Open Access Article
Open Access ArticleRetinal organoid chip: engineering a physiomimetic oxygen gradient for optimizing long term culture of human retinal organoids†
Emma Drabbe ab,
Daniel Pelaez*abc and
Ashutosh Agarwal
ab,
Daniel Pelaez*abc and
Ashutosh Agarwal *acd
*acd
aDepartment of Biomedical Engineering, University of Miami, Coral Gables, FL, USA
bBascom Palmer Eye Institute, Department of Ophthalmology, University of Miami Miller School of Medicine, 1638 NW 10th Ave., Miami, FL 33136, USA. E-mail: dpelaez@miami.edu; Tel: +1 (305) 482 4850
cSylvester Comprehensive Cancer Center, University of Miami Miller School of Medicine, 1951 NW 7th Ave #475, Miami, FL 33136, USA. E-mail: A.agarwal2@miami.edu; Tel: +1 (305) 243 8925
dDesai Sethi Urology Institute, University of Miami Miller School of Medicine, Miami, USA
First published on 3rd December 2024
Abstract
An oxygen gradient across the retina plays a crucial role in its development and function. The inner retina resides in a hypoxic environment (2% O2) adjacent to the vitreous cavity. Oxygenation levels rapidly increase towards the outer retina (18% O2) at the choroid. In addition to retinal stratification, oxygen levels are critical for the health of retinal ganglion cells (RGCs), which relay visual information from the retina to the brain. Human stem cell derived retinal organoids are being engineered to mimic the structure and function of human retina for applications such as disease modeling, development of therapeutics, and cell replacement therapies. However, rapid degeneration of the retinal ganglion cell layers are a common limitation of human retinal organoid platforms. We report the design of a novel retinal organoid chip (ROC) that maintains a physiologically relevant oxygen gradient and allows the maturation of inner and outer retinal cell phenotypes for human retinal organoids. Our PDMS-free ROC holds 55 individual retinal organoids that were manually seeded, cultured for extended periods (over 150 days), imaged in situ, and retrieved. ROC was designed from first principles of liquid and gas mass transport, and fabricated from biologically- and chemically inert materials using rapid prototyping techniques such as micromachining, laser cutting, 3D printing and bonding. After computational and experimental validation of oxygen gradients, human induced pluripotent stem cell derived retinal organoids were transferred into the ROC, differentiated, cultured and imaged within the chip. ROCs that maintained active perfusion and stable oxygen gradients were successful in inducing higher viability of RGCs within retinal organoids than static controls, or ROC without oxygen gradients. Our physiologically relevant and higher-throughput retinal organoid culture system is well suited for applications in studying developmental perturbations to primate retinogenesis, including those driven by inherited traits, fetal environmental exposure to toxic agents, or acquired by genetic mutations, such as retinoblastoma.
Introduction
The retina is a complex multi-cellular stratified tissue responsible for converting photons into electrical signals for the establishment of sight. Human retina derives from the neuroectodermal germ layer during embryonic development in-utero. Retinogenesis is a self-organizing process, directed by biochemical and mechanical cues of the microenvironment. Clonal evolution of the different retinal phenotypes from multipotent retinal progenitor cells is still not entirely understood in developmental biology.1–3 Starting early in retinogenesis and through adulthood, the inner and outer retinas comprise two distinct domains in terms of oxygen perfusion levels,4,5 and respond differently to inspired oxygen variations and vascular permeability modulation.6–8 The inner retina is kept in a hypoxic state, with partial pressures of oxygen maintained constantly below 15 mmHg (or 2% v/v).9 The oxygenation levels of the retina rapidly increase towards the outer retina and Bruch's membrane, reaching a maximum of approximately 140 mmHg (18% v/v) at the choroid (illustrated in Fig. S1†).9,10 Even under hyperoxic ventilation, the hypoxic maintenance mechanisms within the inner retina avoid significant increases in oxygen perfusion, while the outer retina and choroid, devoid of oxygen self-regulation, are allowed to experience oxygenation increases commensurate with the level of oxygen intake.11There is a need for deep comprehension of the physiological and pathological human retinogenesis to explore novel therapeutic approaches for eye disease.10,12 Recent investigations indicate that human induced pluripotent stem cell (iPSC) derived retinal organoids are a promising tool to recapitulate human retinogenesis and therefore increase the understanding of early retinal development.12 A correct temporal and spatial presentation of retinal neuron differentiation is observed, up to and including maturation of photoreceptor outer segments. The retinal ganglion cells (RGCs) at the inner surface of the retina translate visual information received by the photoreceptors into signals for the brain via the optic nerve. These are the first retinal cell phenotypes to appear, both in vivo and in retinal organoids. Initially the RGC development in organoids is similar to in vivo RGC layer formation. However, at later stages of organoid development there is a loss of the RGCs, eventually leading to complete degeneration of the nerve fiber layer.13–15 Degeneration of RGCs in retinal organoids may be related to the exposure to oxygenated microenvironments, since inner retinal cells have been shown to express nuclear HIF1-α, a transcription factor activated by low oxygen concentrations, mediating the metabolic adaptation of cells causing changes in gene expression, impacting migration, proliferation, and metabolism.16 Hence, this limitation makes the use of retinal organoids challenging for in vitro retinal disease models, particularly for the inner retina and optic nerve.
