Open Access Article
Open Access ArticleSmartphones as a platform for molecular analysis: concepts, methods, devices and future potential†
Daina V.
Baker‡
 ,
Jasmine
Bernal-Escalante‡
,
Christine
Traaseth‡
,
Yihao
Wang
,
Michael V.
Tran
,
Seth
Keenan
and
W. Russ
Algar
,
Jasmine
Bernal-Escalante‡
,
Christine
Traaseth‡
,
Yihao
Wang
,
Michael V.
Tran
,
Seth
Keenan
and
W. Russ
Algar
 *
*
Department of Chemistry, University of British Columbia, 2036 Main Mall, Vancouver, BC V6T 1Z1, Canada. E-mail: algar@chem.ubc.ca
First published on 7th February 2025
Abstract
Over the past 15 years, smartphones have had a transformative effect on everyday life. These devices also have the potential to transform molecular analysis over the next 15 years. The cameras of a smartphone, and its many additional onboard features, support optical detection and other aspects of engineering an analytical device. This article reviews the development of smartphones as platforms for portable chemical and biological analysis. It is equal parts conceptual overview, technical tutorial, critical summary of the state of the art, and outlook on how to advance smartphones as a tool for analysis. It further discusses the motivations for adopting smartphones as a portable platform, summarizes their enabling features and relevant optical detection methods, then highlights complementary technologies and materials such as 3D printing, microfluidics, optoelectronics, microelectronics, and nanoparticles. The broad scope of research and key advances from the past 7 years are reviewed as a prelude to a perspective on the challenges and opportunities for translating smartphone-based lab-on-a-chip devices from prototypes to authentic applications in health, food and water safety, environmental monitoring, and beyond. The convergence of smartphones with smart assays and smart apps powered by machine learning and artificial intelligence holds immense promise for realizing a future for molecular analysis that is powerful, versatile, democratized, and no longer just the stuff of science fiction.
1. Introduction
At their time of first release, many classic works of science fiction imagined ambitious and powerful technologies: computers that accepted commands and returned information verbally; handheld devices that processed reams of data on demand; wearable devices for long-range wireless communication; augmented reality eyewear; artificial intelligence that predicted needs and preferences; and portable scanners that measured electromagnetic radiation, vital signs, and molecular composition. Here and now, in the real world, most of these capabilities are genuine features of smartphones, smartphone-linked peripherals, and apps and online tools accessible via smartphone. A notable exception is the scanning of molecular composition, which is neither a capability nor a realistic near-future expectation for smartphones. Nevertheless, smartphones are increasingly useful as detection and readout platforms for assays and sensors that do provide molecular information.Contemporary “lab on a chip” (LOC) devices1–3 hold promise for a future with portable molecular analyses but are nonetheless more aspirational than realized. Microfluidics have done much to advance the LOC concept—including methods for the automation of fluid handling, reduction of sample and waste volumes, faster analyses, precision in mixing, and other benefits from miniaturization—but a key challenge lies beyond the chip, where many microfluidic systems require peripheral equipment for their operation and for the detection of analytes. Examples of such equipment include pumps, pressure generators, power supplies, voltage sequencers, temperature controllers, light sources, microscopes, photodetectors, potentiostats, and other hardware for optical and electrochemical detection. There is a push toward passive microfluidics that eliminate pumping,4 and rapid progress in microelectronics has helped to scale down and lower the cost of hardware, but further reductions in the size, cost, and complexity of off-chip components are still needed.5 LOC systems that require specialized laboratory equipment for their operation are not logistically different than a traditional benchtop analysis.
Laboratories and their specialized equipment are principally located in populous and well-resourced urban centres. These facilities tend to be poorly accessible or inaccessible to residents of rural and remote communities, who represent an underserved 45% of the current global population.6,7 Moreover, laboratory facilities either do not exist at field sites or, when pop-up infrastructure is possible, are necessarily limited in their resources. Examples of field sites where molecular information has high value include sites suspected of a disease outbreak, sites suspected of environmental contamination, farms for agriculture and aquaculture, biomanufacturing plants, workplaces with potential for unsafe conditions, and frontiers of scientific exploration such as the ocean depths and outer space. Even when lab facilities are available in urban centers, restrictions on budget and throughput may hinder the implementation of analyses at the desired scale or frequency—something that is often the case for personalized medicine, environmental surveillance, food and water testing, and process monitoring in manufacturing. Although largely beyond the scope of this review, science education also faces challenges with the costs and access limitations associated with sophisticated scientific instruments and facilities.8,9 In short, there are numerous unmet needs for LOC technologies that offer rapid and quantitative chemical and biological analysis, straightforward and lab-free operation, multitasking between more than one type of analysis, and democratization in the form of being able to be built, distributed, maintained, and utilized without substantial financial or other barriers.
A convergence of microfluidics and many other technologies will be required to create LOC devices and assays that translate to society and industry with global impact. These enabling technologies will originate from electronics, materials science, biotechnology, electrochemistry, optics, photonics, data science, and other fields of research and development. Of course, such a scope is too broad for a single review. Instead, this review focuses on the emergence of smartphones as an enabler of LOC, point-of-care, point-of-need, and other portable devices for chemical and biological analyses. Smartphones already integrate numerous technologies and thereby represent a shortcut to the convergence needed for translatable LOC systems. The review begins by discussing the motivation for adopting smartphones as a portable analysis platform, summarizes their potentially useful features, and highlights technologies and materials that are complementary to those features. It then provides a short tutorial on optical detection methods with a singular focus on use of the smartphone camera. A meta-analysis is presented as a preamble to a categorical and critical summary of the recent literature, including examples of how microfluidic platforms have been advantageously paired with smartphone-based devices. The review closes with a perspective on challenges and opportunities for research and development that will advance the technology from prototypes to authentic tools for molecular analysis. When the global ubiquity and ease of use of smartphones are achievements that LOC technologies aspire to emulate, why not leverage the technology of smartphones to help reach that goal?
2. Why smartphones?
2.1 A global technology
Smartphones are ubiquitous in many parts of the world and global penetration is projected to increase. The number of mobile subscriptions is more than 8B and the number of smartphone users varies from >80% to ≤30% between countries, with greater penetration in wealthier nations.10,11 An estimated 54% of the world's population owns a smartphone,12 and this number increases to ∼70% when basic mobile phones (sometimes called “dumbphones”) are included. Mobile networks of varying technology level are available to 95% of the world's population, with 5G coverage available to 45%.13 To a large extent, modern society has reorganized around the capabilities and pocket presence of smartphones. Middle-income countries are already poised to adopt and deploy mobile devices as alternatives to centralized and costly laboratory facilities.14 There is a trajectory, capacity, and motivation for smartphones to be a truly global technology in the near future.2.2 Market size
Mobile technologies and services generated 5% of the global gross domestic product (GDP) in 2022.15 One of the reasons that smartphone technology advances so quickly is its economy of scale. Annual sales have exceeded 1.3 billion smartphones since 2015 and represent a $500 billion USD market.16 This market size enables the cost of innovation to be amortized across many consumers. Device costs range from $100 to $1200 USD, which are far lower than would be possible with a small market size. Although some of the high purchase price for scientific instruments is attributable to sophistication, much of the price still arises from the need to recover development and manufacturing costs (and profit) from a small customer base. Chemical and biological analysis technologies that leverage mass-produced consumer electronics will be able to have lower purchase costs, more robust supply chains, and more accessible repair options than bespoke instruments.2.3 Integrated package
Smartphones are built to be a complete and integrated technological package (Fig. 1) with seamless and intuitive utilization of their numerous features by users. That is not the case for two other technologies adopted for portable analytical device development: microcontroller units (MCUs; e.g. Arduino) and single-board computers (SBCs; e.g. Raspberry Pi).For a similar set of capabilities, smartphones will better minimize size and weight than custom MCU or SBC devices. Of course, the cost for this benefit is a higher purchase price, a closed hardware system that limits customization, and barriers to replacing individual components rather than the whole device. It contrast, MCUs and SBCs are inexpensive and are designed to be modular and enable custom applications. The open-source options for MCUs and SBCs are also beneficial for democratizing and advancing the technology,17–25 albeit countered by the aforementioned global ubiquity of smartphones.
The technical features (vide infra) and form factors of smartphones are also unmatched. As such, common trade-offs for developing analytical devices around MCUs and SBCs include lower performance from lower-cost components, loss of benefits from the scale of the smartphone market, the need for external power and user interfaces, and the time, effort, expense, and learning curve of reengineering features that are already prepackaged in a smartphone. A smartphone is a more rugged and user-friendly platform, and its many integrated sensors are generally superior to the external options for MCUs and SBCs. Lastly, smartphones will best facilitate the integration of analytical devices and assay outcomes with electronic information systems. These systems are, for example, of ever-growing importance for the delivery of health care, and are a source of big data for gaining new health insights from machine learning (ML) and artificial intelligence (AI).
3. Technical features of smartphones
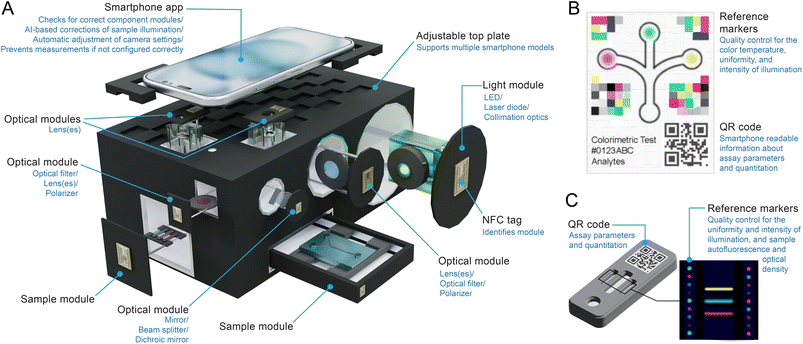
Smartphones have numerous components and features (Fig. 1) that are directly useful for measurements or otherwise support the operation of a portable analytical device.26,273.1 Cameras
Cameras are one of the most heavily marketed and fastest advancing optoelectronic components of a smartphone. Contemporary smartphone models feature multiple camera systems, where each system includes fit-for-purpose lenses, color filters, a CMOS image sensor, and a processing unit that amplifies, digitizes, and processes the sensor data into a digital image (Fig. 2A and B). The specifications that characterize the cameras are summarized in Fig. 1. | ||
| Fig. 2 Important technical components of a smartphone camera. (A) Examples of the exterior view of smartphone camera modules (marked with arrows). (B) Simplified general design of the camera module. (C) Typical arrangements of the Bayer and quad-Bayer color filter arrays (CFAs). (D) An RGB test image decomposed into the corresponding R, G, and B channel images. (E) Typical relative spectral responses of the smartphone camera's R, G, and B color channels. The solid lines in panel E represent the average response across 13 different models and 4 manufacturers of smartphones; the shaded regions reflect the highest and lowest responses at a given wavelength. The response data is from Tominaga et al.28 | ||
The CMOS image sensor generates data from an array of millions of photodetector elements, each corresponding to an individual pixel. More pixels per image sensor improves the nominal resolution, but does not necessarily improve image quality due to the smaller amount of light per pixel. Modern smartphones are now compensating with camera modules that offer pixel binning and one of the multiple camera modules having a larger sensor and pixels.
An infrared (IR)-blocking filter and a Bayer mosaic or similar color filter array (CFA) of red, green (×2), and blue (RGGB) filters are applied over the image sensor to enable color imaging (Fig. 2C and D). A few smartphone camera models were produced with modified CFAs that substituted yellow (Y) pixels for green pixels (i.e. RYYB), added non-selective white (W) pixels (i.e. RGBW), or were monochrome (no CFA), all with the goal of higher light capture in non-scientific photography. For otherwise similar sensor specifications, a study found that a monochrome sensor measured a fluorescence signal that was more than double the signal measured with a color sensor.29 For many smartphones, a quad-Bayer array (Fig. 2C) improves performance at low light levels by supporting pixel binning. To date, the smartphones adopted for portable analyses have almost exclusively had RGB color filters. Most spectroscopists and microscopists would characterize these filters as poor by scientific standards because the transmission spectra overlap and lack a flat top (Fig. 2E). These limitations inform how to optimally use a smartphone camera for optical detection.
Video rates for smartphone cameras range from 24 fps (standard video rate) up to 3840 fps (see Fig. 1). Resolution typically decreases as frame rate increases, but high-end smartphones will support high-definition resolution (720 p or 1080 p) even at hundreds of frames per second. Light capture also decreases as frame rate increases.
3.2 Flashlight
The “flashlight” or “torch” associated with the smartphone camera is another useful component. It is approximately white-light emitting (Fig. 3) and is most often based on the combination of a blue-emitting LED and a phosphor.The flashlight is a useful illumination source for spectroscopic and imaging applications. Peripheral devices with mirrors, optical fibers, and diffusers have been used to direct white light from the flashlight for a variety of brightfield measurements and imaging,30–37 but direct illumination from the flashlight has also been satisfactory for many applications and devices. Other devices have used optical filters to transmit only the blue light from the flashlight for excitation of fluorescence that is imaged in the R and G channels of the camera,38–41 and transmit only red light for SPR measurements.42–45
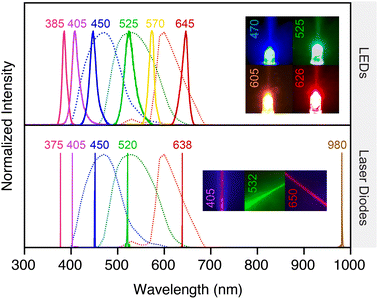
3.3 Display
Almost every contemporary smartphone has a touchscreen display, where each display pixel comprises red, green, and blue (RGB) sub-pixels that emit that color of light (Fig. 4). As little as ten years ago, most smartphones had liquid crystal displays (LCD) displays. This technology is based on a white back light and each sub-pixel utilizes polarizers and a color filter to transmit only R, G, or B wavelengths with intensities controlled by voltage actuation of liquid crystals. At present and for the foreseeable future, organic light-emitting diode (OLED) displays are the predominant technology. Each pixel is made up of R, G, and B sub-pixels (i.e. OLEDs) that emit that color of light directly (Fig. 4B and C). Until recently, OLED displays also incorporated linear polarizers and quarter-wave plates to eliminate reflection of ambient light, but some contemporary displays no longer require these components and thus have better efficiency and higher maximum brightness.In principle, the smartphone display is another source of light for spectroscopy and imaging experiments. The display has, for example, been used as a source of red light for surface plasmon resonance (SPR) measurements,46 a source of blue light for excitation of fluorescence (albeit still in combination with a bandpass filter),47 and as a source of white light for brightfield,48 phase contrast,49 and darkfield imaging.47 A non-smartphone OLED display unit has also been used for multimodal imaging with a smartphone camera, suggesting similar capability for a smartphone display.50 Potential drawbacks of the smartphone display as a light source include a lower intensity than other options and only three pseudo-monochromatic color choices. The spectrum of white light is also non-uniform, although this limitation equally applies to an external white-light LED. Benefits of the display as a light source include the elimination of a component external to the smartphone and the configurability of illumination, the latter of which has been leveraged to enable photochemistry in an array format.51
3.4 Other light-based sensors
Most smartphones have proximity sensors, biometric sensors, and ambient light sensors (ALS), among other sensor types. Proximity sensors use reflected IR light to measure the distance from an object. This feature is used, for example, to turn off the smartphone display when held to a user's ear during a call. Some smartphones include sensors for light detection and ranging (LiDAR) or other time-of-flight (ToF) measurements based on reflected IR light. These features are for the purposes of facial recognition, distance measurements, and supporting augmented reality apps. Although largely untapped for molecular analysis, a study has shown that a smartphone LiDAR sensor is able to measure the viscosity of a liquid droplet, with the potential to analyze blood coagulation and the fat content in milk.52 Smartphone LiDAR has also been proposed for monitoring patient movements.53For biometrics, many smartphone-integrated fingerprint scanners operate based on capacitance or ultrasound imaging, whereas others use a combination of capacitive and optical readout via reflected IR light. Imaging reflected IR light is also the basis for iris recognition on some smartphones. These sensors are not anticipated to be directly useful for molecular analyses, but are a potential means of linking measurement data to relevant identifiers (e.g. patient, analyst) and for data security.
ALS have spectral responses that approximate the photopic response of the human eye. The sensor response is often used to automatically adjust display brightness and apps are available to measure light intensity quantitatively via the ALS. Although this sensor has some latent ability for imaging,54 the capability is far from practical for molecular analyses. To date, the main analytical application of the ALS is non-imaging smartphone-based photometry, where the ALS has a larger dynamic range and bit depth than the smartphone camera. Cuvette-accommodating smartphone attachments have been developed to use the ALS to assess bacterial growth via measurements of turbidity,55 to measure enzyme activity and inhibition by heavy metals through a pH indicator dye,34 and to quantify Cr6+ (aq) in water samples through a chromogenic chelator.56 Smartphone ALS-based devices have also been developed for quantitative analysis of colorimetric dipstick57 and lateral flow assays58 for organophosphate pesticides, and for lateral flow tests of wild-type pseudorabies virus infection,59 blood bilirubin levels,60 prostate-specific antigen (PSA) in patient blood,61 and ampicillin residue in animal milk.61
Multi-channel sensors that measure both the color and intensity of ambient light are the latest generation of ALS technology. These sensors enable automatic adjustment of color temperature for smartphone photography and device displays. This capability will support more robust colorimetric assays under ambient light and help minimize the number of peripheral components (e.g. box, specific light source) needed for reliable smartphone-based measurements.
3.5 Non-optical sensors
Most smartphones include non-optical sensors such as an accelerometer, gyroscope, other orientation and motion sensors, and a magnetometer/geomagnetic field sensor. Collectively, these components provide information about position and movement of a smartphone, and have roles in image stabilization, but are otherwise not anticipated to have direct application to molecular analyses. Every smartphone also has at least one built-in microphone, which is potentially useful for attaching verbal notes to measurement data, monitoring breathing,62–64 and, speculatively, has latent potential for photoacoustic detection for assays. A database of smartphone sensors has been established to support the physical phone experiments (phyphox) education project.653.6 Battery
Smartphone batteries are lithium-ion technology and store up to 4000–6000 mAh of rechargeable energy. The batteries can power not only the phone but also peripheral components and devices. In most cases, up to 10 W of power (5.0 V DC potential and a current up to 2.0 A) can be drawn from the nominal charging port (e.g. USB-C) of the smartphone. This power is sufficient to operate many LEDs, laser diodes, and electric motors, meaning that smartphone-based devices do not necessarily need a separate power supply for components external to the phone. In principle, voltage and current can also be drawn from audio jacks (e.g. tip-ring-ring-sleeve, TRRS). These connectors are increasingly scarce with smartphones and may not reach the same voltage and current levels as a USB-C port. In addition, consumer demand for on-the-go charging of smartphones has led to the mass commercialization of portable power banks that are also useful as a power source in a portable device. Power banks have the potential to provide higher voltages (5–20 V) and currents (up to 5 A), and have longer operating capacities (≥10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 mAh) than the batteries built into smartphones.
000 mAh) than the batteries built into smartphones.
