Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Impact of mucin protein corona on the gastrointestinal behavior and antioxidant activity of food carbon dots extracted from bread crust†
Shichao Mia,
Zimo Liua,
Mingcheng Wanga,
David Julian McClements c,
Chongjiang Cao
c,
Chongjiang Cao a,
Xiao Xu
a,
Xiao Xu *b and
Biao Yuan
*b and
Biao Yuan *a
*a
aDepartment of Food Quality and Safety, National R&D Center for Chinese Herbal Medicine Processing, College of Engineering, China Pharmaceutical University, Nanjing, Jiangsu 211198, China. E-mail: yuanbiao01@gmail.com; yuanbiao@cpu.edu.cn
bSchool of Life Science, Shaoxing University, Shaoxing, Zhejiang 312000, China. E-mail: xiaolexu@126.com
cDepartment of Food Science, University of Massachusetts, Amherst, MA 01003, USA
First published on 7th May 2025
Abstract
Bread products are widely consumed around the globe as they are a staple food in many countries. Recent studies have shown that the elevated temperatures and prolonged times used during bread baking can lead to the formation of appreciable amounts of carbon dots, which may have adverse effects on human health. Consequently, it is important to elucidate the potential fate of these carbon dots within the gastrointestinal tract. In this study, we characterized the properties of bread-derived carbon dots (BDCDs) and then investigated their interactions with mucin. The average diameter of the individual BDCDs was 5.81 nm but they tended to exist as clusters (around 91 nm) in aqueous environments. The BDCDs mainly consisted of carbon (70.9%), oxygen (27.6%), and nitrogen (1.5%). Like other types of carbon dots, the BDCDs were found to exhibit natural fluorescence. In simulated saliva, the carbon dots interacted with mucin to form BDCD–mucin complexes. These interactions caused the zeta potential of the BDCDs to change from −5.97 to −11.07 mV and their hydrodynamic diameters to increase from 91.3 to 122.4 nm. Moreover, these interactions caused changes in the conformation of the mucin. Furthermore, in vitro simulated digestion studies showed that the BDCDs induced conformational changes in pepsin, reducing its enzymatic activity by around 20.8%. However, this effect was mitigated when BDCD–mucin complexes were used instead of BDCDs, highlighting the important role that mucin plays in mediating carbon dot–enzyme interactions. Moreover, the BDCD–mucin complexes were better at attenuating reactive oxygen species generation in Caco-2 cells than BDCDs, which led to higher cell viability. This study provides new insights into the role of mucin coronas in modulating the gastrointestinal behavior of carbon dots, highlighting their potential impact on human health.
1. Introduction
Carbon dots are a class of carbon-based nanoparticles, ranging in size from 4 to 30 nm, that are widely used in many fields due to their high fluorescence, photostability, and solubility.1,2 The preparation of carbon dots for various applications has been a prominent research area for many years. Recently, however, particular attention has been paid to the presence of carbon dots in different food sources, especially regarding their potential health risks.3 Carbon dots have been reported to form during the thermal processing of some foods, especially those that are held at high temperatures for prolonged times, which is the case for products such as bread, hot beverages, and roasted meats.4–6 The high temperatures (175–190 °C) and extended times (20–60 min) used during baking provide optimal conditions for the formation of carbon dots in bread crust, particularly darker colored ones. The carbon dots in bread crust may therefore be readily ingested as part of the daily human diet. Given the widespread presence of carbon dots in bread crust, their potential effects in vivo are non-negligible. Previous studies have shown that carbon dots derived from roasted squid can accumulate in various organs of mice in a time-dependent manner after ingestion, including crossing the blood–brain barrier and entering the brain.7 Furthermore, research has shown that these carbon dots can induce cell membrane damage, cell cycle arrest, mitochondrial dysfunction, and ROS production in cells.8 Consequently, it is important to better understand the potential gastrointestinal fate of carbon dots from bread.Mucin is a highly glycosylated protein that forms a porous gel-like coating around the walls of the gastrointestinal tract, and is present in the gastrointestinal fluids.9 It is secreted by goblet cells to form a protective mucus layer, serving as a barrier that protects intestinal epithelial cells from mechanical and physical damage while allowing nutrient absorption.10 Previous studies have shown that mucin can also adsorb onto the surfaces of nanoparticles to form a coating around them, which is often referred to as the ‘protein corona’ (even though it also contains other constituents), which alters their gastrointestinal fate.11 Zhou et al. reported that the formation of a mucin corona around TiO2 nanoparticles in simulated salivary fluids impacted the dispersion state of TiO2 nanoparticles.12 This change in the oral dispersion state influences their subsequent behavior in the lower regions of the gastrointestinal tract. Other researchers reported that the formation of a mucin corona around transferrin-modified nanoparticles enhanced their endocytosis and transcellular transport.13 In our previous study, we showed that the formation of a whey protein corona around TiO2 nanoparticles altered their aggregation state and surface charge under simulated gastrointestinal conditions.14,15 Despite these studies, there is still a relatively poor understanding of the impact of corona formation on the gastrointestinal fate of different kinds of nanoparticles.16 In this study, we therefore examined the impact of mucin corona formation on the behavior of carbon dots under simulated gastrointestinal conditions.17,18
Previously, it has been shown that nanoparticles can interact with digestive enzymes in the gastrointestinal tract, which may impact their activity. For instance, Song et al. reported that food-derived carbon dots could interact with pepsin to form protein coronas, which decreased the activity of pepsin in simulated gastric fluids.5 These alterations in digestive enzyme activity after interacting with the nanoparticles may be due to changes in the secondary structure of the enzymes after they adsorb onto the particle surfaces.19 Carbon dots with different functional groups have been reported to affect digestive enzyme activity to varying degrees due to the structural modifications induced by these functional groups.20 Interestingly, some researchers have found that interactions between nanoparticles and digestive enzymes actually enhance their activities.21 For instance, the formation of a protein corona consisting of α-amylase and amylopsin around quercetin-protein nanoparticles increased the enzyme activity in simulated salivary and intestinal fluids, respectively, which was attributed to alterations in the secondary structure of these digestive enzymes.22 In contrast, it has been reported that the formation of a pepsin corona around chitin nanowhiskers caused little change in the activity of pepsin, despite variations in its secondary structure.23 Nevertheless, previous studies have overlooked the presence of mucin in the gastrointestinal tract, which may impact the effects of carbon dots on digestive enzymes.
In the present study, bread-derived carbon dots (BDCDs) were first characterized, and then their interactions with mucin were investigated. The impact of the BDCD–mucin interactions on the secondary structure and optical properties of the mucin was also studied. An in vitro simulated digestion model was then used to explore the impact of the mucin corona on the behavior of the BDCDs in the gastrointestinal tract. The impact of BDCDs and BDCD–mucin complexes on the activity of digestive enzymes, particularly pepsin, was also determined, so as to elucidate the role of the mucin. The antioxidant activity of the BDCD–mucin complexes during simulated digestion was studied using radical scavenging assays, as well as a reducing power assay. Furthermore, the effects of the BDCD–mucin complexes on cell viability and reactive oxygen species levels were evaluated using a cell culture model. Overall, our findings reveal that mucin in the gastrointestinal tract modulates the behavior and antioxidant activity of BDCDs through the formation of a corona around them, thereby providing a novel perspective for assessing their safety and functionality.
