Open Access Article
Open Access ArticleBioconversion of a lignin-derived biphenyl dimer into the strategic building block 5-carboxyvanillic acid in Pseudomonas putida KT2440†
Helena
Gómez-Álvarez‡
 *ac,
Carlos
del Cerro-Sánchez‡
*ac,
Carlos
del Cerro-Sánchez‡
 ac,
Pablo
Iturbe
a,
Virginia
Rivero-Buceta
ac,
Juan
Nogales
bc,
Timothy D. H.
Bugg
ac,
Pablo
Iturbe
a,
Virginia
Rivero-Buceta
ac,
Juan
Nogales
bc,
Timothy D. H.
Bugg
 d and
Eduardo
Díaz
d and
Eduardo
Díaz
 *ac
*ac
aDepartment of Biotechnology, Margarita Salas Center for Biological Research, Spanish National Research Council, Ramiro de Maeztu 9, 28040 Madrid, Spain. E-mail: ediaz@cib.csic.es; hgomalv@cib.csic.es
bDepartment of Systems Biology, National Centre for Biotechnology, Spanish National Research Council, Darwin 3, 28049 Madrid, Spain
cInterdisciplinary Platform for Sustainable Plastics towards a Circular Economy-Spanish National Research Council (SusPlast-CSIC), Madrid, Spain
dDepartment of Chemistry, University of Warwick, Coventry CV4 7AL, UK
First published on 27th February 2025
Abstract
The design of new biocatalysts for funneling lignin depolymerization-derived dimers into added-value compounds is nowadays a major challenge in biological lignin valorization. Biphenyl 5,5′-dehydrodivanillate (DDVA) is a model-lignin dimer that contains the C5–C5′ linkage commonly found in lignin depolymerization mixtures. In this work, the metabolic potential of the industrially relevant Pseudomonas putida KT2440 bacterial strain was broadened by expressing synthetic DNA modules encoding selected metabolic and transport steps from the well-characterized DDVA degradation pathway of the Sphingobium lignivorans SYK-6 strain. By employing this heterologous expression strategy, we successfully developed an unprecedented resting cell-based bioprocess to convert DDVA into 5-carboxyvanillic acid (5CVA), a promising building block for the production of innovative bio-based polymers. This proof-of-concept study underscores the essential role of the associated DDVA transport systems. Furthermore, the findings reveal that P. putida KT2440 serves as an effective bacterial chassis for biotechnological processes that require the uptake of substrates through specific TonB-dependent transporters.
Green foundation1. Sustainable management of lignin waste has predominantly focused on valorizing monomers, and limited studies have explored the conversion of lignin-derived dimers. Here, we present a green strategy for producing the key building block 5-carboxyvanillic acid (5CVA) using the biphenyl-type lignin dimer DDVA (biphenyl 5,5′-dehydrodivanillate) as the substrate.2. The process was validated in the Pseudomonas putida KT2440 strain by expressing key metabolic steps of the DDVA degradation pathway from Sphingobium lignivorans SYK-6. DDVA uptake, a major bottleneck in the bioconversion, was addressed by cloning outer (DdvT) and inner (DdvK) membrane transporters, coupled with optimizing resting cell conditions, and a 5CVA yield of 30% was achieved. 3. Further research is required to: (i) optimize bacterial biocatalysts and the process to enhance 5CVA yields; (ii) implement the bioconversion using depolymerized lignin samples as feedstock; and (iii) scale up the bioprocess to meet industrial relevance. |
Introduction
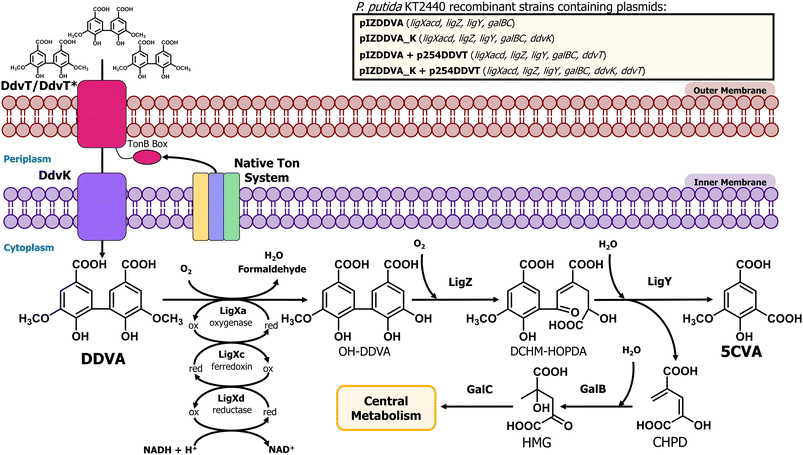
In our societies, there is an urgent need for innovative waste management solutions that contribute to a transition towards a more sustainable circular economy. In particular, due to lignin recalcitrance, lignin recycling constitutes a challenge in the management of lignocellulosic biomass. After cellulose, lignin is considered the second most abundant plant-based biopolymer on Earth accounting for 30% of the organic carbon in the biosphere1 and representing up to 40% of the energy accumulated in lignocellulosic biomass.2 Lignin is extracted in amounts higher than 50 Mt per year3 and is mostly considered a residue, usually burned for its fuel value.4 However, as a massive reservoir of carbon and energy, lignin possesses a high potential value for being valorized through green biotechnology processes. In particular, from a circular economy perspective, the bio-conversion of lignin and its heterogeneous derived compounds to value-added products has emerged as an especially relevant strategy, and genetically modified bacteria have been used for the production of compounds of industrial relevance such as muconic acid,5,6 adipic acid,7 2-pyrone-4,6-dicarboxylic acid,8–10 β-ketoadipic acid,11,12 pyridine dicarboxylic acids,13–15 substituted styrene molecules,16 vanillin,17 or polyhydroxyalkanoates,18–20 among others.21Chemo-catalytic lignin depolymerization methods mostly cleave carbon–oxygen linkages such as β-O-4 aryl ether bonds, thus enriching lignin streams in dimers and oligomers with carbon–carbon linkages.22 One of the most prevalent carbon–carbon linkages between benzene rings in lignin is the C5–C5′ linkage found in biphenyl dimers.23–25 Kraft lignin contains additional biphenyl units caused by radical coupling reactions.26 Therefore, designing biological systems capable of bioconverting recalcitrant lignin-derived biphenyls into value-added products could significantly enhance lignin waste management. This approach would expand current metabolic funneling processes, which are primarily focused on valorizing monomers derived from the cleavage of carbon–oxygen linkages. In particular, 5,5′-dehydrodivanillate (DDVA) has been used as a model of lignin-derived biphenyl compounds and it can be obtained as a result of different lignin depolymerization methods. Thus, this chemical can be generated via fungal depolymerization processes, as in the case of wood decay by Phanerochaete chrysosporium,27,28 as well as by using physico-chemical procedures like the alkaline nitrobenzene oxidation of lignin samples29 or as