Open Access Article
Open Access ArticleA biotin-free diet attenuates the incidence of collagen-induced arthritis and alleviates microbial dysbiosis†
Xiuli
Su‡
b,
Xiaona
Li‡
d,
Xiaojuan
He
ef,
Sijie
Zhou
*a,
Aiping
Lyu
*e and
Zongwei
Cai
 *bc
*bc
aDepartment of Nephrology, The First Affiliated Hospital of Zhengzhou University, Zhengzhou, China. E-mail: fcczhousj@zzu.edu.cn
bEastern Institute of Technology, Ningbo, China. E-mail: zwcai@hkbu.edu.hk
cState Key Laboratory of Environmental and Biological Analysis, Hong Kong Baptist University, Hong Kong, China
dDepartment of Pharmacy, Peking University Third Hospital, Beijing, China
eLaw Sau Fai Institute for Advancing Translational Medicine in Bone and Joint Diseases, School of Chinese Medicine, Hong Kong Baptist University, Hong Kong, China. E-mail: aipinglu@hkbu.edu.hk
fInstitute of Basic Research in Clinical Medicine, China Academy of Traditional Chinese Medicine, Beijing, China
First published on 29th May 2025
Abstract
Emerging evidence has shown that the gut microbiota and its products are important triggers in the pathogenesis of rheumatoid arthritis (RA). Biotin is a diet- and microbiome-dependent metabolite and an immune regulator; however, the role of biotin in RA remains unknown. In this study, we observed abnormal fecal biotin excretion in RA patients, which correlated with microbial alterations. Specifically, biotin content was inversely associated with gut microbial genera enriched in healthy controls, including Roseburia and Dorea. Meanwhile, it positively correlated with Oscillospira, which was highly enriched in RA individuals. Moreover, collagen-induced arthritis (CIA) mice fed a biotin-free diet had attenuated arthritis incidence with depressed differentiation of splenic CD3+ T cells and restored microbial diversities. The biotin-free diet also increased bone mass and protected against inflammation-induced bone loss in CIA mice. Additionally, the biotin-free diet reshaped the host metabolic phenotype of amino acids and microbial composition. Notably, biotin deficiency ameliorated the augmentation of Oscillospira in CIA mice. Collectively, our results suggested a potential link between biotin deficiency, gut microbiota dysbiosis and CIA progression.
1 Introduction
Rheumatoid arthritis (RA) is a chronic, progressive autoimmune disorder marked by synovitis and symmetric, destructive arthropathy.1 While the pathogenesis of RA is not yet fully understood, it is believed to be a combination of genetic and environmental factors.1 One theory regarding the pathophysiology of RA is that the disease initially manifests in mucosal areas due to interactions between mucosal immunity and an abnormal gut microbiome, before progressing to affect the synovial joints.2,3 Evidence from studies on the early stages of the disease supports the hypothesis that these microbiota changes precede the development of RA.4,5 In a cohort study investigating the fecal microbiome of first-degree relatives of RA individuals, subjects in the preclinical stages of RA (defined by the presence of anti-citrullinated protein antibodies, rheumatoid factors or RA-like symptoms) exhibited a significantly altered microbiome compared to asymptomatic individuals without autoantibodies.4 Gut dysbiosis was found in RA individuals and was partly normalized after therapy.5 Moreover, partial depletion of the gut microbiome aggravated arthritis severity in collagen-induced arthritis (CIA) mice.6 Microorganisms can also regulate the differentiation of immunocytes and inflammatory cytokines, changing the permeability and barrier of the mucosa to impact the autoimmunity of RA.7 A hallmark of RA is the infiltration of activated CD4+ T cells into the synovium, particularly Th1 and Th17 subsets, which secrete pro-inflammatory cytokines such as IFN-γ, IL-17, and TNF-α, thereby promoting synovial inflammation and joint destruction.8 In addition, monocytes and macrophages contribute to joint pathology by producing inflammatory mediators.9 Regulatory T cells (Tregs), which normally act to suppress excessive immune activation, are often functionally impaired in RA.4Microbial communities, along with their metabolites and components, are essential for maintaining immunological homeostasis and can impact host vulnerability to various immune disorders.10–12 Metabolites are products and signaling molecules of the gut microbiome, making metabolomics a useful tool for identifying biological responses in patients with rheumatic conditions. A previous study found that tryptophan catabolites produced by gut bacteria can activate the aryl-hydrocarbon receptor in regulatory B cells, which can then suppress arthritis in mice.13 These findings provide insights into the previously unknown connection between the gut microbiome and bone health.
An elevated level of fecal biotin was found in RA patients.14 Since mammals cannot synthesize biotin by themselves, biotin can be obtained only from the diet.15 Additionally, the gut microbiome can de novo synthesize and supply biotin to its host.15 Emerging evidence suggests that biotin also plays a crucial role in regulating immunity.16,17 A biotin-free diet has been associated with impaired T cell proliferation and differentiation, leading to compromised immune responses.18 However, studies on the role of biotin in autoimmune diseases are relatively limited.
We have previously reported that alterations in the gut microbiome and metabolome in RA patients were associated with disease severity.14 The results revealed that elevated levels of fecal biotin in RA patients correlated with alterations in the gut microbiome. We then explored the contribution of diet- and microbiome-dependent biotin to the microbial homeostasis of RA. By using a CIA mouse model, we uncovered the effects of a biotin-free diet on disease development and microbial alterations in CIA mice.