2. Materials and methods
2.1. Materials
The bread crust used in this study was obtained from commercially available toast bread, whose primary ingredients included wheat flour, white granulated sugar, vegetable oil, egg pulp, and milk powder (Shenyang Taoli Bread Co., Ltd, Taoli, Shenyang, China). Dialysis bags with a molecular weight cut-off of 3500 Da and 500 Da were obtained from Viskase (Chicago, USA). Mucin (≥95%), α-amylase (≥95%, from Bacillus licheniformis, exhibits catalytic activity comparable to human salivary α-amylase, albeit with physiologically relevant limitations), pepsin (EC 3.4.23.1, ≥95%, from porcine gastric mucosa), trypsin (EC 3.4.21.4, ≥95%, from porcine pancreas), and iron(II) sulfate heptahydrate (FeSO4·7H2O, ≥99.7%) were obtained from Shanghai Macklin Biochemical Technology Co., Ltd (Shanghai, China). Bile salt (≥95%), benzoyl-L-arginine ethyl ester (C15H22N4O3·HCl, 98%), potassium persulfate (K2S2O8, ≥99.5%), and potassium ferrocyanide trihydrate (K4FeC6N6·3H2O, 98%) were purchased from Aladdin Chemical Reagent Co., Ltd (Shanghai, China). Methanol (CH4O, ≥99.7%) and salicylic acid (C7H6O3, ≥99.7%) was purchased from General Reagent Co., Ltd (Shanghai, China). 3-(4,5-Dimethyl-2-thiazolyl)-2,5-diphenyl-2-H-tetrazolium bromide (MTT, 98%) and a ROS assay kit were purchased from Beyotime Biotechnology (Shanghai, China). Bovine hemoglobin (≥95%) and ABTS (C18H24N6O6S4, 98%) were purchased from Yuanye Biotechnology Co., Ltd (Shanghai, China). Trichloroacetic acid (C2HCl3O2, ≥99.7%) and ferric chloride (FeCl3·6H2O, ≥99.7%) were purchased from Shanghai Hushi Laboratory Equipment Co., Ltd (Shanghai, China). Hydrogen peroxide (H2O2, 30%) was purchased from Nanjing Chemical Reagent Co., Ltd (Nanjing, China). All other chemicals used in this study were of analytical grade.2.2. Isolation of BDCDs
The BDCDs were extracted using a method described in a previous study, with slight modifications.24 Briefly, the bread crust (530 g, wet weight) was first dried in an oven at 60 °C for 2 h, and then the toasted crust edges were collected and ground into a powder (142 g). Next, the bread crust powder was mixed with methanol in a 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 20 ratio and sonicated at 40 kHz for 1 h. The mixture was then centrifuged at 5000g for 30 min, and the resulting supernatant was concentrated to 1/20 of its original volume using vacuum rotary evaporation. After concentration, the concentrated solution was incubated at 4 °C to allow any insoluble matter to sediment, and then the supernatant was collected for further use. The supernatant was dialyzed in a 3500 Da dialysis bag for 48 h, and the outer dialysis solution was collected for further concentration. To remove small water-soluble molecules, the concentrated outer dialysis solution was dialyzed in a 500 Da dialysis bag for 24 h and then filtered through a 0.22 μm filter. The filtrate was freeze-dried to afford 266 mg of BDCDs (yield was calculated based on the wet weight of bread crust). The obtained BDCDs were then stored in a glass desiccator to prevent moisture absorption for further study.
20 ratio and sonicated at 40 kHz for 1 h. The mixture was then centrifuged at 5000g for 30 min, and the resulting supernatant was concentrated to 1/20 of its original volume using vacuum rotary evaporation. After concentration, the concentrated solution was incubated at 4 °C to allow any insoluble matter to sediment, and then the supernatant was collected for further use. The supernatant was dialyzed in a 3500 Da dialysis bag for 48 h, and the outer dialysis solution was collected for further concentration. To remove small water-soluble molecules, the concentrated outer dialysis solution was dialyzed in a 500 Da dialysis bag for 24 h and then filtered through a 0.22 μm filter. The filtrate was freeze-dried to afford 266 mg of BDCDs (yield was calculated based on the wet weight of bread crust). The obtained BDCDs were then stored in a glass desiccator to prevent moisture absorption for further study.
2.3. Characterization of BDCDs
The characterization of the BDCDs was performed based on the methods described in detail in a previous study.5 Briefly, the morphology of the BDCDs was characterized using transmission electron microscopy (TEM) (Thermo Fisher Scientific, Waltham, USA). The chemical composition of the BDCDs was characterized using an X-ray photoelectron spectrometer (XPS) (K-Alpha, Thermo Fisher Scientific, Waltham, USA). Their crystalline structure was recorded using an X-ray diffraction (XRD) instrument (D8 Advanced, Bruker, Karlsruhe, Germany). The fluorescence spectra were determined using a spectrofluorometer (F-7000, Hitachi, Tokyo, Japan). The ultraviolet-visible spectra of BDCD suspensions were recorded using a UV-visible spectrophotometer (Multiskan GO, Thermo Fisher Scientific, Waltham, USA).2.4. BDCD–mucin protein corona formation
BDCD–mucin complexes were prepared according to a method described previously.25 Briefly, BDCDs and mucin were first mixed together in equal volumes, each at the same concentration, and then the resulting mixture was incubated overnight at 37 °C.2.5. Scanning electron microscopy
A scanning electron microscope was used to characterize the morphology of the BDCDs, mucin, and BDCD–mucin complexes.26 These experiments were conducted using a field emission scanning electron microscope (Sigma 360, Zeiss, Oberkochen, Germany). The freeze-dried samples were loaded on conductive tapes and gold-coated prior to SEM observation.2.6. Zeta potential and particle size analysis
The zeta potentials and particle size distributions of the BDCDs, mucin, and BDCD–mucin complexes were measured using a combined particle electrophoresis/dynamic light scattering instrument (Zetasizer Nano, Malvern Instruments, Malvern, Worcestershire, UK).26 All the samples were vortex-dispersed evenly before analysis.2.7. Fourier transform infrared spectroscopy
Freeze-dried BDCDs, mucin, and BDCD–mucin complexes were ground into fine powders. Air was used as a background. The Fourier transform infrared spectra were obtained using a FTIR spectrometer (Nicolet iS50, Thermo Fisher Scientific, Waltham, USA) in the frequency range of 4000 to 400 cm−1.262.8. Circular dichroism spectroscopy
The circular dichroism spectra of mucin and BDCD–mucin complexes were measured using a CD spectropolarimeter (JASCO J-810, JASCO Corporation, Tokyo, Japan), as previously described.27 The samples were prepared by mixing 0.5 mg mL−1 mucin with concentrations of BDCDs ranging from 0.5 to 2.5 mg mL−1, and then the resulting mixtures were incubated for 24 h. The Beta Structure Selection method was used to calculate the secondary structure of the mucin from the spectra.282.9. Ultraviolet–visible spectroscopy
The ultraviolet-visible spectra of BDCDs, mucin, and BDCD–mucin complexes were measured using a UV-visible spectrophotometer (Multiskan GO, Thermo Fisher Scientific, Waltham, USA).29 The concentrations of samples used in this experiment were 0.1 mg mL−1.2.10. Fluorescence spectroscopy
A spectrofluorometer (F-7000, Hitachi, Tokyo, Japan) and microplate reader (SpectraMax M2, Molecular Devices, Sunnyvale, USA) were used to analyze the interactions between BDCDs and mucin, and between BDCD–mucin complexes and digestive enzymes, respectively. To test the interactions between BDCDs and mucin, an excitation wavelength of 280 nm, an emission wavelength range of 300–500 nm, excitation and emission slit widths of 5 nm, and a scanning rate of 1200 nm min−1 were used. To test the interaction between the BDCD–mucin complexes and digestive enzymes, the experiments were conducted by keeping pepsin (2000 U mL−1) and trypsin (100 U mL−1) concentrations constant, while increasing the sample concentrations from 0.5 to 2.5 mg mL−1. The excitation wavelength was set to 280 nm, with an emission wavelength range of 360–500 nm.30 For the detection of three-dimensional fluorescence spectroscopy, the ranges of excitation wavelength and emission wavelength were 210–390 nm and 250–550 nm, respectively. The test was conducted at room temperature.2.11. In vitro simulated digestion
Simulated digestion was performed as previously described.31 All experiments were conducted at 37 °C to simulate the conditions in the gastrointestinal tract. The samples subjected to in vitro simulated digestion were isolated carbon dots of bread crust (BDCDs), not the bread crust. Four sample groups were prepared: control, BDCDs (10 mg mL−1), mucin (10 mg mL−1), and BDCD–mucin complexes (10 mg mL−1). Specifically, simulated salivary fluid (15 mL) containing 150 U mL−1 α-amylase was mixed with 15 mL of the sample and then incubated for 2 min at pH 7. The concentration of BDCDs in the simulated salivary stage was derived from physiological modeling of oral processing for bread crust, accounting for salivary secretion rates and established extraction yields. Subsequently, the resulting mixture (15 mL) was combined with 15 mL of simulated gastric fluid containing 4000 U mL−1 pepsin and incubated for 2 hours at pH 3, with the pH being monitored and adjusted every 30 min to maintain stability.The mixture (15 mL) was then added to 15 mL of simulated intestinal fluid containing 200 U mL−1 trypsin and incubated for 2 hours at pH 7, with the pH monitored and adjusted every 30 min to ensure consistency. After digestion, the digestive enzymes were inactivated by placing the samples in a water bath at 100 °C for 10 min. Simulated digestive fluids without enzymes were used as a negative control.