Despite the capability of the smartphone battery to power external components, most smartphone-based analytical devices still incorporate separate batteries (e.g. 1.5 V AA size, 9 V PP3 size) for this purpose. In some cases, these external batteries provide more direct access to a higher voltage (no need for a voltage converter in a circuit) or current, or might usefully extend the operating time of the smartphone device between recharges (i.e. less drain on the smartphone battery). In other cases, the use of external batteries is a preference for prototyping with no significant functional advantage over the smartphone battery. In both cases, downstream circuit components can modify the electrical output to what is required for an application.
3.7 Wireless connectivity
Most smartphones include technologies for medium-range and short-range (e.g. Bluetooth, near-field communication/NFC) connections and data transmission with other enabled devices in close proximity. These modalities have utility for connecting to and controlling device components external to the smartphone, helping to mitigate any limitations associated with the single physical port on a smartphone.Smartphones also transmit data globally through Wi-Fi and cellular networks based on LTE (long term evolution) and other standards categorized as 3G, 4G, and 5G. For applications in some low resource settings, smartphone-based analytical devices will need to be fully functional in the absence of a network connection (although satellite connectivity for data transmission is reasonably anticipated for smartphones in the future). Connectivity adds processing and storage capacity via cloud services and makes support from human expertise accessible around the globe.
An on-board global positioning system (GPS) and other location services are available to tag data with geographical and temporal information—features that are especially useful for documenting field-based measurements. Most smartphones use assisted (or augmented) GPS (A-GPS) in normal operation, but do contain GPS receivers that function in the absence of cellular service or network connectivity.
3.8 Computation, memory and operating systems
Most contemporary smartphones have tens or hundreds of gigabytes of storage, gigabytes of memory (i.e. RAM), and gigahertz processors—technical specifications that are on par with or better than the minimum requirements that have operated laboratory instruments for decades. Even without considering opportunities for cloud computing, smartphone technology is capable of handling the data acquisition and processing needed for many spectroscopic and imaging applications, and some modest implementations of ML and AI algorithms for data analysis.Smartphones run operating systems (OS) to connect hardware with apps. The two dominant OS are Android (∼71% of the global market) and iOS (∼28%).66 Most researchers adopt Android devices due to the larger market share, and for the more open and flexible ecosystem for app development and distribution. Nonetheless, there are concerns that Android is moving toward a more closed system with increasing reliance on proprietary software and a closed-source version of Android from Google. A closed system like iOS has potential advantages in terms of security and privacy, seamless integration with other devices (e.g. desktop and notebook computers, tablets), and a smaller number of smartphone models with which an app needs to be compatible. Although some researchers code their own apps for smartphone-based measurements, there are pre-existing apps that support research and development. Examples include native and third-party apps that provide users with substantial manual control over camera settings for photography (vide infra), mobile development apps for MATLAB and Python, and support for integration of these codes into custom Android or iOS apps written in Android Studio or Apple Xcode integrated development environments (IDEs).
4. Complementary technologies
On its own, a smartphone is sufficient hardware for only some assays—colorimetric detection being the most frequent example. For other assays, such as those based on fluorescence, a smartphone needs support from external components. There are several technologies that have a recurring role in supporting smartphone-based devices and assays.4.1 3D printing
Three-dimensional (3D) printing fabricates objects through successive deposition of layers of material.67–69 This rapidly proliferating technology is democratizing prototyping via simple-to-operate and consumer-affordable desktop printers. Although there are many classes of 3D printing, the two most relevant classes are fused deposition modelling (FDM)/fused filament fabrication (FFF) printers and photopolymerizable resin-based printers.FDM/FFF printers are available at low to modest cost ($200–5000) and offer print resolution ≥0.2 mm, reasonable print speeds, potentially large build areas, and printing materials such as acrylonitrile butadiene styrene (ABS) and polylactic acid (PLA). These printers have been widely adopted for fabricating the housing of smartphone-based devices, as well as holders, mounts, and supports for optical components, circuit boards, and other peripheral components.
Photopolymerizable resin-based 3D printers are available in stereolithography (SLA), digital light processing (DLP), and masked stereolithography (MSLA; also known as LCD) varieties that use epoxy, acrylic and methacrylic monomers. The purchase prices are similar to FDM/FFF printers. Although post-processing such as UV curing and solvent washes are sometimes needed, the prints are macroscopically smooth, watertight, robust, and (for colorless resins) semi-transparent. The technology is suitable for the fabrication of watertight flow cells and making molds for the soft-lithographic preparation of microfluidic chips.70–72 Lenses can also be fabricated but will tend to require post-print smoothing by sanding or coating procedures to obtain sufficient transparency for practical use.73–75
Collectively, a smartphone, an FDM/FFF 3D printer and filament, a photopolymerization 3D printer and resin, and some basic tools (e.g. sandpaper, knife, rotary tool and bits) have the potential to function as a near-complete kit for prototyping or maintaining a smartphone-based analytical device. Depending on the measurement modality and assay designs, some additional kit may also be warranted: light sources (e.g. LEDs), electronic components (e.g. wire, resistors, connectors, solder, soldering iron), simple optical components (e.g. lenses, mirrors, colored filters), mild solvents (e.g. isopropyl alcohol), and simple plasticware (e.g. tubing, syringes). Although these resources are not household items and not without cost, they are realistic and common for professional and at-home workshops to stock and utilize. Examples of LEGO®-based smartphone devices,76,77 including with deep-learning-based computer vision, further show that analytical function is not predicated on sophisticated hardware external to the smartphone.
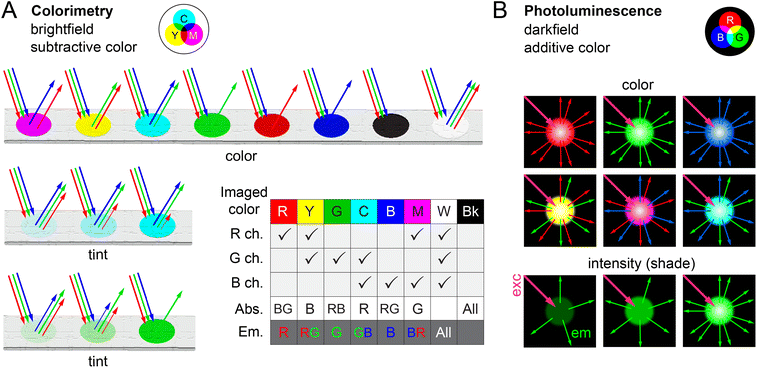
4.2 LEDs and laser diodes
Light-emitting diodes (LEDs) are ubiquitous in consumer electronic devices and have small sizes, low cost ($0.25–100 USD), and long operating lifetimes (≥104 h), making them an ideal peripheral light source for many smartphone-based devices. LEDs with broad-spectrum “white light” emission (similar to a smartphone flashlight) and with nominally monochrome emission (FWHM of 20–70 nm; Fig. 5) from the UV through near-IR regions of the spectrum are available with milliwatt scale powers. With the exception of UV and some violet LEDs, the frequent trade-off for the lower cost of an LED is the loss of access to some or all of the bandwidth of an imaging RGB color channel due to the long-pass optical filter needed to block stray LED light. The inclusion of a narrow bandpass excitation filter in a design mitigates this disadvantage at the cost of lower excitation intensity.78–80Laser diodes are also components in consumer devices (e.g. optical drives, laser pointers, barcode readers, 3D printers, laser printers) and are available at low to high cost ($10–1000) with small size, milliwatt powers, and wavelengths from the UV to the near-infrared. Although cost will generally favor the use of LEDs, the monochromatic output of laser diodes (FWHM ≤ 1 nm; Fig. 5) can be a technical advantage for photoluminescence (PL) measurements—the application where a majority of laser diodes29,81–86 and non-white LEDs have been utilized—because a greater fraction of a smartphone camera RGB channel remains accessible.
Many LEDs and laser diodes are operable with the voltage and current output available from a smartphone USB-C connector, power bank, or common consumer batteries. A simple circuit can step voltage up or down as needed. LED wavelengths have ranged between 285 nm (UVB) and 625 nm (red). Laser diode wavelengths have ranged from 365 nm (UVA) to 638 nm (red), with powers ranging from 20 mW up to 180 mW. The notable exception has been measurements with upconversion materials, which utilize 808 nm or 980 nm wavelengths. For applications with non-trivial illumination requirements (e.g. PL measurements, microscopy, spectroscopy), laser diodes have been adopted as the excitation source about 15% of the time, whereas LEDs have been adopted about 65% of the time (or 70% when including the smartphone flashlight, vide infra).
4.3 Microcontrollers
SBCs such as Raspberry Pi compete with smartphones as a technology for operating portable analytical instruments. Certain models of SBC have some similar technical capability (GHz processors, up to 8 GB RAM, Bluetooth, Wi-Fi, Linux OS) but lack the display, user input, power, and sensor (e.g. camera) components that are built into a smartphone. Potential advantages of these SBCs over smartphones are their lower cost ($50–100 USD) and on-board pins for digital input/output (GPIO; 3.3 V, 16 mA) and power out (5 V, 1A), albeit that similarly functioning USB-C-connected external modules for GPIO can be connected to a smartphone. High-end external camera modules ($50 USD) for Raspberry Pi are available with adequate specifications and RAW support. The overall capabilities and features for smartphones and SBCs are generally too similar for both technologies to be logically used in tandem, and smartphones have an edge in several categories (Fig. 6).In contrast to SBCs, MCUs have the potential to complement smartphones as a technology for operating portable analytical instruments. The open-source Arduino boards are popular for rapid prototyping,87 and their combination with 3D printing has been reported to reduce costs by an order of magnitude for scientific technology.88 Arduinos integrate features like a modest central processing unit (8–84 MHz), memory (up to 8 MB RAM), wireless communication (e.g. Wi-Fi, Bluetooth), and 5 V pins for digital input/output, analog output, and pulse-width modulation output (a digital means of mimicking analog output). These devices require external power, do not have built-in user interfaces, and do not utilize an OS, but there is software for writing and uploading code (e.g. Arduino IDE, based on C++, or MicroPython). The Arduino hardware catalog also includes external “shields” that interface with the main boards to provide additional features (e.g. motor drivers). Some non-Arduino MCUs, such as ESP32 and Teensy models, have higher processing power (up to 240 MHz and more RAM) but lower output voltage (3.3 V). With prices that range from $10–100 USD and minimal duplication of smartphone capability and features, MCUs add versatility by supplementing smartphone-to-peripheral connections with smartphone-to-MCU-to-peripheral connections. Several devices reported in the literature have paired smartphones with MCUs.89–97
4.4 Microfluidics
Microfluidic and nanofluidic devices manipulate small volumes (μL to pL) of fluid as continuous flows in channels and as discrete droplets in multiple formats. Fluid is transported via the application of pressure, electrowetting,98,99 capillary forces, light,100 and more.101,102 Smartphone devices have the potential to interface with microfluidic devices as optical detectors, power supplies, controllers, data handlers, and user interfaces.103 Ideally, the smartphone replaces a laboratory instrument for detection and the microfluidic chip replaces the benchtop sample preparation and assay steps done by a lab technician.Common materials for microfluidic chips include glass, polymers, and even paper.104 Although long associated with fabrication in clean rooms, channel microfluidics are now broadly accessible through commercially available glass capillaries, 3D printing of molds for polydimethylsiloxane (PDMS)-based soft lithography, and 3D printing of complete chips.70–72 Do-it-yourself hardware is increasingly common: syringe pumps and peristaltic pumps that drive fluid flow have been built using 3D-printed components and low-cost motors that can be powered by batteries or spring loading,105–112 and a low-cost 3D printer kit has been transformed into multiple syringe pumps.113 Alternatively, microfluidic paper analytical devices (μPADs)114 and capillaric chips115 do not need pumps because flow is driven by capillary action—an operational simplicity that is ideal for portability. Paper also has the advantages of being lightweight, inexpensive, biodegradable, incinerable (e.g. if samples are biohazardous), and capable of sample cleanup via filtering action. Despite the discontinuation of commercial wax ink office printers, wax printing remains the most common method for patterning channels and zones on paper, but alternative methods of creating hydrophobic barriers have been developed.116,117
Beyond channels, digital microfluidics based on electrowetting are promising because of the complex operations that are possible in this format, and the potential to miniaturize the driving electronics and use optical actuation.118–123
For smartphone-based analyses, a microfluidic chip must have optical characteristics and a design that are suitable for the desired type of measurement. For example, reflection colorimetry often needs a white background, such as from a paper substrate or μPAD. PL detection benefits from transparent optical paths for emission and excitation light, avoidance of strong camera-directed reflections of excitation light, and low autofluorescence from the chip material. Nominally transparent polymers such as poly(methyl methacrylate),124 polycarbonate, polyethylene terephthalate,125–127 cyclic olefin copolymer,128,129 and PDMS are useful materials for microfluidic chips.130 These polymers transmit wavelengths longer than 400 nm, but transmission is usually lost between 300–400 nm, which has implications for excitation in PL measurements. The nominally transparent resins for photopolymerization 3D printing have a narrower range of transmission, with short-wavelength cut-offs nearer to ca. 430 nm, and often require polishing to be more transparent than translucent. Microfluidic chips made entirely of glass or with a glass face (e.g. PDMS-on-glass chips) typically provide the widest range of wavelength transmission. Autofluorescence and the scattering and reflection of light are sometimes challenging to address with smartphone-based devices, where considerations such as cost, robustness, and compact design may restrict the practical range of solutions to these problems.
5. Complementary materials
Smartphone cameras are capable imagers but nonetheless have technical limitations intrinsic to their design and intended use. The analytical performance of a smartphone-based device will almost always be less than what is possible with in-lab sample handling and a modern laboratory instrument, making it advantageous to adopt materials that help mitigate these limitations or otherwise simplify detection.5.1 Indicator dyes
Indicator dyes undergo a spontaneous change or restriction in their molecular structure or conformation in response to an analyte, with a concomitant change in their visible color or fluorescence properties (e.g. brightness). Numerous indicator dyes exist for pH and the detection of ions, single-stranded DNA, double-stranded DNA, RNA, and more. These materials facilitate “mix-and-measure” assays that do not require washes or other signal development steps, and produce signals that can be followed in real time. The simplicity is ideal for a portable diagnostic test with smartphone-based readout. A potential drawback is that the typical absorption coefficients and fluorescence brightness for molecular dyes (104–105 M−1 cm−1) are often modest compared to those for some nanomaterial chromophores and fluorophores, potentially resulting in less favourable detection limits and sensitivity.131 The excitation and emission spectra of many fluorescent dyes will also tend to restrict usage of one or more smartphone camera RGB color channels.Under approximate white-light illumination, molecular dyes produce color in smartphone images based on the wavelengths of light absorbed relative to the RGB color channel responses (Fig. 7A). In the dark, the imaged color of a fluorescent dye corresponds to the wavelengths of light emitted relative to the RGB channel responses (Fig. 8A).
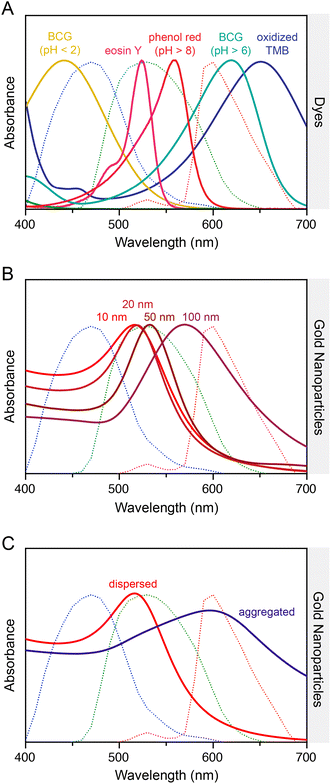 | ||
| Fig. 7 Normalized extinction spectra of selected materials. (A) Examples of common dyes: oxidized TMB,132 phenol red,133 eosin Y,133 bromocresol green (BCG). (B) Different diameters of AuNPs. (C) Dispersed and aggregated AuNPs. The spectra are superimposed on the typical RGB channel responses of a smartphone camera (dotted lines). Note: the spectrum for oxidized TMB is for its charge transfer complex with TMB, which is usually the first colored product from the catalyzed oxidation of TMB. Uncomplexed oxidized TMB has an absorption maximum at 450 nm (not shown) and is usually isolated by the addition of acid. The source data for dye spectra are cited. AuNP and BCG spectra were measured for this review. | ||
 | ||
| Fig. 8 PL emission spectra of selected materials: (A) common fluorescent dyes (DAPI = 4′,6-diamidino-2-phenylindole, fluorescein, TAMRA = carboxytetramethylrhodamine, Alexa Fluor 647, and the emissive product from turnover of furimazine by NanoLuc (dye spectral data from http://fpbase.org/); (B) different sizes of QDs; and (C) some trivalent lanthanide ions (data replotted form ref. 134). These spectra are superimposed on the typical RGB channel responses of a smartphone camera (dotted lines). | ||
5.2 Nanozymes
When sufficiently robust and selective, catalysts that are organic, inorganic, or biomolecular in nature are a useful means of producing amplified signals from individual binding events. Enzymatic amplification in molecular analysis is well-established in the form of enzyme-linked immunosorbent assays (ELISAs) and real-time polymerase chain reaction (PCR) to enhance detection limits with laboratory instruments. The catalytic amplification of chromogenic and fluorogenic reactions (typically using molecular dyes) has equal or greater value in mitigating the modest sensitivity of smartphone cameras compared to research-grade cameras and photodetectors. The potential trade-off is that these reactions add washing and signal development steps to assays, which conflicts with the oft-desired minimization of user actions.Borrowing directly from classic ELISA methods, the enzyme-catalyzed (e.g. horseradish peroxidase, HRP) and nanomaterial-catalyzed oxidation of 3,3′,5,5′-tetramethylbenzidine (TMB) to yield a blue color has been the most widely adopted chromogenic reaction for smartphone-based assays. Peroxidase mimics and other nanoparticle (NP) catalysts that are able to turnover a chromogenic substrate under ambient conditions (a.k.a. nanozymes) are of interest because of their potentially greater robustness and shelf life, lower manufacturing cost, and ease of modification versus conventional enzymes.135,136 Analogous to enzymes, nanozymes are not immune to inhibition by components of a sample matrix, and inhibitory effects are sometimes useful for the detection of certain analytes. Nanozyme-based amplification will benefit from the microfluidic automation of fluid handling to reduce multiple assay steps to a single user step.
5.3 Metal NPs
Gold nanoparticles (AuNPs; ca. 10–100 nm diameter) are a popular choice as a colorimetric label for smartphone-based assays. The plasmon resonance of these materials (Fig. 7B) generates a deep red color with very large per-particle extinction coefficients (107–1011 M−1 cm−1), and there is a substantial spectral change in optical extinction between dispersed and aggregated states, with color turning from red (strong green absorption) to blue/purple (broad absorption from yellow to red; Fig. 7C). AuNPs are most frequently paired with smartphone cameras for brightfield imaging, and occasionally for darkfield imaging. The main advantage in both cases is the sensitivity boost from a high optical contrast per small amount of material. Unlike many other NPs, where light absorption is the dominant contribution to extinction, light scattering is also very strong for metal NPs. Moreover, metal NPs can be leveraged for metal-enhanced fluorescence. The robust chemical stability of AuNPs is why these materials are much more prevalent in assays than other metal NPs with similar plasmonic properties.In most applications, AuNPs are adopted as a label for generating contrast in binding assays. Silver enhancement—the reductive growth of a thick layer of silver on AuNPs—has been used to generate even greater optical contrast, but the post-assay development steps are not ideal for a portable analysis without on-chip automation. The aggregative behavior of AuNPs also functions as a “mix-and-measure” sensor, but ensuring robust selectivity sometimes requires more careful and challenging design than for molecular indicator dyes.