2 Materials and methods
2.1 Collagen-induced arthritis in mice
DBA/1 mice (male, 7–8 week old) were obtained from Charles River Laboratories (Beijing, China). Arthritis was induced by intradermal injection of chick type II collagen emulsified with complete Freund's adjuvant at the base of the tail on day 1, followed by a booster immunization on day 21. Starting on day 28, disease severity was evaluated twice weekly. The severity of arthritis was graded by assessing the four limbs of the CIA mice based on established criteria.19 AIN-93 purified diets with and without biotin were purchased from Trophic Animal Feed High-tech Co., China. All protocols used in the mouse study received approval from the Research Ethics Committee of the Institute of Basic Theory of Chinese Medicine, China Academy of Chinese Medical Sciences.2.2 Targeted metabolomics of biotin and amino acids
We performed targeted metabolomics of 22 amino acids and biotin in serum and fecal samples. Two hundred microliters of methanol containing internal standards were added to 50 μL of serum to extract the metabolites, and a vortex was applied. Thirty-five grams of feces were homogenized in 500 μL of 80% cold methanol with internal standards. The internal standards included 4-chloro-phenylalanine, D3-methionine, D5-tryptophan and D4-biotin. The supernatants were lyophilized after centrifugation. The resulting residues were reconstituted in 85% acetonitrile with 0.1% formic acid. Targeted metabolomics was conducted using a previously described method.202.3 Mouse microbiota analysis
Feces from DBA/1 mice were collected and promptly frozen in liquid nitrogen. Feces were stored at −80 °C until analysis. High-quality total DNA for sequencing the gene encoding 16S rRNA was extracted from the frozen stool samples using an E.Z.N.A.® Stool DNA Kit. Qualified libraries were sequenced on a HiSeq System (HiSeq SBS Kit V2, Illumina).2.4 Subpopulation of T lymphocyte analysis
A single-cell suspension was harvested from the mouse spleen in a cell staining buffer. After lysing red blood cells, the cell surface was stained with a Mouse T Lymphocyte Subset Antibody Cocktail (PE-Cy™7 CD3e, PE CD4, FITC CD8; BD Biosciences) according to the manufacturer's instructions. Then samples were measured using an Accuri™ C6 flow cytometer (BD Biosciences, San Jose, CA).2.5 Cytokine analysis
Serum levels of IL-1β, TNF-α, IFN-γ, IL-6, IL-4, IL-10 and IL-17A in model mice were measured using Bio-Plex (Bio-Rad) and a Luminex instrument according to the manufacturer's instructions.2.6 Micro-computed tomography
Micro-computed tomography (micro-CT) imaging and analysis were performed using a vivaCT 40 system (Scanco Medical, Switzerland). Imaging was conducted at a voxel size of 17.5 μm, with a voltage of 70 kVp, a current of 114 μA, and an integration time of 200 ms. The region of interest in the tibial proximal metaphysis started 0.25 mm distal to the growth plate. Additionally, cortical bone from the midshaft of the tibia was analyzed.2.7 Statistical analysis
Statistical analysis was conducted using R and SPSS. Data distribution was examined by using the Shapiro–Wilk test. Associations between metabolites and the gut microbiome were assessed by Spearman's rank correlation test. For the mouse study, one-way ANOVA was performed to assess the significance of differential abundance. Statistical significance was considered at p < 0.05.3 Results
3.1 Elevated biotin in RA patients correlates with the gut microbiome
Emerging research suggested that the gut microbiota and its products may contribute to the pathogenesis of RA.5,13,21 Fecal biotin levels in RA individuals of the discovery set14 were higher than those in healthy control subjects, whereas serum biotin levels did not show significant differences between the two groups (Fig. 1A). We also found that bacterial genes related to biotin metabolism were also increased in RA individuals (Fig. 1B). In addition, fecal biotin and bacterial genes related to biotin metabolism were significantly positively associated with the phylum Bacteroidetes (r = 0.405 and r = 0.709, respectively, p < 0.01) but negatively associated with Firmicutes (r = −0.576, p < 0.01) (Fig. 1C), which was consistent with the distribution of genes related to biotin biosynthesis.22 Therefore, we assumed that the upregulation of fecal biotin in RA patients resulted from a higher proportion of bacterial biosynthesis.To investigate the association between the altered microbiome and biotin, we performed Spearman rank correlation analysis (Fig. 1D). Specifically, biotin was inversely associated with control-enriched gut microbial species, including Dorea (r = −0.359, p = 0.001) (Table S1†). In contrast, biotin content was positively correlated with RA-enriched gut microbiome constituents, such as Oscillospira (r = 0.234, p = 0.028), Bosea (r = 0.266, p = 0.016), Bilophila (r = 0.262, p = 0.014), and so on. We also found that biotin metabolism was inversely related to Dorea (r = −0.406, p < 0.001) and Roseburia (r = −0.383, p < 0.001). Besides, biotin metabolism was positively correlated with RA-enriched gut microbiome constituents, including Bosea (r = 0.360, p < 0.001) and Bilophila (r = 0.405, p < 0.001). Among these genera, Roseburia was negatively correlated with the levels of the erythrocyte sedimentation rate (ESR) (Fig. S1†), a diagnostic marker of inflammation in the body.1 In addition, the genus Oscillospira was highly enriched in RA patients and positively correlated with ESR levels (Fig. S1†).