2.12. Pepsin activity assay
The pepsin activity was quantified as described previously, with slight modifications.32 Initially, a 0.1 mL aliquot of simulated gastric fluid was added into 0.5 mL of 2% (w/v) bovine hemoglobin solution (pH 2) and incubated at 37 °C for 30 min. Then, the mixture was combined with 1 mL of 5% (w/v) trichloroacetic acid and centrifuged at 12![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000g for 10 min at 4 °C using a centrifuge (FC5515R, Ohaus, New Jersey, USA). The absorbance of the supernatant at 280 nm was measured.
000g for 10 min at 4 °C using a centrifuge (FC5515R, Ohaus, New Jersey, USA). The absorbance of the supernatant at 280 nm was measured.
2.13. Trypsin activity assay
The trypsin activity assay was performed as previously described, with slight modifications.33 The simulated intestinal fluid was diluted 10-fold and then added to the benzoyl-L-arginine ethyl ester solution (3.1 mL, 0.25 mM). The absorbance of the mixture at 253 nm was then recorded every minute for a duration of 5 min.2.14. ABTS radical scavenging assay
The ABTS stock solution was prepared by mixing 10 mL of 7 mM ABTS solution with potassium persulfate solution (178 μL, 140 mM) and incubating in the dark overnight.14 The stock solution was then diluted with dH2O to achieve an absorbance of 0.7 at 734 nm for further use. The absorbance was measured at 734 nm after incubating the ABTS reaction solution and digestive fluids at room temperature for 1 h.2.15. Hydroxyl radical scavenging assay
The hydroxyl radical scavenging assay was conducted as previously described, with a slight modification.14 Equal volumes (0.5 mL each) of digestive fluids, 6 mM FeSO4, 6 mM H2O2, and 6 mM salicylic acid–ethanol solution were mixed and incubated at 37 °C for 1 h in the dark. The absorbance was measured at 510 nm. Additionally, the volume of digestive fluid was optimized based on its hydroxyl radical scavenging activity.2.16. Total reducibility assay
The total reducibility assay was performed as described previously.14 Briefly, 200 μL of digestive fluids, 200 μL of phosphate buffer solution (200 mM, pH 6.6), and 200 μL of 1% (w/v) potassium ferricyanide were mixed and then incubated at 55 °C for 20 min. Then, 200 μL of 10% (w/v) trichloroacetic acid was added, and the mixture was centrifuged at 1000g for 15 min. Next, the supernatant (500 μL) was mixed with 500 μL of dH2O and 100 μL of 0.17% (w/v) ferric chloride, and incubated at room temperature for 10 min. The absorbance was then measured at 700 nm using a UV-visible spectrophotometer.2.17. Cell viability assay
Cell viability was assessed using the MTT (3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide) assay.34 Briefly, Caco-2 cells in a 96-well plate were treated with 200 μL of serum-free medium containing various concentrations (from 6.25 to 1600 μg mL−1) of BDCDs, mucin, and BDCD–mucin complexes for 24 h. After treatment, cells were incubated with MTT for 4 h, and the formazan crystals were dissolved in DMSO. The absorbance at 570 nm was measured using a microplate reader (Thermo Fisher Scientific, Waltham, USA).2.18. Reactive oxygen species measurement
The measurement of reactive oxygen species (ROS) was carried out as previously described.34 Caco-2 cells were seeded in a 12-well plate and treated with BDCDs, mucin, and BDCD–mucin complexes at concentrations of 100 and 400 μg mL−1 for 24 h. The concentration of BDCDs used in this experiment was calculated based on the mass of bread crust consumed and the total volume of digestive fluids secreted. Then, the cells were incubated with DCFH-DA for 20 min, and images of cells with high ROS levels were captured using a fluorescence microscope (DMi8, Leica, Wetzlar, Germany).2.19. Statistical analysis
Statistical analysis was conducted by one-way analysis of variance (ANOVA) using OriginPro 2024b software. Results are expressed as mean ± SD. Significance analysis was performed using the Tukey test and Student's t-test, with p < 0.05 considered statistically significant.3. Results and discussion
3.1. Characterization of BDCDs
Previous studies have demonstrated that carbon dots are formed during bread baking and exhibit cytotoxicity toward human mesenchymal stem cells in vitro.35 Additionally, it has been reported that nanoparticles can adsorb biomacromolecules from bodily fluids, forming “protein” coronas that play a crucial role in influencing their biological effects.36 Furthermore, the mucus barrier, which is primarily composed of mucin proteins, serves as the first line of defense in the gastrointestinal tract, protecting the intestinal epithelium from external damage. Once carbon dots enter the gastrointestinal tract, they therefore interact with mucin in the intestinal mucosa. Therefore, we investigated the impact of mucin corona formation on the gastrointestinal fate of BDCDs.Initially, the carbon dots were extracted from burnt bread crust and then characterized (Fig. 1A). Fig. 1B shows the photographs of BDCD solutions (1 mg mL−1) when exposed to either visible light or ultraviolet light (365 nm). The BDCD solutions were clear yellowish liquids under visible light but emitted blue fluorescence upon ultraviolet light, indicating that they were fluorescent carbon dots with good water solubility. The morphology of the BDCDs was investigated using transmission electron microscopy (Fig. 1C–E). The regular TEM images showed that the BDCDs consisted of small spheroidal particles with an average diameter of around 5.81 nm, which assembled into clumps of around 100 nm. The high-resolution TEM images showed that the interplane distances within the carbon dots were 0.246 and 0.422 nm. Previous researchers have extracted carbon dots with an average diameter of 2.67 nm from breadcrumbs fried at 180 °C.37 Thus, the dimensions of the BDCDs extracted from bread crust baked at a similar temperature were on the same order of magnitude as these.