We sought to develop a large volume microfluidic system that recapitulates the in vivo like microenvironment of the human retina, maintaining a stable oxygen concentration gradient for the entire extended culture time of developing retinal organoids. Current microfluidic systems that induce oxygen gradients often do not have the clearances to hold up to 1000 μm diameter organoids. Further, they typically employ polydimethylsiloxane (PDMS), which can absorb gases and small molecules like drugs, and hence, is not a preferable material for developing organs on chips meant for disease modeling and drug discovery.17 Particularly for retinal organoids, culture platforms are often designed for research involving the retinal outer segments, and typically do not include RGCs, and employ different cell sources merged in a multilayered system, which does not replicate retinal development.18–23 Additionally, despite the existence of culture systems incorporating an oxygen gradient for other research purposes, none of the current systems are suitable for retinal organoid culture over extended periods of time because of the large size of organoids (up to 1000 μm), and extremely long culture times (several months).24 We describe a PDMS-free retinal organoid chip (ROC) that holds 55 individual retinal organoids that were manually seeded, cultured for extended periods (multiple months), imaged, and retrieved for downstream analytical processing. ROC is designed from first principles of liquid and gas mass transport, fabricated from biologically- and chemically-inert materials using rapid prototyping techniques such as micromachining, laser cutting, 3D printing, and bonding. After experimental verification of oxygen gradients, human iPSC derived retinal organoids are transferred into the ROC, and differentiated, cultured, and imaged within the ROC. ROCs that maintained active perfusion and stable oxygen gradients were successful in inducing higher viability of RGCs within retinal organoids than static controls, or ROC without oxygen gradients.
Materials and methods
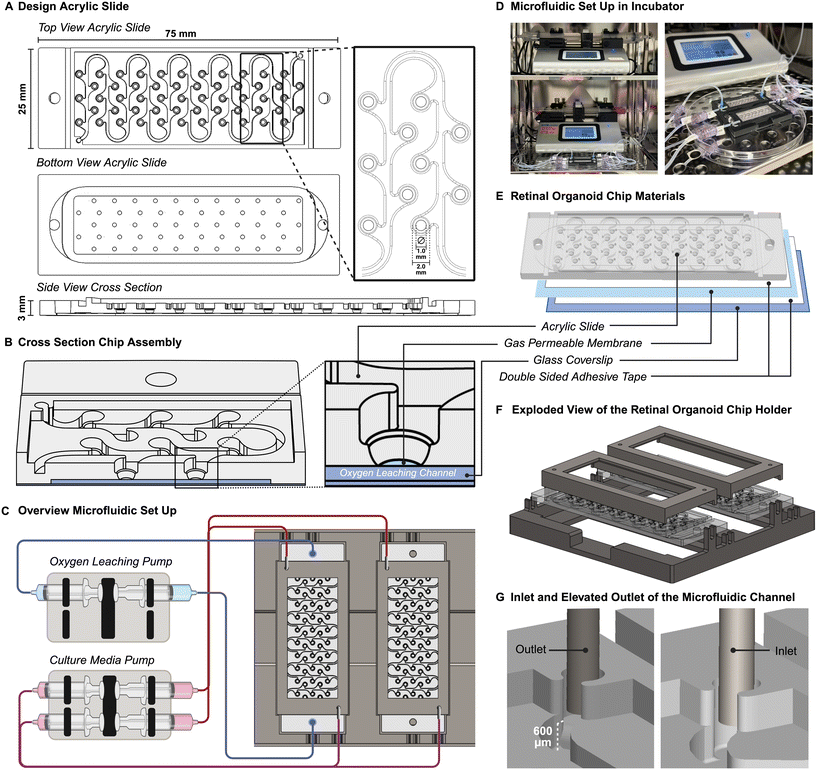
Retinal organoid Chip (ROC) design, fabrication and assembly
ROC was composed from three different materials, bonded together by double-adhesive optically clear mounting tape. The top component was made of 1/8′′ thick acrylic (polymethyl methacrylate or PMMA (McMaster-Carr, Elmhurst, IL)) and contains the structure for the microfluidic channel and culture wells for the retinal organoids. This part was designed in computer aided design software Solidworks® (SolidWorks, Dassault Systèmes), and then fabricated out of a 3 mm thick slide of acrylic using a computer numerical control milling machine (MODELA MDX-540, Roland) and a 30 W CO2 laser machine (Legend Helix, Epilog Laser). Optically clear double-adhesive mounting tape (7602A56, McMaster-Carr, USA) was cut into shape to fit around the bottom of the culture wells and applied to acrylic. On the other side of the tape the clear durable PFA sheet with 0.25 μm thickness (84955K22, McMaster-Carr, USA) was applied. Excess PFA is removed with a scalpel. A second double-sided adhesive tape (PS-1340, Polymer Science) was cut to fit the outer contour of the ROC and applied around the bottom channel. Finally, a 75 × 25 mm glass coverslip was applied to the other side of the adhesive to seal the bottom channel. The assembled ROC was then placed between two microscopic glass slides (VWR, 48311-703) to prevent deformation and was autoclaved for 20 minutes at 121 °C. The fully fabricated and sterilized ROC was then coated with anti-adherence solution (Stemcell Technologies, cat# 07010). The holder plates and lids for the ROC were designed in Solidworks® and 3D printed using the Bambu Lab® 3D printer and PLA-CF filament (Bambu, 14100). The glass top of the lid is a commercially available cover glass 50 × 22 mm (Corning®, 2980-225) glued to the PLA-CF structure with medical grade instant adhesive (Loctite 4011®, 50-949-057).Fluidic system set-up