an oxidation product of lignosulfonates.30 Alternatively, DDVA can be obtained through enzymatic oligomerization of vanillic acid, another lignin-derived product, using fungal laccases.31 The DDVA degradation pathway has been well characterized in the bacterium Sphingobium lignivorans SYK-6.32 The first step of DDVA catabolism includes an O-demethylation carried out by a multicomponent oxygenase composed of LigXa (oxygenase), LigXc (ferredoxin) and LigXd (ferredoxin reductase) subunits, generating 2,2′,3-trihydroxy-3′-methoxy-5,5′-dicarboxybiphenyl (OH-DDVA) (Fig. 1).33,34 In the second step, the extradiol dioxygenase LigZ is able to cleave one of the aromatic rings generating a meta-ring cleavage compound (4,11-dicarboxy-8-hydroxy-9-methoxy-2-hydroxy-6-oxo-6-phenylhexa-2,4-dienoate) (DCHM-HOPDA) that is subsequently hydrolyzed by the LigY hydrolase to yield 5-carboxyvanillate (5CVA) and 4-carboxy-2-hydroxypenta-2,4-dienoate (CHPD)35,36 (Fig. 1). 5CVA is converted to vanillate by the LigW and LigW2 decarboxylases37,38 and then redirected to the central metabolism through the protocatechuate 4,5-cleavage pathway.39
Interestingly, 5CVA, an intermediate in the DDVA pathway, is a structural analogue of isophthalic acid but with enhanced oxygen functionalities. This makes it a highly promising building block for the development of innovative bioplastics. In fact, using artificial intelligence-based approaches, it has been recently demonstrated that 5CVA presents optimal characteristics as a novel building block for the production of bio-based polymers. Notably, this diacid can be polymerized into poly(ethylene 5-carboxyvanillate) (PEC), a polyester that exhibits superior properties compared to polyethylene terephthalate (PET).40 With the growing concern over the environmental impact of petrochemical-based plastics,41 the development of bioplastics from renewable feedstocks offers a crucial and sustainable alternative. In support of this goal, our study introduces a novel biotechnological approach for the bioconversion of a lignin-derived aromatic dimer, DDVA, into the valuable 5CVA building block.
P. putida KT2440 is a non-pathogenic industrially relevant bacterium that has been extensively used for lignin valorization.6,10,12,15,20,21,42–44 In this work, a genetically engineered P. putida KT2440 strain was developed to express a specific set of genes from S. lignivorans SYK6, enabling the bioconversion of DDVA into 5CVA. A successful growth-independent biotransformation, known as a resting cell process, was established as a novel procedure for the production of 5CVA.
Experimental
Chemicals, bacterial strains, plasmids and growth conditions
All chemicals were purchased from Merck unless otherwise stated. DDVA and 5CVA were purchased from Biosynth and APIN Chemicals Ltd, respectively. DDVA and 5CVA 0.05 M stock solutions were prepared in water after neutralization with equimolar amounts of NaOH and stored at −20 °C. The bacterial strains and plasmids used in this work are detailed in Table 1. P. putida KT2440 strains and S. lignivorans SYK-6 were cultivated in liquid lysogeny broth (LB) medium45 at 30 °C and 200 rpm agitation. M63 minimal medium was also used to grow P. putida KT2440 strains.46Escherichia coli strains were cultivated in LB medium at 37 °C and 200 rpm agitation. When required, gentamicin (10 μg ml−1) or kanamycin (50 μg ml−1) was added to the growth medium.| Relevant genotype | Reference or source | |
|---|---|---|
| Bacterial strains | ||
| E. coli DH10B | F-, mcrA Δ(mrr-hsdRMS-mcrBC) ϕ80dlacZΔM15 ΔlacX74 deoR recA1 araD139 Δ(ara-leu)7697 galU galK rpsL endA1 nupG | Invitrogen |
| P. putida KT2440 | Wild-type strain | Bagdasarian et al.47 |
| S. lignivorans SYK-6 | Wild-type strain | Masai et al.32 |
| Plasmids | ||
| pIZ1016 | GmR, broad-host range expression vector bearing lacIq and Ptac, and the pBBR1MCS replication origin | Moreno-Ruiz et al.48 |
| pSEVA254 | KmR, broad-host range expression vector bearing lacIq and Ptrc, and the RSF1010 replication origin | Silva-Rocha et al.49 |
| pSEVA234 | KmR, broad-host range expression vector bearing lacIq and Ptrc, and the pBBR1 replication origin | Silva-Rocha et al.49 |
| pSEVA238 | KmR, broad-host range expression vector bearing xylS and Pm, and the pBBR1 replication origin | Silva-Rocha et al.49 |
| pIZDDVA | GmR, pIZ1016 derivative expressing ligXaXcXd, ligZ, ligY, galBC under the lacIq/Ptac regulatory couple. | This work |
| pIZDDVA_K | GmR, pIZ1016 derivative expressing ligXaXcXd, ligZ, ligY, ddvK, galBC under the lacIq/Ptac regulatory couple | This work |
| p254DDVT | KmR, pSEVA254 derivative expressing ddvT under the lacIq/Ptrc regulatory couple | This work |
| p254DDVTB | KmR, pSEVA254 derivative expressing a chimeric ddvT* gene with the consensus KT2440 TonB box “LELPATVITA” under the lacIq/Ptrc regulatory couple | This work |
Molecular biology techniques
Standard molecular biology techniques were performed as previously described.45 Plasmid DNA extractions were performed using a High Pure Plasmid Isolation Kit (Roche Applied Science). DNA fragments were purified from agarose gels or PCR mixtures with a Gene Extraction Kit or a PCR Purification Kit (Qiagen), respectively. Gibson Assembly reactions were performed using an NEBuilder HiFi DNA Assembly Cloning Kit (New England Biolabs). The oligonucleotides employed (see Table S1 of the ESI†) were supplied by IDT. All cloned inserts and DNA fragments were confirmed by DNA sequencing in an ABI Prism 377 automated DNA sequencer. Transformation of E. coli cells was carried out by using the RbCl method.45 Plasmids were transferred to P. putida KT2440 cells by electroporation (Gene Pulser, Bio-Rad).45Construction of plasmids pIZDDVA, pIZDDVA_K, p254DDVT, and p254DDVTB
The original signal peptide of the DdvT protein has been maintained for its heterologous expression in P. putida KT2440.