XRD analysis was used to determine the physical state of the BDCDs. The XRD patterns revealed the presence of a single peak at 2θ = 20.7°, which suggested that the BDCDs were amorphous (rather than crystalline) (Fig. 1F). According to the XPS analysis, the surface chemical composition of the BDCDs contained C, N, and O constituents, which accounted for 70.9%, 1.5%, and 27.6%, respectively (Fig. 1G and H). Previous studies have reported that nitrogen can increase the quantum yield and enhance the fluorescence of carbon dots,38,39 which might explain the blue fluorescence of the BDCDs observed in our study. The detailed XPS spectra of C 1s, N 1s, and O 1s were measured to identify the surface functional groups of the BDCDs (Fig. 1I–K). The characteristic peaks at 284.80, 286.39, and 288.29 eV in the C 1s spectra corresponded to C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C/C–C, C–O, and C
C/C–C, C–O, and C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O bonds, respectively. In the N 1s spectra, the peaks observed at 399.61, 399.93, and 401.70 eV correspond to pyridinic N, pyrrolic N, and N–H groups, respectively. In the O1s spectra, the peaks observed at 532.12, 532.60, and 534.86 eV correspond to O
O bonds, respectively. In the N 1s spectra, the peaks observed at 399.61, 399.93, and 401.70 eV correspond to pyridinic N, pyrrolic N, and N–H groups, respectively. In the O1s spectra, the peaks observed at 532.12, 532.60, and 534.86 eV correspond to O![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C–O, C–O, and O
C–O, C–O, and O![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C–O* functional groups, respectively.40,41
C–O* functional groups, respectively.40,41
Furthermore, the optical properties of the BDCDs were investigated using both fluorescence and UV-visible spectrophotometry. The fluorescence emission spectra of the BDCDs were measured at different excitation wavelengths, revealing a maximum excitation/emission wavelength of 344/437 nm. BDCDs exhibited a quantum yield of 1.03% when excited at 344 nm, along with a decay time of 5.75 ns (Fig. S1†). Additionally, the emission wavelength exhibited a redshift as the excitation wavelength increased, a phenomenon commonly observed in carbon dots (Fig. 1L). Mechanistically, the formation of complex surface states in BDCDs during the bread-baking process may result in this bathochromic emission phenomenon.5 Moreover, the fluorescence intensity of BDCDs excited by the excitation wavelength of 344 nm decreased with decreasing BDCD concentration (Fig. 1M). However, compared to the fluorescence intensity of BDCDs at 500 μg mL−1, the intensity at 1000 μg mL−1 was lower, which could be due to the agglomeration of BDCDs at higher concentrations (as seen in the TEM images), leading to the masking of the fluorophores. The UV-visible spectra of BDCDs had an absorption peak at 276 nm, and the absorbance decreased with decreasing concentration of BDCDs (Fig. 1N). The absorption peak may result from the π–π* conjugation of C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C double bonds in the surface functional groups of BDCDs.7
C double bonds in the surface functional groups of BDCDs.7
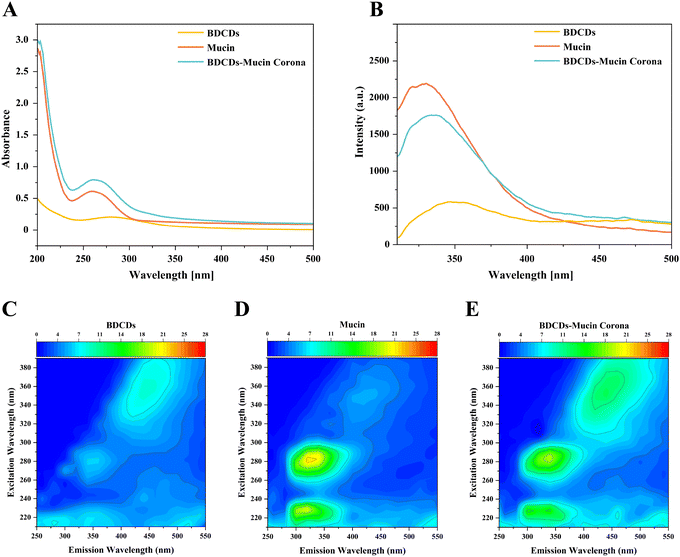
3.2. Interactions between BDCDs and mucin
The interactions between the carbon dots derived from bread crust and mucin were characterized using a variety of analytical methods.The hydrodynamic diameters of the BDCDs, mucin, and BDCD–mucin complexes were measured by dynamic light scattering. The BDCDs had an average diameter of around 91.28 nm, which was consistent with the dimensions of the particles observed in the TEM images discussed earlier. This size was considerably larger than the size of the individual BDCDs, which again highlights the fact that they tended to exist as clusters in solution. The mucin had an average diameter of around 78.82 nm, which was again consistent with the TEM images, and suggests that the mucin molecules existed as clumps in solution. The average diameter of the BDCD–mucin complexes (122.40 nm) was larger than either the BDCDs or the mucin. This effect may have been because the incorporation of the BDCDs into the clumps of mucin led to an increase in their average size, which was consistent with the SEM images. Other researchers have also reported that the formation of protein–nanoparticle complexes leads to an increase in diameter.27 Interestingly, however, it has been reported that the formation of a mucin corona can alter the agglomeration state of TiO2, while only leads to minimal changes in the particle size measured by light scattering.12 This result suggests that the impact of mucin depends on the nature of the nanoparticles involved.
![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C group.30 Meanwhile, the peaks at 1220 and 1059 cm−1 corresponded to the C–C stretching vibrations and aromatic alkoxy bonds, respectively.5 Compared to mucin and the BDCD–mucin complexes, the BDCDs exhibited only small or no peaks at 2919, 2851, 1628, 1537, and 1220 cm−1. After the formation of the BDCD–mucin complexes, a redshift of the –NH2 and –OH stretching vibration peaks of mucin was observed, suggesting hydrogen bonding and hydrophobic interactions between the BDCDs and mucin.30 Moreover, compared to the mucin group, the peak at 1059 cm−1 in the BDCD–mucin complex group exhibited a significant blueshift, suggesting that the BDCDs altered the conformation of mucin. It is worth noting the similarity in the FTIR spectra of mucin and BDCD–mucin complexes, which suggested that mucin dominated the spectra of the complexes. Previous researchers have reported that the presence of a strong amide group peak in the FTIR spectra of nanoparticles was consistent with the adsorption of proteins onto their surfaces.45
C group.30 Meanwhile, the peaks at 1220 and 1059 cm−1 corresponded to the C–C stretching vibrations and aromatic alkoxy bonds, respectively.5 Compared to mucin and the BDCD–mucin complexes, the BDCDs exhibited only small or no peaks at 2919, 2851, 1628, 1537, and 1220 cm−1. After the formation of the BDCD–mucin complexes, a redshift of the –NH2 and –OH stretching vibration peaks of mucin was observed, suggesting hydrogen bonding and hydrophobic interactions between the BDCDs and mucin.30 Moreover, compared to the mucin group, the peak at 1059 cm−1 in the BDCD–mucin complex group exhibited a significant blueshift, suggesting that the BDCDs altered the conformation of mucin. It is worth noting the similarity in the FTIR spectra of mucin and BDCD–mucin complexes, which suggested that mucin dominated the spectra of the complexes. Previous researchers have reported that the presence of a strong amide group peak in the FTIR spectra of nanoparticles was consistent with the adsorption of proteins onto their surfaces.45| System | β-Sheet (%) | β-Turn (%) | Random coil (%) |
|---|---|---|---|
| Mucin | 42.0 | 23.7 | 34.3 |
BDCDs![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) mucin = 1 mucin = 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 1 |
38.2 | 25.7 | 36.1 |
BDCDs![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) mucin = 2 mucin = 2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 1 |
43.7 | 21.9 | 34.3 |
BDCDs![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) mucin = 3 mucin = 3![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 1 |
39.7 | 23.2 | 37.1 |
BDCDs![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) mucin = 4 mucin = 4![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 1 |
44.4 | 23.9 | 31.7 |
BDCDs![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) mucin = 5 mucin = 5![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 1 |
41.8 | 22.1 | 36.1 |
The samples were therefore excited using an excitation wavelength of 280 nm and the resulting maximum emission peak around 330 nm was recorded. Compared to mucin alone, there was a red shift of the maximum emission peak of mucin after interacting with BDCDs, which indicated that the presence of the nanoparticles enhanced the polarity of the tryptophan residues in the mucin, suggesting that they altered their microenvironment. Previously, it has been reported that an increase in the polarity of the microenvironment surrounding the aromatic amino acids in the proteins of curcumin–myofibrillar protein complexes led to a red shift in their maximum fluorescence intensity during in vitro digestion.48 Moreover, the fluorescence intensity of the BDCD–mucin complexes was lower than mucin alone, further suggesting the structural changes in the mucin (Fig. 3B). It is important to note that these changes in the microenvironment of the amino acid residues in mucin may disrupt its normal physiological function. For instance, the interaction between black carbon nanoparticles and mucin has been reported to alter the selective permeability of mucin, increasing the translocation of cationic molecules across the mucin barrier.49
In the presence of BDCDs, the fluorescence intensity of the mucin at excitation wavelengths of 230 nm and 280 nm decreased, which indicated that both the polypeptide skeleton structure and the tryptophan and tyrosine residues of mucin were involved in its interaction with BDCDs.50 Moreover, the increased fluorescence intensity observed at an excitation wavelength of 350 nm in the BDCD–mucin complexes also suggests that the BDCDs altered the structure of the proteins in the mucin following their interaction (Fig. 3E). Overall, these results demonstrated that the presence of BDCDs induced a conformational change in the mucin.
