 Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Gram-scale production of 4-vinyl guaiacol in the fast-growing phototrophic cyanobacterium Synechococcus sp. PCC 11901†
Thomas
Rohr
 and
Florian
Rudroff
and
Florian
Rudroff
 *
*
Institute of Applied Synthetic Chemistry, TU Wien, Getreidemarkt 9/OC-163, 1060 Vienna, Austria. E-mail: florian.rudroff@tuwien.ac.at
First published on 10th April 2025
Abstract
Whole-cell catalysis in cyanobacteria allows the transformation of light energy into chemical energy by co-factor recycling and in situ production of oxygen by photosynthesis, requiring only light, CO2, water, and a few minerals. Despite these benefits, cyanobacteria have not been deployed on a large scale due to low yields, comparably slow growth, and low biomass accumulation. Additionally, previous works on whole-cell catalysis in cyanobacteria indicate higher yields at high CO2 concentrations, highlighting the need for a source of inorganic carbon to balance photosynthesis and prevent photorespiration. Here, we addressed these problems by combining a fast-growing, high biomass-accumulating strain, Synechococcus sp. PCC 11901, with a CO2-releasing enzymatic reaction, a phenolic acid decarboxylase. After identifying the product toxicity as the limiting factor, we were able to achieve a final product concentration of 80 mM from ferulic acid to 4-vinyl guaiacol with a two-phasic system with diisononyl phthalate and also describe the first use with cyanobacteria of the environmentally benign alternative isopropyl myristate, thus converting a lignin-derived waste product into a valuable precursor molecule for bioplastics and fragrances. We were able to scale the reaction up by employing an inexpensive cultivation system to a final yield of 1.19 g (97% yield) with 100 mL cell suspension and a simple extraction method. This configuration could enable continuous, photosynthetic oxygen production during large-scale cyanobacterial whole-cell catalysis without requiring the addition of an external carbon source.
Green foundation1. We introduce a carbon-negative whole-cell catalysis platform using a fast-growing cyanobacterium (Synechococcus sp. PCC 11901). Its streamlined photosynthesis harnesses light for cofactor replenishment and water splitting, overcoming oxygen limitations and boosting biomass yield.2. Our system fully converts up to 80 mM lignin-derived ferulic acid into valuable 4-vinyl guaiacol within just 8 hours, which is the highest reported whole cell biocataylsis in cyanobacteria. It has been scaled to produce >1 g 4-vinyl guaiacol using cost-effective equipment. 3. Optimizing reaction conditions, scaling production further, and integrating exhaust gas as a CO2 source could make the process even greener and more industrially viable. |
Introduction
Greenhouse gases, such as CO2, are the main driver of anthropogenic climate change, which poses the greatest threat to humanity.1 Biocatalysis allows the mitigation of greenhouse gas emissions to a more environmentally benign approach in producing bulk and fine chemicals compared to classical chemical synthesis. The advantages include lower temperatures and pressures, less solvents and less need for heavy metal catalysts.2,3 Whole-cell catalysis takes up a special role, as an intact cell provides a more stable environment for the enzyme(s) of interest in terms of salinity or pH and offers a system for co-factor recycling, such as adenosine triphosphate (ATP) or nicotinamide adenine dinucleotide (phosphate) (NAD(P)H).2 Established platform organisms for whole-cell catalysis are commonly heterotrophic prokaryotes or unicellular fungi, such as Escherichia coli, Saccharomyces cerevisiae, or Pichia pastoris, which require carbohydrates, e.g., glucose, for growth and co-factor regeneration, thereby increasing costs of production and competing for resources with the food industry. Moreover, the metabolic activities of these organisms consume oxygen, which can impede upscaling in oxygenation reactions. Cyanobacteria, ancient prokaryotes capable of oxygenic photosynthesis, offer a potential solution to this limitation.4,5 These organisms have garnered significant interest in biotechnology due to their efficient photosynthetic apparatus, rapid growth compared to land plants, and amenability to genetic engineering. Cyanobacteria have demonstrated the ability to convert CO2 into bulk and fine chemicals,6 with recent studies highlighting their potential as whole-cell catalysts.4,5 Particularly, their use in enzyme-mediated redox-reactions, including those involving enoate reductase,7 cytochrome P450 monooxygenases,8 Baeyer–Villiger monooxygenases9 and others,4,5 has shown highly promising results and has already achieved gram-scale yields.8,10Despite advancements, a critical aspect is frequently neglected: the production of oxygen by cyanobacteria depends not only on light but also on the presence of inorganic carbon (Ci). Most reactions are conducted in closed vessels, leading to rapid Ci depletion, stalling oxygen production, and reducing the pool of co-factors like NADPH. The lack of the carbon assimilation reactions, a major electron sink, forces the cyanobacterium to find alternative electron sinks or reduce the light reaction of photosynthesis, thus limiting electron release from water-splitting.11 Optimal yields and reaction rates are achieved under conditions of high light and high Ci availability, such as in a continuous flow reactor with a steady influx of a carbonate-supplemented medium.12,13 Alternatively, a reaction, that releases CO2 could bypass this limitation.