DDVA bioconversion in P. putida resting cell assays
1 mL of an overnight seed culture of the different P. putida KT2440 recombinant strains, containing different combinations of plasmids pIZ1016, pSEVA254, pIZDDVA, pIZDDVA_K, p254DDVT or p254DDVTB and grown in LB with kanamycin and gentamycin, was used to inoculate 200 mL of fresh LB medium without antibiotics. They were incubated for 3 h at 30 °C. 1 mM IPTG was then added to induce expression of galBC, ligXacdYZ, ddvT, and ddvK genes, followed by an additional 24 h incubation period. Cells (equivalent to an OD600 of 70 or to 0.28 ± 0.05 g dry cell weight) were collected at 8500 rpm for 15 minutes at 15 °C and then resuspended in 10 mL of the resting cell medium, unless otherwise stated: 50 mM sodium phosphate buffer pH 7.5 supplemented with the aromatic substrate (1 mM DDVA) and 0.1% glucose. For biotransformations performed with cells grown in M63 mineral medium, this was supplemented with 1 mM MgSO4, trace elements and 0.034 mM EDTA as previously described50 and either 20 mM glucose or 15 mM sodium octanoate (carbon/nitrogen ratio equal to 4 (mol/mol) in both cases) as carbon sources. When required, resting cell experiments were also performed in sodium phosphate buffer pH 6 or with higher amounts of the substrate (5 mM DDVA). These resting cell mixtures were then incubated at 30 °C and 200 rpm agitation in a 50 mL flask. A 1 mL sample was taken at different time points, filtered through 0.45 μm pore size filters and stored at −20 °C for further analysis of metabolites accumulated in the supernatant.Metabolite analyses
To monitor 5CVA production from DDVA, both DDVA and 5CVA stock solutions and the supernatant of the samples collected were diluted in 0.1 M sodium phosphate buffer pH 6.5 and analyzed by HPLC-MS (high-performance liquid chromatography coupled to mass spectroscopy) using 1260 Infinity II and Infinity Lab LC/MSD XT systems (both from Agilent) and a Poroshell 120 EC-C18 (4.6 × 100 mm, 4 μm, Agilent) reverse phase column. DDVA and 5CVA were detected and quantified at 230 nm in a diode array type of UV/VIS detector (DAD). The solvents used for HPLC analyses were water/0.1% formic acid (solvent A) and acetonitrile/0.1% formic acid (solvent B). The applied gradient for analysis was 0–40% of solvent B for 12 min followed by a 3 min column re-equilibration step in which solvent B is restored to 0%. The flow rate was always kept at 1 mL min−1. Chromatograms generated from the samples were compared to commercial standards. The identity of DDVA and 5CVA was confirmed by extracted ion analysis for fragments 334 and 212 m/z, respectively, in positive ion mode, and they were compared with the same standard-generated profiles.In silico identification of a TonB box
P. putida KT2440 TonB-dependent transporters annotated by Cornelis & Bodilis51 were considered for TonB box detection. The first 50 amino acids of all 27 receptors were aligned using MUSCLE52 excluding signal peptides. Alignments of the putative TonB region were corrected manually using the corresponding sequence of the characterized BtuB receptor of P. aeruginosa.53 11 P. putida KT2440 putative TonB box sequences were selected due to their similarity to DdvT and BtuB sequences and the resulting consensus sequence was obtained.Results and discussion
Construction of a P. putida KT2440 recombinant biocatalyst for the bioconversion of DDVA into 5CVA
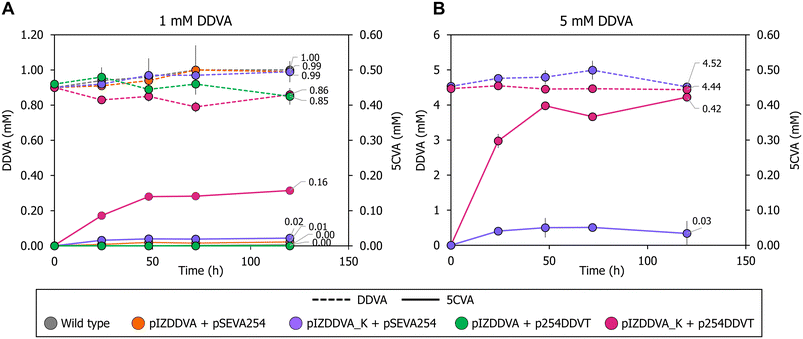
As indicated in the Introduction, S. lignivorans SYK-6 can degrade DDVA with the formation of 5CVA, but then this intermediate is further channeled to the central metabolism of the cell and it does not accumulate as the final product.54 Thus, to develop a bacterial biocatalyst able to accumulate 5CVA from DDVA, we have engineered the wild-type P. putida KT2440 strain, which is unable to degrade or modify 5CVA, by expressing the SYK-6 genes required to convert DDVA into 5CVA. To accomplish this, P. putida KT2440 was transformed with plasmid pIZDDVA that contains the ligXa, ligXc, ligXd, ligZ, and ligY genes from strain SYK-6 (Fig. 1) under control of the lacIq/Ptac regulatory couple, allowing gene induction when required by the addition of IPTG. During DDVA metabolism, the activity of the LigX, LigZ and LigY enzymes produces not only 5CVA but also 4-carboxy-2-hydroxypenta-2,4-dienoate (CHPD) (Fig. 1),35 a molecule that has been proposed to be metabolized to pyruvate by the GalB hydratase and the GalC aldolase, two enzymes of the gallate degradation pathway in P. putida KT2440 and that are functionally equivalent to LigJ and LigK from strain SYK6, respectively.55,56 Therefore, the galB and galC genes were also cloned into pIZDDVA (Fig. 1), to promote CHPD consumption and avoid a potential product inhibition of LigY, caused by the accumulation of CHPD, hence optimizing 5CVA production.When the recombinant P. putida KT2440 (pIZDDVA) strain was tested, we did not observe growth in minimal medium containing 5 mM DDVA as the sole carbon source. Moreover, when resting cells of the same strain were assayed in the presence of 1 mM DDVA, only a very minor amount (0.01 mM) of 5CVA was detected after 120 h of incubation (Fig. 2). In the wild-type KT2440 strain, the 5CVA production was completely undetectable at any time (Fig. 2). These results suggested that the heterologous expression of the genetic determinants that encode the enzymes needed for the conversion of DDVA into 5CVA was not enough to allow 5CVA production in the recombinant P. putida cells.