3.3. Interactions between BDCD–mucin complexes and digestive enzymes during in vitro digestion
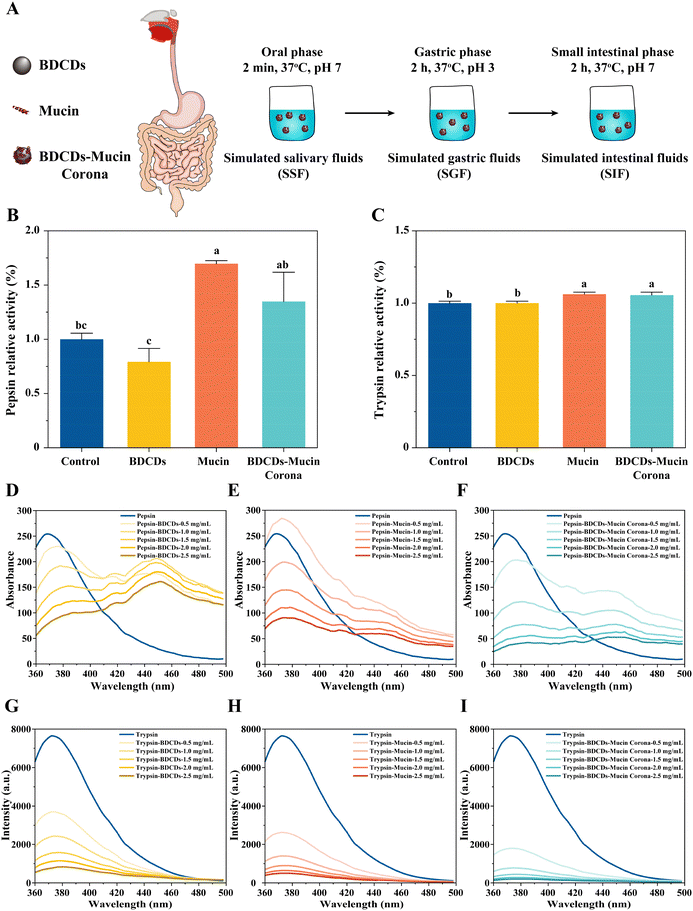
In this series of experiments, an in vitro digestion model was used to investigate the interactions between the BDCD–mucin complexes and digestive enzymes (Fig. 4A). In general, these in vitro simulated digestion models provide a rapid and highly reproducible means of studying the gastrointestinal behavior of foods.51In summary, these results suggest that mucin may help protect the gastrointestinal digestive function from being impaired by carbon dots in foods.
The fluorescence intensity of trypsin displayed a trend similar to that of pepsin after interacting with BDCDs, mucin, and the BDCD–mucin complexes (Fig. 4G–I). However, the key difference was the absence of a fluorescence peak at 450 nm. These results suggest that the presence of our samples altered the structure of trypsin, although they had little effect on its activity. Wang et al. reported that epigallocatechin gallate-resistant starch nanoparticles altered the conformation and microenvironment of trypsin, while leaving its activity unaffected.30 Again, the above results suggest that BDCDs have little effect on the active site of trypsin, while the presence of mucin or BDCD–mucin complexes may even activate trypsin. Furthermore, the redshift of the trypsin peak was slightly smaller than that of pepsin when treated with BDCDs, mucin, and the BDCD–mucin complexes, which may correspond to the smaller changes in trypsin activity observed for these treatments.
The quenching mechanisms of pepsin and trypsin were investigated using the Stern–Volmer equation (Fig. S3†). According to the F0/F curve, a linear correlation was observed with BDCDs, mucin, and BDCD–mucin complexes at concentrations ranging from 0.5 to 2.5 μg mL−1, which suggests that the Stern–Volmer equation was appropriate for interpreting the data. Furthermore, the slight upward curvature observed in the F0/F curve suggests the coexistence of static and dynamic quenching. These energy results suggest that BDCDs, mucin, and BDCD–mucin complexes could interact with pepsin and trypsin to form non-fluorescent complexes, accompanied by a charge or transfer between them.56–58 Compared to BDCDs alone, the BDCD–mucin complexes exhibited a weaker linear relationship with pepsin and trypsin, suggesting that reduced non-fluorescent complexes formed between the complexes and these enzymes.
3.4. Changes in the antioxidant activity of BDCD–mucin complexes induced by in vitro digestion
It has been reported that free radicals are generated during aerobic metabolism and can influence the physiological functions and metabolism of the human body.59 Studies have shown that mucins can protect DNA from hydroxyl radical damage and that sialic acid in mucin is an essential component of its antioxidant activity.60 Therefore, the antioxidant activity of BDCDs, mucin, and BDCD–mucin complexes during in vitro simulated digestion was measured using three assays: the ABTS radical scavenging, hydroxyl radical scavenging, and total reducibility assays. The results of the ABTS radical scavenging assay indicated that the formation of the BDCD–mucin complexes increased the ability of the BDCDs to scavenge ABTS radicals during the simulated salivary and intestinal stages (Fig. 5A–D). Notably, the mucin itself had a relatively low ABTS radical scavenging ability during the simulated gastric stage. After exposure to simulated gastric conditions, the ABTS radical scavenging ability of the BDCD–mucin complexes was lower than that of the BDCDs, which may be because the carbon dots were trapped inside mucin clumps. Furthermore, the ABTS radical scavenging ability of BDCDs, mucin, and BDCD–mucin complexes remained stable, with only a slight decline at each stage of digestion, indicating their relative resistance to digestive processesTypically, an increase in the antioxidant activity of proteins during simulated digestion occurs due to the generation of antioxidant peptides after hydrolysis by proteases. Previous studies have shown that mucin is relatively resistant to digestion by digestive enzymes in the gastrointestinal tract, which is consistent with the stable ABTS radical scavenging activity of mucin and the BDCD–mucin complexes when exposed to different digestion regions.61
Interestingly, the results of the hydroxyl radical scavenging assay were similar to those of the ABTS radical scavenging assay (Fig. 5E–H). The hydroxyl radical scavenging ability of mucin remained high during the simulated gastric phase. Moreover, the hydroxyl radical scavenging ability of the BDCDs significantly decreased during the simulated gastric and intestinal phases as digestion progressed, suggesting that digestive enzymes may interact with BDCDs to form protein coronas, thereby masking the reducing groups on their surfaces.5 However, in simulated gastric fluids, the hydroxyl radical scavenging ability of the BDCD–mucin complexes remained unchanged, suggesting that the formation of the mucin protein corona prevented further interaction between the BDCDs and digestive enzymes. It should be noted that the hydroxyl radical scavenging activity of the BDCD–mucin complexes decreased significantly during the simulated intestinal phase, although it remained higher than that of the BDCDs at the end of digestion. One possible explanation is that the structure changes of the BDCD–mucin complexes during the simulated intestinal phase resulted in masking of the reducing groups on their surfaces.
In the total reducibility assay, mucin exhibited a low reducing ability throughout the in vitro simulated digestion, whereas the BDCDs retained a relatively high reducing ability. As shown in Fig. 5J–L, it is clear that the formation of the BDCD–mucin complexes enhanced the reducing ability of the BDCDs during simulated digestion. Previous studies have reported that the formation of a whey protein corona around TiO2 nanoparticles enhanced their antioxidant properties. However, TiO2 nanoparticles may also reduce the antioxidant activity of whey by adsorbing some of the proteins and peptides onto their surfaces.14 Furthermore, the reducing ability of the BDCDs, mucin, and BDCD–mucin complexes decreased slightly after each stage of digestion, which was consistent with the results of the ABTS radical scavenging assay. These results suggest that the BDCD–mucin complexes remain relatively stable in the gastrointestinal tract. It should be noted, however, that compared to the antioxidant activity of simulated digestive fluid without digestive enzymes, the presence of digestive enzymes enhanced the antioxidant activity of the digestive fluids (Fig. 5).
Based on the antioxidant activity results, it can be speculated that the BDCD–mucin complexes remained relatively stable during simulated digestion, which may allow them to play a role in the colon.
Interestingly, although the antioxidant activity of the BDCDs, mucin, and BDCD–mucin complexes exhibited some fluctuations during in vitro simulated digestion, the BDCD–mucin complexes demonstrated higher antioxidant activity than the BDCDs but lower than the mucin at the end of the digestion process, particularly for the ABTS and hydroxyl radical scavenging capacities. A plausible explanation for this effect is that the reductive peptides of mucin were adsorbed onto the surface of BDCDs, resulting in a reduced radical scavenging activity compared to that of free mucin.14 Additionally, the presence of BDCDs may modulate the diversity and abundance of peptides generated during simulated digestion.62 In comparison with BDCDs alone, the enhanced antioxidant activity of the BDCD–mucin complexes could be attributed to the surface chemistry modification of the BDCDs by mucin, which may further shield BDCDs from interactions with other gastrointestinal components, such as digestive enzymes, thereby maintaining a relatively high antioxidant activity.