Cyanobacteria also have other hurdles to overcome before replacing heterotrophic microbes for whole-cell catalysis. One is their slow growth and limited maximum cell density compared to the established platform organisms mentioned above. Bioreactor design substantially improved growth rate and biomass maxima in cyanobacteria and microalgae, for example with an internal illumination system14 or by improving mass transfer with a gas-permeable membrane,15 but are still orders of magnitudes smaller compared to commonly used bacteria and yeasts.16 In 2020, the marine cyanobacterium Synechococcus sp. PCC 11901 was described, showing growth rates comparable to yeasts (doubling times of ∼2 h) and can reach a cell density comparable to heterotrophic bacteria of up to 33 gDCW L−1, the highest cell density reached in any cyanobacterium.17 Since its first description, several groups have developed an elaborate genetic toolbox for this strain, facilitated by its natural transformability, which includes a suite of inducible and constitutive promoters, CRISPR-based systems for genomic manipulation as well as vitamin B12 independent mutants and potential self-replicating plasmids, and the first genome-scale model has also been described. With these tools at hand, the strain is posed for metabolic engineering, but only limited examples have been described yet, currently restricted to the production of glycosyl glycerol and free fatty acids.17–22 Despite its highly efficient and streamlined photosynthesis apparatus, this strain has not yet been described for whole-cell catalysis approaches.20
The goal of this work (Scheme 1) was to establish Synechococcus sp. PCC 11901 as a host for whole-cell catalysis and circumventing carbon depletion by using an enzymatic reaction, which releases CO2, namely the non-oxidative decarboxylation of aromatic compounds by a phenolic acid decarboxylase (PAD) from Bacillus coagulans. This enzyme converts cinnamic acid derivatives into hydroxy-styrenes, e.g. ferulic acid (1a) into 4-vinyl guaiacol (1b).23 These compounds are of interest as aromatic fragrances or bases for biobased plastics. Additionally, they can be further reacted to aldehydes (such as Vanillin) or other biobased plastic precursors.23,24 In a future application, this reaction could be implemented in a cascade reaction with oxidases and thereby evade the need for additional Ci supplementation and so stabilize the phototrophic metabolism during whole-cell catalysis. We identified the toxicity of the product as a limiting factor and employed a two-phasic system for in situ product removal and achieved unprecedented yields for whole-cell catalysis in cyanobacteria. Finally, we highlight the applicability of this strain for biotechnological applications by up-scaling the reaction to gram-scale using inexpensive equipment and using an ecologically benign solvent alternative, namely isopropyl myristate.
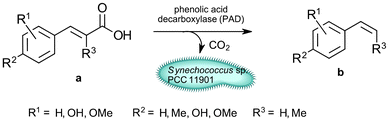 | ||
| Scheme 1 Biocatalytic production of styrene derivatives by phenolic acid decarboxylase expressing Synechococcus sp. PCC 11901. | ||
Results and discussion
Substrate scope of phenolic acid decarboxylase in S. PCC 11901
The enzyme phenolic acid decarboxylase (PAD) from Bacillus coagulans DSM1123 (PDB entry: 2P8G) was integrated into the genome of S. PCC 11901 under the control of an IPTG-inducible promoter (Pclac143) into the fadA locus with Kanamycin as a selection marker, as described before.17 Genomic integration of PAD was confirmed by colony PCR and subsequent sequencing of the amplicon (Microsynth, Balgach, Switzerland, ESI Fig. 1†), and expression was visible by SDS-PAGE (see ESI Fig. 2†). When protein expression was induced with 1 mM IPTG, the strain successfully converted 1a into 1b (Table 1).| Number | Substrate | Consumptiona [%] |
|---|---|---|
| a Based on substrate consumption of 3 replicates. b Polymerization of product prevented direct product detection. | ||
| 1a ferulic acid |
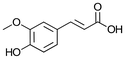
|
81 ± 8 |
| 2a p-coumaric acid |
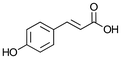
|
93 ± 8 |
| 3a caffeic acid |

|
100b |
| 4a cinnamic acid |

|
n.c. |
| 5a 3,4-dimethoxy cinnamic acid |

|
n.c. |
| 6a α-methyl cinnamic acid |

|
n.c. |
| 7a 4-methoxy cinnamic acid |

|
n.c. |
| 8a 4-methyl cinnamic acid |

|
n.c. |
The substrate scope tested, see Table 1, included 8 derivatives of cinnamic acid.