A microfluidic tubing network was established using two Legato® dual syringe pumps (KD Scientific, 788270) placed in the incubator, with syringes connected to 1/32′′ Tygon® PTFE tubing (Cole Parmer) with 1/32′′ barbed Luer connectors (Qosina). The ends of the tubing were connected to 20-gauge stainless steel 90° angled needles (Fisnar, 8001169) which were placed in the inlet and outlet ports of the ROC. The syringe pumps held 30 mL syringes (BD Scientific) with the corresponding culture media, and 60 mL syringes with a 3 mM sodium sulfite (Sigma-Aldrich, 239321) oxygen leaching solution in PBS (Gibco).Finite element modeling of oxygen transport and fluid flow
Computational modeling of the fluid and gas dynamics was performed using finite element modeling software COMSOL Multiphysics 5.0. The fluidic area in the ROC was obtained using an inversion of the Solidworks® design of the ROC and imported as a Parasolid file. Computational modeling of the oxygen transport was done with the transport of diluted species physics modules governed by the Fick's law of diffusion. An excess sodium sulfite concentration (>1 mM) (sodium sulfite MW: 126.04 g mol−1) removes all oxygen from the liquid, therefore oxygen concentration along the syringe and tubing was assumed to be constant, which was experimentally confirmed by testing the oxygen concentrations at the outlets with PreSens oxygen sensors.Microfluidic fluid flow was modeled with the laminar flow physics module to solve for oxygen diffusion and fluid velocity within the culture media channel. Fluid flow was modeled as the incompressible flow of water (density = 1000 kg m−3 and dynamic viscosity = 0.001 Pa s) using Navier–Stokes and continuity equations. With a flow rate of 50 μL per hour the Reynolds number was small enough to assume a laminar flow. All modeling parameters are listed in Table S1.†
Experimental oxygen concentration gradient validation
Validation of theoretical results was done using PreSens Profiling Oxygen Microsensor PM-PSt7 (PreSens, Germany) secured in a micromanipulator connected to multi-channel trace O2 meter OXY-4 ST operated with PreSens Measurement Studio 2 software. After running the ROC system for a few hours for equilibration purposes, the sensor was carefully placed on the bottom of the culture well, lightly touching the PFA membrane. After 60 seconds of stabilization the oxygen concentration was measured, and the sensor was moved up in steps of 50 μm, each separated by 60 seconds wait time to let the concentrations equilibrate again. These measurements were repeated over two different chips and eight different culture wells.iPSC culture and retinal organoid differentiation
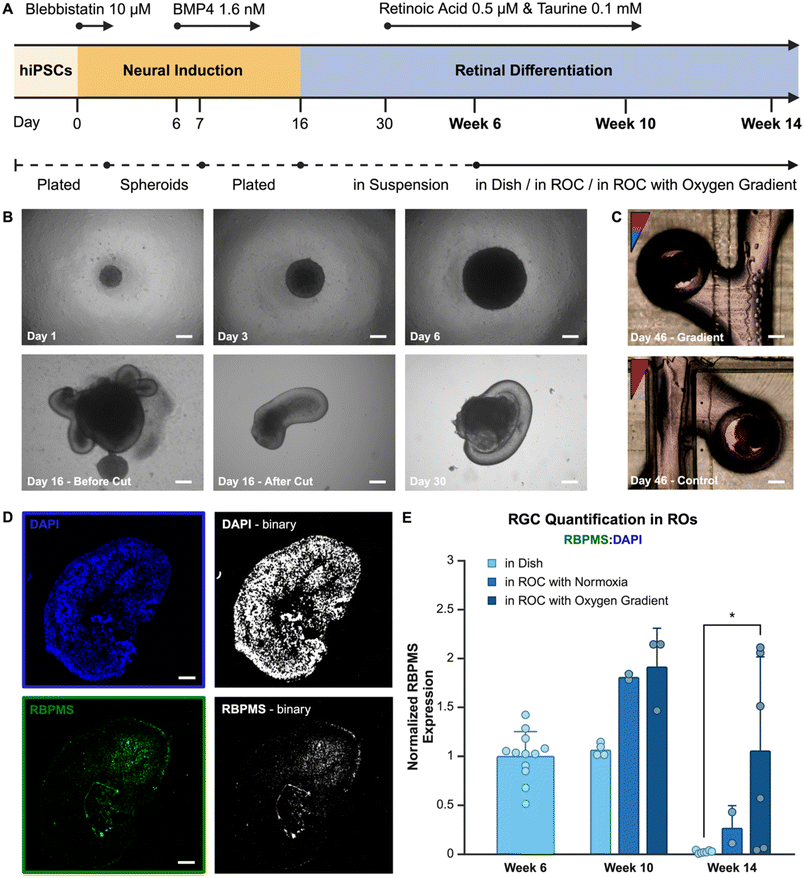
Human iPSCs were previously created using a dental pulp stem cell (DPSC) line with the CytoTune™-IPS Sendai Reprogramming Kit (Invitrogen, A16517) to induce pluripotency. Pluripotency of the DPSC-iPSC line was verified with immunofluorescence staining for pluripotency markers (Fig. S2†). The created DPSC-iPSC line has been used between passages 28–38 and was cultured on Cultrex™ (R&D Systems, 3433-005-01) coated plates in supplemented mTESR1™ (Stemcell Technologies, cat# 85850). Media was changed every day and until the cells were 80–90% confluent, at which time they were passaged using ReLeSR™ (Stemcell Technologies, cat# 100-0484). After passaging, 10 μm Y-27632 Rock Inhibitor (SelleckChem, S1049) was added to the cell suspension for a maximum of 24 hours.An optimized retinal organoid formation protocol was adapted from previously published protocols.14,25 Starting on day 0, 5000 cells per well were seeded in Corning® Costar® ultra-low attachment 96 well plates (Sigma-Aldrich, St. Louis, USA) in mTESR1™ supplemented with 10 μm blebbistatin (Stemcell Technologies, 72402). The first three days cells were weaned onto Neural Induction Media (Table S2†). Then, 1.6 nM BMP4 was added at day 6, which is tapered down until day 10 of differentiation. On day 7, the formed spheroids were plated onto Cultrex™ coated (R&D Systems, 3433-005-01) 6 well plates. At day 16, the optic cups were separated from the organoids and cultured in Corning® Costar® ultra-low attachment 96 well plates with retinal differentiation medium (Table S2†). All cells were cultured in normoxia (18.6% O2) until week 6 of differentiation, then retinal organoids were added to the ROC, pre-treated with anti-adherence solution, or continued to grow in culture plates as control. Starting at day 30, the retinal differentiation medium was supplemented with 200 μM taurine and from day 30 to day 120, 1 μm all-trans retinoic acid (Stemcell Technologies, 72262) was added to the medium.
