acids |
| SFAs | Saturated fatty acids |
Data availability
The datasets used and analyzed in the present study are available from the corresponding authors upon reasonable request. The datasets generated and/or analyzed during the current study are available in the GWAS (https://gwas.mrcieu.ac.uk/) database.Conflicts of interest
The authors declare no competing interests.References
- P. Zhuang, Y. Ao, X. Liu, H. Ye, H. Li, X. Wan, Y. Zhang and J. Jiao, Circulating fatty acids and risk of severe non-alcoholic fatty liver disease in the UK biobank: a prospective cohort of 116 223 individuals, Food Funct., 2024, 15(20), 10527–10538, 10.1039/d4fo01182a.
- P. Sekula, M. F. Del Greco, C. Pattaro and A. Köttgen, Mendelian Randomization as an Approach to Assess Causality Using Observational Data, J. Am. Soc. Nephrol., 2016, 27(11), 3253–3265, DOI:10.1681/asn.2016010098.
- A. K. Leamy, R. A. Egnatchik and J. D. Young, Molecular mechanisms and the role of saturated fatty acids in the progression of non-alcoholic fatty liver disease, Prog. Lipid Res., 2013, 52(1), 165–174, DOI:10.1016/j.plipres.2012.10.004.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d4fo05002a |
| This journal is © The Royal Society of Chemistry 2025 |