The only three substrates converted had a hydroxy-group in para-position, such as 1a, 2a, and 3a, which is consistent with the literature.25 In Table 1 10 mM of substrate were tested, and conversion was achieved for 1a, 2a, and 3a, whereas conversion of 3a was measured on substrate consumption, as the product, 3,4-dihydroxystyrene (3b), was not quantifiable due to polymerization, but strong dark discoloration indicated product formation (see ESI Fig. 3†).
During the screening of the substrate conversions, a loss of green pigments of the cells was observed 24 h after adding 1a, resulting in a yellowish color (see ESI Fig. 4†). It was hypothesized, that the discoloration was due to toxicity of either the substrate or the product, which could be the cause for the low product concentration, compared to already published results.23 Substrate and product toxicity was so further investigated as limiting factor for conversion.
Product inhibition and effect on photosynthetic oxygen production
As mentioned above, adding 1a to the PAD expressing strain caused strong discoloration in S. PCC 11901, and the Chlorophyll a (Chl a) content decreased under the detection limit. At the same time, 1a did not affect Chl a content (100% ± 5 compared to before the 1a addition). The same discoloration was observed when 10 mM of the product, 1b, was added, but not, when the substrate 1a was added to the WT. This is a strong indicator of product toxicity. The effect of substrate and product on photosynthetic oxygen production was investigated next to elucidate if photosynthetic activity was compromised.The effect of substrate and product on the photosynthetic activity of Synechococcus sp. PCC 11901 was investigated by simulating a carbon-limited environment and monitoring changes in dissolved oxygen concentration. In a closed vessel, CO2 concentration can be depleted by the cyanobacteria, causing a shift from photosynthesis to photorespiration, which can be seen by a drop in oxygen concentration. The addition of carbonate causes an increase in oxygen concentration, as the cyanobacterium switches away from photorespiration. This allows the close observation of photosynthetic activity during the reaction.
In Fig. 1A, S. PCC 11901 shows robust oxygen production after adding 10 mM carbonate, and no significant changes are observed after adding 10 mM of substrate 1a. When 1a was added to Synechococcus sp. PCC 11901 fadA::pad, an increase in oxygen concentration was observed, as would be expected, due to the release of CO2, but oxygen production ceased after a few minutes (Fig. 1B). A similar effect was observed when 1b was added to the WT strain, where oxygen production halted directly after adding 10 mM 1b.
The inhibition of oxygen evolution, in combination with loss of pigments (Chl a and carotenoids), caused by 1b, indicated a strong toxic effect on S. PCC 11901. In the next section, we investigated whether this effect was also inhibitory on the enzymatic reaction.
In situ product removal (ISPR) significantly increased the yield and final product concentration
An in situ product removal might circumvent the problem of product toxicity. Three solvents have been described to work with cyanobacteria, namely dodecane,26 ethyl oleate, and diisononyl phthalate (DINP).27 DINP was the only appropriate solvent for 1b due to its solubility, additionally, more environmentally benign alternatives to DINP were also evaluated, focusing on solvents that fit the criteria of ‘green solvents’.28 The compatibility of the solvents with the model strain was tested by incubating the WT in the presence of the potential solvents and comparing their Chl a and carotenoid content to the control without solvent after 24 h at the reaction conditions (500 μmol photons per m2 per s, 37 °C, constant shaking). DINP and isopropyl myristate (IPM) had the least effect on pigment content within the cells (a decrease of ∼13% in Chl a and <2% in carotenoids) while the other tested solvents, cyclopentyl methyl ether (CPME), ethyl acetate, limonene, p-cymene, and methyl tert-butyl ether (MTBE), decreased the Chl a content by at least 82%, while carotenoids were reduced by at least 59% (Fig. 2A). Consequently, DINP and IPM were chosen for further testing as an organic phase overlay. IPM is the propyl ester of myristic acid, is considered environmentally harmless, and is used in skincare products, while DINP is mainly produced from fossil fuels and has been shown to affect the endocrine system of vertebrates.29,30Full conversion of 10 mM 1a was observed in the presence of DINP and IPM in a ratio of 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 of aqueous to organic phase. Motivated by the results, we then tested higher concentrations of 1a under strong mixing and determined the conversion (Fig. 2B). Full conversion was reached up to a substrate concentration of 40 mM, irrespective of light exposure. At higher substrate amounts, unreacted 1a remained in the aqueous phase. The reasons for this specific limit remain unknown and could be either attributed to substrate toxicity, or 1b accumulates faster than it is removed by the organic phase. PAD remained active enough to fully convert another 40 mM of substrate that was subsequently fed to the culture, increasing the total amount of product to ∼80 mM.