Immunofluorescence imaging, quantification and statistical analyses
Retinal organoids used for imaging were taken out of the plates or ROC and washed in PBS before fixation in 4% PFA. Samples were then embedded in paraffin and sliced into 10 μm thick slices with a microtome (Leica). Slices were deparaffinized and stained with primary antibody RBPMS (Invitrogen, PA5-31231), which is a phenotypic marker for RGCs, and immunofluorescence secondary antibody Alexa 488 (Abcam, ab150077). Samples were then incubated with DAPI (Thermo Scientific, 62248) before mounted on microscopic slides with Fluoromount-G™ (Electron Microscopy Science, cat# 17984-25). Immunofluorescence imaging was done with Leica THUNDER (Leica) with a 20× objective. Images were taken with the exact same image settings and then imported in ImageJ as .lif file using the bio formats importer plugin.An ImageJ macro was created to separate the DAPI and RBPMS channels of each image and converting them into binary images by running command “Convert to Mask”. Finally, the white pixel area of each binary image was measured by running command “Measure” for area. The pixel count of all images was summarized in a table and exported to Excel. To account for organoid size, the ratio of RBPMS/DAPI was calculated for each image and which represented the ratio of cells positive for the RGC marker RBPMS. A schematic of the RBPMS quantification workflow is shown in Fig. S3.† These ratios were normalized to the images of the day the organoids were seeded into the ROC. Values were plotted in bar graphs using Biorender and were tested for normal distribution with Shapiro Wilk test and analyzed with Welch's t-test for significance.
Results and discussion
ROC design and fabrication
The general design of the ROC was created in computer aided design software Solidworks® and was fabricated using computer numerical controlled machining techniques on acrylic slides and double adhesive tapes, which were then combined with a gas permeable membrane and glass coverslip to complete the ROC. This method of fabrication is ideal to test different iterations of design and make small adjustments during the process. Whereas PDMS chips require the development of a new mask for each iteration using an expensive and time-consuming cleanroom-based photolithography method. The design of the acrylic part of the ROC is a single microfluidic channel connecting 55 round bottom wells for retinal organoids to each other (Fig. 1A). The wells are connected in leaf-like structures to capture the organoids in their designated wells once placed and prevent them from flowing through the microfluidic channel. This allows individual tracking of each organoid over time with microscopic imaging. The bottom of the chip contains a thin chamber between the glass coverslip and the perfluoroalkoxy alkane (PFA) membrane that allows the oxygen scavenger solution containing sodium sulfite, to flow through, while keeping a small (<200 μm) and transparent working distance for live imaging of the organoids (Fig. 1B).The microfluidic set up was completed with push and pull syringe pumps, of which one supplies an oxygen leaching solution to the bottom channel and the other the culture media to the culture media channel. The ROCs are connected to the pumps with silicone tubing and 90 degree angled stainless steel injection needles. Due to the open-well design of the ROC, there was no need for pressure control with pumps. To test the effect of the oxygen concentration gradient on the RO culture, only one ROC was perfused with the oxygen leaching solution creating the oxygen gradient, and a second ROC functioned as control with a normoxic culture environment (Fig. 1C). The ROC had a culture media supply in the syringe pushing at a rate of 50 μL per hour, and waste media was collected on the pulling side of the syringe pump. The microfluidic system ran for 21 days at a time independently, after which both fresh and waste media syringes were replaced. Compared to static culture, 50% less culture media was needed throughout the duration of the retinal organoid differentiation process. The whole set-up was placed inside the incubator for 110 days (Fig. 1D).
To precisely control the oxygen concentrations, non-permeable materials were used for the ROC and microfluidic system. In the microfluidic system, the syringes were made of polypropylene which has low O2 permeability,26 and the Tygon® tubing has lower gas permeability compared to regular silicone tubing. The ROC components, glass and PMMA are both non-permeable to oxygen (PMMA has an oxygen diffusivity index of 2.7 × 10−8 cm2 s−1, whereas commonly used PDMS has a 3.4 × 10−5 cm2 s−1 diffusivity to oxygen) (Fig. 1E). Using the size of a microscopic glass cover slide allowed using commercially available materials and reduced complexity of ROC production.
Due to long-term culture, the ROCs were for single use and sterilized thoroughly before the start of culture. It was possible to autoclave the entire ROC between two glass cover slides to prevent deformation. To facilitate the ease of use and ensure sterility during long-term culture, a holder was designed and fabricated to ensure a closed system keeping the microfluidic tubing in place, while the ROCs were still accessible for imaging and taking out retinal organoids at different time points (Fig. 1F).
The height of culture media in the chip is an important consideration for the oxygen concentration gradient. Therefore, the liquid height was controlled with the relative positioning heights of the inlet and outlet ports of the microfluidic channel. The inlet was leveled with the bottom of the channel, while the outlet was on a 600 μm raised platform, which was the height of the intended liquid column level (Fig. 1G). While the syringe pumps pushed and pulled culture media to and from the ROC at the exact same rate, the microfluidic channels would remain filled at the same height, and the elevated outlet prevented the channels from drying out due to evaporation.