It is known that the Major Facilitator Superfamily (MFS) inner membrane transporter DdvK and the Ton system-dependent outer membrane transporter DdvT are required for efficient uptake and catabolism of DDVA in S. lignivorans SYK-6.57–59 Unlike DdvK, which seems completely essential for DDVA uptake independently of the compound concentration in the culture medium, DdvT appears to be partially replaceable by unknown outer membrane porins or transporters in the presence of higher amounts (1 to 5 mM) of DDVA.57,58 Since neither the DdvK nor the DdvT ortholog is present in P. putida KT2440, we checked the contribution of these two DDVA transport elements in the previously engineered P. putida KT2440 strain expressing the ligXYZgalBC genes. For this purpose, ddvK was engineered in cis with the ligXYZgalBC genes yielding plasmid pIZDDVA_K (Fig. 1). For ddvT expression under control of the lacIq/Ptrc regulatory couple, plasmid p254DDVT was constructed (Fig. 1). As shown in Fig. 2A, ddvK co-expression with the catabolic genes turned into an earlier 5CVA generation in resting cells, although the biotransformation efficiency was still low (production of 0.02 mM 5CVA after 48 hours). In contrast, the expression of the catabolic genes and ddvT did not allow the detection of 5CVA (Fig. 2A). However, the simultaneous expression of ddvK, ddvT, and the catabolic genes in the P. putida KT2440 (pIZDDVA_K, p254DDVT) strain boosted DDVA utilization, reaching up to 0.16 mM 5CVA production after 48 h of resting cell assay (Fig. 2A). Therefore, these results show for the first time that bacterial biocatalysts can be successfully engineered for the bioconversion of DDVA into 5CVA.
The production of 5CVA by P. putida KT2440 (pIZDDVA_K, p254DDVT) resting cells increased to 0.42 mM when 5 mM DDVA was used as the substrate (Fig. 2B). This suggests that the enzymatic machinery responsible for converting DDVA into 5CVA is not yet saturated at a substrate concentration of 1 mM DDVA. The relatively low production yield of 5CVA (approximately 16% in the presence of 1 mM DDVA), which results in a significant amount of DDVA substrate remaining in the culture medium, combined with the absence of detectable metabolic intermediates from DDVA degradation other than 5CVA (Fig. S2†), strongly suggests that the primary limitation in using P. putida as a biocatalyst for 5CVA production is the uptake of DDVA into the bacterial cells, rather than the enzymatic activity responsible for its degradation.
DdvT is recognized by the native Ton system of P. putida KT2440
The results presented above demonstrate that active DDVA uptake through the outer membrane of P. putida is crucial for its subsequent bioconversion to 5CVA. This implies that the native inner membrane Ton system in P. putida KT2440 can, at least to some extent, interact with the outer membrane DdvT transporter from S. lignivorans SYK-6. However, since DDVA transport efficiency appears to be a significant bottleneck in the bioconversion of DDVA to 5CVA, we hypothesized that DdvT may not operate optimally with the native Ton machinery of KT2440. The genome of KT2440 contains annotations for at least one Ton system (PP_5306, PP_5307, and PP_5308) and several TonB-like genes (PP_0696, PP_1221, PP_1512, and PP_4994). Consequently, we sought to optimize this transport step. Initially, we attempted to clone the DdvT-associated Ton system, which has been identified as essential for SYK-6 survival and DDVA uptake.58 The tonB1-exbB1-exbD1-exbD2 genes were PCR-amplified from the SYK-6 genome and they were tried to be cloned into the p254DDVT plasmid under the control of the lacIq/Ptrc regulatory system. However, no stable plasmid constructions were obtained, suggesting that the expression of these membrane proteins together with DdvT might disrupt cellular functionality in E. coli when using a pSEVA254 vector. Alternatively, a second approach involved replacing the reported TonB box of DdvT with a synthetic TonB box designed to be more compatible with the native inner membrane Ton system of KT2440. To achieve this, the DdvT TonB box motif (I-I-V-T)58 was aligned with the corresponding region of 27 different TonB-dependent transporters annotated in KT2440.51 From this alignment, a TonB box containing the L-X-X-X-X-V/I-T motif was identified in 11 of these outer membrane transporters, as well as in the well-characterized BtuB receptor of P. aeruginosa53 (Fig. S3A†). Notably, the alignment revealed a fully conserved leucine residue that was absent in the TonB box of DdvT from strain SYK-6 (Fig. S3A†). To determine whether this leucine was crucial for establishing an efficient interaction between DdvT and the native Ton system of KT2440, a synthetic consensus TonB box (L-E-L-P-A-T-V-I-T-A amino acid sequence) was introduced to replace the corresponding residues in DdvT, generating a DdvT* chimeric protein in plasmid p254DDVTB (Fig. 1). P. putida KT2440 (pIZDDVA_K, p254DDVTB) resting cell assays revealed that the chimeric DdvT* transporter was functional and DDVA was converted to 5CVA (Fig. S3B†). However, the 5CVA production yield using the DdvT* chimeric transporter was not higher than that obtained with the wild-type DdvT protein (Fig. 2A). This finding suggests that the bottleneck in DDVA uptake is unlikely to result from a deficient interaction between the SYK-6 DdvT transporter and the KT2440 inner membrane Ton machinery. Furthermore, these results demonstrate that P. putida KT2440 can serve as a versatile microbial platform for the functional expression of heterologous TonB-dependent transporters, even those with specificity closely aligned to their native inner membrane Ton machinery.Enhancing 5CVA production