1 of aqueous to organic phase. Motivated by the results, we then tested higher concentrations of 1a under strong mixing and determined the conversion (Fig. 2B). Full conversion was reached up to a substrate concentration of 40 mM, irrespective of light exposure. At higher substrate amounts, unreacted 1a remained in the aqueous phase. The reasons for this specific limit remain unknown and could be either attributed to substrate toxicity, or 1b accumulates faster than it is removed by the organic phase. PAD remained active enough to fully convert another 40 mM of substrate that was subsequently fed to the culture, increasing the total amount of product to ∼80 mM.
The ISPR thus strongly increased 1a conversion, showing the toxicity of 1b as a limiting factor. To the best of our knowledge, this is the highest yield achieved so far for whole-cell catalysis in cyanobacteria. Additionally, we added IPM to the collection of tolerable solvents for cyanobacteria.
Gram-scale production of 4-vinyl guaiacol in S. PCC 11901 fadA::pad
The fast growth and high biomass accumulation in combination with the fast conversion of S. PCC 11901 fadA::pad renders this strain an interesting candidate for industrial biotechnological applications, but upscaling of reactions involving cyanobacteria is challenging.4 Therefore, we next examined the scalability of the enzymatic, non-oxidative decarboxylation of 1a into 1b by S. PCC 11901 fadA::pad. We used the CellDEG system with carbonate buffer as a Ci source, separated from the culture vessel by a hydrophobic but gas-permeable membrane (CellDEG, Berlin, Germany).15 This system is an inexpensive alternative to typically rather costly photobioreactors and can be used in combination with an LED light source and an orbital shaker, which could increase the acceptance of cyanobacteria in the field of biotechnology research by reducing investment costs. After 96 h of incubation, at a cell density of ∼3.9–5.2 gDCW L−1 in 100 mL MAD2 medium, the cells were centrifuged and resuspended in YBG11 medium to a final cell density of 3.9 gDCW L−1. The switch from the saltwater medium MAD2 to the YBG11 was done to avoid high NaCl concentrations while providing the necessary nutrients for cell survival. The cell suspension was transferred to the reaction vessel (Media composition can be found in the ESI Table 1†). The reaction was prepared in a 500 mL glass bottle (SCHOTT, Germany) with 100 mL DINP and a magnetic stirrer, at 37 °C, 300 rpm, 500 μmol photons per s per m2. The substrate was added in two portions of 0.78 g each, at 0 h and 4 h. After 8 h, the reaction was quenched by removing the organic phase and adding 100 mL of ethyl acetate to the cell suspension to remove any residual 1b from the aqueous phase. The organic phase and ethyl acetate extract were then mixed. The product was extracted from the organic phase by adding 100 mL of 2 N NaOH to deprotonate the phenolic group, which was then separated from the organic phase and neutralized with 2 N HCl. The product was again extracted with ethyl acetate, which was then evaporated.This basic extraction procedure yielded 1.19 g 1b, 97% of the theoretical yield. The product was confirmed to be pure by gas-chromatography and NMR, while NMR showed no signs of polymerization (ESI Fig. 5†). The reaction was repeated under the same conditions with IPM instead of DINP. 1a was fully converted, and basic extraction yielded 1.10 g of 1b but with 9% IPM impurity, which required removal via column chromatography. The final concentration of the reaction of 5.5–5.6 g L−1 is the highest described yet for cyanobacteria, and the possibility of producing >1 g with simple, inexpensive equipment is a promising first step to show the scalability of this new cyanobacterial strain. Further optimization may allow an even higher efficiency of this process, for example, optimized lightning or controlled CO2 atmosphere, instead of an unsteady flow from a carbonate buffer.