It is worth emphasizing that biotin is involved in pathways of lipids, amino acids and carbohydrates16 evidenced by the correlation between bacterial genes for biotin metabolism and other pathways (Fig. 1E). Genes for biotin metabolism were positively related to some lipid pathways, including glycosphingolipid biosynthesis, linoleic acid metabolism and sphingolipid metabolism, but negatively correlated with glycerophospholipid metabolism and glycerolipid metabolism. The first-ranking bacterial genes related to D-alanine metabolism may be associated with the decreased bacterial richness in RA patients.23 Furthermore, biotin metabolism was positively associated with histidine metabolism, glycine, serine and threonine metabolism, and valine, leucine and isoleucine degradation. The close association between biotin and the amino acid-related pathway indicated that biotin played an important role in the process of amino acid metabolism. Taken together, our results suggested that the increased biotin may result from the increased proportion of biotin-generated microbiome in RA individuals, which further perturbed the host metabolic phenotype, such as amino acid metabolism.
3.2 Biotin-free diet attenuates the incidence of collagen-induced arthritis
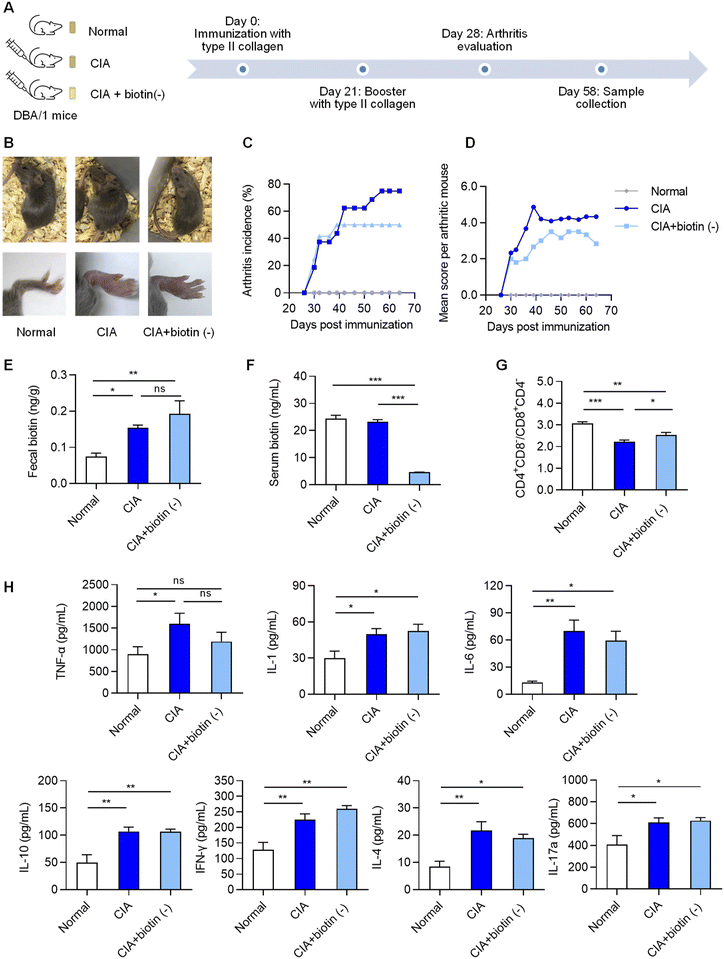
Based on the metabolomics study of clinical samples, biotin was identified as a key perturbed metabolite altered by the gut microbiome under RA inflammation. Since biotin is a diet- and microbiome-dependent metabolite and an immune regulator,24 we hypothesized that biotin metabolism linked to the symbiotic gut microbiota might contribute to the pathogenesis of RA. To investigate the function of biotin in RA, the most commonly used autoimmune model of RA, the CIA model, was used. We removed the biotin intake of CIA mice by providing a biotin-free diet (Fig. 2A). CIA mice with deprivation of dietary biotin did not show any signs of body weight loss, growth defects and alopecia compared to CIA mice on a normal diet (Fig. 2B and Fig. S2A†). Notably, the arthritic incidence and the severity of inflammation were lower than those of the CIA model group on a normal diet (Fig. 2C and D). Biotin was targeted and monitored to assess the biotin-deficient state in vivo. Fecal biotin content in CIA mice was significantly increased compared to that in normal mice (Fig. 2E), which was in accordance with the results of the clinical study. The biotin-free diet reduced serum biotin but did not significantly change the fecal biotin amount in CIA mice compared with that in mice fed a normal diet (Fig. 2E and F). This suggested that the biotin-free diet promoted a biotin-deficient state in the host, which may contribute to a lower incidence of arthritis.3.3 Biotin-free diet affects host immunity
CD4+ T cells and CD8+ T cells are central mediators in the pathological process of RA, clonally expanding and differentiating into cytokine-producing effector cells.25,26 To assess the effect of a biotin-free diet on host immunity, splenic CD4+ Th cells and CD8+ Tc cells were determined. There was a decrease in the CD4+/CD8+ T cell ratio in CIA mice compared to normal mice (Fig. 2G). The biotin-free diet restored the splenic CD4+/CD8+ T cell ratio of CIA mice (Fig. 2G), suggesting that the biotin-free diet may affect the immune system of CIA mice. Further analysis revealed decreases in the splenic CD3+CD4+ cell percentage and splenic CD3+ cell percentage in CIA mice fed a normal diet compared with those in normal mice (Fig. S2B†). The finding suggested that the subpopulations of naive T cells were altered under the rheumatic state, which may contribute to autoimmune conditions. Apparently, a biotin-free diet recovered the CD3+ and CD3+CD4+ cell percentages of CIA mice (Fig. S2B†). The result indicated that a biotin-free diet may change the T cell subpopulations, leading to depressed autoimmune activity.To assess the relationship between a biotin-free diet and inflammation, we detected several inflammatory cytokines in serum, which have been presumed to be critical players in the pathogenesis of RA, such as TNF-α and IL-6.1 The levels of inflammatory cytokines increased in CIA mice fed a normal diet compared to those in normal mice due to the activation of autoimmunity (Fig. 2H). However, the expression of inflammatory cytokines was not significantly changed in CIA mice fed a biotin-deficient diet compared to that in CIA mice fed a normal diet.