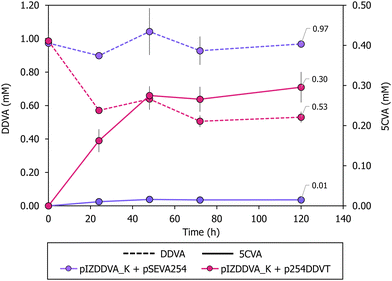
The initial experiments demonstrated a low yield of 5CVA. To assess whether the culture medium used for biocatalyst growth affects 5CVA production efficiency during the resting cell process, we compared the results obtained when cultivating P. putida KT2440 (pIZDDVA_K, p254DDVT) cells in LB medium and minimal media containing two different carbon sources, i.e., glucose and octanoate, leading to biocatalysts with varying energy and reducing power levels.60 The efficiency of 5CVA production was similar when using cells grown in LB medium or glucose-containing minimal medium. However, cells grown in octanoate-containing minimal medium appear to lead to a lower production yield (Fig. S4†). These findings indicate that the choice of carbon source during cell cultivation is crucial in determining the 5CVA production yield, with LB medium being a suitable choice for growing the P. putida KT2440 (pIZDDVA_K, p254DDVT) biocatalyst.We then investigated whether modifications in the resting cell conditions could enhance 5CVA production. The results previously presented indicate that the outer membrane DdvT transporter is functional in P. putida KT2440, and hence it is likely that the transport bottleneck limiting DDVA bioconversion to 5CVA is due to the inner membrane Ton system and/or to the DdvK MFS transporter.57 MFS transporters responsible for the uptake of lignin-derived aromatic compounds belong to either the aromatic acid/H+ symporter (AAHS) or the metabolite/H+ symporter (MHS) families, which rely on a proton gradient across the inner membrane.61 Similarly, the proton motive force also powers the activity of TonB-dependent transporters.61 To enhance DDVA transport across the membrane, we conducted resting cell assays at lower pH levels to increase the proton concentration in the periplasm,62 thereby boosting the proton motive force required for the TonB-dependent DdvT activity and for the DdvK activity.63–65 At pH 6, P. putida KT2440 (pIZDDVA_K, p254DDVT) resting cells achieved a 5CVA production of 0.3 mM from 1 mM DDVA substrate (Fig. 3), representing a 30% yield, higher than the 16% yield observed at pH 7.5 (Fig. 2A). Notably, no increase in 5CVA production was observed at pH 6 when using P. putida KT2440 (pIZDDVA_K, pSEVA254) resting cells (Fig. 3), reinforcing the notion that passive diffusion of non-charged (protonated) DDVA across cell membranes is an inefficient transport system and that both DdvK and DdvT are crucial for effective active DDVA uptake. In summary, lowering the pH in resting cell assays proved to be an effective strategy to overcome DDVA transport limitations, leading to a substantial improvement in 5CVA production yields.
Although lowering the pH during resting cell incubation improved the 5CVA production yield, further optimization of the DDVA bioconversion process would be necessary to achieve higher 5CVA production levels. DDVA transport system may still be underperforming; and therefore, further optimization is advisable. For instance, optimizing the signal peptide of DdvT to ensure proper recognition by the P. putida secretion machinery could enhance transport efficiency.66 On the other hand, cellular toxicity of certain DDVA pathway products, e.g., formaldehyde, and intermediates, e.g., DCHM-HOPDA (Fig. 1),35 should be mitigated. In this sense, we observed that DDVA bioconversion was undetectable without glucose supplementation in the resting cell medium. This observation aligns with previous studies demonstrating that adding glucose increases intracellular NAD(P)H levels, which helps counteract the toxicity of compounds generated during the O-demethylation of aromatic substances.10,67,68 Enhancing DDVA bioconversion efficiency can also be achieved by optimizing enzyme dosage and functionality. Strategies such as codon-usage optimization, employing alternative transcription and translation signals, and rational enzyme dosing (an optimal ratio of 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 30
30![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2 has been proposed for LigXa
2 has been proposed for LigXa![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) LigXc
LigXc![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) LigXd),33 are promising. To mitigate potential metabolic burdens associated with the DDVA pathway in recombinant P. putida biocatalysts, chromosomal integration of the synthetic gene cassette followed by adaptive laboratory evolution (ALE) could also be beneficial. Finally, the reuse of the optimized recombinant biocatalyst could further increase the 5CVA production levels.
LigXd),33 are promising. To mitigate potential metabolic burdens associated with the DDVA pathway in recombinant P. putida biocatalysts, chromosomal integration of the synthetic gene cassette followed by adaptive laboratory evolution (ALE) could also be beneficial. Finally, the reuse of the optimized recombinant biocatalyst could further increase the 5CVA production levels.