Conclusion
In this study, a phenolic acid decarboxylase was successfully expressed in the fast-growing cyanobacterium Synechococcus sp. PCC 11901. It was shown that the product 1b inhibits enzymatic activity, suppresses oxygen evolution, and causes bleaching. A two-phase system allowed in situ product removal and prevented the detrimental effects, resulting in a total product concentration of up to 80 mM 1b when administered in a semi-fed batch approach, with 2 administrations at least 4 h apart. It was possible to scale this process up by applying the low-cost CellDEG system (CellDEG GmbH, Berlin, Germany), as opposed to a high-cost photobioreactor, to produce >1 g of 1b in 100 mL cell suspension and simple extraction from the organic phase. We also highlighted the applicability of green solvents for ISPR with cyanobacteria. In total, we yielded a product concentration of 5.5–5.6 g L−1, the highest achieved by whole-cell catalysis with cyanobacteria, to the best of our knowledge.Cyanobacteria are promising platform organisms for whole-cell catalysis but are hindered by their comparably slow growth and low biomass accumulation. With this work, we highlighted the use of a fast-growing and high-biomass accumulating strain and a way to circumvent Ci depletion during the reaction in a closed vessel while also showing that smart design of cultivation systems can reduce production costs significantly. Previously described gram-scale whole-cell catalysis with the commonly used cyanobacterium Synechocystis sp. PCC6803 required longer incubation times, sometimes followed by resuspension in smaller volumes to reach a cell density of ∼1 gDCW L−1 with expensive and labor-intensive photobioreactors, while here, we were able to reach higher cell densities in less time and with inexpensive equipment.10,12 It should be mentioned that by optimizing growth conditions, reactor design, and medium composition, maximum biomass can be significantly increased.4,16,31 The CellDEG system currently operates at a maximum of 2 L total volume, which is too little for industrial applications. A broad acceptance of cyanobacteria in whole-cell catalysis requires further basic research.
With 1a as a readily available and common phenolic acid, which can easily be extracted from agro-waste, it can be used as a feedstock in a circular economy.32,33 While most research on phenolic acid decarboxylases focuses on the enzyme itself, with highly promising results and constant development of new high-throughput systems, using cyanobacteria as a platform to produce these enzymes, the production becomes even more sustainable, although the final product concentration is still comparably smaller.24,34,35
Methods and materials
Strain cultivation and genomic integration
Synechococcus sp. PCC 11901 was acquired via the Pasteur Culture Collection of Cyanobacteria (Institut Pasteur, Paris, France). The strain was commonly cultivated at 39 °C, 150 rpm, 1% CO2, and 500 μmol photons per m2 per s (light source: Samsung quantum boards LM301B full spec, 3500 K) in a high cell density medium MAD2 (described in ref. 17, see ESI Table 1†). Cryo-cultures were prepared by centrifugation of culture in the linear growth phase and resuspending in AD7 medium supplemented with 1% DMSO and stored at −80 °C. Culture density was measured as absorbance at 750 nm (OD750) in a NanoDrop One (Thermo Fisher Scientific Inc., Waltham, USA) with a factor of 0.24 gDCW L−1 OD750−1. Cell dry weight was determined by preparing cultures at different cell densities, centrifugation (2500g for 10 min), and subsequent resuspension in deionized water three times and transferring the remaining culture into a 30 mL glass vial and letting it dry at 75 °C for 48 h. The conversion factor was calculated from a regression line across 5 different OD750 values (see ESI Fig. 6†).The plasmids used in this work were constructed using the NEBuilder HiFi DNA Assembly system (New England Biolabs, Ipswich, USA). In short, the sequence for the gene of interest, the Phenolic Acid Decarboxylase, PAD, from Bacillus coagulans DSM11, was codon optimized for Synechococcus (Gensmart codon optimizer, Version 28.01.21), inserted into PSZT025, containing the fadA flanking region and the IPTG-inducible Pclac143 promoter (purchased via Addgene, Watertown, USA, plasmid #140033). DNA sequences, Plasmids, and primer used in this study can be found in the ESI.†
The strain Synechococcus sp.PCC 11901 fadA::pad was constructed via natural transformation. A culture of 25 mL in a 200 mL shaking flask with AD7 medium at 30 °C, 150 rpm, ambient air, 100 μmol photons per m2 per s was grown to OD750 ∼ 1, then 1 mL of culture was transferred into a 50 mL centrifugation tube, mixed with 1 μg DNA (containing the gene of interest and the flanking region, amplified via PCR) and incubated for 24 hours at 100 μmol photons per m2 per s at 39 °C. Cells were then transferred to an agar plate containing AD7 agar (same composition as the minimal medium and supplemented with 1.5% bacto-agar and 1 g L−1 thiosulfate) and further incubated at 250 μmol photons per m2 per s at 1% CO2. Successful integration and complete segregation were verified via colony PCR and subsequent sequencing (Microsynth Austria, Vienna, Austria).