3.4 Biotin-free diet prevents inflammation-induced bone loss
To determine whether a biotin-free diet affects bone mass, we analyzed bone mass in the trabecular and cortical parts of the tibias. Micro-computed tomography (micro-CT) analysis showed that biotin-free diet treatment significantly reduced the bone erosion in the hind paws together with increased bone mass as shown by increased trabecular thickness and trabecular bone volume per tissue volume in CIA mice (Fig. 3A and B). In addition, obvious effects on bone mass and bone mineral density in cortical bone were observed in CIA mice treated with a biotin-free diet (Fig. 3C), suggesting that the biotin-free diet had protection from inflammation and inflammation-induced bone loss.3.5 Biotin-free diet affects the gut microbiome
We also analyzed gut microbiome compositions and proportions to investigate the gut microbiome alteration under biotin-deficient conditions. The results showed that the biotin-free diet significantly increased the microbial diversity of CIA mice, including ACE diversity, observed diversity and Fisher diversity (Fig. 4A). Principal component analysis (PCA) at the OTU level suggested a heterogeneous microbial community structure among the three groups (Fig. 4B and C). The results demonstrated the specific and predominant genera of different groups of mice, revealing that Oscillospira was predominant in the CIA group and was decreased in CIA mice on a biotin-free diet (Fig. 4D). The beneficial genera Roseburia and Dorea, and inflammation-associated gut commensals, were predominant under biotin-deficient conditions, while the level of RA-enriched Prevotella was reversed.We performed PICRUSt2-based functional prediction using our 16S rRNA sequencing data to assess the relative abundance of microbial genes related to biotin metabolism and biotin biosynthesis (bioA, bioB, bioC, and bioF). Interestingly, we did not observe significant differences in the predicted abundance of these genes between the biotin-free diet group and the control group (Fig. S3A†). These findings suggest that while the biotin-free diet altered the gut microbiota composition, it did not appear to significantly affect the microbial community's overall potential for biotin metabolism and biosynthesis.
3.6 Biotin-free diet affects microbial and host metabolism
To investigate the effects of biotin deficiency on the gut microbiota and its subsequent impact on host metabolism, we employed an untargeted metabolomics study on mouse feces and serum samples as a comprehensive approach for capturing the dynamic metabolic shifts. Score plots of orthogonal partial least squares discriminant analysis (OPLS-DA) revealed metabolic shifts occurring within both the microbiome and host under biotin-free dietary conditions (Fig. 5A and B). Pathway analysis of fecal metabolites highlighted tryptophan metabolism as a key affected pathway (Fig. 5C), which was associated with inflammation.27 Targeted metabolomics of fecal samples demonstrated that biotin deficiency notably downregulated the bacterial synthesis of kynurenine, tyrosine, 3-indolepropionic acid (IPA), and their precursor metabolites, tryptophan and phenylalanine (Fig. 5D). Additionally, biotin deficiency significantly restored serum IPA levels (Fig. 5E), implying a reduction in inflammation.28Biotin is also involved in amino acid metabolism responding to host immunity.29 Pathway analysis of fecal and serum metabolites revealed significant alterations in amino acid-related pathways, including tryptophan metabolism, arginine biosynthesis, and glycine, serine, and threonine metabolism (Fig. 5C and Fig. S3B†). We detected 22 amino acids in serum and fecal samples from normal mice, CIA mice and biotin-deficient CIA mice to investigate their changes under rheumatic and biotin-deficient conditions. The serum levels of certain amino acids decreased in CIA model mice, while they increased in biotin-deficient-fed mice and were close to the levels of normal mice (Fig. 5F). For instance, glycine (FC = 0.84, p = 0.104), serine (FC = 0.75, p = 0.033), alanine (FC = 0.73, p = 0.008) and threonine (FC = 0.78, p = 0.022), which are upstream products of pyruvate, were downregulated in serum of CIA mice on a normal diet compared to normal mice. The decreasing tendency of these amino acids was consistent with a previous study and indicated the accumulation of pyruvate and a high rate of glycolysis in CIA mice, activating immune cells.30,31 The serum levels of glycine (FC = 1.07, p = 0.031), serine (FC = 1.02, p = 0.023), alanine (FC = 1.09, p = 0.001) and threonine (FC = 1.04, p = 0.01) were restored in biotin-deficient CIA mice compared to normal-diet CIA mice. For fecal levels of amino acids, most of them decreased under biotin-deficient conditions, except for serine, alanine, threonine and glutamate (Fig. 5F). Microbial amino acid metabolism was perturbed by the biotin-deficient diet despite no change in fecal biotin levels, suggesting that not only gut microbiome-derived biotin but also dietary biotin is implicated in the amino acid metabolism of the gut microbiome. These findings further supported the idea that biotin-free diet-induced biotin deprivation influences both microbial composition and host metabolic pathways.