5.4 Quantum dots
Quantum dots (QDs) are colloidal semiconductor nanocrystals (2–10 nm diameter137) that are brightly photoluminescent when prepared as high-quality core/shell materials.138 The emission wavelength is tunable across the visible spectrum via nanocrystal size and composition. For monodisperse CdSe/ZnS, CdTe/ZnS and related QD materials, the emission is also symmetric and spectrally narrow (FWHM 25–35 nm; Fig. 8B). Other QD materials, such as InP/ZnS, have visible emission that is less narrow (FWHM 35–50 nm) but still symmetric. QDs are also characterized by spectrally broad absorption and higher brightness versus organic fluorophores. The latter arises from larger molar absorption coefficients (105–107 M−1 cm−1), competitive emission quantum yields, and superior resistance to photobleaching. QDs have been shown to have a clear brightness advantage over fluorescent dyes and proteins, especially when blue light filtered from the smartphone flash is used for excitation.38The PL properties of QDs are an ideal match to smartphone cameras: high brightness mitigates the modest sensitivity of the image sensor versus research-grade cameras and photodetectors; the resistance to photobleaching enables longer image exposure times to further enhance signals; the narrow PL emission can be aligned to one of the RGB color channels to maximize sensitivity while minimizing crosstalk with other channels; and the broad absorption enables all colors of QD to be optimally excited with violet or UV light, leaving all of the camera RGB channels available for PL emission measurements. Most organic fluorescent dyes do not offer the same capability. At present, QDs are the material that most readily enables up to three-color multiplexed detection with a smartphone camera.139 QDs also function as “mix-and-measure” sensors for smartphone-based analyses through processes such as Förster resonance energy transfer (FRET),139,140 where these sensors can be sufficiently bright for detection directly in whole blood.141
The main drawback of QDs is the frequent inclusion of cadmium in the best-in-class materials. This inclusion is often framed as a toxicity issue, but such framing is an oversimplification.142–144 Toxicity has limited relevance to in vitro analyses, where measurements of biological samples are ex vivo, minute quantities of material are needed per test, and some assay formats will already include some level of containment for the sample. Rather, the more relevant challenge is one of environmentally conscious or regulated disposal in low-resource settings and, ultimately, sustainability for use at a global scale.
5.5 Semiconducting polymer dots
Semiconducting polymer dots (Pdots) and conjugated polymer NPs are two classes of NP (20–100 nm diameter) with a high mass percentage (>50% w/w) of fluorescent semiconducting polymer in their composition. These materials have very large molar absorption coefficients for UV and blue light (106–109 M−1 cm−1), competitive emission quantum yields, and photobleaching rates that are often slower than for fluorescent dyes. The semiconducting polymer emission is spectrally broad (FWHM 40–100 nm) but can be narrowed by incorporating suitable fluorophores into the polymer structure. The substantial advantage of Pdots is brightness that tends to be the highest per particle among the different types of fluorescent NPs. For example, Pdots have been shown to provide much lower detection limits than QDs in lateral flow-style binding assays, albeit outperformed by QDs for two-color multiplexing between the R and G smartphone camera channels.145 The brightness was even sufficient to measure subcutaneous glucose and NADH in mouse models using red-fluorescent Pdot-based sensors with smartphone-based imaging under a UV lamp.146,1475.6 Upconversion nanoparticles
Lanthanide-doped upconversion NPs (UCNPs; 20–150 nm diameter) leverage the electronic transitions that are possible within the 4f electron shells of lanthanide ions to generate visible luminescence from infrared excitation.148,149 As is characteristic of trivalent lanthanide ions, the PL emission consists of narrow lines (FWHM 15–30 nm) at specific wavelengths and with relative intensities that depend on the composition of the UCNPs. Several lanthanide emitters have emission lines in the visible spectrum (Fig. 8C). Excitation of UCNP PL is via sequential two-photon absorption, typically using laser diode wavelengths of either 808 nm or 980 nm.The main advantage of UCNPs with smartphone cameras is the near-IR excitation, which is a spectral region that is already blocked by the IR filter in the smartphone camera assembly and induces negligible autofluorescence from most samples and materials. UCNPs thereby enable imaging without an emission filter, with very low background, and with potentially less pre-processing of crude samples. For assay formats with short optical path lengths through samples, there is significant potential for the suppression of autofluorescence background from the assay vehicle (e.g. lateral flow membranes, paper test strips, microfluidic chips) to be a greater benefit than the suppression of autofluorescence from the sample itself.
A potential disadvantage of UCNPs is their modest brightness from relatively low absorption coefficients, modest upconversion quantum yields, and low emission rates. The emerging technology of organic-based triplet-triplet annihilation (TTA) upconversion NPs has the potential to provide higher brightness, but color multiplexing will typically be limited by the broad emission spectra.131 Although some UCNP compositions have an narrow emission line that is well aligned with an RGB color channel, other compositions have lines that are detected in multiple color channels. Results with smartphone-based imaging of luminescent lanthanide complexes (LLCs) suggest that the combination of Tb(III) and Eu(III) emitters is effective for two-color multiplexing between the G and R channels,150 and there is potential for Tm(III) to leverage the blue channel as well. With their typical UV excitation, molecular luminescent lanthanide complexes are arguably less appealing for smartphone imaging than UCNPs because of the technical need for a time-gated imaging mechanism to supress autofluorescence, which is more demanding than simple inclusion of a NIR laser diode. Otherwise, LLCs are expected to perform similarly to lanthanide-based UCNPs, with the possible added benefit of being directly substitutable for fluorescent dyes in assays.
5.7 Persistent-luminescence NPs
Persistent-luminescent NPs (PLNPs), also known as “afterglow” NPs, are materials that transiently trap and store energy from the absorption of UV-visible light or X-rays. This energy is gradually released as luminescence over a period of minutes, hours, or even days after the excitation source is turned off.151 The emission centers tend to be lanthanide ions, transition metal ions, or main group/post-transition metal ions, with peak emission wavelength and bandwidths that correspond to these origins.The primary advantage of PLNPs is the temporal separation of excitation and detection. An imaging camera is able to be turned off when the excitation light is turned on, and vice versa. This capability eliminates background from stray excitation light and sample autofluorescence. Persistent luminescence makes it possible for device designs to omit an optical filter (something that is essential for conventional fluorescence measurements) and to avoid background from autofluorescence. The potential drawbacks of PLNPs are concerns related to some of their metal-based compositions, the lower brightness that is an outcome of their slow emission rate, and additional parameters to keep constant for intensity-based calibration (e.g. excitation duration, time between the end of excitation and the start of imaging). In principle, the most suitable applications for PLNPs are those where the decrease in background more than offsets the decrease in signal compared to another type of luminescent material. In practice, it is difficult to fully benchmark the advantages and disadvantages of PLNPs because of the few studies with smartphone-based detection.
6. Measurement modalities and technical approaches
6.1 Role of the smartphone
Smartphones are utilized as components of portable analytical devices in three main ways: linked, dongle, and integrated (Fig. 9). Smartphone-linked devices are standalone devices in every way other than a wireless connection to a smartphone. Dongle devices require a physical connection to a smartphone, which is principally a user interface and power supply but is not directly active in analyte detection. Smartphone-integrated devices make direct use of the smartphone camera or another on-board feature (e.g. ALS, flashlight) for analyte detection. Integrated devices will frequently assemble dongle-like, wirelessly connected, and unconnected peripheral components around the smartphone to provide the full technical capability needed for a measurement. | ||
| Fig. 9 Conceptualization and characteristic features of (A) linked, (B) dongle, and (C) integrated devices that utilize smartphones. | ||
Linked and dongle devices are often bespoke and therefore adopt designs and components that are optimized for the analytical measurement and user context. The smartphone functions as a user interface and as a platform for data processing, storage, and sharing. Compared to other benchtop and portable instrument systems, linked and dongle devices benefit from offloading bulkier components (e.g. power, display, user input) to the phone. This approach decreases the size and cost of the analytical device when an analyst already has access to a smartphone. The anticipated worldwide standardization of smartphone charging ports to USB-C (already implemented in European Union152) will also enable linked and dongle devices to function independent of the make and model of smartphone. Indeed, dongle devices are an inevitable future for portable electrochemical analysis. The future for optical analysis is less certain because cameras and light sources are built into smartphones.
Given their bespoke designs and mere “connected” role for the smartphone, linked and dongle devices for molecular analysis are not discussed further in this review. Rather, the focus is on integrated devices that adopt, at minimum, the smartphone camera as an essential component. This approach requires fewer external components and less secondary engineering than linked and dongle devices, leading to simpler device prototyping (especially by non-engineers) and greater potential for democratization. The highly competitive consumer market also ensures that smartphones provide access to best-in-class image sensors, regularly upgraded technology, and opportunities to utilize non-imaging features to enhance measurements or data collection.
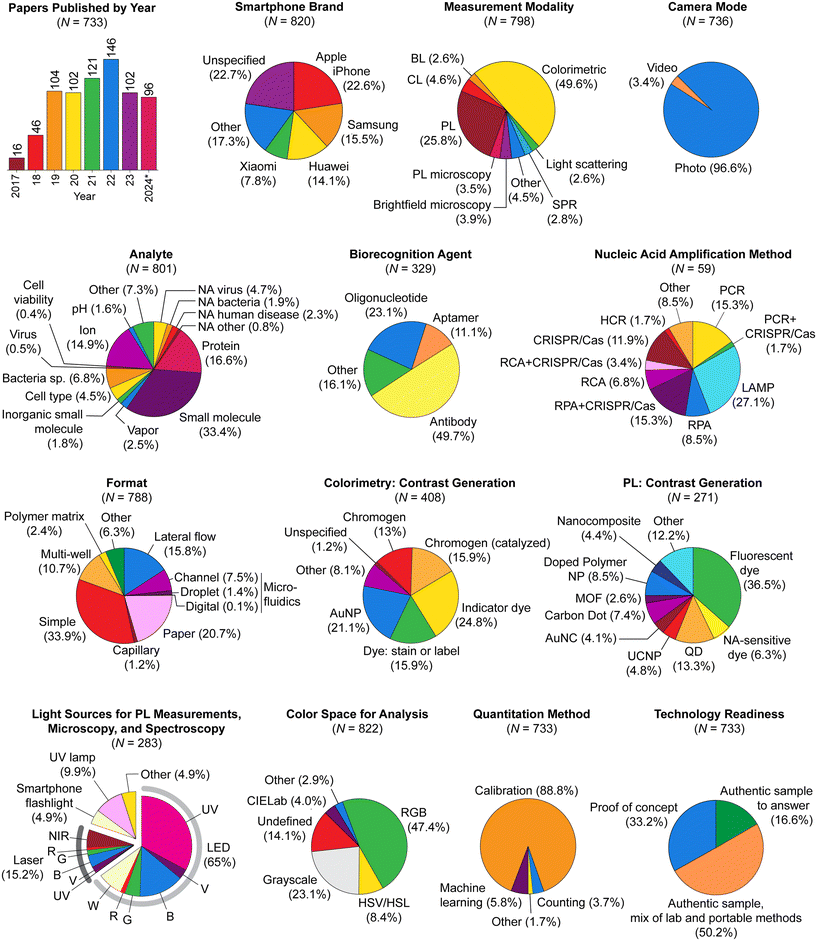
6.2 Colorimetry
This detection modality uses a smartphone camera to identify and quantitate changes in color, where both transmitted- and reflected-light configurations are useful for these measurements. Most measurements are under white light and the observed color is best predicted by subtractive color theory (Fig. 10A), with cyan (red absorbing), magenta (green absorbing), and yellow (blue absorbing) primary colors. Common signaling motifs include analyte-induced transitions between colorless and colored states, transitions between two different colors, and analyte-induced changes in color patterns for an array of chromophores. These signals most frequently originate from changes in the efficiency or dominant wavelength of light absorption associated with indicator dyes and chromogenic reactions, and from the retention of chromophores in binding assays (vide infra for examples). Changes in Bragg diffraction (e.g. photonic crystal sensors153) or thin film interference (e.g. etalon-based sensors154) would also produce analytically useful color changes but, to our knowledge, remain mostly unexplored with smartphone-based detection.Potential advantages of colorimetric methods include operation in ambient light, simplification or elimination of hardware peripheral to the smartphone, and human vision as a redundancy and quality control measure for camera-obtained results—all of which contribute to the status of colorimetry as the most popular detection modality with smartphone-based devices. Potential disadvantages include less favorable detection limits than luminescence methods, interference from samples that are naturally colored, and a functional dependence on the color temperature or spectrum of the illumination. Approaches that have been used to address the last challenge are the use of a box155–159 or an attachment160 that excludes ambient light and provides reproducible and optimized illumination from a built-in source, and the use of color standards to calibrate ambient lighting.161–165
Most devices for colorimetric detection have been designed for use with cuvettes, lateral flow membranes, μPADs, and other test strips. Additionally, some devices for absorbance measurements have been designed for 96-well plates. One technical approach is to use an array of 96 optical fibers to deliver light transmitted through each well to a defined spatial position on the smartphone camera image. Examples of light sources for this type of configuration include an array of blue LEDs166 and an electroluminescent plate as a white light source.167 An alternate approach is to use a large-diameter convex lens for concurrent imaging of an 8 × 8 array of wells without significant optical distortion.168
6.3 Light scattering
Elastic light scattering assays measure light intensity at a position that is not along the optical axis of sample illumination or its reflection, with a dark field of view in the absence of scatter. The necessary optical arrangement is detection at an orthogonal or large oblique angle to a light beam through a cuvette or glass capillary. In practice, light scattering measurements also tend to differ from colorimetric measurements in their use of a light source that is monochromatic (e.g. laser diode) or pseudo-monochromatic (e.g. single-color LED) rather than white. Nonetheless, there are analyses that utilize AuNPs with white-light illumination and analyze the scattered light via a color ratio.169 Light scattering assays require very efficient scatters and large analyte-induced changes in scattering efficiency to be practical and robust, such as in the case of analyte-dependent dissolution of NPs.170,1716.4 Luminescence
This detection modality uses the smartphone camera to measure the intensity or color of light emitted against a dark background (Fig. 10B). The observed color is best predicted by additive color theory, with blue, green, and red primary colors. Relevant mechanisms include photoluminescence (PL), chemiluminescence (CL), bioluminescence (BL), and electrochemiluminescence (ECL). The main advantages of luminescence detection are higher sensitivity and lower detection limits than other modalities, and capacity for color-based multiplexed detection within the same measurement volume. In some cases, luminescence methods will also better tolerate strong sample coloration.The main drawback of luminescence detection is the additional components that are required for a functional device. A box or compartment is needed to exclude ambient light and provide a dark imaging environment. An excitation light source is also needed for PL measurements, along with an emission filter to prevent reflected or scattered excitation light from reaching the smartphone camera. External single-color LEDs and laser diodes are common excitation light sources. Violet and UV wavelengths enable full utilization of the camera's RGB channels, whereas longer wavelengths preclude imaging in one or more color channels. Excitation filters are needed to isolate a band of wavelengths when the smartphone flash or another white-light source is used for PL excitation.38,41,145,172 Although infrequent, narrow bandpass filters have also been used with single-color LEDs to minimize the leakage of excitation light into the camera color channel to be used for detection.173,174 This approach avoids reducing the range of wavelengths useful for PL detection, which can sometimes be a consequence of using an emission filter that blocks the full spectrum of an excitation LED. Such filtering also has the side-effect of reducing the excitation intensity. Monochromatic laser diodes do not pose the same challenge because long-pass and notch filters are able to block the excitation light from reaching the camera while maximizing the remaining wavelength range for PL detection.
Although of minimal importance for brightfield colorimetric imaging, the distance between the sample and the smartphone camera (or another primary imaging lens) is a relevant design consideration because PL, CL and BL exhibit isotropic emission. The measured intensity thus scales as the inverse square of this distance and greatly impacts assay sensitivity.
An additional consideration for PL detection is that components in the path of the excitation light and in the field of view of the camera must not be autofluorescent, and should be transparent to the excitation wavelength to maximize sensitivity. These requirements are sometimes more challenging than anticipated when working with violet and ultraviolet excitation: some paper,175 plastic,176,177 and (more rarely) glass materials generate significant levels of autofluorescence. Some low-cost colored glass filters will also exhibit autofluorescence when incident excitation light (e.g. from a reflection) is too intense.178 Common clear plastics and glasses are transparent to NIR light (at 808 nm and 980 nm) and, as noted earlier, the use of UCNPs and PLNPs will avoid autofluorescence from samples and device materials alike. Nonetheless, UCNP users should also be cognizant of materials that could burn from extended exposure to the IR laser. UV light sources and NIR lasers suitable for upconversion PL both pose safety risks to skin and eyes that will need to be mitigated by suitable engineering of any device.
In contrast to PL detection, CL, BL, and ECL detection do not need an excitation source and optical filters. Nevertheless, there is intricacy associated with the temporal decay of CL and BL signals as a reaction progresses. In many cases, a greater number of reagents and assay steps are needed to generate signal. ECL provides a greater level of spatial and temporal control because the luminescence is restricted to an electrode interface and requires an applied voltage to be initiated.179,180 ECL emitters can sometimes also be regenerated on the fly. In general, the emission rates for CL, BL, and ECL have a lower ceiling than with fluorescence, such that competitive detection limits are predicated on the background being darker than what is possible with PL, whether due to technical considerations or sample autofluorescence.