Additional information about the stability of the BDCD–mucin complexes during the digestion process was obtained by measuring changes in their fluorescence and UV-visible absorption spectra after being exposed to different pH conditions, which were designed to simulate the gastrointestinal environment (Fig. S4†). These results showed that there were some modest changes in the fluorescence and UV-visible absorption spectra after exposure of the BDCD–mucin complexes to simulated gastrointestinal conditions, which suggests that there were some alterations in protein conformation and/or the aggregation state of the complexes when exposed to different digestion conditions. These alterations may account for the changes in the antioxidant properties of the complexes in different gastrointestinal regions discussed earlier.
3.5. Biological effects of the BDCD–mucin complexes on intestinal epithelial cells
Finally, the potential biological effects of the carbon dot–mucin complexes were determined by measuring their impact on the viability and oxidation state of model intestinal epithelial cells.4. Conclusion
In summary, this study characterized the carbon dots isolated from bread crust and then investigated their interactions with mucin. The bread-derived carbon dots (BDCDs) were found to interact with mucin and form BDCD–mucin complexes, where clusters of carbon dots were embedded within mucin clumps. The formation of these complexes led to changes in zeta potential and particle size of the BDCDs, as well as alterations in the secondary structure of the mucin. In vitro simulated digestion showed that BDCDs reduced pepsin activity by altering its conformation, as evidenced by changes in the fluorescence spectrum. Furthermore, the presence of mucin reduced the ability of the BDCDs to inactivate pepsin, and actually increased the activity of trypsin. The antioxidant activity of BDCDs during simulated digestion was found to be enhanced by the formation of the carbon dot–mucin clusters. Additionally, compared to the BDCDs, the BDCD–mucin complexes enhanced the viability of Caco-2 cells by reducing cellular ROS levels. Our results highlight the impact of mucin on the gastrointestinal behavior of food-derived carbon dots, which may be important in understanding their potential adverse effects on the healthiness of foods.Author contributions
Mi Shichao: writing – original draft, methodology, formal analysis, investigation, and data curation. Liu Zimo: methodology, formal analysis, and data curation. Wang Mingcheng: methodology and formal analysis. David Julian McClements: formal analysis and review & editing. Cao Chongjiang: conceptualization and validation. Xu Xiao: formal analysis, visualization, supervision, validation, and review & editing. Yuan Biao: funding acquisition, conceptualization, supervision, review & editing, and project administrationData availability
The data will be made available from the corresponding author upon reasonable request.Conflicts of interest
There are no conflicts of interest to declare.Acknowledgements
This work was supported by the National Natural Science Foundation of China (32472355, 31901699 and 32302040), the Fundamental Research Funds for the Central Universities (2632021ZD01 and 2632024HL03), the China Postdoctoral Science Foundation (2020T130138ZX), the Jiangsu Planned Projects for Postdoctoral Research Funds and the National Innovation and Entrepreneurship Training Program for Undergraduate.References
- D. H. H. Nguyen, H. El-Ramady and J. Prokisch, Food safety aspects of carbon dots: a review, Environ. Chem. Lett., 2025, 23, 337–360 CrossRef CAS.
- H. E. Emam, Carbon quantum dots derived from polysaccharides: Chemistry and potential applications, Carbohydr. Polym., 2024, 324, 121503 CrossRef CAS PubMed.
- H. Wang, W. Su and M. Tan, Endogenous fluorescence carbon dots derived from food items, Innovation, 2020, 1, 100009 CAS.
- B. Zhang, X. Fan, H. Du, M. Zhao, Z. Zhang, R. Zhu, B. He, Y. Zhang, X. Li, J. Li and N. Gu, Foodborne carbon dot exposure induces insulin resistance through gut microbiota dysbiosis and damaged intestinal mucus layer, ACS Nano, 2023, 17, 6081–6094 CrossRef CAS PubMed.
- X. Song, Y. Song, Z. Guo and M. Tan, Influence of protein coronas between carbon nanoparticles extracted from roasted chicken and pepsin on the digestion of soy protein isolate, Food Chem., 2022, 385, 132714 CrossRef CAS PubMed.
- H. B. Ahmed, M. El-Shahat, A. K. Allayeh and H. E. Emam, Maillard reaction for nucleation of polymer quantum dots from chitosan-glucose conjugate: Antagonistic for cancer and viral diseases, Int. J. Biol. Macromol., 2023, 224, 858–870 CrossRef CAS PubMed.
- R. Liu, K. Liu and M. Tan, Nanocorona formation between foodborne nanoparticles extracted from roast squid and human serum albumin, J. Agric. Food Chem., 2019, 67, 10470–10480 CrossRef CAS PubMed.
- R. Liu, K. Liu, G. Cui and M. Tan, Change of cell toxicity of food-borne nanoparticles after forming protein coronas with human serum albumin, J. Agric. Food Chem., 2022, 70, 1261–1271 CrossRef CAS PubMed.
- C. Song, Z. Chai, S. Chen, H. Zhang, X. Zhang and Y. Zhou, Intestinal mucus components and secretion mechanisms: what we do and do not know, Exp. Mol. Med., 2023, 55, 681–691 CrossRef CAS PubMed.
- N. Reznik, A. D. Gallo, K. W. Rush, G. Javitt, Y. Fridmann-Sirkis, T. Ilani, N. A. Nairner, S. Fishilevich, D. Gokhman, K. N. Chacón, K. J. Franz and D. Fass, Intestinal mucin is a chaperone of multivalent copper, Cell, 2022, 185, 4206–4215 CrossRef CAS PubMed.
- X. Rui, K. Fu, H. Wang, T. Pan and W. Wang, Formation mechanisms of protein coronas on food-related nanoparticles: their impact on digestive system and bioactive compound delivery, Foods, 2025, 14, 512 CrossRef CAS PubMed.
- H. Zhou, J. K. Pandya, Y. Tan, J. Liu, S. Peng, J. L. Muriel Mundo, L. He, H. Xiao and D. J. McClements, Role of mucin in behavior of food-grade tio2 nanoparticles under simulated oral conditions, J. Agric. Food Chem., 2019, 67, 5882–5890 CrossRef CAS PubMed.
- D. Yang, Y. Feng, Y. Yuan, L. Zhang, Y. Zhou, A. C. Midgley, Y. Wang, N. Liu, G. Li, X. Yao and D. Liu, Protein coronas derived from mucus act as both spear and shield to regulate transferrin functionalized nanoparticle transcellular transport in enterocytes, ACS Nano, 2024, 18, 7455–7472 CrossRef CAS PubMed.
- H. Shan, Y. Guo, J. Li, Z. Liu, S. Chen, B. Dashnyam, D. J. McClements, C. Cao, X. Xu and B. Yuan, Impact of whey protein corona formation around tio2 nanoparticles on their physiochemical properties and gastrointestinal fate, J. Agric. Food Chem., 2024, 72, 4958–4976 CrossRef CAS PubMed.
- G. Lee, Y.-J. Jhang, Y.-T. Jhang, Y.-C. Chang, H.-W. Chang, C.-Y. Chuang, Y.-K. Chuang, C.-W. Lin and I. L. Hsiao, Artificial digestion represents the worst-case scenario for studying nanoplastic fate in gastrointestinal tract, J. Hazard. Mater., 2025, 485, 136809 CrossRef CAS PubMed.
- R. Liu, D. Yu, A. M. Abd El-Aty and M. Tan, Protein corona of food nanoparticles: Implications for biological responses and future research directions, Trends Food Sci. Technol., 2023, 141, 104179 CrossRef CAS.
- Y. Wang, Y. L. Mo, Y. W. Sun, J. Li, Y. An, N. P. Feng and Y. Liu, Intestinal nanoparticle delivery and cellular response: a review of the bidirectional nanoparticle-cell interplay in mucosa based on physiochemical properties, J. Nanobiotechnol., 2024, 22, 669 CrossRef PubMed.