4 Discussion
Gut dysbiosis-driven metabolic shifts and immune system dysfunction during RA progression have attracted increasing attention.5,13 RA status was associated with microbial imbalances, including abatement of some beneficial bacteria (i.e., Roseburia and Dorea) and augmentation of other bacteria (i.e., Oscillospira and Prevotella).14,32–35 The mutual interactions between the gut microbiota and RA have been increasingly recognized. On one hand, RA-related inflammation can alter gut permeability and immune signaling, thereby reshaping the microbial ecosystem.36–38 On the other hand, the altered gut microbiota may influence host immune responses through microbial metabolites (e.g., SCFAs and lipopolysaccharides), antigen mimicry, and modulation of T cell differentiation.13,39,40We found that microbiome-associated biotin content was increased in the individual feces in the RA state. The phyla Bacteroidetes, Fusobacteria and Proteobacteria contain essential genes for biotin biosynthesis, but the synthesis of biotin occurs rarely in the Firmicutes phylum.22 Most Actinobacteria genomes lack the complete pathway for biotin biosynthesis but retain biotin transport systems such as BioY, suggesting a reliance on exogenous biotin.22 The gut microbiota (e.g. Bacteroides fragilis and Escherichia coli) can synthesize biotin and secrete it into the environment, while certain bacteria cannot synthesize biotin themselves but possess biotin transporters to acquire biotin from the surroundings.22,41 Compared to dietary biotin, gut microbiome-produced biotin is predominantly absorbed in the colon.42 The amount of biotin uptake sometimes is more than that of dietary biotin, indicating that gut microbiome-derived biotin participates in host metabolism.43
Emerging research has proved that dietary restrictions, i.e., fasting, methionine restriction and caloric restriction, can modulate host innate immunity and affect the pathological process through metabolic regulation and the gut microbiome.44–46 Previous studies have reported that biotin impacted the functions of adaptive immune T and NK cells.47 Moreover, biotin deficiency enhanced the inflammatory response via elevated levels of cytokines in normal cell models, such as TNF-α, IFN-γ and IL-17.18,47 In this study, a biotin-free diet led to a deficient state of host biotin and a considerable decrease in the arthritis incidence of CIA mice by changing the differentiation of naive T cells. Biotin deprivation attenuated the subpopulations of spleen lymphocytes in BALB/cAnN mice.48 Immune responses to antigens were depressed by biotin deficiency, resulting in the inhibition of the development of experimental allergic encephalomyelitis in rats.49 That is to say, biotin deprivation restrained immunologic responses in experimentally induced autoimmunity, which was similar to our study. Although biotin deficiency markedly altered the gut microbial composition, functional prediction did not reveal significant differences in the abundance of microbial genes related to biotin biosynthesis (e.g., bioA, bioB, bioC, and bioF). This indicates that the microbial community's capacity for biotin production may not be the primary factor linking dietary biotin deficiency to arthritis attenuation. Instead, the beneficial effects may stem from broader shifts in microbial taxa and the host immune response, rather than direct modulation of microbially derived biotin.
Biotin is critical for amino acid metabolism, glycolysis and fatty acid metabolism responding to host immunity.24,29,50 Bacteria, such as Bacteroidetes, Fusobacteria, and Proteobacteria, e.g., E. coli, supply biotin to the host under biotin-scarce conditions.41 Interestingly, while biotin deficiency suppressed certain microbial metabolites, it also significantly restored serum IPA levels, implying a potential compensatory response to reduce inflammation in the host. This suggested that biotin deficiency disrupted microbial metabolism and triggered adaptive mechanisms in the host. Although fecal biotin levels were not affected, a biotin-free diet reshaped the metabolic phenotype of amino acids and the inflammation-associated gut commensals in CIA mice, such as Roseburia and Oscillospira. However, the underlying molecular mechanism by which biotin deficiency influences the homeostasis or function of immune cells is not yet known; previous research and our study suggest that the activation of the mTOR signaling pathway played a role in that.18 In addition, future studies are required to include both biotin-supplemented groups and direct synovial tissue analyses to comprehensively evaluate the dose-dependent and local joint effects of biotin in arthritis progression. In a nutshell, our study revealed that biotin may modulate the immune homeostasis and susceptibility of the host to RA through the gut microbiome and metabolic regulation. The findings provided a better understanding of how the crosstalk between the microbiota and its derived metabolites influences the host immune system under RA inflammation.