Conclusions
Previous studies have demonstrated that recombinant strains of P. putida KT2440 are effective hosts for the valorization of various lignin-derived monoaromatics.6,10,42–44 More recently, it has been shown that a recombinant P. putida KT2440 strain expressing the LsdE and LsdA enzymes from Novosphingobium aromaticivorans DSM12444 acquired the ability to catabolize the dimer erythro-DGPD (1,2-diguaiacylpropane-1,3-diol), and this pathway has been further engineered to produce cis,cis-muconic acid.69This study broadens the current understanding of P. putida KT2440 as a versatile bacterial chassis for lignin valorization by demonstrating that the heterologous expression of the ligXacdZY genes from S. lignivorans SYK6 enables the successful bioconversion of the biphenyl-type lignin dimer DDVA into the strategic building block 5CVA. Unlike erythro-DGPD, which does not appear to require a specific transport system in P. putida, DDVA uptake depends on a dedicated active transport system involving an outer membrane TonB-dependent transporter (DdvT) and an inner membrane MFS transporter (DdvK). This transport system has been identified as a major bottleneck in the conversion of DDVA to 5CVA, but optimizing resting cell conditions—such as lowering the pH—can help overcome this limitation.
Overall, this work highlights the importance of engineering not only metabolic pathways but also transport systems for the efficient valorization of certain lignin-derived compounds in heterologous hosts. Additionally, it reveals for the first time that P. putida KT2440 is a promising bacterial chassis for biotechnological processes requiring the uptake of substrates via a specific TonB-dependent transporter. This proof-of-concept study also lays the foundation for developing a sustainable strategy to produce 5CVA, a strategic building block for the synthesis of novel bio-based polymers with enhanced properties.40
Author contributions
Helena Gómez-Álvarez: conceptualization, investigation, formal analysis, writing – original draft, and writing – review & editing; Carlos del Cerro-Sánchez: conceptualization, investigation, formal analysis, writing – original draft, writing – review & editing, and funding acquisition; Pablo Iturbe: investigation; Virginia Rivero-Buceta: methodology; Juan Nogales: conceptualization and writing – review & editing; Timothy D. H. Bugg: conceptualization, writing – review & editing, and funding acquisition; and Eduardo Díaz: conceptualization, writing – original draft, writing – review & editing, and funding acquisition.Data availability
All the generated data are available within this manuscript and its ESI.†Conflicts of interest
The authors declare no conflict of interest.Acknowledgements
We thank Prof. E. Masai for kindly donating the Sphingobium lignivorans SYK-6 strain and A. Valencia for technical assistance. Funding was provided by grant PID2022-142540OB-I00 of the Spanish Ministry of Science, Innovation and Universities MICIU/AEI/10.13039/501100011033, by ERA CoBiotech grant PCI2019-111833-2 (MILIMO), and by the RELAY project supported by a fellowship from the Fundación General CSIC's ComFuturo program which has received funding from the European Union's Horizon 2020 research and innovation program under the Marie Skłodowska-Curie grant agreement no. 101034263.References
- W. Boerjan, J. Ralph and M. Baucher, Annu. Rev. Plant Biol., 2003, 54, 519–546 CrossRef PubMed.
- J. Zakzeski, P. C. A. Bruijnincx, A. L. Jongerius and B. M. Weckhuysen, Chem. Rev., 2010, 110, 3552–3599 CrossRef CAS PubMed.
- D. S. Bajwa, G. Pourhashem, A. H. Ullah and S. G. Bajwa, Ind. Crops Prod., 2019, 139, 111526 CrossRef CAS.
- W.-J. Liu, H. Jiang and H.-Q. Yu, Green Chem., 2015, 17, 4888–4907 RSC.
- Y. Chen, B. Fu, G. Xiao, L.-Y. Ko, T.-Y. Kao, C. Fan and J. Yuan, ACS Food Sci. Technol., 2021, 1, 382–387 CrossRef CAS.
- C. W. Johnson, D. Salvachúa, P. Khanna, H. Smith, D. J. Peterson and G. T. Beckham, Metab. Eng. Commun., 2016, 3, 111–119 CrossRef PubMed.
- D. R. Vardon, M. A. Franden, C. W. Johnson, E. M. Karp, M. T. Guarnieri, J. G. Linger, M. J. Salm, T. J. Strathmann and G. T. Beckham, Energy Environ. Sci., 2015, 8, 617–628 RSC.
- Y. Suzuki, Y. Okamura-Abe, M. Nakamura, Y. Otsuka, T. Araki, H. Otsuka, R. R. Navarro, N. Kamimura, E. Masai and Y. Katayama, J. Biosci. Bioeng., 2020, 130, 71–75 CrossRef CAS PubMed.
- J. M. Perez, W. S. Kontur, M. Alherech, J. Coplien, S. D. Karlen, S. S. Stahl, T. J. Donohue and D. R. Noguera, Green Chem., 2019, 21, 1340–1350 RSC.
- S. Notonier, A. Z. Werner, E. Kuatsjah, L. Dumalo, P. E. Abraham, E. A. Hatmaker, C. B. Hoyt, A. Amore, K. J. Ramirez, S. P. Woodworth, D. M. Klingeman, R. J. Giannone, A. M. Guss, R. L. Hettich, L. D. Eltis, C. W. Johnson and G. T. Beckham, Metab. Eng., 2021, 65, 111–122 CrossRef CAS PubMed.
- T.-M. Lo, I. Y. Hwang, H.-S. Cho, R. E. Fedora, S. H. Chng, W. J. Choi and M. W. Chang, Front. Microbiol., 2021, 12, 663642 CrossRef PubMed.
- A. Z. Werner, W. T. Cordell, C. W. Lahive, B. C. Klein, C. A. Singer, E. C. D. Tan, M. A. Ingraham, K. J. Ramirez, D. H. Kim, J. N. Pedersen, C. W. Johnson, B. F. Pfleger, G. T. Beckham and D. Salvachúa, Sci. Adv., 2023, 9, eadj0053 CrossRef CAS PubMed.