6.5 Surface Plasmon resonance
Two main optical approaches have been used to implement surface plasmon resonance (SPR) measurements with a smartphone camera. In an optical fiber format (Fig. 11A), one end of the fiber is coupled to a light source and the other end of the fiber is imaged by the camera. A segment along the length of the fiber is coated with a gold film and integrated into a flow cell, where the plasmon is excited by light coupled into the fiber by total internal reflection. The light source has been the flash on a smartphone (filtered to select red light)42–45 or a standalone red LED.44,181 The other common format is to use a non-fiber waveguide with a planar gold film in a flow cell. Examples have included a 3D-printed clear-plastic optical coupler that totally internally reflects red light from the phone display to the front-facing smartphone camera;46,182 Kretschmann configurations that use the rear camera and a polarizer with either a red LED183 or the phone flash (plus a red bandpass filter; Fig. 11B);45 and a reverse Kretschmann configuration for SPR-coupled emission with a hemispherical prism.184–187 Although a niche measurement modality at present, SPR has the prospective advantages of label-free and indicator-free detection of binding.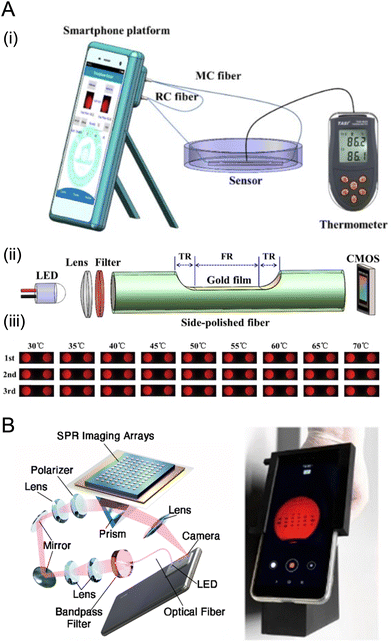 | ||
| Fig. 11 Examples of the two main optical approaches for SPR with a smartphone. (A) An optical fiber format: (i) measurement setup; (ii) optical axis; (iii) examples of smartphone images of the reference (RC) and measurement (MC) fibers. Other abbreviations: TR, transition region; FR, flat region. Figure adapted with permission from ref. 44 Copyright 2019 Optical Society of America. (B) A Kretschmann configuration with a planar gold film on a chip in a flow cell (left) and an example of a smartphone-captured SPR image of the chip. Figure adapted from ref. 45 Copyright 2022 Royal Society of Chemistry. | ||
6.6 Microscopy
Devices for smartphone microscopy typically incorporate lenses that are external to the camera to achieve micron-scale image resolution. In practice, these external lenses are low-cost objective lenses or bespoke combinations of individual lenses that serve the same purpose. Magnification has also been increased with attachment of the lens from the laser pickup head of a CD player over the smartphone camera.188 Ball lenses have also proven useful for magnification with simple and compact designs.189,190 With some smartphone cameras now offering up to 10X optical zoom, some low-magnification applications may bypass the need for an external lens. Multiple camera units on the back of the smartphone will also support optics for stereomicroscopy.191Beyond external magnification lenses, the components utilized for smartphone-based microscopy are similar to the components utilized for non-microscopy measurements. Devices for brightfield microscopy will incorporate a peripheral white LED source or redirect the smartphone flash for sample illumination because ambient light is not sufficiently intense for high magnification.192 Although most microscope devices have been built around a single smartphone, there have been instances of using two smartphones—one for the flashlight to illuminate the sample and one for the camera to image the sample—to simplify optical design and avoid any circuits.193
Although uncommon, the smartphone display has been used as a light source for microscopy. Examples include the use of a liquid light guide to collect light from the display for downstream modulation by a digital micromirror device for hybrid illumination (HiLo) optical sectioning with 12 μm axial resolution,194 and the use of the display as a dynamic and programmable light source to replace the typical two-dimensional LED array for Fourier ptychographic microscopy to obtain half-pitch resolution of 0.87 μm with a field of view of 3 mm2 (Fig. 12).195 An arguably simpler design used a separate OLED display, operated by a microcontroller, to act as a programmable light source for smartphone microscopy, demonstrating six different imaging modes: bright-field, dark-field, oblique illumination, Rheinberg, differential phase contrast, and fluorescence.50 More commonly, single-color LEDs and laser diodes are used with appropriate optical filters for PL microscopy. Theater stage lighting gel films and acrylic films have been used as a low-cost alternative to conventional scientific-grade emission filters.196,197 A related advance has been the development of colored polymer lenses that mount directly on a smartphone camera to concurrently magnify and block excitation light in PL imaging.198 Another example of custom imaging is a tunable optofluidic design, based on hydraulic deformation of an elastomeric membrane, that offers an adjustable focus.199 Simple mechanical approaches to focus adjustment have included spring-loaded screws to attach a phone cradle as the lid of a dark box,84 and a screw-actuated 3D-printed seesaw mechanism that offers as small as 5 μm adjustments.200
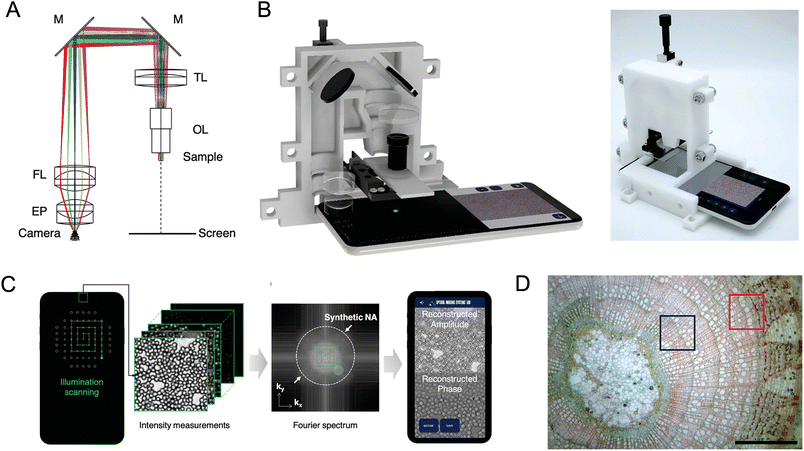 | ||
| Fig. 12 A smartphone-based Fourier ptychographic microscope using the display screen for illumination. (A) Optical layout and (B) rendering and photograph of the microscope. Diagram abbreviations: OL, objective lens; TL, tube lens; M, mirror; FL, field lens, EP, eyepiece lens. (C) Working principle of the microscope. (D) Full field-of-view color reconstruction result of cross-section of a Tilia stem. (The blue and red boxes indicate image regions that are enlarged in the original publication). Figure adapted with permission from ref. 195 Copyright 2021 American Chemical Society. | ||
6.7 Spectroscopy
Smartphone-based spectroscopy comprises absorbance, light scattering, luminescence, and SPR measurements with wavelength resolution. It is defined by the introduction of a dispersive element (e.g. prism, grating) as a peripheral component to the smartphone. The camera images the dispersion of the incoming light, obtaining wavelength-resolved intensity information (i.e. spectra) as a function of pixel address. The RGB color channels are inherent to the analysis but are secondary to the spatial encoding of wavelength. That said, the increasingly common presence of multiple rear cameras on smartphones may provide a technical solution to parallel spectral measurements and conventional imaging of the same sample.Most smartphone-based absorption spectrometry devices have been designed to utilize cuvettes. A plurality of designs use a piece of a DVD201–203 or CD204,205 as a proxy for a made-for-purpose diffraction grating. The grating is typically used in a transmission format, although some reflection designs have been reported. Low-cost thin film gratings designed for classroom education have also been adopted,206–208 as have research-grade transmission209 and reflection gratings.210 When reported, spectral resolutions have been <1 nm per pixel.204,209,211 Although most devices have 3D-printed housing, some devices have been made with laminated paper,201 cardboard and wood,203 and Plexiglas.202 The most common light source is a white LED external to the smartphone,201 although some designs have utilized a tungsten halogen lamp (i.e. incandescent bulb),203,209 a flashlight-sized xenon bulb,212 and redirection of the phone flash via a light guide.211,213 Two versatile designs implemented a bifurcated optical fiber to deliver light from the flash to the sample and to send reflected light back to the grating assembly and smartphone camera.32,208 Other interesting device designs have included a double-beam system,207 an eight-channel spectrometer based on a prism array for working with 96-well plates212 and 8-well strips,214 and endoscope designs.213,215 Two designs have also omitted a light source, once for flexibility in source selection216 and once for use with the Sun.204 The Sun offers the advantage of a broader and more uniform spectrum than an LED or lamp, but poses challenges with ensuring consistent incident light intensity between samples and blanks, and with weather-dependent utility.
Regarding the smartphone, a study evaluated more than 60 models for spectral measurements and highlighted the importance of manual control over image acquisition and processing settings, the trade-off between higher signal-to-noise ratios from larger camera pixel sizes and higher wavelength resolution from more pixels, and the generally superior analytical performance of newer phones versus older phones.211
The advantage of smartphone-based spectroscopy over imaging is wavelength-resolved data that is more analogous to lab-based spectrometers. In principle, this resolution will support more robust or higher levels of spectral multiplexing and facilitate differentiation of analytical signals from background. In practice, smartphone spectrometers are often better suited to education and citizen science than for chemical and biomolecular analyses. The built-in RGB channels of the smartphone camera intrinsically provide some wavelength-related information, and few studies to date have demonstrated an application where the extra resolution from a grating provides essential new information.
6.8 Image processing and analysis
A full discussion of image compression algorithms are beyond the scope of the review and can be found elsewhere.217–219 In brief, JPEG is a lossy format that simplifies the pixel data in the original image based, in part, on the limits of human vision and color perception. Nominally redundant data in the form of less perceivable detail is eliminated to conserve memory. Pixels in JPEG images can, in principle, adopt 28 = 256 different (i.e. 8-bit) levels of red, green, and blue (R, G, and B), for a total of 224 = 16![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 777
777![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 216 (i.e. 24-bit) colors. Compared to the JPEG format, the HEIC format uses less memory to store higher quality images, with the potential for up to 16-bit depth for each RGB color channel.
216 (i.e. 24-bit) colors. Compared to the JPEG format, the HEIC format uses less memory to store higher quality images, with the potential for up to 16-bit depth for each RGB color channel.
For both JPEG and HEIC images, an acquired raw image is first processed by demosaicing. This process uses pixel interpolation and correlation algorithms to assign a full triplet of R, G, and B values to an image pixel, even though each image sensor pixel was sensitive to only one of these colors. Subsequent processing includes color space conversion, white balance or color temperature adjustment, and file compression steps. Notably, the color processing steps alter the quantitative data from the image sensor. Most image sensors have a linear response to light intensity and the pixel signals are typically digitized on 10-bit up to 14-bit scales. In addition to reducing the bit depth in a stored image, standard JPEG or HEIC conversion will also apply a gamma correction that transforms the original linear correlation between pixel value and light intensity to a non-linear one that better mimics human vision. Increasingly, there are also other behind-the-scenes actions by the smartphone camera app: various computational methods are used to further enhance and optimize image quality, thereby better mitigating the hardware compromises necessary for a small form factor, user friendliness, and versatility. Although these operations add value to the popular uses of smartphone cameras, autonomous image processing algorithms may not be well calibrated to scenarios encountered in scientific imaging and measurements. For example, many smartphone cameras and their default apps will attempt to make dim light brighter by altering both the hardware parameters for image capture and the downstream digital image processing. Such processing risks a low luminescence signal from a low concentration of analyte falsely appearing to be high signal from a high concentration of analyte. In short, undefined image processing algorithms are anathema to quantitative analysis and are to be avoided.
One strategy for avoiding non-quantitative data is to use smartphone camera apps that offer control over important settings used for image capture and processing. Examples of such settings include the focus, exposure, ISO level, and white balance/color temperature. Manual control provides a level of consistency between images and is sometimes adequate for quantitative analysis even when storing images or videos in compressed formats. An alternative to JPEG and HEIC is to store images in RAW format. The RAW format provides a digital image with the highest fidelity to what the image sensor recorded, including greater bit depth, with the trade-off of requiring more memory. RAW files ideally retain the linear response of the image sensor and are minimally processed. Acquiring RAW images will thus be the best approach to any quantitative analysis that is based on the light intensity measured in one or more of the camera's RGB color channels. Numerous apps are available to enable RAW image acquisition with manual control over the aforementioned camera settings, including the advanced or “pro” modes of some default camera apps for newer smartphones. Examples of non-default apps include Adobe Lightroom, OpenCamera, Camera FV5, Halide Mark II, and several apps that have “Pro Camera” (or similar) in their name. Apps capable of RAW video acquisition have been rare but do exist (e.g. MotionCam). There are also tools available for researchers to code their own custom camera apps using application programming interfaces (APIs) and software development kits (SDKs) that are available for smartphone operating systems such as Android, iOS, and HarmonyOS.
With smartphones, the precise details of the processing applied to the exported RAW image (e.g. digital negative, DNG, file format) depends on the device and app, but typically includes a demosaicing (a.k.a. debayering) process to obtain a full color image. Note that some image processing and analysis programs may require that the bit depth of an imported RAW image (e.g. 10-bit) be downscaled or upscaled to a supported value (e.g. 8-, 16-, 32-bit). Downscaling will reduce image detail and dynamic range, and add quantization noise. Upscaling will add gaps in the pixel intensity histogram of an image but otherwise retains the original quality of the data. Upscaling cannot undo the effects of a prior downscaling.
Color Spaces. The additive RBG color space is the natural default for smartphone camera images because both the camera and the smartphone display model color as a combination of red, green, and blue values. These primary colors combine in pairs to make yellow, cyan, and magenta. A full contribution from all three primary colors approximates white light and the full absence of all three colors produces black. When each primary color is plotted on an orthogonal axis, the result is a cubic color space (Fig. 13A). For data analysis, the RGB color space is intuitive in how it mimics the operational principles of the smartphone camera. It is arguably non-intuitive in that, from a visual perspective, the identity of a color (i.e. hue) and its tint (for brightfield imaging with a white background) or shade (for darkfield imaging) are not independent parameters. This aspect of the RGB model sometimes motivates the use of other color models.
The hue-saturation-value color space (HSV; also called hue-saturation-brightness, HSB; Fig. 13B) is an alternative model to the RGB color space. HSV space uses a single coordinate, hue (H), to identify colors, where H is expressed in degrees (0–360°) on a cone or cylinder. For example, the RGB hues of red (255, 0, 0; 8-bit RGB coordinates), yellow (255, 255, 0), green (0, 255, 0), cyan (0, 255, 255), blue (0, 0, 255), and magenta (255, 0, 255) are at 60° intervals starting at 0° for red and ending at 300° for magenta, with brightness and saturation at 100%. The saturation (S) is expressed as a radial coordinate (0–100%) and quantifies the amount of a given hue versus a neutral white. The value (V) is expressed as the axial coordinate (0–100%) and is a measure of the perceived brightness of the hue. White has 8-bit RGB coordinates of (255, 255, 255) and HSV coordinates of (H, 0%, 100%). The corresponding coordinates for black are (0, 0, 0) and (H, S, 0%). There are defined equations for converting between the RGB and HSV color spaces.
Other examples of color spaces are the International Commission on Illumination (CIE) Lab model, its still-common predecessor, the CIE XYZ model, and two-dimensional representations commonly referred to as CIE 1931 (xy) and CIE 1976 (u′v′) (Fig. 13C). These models are perhaps less intuitive than the HSV color space, but are better mimics of the human perception of color. The CIE XYZ model defines X, Y, and Z coordinates as tristimulus values that nominally reflect the degree to which different wavelengths of light stimulate the three types of cone cells in an average human eye. Equations are defined to convert RGB coordinates to XYZ coordinates. In turn, equations are defined to convert the XYZ coordinates to the (x, y) and (u′, v′) coordinates for two-dimensional representation, and to convert XYZ to the L*a*b* coordinates of the CIE Lab model. This latter and newer model aligns with how the average human actually experiences four pure color stimuli (red, green, blue, yellow) due to cone-opponent neurons that combine signals from the three types of cone cells. The L* coordinate is the lightness of a color. The a* and b* coordinates reflect the redness-greenness and yellowness-blueness of a color, respectively, where a positive value indicates a predominance of the first color in a pair and a negative value indicates a predominance of the second color.
RGB is the natural color space for smartphone image analysis and has been adopted in close to half of published studies (vide infra). Many studies have also used HSV and L*a*b* coordinates, where a few of these studies have advanced a notion that these color spaces have the potential to enhance the data. This idea is a misconception: the HSV and L*a*b* coordinates are calculated directly from the original RGB values and thus cannot improve the quality of the data. Rather, conversion to these alternate color spaces has the potential to function as a form of data reduction, enabling fewer coordinates to reflect trends in the data and better correlating data with human perception. Grayscale conversion is likewise a data reduction mechanism because it is a weighted average of the RGB triplet values. Ad hoc color space conversions are not a substitute for knowledge of the optical spectroscopy relevant to a smartphone-based assay. The optimum approach to data analysis will emerge from careful consideration of the sources of signal, background and noise in an experiment, the sensitivity of the camera's RGB pixels to each source, and the contributions of each source to the coordinates of an image color space. Coordinates that maximize signal-to-background and signal-to-noise ratios will maximize analytical performance.
Signaling between low and high contrast states usually takes the form of a colorless to colored transition for colorimetric measurements, or a transition from dark to bright for luminescence measurements. For luminescence assays with a single luminophore, the transition is simply and effectively quantified via changes in the magnitude of the RGB channel values. The relevant channels will be determined by the overlap between the emission spectrum of the luminophore and the RGB channel responses, where the channel that provides the highest signal-to-background ratio will most often provide the highest quality data. Image analysis programs and coded scripts will output RGB coordinate values for pixels of interest and will split a color image into a separate monochrome image for each of the three channels. Background subtraction is important and should not be neglected.
Despite being adopted for data analysis in almost one quarter of studies (vide infra), the best-case scenario is that grayscale conversion has a neutral effect on data quality for colorimetric and luminescence measurements. It is equally likely to have a detrimental effect from averaging in color channels with inferior signal-to-background ratios. It should also be noted that data from an image is unreliable for quantitative analysis if a significant number of pixels are saturated in a color channel (i.e. an 8-bit value close to 255). Gross saturation is frequently observed as a white-hot glow for an imaged color of luminescence, but pixel data can be adversely affected before this effect is visually apparent.
For a transition between a colorless and colored state under approximate white light, the relevant color coordinate will be determined by the overlap between the absorbance spectrum of the chromophore and the RGB channel responses. The RGB channel that covers the largest fraction of the area of the absorbance spectrum will be most sensitive to the color transitions. Unlike in darkfield luminescence imaging, where, for primary colors, the visual color tends to match the optimum RGB channel, the visual color for brightfield imaging under approximate white light will tend to be complementary to the color of the optimum RGB channel. For example, red luminescence is best quantified in the R channel, whereas a red color in brightfield imaging is often best quantified in the G channel. The S value in HSV space is also an intuitive and suitable parameter for quantifying colorless to colored transitions, with the caveat that there is still potential for some of the data variance to be captured by the H and V coordinates. The data reduction from grayscale conversion has the potential to increase dynamic range from its averaging in of less-responsive RGB color channels, but risks a reduction in sensitivity for vivid colors.
Another common scenario is quantifying the degree of a transition between two color extremes, such as a blue-to-yellow transition or a green-to-red transition. A ratio of values between two RGB channels will often suffice to produce a monotonic trend in both darkfield luminescence and brightfield color imaging. The specific RGB channels will be those that align with the majority of the absorbance or emission that produces the two colors. A ratio between the sum of two channels and the third channel (or the mathematical inverse) also has the potential to be useful in the case of color changes associated with large shifts in broad spectra. Alternatively, the H coordinate of the HSV color space is a good conceptual match to this scenario and often utilized for this purpose. Although sometimes utilized, ML methods are generally unnecessary for quantifying a simple transition between two colors. Rather, the value of ML in this situation is accounting for significant variation in the illumination or in the optical properties of sample matrices (i.e. inconsistent background and noise). ML methods have similar utility when applied to colorless to colored transitions or dark to bright transitions in luminescence.
The last common scenarios for data analysis are color recognition and color pattern recognition, and it is here that ML methods are excellent for signal processing. When illumination is not well controlled, ML will have the dual role of color identification and correction for variation in conditions. Examples of ML for color recognition include support vector machines (SVM) for brightfield220–222 and luminescence85,223 imaging, and neural networks (NNs) for brightfield.165,224,225 NNs, hierarchical cluster analysis (HCA), and linear discriminant analysis (LDA), and principle components analysis (PCA) have been used to discriminate between color patterns from array-based assays.226–232 The data processing usually requires a calibration or training data set that spans a broad scope of possible assay inputs, color outcomes, and imaging conditions. Since most of these ML methods intrinsically perform a data reduction or transformation, there is little or no value to a prior conversion from RGB to another color space.