- J. Wang, Z. Zhang, Z. Zhang, Z. Zou, Y. Zhuo, C. Liu, D. Nie, Y. Gan and M. Yu, Enhanced gut-to-liver oral drug delivery via ligand-modified nanoparticles by attenuating protein corona adsorption, ACS Nano, 2024, 18, 35310–35324 CrossRef CAS PubMed.
- Q. Peng, Y. Ma, Z. Wang and J. Wang, Inhibition mechanism of different structural polyphenols against α-amylase studied by solid-state NMR and molecular docking, Int. J. Biol. Macromol., 2024, 275, 133757 CrossRef CAS PubMed.
- C. Lei, M. Tao, L. Xu, L. Yue, X. Cao, B. Cheng, C. Wang and Z. Wang, Different functional groups of carbon dots influence the formation of protein crowns and pepsin characteristic in vitro digestion, Food Chem., 2024, 440, 138224 CrossRef CAS PubMed.
- Y. Chen, Q. Liu, H. Yu, Y. Guo, Y. Cheng, H. Qian, Y. Xie and W. Yao, Protein corona formed on the TiO2 nanoparticles promotes the hydrolysis of collagen in simulated gastrointestinal fluids, Food Biosci., 2023, 53, 102786 CrossRef CAS.
- Y.-R. Wu, Q. Zhou, J. Li, W. Wang, Y.-B. Zhou and K. Liu, The formation of protein coronas and its effect on the quercetin-edible dock protein nanoparticles, Food Hydrocolloids, 2024, 157, 110432 CrossRef CAS.
- Y. Wang, L. Zhou, Y. Sun, H. Mu, X. Li, Y. Wang and Q. Sun, Formation of protein corona on interaction of pepsin with chitin nanowhiskers in simulated gastric fluid, Food Chem., 2022, 383, 132393 CrossRef CAS PubMed.
- Q. Huang, L. Yan, H. Wei, X. Ye, W. Wang, C. Yang, X.-P. Yan and Y. Zhang, Extraction, characterization and toxicity evaluation of fluorescent carbon nanoparticles in bamboo charcoal peanut, J. Food Compos. Anal., 2023, 121, 105357 CrossRef CAS.
- K. Liu, Y. Song and M. Tan, Toxicity alleviation of carbon dots from roast beef after the formation of protein coronas with human serum albumin, J. Agric. Food Chem., 2020, 68, 9789–9795 CrossRef CAS PubMed.
- Q. Zhao, H. Shan, Y. Li, B. Jiang, X. Xu, D. Julian McClements, C. Cao and B. Yuan, Investigation of the interactions between food plant carbohydrates and titanium dioxide nanoparticles, Food Res. Int., 2022, 159, 111574 CrossRef CAS PubMed.
- H. Shan, Q. Zhao, Y. Guo, M. Gao, X. Xu, D. J. McClements, C. Cao and B. Yuan, Impact of pH on the formation and properties of whey protein coronas around TiO2 nanoparticles, J. Agric. Food Chem., 2023, 71, 5756–5769 CrossRef CAS PubMed.
- A. Micsonai, É. Moussong, F. Wien, E. Boros, H. Vadászi, N. Murvai, Y.-H. Lee, T. Molnár, M. Réfrégiers, Y. Goto, Á. Tantos and J. Kardos, BeStSel: webserver for secondary structure and fold prediction for protein CD spectroscopy, Nucleic Acids Res., 2022, 50, W90–W98 CrossRef CAS PubMed.
- B. Jiang, Q. Zhao, H. Shan, Y. Guo, X. Xu, D. J. McClements, C. Cao and B. Yuan, Impact of heat treatment on the structure and properties of the plant protein corona formed around TiO2 nanoparticles, J. Agric. Food Chem., 2022, 70, 6540–6551 CrossRef CAS PubMed.
- S. Wang, Z. Duan, L. Zheng, Y. Yang, X. Zheng, D. Xiao, B. Ai, M. Wang and Z. Sheng, Digestive enzyme corona formed in simulated gastrointestinal tract and its impact on EGCG release from banana resistant starch nanoparticles, Food Hydrocolloids, 2024, 146, 109267 CrossRef CAS.
- H. Yang, P. Wilde, R. J. Wang, Q. Meng, H. Y. Shi, H. Yu, Z. J. Zhou, J. Z. Han and W. L. Liu, Effect of complexation with different molecular weights of oat β-glucan and sea buckthorn flavonoid on the digestion of rice flour, J. Agric. Food Chem., 2024, 72, 23567–23579 CrossRef CAS PubMed.
- J. Zhang, Q. Li, X. Jiang, X. Li, P. Dong, J. Li, M. Komiyama and X. Liang, Effect of sulfated polysaccharides on the digestion of DNA by pepsin under simulated gastric juice in vitro, Food Funct., 2020, 11, 1790–1797 RSC.
- M. Sun, Z. Cai, C. Li, Y. Hao, X. Xu, K. Qian, H. Li, Y. Guo, A. Liang, L. Han, H. Shang, W. Jia, Y. Cao, C. Wang, C. Ma, J. C. White and B. Xing, Nanoscale ZnO improves the amino acids and lipids in tomato fruits and the subsequent assimilation in a simulated human gastrointestinal tract model, ACS Nano, 2023, 17, 19938–19951 CrossRef CAS PubMed.
- S. Mi, M. Shen, Z. Liu, Y. Yu, H. Shan, J. Cao, D. J. McClements, C. Cao, X. Xu and B. Yuan, A glutenin protein corona ameliorated TiO2 nanoparticle-induced gut barrier dysfunction and altered the gut microbiota composition, Food Funct., 2024, 15, 12101–12117 RSC.
- A. M. Al-Hadi, V. S. Periasamy, J. Athinarayanan, A. S. Al-Khalifa and A. A. Alshatwi, Extraction of ultrafine carbon nanoparticles from samooli Bread and evaluation of their in vitro cytotoxicity in human mesenchymal stem cells, Process Biochem., 2017, 52, 250–258 CrossRef CAS.
- H. Tang, Y. Zhang, T. Yang, C. Wang, Y. Zhu, L. Qiu, J. Liu, Y. Song, L. Zhou, J. Zhang, Y. K. Wong, Y. Liu, C. Xu, H. Wang and J. Wang, Cholesterol modulates the physiological response to nanoparticles by changing the composition of protein corona, Nat. Nanotechnol., 2023, 18, 1067–1077 CrossRef CAS PubMed.
- J. Yin, K. Liu, S. Yuan, Y. Guo, H. Yu, Y. Cheng, Y. Xie, H. Qian and W. Yao, Carbon dots in breadcrumbs: Effect of frying on them and interaction with human serum albumin, Food Chem., 2023, 424, 136371 CrossRef CAS PubMed.
- K. Chan and A. Zinchenko, Aminolysis-assisted hydrothermal conversion of waste PET plastic to N-doped carbon dots with markedly enhanced fluorescence, J. Environ. Chem. Eng., 2022, 10, 107749 CrossRef CAS.
- M. M. Mikhail, H. B. Ahmed, A. E. M. Abdallah, M. El-Shahat and H. E. Emam, Surface passivation of carbon dots for tunable biological performance, J. Fluoresc., 2024 DOI:10.1007/s10895-024-03806-6.
- M. Esmaeili Koutamehr, M. Moradi, H. Tajik, R. Molaei, M. Khakbaz Heshmati and A. Alizadeh, Sour whey-derived carbon dots; synthesis, characterization, antioxidant activity and antimicrobial performance on foodborne pathogens, LWT, 2023, 184, 114978 CrossRef CAS.
- N. Rahmatian, S. Abbasi, N. Abbasi and M. Tavakkoli Yaraki, Alginate carbon dot nanocomposite: A green approach towards designing turn-on aptasenor for Candida albicans fungus, Int. J. Biol. Macromol., 2024, 282, 137315 CrossRef CAS PubMed.
- Y. Qu, S. Zhao, J. Ni, L. Jiao, X. Zhang, S. Benjakul, Z. Liu, X. Chen and B. Zhang, In situ formation mechanism of endogenous fluorescent carbon dots during the roasting process of small yellow croaker (Larimichthys polyactis), Food Chem.: X, 2025, 25, 102187 CAS.