Whole-cell catalysis with S. PCC 11901
Whole-cell catalysis in Synechococcus sp. PCC 11901 fadA::pad was performed as follows. At first, 50 mL MAD2 medium was inoculated either with 1 mL of fully grown cells (not older than 2 weeks) or directly from a cryo-culture to a start OD750 or ∼0.05–0.1. Cells were then grown at 39 °C, 150 rpm, 1% CO2, and 500 μmol photons per m2 per s for 24 h, and protein expression was induced with 1 mM IPTG (OD750 ∼ 5–8) and incubation continued for another 24 h. Cells were harvested by centrifugation (3500g, 10 min) and resuspended in YBG11 medium supplemented with 100 mM HEPES (pH 7.4) as a buffer to a cell density of 3.6 g DCW L−1. The minimal sweet-water medium was selected to prevent the effects of excessive salt. The reactions were performed in 30 mL glass vials with screw caps, with 5 or 10 mL culture. For the in situ product removal, the organic phase was added in a 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 ratio to the cell suspension. Stirring was done with a magnetic stirrer bar to ensure an emulsion of organic and aqueous phases.
1 ratio to the cell suspension. Stirring was done with a magnetic stirrer bar to ensure an emulsion of organic and aqueous phases.
Compound toxicity assessment via oxygen measurement
The toxicity assays were performed in a 30 mL flat bottom flask with 10 mL of cell culture at 55 rpm and 37 °C. Oxygen was measured using the firesting OXROB10 probes (Pyroscience, Aachen, Germany) fixed through the screw caps. Data analysis was done using Python 3.0. The oxygen production was measured by letting the oxygen concentration level at ∼200–240 μmol L−1 and adding 10 mM NaHCO3 or KHCO3 from a 500 mM stock solution to the cells via a syringe. Chlorophyll a concentration was measured via methanol extraction.36 In brief, 100 μL cells were centrifuged for 7 min at 12![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000g, resuspended in 1 mL cold methanol, incubated at 4 °C in the dark for 20 min, and again centrifuged. The supernatant was measured for absorbance at 470, 655, and 720 nm in a microwell-plate reader (Tecan Spark, Tecan, Switzerland). Pure Methanol was used as blank and concentration was calculated as follows:
000g, resuspended in 1 mL cold methanol, incubated at 4 °C in the dark for 20 min, and again centrifuged. The supernatant was measured for absorbance at 470, 655, and 720 nm in a microwell-plate reader (Tecan Spark, Tecan, Switzerland). Pure Methanol was used as blank and concentration was calculated as follows:| Chlorophyll a (Chla) [μg mL−1] = 12.9447(A665 − A720) |
Analytics
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 ration within 12 min). Samples were diluted 1
1 ration within 12 min). Samples were diluted 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 20 or 1
20 or 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 40 in Acetonitrile and filtered (0.2 μm pore size).
40 in Acetonitrile and filtered (0.2 μm pore size).
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 20 or 1
20 or 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 40 in ethyl acetate with 1 mM methyl benzoate as internal standard.
40 in ethyl acetate with 1 mM methyl benzoate as internal standard.
Author contributions
T. R.: conceptualization, investigation, methodology, writing – original draft. F. R.: conceptualization, supervision, writing – review & editing.Data availability
The data supporting this article have been included as part of the ESI.†Conflicts of interest
There are no conflicts to declare.Acknowledgements
Financial support by the doctoral school Bioactive, the TU Wien and the EU Interreg. Project GreenChemForCE (CE0200857) is gratefully acknowledged.References