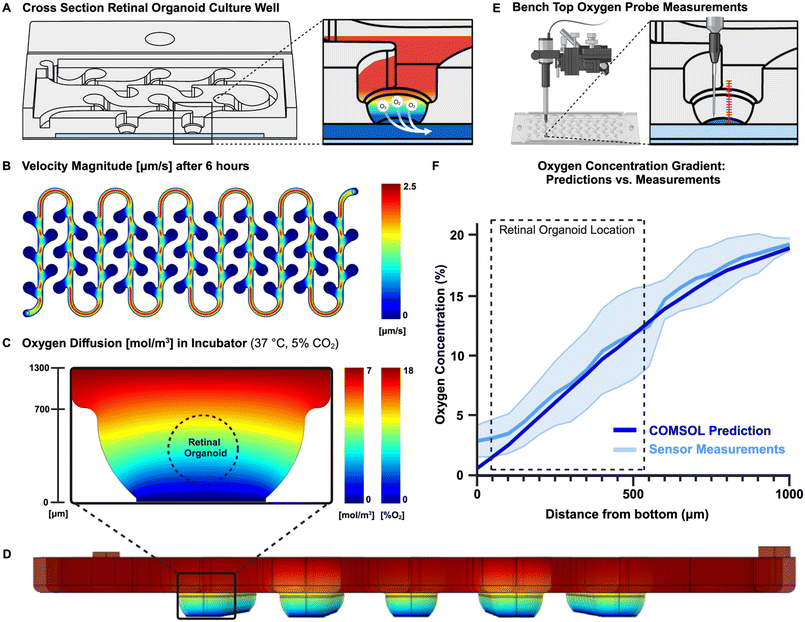
Computational prediction of system boundaries
Due to the open-well structure of the ROC and the gas permeable membrane separating the well and the oxygen leaching channel, the principle of simple gas diffusion would act on the oxygen concentration, which moved oxygen from the oxygenated culture media towards the oxygen leaching channel (Fig. 2A). The oxygen concentration gradient established due to these physics is the most important feature of the ROC and was defined using theoretical and experimental methods. As retinal organoids are sensitive and harmed by shear stress, the velocity of culture media through the ROC was an important factor as well. The combination of Solidworks® for computer aided design and Multiphysics simulation of COMSOL, provides a powerful tool for product design, evaluation, and optimization. However, computational predictions might be slightly different than reality, which is why the computational theoretical predictions were validated by experimental measurements. Finite Element Modeling software COMSOL was used to predict transport in the ROC with this geometry, dimensions, and pump settings. An optimal flow velocity was determined to set as culture media inflow on the syringe pumps. This was optimized by comparing the total volume of the well, with a rough estimate of the syringes volume, to determine how long they last. Even though theoretically evaporation should not happen within the moisturized ROC, realistically there is still some loss of liquid. Therefore, the flow rate had to be high enough to change the total volume of media at least twice a day. Since the total system volume is approximately 530 μL, the velocity was set at 50 μL per hour (or 1200 μL per day), which represented 2.2 times of system volume change every day (Fig. 2B).Oxygen gradient validation
Computational modeling software COMSOL Multiphysics® has predicted the oxygen transport throughout the ROC in combination with the microfluidic flow of culture medium. A two-dimensional model was created to define the oxygen concentration gradient within the culture well (Fig. 2C) and a three-dimensional model showed the oxygen concentrations throughout the ROC (Fig. 2D). In addition, oxygen sensors have measured an established oxygen gradient within the culture wells of the ROC, which is the most important feature of this novel ROC. The oxygen concentrations in the ROC were measured with PreSens oxygen sensors starting from the bottom of the well going up with increments of 50 μm using a micromanipulator (Fig. 2E). Since measurements were performed on a bench top, the values will be plotted against oxygen concentrations predicted with the same benchtop environment (Fig. 2F). The COMSOL Multiphysics® calculations for the bench top environment closely mimicked the sensor measurements, therefore the gas diffusion model was validated and could be used to predict the oxygen gradient concentrations within the incubator environment. The oxygen concentrations in the culture wells of the ROC approximately range from 3% to 18% O2, which closely mimics the in vivo retinal oxygen concentrations from 2% to 18% O2.The measurements done with PreSens oxygen sensors were hard to align perfectly with the bottom of the membrane, as the sensor tip location had to be estimated and could break when touching solid materials such as the membrane. Therefore, we believe the high standard deviation and higher start at 0 μm is due to practical limitations of manual measurements.
Retinal organoids in ROC
The chip and its gradient were specifically designed for retinal organoid culture. Retinal organoids were cultured using an optimized protocol adapted from Zhong et al.14 Human dental pulp stem cell (DPSC) derived iPSCs were used to create spheroids, which differentiated with neural induction media. After optic cups had formed, they were harvested and individually placed in low-adherence wells with retinal differentiation media. At week 6 of differentiation the organoids were manually selected based on their phenotype (Fig. 3A) and each organoid was placed into a well in the microfluidic ROCs. Over extended culture time organoids stayed in place due to the organoid capture design structure of the microfluidic channel and culture wells (Fig. 3B). At week 10 and 14, organoids were removed for fluorescent image analysis of RGCs (Fig. 3C).RBPMS is an RGC specific marker used in vertebrate retina labeling of RGCs for quantitative analysis.27 Retinal organoids were embedded and sliced into sections for further analyses. The image surface area expressing RBPMS in each organoid was compared to the surface area with cell nuclei stained with DAPI (representative images in Fig. 3D), the ratio between these two was used to express relative quantitation of RGCs. Using this method, results from different size organoids can be compared. However, a major limitation of this method is the loss of knowledge on the orientation of the retinal organoid within the oxygen gradient in the microfluidic chip.
All results were normalized to week 6, which is the time point when organoids are added to the chip. The results show a relatively high standard deviation for the oxygen gradient condition compared to the other conditions. This is likely due the heterogeneous microenvironment of the organoids, of which slices in uncertain orientations are used for analysis. In the homogeneous control conditions the same cell composition in all orientations would be expected, whereas in the oxygen gradient a higher RGC population in the hypoxic area is hypothesized with a lower RGC population in normoxia. Overall, the results (Fig. 3E) show a higher RGC population in the organoids grown in an oxygen gradient, which remains present over a longer period of time. The results indicate that, at week 10 of differentiation, the presence of continuous microfluidic flow improves the RGC quantity compared to the standard retinal organoid differentiation protocol. At week 14 of differentiation, the presence of an oxygen gradient appears to contribute positively to RGC survival. There is a significantly (p = 0.045) higher amount of RGCs present in retinal organoids grown in the ROC with the oxygen gradient compared to the control group grown with the standard organoid differentiation method in a culture dish. Having the RGCs present through the maturation of photoreceptor outer segments of the retina is important for physiological relevance of the platform and accurate disease modeling purposes, where all retinal cell phenotypes are potentially involved, such as retinoblastoma.