- C. W. Johnson, D. Salvachúa, N. A. Rorrer, B. A. Black, D. R. Vardon, P. C. St John, N. S. Cleveland, G. Dominick, J. R. Elmore, N. Grundl, P. Khanna, C. R. Martinez, W. E. Michener, D. J. Peterson, K. J. Ramirez, P. Singh, T. A. VanderWall, A. N. Wilson, X. Yi, M. J. Biddy, Y. J. Bomble, A. M. Guss and G. T. Beckham, Joule, 2019, 3, 1523–1537 CrossRef CAS.
- E. M. Spence, L. Calvo-Bado, P. Mines and T. D. H. Bugg, Microb. Cell Fact., 2021, 20, 15 CrossRef CAS PubMed.
- H. Gómez-Álvarez, P. Iturbe, V. Rivero-Buceta, P. Mines, T. D. H. Bugg, J. Nogales and E. Díaz, Bioresour. Technol., 2022, 346, 126638 CrossRef PubMed.
- J. J. Williamson, N. Bahrin, E. M. Hardiman and T. D. H. Bugg, Biotechnol. J., 2020, 15, e1900571 CrossRef PubMed.
- P. D. Sainsbury, E. M. Hardiman, M. Ahmad, H. Otani, N. Seghezzi, L. D. Eltis and T. D. H. Bugg, ACS Chem. Biol., 2013, 8, 2151–2156 CrossRef CAS PubMed.
- D. Salvachúa, T. Rydzak, R. Auwae, A. De Capite, B. A. Black, J. T. Bouvier, N. S. Cleveland, J. R. Elmore, A. Furches, J. D. Huenemann, R. Katahira, W. E. Michener, D. J. Peterson, H. Rohrer, D. R. Vardon, G. T. Beckham and A. M. Guss, Microb. Biotechnol., 2020, 13, 290–298 CrossRef PubMed.
- H. Tang, M.-J. Wang, X.-F. Gan and Y.-Q. Li, Bioresour. Technol., 2022, 362, 127837 CrossRef CAS PubMed.
- M.-T. Manoli, Á. Gargantilla-Becerra, C. del Cerro Sánchez, V. Rivero-Buceta, M. A. Prieto and J. Nogales, Cell Rep., 2024, 43, 113979 CrossRef CAS PubMed.
- F. Weiland, M. Kohlstedt and C. Wittmann, Metab. Eng., 2022, 71, 13–41 CrossRef CAS PubMed.
- M. V. Galkin and J. S. M. Samec, ChemSusChem, 2016, 9, 1544–1558 CrossRef CAS PubMed.
- E. A. Capanema, M. Y. Balakshin and J. F. Kadla, J. Agric. Food Chem., 2004, 52, 1850–1860 CrossRef CAS PubMed.
- M. Sette, R. Wechselberger and C. Crestini, Chem. – Eur. J., 2011, 17, 9529–9535 CrossRef CAS PubMed.
- H. Hirayama, T. Akiyama, A. Tamai, D. S. Nawawi, W. Syafii, T. Yokoyama and Y. Matsumoto, Holzforschung, 2019, 73, 569–578 CrossRef CAS.
- C. Crestini, H. Lange, M. Sette and D. S. Argyropoulos, Green Chem., 2017, 19, 4104–4121 RSC.
- M. A. Goñi, B. Nelson, R. A. Blanchette and J. I. Hedges, Geochim. Cosmochim. Acta, 1993, 57, 3985–4002 CrossRef.
- C. L. Chen, H. M. Chang and T. Kent Kirk, Holzforschung, 1982, 36, 3–9 CrossRef CAS.
- A. Tamai, H. Goto, T. Akiyama and Y. Matsumoto, Holzforschung, 2015, 69, 951–958 CrossRef CAS.
- I. A. Pearl, J. Am. Chem. Soc., 1956, 78, 5672–5674 CrossRef CAS.
- J.-M. Bollag, S.-Y. Liu and R. D. Minard, Soil Biol. Biochem., 1982, 14, 157–163 CrossRef CAS.
- E. Masai, Y. Katayama, S. Nishikawa, M. Yamasaki, N. Morohoshi and T. Haraguchi, FEBS Lett., 1989, 249, 348–352 CrossRef CAS PubMed.
- T. Yoshikata, K. Suzuki, N. Kamimura, M. Namiki, S. Hishiyama, T. Araki, D. Kasai, Y. Otsuka, M. Nakamura, M. Fukuda, Y. Katayama and E. Masai, Appl. Environ. Microbiol., 2014, 80, 7142–7153 CrossRef PubMed.
- T. Sonoki, T. Obi, S. Kubota, M. Higashi, E. Masai and Y. Katayama, Appl. Environ. Microbiol., 2000, 66, 2125–2132 CrossRef CAS PubMed.
- E. Kuatsjah, H. Chen, S. G. Withers and L. D. Eltis, FEBS Lett., 2017, 591, 1001–1009 CrossRef CAS PubMed.
- X. Peng, T. Egashira, K. Hanashiro, E. Masai, S. Nishikawa, Y. Katayama, K. Kimbara and M. Fukuda, Appl. Environ. Microbiol., 1998, 64, 2520–2527 CrossRef CAS PubMed.
- X. Peng, E. Masai, H. Kitayama, K. Harada, Y. Katayama and M. Fukuda, Appl. Environ. Microbiol., 2002, 68, 4407–4415 CrossRef CAS PubMed.
- X. Peng, E. Masai, D. Kasai, K. Miyauchi, Y. Katayama and M. Fukuda, Appl. Environ. Microbiol., 2005, 71, 5014–5021 CrossRef CAS PubMed.
- E. Masai, Y. Katayama and M. Fukuda, Biosci. Biotechnol. Biochem., 2007, 71, 1–15 CrossRef CAS PubMed.
- A. N. Wilson, P. C. St John, D. H. Marin, C. B. Hoyt, E. G. Rognerud, M. R. Nimlos, R. M. Cywar, N. A. Rorrer, K. M. Shebek, L. J. Broadbelt, G. T. Beckham and M. F. Crowley, Macromolecules, 2023, 56, 8547–8557 CrossRef CAS PubMed.