- K. Calvin, D. Dasgupta, G. Krinner, A. Mukherji, P. W. Thorne, C. Trisos, J. Romero, P. Aldunce, K. Barrett, G. Blanco, W. W. L. Cheung, S. Connors, F. Denton, A. Diongue-Niang, D. Dodman, M. Garschagen, O. Geden, B. Hayward, C. Jones, F. Jotzo, T. Krug, R. Lasco, Y.-Y. Lee, V. Masson-Delmotte, M. Meinshausen, K. Mintenbeck, A. Mokssit, F. E. L. Otto, M. Pathak, A. Pirani, E. Poloczanska, H.-O. Pörtner, A. Revi, D. C. Roberts, J. Roy, A. C. Ruane, J. Skea, P. R. Shukla, R. Slade, A. Slangen, Y. Sokona, A. A. Sörensson, M. Tignor, D. Van Vuuren, Y.-M. Wei, H. Winkler, P. Zhai, Z. Zommers, J.-C. Hourcade, F. X. Johnson, S. Pachauri, N. P. Simpson, C. Singh, A. Thomas, E. Totin, P. Arias, M. Bustamante, I. Elgizouli, G. Flato, M. Howden, C. Méndez-Vallejo, J. J. Pereira, R. Pichs-Madruga, S. K. Rose, Y. Saheb, R. Sánchez Rodríguez, D. Ürge-Vorsatz, C. Xiao, N. Yassaa, A. Alegría, K. Armour, B. Bednar-Friedl, K. Blok, G. Cissé, F. Dentener, S. Eriksen, E. Fischer, G. Garner, C. Guivarch, M. Haasnoot, G. Hansen, M. Hauser, E. Hawkins, T. Hermans, R. Kopp, N. Leprince-Ringuet, J. Lewis, D. Ley, C. Ludden, L. Niamir, Z. Nicholls, S. Some, S. Szopa, B. Trewin, K.-I. Van Der Wijst, G. Winter, M. Witting, A. Birt, M. Ha, J. Romero, J. Kim, E. F. Haites, Y. Jung, R. Stavins, A. Birt, M. Ha, D. J. A. Orendain, L. Ignon, S. Park, Y. Park, A. Reisinger, D. Cammaramo, A. Fischlin, J. S. Fuglestvedt, G. Hansen, C. Ludden, V. Masson-Delmotte, J. B. R. Matthews, K. Mintenbeck, A. Pirani, E. Poloczanska, N. Leprince-Ringuet and C. Péan, in IPCC, 2023: Climate Change 2023: Synthesis Report. Contribution of Working Groups I, II and III to the Sixth Assessment Report of the Intergovernmental Panel on Climate Change, ed. Core Writing Team, H. Lee and J. Romero, Intergovernmental Panel on Climate Change (IPCC), Geneva, Switzerland, 1st edn, 2023 Search PubMed.
- M. Schrewe, M. K. Julsing, B. Bühler and A. Schmid, Chem. Soc. Rev., 2013, 42, 6346–6377, 10.1039/C3CS60011D.
- B. Lin and Y. Tao, Microb. Cell Fact., 2017, 16, 106, DOI:10.1186/s12934-017-0724-7.
- J. Jodlbauer, T. Rohr, O. Spadiut, M. D. Mihovilovic and F. Rudroff, Trends Biotechnol., 2021, 39(9), 875–889, DOI:10.1016/j.tibtech.2020.12.009.
- L. Malihan-Yap, H. C. Grimm and R. Kourist, Chem. Ing. Tech., 2022, 94, 1628–1644, DOI:10.1002/cite.202200077.
- J. Kumar, D. Singh, M. B. Tyagi and A. Kumar, Cyanobacteria: Applications in Biotechnology, Elsevier Inc., 2018, vol. 7421 Search PubMed.
- M. Hall, C. Stueckler, H. Ehammer, E. Pointner, G. Oberdorfer, K. Gruber, B. Hauer, R. Stuermer, W. Kroutil, P. Macheroux and K. Faber, Adv. Synth. Catal., 2008, 350, 411–418, DOI:10.1002/adsc.200700458.
- A. Hoschek, J. Toepel, A. Hochkeppel, R. Karande, B. Bühler and A. Schmid, Biotechnol. J., 2019, 14, 1–10, DOI:10.1002/biot.201800724.
- S. Böhmer, K. Köninger, Á. Gómez-Baraibar, S. Bojarra, C. Mügge, S. Schmidt, M. Nowaczyk and R. Kourist, Catalysts, 2017, 7, 240, DOI:10.3390/catal7080240.
- A. Tüllinghoff, H. Djaya-Mbissam, J. Toepel and B. Bühler, Plant Biotechnol. J., 2023, 21, 2074–2083, DOI:10.1111/pbi.14113.
- S. C. Holland, A. D. Kappell and R. L. Burnap, Biochim. Biophys. Acta, Bioenerg., 2015, 1847, 355–363, DOI:10.1007/s11120-021-00835-1.
- A. Tüllinghoff, M. B. Uhl, F. E. H. Nintzel, A. Schmid, B. Bühler and J. Toepel, Front. Catal., 2022, 1, 780474, DOI:10.3389/fctls.2021.780474.
- A. Hoschek, I. Heuschkel, A. Schmid, B. Bühler, R. Karande and K. Bühler, Bioresour. Technol., 2019, 282, 171–178, DOI:10.1016/j.biortech.2019.02.093.
- M. Hobisch, J. Spasic, L. Malihan-Yap, G. D. Barone, K. Castiglione, P. Tamagnini, S. Kara and R. Kourist, ChemSusChem, 2021, 14, 3219–3225, DOI:10.1002/cssc.202100832.
- L. Bähr, A. Wüstenberg and R. Ehwald, J. Appl. Phycol., 2016, 28, 783–793 CrossRef.
- S. N. Chanquia, G. Vernet and S. Kara, Eng. Life Sci., 2022, 22, 712–724, DOI:10.1002/elsc.202100070.