For research and development, an offboard approach is common for the implementation of data analysis methods. Familiar computer-based tools (e.g. ImageJ/Fiji, Adobe Photoshop, Icy) and programming tools (e.g. MATLAB, Python) are utilized for extraction and processing of data from smartphone-acquired images. For onboard analysis, whether for prototype development or downstream applications and end-users, the norm is development of a custom smartphone app. There are numerous online resources to assist novices in coding smartphone apps, inclusive of leveraging code already generated in, for example, MATLAB and Python.
7. Analytes and assays
7.1 Metanalysis
Fig. 14 categorizes and summarizes some of the recent trends in the literature on smartphone-based detection and devices. This analysis surely did not capture the totality of studies published over the past 7 years, but is enough of a sampling to identify trends. Prevalent trends include colorimetric assays, static imaging, the use of antibodies as biorecognition agents for analytes, standard calibration, and portable detection without fully portable sample processing. Although some of these trends are likely to continue over the next several years, the implementation of ML and AI methods is rapidly increasing. We anticipate that the percentage of studies using these tools will be notably larger even within a year or two.An opportunity for improvement in the field is for more studies to provide full technical details about the components of their smartphone-based device, which are sometimes lacking in full or in part. Common examples of omissions include the power of light sources and wavelength information beyond a nominal color (e.g. peak and bandwidth), specifications for lenses and other optical components, the name of the app and full imaging settings (e.g. exposure time/shutter speed, ISO value, color temperature), and details around the extraction of quantitative data from images. Nevertheless, the meta-analysis data highlights the prevalence of some of the technical approaches discussed in the preceding sections and the diversity of the assay designs discussed in the following sections.
7.2 Colorimetric analyses
Smartphone cameras have led to renewed interest in methods of analysis based on color changes. The RGB image from the camera enables color identification to be quantitative rather than qualitative, and for analyte quantity to be determined by calibration rather than titration to an end point. Almost 400 papers published in past 7 years have reported different variations on the colorimetric detection of analytes, making it the most popular detection modality with a smartphone camera. Collectively, indicator dyes, stains, and other molecular chromogens have been used for color generation in about two thirds of published studies, with AuNP adopted in about one in five studies (Fig. 14). | ||
| Fig. 15 Colorimetric RT-LAMP assay with an indicator dye and smartphone-based color measurement. (A) Assay work flow. (B) Color change induced by mixing a drop of SYBR green I in the lid of the tube after iLAMP reaction with different copy numbers of synthetic viral RNA. (C) Hue measurement of different sample pools after amplification. Samples from healthy individuals are marked in blue; samples for SARS-CoV-2-positive patients are marked in green. Adapted from ref. 241 under a Creative Commons Attribution 4.0 International License. | ||
In other cases, analytes have reacted with colorless reagents to form a new colored species. Examples include the reactions of 2,4,6-trinitrotoluene with an amine to generate a brown Meisenheimer complex,258 formaldehyde with acetylacetone and ammonium to generate a yellow product,259 furfural vapour from gasoline with aromatic amines to generate a pink product,260 and phenolics (e.g. capsaicinoid analytes261) with Gibbs reagent and with Folin–Ciocalteu assay reagents to generate a blue color.262 Further examples include the detection of creatinine in urine via its reaction with picric acid (Jaffé method; orange color),263 nitrite via its reaction with Griess reagents (pink color),264 a cannabinoid via its reaction with Ehrlich reagent (pink color),265 and gamma-hydroxybutyrate (a recreational drug) via its reaction with hydroxylamine and complexation of aqueous Fe3+ (purple color).266
Clearly, there is no shortage of chromogenic reactions that a smartphone camera is equipped to measure; however, their practicality for portable detection depends on the degree to which reagents must be handled by a user and the associated hazards. Most of the procedures for the detection of the solution-phase analytes noted above were at small scale and used a smartphone for quantitative readout, but were otherwise not substantially different from a standard benchtop laboratory protocol. This aspect is a potential barrier to successful translation to non-laboratory settings.
When an analyte is neither a substrate nor an inhibitor of an enzyme, then HRP-conjugates are frequently adopted to enable chromogenic detection. Examples of blot-style assays include the aptamer-based detection of Mycobacterium tuberculosis with TMB chromogen,274 and the immunodetection of the SARS-CoV-2 spike and nucleocapsid antigens (LOD 2 pM) via a chromogenic HRP-induced silver metallization reaction.275 Competitive enzyme-linked immunosorbent assays (ELISAs) have been ported to paper substrates for the smartphone-based detection of tetracycline (LOD 0.5 ng mL−1) and chloramphenicol (0.05 ng mL−1),276 and ketamine in clinical oral samples (0.3 ng mL−1).277 Color generation in ELISAs has been enhanced by NP scaffolds that carry multiple copies of HRP, as in the cases of AuNPs in an assay for saxitoxin (0.4 ng mL−1; Fig. 16)278 and graphene oxide in an assay for okadaic acid (0.02 ng mL−1).279 To make the ELISA more user friendly for portable detection, capture antibodies were immobilized on microbeads within wells that had a membrane bottom with pores smaller than the microbead diameter. The wells were placed on top of an adsorbent pad to facilitate washing through the pores while retaining the microbeads for detection for SARS-CoV-2 nucleocapsid protein (LOD 7.5 pg mL−1).167 As another example of a strategy for simpler execution of an ELISA, capture antibodies for FXYD3 (a urothelial cancer biomarker) were immobilized on the inner surface of the lid of a glass vial.280 The lid was moved stepwise between vials with the different assay reagents and shook, while also adopting AuNP-HRP conjugates for extra amplification per binding event.
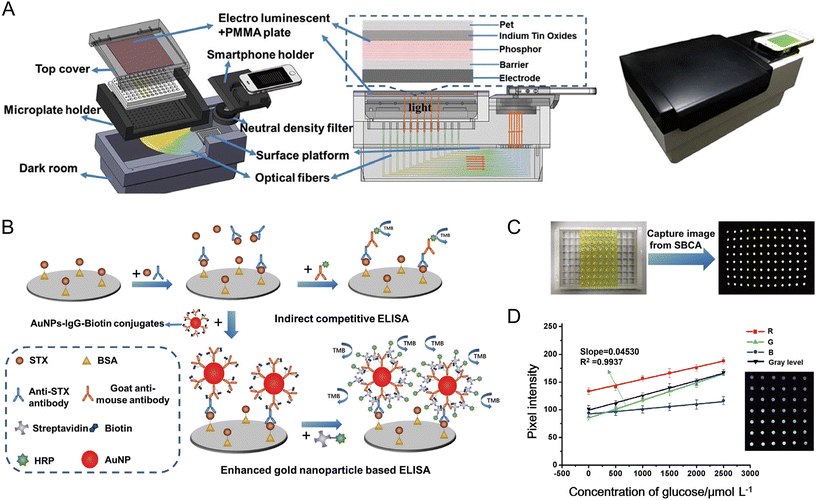 | ||
| Fig. 16 Smartphone-based colorimetric analyzer for an AuNP-enhanced ELISA. (A) Device design. (B) Assay method. (C) Smartphone image (right) derived from the multi-well plate (left). (D) Plot of pixel intensity versus glucose concentration from RGB and grayscale image analysis. Abbreviations: STX, saxitoxin; BSA, bovine serum albumin. Also refer to the Abbreviations list. Adapted with permission from ref. 278 Copyright 2019 Springer Nature. | ||
Although colorimetric detection of nucleic acids is rare, one study leveraged the decrease in pH from polymerase activity during LAMP amplification to detect a SARS-CoV-2 gene (LOD < 50 copies/reaction) using the color change of phenol red pH indicator.281 Another study combined RT-LAMP, CRISPR/Cas9, and a DNAzyme-catalyzed chromogenic reaction for the detection of SARS-CoV-2 in clinical samples.282
Other colorimetric assays with NPs have been special cases where an analyte modulates the catalytic activity of the NP. For example, serum glutathione (LOD 0.3 μM) was detected through its inhibition of the oxidation of TMB by zeolitic imidazolate framework (ZIF-8)/AuNP materials.289 Conversely, aqueous Hg2+ (LOD < 8 ng mL−1)290 and Cr6+ (LOD 0.05 nM)291 were detected via their enhancement of the catalytic activity of AuNPs toward oxidation of TMB and methylene blue, respectively.
For the parallel detection of multiple analytes, each chromogen nominally responds to only one analyte. For example, a wearable patch was developed for detecting pH, urea, and Ca2+ (aq) in human sweat via separate spots with a cocktail of three pH indicator dyes, p-dimethylaminobenzaldehyde (Ehrlich reaction with urea), and o-methyl phenolphthalein indicator (Fig. 17).164 Another wearable patch was reported for the AI-assisted analysis of pH, vitamin C, Ca2+ (aq), and protein levels in tears using a cocktail of indicator dyes and three chromogenic systems.230 A trio of indicator dyes was used on a paper substrate for the detection of gaseous ammonia, formaldehyde, and hydrogen sulfide,294 and chromogenic inks were printed in a QR code pattern for dosimeter-like response to relative humidity, temperature, and gaseous H2S, NH3, and CO2.295 NO3− (aq), Mg2+ (aq), Ca2+ (aq), and NH4+ (aq) were detected as macronutrients of interest through a μPAD with a combination of the Greiss method, three indicator dyes, and the salicylate method.296 A conceptually similar mix of indicator dyes and chromogenic reactions was used to develop a μPAD for the detection of Fe3+, Ni2+, Cr6+, Cu2+, Al3+, and Zn2+ (aq).297 NNs enabled an array of five chromogenic reactions to distinguish between seven illicit drugs,231 and enabled a linear barcode of 20 different compositions of a halochromic dye, chitosan NPs, and cellulose acetate to predict meat freshness from its volatile amines.229
 | ||
| Fig. 17 Stretchable patch for colorimetric sweat sensing directly from skin. (A) Design of the patch. (B) Photograph of the patch with six reference colors and indicator spots for detecting pH, urea, and Ca2+ (aq). (C) Smartphone imaging of the patch for in situ colorimetric analysis. Adapted with permission from ref. 164 Copyright 2024 American Chemical Society. | ||
Enzyme-catalyzed chromogenic reactions have also been multiplexed. One example is a plate-based assay for the detection of glucose, uric acid, cholesterol, and triglycerides via oxidase-peroxidase coupled reactions ending in the chromogenic oxidation of 4-aminophenazone, supplemented with a classic biuret reaction and indicator dye for the additional detection of total protein and albumin (Fig. 18).298 A related example is an injectable “ink” for pH measurement (via a cocktail of three indicator dyes), glucose (via the glucose oxidase–peroxidase-TMB chromogenic system), and albumin (via a protein-binding indicator dye).299
 | ||
| Fig. 18 Colorimetric assay for the simultaneous determination of glucose (GLU), total protein, human serum albumin, uric acid, total cholesterol, and triglycerides in plasma samples. (A) Smartphone image of the multi-well layout of the colorimetric array (each row) and its response to different samples (each column). (B) Comparison of calibration curves for glucose measured by smartphone (left y-axis) and by conventional spectrophotometry (right y-axis). Adapted with permission from ref. 298 Copyright 2024 Springer Nature. | ||
In aggregative detection mechanisms, an analyte compromises the colloidal stability of the AuNPs. However, loss of colloidal stability is also a potential outcome from changes in temperature, ionic strength, and pH, as well as from non-specific binding interactions with molecules in a complex sample matrix. Although some aggregative mechanisms have been functional with authentic samples, a majority of studies have used lab-prepared solutions of analyte for proof of concept or relied on substantial dilution of authentic samples. Many of these approaches lack a defined and robust mechanism for selectivity and are not discussed in this review. Nevertheless, there have been examples of aggregative detection with a selective component: the nominal trypsin activity in pancreatitis samples was detected via a color change when the bovine serum albumin molecules stabilizing AuNPs were hydrolyzed to cause particle aggregation;300 and a five-material array of AuNPs synthesized with different ligands and reducing agents was able to discriminate between eight different pesticides in washes of fruit samples.301 In the latter case, the color changes were related to aggregation dependent on the level of inhibition of acetylcholine esterase activity by the pesticide.
Assembly-based assays incorporate a component for selective recognition of analyte that induces or disrupts a close proximity between AuNPs. As a simple and elegant example of this approach, AuNP–antibody conjugates bound to the spike protein on SARS-CoV-2 virus with sufficient density to generate a color change from red to blue (LOD 0.3 PFU mL−1; Fig. 19).302 Analogous detection of Listeria monocytogenes was also demonstrated,303 and the concept was extended to the detection of a nucleic acid reporter sequence from a magnetic pull-down immunoassay for creatine kinase-MB (CK-MB; LOD 0.8 pM).304 In the later example, the reporter was amplified by rolling circle amplification (RCA) and the proximity needed for a color change arose from binding of AuNPs (less sensitive) and AuNP-tetrahedra (more sensitive) along the repeating sequence of the linear product.304 In another case, hybridization-mediated assembly between AuNP–oligonucleotide conjugates generated signal in a proteolysis-responsive transcription assay for matrix metalloproteinase-2 activity (LOD 5.7 pM) in serum, which distinguished between healthy persons and cancer patients.305 Conversely, the trans-nuclease activity of CRISPR/Cas12a was used to prevent the assembly of AuNP–oligonucleotide conjugates for the detection of SARS-CoV-2 (LOD 1 copy per μL).306
 | ||
| Fig. 19 Colorimetric detection of SARS-CoV-2 analysis in saliva samples using a smartphone. (A) Assay concept. (B) Change in color, measured via a B/R ratio, for samples with and without SARS-CoV-2 virus. The inset shows the change in the color of solutions with different amounts of virus (measured in plaque-forming units). Adapted with permission from ref. 302 Copyright 2022 American Chemical Society. | ||
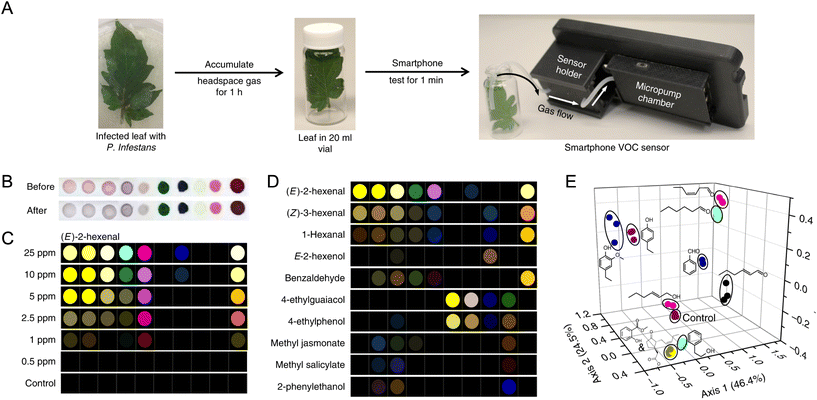 | ||
Fig. 20 Analysis of plant volatiles using a portable smartphone-based device and colorimetric array. (A) Assay work flow and photograph of the device. (B) Smartphone image of the colorimetric array before and after exposure to headspace gases. (C) RGB difference profiles for the array exposed to different concentrations of (E)-2-hexenal (0.5–25![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) ppm). (D) RGB difference profiles of ten representative plant volatiles at 10 ppm). (D) RGB difference profiles of ten representative plant volatiles at 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) ppm. (E) PCA score plot showing differentiation between multiple volatiles. Adapted with permission from ref. 310 Copyright 2019 Springer Nature Limited. ppm. (E) PCA score plot showing differentiation between multiple volatiles. Adapted with permission from ref. 310 Copyright 2019 Springer Nature Limited. | ||
The reactivity of NPs has also been leveraged for the detection of solution-phase analytes. Bromide (LOD 0.5 mg dL−1) and glucose (LOD 1 mg dL−1) were detected via their direct and indirect (H2O2 production by glucose oxidase) etching of silver triangular nanoprisms;225 Hg2+ (LOD 50 μM) was detected in tap water via its color-fading amalgamation with AgNPs;311 and the activity of acid phosphatase (LOD of 0.97 U L−1) in human serum was quantified through a set of coupled reactions that ultimately produced triiodide to etch gold nanorods (AuNRs).312 As another example, cyanide was detected by its ability to etch Au@Au–Ag NPs, where three different sizes of the NPs (and thus starting colors) were used in an array format for greater quantitative reliability.313
A more exotic approach to detection has been the growth of NPs to generate color as the readout of immunoassays. A competitive immunoassay for tyramine utilized an alkaline phosphatase conjugate of the antibody to convert ascorbic acid phosphate to ascorbic acid to enable the formation of gold nanostars from gold salt.314 A similar approach grew silver on AuNR seeds for the detection of acrylamide, albeit requiring chemical derivatization of the analyte as the first step in the immunoassay.315
The principal motivation for smartphone-based imaging of LFIAs is quantitation based on the color intensity of the test line. Whereas qualitative LFIAs are useful for binary scenarios (e.g. pregnant or not, infected with virus or not), quantitative LFIAs enable assays of analytes that are naturally present in a sample to determine if levels are too high or too low. Quantitative smartphone readout of LFIA test lines has been shown to be well correlated with lab-based CL immunoassays.324 When applied with LFIAs with multiple test lines for the same analyte—a configuration that nominally renders the assays pseudo-quantitative by visual inspection—smartphone readout provided a modest improvement in detection limit.325
In general, any strategy that improves LFIA sensitivity by visual inspection also benefits readout by smartphone. Examples of strategies include compression of a zone of the membrane to slow flow at the test line,326 narrowing of the membrane at the test line to increase the density of analyte binding,327 and electrokinetic preconcentration based on ion concentration polarization with a commercial 9 V battery (2.7-fold improvement in LOD).328 Assembly of a cluster-like network of AuNPs, without a change in their plasmon resonance (i.e. red color), decreased the detection limit for an LFIA for enrofloxacin (LOD 10–1 μg L−1) by three orders of magnitude compared to single AuNPs.329
Peripheral hardware for smartphone-based colorimetric LFIAs has been wide ranging. At the extreme of simplicity, hardware is absent and the assay relies on ambient light and manual positioning for image acquisition. In other cases, there are attachments and boxes to position the smartphone and LFIA strip and provide consistent illumination via an internal light source, whether the source is the smartphone flash77,330 or LEDs external to the phone.31,316,331,332 It is also possible to combine a box with ambient lighting for readout.317 At the technical extreme, a multi-step LFIA was semi-automated by means of a device with a motorized rotor (Fig. 21).333 The rotor had four diametric pairs of sample pads (for adding chromogenic signal development reagents) and absorbent pads (for driving fluid flow) that moved in sequence to bracket a central test zone membrane. A less technical approach for two-step development of signal in an LFIA was a two-tiered LFA with a slow-dissolving polymer barrier to delay the arrival of reagents for amplification (by NP growth) until after the initial binding of analyte and AuNP-antibody conjugates at the test line.332
 | ||
| Fig. 21 Rotary device for an enzyme-linked LFIA with a colorimetric detection. (A) A photograph and illustration of the rotary device containing the nitrocellulose (NC) membrane, the sample pads and absorbent pads. The top layer is rotated via a motor controlled by the smartphone via Bluetooth. (B) Assay steps (a–e). Each step takes 5–10 min. (C) Smartphone images showing (top) the signal generation from the chromogenic turnover of 3,3′-diaminobenzidine (DAB) with increasing concentrations of the target E. coli bacteria, and (bottom) the selectivity for E. coli over other bacteria species. Figure adapted from ref. 333 under a Creative Commons CC BY 4.0 license. | ||
Image processing for LFIAs have been as straightforward as color analysis of regions of interest in the RGB, HSV, and CIE spaces discussed earlier, as well as grayscale, but has also incorporated computer vision and ML for the recognition of test cartridge and line positions, and for image calibration.334,335 Examples include the application of statistical image analysis,336 support vector machines,337,338 convolutional NNs,339,340 and other deep NNs.341
Smartphone cameras have also benefited multiplexing with LFIAs. Some approaches, like bifurcated membranes, derive only the aforementioned quantitation advantage.342 Other multicolor approaches leverage the RGB information available. In one case, a single competitive LFIA was able to distinguish between food contamination with one of two different types of mycotoxin (aflatoxin B1 and type-B fumonisins; LODs of 1 and 50 ng mL−1) through a pair of AuNP–antibody conjugates that roughly corresponded to red (30 nm diameter) and blue (75 nm) colors.343 In another case, a sandwich LFIA featured a combination of two test lines, each with two co-immobilized capture antibodies, plus two different reporter antibody conjugates of each of blue and red latex microparticles, to detect and discriminate between serogroups of Salmonella (Fig. 22).344 The position (first or second line) and color (blue or red) of test-line binding indicated the specific serogroup. In both cases, the camera provided a non-subjective means of color recognition. Another approach utilized an LFIA with three test lines for three mycotoxins and red, green, and blue colors of reporter antibody–latex microsphere conjugates.345 Here, the color and line location provided a level of redundancy.