Retinogenesis, retinal organoid differentiation, and ROC
Extensive knowledge of human retinal development and pathogenesis remains lacking due to physiological differences between species. Mouse retinogenesis is wildly divergent from primate retinogenesis in temporal and spatial distribution of nascent differentiated cells. Similarly, mouse retinas lack a fovea, the structure responsible for high-acuity vision in primates with the highest density of photoreceptor cells. Furthermore, the mouse genome does not contain the NBPF family of genes, which are critical for structural development of the primate central nervous system,28 including the retina. A great benefit of human retinal organoids is their ability to mimic the in vivo development of human retinas, allowing the investigation of early-stage retinal development and disease. Because organoids are derived from iPSCs, genetic manipulations can be implemented stably to create reporters for individual genes or to model genetic disease.Development of the human retina and the differentiation of retinal organoids share several similarities but also have distinct differences. The human retina begins to form early in embryogenesis from the optic vesicle, which is an outgrowth of the forebrain (Fig. 4A). Retinal progenitor cells differentiate into various retinal cell types, including RGCs in a highly regulated sequence, and finally, retina develops into a laminated structure with distinct layers, each containing specific cell types. Similar to human retina, in vitro development of retinal organoids may lead to laminated structures with photoreceptors, ganglion cells, and other retinal cell types (Fig. 4B). However, the efficiency and maturity of these layers can vary depending on the protocol used. We attempted to compare and overlay the presence of RGCs in retinal organoids during development using the three different culture methods (Fig. 4C). All methods employed the same differentiation protocol up to week 6. Subsequently, the retinal organoids were allocated to one of three conditions: a static culture well, a microfluidic ROC without the gradient or a ROC including the oxygen concentration gradient. The organoids in the first two conditions are exposed to normoxia, and the third condition to an oxygen concentration gradient from day 30 throughout the duration of the experiment. The overlay of the bar graph results from Fig. 3E show the presence of RGCs in retinal organoids throughout the early phase retinogenesis. The presence of RGCs at the end of the RGC differentiation window is important in establishing a retinal organoid model with both inner and outer retinal cell phenotypes present, as the RGC development stops going into the late-phase retinogenesis.
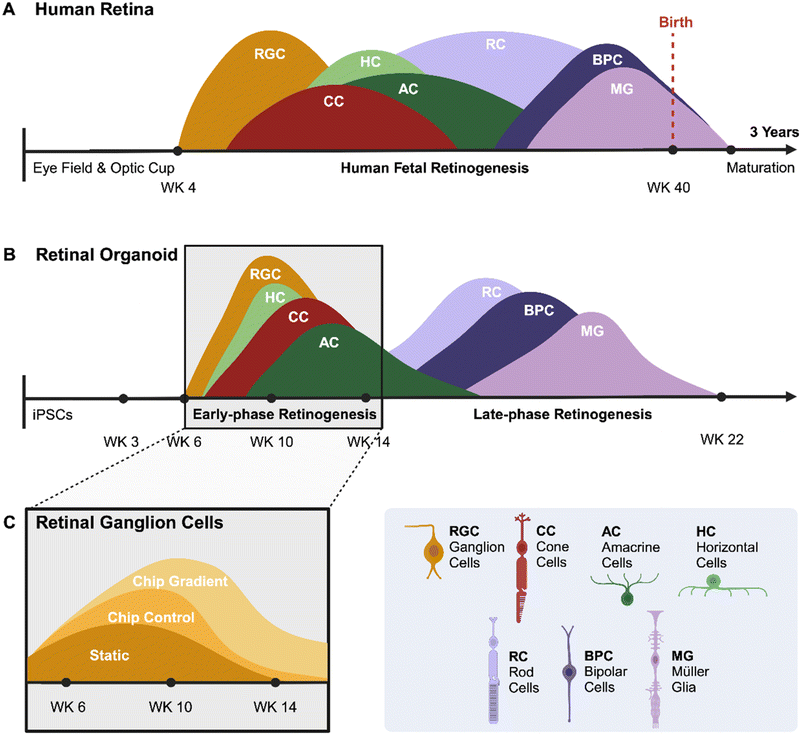 | ||
| Fig. 4 Overview of retinogenesis. A developmental comparison between the embryogenesis of (A) the human retina and the differentiation process of (B) a retinal organoid. Peaks show the window of differentiation of progenitor cells into the specific retinal cell phenotypes (adapted from O'Hara-Wright and Gonzalez-Cordero (2020)29). (C) A schematic diagram of the results from Fig. 3E show the presence of RGCs in retinal organoids in early-phase retinogenesis cultured with three different methods. | ||
Conclusion
Here we describe a novel organ on chip design that has implemented a vertical oxygen concentration gradient ranging from 3% to 18% O2, while maintaining the capability to access and image tissues throughout culture. There is a need for fine oxygen concentration control within human physiological and pathological tissue models of the retina, but addressing this need can be challenging depending on the platform requirements.30 RGC degeneration in human retinal models has been an ongoing limitation in the retinal organoid research field. Although the cause of RGC degeneration is still unknown, there are different speculations on the reasons for this phenomenon aside from the lack of exposure to a physio mimetic microenvironment with physiological oxygen concentrations. Other studies imply several different reasons for RGC degeneration. For example, microglia, astrocytes and Muller glia are cell types present in the retina that maintain the retina's homeostasis. Astrocytes play a role in the maturation of ganglion cells and are therefore thought to have the ability to help RGC survival.31 Research has shown improved RGC survival when the role of these cell types was promoted.31,32 Another implication is the lack of neurotrophic feedback from the visual centers in the brain to the nascent RGCs. Finally, it is suggested by recent studies that primary cilia in retinal organoids are related to the survival of RGCs.33Previous research has shown the improvement of retinal organoid development and long-term culture using a microfluidic culture system.22,23 Our system combines continuous microfluidic perfusion with physio mimetic oxygen concentration values, which seems promising for enhancing the RGC development process and preventing complete cell death. Compared to existing retinal organoid systems, the benefit of this system is the ease of use due to the open well design, the absence of a pressure control system and quickly adjustable design for different applications. Although we are focused on improved survival of the ganglion cells in the retina, to verify the differences between the control and gradient condition, the goal is to have a complete representation of all retinal cell phenotypes, and therefore organoids should be stained for photoreceptors and other retinal cell phenotypes as well, to verify how the presence of ganglion cells will affect the development of rods and cones. Second, taking out the retinal organoids from the ROC at different timepoints makes it impossible to retain information regarding the orientation of the organoid within the oxygen concentration gradient. Therefore, slices of the fixed tissue have random orientations and cell phenotype ratios. When all organoids are sacrificed at the same time, the fixation and embedding process can be done in the chip itself, which retains the orientation of the organoids. Additionally, all ROCs have to be fabricated with computer numerical controlled machining techniques, and manually assembled. This might create slight differences between ROCs and is time consuming. For future use of this design, an injection molding technique can be adopted to reduce fabrication labor and variability. In addition, the ROC materials can be assembled using more advanced techniques such as heat bonding, to ensure reproducibility. Furthermore, we note that the microfluidic channel connects the culture wells in a series configuration. Given that the culture media is refreshed twice daily, the positional differences of organoids within the channel are generally considered negligible. However, the implications of paracrine signaling factors and the delivery of compounds have not yet been investigated. Future research is essential to elucidate the effects of culturing organoids in a series arrangement. Additionally, each microfluidic channel is supplied by a syringe linked to a singular syringe pump, which constrains the incorporation of different biological groups that require different culture media compositions, as this is limited by the number of available syringe pumps.