- A. Wlodarczyk, T. T. Selão, B. Norling and P. J. Nixon, bioRxiv, DOI:10.1101/684944.
- L. Mills, J. Moreno-Cabezuelo, A. Włodarczyk, A. Victoria, R. Mejías, A. Nenninger, S. Moxon, P. Bombelli, T. Selão, A. McCormick and D. Lea-Smith, Biomolecules, 2022, 12, 872, DOI:10.3390/biom12070872.
- S. Ravindran, N. Hajinajaf, P. Kundu, J. Comes, D. R. Nielsen, A. M. Varman and A. Ghosh, ACS Synth. Biol., 2024, 13(10), 3281–3294, DOI:10.1021/acssynbio.4c00379.
- A. J. Victoria, T. T. Selão, J. Á. Moreno-Cabezuelo, L. A. Mills, G. A. R. Gale, D. J. Lea-Smith and A. J. McCormick, 2023, preprint, DOI:10.1101/2023.08.04.552009.
- B. A. Cho, J. Á. Moreno-Cabezuelo, L. A. Mills, E. A. Del Río Chanona, D. J. Lea-Smith and D. Zhang, Algal Res., 2023, 70, 102997, DOI:10.1016/j.algal.2023.102997.
- T. Zhang, S. Li, L. Chen, T. Sun and W. Zhang, 2023, preprint, DOI:10.1101/2023.08.23.554402.
- J. Ni, Y. T. Wu, F. Tao, Y. Peng and P. Xu, J. Am. Chem. Soc., 2018, 140, 16001–16005, DOI:10.1021/jacs.8b08177.
- P. Petermeier, J. P. Bittner, T. Jonsson, P. Domínguez De María, E. Byström and S. Kara, Commun. Chem., 2024, 7, 57, DOI:10.1038/s42004-024-01138-x.
- J. Ni, Y.-Y. Gao, F. Tao, H.-Y. Liu and P. Xu, Angew. Chem., 2018, 130, 1228–1231 CrossRef.
- F. K. Davies, V. H. Work, A. S. Beliaev and M. C. Posewitz, Front. Bioeng. Biotechnol., 2014, 2 DOI:10.3389/fbioe.2014.00021.
- A. Hoschek, B. Bühler and A. Schmid, Biotechnol. Bioeng., 2019, 116, 1887–1900, DOI:10.1002/bit.27006.
- D. Prat, O. Pardigon, H.-W. Flemming, S. Letestu, V. Ducandas, P. Isnard, E. Guntrum, T. Senac, S. Ruisseau, P. Cruciani and P. Hosek, Org. Process Res. Dev., 2013, 17, 1517–1525, DOI:10.1021/op4002565.
- R. N. Vadgama, A. A. Odaneth and A. M. Lali, Biotechnol. Rep., 2015, 8, 133–137, DOI:10.1016/j.btre.2015.10.006.
- I. Forner-Piquer, S. Santangeli, F. Maradonna, A. Rabbito, F. Piscitelli, H. R. Habibi, V. Di Marzo and O. Carnevali, Environ. Pollut., 2018, 241, 1–8, DOI:10.1016/j.envpol.2018.05.007.
- P. Van Alphen, H. Abedini Najafabadi, F. Branco Dos Santos and K. J. Hellingwerf, Biotechnol. J., 2018, 13, 1700764, DOI:10.1002/biot.201700764.
- A. Tüllinghoff, H. Sträuber, F. C. F. Baleeiro, A. Aurich, M. C. Morejón, K. Meisel, K.-F. Cyffka, F. Harnisch, K. Bühler and D. Thrän, In Review, 2024, preprint, DOI:10.21203/rs.3.rs-5460981/v1.
- E. Valanciene, I. Jonuskiene, M. Syrpas, E. Augustiniene, P. Matulis, A. Simonavicius and N. Malys, Biomolecules, 2020, 10, 874, DOI:10.3390/biom10060874.
- H. Terholsen, K. Myrtollari, M. Larva, C. Möller, A. Taden, R. Kourist, U. T. Bornscheuer and D. Kracher, ChemBioChem, 2023, 24, e202300207, DOI:10.1002/cbic.202300207.
- L. Li, L. Long and S. Ding, Front. Microbiol., 2019, 10, 1798, DOI:10.3389/fmicb.2019.01798.
- T. Zavrel, M. Sinetova and J. Cerven, Bio-Protoc., 2015, 5(9) DOI:10.21769/BioProtoc.1467.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d5gc00905g |
| This journal is © The Royal Society of Chemistry 2025 |