- J. Bing, X. Xiao, D. J. McClements, Y. Biao and C. Chongjiang, Protein corona formation around inorganic nanoparticles: Food plant proteins-TiO2 nanoparticle interactions, Food Hydrocolloids, 2021, 115, 106594 CrossRef CAS.
- Z. Zhong, S. Fang, Y. Li, Y. Huang, Y. Zhang, H. Chen, J. Zhang, H.-X. Wang, H. Xiong, Q. Zou and S. Wang, Quantitative analysis of protein corona on precoated protein nanoparticles and determined nanoparticles with ultralow protein corona and efficient targeting in vivo, ACS Appl. Mater. Interfaces, 2021, 13, 56812–56824 CrossRef CAS PubMed.
- G. Mummaleti, J. Feng, A. Mohan, J. Suh, Z.-L. Kong and F. Kong, Microplastics interactions and transformations during in vitro digestion with milk, Food Res. Int., 2024, 197, 115247 CrossRef CAS PubMed.
- N. Barbero, M. Coletti, F. Catalano and S. Visentin, Exploring gold nanoparticles interaction with mucins: A spectroscopic-based study, Int. J. Pharm., 2018, 535, 438–443 CrossRef CAS PubMed.
- N. Archipowa, L. Wittmann, J. Köckenberger, F. J. Ertl, J. Gleixner, M. Keller, M. R. Heinrich and R. J. Kutta, Characterization of fluorescent dyes frequently used for bioimaging: photophysics and photocatalytical reactions with proteins, J. Phys. Chem. B, 2023, 127, 9532–9542 CrossRef CAS PubMed.
- C. Wu, H. Dong, P. Wang, M. Han and X. Xu, Sequential changes in antioxidant activity and structure of curcumin-myofibrillar protein nanocomplex during in vitro digestion, Food Chem., 2022, 382, 132331 CrossRef CAS PubMed.
- M. Marczynski, T. M. Lutz, R. Schlatterer, M. Henkel, B. N. Balzer and O. Lieleg, Contamination with black carbon nanoparticles alters the selective permeability of mucin hydrogels: implications for molecular transport across mucosal barriers, ACS Appl. Nano Mater., 2022, 5, 16955–16970 CrossRef CAS.
- X. Qiao, L. Yang, J. Gu, Y. Cao, Z. Li, J. Xu and C. Xue, Kinetic interactions of nanocomplexes between astaxanthin esters with different molecular structures and β-lactoglobulin, Food Chem., 2021, 335, 127633 CrossRef CAS PubMed.
- M. Minekus, M. Alminger, P. Alvito, S. Ballance, T. Bohn, C. Bourlieu, F. Carrière, R. Boutrou, M. Corredig, D. Dupont, C. Dufour, L. Egger, M. Golding, S. Karakaya, B. Kirkhus, S. Le Feunteun, U. Lesmes, A. Macierzanka, A. Mackie, S. Marze, D. J. McClements, O. Ménard, I. Recio, C. N. Santos, R. P. Singh, G. E. Vegarud, M. S. J. Wickham, W. Weitschies and A. Brodkorb, A standardised static in vitro digestion method suitable for food-an international consensus, Food Funct., 2014, 5, 1113–1124 RSC.
- H.-j. Zeng, J. You, H.-l. Liang, T. Qi, R. Yang and L.-b. Qu, Investigation on the binding interaction between silybin and pepsin by spectral and molecular docking, Int. J. Biol. Macromol., 2014, 67, 105–111 CrossRef CAS PubMed.
- X. Li, R. Xu, L. Shi and T. Ni, Design of flavonol-loaded cationic gold nanoparticles with enhanced antioxidant and antibacterial activities and their interaction with proteins, Int. J. Biol. Macromol., 2023, 253, 127074 CrossRef CAS PubMed.
- C. Yang, G. Xu, C. Hou and H. Zhang, Cobalt oxyhydroxide nanoflakes enable ratiometric fluorescent assay of gallic acid, Food Chem.: X, 2024, 24, 101843 CAS.
- Y. Wang, Y. Sun, J. Yang, L. Dai, N. Ji, L. Xiong and Q. Sun, Interactions of surface-functionalized starch nanoparticles with pepsin and trypsin in simulated gastrointestinal fluids, J. Agric. Food Chem., 2020, 68, 10174–10183 CrossRef CAS PubMed.
- H. B. Ahmed and H. E. Emam, Environmentally exploitable biocide/fluorescent metal marker carbon quantum dots, RSC Adv., 2020, 10, 42916–42929 RSC.
- T. Sasikumar, J. S. Packialakshmi, S. J. Hong, S. Y. Ha, G. H. Shin and J. T. Kim, Multifunctional green-emitting fluorescent carbon dots: A versatile fluorometric probe for glyphosate detection and applications in food, J. Environ. Chem. Eng., 2024, 12, 113356 CrossRef CAS.
- H. Huang, H. Huang, J. Yang, H. Yang, J. Dai, Z. Li, W. Yao and X. Guo, Synthesis of P, N-dopped carbon nanosheets for highly sensitive fluorescence analysis of nitrofuran antibiotics in fish, Food Chem., 2024, 459, 140445 CrossRef CAS PubMed.
- Z. Zhang, F. Kong, H. Ni, Z. Mo, J.-B. Wan, D. Hua and C. Yan, Structural characterization, α-glucosidase inhibitory and DPPH scavenging activities of polysaccharides from guava, Carbohydr. Polym., 2016, 144, 106–114 CrossRef CAS PubMed.
- Y. Ogasawara, T. Namai, F. Yoshino, M.-C.-i. Lee and K. Ishii, Sialic acid is an essential moiety of mucin as a hydroxyl radical scavenger, FEBS. Lett., 2007, 581, 2473–2477 CrossRef CAS PubMed.
- A. Abodinar, K. Tømmeraas, E. Ronander, A. M. Smith and G. A. Morris, The physicochemical characterisation of pepsin degraded pig gastric mucin, Int. J. Biol. Macromol., 2016, 87, 281–286 CrossRef CAS PubMed.
- B. H. Sarmadi and A. Ismail, Antioxidative peptides from food proteins: a review, Peptides, 2010, 31, 1949–1956 CrossRef CAS PubMed.
- A. W.-S. Zongo, D. Zogona, M. Youssef, S. Ye, F. Zhan, J. Li and B. Li, Senegalia macrostachya seed polysaccharides attenuate inflammation-induced intestinal epithelial barrier dysfunction in a Caco-2 and RAW264.7 macrophage co-culture model by inhibiting the NF-κB/MLCK pathway, Food Funct., 2022, 13, 11676–11689 RSC.
- Y. Wu, X. Song, N. Wang, S. Cong, X. Zhao, R. Rai and M. Tan, Carbon dots from roasted chicken accumulate in lysosomes and induce lysosome-dependent cell death, Food Funct., 2020, 11, 10105–10113 RSC.
- Y.-t. Li, K.-C. Mei, R. Liam-Or, J. T.-W. Wang, F. N. Faruqu, S. Zhu, Y.-l. Wang, Y. Lu and K. T. Al-Jamal, Graphene Oxide Nanosheets Toxicity in Mice Is Dependent on Protein Corona Composition and Host Immunity, ACS Nano, 2024, 18, 22572–22585 CrossRef CAS PubMed.
- Y. Ji, Y. Wang, D. Shen, Q. Kang and L. Chen, Mucin corona delays intracellular trafficking and alleviates cytotoxicity of nanoplastic-benzopyrene combined contaminant, J. Hazard. Mater., 2021, 406, 124306 CrossRef CAS PubMed.
- D. Yang, D. Liu, M. Qin, B. Chen, S. Song, W. Dai, H. Zhang, X. Wang, Y. Wang, B. He, X. Tang and Q. Zhang, Intestinal mucin induces more endocytosis but less transcytosis of nanoparticles across enterocytes by triggering nanoclustering and strengthening the retrograde pathway, ACS Appl. Mater. Interfaces, 2018, 10, 11443–11456 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d5fo00088b |
| This journal is © The Royal Society of Chemistry 2025 |