 | ||
| Fig. 22 Two-line two-color LFIA for serotyping Salmonella. (A) Assay design (left), smartphone images of LFIA results for four different serotypes (O:2, O:3, O:7, O:9, top-right), and position of the blue and red latex microspheres (LMs) on an HSB color space projection. (B) Smartphone images of LFIA test strip response to different concentrations of O:2 standard strain and O:3 standard strain. Adapted from ref. 344 with permission. Copyright 2023 Elsevier. | ||
The vast majority of LFIAs have, to date, utilized AuNPs to generate contrast at test and control lines, and smartphone-based approaches are no exception. As noted above, dyed latex microparticles—another classic contrast label for LFIAs—have also been utilized with smartphone readout.331,344 Nevertheless, numerous papers have evaluated alternative materials: blue-colored iridium (VI) oxide NPs,346 which provided approximately double the contrast compared to AuNPs; rhodamine B-labeled polymersomes;347 polydopamine NPs;334 composite materials derived from hyperbranched AuNPs and polydopamine;348 carbon black NPs;349 and NPs derived from cuttlefish ink.350 A caveat in comparing the contrast from different NP materials is that extinction coefficients tend to increase with increasing size. It is therefore necessary to consider both the type and size of a material when choosing a colorimetric label. Size is also a potential consideration in so far as it affects NP transport on the LFIA membrane, non-specific binding, and blocking of membrane and pads.
Other LFIAs have adopted nanozymes and other NPs that catalyze chromogenic reactions for signal amplification with LFIAs. Examples of such materials include PtPd NPs with DTNB chromogen;58 Au@Pt NPs,351 CuCo NPs,352 Rh NPs (50-fold improvement in LOD versus standard AuNP usage),353 Au@PtPd NPs with TMB and peroxide;31 and cerium oxide NPs with TMB (no peroxide; two-fold improvement in sensitivity versus AuNPs).354 The gain in signal comes at the cost of an extra processing step and additional reagents.
Although not very common, the lateral flow assay format with smartphone readout has been adapted to the detection of nucleic acids. A simple adaptation utilized a test line with streptavidin, a control line with an oligonucleotide probe, and AuNP–oligonucleotide conjugates to detect biotinylated amplicons from recombinase polymerase amplification (RPA) of a Salmonella gene (LOD 19 CFU mL−1).355 Biotinylated and fluorescein-labeled LAMP amplicons for SARS-CoV-2 detection (LOD 4000 copies per mL) were likewise detected with a streptavidin test line and AuNP–anti-fluorescein–antibody conjugates.356 Tilapia lake virus RNA was detected through RT-RPA amplification followed by CRISPR/Cas12a amplification with a biotin–oligonucleotide–fluorescein reporter and AuNP–anti-fluorescein conjugates (LOD 200 copies/reaction).357 A similar approach was adopted to screen for six genes of Bacillus cereus in milk and rice samples (LOD 10−4 ng mL−1),358 and the CaMV35S promoter sequence relevant to genetically modified organisms was detected (LOD 10 aM) with substitution of RCA for RPA.359 A lateral flow assay for detection of a Salmonella gene (LOD 1 CFU mL−1) in food samples used a combination of RPA, CRISPR/Cas12a, and chromogenic oxidation of TMB.360 Assays of this nature have notably compromised simplicity and portability for enhancement of detection in an LFIA format.
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 cells);362 a paper substrate with WST-8 dye (colorless to orange from reduction) for screening MCF-7, L-02, and HepG2 cells responses to cytotoxic drugs;363 and a gel-filled poly(methyl methacrylate) through-hole array with resazurin (Alamar blue) for screening HeLa cell response to drugs.364 Calibration relates viable cell count to color, where image analysis for these measurements has utilized both the RGB and HSV color spaces (analyzing specific color channels and saturation values) and grayscale conversion.
000 cells);362 a paper substrate with WST-8 dye (colorless to orange from reduction) for screening MCF-7, L-02, and HepG2 cells responses to cytotoxic drugs;363 and a gel-filled poly(methyl methacrylate) through-hole array with resazurin (Alamar blue) for screening HeLa cell response to drugs.364 Calibration relates viable cell count to color, where image analysis for these measurements has utilized both the RGB and HSV color spaces (analyzing specific color channels and saturation values) and grayscale conversion.
Other approaches for smartphone-based cell counting rely on the direct imaging of cells. As a unique example of bulk imaging, L929 cells were counted with the assistance of a trained convolutional NN through brightfield imaging of a centrifuged pellet of cells in a plastic tube.365 However, most approaches resolve individual cells to generate counts. Stains have sometimes been used to enhance image contrast or to help identify target cells, as in the cases of counting red blood cells in a Leishman-stained blood sample,366 and counting Cryptosporidium and Giardia (oo)cysts in vegetable wash water and river water samples with Lugol's iodine-staining.367
Some cell-counting approaches have also bypassed staining. A method of counting CD4+ T cells for HIV monitoring relied on immunocapture of the target cells (from whole blood) within a microfluidic chip.368 A battery-powered clip-on device utilized a broadband LED for illumination and a pair of lenses from a DVD drive to supplement the smartphone camera for brightfield imaging. Individual cells counted in the field of view were converted to a sample cell count. A similar technical approach was used to count sickled cells (i.e. monitor sickle cell disease) from whole blood samples.369 The main difference was that immunocapture was not necessary since the image analysis algorithm distinguished round (healthy) cells from misshapen (sickle) cells.
Whereas most cell counting approaches have utilized a sample preparation with static cells, a smartphone-based brightfield imaging flow cytometer has been reported. The device used a high-power LED illumination source, a simple PDMS-on-glass microfluidic chip, and objective and eyepiece lenses borrowed from a conventional microscope.370 The prototype sacrificed some portability for performance, requiring an outlet-connected power supply for the LED source, but was able to image cells with <700 nm spatial resolution at real-time rates up to 100 cells per second and post-processing rates of 67![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 cells per second. The high-power LED was necessary to produce sufficient illumination intensity for imaging the fast-flowing cells at a high frame rate to avoid motion blur. A convolutional NN analyzed cell size and morphological features to distinguish Jurkat and EL4 cells.
000 cells per second. The high-power LED was necessary to produce sufficient illumination intensity for imaging the fast-flowing cells at a high frame rate to avoid motion blur. A convolutional NN analyzed cell size and morphological features to distinguish Jurkat and EL4 cells.
Tissue specimens have also been analyzed by smartphone microscopy. The assessment of nonalcoholic fatty liver disease lesions from hematoxylin-and-eosin-stained liver allograft biopsies,371 and the evaluation of allograft kidney biopsy specimens,372 are two examples of applications. A growing trend in smartphone microscopy is the use of deep learning (i.e. NNs) for the analysis of acquired images. Benefits of this approach include automation, elimination of the need for a trained expert for image interpretation, mitigation of potential limitations in image quality, and additional informing power that avoids the need for further analysis (e.g. immunohistochemistry). Examples of deep-learning-supported imaging have included screening for sickle cell disease from patient blood smears (Fig. 23),89 screening for cervical cancer from liquid cytology samples,373,374 classification of skin lesions as malignant or benign,375 identification of five species of an invasive insect (genus Liriomyza),376 early detection of mold on bread,377 and prediction of the presence of asbestos in building materials.378
 | ||
| Fig. 23 Smartphone-based brightfield microscope for sickle cell identification. (A) Photograph (left) and design (right) of the microscope device. (B) Close-up of the optical path. (C) Workflow from image acquisition to identification of sickle cells. Adapted from ref. 89 under a Creative Commons Attribution 4.0 International License. | ||
In general, the above methods utilized clip-on attachments for a smartphone to enable brightfield microscopy with suitable magnification, or had 3D printed a small smartphone-based microscope device for the benchtop. One noteworthy clip-on attachment was only 0.15 cm3 in volume, 0.5 g in weight, and $10 in cost, with a reported order of magnitude improvement in depth of field compared to a conventional microscope. It was combined with deep learning for classifying skin moisture levels.379 At the other extreme of device design, microscopy has been approached by mounting a smartphone on a scientific benchtop microscope that would otherwise not be equipped with a digital camera. This approach has been implemented with deep learning for determining the risk of malignancy for thyroid fine-needle aspiration biopsy samples,380 for subtyping of cancer cells and detecting lymph node metastasis from hematoxylin-and-eosin-stained pathology tissue slices,381 and for evaluating the morphology of embryos for in vitro fertilization.382 A third viable platform for smartphone microscopy has been the low-cost, commercially-available “foldscope” paper microscope, which was adopted to screen antifungal agents for their effect on the C. albicans pathogen,383 and for imaging prepared slides of mammalian endocrine glands in a classroom setting.384
As a brightfield microscopic approach to molecular detection, polystyrene microspheres were combined with magnetic NPs to enable a digital competitive immunoassay for aflatoxin B1.385 To report on a plate-based immunoassay, the secondary antibody for detection was conjugated with alkaline phosphatase to produce ascorbic acid, which then initiated an azide-alkyne click reaction that coupled magnetic nanoparticles to microspheres in the assay supernatant. The analyte was quantified through magnetic capture and AI-assisted counting of the microspheres. In principle, this strategy is adaptable to a variety of immunoassays and coupling or ligation reactions.
As described, all of the assays noted above would have been possible with conventional RGB imaging with a smartphone camera. Equivalent analytical performance would also be anticipated for conventional imaging, particularly when aided by modern ML methods. A smartphone spectrometer will be most useful when there is a need for detailed or continuous spectral data over color information, such as when two signals to be unmixed do not make significantly different contributions to two different RGB channels. This scenario is rare and thus the spectral resolution will often not be worth the technical costs.
7.3 Light scattering analyses
A variety of smartphone assays have been developed around imaging of light orthogonally scattered from the beam of a red laser by NPs. In each case, analyte directly or indirectly induces the aggregation or formation of metal NPs (e.g. gold, silver) to increase light scatter, or induces the dissolution of the NPs for a decrease in light scatter. Analytes have included cocaine and a protein biomarker,387 creatinine,388 glyphosphate,389 and aqueous metal ions (Fe3+, Hg2+, Ag+, Cu2+).390–395 A conceptually similar light scattering assay with AuNPs and aptamers utilized illumination with a green LED for the detection of certain antibiotics (tobramycin, kanamycin, alternariol; LODs < 200 ng mL−1).396In addition to metal NPs, materials such as metal–organic frameworks and cobalt oxyhydroxide nanoflakes have been used as scattering reporters. Scattering assays without synthetic reporters are also possible in certain cases: bacteria were detected in suspension through the loss of light scattering as a bacteriophage lysed the cells (and provides selectivity);397 and SARS-CoV-2 genes (LOD 1 copy per μL) were detected using a water-in-oil emulsion implementation of LAMP, where the production of amplicons decreased the interfacial tension, decreasing the droplet size and scattering intensity as the amplification progressed.398
Smartphone imaging formats have also been developed around light scattering. In one example, a smartphone-based device with built-in polarizers and a green laser imaged backscattered light from bacterial colonies in a petri dish (Fig. 24).399 Images features were algorithmically extracted, and the type of bacteria was identified through a support vector machine learning analysis of the colony morphology. A related example is the application of ML to count “DNA colonies” that were formed by LAMP reactions confined within a polyacrylamide hydrogel (Fig. 25).400 The production of pyrophosphates during the amplification led to the formation of magnesium pyrophosphate precipitates, which scattered light and were highly visible by reflection imaging (LOD 1 copy/reaction). A darkfield configuration based on total internal reflection of white-light was used to image light scattered from metal precipitates to identify Pb2+ and Hg2+ in drinking water, where the color of the precipitate provided information about the identity of the metal.401
 | ||
| Fig. 24 Smartphone imaging and identification of bacterial colonies via elastic light scattering and machine learning. (A) Design of the smartphone-based device. (B) Examples of backscatter patterns from colonies of several species of bacteria. Abbreviations: BS, beam splitter; PCX, Plano-convex. Figure reproduced from ref. 399 under a Creative Commons CC BY 4.0 licence. | ||
 | ||
| Fig. 25 Label-free detection of DNA via light scattering. (A) Assay methodology (PAM = polyacrylamide monomers). (B) Smartphone images of “DNA colonies” (precipitates of magnesium pyrophosphate) in a hydrogel. The colonies are ultimately counted by a machine learning algorithm. Adapted with permission from ref. 400 Copyright 2024 American Chemical Society. | ||
7.4 PL analyses
PL-based assays are the second most abundant class of assay after colorimetric assays. Molecular dyes are the most popular source of fluorescence, but there is significant adoption of luminescent NPs. LEDs are the most common excitation source, and there is frequent use of laser diodes and UV lamps, but only limited adoption of the light sources on-board the smartphone (Fig. 14).Fluorescent dyes have also been developed for interaction or reaction with an analyte, including dyes applied to the smartphone-based detection of malononitrile403 (a cyanogen) and biogenic amines404 (for assessing food freshness) through changes in emission color, and the detection of gamma-hydroxybutyric acid (the date rape drug) in carbonated and alcoholic drinks through an increase in emission intensity.405 A thiol-responsive AIEgen (a dye that undergoes aggregation induced emission) was developed for the detection of an organophosphorous pesticide in hydrolyzed leaf extracts.406 Dipicolinic acid (an anthrax biomarker) was detected through a competitive coordination interaction that activated the fluorescence from Tb3+ ion.407 Volatile amines have also been identified through a combined analysis of the absorbance color and fluorescence color of a specially synthesized BODIPY dye.408
Examples of smartphone-based PL assays with enzyme-catalyzed fluorogenic reactions include measurement of the activity of β-glucosidase using a fluorogenic substrate,409 and the detection of glucose in wound exudate through the coupled reactions of glucose oxidase and HRP to convert dichlorofluorescein-diacetate to its fluorescent form.410
Arrays of fluorophores have also been used to detect and identify analytes. A paper-based array of poly(arylene ethynylene) fluorophores identified 10 different polyaromatic hydrocarbons through changes in the RGB emission color of the polymers combined with principal component analysis and discriminant analysis.226 Similarly, an array of six different fluorescent dyes distinguished between five types of amyloid fibrils, including with diluted plasma and artificial cerebrospinal fluid samples, through ML analysis of changes in fluorescence intensity and RGB color.232
The reactivity of NP materials has also been exploited for the detection of analytes. Examples include the quenching effects of Cu2+ (aq) on carbon dots,414 Hg2+ (aq) and S2− (aq) on CdZnTe QDs,415 Hg2+ (aq) on gold nanoclusters,416 and water in ethanol on perovskite QDs.417 Quenching of luminescent metal organic frameworks have also been utilized for detection of chlortetracycline in seafood homogenate.418
Various photoluminescent NPs have been adopted for smartphone-based readout of LFIAs. QDs are one of the more popular materials and have been utilized for the detection of Influenza A nucleoprotein in simulated nasal matrix (LOD ≤ 3 fmol),421 ciliary neurotrophic factor (biomarker for glaucoma; LOD 6 pg mL−1),425 and antibodies toward the Taenia solium rT24H antigen with samples from patients with neurocysticercosis.426 Other LFIAs have amplified test-line signals by using silica or polymer particles doped with many QDs. Examples include the detection of PSA in human serum samples (LOD 0.14 ng mL−1);427 interleukin-6,428 ciprofloxacin (LOD 0.05 ng mL−1),429 and Zika virus non-structural protein 1 in serum (LOD 0.15 ng mL−1).430
In addition to their high brightness, the spectrally narrow emission of QDs has enabled a novel detection approach for LFIAs: signaling of analyte through a change in the color of the test line rather than a change in its single-color fluorescence intensity. The analyte-induced transition is from green to red QD PL at the test line, with intermediate hues of yellow and orange. This color-changing approach is less dependent on illumination conditions and has better interpretability to the naked eye. It has been adopted for competitive assays of aflatoxin M1 (LOD 1 pg mL−1)423 and benzothiostrobin (LOD < 1 ng mL−1),431 and for sandwich assays for heart-type fatty acid binding protein (LOD 0.21 ng mL−1; Fig. 26)432 and the SARS-CoV-2 nucleocapsid protein (LOD 0.1 ng mL−1).433 Beyond QDs, there is also an example of this strategy with red-fluorescent ruthenium(II)-complex doped NPs and green-fluorescent gold nanoclusters.434
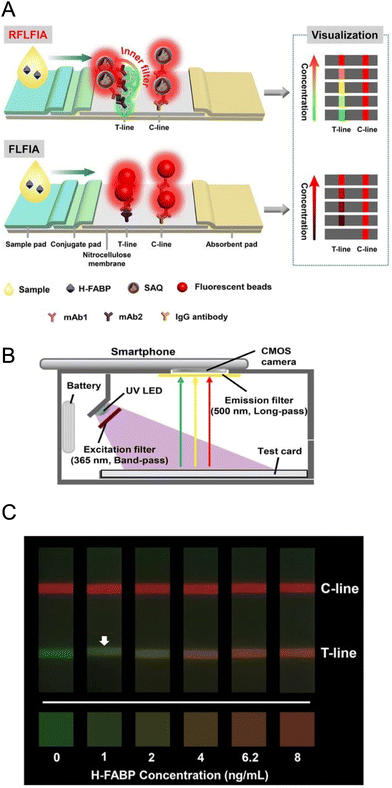 | ||
| Fig. 26 A ratiometric fluorescence LFIA (RFLFIA). (A) Assay design and comparison to a conventional non-ratiometric fluorescence LFIA (FLFIA). Abbreviations: H-FABP, heart-type fatty acid binding protein; SAQ, silica nanosphere loaded with AuNPs and red-light emitting CdSe/CdS/ZnS QDs; mAb, monoclonal antibody. (B) Smartphone-based device for imaging the RFLFIA. (C) Examples of smartphone images of the RFLFIAs with increasing concentration of the heart-type fatty acid binding protein analyte. Adapted from ref. 432 with permission. Copyright 2021 John Wiley and Sons. | ||
The PL colors of QDs also have potential for multiplexed LFIAs. One strategy has been to use LFIAs with two test lines, either on the same strip (Fig. 27)78 or on separate arms of a bifurcated membrane,435 where immunoconjugates of green and red QDs (or QD assemblies) bind to separate lines for capture of each analyte. Because of the spatial separation of the two test lines, two different immunoconjugates of a single color of QDs would still be useful for these assays, albeit that the potential benefit from two colors is redundancy to guard against non-specific binding to the wrong test line. A format where the two analytes are captured on the same test line requires the two distinct colors of QD and a color-based analysis, as was the case for an LFIA of free and complexed forms of PSA.436
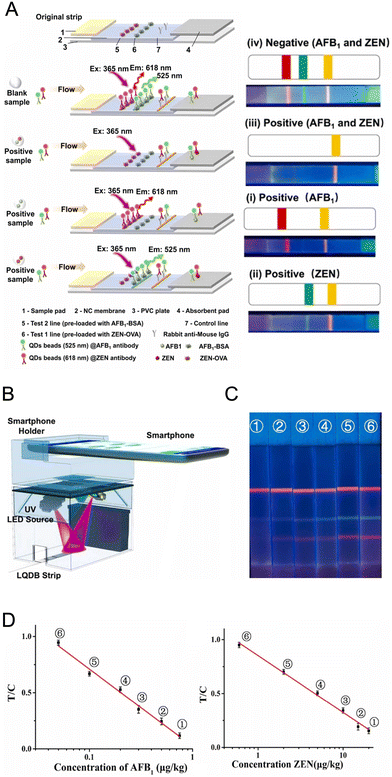 | ||
| Fig. 27 Two-color, two-line multiplexed competitive LFIA with fluorescence detection. (A) Assay design and smartphone images of assay outcomes. (B) Design of the smartphone-based device used to image the LFIAs. (C) Smartphone images of LFIAs and (D) the corresponding calibration curves for each analyte, aflatoxin B1 (AFB1) and zearalenone (ZEN). The y-axis in panel D is the ratio of signal between the test and control lines. Adapted from ref. 78 with permission from Elsevier. | ||
Many other luminescent NPs have been adopted in LFIAs. Polymer microspheres and nanoparticles doped with Eu(III) complexes were used as labels to detect thyroid stimulating hormone in diluted serum (LOD 0.02 μIU mL−1);422 lung-cancer relevant plasma extracellular vesicles (LOD 105 mL−1);437 ochratoxin A in wheat samples (LOD 0.32 μg kg−1);438 Hepatitis B virus in serum samples;439 and five different classes of mycotoxin, spanning 20 individual compounds, with a five-line LFIA.440 Latex NPs conjugated with phycocyanobilin have also been utilized for LFIAs,80 and lanthanide-doped UCNPs were labels to detect ochratoxin A, Hg2+ (aq), Salmonella, hepatitis B virus nucleic acids, and protein ST-2,441 as well as C-reactive protein (LOD 0.05 ng mL−1).442 Two-color two-test-line LFIAs have also been demonstrated with Er3+ (green) and Tm3+ (blue) emitters for PSA and ephrin type-A receptor 2 (EphA2),443 and for brain natriuretic peptide and suppression of tumorigenicity 2 antigen in serum.444 PLNPs have been adopted for LFIAs of hCG (LOD 45 pg mL−1),35 17β-estradiol in milk,445 and T-2 toxins (0.025 ng mL−1) in food samples,446 plus two-test-line LFIA with green and blue colors for PSA and hCG.447 Other examples of materials include a red-emitting gold nanocluster-doped zeolitic imidazolate framework as a label in an LFIA of olaquindox (LOD 2.5 μg L−1),448 and AIEgen-doped MOF as a label in an LFIA of aflatoxin B1.449
Any comparisons of analytical performance between different luminescent NPs will need to account for differences in excitation intensity and detection efficiencies between different smartphone-based devices. The most reliable comparisons will be head-to-head with the same device, and there are few instances of such studies in the literature.