- R. Geyer, J. R. Jambeck and K. L. Law, Sci. Adv., 2017, 3, e1700782 CrossRef PubMed.
- M. Kumar, S. You, J. Beiyuan, G. Luo, J. Gupta, S. Kumar, L. Singh, S. Zhang and D. C. W. Tsang, Bioresour. Technol., 2021, 320, 124412 CrossRef CAS PubMed.
- A. Weimer, M. Kohlstedt, D. C. Volke, P. I. Nikel and C. Wittmann, Appl. Microbiol. Biotechnol., 2020, 104, 7745–7766 CrossRef CAS PubMed.
- A. L. Yaguchi, S. J. Lee and M. A. Blenner, Trends Biotechnol., 2021, 39, 1037–1064 CrossRef CAS PubMed.
- J. Sambrook and D. W. Russell, Molecular cloning: a laboratory manual, Cold Spring Harbor Laboratory Press, Cold Spring Harbor, New York, 2001 Search PubMed.
- J. H. Miller, Experiments in Molecular Genetics, Cold Spring Harbor Laboratory, Cold Spring Harbor, New York, 1972 Search PubMed.
- M. Bagdasarian, R. Lurz, B. Rückert, F. C. H. Franklin, M. M. Bagdasarian, J. Frey and K. N. Timmis, Gene, 1981, 16, 237–247 CrossRef CAS PubMed.
- E. Moreno-Ruiz, M. J. Hernáez, O. Martínez-Pérez and E. Santero, J. Bacteriol., 2003, 185, 2026–2030 CrossRef CAS PubMed.
- R. Silva-Rocha, E. Martínez-García, B. Calles, M. Chavarría, A. Arce-Rodríguez, A. de las Heras, A. D. Páez-Espino, G. Durante-Rodríguez, J. Kim, P. I. Nikel, R. Platero and V. de Lorenzo, Nucleic Acids Res., 2013, 41, D666–D675 CrossRef CAS PubMed.
- M. T. Manoli, J. Nogales and A. Prieto, mBio, 2022, 13, e01794–e01721 CrossRef CAS PubMed.
- P. Cornelis and J. Bodilis, Environ. Microbiol. Rep., 2009, 1, 256–262 CrossRef CAS PubMed.
- R. C. Edgar, Nucleic Acids Res., 2004, 32, 1792–1797 CrossRef CAS PubMed.
- J. S. Oeemig, O. H. S. Ollila and H. Iwaï, PeerJ, 2018, 6, e5412 CrossRef PubMed.
- X. Peng, E. Masai, Y. Katayama and M. Fukuda, Appl. Environ. Microbiol., 1999, 65, 2789–2793 CrossRef CAS PubMed.
- S. Mazurkewich, A. S. Brott, M. S. Kimber and S. Y. K. Seah, J. Biol. Chem., 2016, 291, 7669–7686 CrossRef CAS PubMed.
- J. Nogales, Á. Canales, J. Jiménez-Barbero, B. Serra, J. M. Pingarrón, J. L. García and E. Díaz, Mol. Microbiol., 2011, 79, 359–374 CrossRef CAS PubMed.
- K. Mori, K. Niinuma, M. Fujita, N. Kamimura and E. Masai, Appl. Environ. Microbiol., 2018, 84, e01314–e01318 CrossRef CAS PubMed.
- M. Fujita, K. Mori, H. Hara, S. Hishiyama, N. Kamimura and E. Masai, Commun. Biol., 2019, 2, 432 CrossRef CAS PubMed.
- M. Fujita, S. Yano, K. Shibata, M. Kondo, S. Hishiyama, N. Kamimura and E. Masai, Sci. Rep., 2021, 11, 22444 CrossRef CAS PubMed.
- K. Olavarria, M. P. Marone, H. da Costa Oliveira, J. C. Roncallo, F. N. da Costa Vasconcelos, L. F. da Silva and J. G. C. Gomez, FEBS Open Bio, 2015, 5, 908–915 CrossRef CAS PubMed.
- I. Mutanda, J. Sun, J. Jiang and D. Zhu, Biotechnol. Adv., 2022, 59, 107952 CrossRef CAS PubMed.
- J. C. Wilks and J. L. Slonczewski, J. Bacteriol., 2007, 189, 5601–5607 CrossRef CAS PubMed.
- M. A. Whooley and A. J. Mcloughlin, J. Gen. Microbiol., 1983, 129, 989–996 CAS.
- M. Salema, J. S. Lolkema, M. V. San Romão and M. C. Lourero Dias, J. Bacteriol., 1996, 178, 3127–3132 CrossRef CAS PubMed.
- H. Celia, N. Noinaj, S. D. Zakharov, E. Bordignon, I. Botos, M. Santamaria, T. J. Barnard, W. A. Cramer, R. Lloubes and S. K. Buchanan, Nature, 2016, 538, 60–65 CrossRef CAS PubMed.
- S. Grasso, V. Dabene, M. M. W. B. Hendriks, P. Zwartjens, R. Pellaux, M. Held, S. Panke, J. M. van Dijl, A. Meyer and T. van Rij, ACS Synth. Biol., 2023, 12, 390–404 CrossRef CAS PubMed.
- L. M. Blank, G. Ionidis, B. E. Ebert, B. Bühler and A. Schmid, FEBS J., 2008, 275, 5173–5190 CrossRef CAS PubMed.
- J. A. Lee, S. Stolyar and C. J. Marx, Front. Microbiol., 2022, 13, 849573 CrossRef PubMed.
- G. N. Presley, A. Z. Werner, R. Katahira, D. C. Garcia, S. J. Haugen, K. J. Ramirez, R. J. Giannone, G. T. Beckham and J. K. Michener, Metab. Eng., 2021, 65, 1–10 CrossRef CAS PubMed.
Footnotes |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d4gc06537a |
| ‡ These authors contributed equally to this work. |
| This journal is © The Royal Society of Chemistry 2025 |