In addition to optimizing the generation of complex retinal models, this platform allows us to study the effects of developmental perturbations to primate retinogenesis, such as those introduced by acquired genetic mutations, inherited traits, or fetal environmental exposure to toxic agents. As a malignant tumor of the developing retina, retinoblastoma is the prototypical disease that can be modeled using this retinal organoid platform. Retinoblastoma tumors arise in the outer retina and are exquisitely oxygen dependent early in their formation, only growing close to the retinal vasculature. However, most retinoblastomas undergo a phenotypic switch wherein they become resistant to hypoxia and commence disseminating throughout the more hypoxic areas of the eye, including the vitreous and optic nerve. Therefore, the presentation of correct oxygenation profiles is important in retinal organoid differentiation and retinoblastoma disease studies. The use of animals is unsuited for retinoblastoma disease origination and progression, as the mouse retinal photoreceptor pool is predominantly made of rod photoreceptors, while a cone photoreceptor precursor is thought to be the cell-of-origin for human retinoblastoma tumors.34 Using the ROC as retinoblastoma disease model can reveal more on prenatal appearance of the tumor and hypoxic adaptation in the malignant progression of retinoblastoma lesions. We know that RB1 mutations are responsible for retinoblastoma initiation, yet these alone do not lead to malignant disease. Retinal organoids made from inducible RB1 knock out iPSCs can shed light on the origin, progression, and metastasis of retinoblastoma.
In conclusion, the ROC is user-friendly and enables the exposure of three-dimensional cellular constructs to a continuous microfluidic flow and an oxygen concentration gradient. This capability may extend to the development of various other organoids of tissues beyond the human retina. During embryogenesis, physiological oxygen concentration levels are significantly lower, primarily driving cellular proliferation. This suggests that iPSC-derived organoids may benefit form spatial and temporal presentation of different oxygen tensions to mimic human embryogenesis.35 Furthermore, modifications to the acrylic slide can be easily implemented, allowing for the creation of channels or culture wells of varying sizes, thereby enhancing the ROC's suitability for applications such as drug testing. Lastly, the ROC can facilitate the establishment of a pathological environment, enabling the study of patient-derived tumor organoids within a tumor-like microenvironment for the purposes of personalized medicine.
Data availability
The data supporting the computational model have been included as part of the ESI.† We include raw images and analyses at https://miami.box.com/s/1xel8d9mh44m28d9zp8166vyj623b6ez. Other experimental data for this article, including image analyses software are available at public data repository maintained by the University of Miami Libraries https://guides.library.miami.edu/biomedicaldata.Conflicts of interest
None.Acknowledgements
Funding is acknowledged from NIH-NCI (R01CA248890 to DP), NIH-NEI (Center Core Grant P30EY014801), and Research to Prevent Blindness – Unrestricted Grant (GR004596-1).References
- Z. Chen, X. Li and C. Desplan, Deterministic or stochastic choices in retinal neuron specification, Neuron, 2012, 75(5), 739–742 CrossRef CAS.
- J. He, et al., How variable clones build an invariant retina, Neuron, 2012, 75(5), 786–798 CrossRef CAS PubMed.
- F. L. Gomes, et al., Reconstruction of rat retinal progenitor cell lineages in vitro reveals a surprising degree of stochasticity in cell fate decisions, Development, 2011, 138(2), 227–235 CrossRef CAS PubMed.
- S. J. Cringle, et al., Oxygen distribution and consumption in the developing rat retina, Invest. Ophthalmol. Visual Sci., 2006, 47(9), 4072–4076 CrossRef.
- N. D. Wangsa-Wirawan and R. A. Linsenmeier, Retinal oxygen: fundamental and clinical aspects, Arch. Ophthalmol., 2003, 121(4), 547–557 CrossRef PubMed.
- R. A. Linsenmeier, Electrophysiological consequences of retinal hypoxia, Graefes Arch. Clin. Exp. Ophthalmol., 1990, 228(2), 143–150 Search PubMed.
- N. Ruzafa, et al., Effect of hypoxia on the retina and superior colliculus of neonatal pigs, PLoS One, 2017, 12(4), e0175301 Search PubMed.