Author contributions
XS: conceptualization, investigation, methodology, and writing – original draft. XL: conceptualization, investigation, and writing – review & editing. XH: methodology. ZC, AL and SZ: supervision, writing – review & editing, and funding acquisition.Data availability
The data supporting this article have been included as part of the ESI.†Conflicts of interest
The authors declare that they have no conflict of interest.Acknowledgements
This research was funded by the Interdisciplinary Research Matching Scheme (IRMS) of Hong Kong Baptist University (Project ID RC-IRMS/13-14/03).References
- J. S. Smolen, D. Aletaha and I. B. McInnes, Rheumatoid arthritis, Lancet, 2016, 388, 2023–2038 Search PubMed.
- Y. Tong, T. Marion, G. Schett, Y. Luo and Y. Liu, Microbiota and metabolites in rheumatic diseases, Autoimmun. Rev., 2020, 19, 102530 Search PubMed.
- J. U. Scher and S. B. Abramson, The microbiome and rheumatoid arthritis, Nat. Rev. Rheumatol., 2011, 7, 569 Search PubMed.
- M. M. Zaiss, H.-J. Joyce Wu, D. Mauro, G. Schett and F. Ciccia, The gut-joint axis in rheumatoid arthritis, Nat. Rev. Rheumatol., 2021, 17, 224–237 Search PubMed.
- X. Zhang, D. Zhang, H. Jia, Q. Feng, D. Wang, D. Liang, X. Wu, J. Li, L. Tang and Y. Li, The oral and gut microbiomes are perturbed in rheumatoid arthritis and partly normalized after treatment, Nat. Med., 2015, 21, 895–905 Search PubMed.
- I. Dorozynska, M. Majewska-Szczepanik, K. Marcinska and M. Szczepanik, Partial depletion of natural gut flora by antibiotic aggravates collagen induced arthritis (CIA) in mice, Pharmacol. Rep., 2014, 66, 250–255 Search PubMed.
- V. Taneja, Microbiome in 2016: T follicular helper cells and the gut microbiome in arthritis, Nat. Rev. Rheumatol., 2017, 13, 72–74 Search PubMed.
- H. Evans-Marin, R. Rogier, S. B. Koralov, J. Manasson, D. Roeleveld, P. M. van der Kraan, J. U. Scher, M. I. Koenders and S. Abdollahi-Roodsaz, Microbiota-dependent involvement of Th17 cells in murine models of inflammatory arthritis, Arthritis Rheumatol., 2018, 70, 1971–1983 Search PubMed.
- C. M. Weyand and J. J. Goronzy, The immunology of rheumatoid arthritis, Nat. Immunol., 2021, 22, 10–18 Search PubMed.
- M. S. Donia and M. A. Fischbach, Small molecules from the human microbiota, Science, 2015, 349, 1254766 Search PubMed.
- M. G. Rooks and W. S. Garrett, Gut microbiota, metabolites and host immunity, Nat. Rev. Immunol., 2016, 16, 341–352 Search PubMed.
- P. D. Cani, M. Van Hul, C. Lefort, C. Depommier, M. Rastelli and A. Everard, Microbial regulation of organismal energy homeostasis, Nat. Metab., 2019, 1, 34–46 Search PubMed.
- E. C. Rosser, C. J. M. Piper, D. E. Matei, P. A. Blair, A. F. Rendeiro, M. Orford, D. G. Alber, T. Krausgruber, D. Catalan, N. Klein, J. J. Manson, I. Drozdov, C. Bock, L. R. Wedderburn, S. Eaton and C. Mauri, Microbiota-derived metabolites suppress arthritis by amplifying aryl-hydrocarbon receptor activation in regulatory B cells, Cell Metab., 2020, 31, 837–851 Search PubMed.
- X. Su, X. Li, Y. Bian, Q. Ren, L. Li, X. Wu, H. Luan, B. He, X. He and H. Feng, Microbiome-derived bile acids contribute to elevated antigenic response and bone erosion in rheumatoid arthritis, arXiv, 2023, preprint, arXiv:2307.08848, DOI:10.48550/arXiv.2307.08848.
- H. M. Said, Cell and molecular aspects of human intestinal biotin absorption, J. Nutr., 2009, 139, 158–162 CrossRef CAS PubMed.
- A. León-Del-Río, V. Valadez-Graham and R. A. Gravel, Holocarboxylase synthetase: a moonlighting transcriptional coregulator of gene expression and a cytosolic regulator of biotin utilization, Annu. Rev. Nutr., 2017, 37, 207–223 Search PubMed.
- J. Skupsky, S. Sabui, M. Hwang, M. Nakasaki, M. D. Cahalan and H. M. Said, Biotin supplementation ameliorates murine colitis by preventing NF-κB activation, Cell. Mol. Gastroenterol. Hepatol., 2020, 9, 557–567 Search PubMed.
- A. Elahi, S. Sabui, N. N. Narasappa, S. Agrawal, N. W. Lambrecht, A. Agrawal and H. M. Said, Biotin deficiency induces Th1-and Th17-mediated proinflammatory responses in human CD4+ T lymphocytes via activation of the mTOR signaling pathway, J. Immunol., 2018, 200, 2563–2570 Search PubMed.