To date, the majority of published studies on smartphone-based detection of nucleic acids have focused on amplification of the target sequence and detection and quantification of the amplified targets. As such, sample preparation has tended to follow standard laboratory protocols using commercial kits for nucleic acid extraction, and mixing of sample with the reagent cocktails required for reverse transcription (for RNA targets) and the amplification method of choice. Only a few studies have streamlined the sample processing steps alongside smartphone-based detection. One unique approach used a patch-like microneedle array (made from polyvinyl alcohol) to extract RNA by pressing it onto a plant leaf and releasing the RNA by simple rinsing of the array with buffer.450 The extracted RNA was ultimately used to detect plant infections, including Phytophthora infestans and tomato wilt virus (LOD of 1 pg; Fig. 28). With similar simplicity, hepatitis C viral DNA (LOD 5 pmol) was detected in clinical samples without an amplification step using a folded paper test square with immobilized peptide nucleic acid probes and an absorbent pad underneath.451 Sample was added, washed, and fluorescently stained in successive steps that consisted of dropping microliters of solution onto the test square. These examples highlight how user actions can be simplified and minimized, to various degrees, despite the frequent requirement for numerous reagents and multiple steps in a nucleic acid analysis that goes from sample to answer.
 | ||
| Fig. 28 Microneedle extraction of nucleic acid and on-chip LAMP with smartphone-based detection. (A) Photograph of a microneedle (MN) patch and work flow for the nucleic acid extraction. (B) Schematic of the LAMP sample cassette and cross-sectional view of smartphone-based reader device. (C) Representative smartphone fluorescence images of the parallel LAMP assay chip (top) for different nucleic acid sequences (I–IV) and photographs of healthy and infected leaf samples (bottom). (D) Real time amplification curves for P. infestans DNA obtained via the smartphone-based LAMP platform. (E) Comparison between the smartphone device and a conventional laboratory instrument. Adapted with permission from ref. 450 Copyright 2021 Elsevier B.V. | ||
With respect to amplification, polymerase chain reaction (PCR) coupled with a fluorescence detection method is the gold standard for nucleic acid diagnostics. This amplification method requires that thermocycling be implemented in a portable device. One approach to both conventional and digital PCR with smartphone-based fluorescence detection is reproduction of the classic temperature programming of PCR via a Peltier (i.e. thermoelectric) module, as done for the detection of the CD147 gene and cancer biomarker.94 Convective thermocycling PCR, whether separate435,452 or interfaced453 with the detection device, has been an alternative approach that enables a heating element to be maintained at a constant temperature. It has been utilized with smartphone-based devices to detect lambda DNA (LOD 3000 copies),452 genes diagnostic of methicillin-sensitive and methicillin-resistant Staphylococcus aureus (LOD 5000 copies),435 and influenza A (H1N1) virus (LOD of 1 tissue culture infectious dose per mL).453
Another implementation that utilizes a heater at fixed temperature is continuous flow PCR, where, for example, one face of a trigonal helix of sample-carrying tubing was in contact with a constant-temperature heater to produce alternating hot and warm temperature zones that functioned for thermocycling.454 Another example doped the PDMS of a microfluidic chip with gold nanorods for photothermal heating under illumination from an 808 nm laser.91 Thermocycling arose from a circular geometry from a serpentine channel that moved in and out of the hot zone produced from the radial intensity profile of the laser spot. In these two examples, the analytes were H7N9 avian influenza DNA (LOD 104 copies per μL) and hepatitis B virus DNA (LOD 12 copies per μL, 40 min assay time).91 From the standpoint of device engineering, the difference between temperature programming and oscillating sample along a temperature gradient is minimal. Both formats regulate temperature using feedback from an integrated temperature sensor. Temperature programming devices more frequently add a cooling fan, but more readily accommodate array-based multiplexing since samples can be compartmentalized in a small area.
As an alternative to the thermocycling of PCR, loop-mediated isothermal amplification (LAMP) methods are frequently paired with smartphone-based detection. Some reported smartphone-based devices have had integrated heaters to obtain the 60–65 °C typical of the method,450,455 another device had a distinct compartment dedicated to the heating step of the assay,456,457 and some assays have relied upon benchtop heating instruments.456–458 Analytes were lambda DNA (LOD 1 copy per μL),458 SARS-CoV-2 RNA (LOD 10 copies per μL, 60 min assay time),456 and human papillomavirus (HPV; 30 min assay time).457 Another common isothermal method (normally 37–42 °C, less efficient at room temperature) is recombinase polymerase amplification (RPA). Examples of targets amplified by RPA have included HIV-1 RNA (LOD 67 copies), where a consumer coffee warming device was used as the heater,459 and HIV and hepatitis B genes (LOD 1000 copies per mL).81 The latter example was a small-scale suspension array that used the smartphone camera to recognize QD-doped bead colors corresponding to specific nucleic acid targets. Many other targets have also been measured through pairing of RPA with CRISPR/Cas signal amplification (vide infra).
Nearly all smartphone-based PL assays for nucleic acid detection have used fluorescent dyes for signal generation. Fluorescein is widely used as a dye label on oligonucleotide primers and reporters, likely due to is combination of low cost and high brightness relative to other dyes, optimal excitation by widely available blue LEDs, and green emission that overlaps well with the G channel response of smartphone cameras. Alexa Fluor 647 has also been utilized for its compatibility with orthogonal interrogation with QD-based microbead barcodes for optically multiplexed detection of DNA associated with infectious diseases such as human immunodeficiency virus (HIV) and hepatitis B and C (Fig. 29).81 Alternatively, intercalators and DNA-binding dyes (e.g. TB green, EvaGreen, SYBR Green) that switch from dark to bright PL upon interaction with double-stranded DNA have been utilized for readout from both PCR and LAMP amplifications.450,454,455,460,461
 | ||
| Fig. 29 Portable device for amplification and suspension-array-based detection of nucleic acid sequences. (A) Barcode detection of target DNA from patient samples by quantum dots. (B) Typical microwell chip containing different barcodes in each well. (C) Smartphone camera captures the image of four different quantum dot barcodes arrayed on the surface of the chip. (D) Schematic and real images of the smartphone device. (E) Visual representation of the envisioned final device, with the different compartments. (F) Yellow, green, and red barcodes are deposited on the chip and imaged using the device. (G) Sensitivity curves for genetic biomarkers for the blood-borne viruses human immunodeficiency virus (HIV), hepatitis B virus (HBV), and hepatitis C virus (HCV). (H) Comparison between the average combined statistics of all subjects of the HIV-negative group (3 subjects) and HIV-positive group (10 patients). Adapted with permission from ref. 81 Copyright 2015 American Chemical Society. | ||
The smartphone-implemented fluorescence detection methods paired with nucleic acid amplification methods have included commercial TaqMan probes,94,453 molecular beacons,462 magnetic pull-down binding assays (PCR, fluorescein, phycoerythtrin),452,463 and lateral flow binding assays (QDs with PCR;435 QDs with RCA amp;41 QDs with RTF-EXPAR, 10 aM, for multiple QDs per target;464 UCNPs with PCR amplicons465). A smartphone-based microchip capillary electrophoresis unit has even been proposed for the detection of fluorescently labeled PCR amplicons.124
In several other studies, the target nucleic acid sequence triggers the trans cleavage activity of CRISPR/Cas12a or CRISPR/Cas13a to hydrolyze the reporter with loss of FRET and recovery of the dye fluorescence. Fluorescein is by far the most common dye in such fluorophore-quencher reporter systems, although carboxyrhodamine (ROX), carboxytetramethylrhodamine (TAMRA), and cyanine 5 (Cy5) have also been adopted. Examples of assays have included the detection of genes diagnostic of African Swine fever virus (LODs of 8 copies per μL),466,467 Frog virus 3 (LOD 10 aM),468 and SARS-CoV-2 (LOD 100 copies per μL).469 Although these examples utilized a standard lab-based nucleic acid extraction, another study reported a small and low-cost device for simple isolation of RNA from saliva samples in one step and detection in a second step (Fig. 30).470 The first step isolated RNA on a polyethersulfone (PES) membrane disc with pre-loaded lysis reagents and provided 95 °C heat for inactivation of nucleases. The second step transferred the membranes to a reaction chamber that was pre-loaded with freeze-dried assay reagents and provided 37 °C heat for RPA and Cas-mediated detection (LOD 1000 cp per mL; 60 min assay time).470 Alternatively, multiple CRISPR/Cas methods reported for the detection of SARS-CoV-2 have returned results without extraction from patient samples, with assay detection limits in range of 100–1000 copies per mL with assay times of 15–50 min.82,471,472 One such method used a laser-based smartphone device for readout of fluorescence from fluorophore-quencher reporter oligonucleotides (Fig. 31).82
 | ||
| Fig. 30 Sample-to-answer device for CRISPR-based detection SARS-CoV-2 and variant nucleic acids. (A) Design of the device, including sample processing (left) and assay (right) modules. (B) Photographs of the device, which is capable of two-plex detection. (C) Smartphone-acquired fluorescence images of samples. Figure reproduced from ref. 470 under a Creative Commons CC BY 4.0 license. | ||
 | ||
| Fig. 31 CRISPR/Cas method for detection of viral RNA from SARS-CoV-2. (A) Workflow of the assay. (B) Design of a smartphone-based fluorescence reader. (C) Smartphone-acquired image of fluorescence from a multi-well chip preloaded with assay reagents. (D) Calibration curve for measured fluorescence intensity versus the copy number of viral RNA. (E) Comparison of results between the smartphone-based device and RT-qPCR as a standard laboratory method. Figure reproduced from ref. 82 under a Creative Commons CC BY 4.0 license. | ||
Whereas the foregoing studies combined CRISPR/Cas12a with RPA, another approach has relied only on the signal amplification intrinsic to the multiple turnover capability of Cas12a for the detection of genes diagnostic of hepatitis B virus and HPV (LODs of 200 pM for a conventional assay and 5 fM as a digital assay).473 Other signal amplification approaches for smartphone-based assays have included the hybridization chain reaction for detection of cancer relevant miRNA-224 from plasma,474 Exo-III-assisted amplification for detection of miRNA-155 (LOD 42 aM), 16S rRNA of S. aureus (LOD 18 CFU mL−1), and a gene from COVID-19 pseudovirus RNA (LOD 87 copies per μL).475
The predominant use of fluorescent dyes for detection likely arises from four considerations: the widespread commercial availability of dye-labeled oligonucleotides; status as the gold-standard class of material for fluorescent labeling in lab-based nucleic acid assays; tolerance of dye labels by the enzymes leveraged for nucleic acid amplification and hydrolysis; and, in some cases, intrinsic sensitivity of dye fluorescence to nucleic acid structure. Likewise, these considerations almost certainly underlie the almost uniform adoption of homogeneous solution-phase assay formats. Alternative fluorescent materials such as QDs have a higher ceiling for sensitive detection, but their productive combination with relevant enzymes—though possible, including in the context of smartphone-based nucleic acid detection476 (amplification-free LOD of 50–100 pM)—is not as turnkey as it is for dyes. Nucleic acid assays with QDs are therefore more likely to be in the form of post-amplification binding assays that require wash steps, albeit avoidable with a lateral flow format, as demonstrated for the detection of DNA from methicillin-resistant S. aureus (LOD 4700 copies post-convective PCR).435 Perhaps the best aspiration for ultra-bright fluorescent NPs is the amplification-free detection of clinically-relevant amounts of target nucleic acid with a smartphone device. A study has already demonstrated that plasmonic DNA nanostructures with silver NPs—in conjunction with a high numerical aperture objective and an atypically powerful (180 mW) laser diode—enable single-molecule fluorescence detection in DNA hybridization assays with a smartphone camera (Fig. 32).83 Fluorescent NPs have the opportunity to lower the technical requirements for such performance.