- A. Shakoor, et al., Chorioretinal vascular oxygen tension changes in response to light flicker, Invest. Ophthalmol. Visual Sci., 2006, 47(11), 4962–4965 CrossRef.
- D.-Y. Yu and S. J. Cringle, Oxygen Distribution and Consumption within the Retina in Vascularised and Avascular Retinas and in Animal Models of Retinal Disease, Prog. Retinal Eye Res., 2001, 20(2), 175–208 CrossRef CAS PubMed.
- S. Jeon and I. H. Oh, Regeneration of the retina: toward stem cell therapy for degenerative retinal diseases, BMB Rep., 2015, 48(4), 193–199 CrossRef CAS PubMed.
- D. Y. Yu, S. J. Cringle and E. N. Su, Intraretinal oxygen distribution in the monkey retina and the response to systemic hyperoxia, Invest. Ophthalmol. Visual Sci., 2005, 46(12), 4728–4733 CrossRef PubMed.
- J. Kang, et al., Application of Human Stem Cell Derived Retinal Organoids in the Exploration of the Mechanisms of Early Retinal Development, Stem Cell Rev. Rep., 2023, 19(6), 1755–1772 Search PubMed.
- K. Kruczek and A. Swaroop, Pluripotent stem cell-derived retinal organoids for disease modeling and development of therapies, Stem Cells, 2020, 38(10), 1206–1215 CrossRef PubMed.
- X. Zhong, et al., Generation of three-dimensional retinal tissue with functional photoreceptors from human iPSCs, Nat. Commun., 2014, 5, 4047 CrossRef CAS.
- K. C. Eldred and T. A. Reh, Human retinal model systems: Strengths, weaknesses, and future directions, Dev. Biol., 2021, 480, 114–122 CrossRef CAS.
- L. Cheng, et al., Hypoxia-Inducible Factor-1alpha Target Genes Contribute to Retinal Neuroprotection, Front. Cell. Neurosci., 2017, 11, 20 Search PubMed.
- C. G. Alver, et al., Roadblocks confronting widespread dissemination and deployment of Organs on Chips, Nat. Commun., 2024, 15(1), 5118 CrossRef CAS PubMed.
- K. Achberger, et al., Merging organoid and organ-on-a-chip technology to generate complex multi-layer tissue models in a human retina-on-a-chip platform, eLife, 2019, 8, e46188 CrossRef PubMed.
- L. J. Chen, et al., Microfluidic co-cultures of retinal pigment epithelial cells and vascular endothelial cells to investigate choroidal angiogenesis, Sci. Rep., 2017, 7(1), 3538 CrossRef PubMed.
- M. Chung, et al., Wet-AMD on a Chip: Modeling Outer Blood-Retinal Barrier In Vitro, Adv. Healthcare Mater., 2018, 7(2), 1700028 CrossRef.
- J. Yeste, et al., A compartmentalized microfluidic chip with crisscross microgrooves and electrophysiological electrodes for modeling the blood-retinal barrier, Lab Chip, 2017, 18(1), 95–105 RSC.
- J. Gong, et al., A controllable perfusion microfluidic chip for facilitating the development of retinal ganglion cells in human retinal organoids, Lab Chip, 2023, 23(17), 3820–3836 RSC.
- Y. Xue, et al., Retinal organoids on-a-chip: a micro-millifluidic bioreactor for long-term organoid maintenance, Lab Chip, 2021, 21(17), 3361–3377 RSC.
- K. R. Rivera, et al., Measuring and regulating oxygen levels in microphysiological systems: design, material, and sensor considerations, Analyst, 2019, 144(10), 3190–3215 CAS.
- E. E. Capowski, et al., Reproducibility and staging of 3D human retinal organoids across multiple pluripotent stem cell lines, Development, 2019, 146(1), dev171686 Search PubMed.
- V. S. Shirure, et al., Quantitative design strategies for fine control of oxygen in microfluidic systems, Lab Chip, 2020, 20(16), 3036–3050 CAS.
- J. M. Kwong, J. Caprioli and N. Piri, RNA binding protein with multiple splicing: a new marker for retinal ganglion cells, Invest. Ophthalmol. Visual Sci., 2010, 51(2), 1052–1058 Search PubMed.
- F. Zimmer and S. H. Montgomery, Phylogenetic Analysis Supports a Link between DUF1220 Domain Number and Primate Brain Expansion, Genome Biol. Evol., 2015, 7(8), 2083–2088 CrossRef CAS PubMed.
- M. O'Hara-Wright and A. Gonzalez-Cordero, Retinal organoids: a window into human retinal development, Development, 2020, 147(24), dev189746 CrossRef.
- J. N. Mazerik, S. Becker and P. A. Sieving, 3-D retina organoids: Building platforms for therapies of the future, Cell Med., 2018, 10, 2155179018773758 CrossRef.
- K. B. VanderWall, et al., Astrocytes Regulate the Development and Maturation of Retinal Ganglion Cells Derived from Human Pluripotent Stem Cells, Stem Cell Rep., 2019, 12(2), 201–212 CAS.
- K. A. Wong and L. I. Benowitz, Retinal Ganglion Cell Survival and Axon Regeneration after Optic Nerve Injury: Role of Inflammation and Other Factors, Int. J. Mol. Sci., 2022, 23(17), 10179 CrossRef CAS PubMed.
- K. Ning, et al., Characterization of Primary Cilia Formation in Human ESC-Derived Retinal Organoids, Stem Cells Int., 2023, 2023, 6494486 Search PubMed.
- E. Bogenmann, M. A. Lochrie and M. I. Simon, Cone cell-specific genes expressed in retinoblastoma, Science, 1988, 240(4848), 76–78 CAS.
- S. Fathollahipour, P. S. Patil and N. D. Leipzig, Oxygen Regulation in Development: Lessons from Embryogenesis towards Tissue Engineering, Cells Tissues Organs, 2018, 205(5–6), 350–371 CAS.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d4lc00771a |
| This journal is © The Royal Society of Chemistry 2025 |