- D. D. Brand, K. A. Latham and E. F. Rosloniec, Collagen-induced arthritis, Nat. Protoc., 2007, 2, 1269–1275 Search PubMed.
- X. Su, X. Li, H. Wang and Z. Cai, Simultaneous determination of methionine cycle metabolites, urea cycle intermediates and polyamines in serum, urine and intestinal tissue by using UHPLC-MS/MS, Talanta, 2021, 224, 121868 Search PubMed.
- J. U. Scher, A. Sczesnak, R. S. Longman, N. Segata, C. Ubeda, C. Bielski, T. Rostron, V. Cerundolo, E. G. Pamer and S. B. Abramson, Expansion of intestinal Prevotella copri correlates with enhanced susceptibility to arthritis, eLife, 2013, 2, e01202 Search PubMed.
- S. Magnúsdóttir, D. Ravcheev, V. de Crécy-Lagard and I. Thiele, Systematic genome assessment of B-vitamin biosynthesis suggests co-operation among gut microbes, Front. Genet., 2015, 6, 148 Search PubMed.
- W. Qiu, X. Zheng, Y. Wei, X. Zhou, K. Zhang, S. Wang, L. Cheng, Y. Li, B. Ren and X. Xu, d–Alanine metabolism is essential for growth and biofilm formation of Streptococcus mutans, Mol. Oral Microbiol., 2016, 31, 435–444 CrossRef CAS PubMed.
- J. Zempleni and T. Kuroishi, Biotin, Adv. Nutr., 2012, 3, 213–214 Search PubMed.
- D. A. Rao, M. F. Gurish, J. L. Marshall, K. Slowikowski, C. Y. Fonseka, Y. Liu, L. T. Donlin, L. A. Henderson, K. Wei, F. Mizoguchi, N. C. Teslovich, M. E. Weinblatt, E. M. Massarotti, J. S. Coblyn, S. M. Helfgott, Y. C. Lee, D. J. Todd, V. P. Bykerk, S. M. Goodman, A. B. Pernis, L. B. Ivashkiv, E. W. Karlson, P. A. Nigrovic, A. Filer, C. D. Buckley, J. A. Lederer, S. Raychaudhuri and M. B. Brenner, Pathologically expanded peripheral T helper cell subset drives B cells in rheumatoid arthritis, Nature, 2017, 542, 110–114 Search PubMed.
- R. Winchester, J. T. Giles, S. Nativ, K. Downer, H. Z. Zhang, A. Bag-Ozbek, A. Zartoshti, S. Bokhari and J. M. Bathon, Association of elevations of specific T cell and monocyte subpopulations in rheumatoid arthritis with subclinical coronary artery atherosclerosis, Arthritis Rheumatol., 2016, 68, 92–102 Search PubMed.
- H. M. Roager and T. R. Licht, Microbial tryptophan catabolites in health and disease, Nat. Commun., 2018, 9, 3294 Search PubMed.
- D. Dodd, M. H. Spitzer, W. V. Treuren, B. D. Merrill, A. J. Hryckowian, S. K. Higginbottom, A. Le, T. M. Cowan, G. P. Nolan and M. A. Fischbach, A gut bacterial pathway metabolizes aromatic amino acids into nine circulating metabolites, Nature, 2017, 3, e00438 Search PubMed.
- J. R. Brestoff and D. Artis, Commensal bacteria at the interface of host metabolism and the immune system, Nat. Immunol., 2013, 14, 676–684 Search PubMed.
- J. Zhou, J. Chen, C. Hu, Z. Xie, H. Li, S. Wei, D. Wang, C. Wen and G. Xu, Exploration of the serum metabolite signature in patients with rheumatoid arthritis using gas chromatography-mass spectrometry, J. Pharm. Biomed. Anal., 2016, 127, 60–67 Search PubMed.
- R. Garcia-Carbonell, A. S. Divakaruni, A. Lodi, I. Vicente-Suarez, A. Saha, H. Cheroutre, G. R. Boss, S. Tiziani, A. N. Murphy and M. Guma, Critical role of glucose metabolism in rheumatoid arthritis fibroblast–like synoviocytes, Arthritis Rheumatol., 2016, 68, 1614–1626 Search PubMed.
- Y. Li, S.-X. Zhang, X.-F. Yin, M.-X. Zhang, J. Qiao, X.-H. Xin, M.-J. Chang, C. Gao, Y.-F. Li and X.-F. Li, The gut microbiota and its relevance to peripheral lymphocyte subpopulations and cytokines in patients with rheumatoid arthritis, J. Immunol. Res., 2021, 2021, 6665563 Search PubMed.
- Y. Sun, Q. Chen, P. Lin, R. Xu, D. He, W. Ji, Y. Bian, Y. Shen, Q. Li and C. Liu, Characteristics of gut microbiota in patients with rheumatoid arthritis in Shanghai, China, Front. Cell. Infect. Microbiol., 2019, 9, 369 Search PubMed.
- Z. Liu, Y. Wu, Y. Luo, S. Wei, C. Lu, Y. Zhou, J. Wang, T. Miao, H. Lin, Y. Zhao, Q. Liu and Y. Liu, Self-balance of intestinal flora in spouses of patients with rheumatoid arthritis, Front. Med., 2020, 7, 538 Search PubMed.
- V. K. Gupta, K. Y. Cunningham, B. Hur, U. Bakshi, H. Huang, K. J. Warrington, V. Taneja, E. Myasoedova, J. M. Davis and J. Sung, Gut microbial determinants of clinically important improvement in patients with rheumatoid arthritis, Genome Med., 2021, 13, 149 Search PubMed.
- R. Audo, P. Sanchez, B. Rivière, J. Mielle, J. Tan, C. Lukas, L. Macia, J. Morel and C. I. Daien, Rheumatoid arthritis is associated with increased gut permeability and bacterial translocation that are reversed by inflammation control, Rheumatology, 2022, 62, 1264–1271 Search PubMed.
- C. Heidt, U. Kämmerer, M. Fobker, A. Rüffer, T. Marquardt and M. Reuss-Borst, Assessment of Intestinal Permeability and Inflammation Bio-Markers in Patients with Rheumatoid Arthritis, Nutrients, 2023, 15, 2386 Search PubMed.
- Y. Maeda and K. Takeda, Host-microbiota interactions in rheumatoid arthritis, Exp. Mol. Med., 2019, 51, 1–6 Search PubMed.
- S. Lucas, Y. Omata, J. Hofmann, M. Böttcher, A. Iljazovic, K. Sarter, O. Albrecht, O. Schulz, B. Krishnacoumar, G. Krönke, M. Herrmann, D. Mougiakakos, T. Strowig, G. Schett and M. M. Zaiss, Short-chain fatty acids regulate systemic bone mass and protect from pathological bone loss, Nat. Commun., 2018, 9, 55 Search PubMed.
- J. He, Y. Chu, J. Li, Q. Meng, Y. Liu, J. Jin, Y. Wang, J. Wang, B. Huang, L. Shi, X. Shi, J. Tian, Y. Zhufeng, R. Feng, W. Xiao, Y. Gan, J. Guo, C. Shao, Y. Su, F. Hu, X. Sun, J. Yu, Y. Kang and Z. Li, Intestinal butyrate-metabolizing species contribute to autoantibody production and bone erosion in rheumatoid arthritis, Sci. Adv., 2022, 8, eabm1511 Search PubMed.
- C. Neophytou and C. Pitsouli, Biotin controls intestinal stem cell mitosis and host-microbiome interactions, Cell Rep., 2022, 38, 110505 CrossRef CAS PubMed.
- J. G. LeBlanc, C. Milani, G. S. de Giori, F. Sesma, D. van Sinderen and M. Ventura, Bacteria as vitamin suppliers to their host: a gut microbiota perspective, Curr. Opin. Biotechnol., 2013, 24, 160–168 Search PubMed.
- M. G. Langille, C. J. Meehan, J. E. Koenig, A. S. Dhanani, R. A. Rose, S. E. Howlett and R. G. Beiko, Microbial shifts in the aging mouse gut, Microbiome, 2014, 2, 50 CrossRef PubMed.
- Z. Wu, M. Isik, N. Moroz, M. J. Steinbaugh, P. Zhang and T. K. Blackwell, Dietary restriction extends lifespan through metabolic regulation of innate immunity, Cell Metab., 2019, 29, 1192–1205 Search PubMed.
- C. Barcena, P. M. Quiros, S. Durand, P. Mayoral, F. Rodriguez, X. M. Caravia, G. Marino, C. Garabaya, M. T. Fernandez-Garcia, G. Kroemer, J. M. P. Freije and C. Lopez-Otin, Methionine restriction extends lifespan in progeroid mice and alters lipid and bile acid metabolism, Cell Rep., 2018, 24, 2392–2403 Search PubMed.
- M. C. Dao, A. Everard, J. Aron-Wisnewsky, N. Sokolovska, E. Prifti, E. O. Verger, B. D. Kayser, F. Levenez, J. Chilloux and L. Hoyles, Akkermansia muciniphila and improved metabolic health during a dietary intervention in obesity: relationship with gut microbiome richness and ecology, Gut, 2016, 65, 426–436 Search PubMed.
- S. Agrawal, A. Agrawal and H. M. Said, Biotin deficiency enhances the inflammatory response of human dendritic cells, Am. J. Physiol.: Cell Physiol., 2016, 311, C386–C391 CrossRef PubMed.
- A. Báez-Saldaña, G. Díaz, B. Espinoza and E. Ortega, Biotin deficiency induces changes in subpopulations of spleen lymphocytes in mice, Am. J. Clin. Nutr., 1998, 67, 431–437 Search PubMed.
- B. S. Rabin, Inhibition of experimentally induced autoimmunity in rats by biotin deficiency, J. Nutr., 1983, 113, 2316–2322 Search PubMed.
- J. K. Nicholson, E. Holmes, J. Kinross, R. Burcelin, G. Gibson, W. Jia and S. Pettersson, Host-gut microbiota metabolic interactions, Science, 2012, 336, 1262–1267 Search PubMed.
Footnotes |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d5fo01457c |
| ‡ Both authors contributed equally to this work. |
| This journal is © The Royal Society of Chemistry 2025 |