 | ||
| Fig. 32 Single-molecule fluorescence detection on a smartphone using plasmonic nanostructures for metal-enhanced fluorescence. (A) Schematic of the DNA–silver NP nano antennas with cleared hot spots (NACHOS). (B) Diagram (top) and confocal fluorescence images (bottom) images of the NACHOS with three capture strands in the hotspot and a green-fluorescent reference dye for labeling of the DNA origami. (C) Diagram of the portable smartphone-based microscope (left) and a photograph of the display during imaging (right). (D) Top: Fluorescence enhancement histogram of the sandwich assay in buffer solution (light blue) and in serum (dark blue). Bottom: Binding yield obtained for the full sandwich assay with and without added DNA target. Figure reproduced from ref. 83 under a Creative Commons CC BY 4.0 license. | ||
Fluorescence detection has also been used for the imaging and counting of individual cells. Unlike brightfield microscopy, cell staining is a requirement for these measurements. Nuclear (i.e. DNA) stains are one of the most common methods for generating fluorescence contrast from cells. Examples include the use of SYTO16 for imaging (with174 and without479 ML analysis) of leukocytes extracted from human blood, and acridine staining for both total and differential white blood cell counts from blood samples (where the differential count used an R/G ratio in image analysis to distinguish between granulocytes and agranulocytes).480 Other fluorescent stains have included Calcofluor White M2R and Solophenyl Flavine 7GFE 500 for the selective staining of Nosema spores in honey bee midgut samples (LOD 5 × 105 spores per bee),481 Calcofluor White or Blankophor for proof-of-concept imaging of cultured dermatophytes,482 and a newly-synthesized dye for staining microplastic particles.97
Fluorescent immunolabeling is also a viable approach to cell detection with smartphone-based microscopy. A compact clip-on device was designed for smartphone microscopy via ultraviolet surface excitation, demonstrating two-color imaging of brain tissue slices using an Alexa Fluor 488-labeled antibody (green fluorescence) and propidium iodide nuclear stain (red fluorescence), and was also suitable for imaging a combination of DAPI (blue) and Rhodamine B (orange) staining of tissue (Fig. 33).483 A much larger 3D-printed benchtop microscope was used for imaging T. cruzi parasites stained with fluorescein-conjugated antibodies.484
 | ||
| Fig. 33 Compact smartphone mounted microscope. (A) Design of the microscope attachment, which uses ultraviolet surface excitation. (B) Fluorescence image of a mouse kidney section stained with rhodamine B and DAPI dyes. Figure adapted from ref. 483 under a Creative Commons CC BY 4.0 license. | ||
As much as QDs have been superior labels versus dyes for smartphone imaging, nanoscale assemblies of many QDs have outperformed individual QDs. For example, immunoconjugates of both supra- and super-particle assemblies of QDs have been developed for cell detection and imaging with smartphone-based devices. In one study, assemblies of magnetic particles and QDs were used for immunomagnetic isolation and fluorescent labeling of a target breast cancer cell line, which was then imaged and counted in a simple plastic chamber slide (Fig. 34).84 Supra-QD assemblies with a silica NP scaffold have also been used for cell immunolabeling and imaging, providing an order of magnitude signal enhancement over conventional QDs with smartphone-based devices (Fig. 35A–D).485 All these materials also utilized tetrameric antibody complexes, which enabled spontaneous immunoconjugation with speed and efficiency suitable for chemistry-free on-demand preparation. The supra-QD materials were further adopted for immunofluorescent flow cytometry (Fig. 36).85 The high brightness of the materials provided the sensitivity necessary for the detection of cell-surface antigens with video rate imaging. A subsequent study showed that designer colors of supra-QD were able to be prepared from mixtures of R-, G-, and B-emitting QDs, with the potential for machine-learning-assisted detection of up to 14 colors with a smartphone camera (Fig. 35E).223 The next generation of labels are super-particle assemblies, which may provide even larger signal enhancements than supra-QDs (Fig. 35F).486
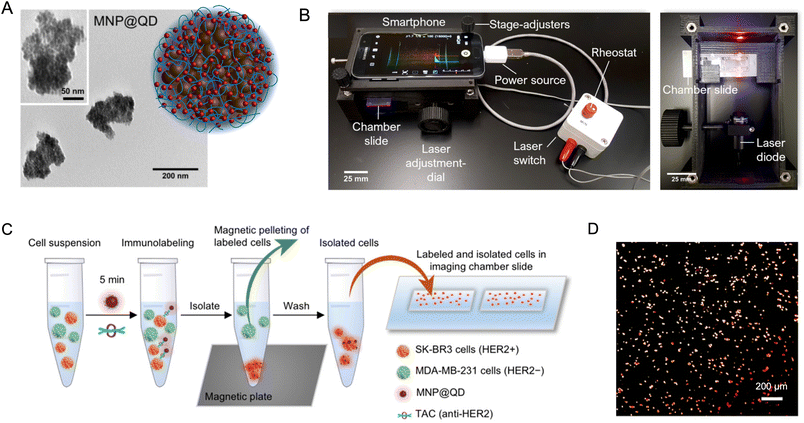 | ||
| Fig. 34 Magnetic NP-QD assemblies for cellular isolation, imaging and enumeration with a smartphone-based device. (A) Cartoon and TEM of cluster-like assemblies of iron oxide magnetic NPs and QDs (MNP@QD). (B) Exterior and interior photographs of the smartphone-based device used for cell imaging. The laser diode is powered by the smartphone battery. (C) Workflow for cell isolation. (D) Processed smartphone PL image of isolated target cells for counting. Figure adapted with permission from ref. 84 Copyright 2019 American Chemical Society. | ||
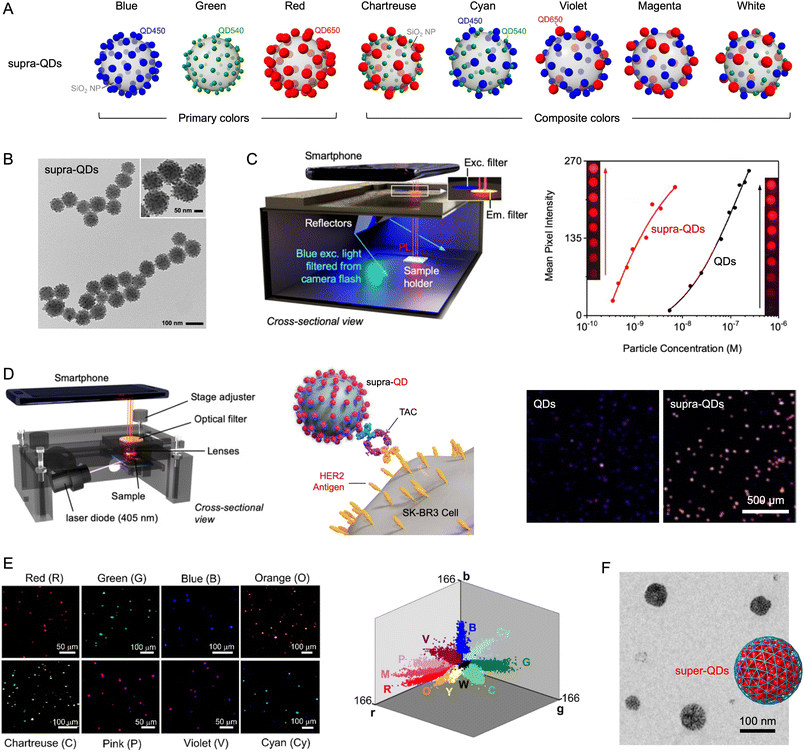 | ||
| Fig. 35 QD-based assemblies for cellular imaging and enumeration with smartphone-based devices. (A) Illustrations of supra-QD assemblies prepared with QDs that emitted primary colors of PL (RGB). Supra-QDs with composite colors of PL are prepared with mixtures of primary-color emitting QDs on the same silica NP scaffold. (B) TEM image of supra-QDs. (C) Rendering (left) of a smartphone-based device that uses blue light filtered from the smartphone flashlight to excite PL. With this device, the supra-QDs provide an order of magnitude increase in detection limit (right). (D) Smartphone-based device with laser-diode excitation of PL from labeled cells in a plastic chamber slide (left). Diagram of TAC-based immunolabeling of the cells (middle). Cells labeled by supra-QDs are far brighter in smartphone images than the cells labeled with QDs. (E) Smartphone images of cell-sized objects labeled with primary and composite colors of supra-QD (left). Each color maps out a trajectory in the RGB color space (right). (F) TEM image and diagram of super-QDs, which are recently developed as potentially brighter successor material to supra-QDs. The internal volume of super-QDs is made up of many individual QDs. Panels A and E are adapted with permission from ref. 223 Copyright 2023 American Chemical Society. Panels B–D are adapted with permission from ref. 485 Copyright 2020 American Chemical Society. Panel F is adapted with permission from ref. 486 Copyright 2023 American Chemical Society. | ||
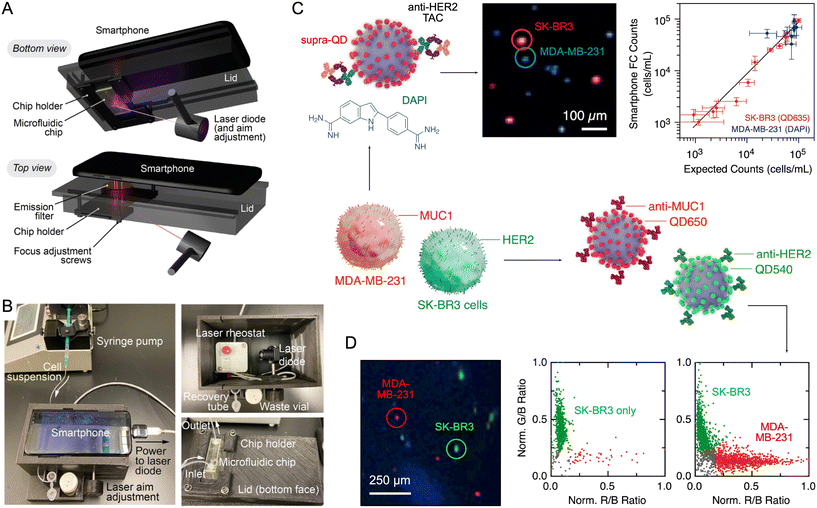 | ||
| Fig. 36 Smartphone-based flow cytometry with immunofluorescent detection via supra-QDs. (A) Design and (B) photographs of the device. (C) Non-selective labeling of all cells (including non-target MDA-MB-231) with DAPI nuclear stain and immunolabeling of target cells (SK-BR3) with supra-QDs. An example of a zoomed smartphone image (left) is shown alongside validation of cell counting data (right). (D) Two-color immunolabeling of two cell lines (SK-BR3 and MDA-MB-231) using two different colors of supra-QD immunoconjugates. A zoomed smartphone image (left) is shown alongside flow cytometry data (right) for samples with a constant quantity of SK-BR3 cells, without and with MDA-MB-231 cells. Figure adapted from ref. 85 under a Creative Commons CC BY 4.0 license. | ||
Many of the devices used for cytometry and microscopy have been custom designs made via 3D printing with simple lens sets or conventional microscope objectives for magnification. Alternatively, there are several examples of smartphone imaging with commercially available microscope attachments for a smartphone. In each case, commercial dye-doped polymer nanoparticles were used for imaging assays of analytes such as cellular CD56 antigen expression,487 bacteria (non-selective),488 norovirus,172,489 ROR1-antigen-positive cancer cells,490 SARS-CoV-2,491 α-amanitin mushroom toxin,492 tetrahydrocannabinol,493 and B. burgdorferi antigen (the marker for Lyme disease).494 The assay format and details of the measurement and analysis varied between analytes, but generally involved a simple wax-patterned paper substrate and direct or indirect counting of cells labeled with the polymer nanoparticles, analyte-induced aggregates of the polymer nanoparticles, or particles bound in a test zone.
Time-gated imaging leverages luminophores with long PL lifetimes to insert a time delay between an excitation pulse and imaging. It differs from persistent luminescence in that the timescale is only microseconds to milliseconds, but has the same effect of eliminating background from excitation light and autofluorescence. Time-gated imaging has been implemented with smartphones using motor-driven choppers. One design used a single chopper, an optical switch component, and other electronics to enable excitation pulses and time-gated imaging, including proof-of-concept imaging of a Eu(III) chelate on a mouse tissue section (using the smartphone in tandem with an FPGA),495 and for detection of tetracycline directly in 50% milk samples (using an analog circuit).496 As a fully mechanical alternative, two coaxial and out-of-phase choppers, with timing controlled by the rotation speed, were used for time-gated imaging of cells non-specifically stained with a Tb(III) complex, and for proof-of-concept of a time-gated FRET sensor for a target DNA oligonucleotide (Fig. 37).150 The same study demonstrated that nanomolar and picomolar levels of detection of Tb(III) complexes and Eu(III)-doped NPs were possible in serum, and that quantitative two-color multiplexing of Tb(III) and Eu(III) complexes was possible using both the RGB color channels of the smartphone and two different chopper rotation speeds that exploited the difference in PL lifetime between the two complexes.
 | ||
| Fig. 37 Smartphone-based time-gated imaging with a dual chopper system. (A) Design of the dual chopper and features that control (B) the timing of excitation pulses, the gate delay, and PL signal acquisition. (C and D) diagrams and photograph of the device abbreviations: exc = excitation; em = emission; LOS = line of sight. (E) Comparison of prompt and time-gated smartphone images of samples with short PL lifetimes (1, 2) and long PL lifetimes (4, 5), and without (3–6) and with (7–9) autofluorescent sample matrices. (F) Prompt and time-gated images of cells stained with a Tb(III) complex with an autofluorescent tissue phantom matrix. Figure adapted with permission from ref. 150 Copyright 2023 American Chemical Society. | ||
Non-mechanical approaches to time-gating with smartphones have been developed as well. One platform used an Arduino MCU to control the timing of excitation pulses and image acquisition with a smartphone camera.497 An extra layer of background suppression was implemented through the use of upconversion PL excited by a NIR laser. A mixture of UCNP-antibody conjugates (anti-EpCAM, anti-CD44, and anti-Trop2) were used to stratify clinical cervical cell samples into high-risk, low-risk, and benign categories. In another study, a virtual chopper design (i.e. no physical chopper) for time-gated imaging with a smartphone was reported.498 An LED was pulsed while a video was recorded by the phone, after which a convolutional NN reconstructed the video frames into an order that represented the PL decay of Eu(III) phosphors.
Smartphone fluorescence spectrometers are most likely to be useful in situations where spectral information is valuable, such as analyzing tissue autofluorescence, separating background autofluorescence from analytical signal, and multiplexing with more than three spectrally narrow emitters. Cuvette-based assays with a single dye, such as proof-of-concept detection of a miR-21 target as a potential biomarker for cardiovascular disease, are interesting but do not practically need a spectrometer.501 In contrast, a smartphone-based endoscope for analyzing the auto-fluorescence of cervical tissue did require spectral resolution. The endoscope design was capable of both reflectance-based absorption spectrometry and fluorescence spectrometry, incorporating beam splitters so that two light sources (a white LED and 405 nm laser diode) were able to utilize the same light paths to the sample and camera. Deep learning algorithms with spectral data were used to classify different grades of cervical cancer.506 A similar design measured diffuse reflectance spectra alongside fluorescence imaging of oral tissues (Fig. 38).215
 | ||
| Fig. 38 Smartphone-based spectroscopic endoscope. (A) Diagram and photograph of the endoscope, which is capable of both diffuse reflectance spectroscopy and high-resolution fluorescence imaging. (B) Smartphone images of lamps for wavelength calibration. (C) Reflectance spectra from an in vivo study of healthy oral tissue (left) and corresponding fluorescence images (right). Abbreviations: BP, bandpass filter; CL, condenser lens; DBS, dichroic beam-splitter; EP, eyepiece; OBJ, objective lens. Figure adapted from ref. 215 under a Creative Commons CC BY 4.0 license. | ||
7.5 CL, BL, and ECL analyses
Collectively, CL and BL are the third most common modality for the detection. Mirroring the broader literature on CL assays, the majority of smartphone-based assays utilize luminol and its blue emission. A simple dark box tends to be the only peripheral component of the smartphone-based device. Analytes have included microRNA,507–510 oligonucleotides and DNA amplicons,511,512 protein biomarkers,128,513–518 pathogenic bacteria,519–521 alkaline phosphatase,522 drugs,523 pesticides,524,525 an explosive,526 phenolics,527 mycotoxins,528,529 catechols,530 and sugars and other metabolites.531–533 For the most part, the selective detection mechanism in these assays are nucleic acid hybridization, immunocomplexation, or inhibition or catalysis of the CL reaction.Many published reports represent proof of concept for smartphone-based CL detection, but have not yet simplified the CL assay procedure to a level that is optimal for non-laboratory settings, still requiring users to execute multiple wet chemical or wash steps. One approach to reducing steps has been the adoption of the LFIA format, which ideally reduces the assays to two steps: addition of sample to the sample pad and subsequent addition of a CL or BL reagent to the test line zone. Such LFIAs have been developed for the detection of SARS-CoV-2 with bis(2,4,6-trichlorophenyl) oxalate “glowstick” chemistry with dye-doped NPs,534 and luminol with a peroxidase enzyme535 or mimic.514 A CRISPR/Cas13a-coupled lateral flow nucleic acid assay has also been reported with a luminol system and chemiluminescence resonance energy transfer (CRET) to an AuNP.521
BL assays with smartphone detection have, to some degree, followed the same trends as CL assays. The most commonly utilized enzyme is the NanoLuc luciferase, which generates blue emission, usually from either furimazine or coelenterazine as the substrate. As with CL, smartphone-based devices are potentially as simple as a dark box. Analytes have included microRNA,536–538 thrombin (Fig. 39) and other protein biomarkers,539–542 pesticides,543 disinfectant,544 drugs,545,546 other small molecules,547,548 Hg2+ (aq) (Fig. 40) and other metal ions.549,550 Immuno-complexation and nucleic acid hybridization remain common mechanisms for selective detection, and there have been several instances of detection that leverage bioluminescence resonance energy transfer (BRET)536,539–541,547 and cells or bacteria encoded with reporter genes to express analyte-sensitive bioluminescent proteins.545,549,551,552 An LFIA for imidaclothiz (an insecticide) has also been reported with NanoLuc as a BL system.543
 | ||
| Fig. 39 BRET-based detection via smartphone. (A) Binding of aptameric probes to thrombin triggers, via two kinetically trapped DNA hairpins (BH1, BH2), the assembly of a DNA-templated BRET construct with luciferase (nLZ-2) and green fluorescent protein (MGA-1). (B) Linear relationship between the G and B smartphone image channels and the thrombin concentration. (C) Smartphone image of the bioluminescent response to various concentrations of human thrombin. Adapted with permission from ref. 540 Copyright 2018 American Chemical Society. | ||
 | ||
| Fig. 40 Smartphone-based device for a dual colorimetric and bioluminescent detection of Hg2+ (aq). Photographs of (A) the device and (B) the three-leaf test strip system. The test used a chromogenic reaction catalyzed by β-galactosidase and two bioluminescent bacterial strain reporters (E. coli and A. fisheri). (C) Brightfield (COL) and BL images of the test strip responses to Hg2+, Cd2+ and low pH. Adapted with permission from ref. 549 Copyright 2021 Elsevier B.V. | ||
Both BRET and engineered cells are strategies that avoid wash steps in assays and thus minimize user actions. Signaling in the BRET assay is generally a color change of the emission from blue to green, where tracking of the change in the B or G signal, or the G/B ratio, is useful for quantitation. Signaling with reporter-encoded cells is based on analyte-induced luciferase expression and more intense BL from higher analyte levels. Some of these systems use NanoLuc, whereas others use luciferases that generate green or red emission from their substrate. There are two consequences to the functional requirement for the cells to be alive: the assays must incorporate analyte-insensitive bioluminescent cells as viability controls, and the scope of possible field applications is more limited.
Similar to CL assays, efforts have been made to simplify assay procedures by drying reagents onto paper. For example, a chip was developed to contain spheroids encoded with green- and red-emitting luciferases.551 The green emission was a viability control and the red emission increased with the concentration of tumor necrosis factor (TNFα; LOD 0.2 ng mL−1). The assay had simple but separate steps for the addition of luciferase substrate. Another study developed a bioluminescent reporter-encoded E. coli system to detect mercuric ion (parts per billion LOD).549 The bacteria were pre-loaded on a paper layer on which sample was added, and luciferase substrate was pre-lyophilized on a second paper layer that was applied to the first layer at a defined time after sample addition. Similarly, for a BRET-based microRNA assay (LOD of 2 fM), dried reagents for rolling circle nucleic acid amplification and BRET sensing were split between two paper discs that were added sequentially with sample in a microplate well.536 In another case, a paper stack was designed to have two twisted-together threads sewn into one of the layers (Fig. 41).541 One thread contained dried-on luciferase substrate and the other thread contained dried-on BRET sensor, where the latter was a fusion of NanoLuc and mNeonGreen fluorescent protein with an epitope-containing linker to detect target antibody biomarkers (LODs from 2–15 nM). The addition of sample and hydration of the threads reconstituted and mixed the sensors and luciferase substrate.
 | ||
| Fig. 41 Thread-based BRET detection of target antibodies to avoid user steps after the addition of sample. (A) Design of the threaded paper stack, with luciferase and its substrate dried on separate threads. (B) Assay workflow. (C) Plot of smartphone-measured luminescence hue versus analyte concentration. (D) Smartphone-acquired BL images for different concentrations of the anti-HIV antibody analyte. (E) Photographs of the smartphone-based imaging device. Figure adapted with permission from ref. 541 Copyright 2020 American Chemical Society. | ||
Although CL and BL methods are logical for the detection of analytes that are reactants or inhibitors of these mechanisms, the selection of CL or BL for many other assays is less intuitive. To date, we have not encountered a study with a head-to-head comparison of either CL or BL with PL. It is thus an open question as to whether the anticipated lower backgrounds of CL and BL methods ultimately provide better signal-to-background ratio than the anticipated greater signal of PL methods. The outcome of such a comparison will likely be very dependent on the nature of the sample and the fluorophore selection.
Examples of ECL detection with smartphones include proof-of-concept assays for glucose via a glucose oxidase-mediated ECL reaction with luminol and peroxide,553,554 an analogous assay for choline via choline oxidase,553 and an immunoassay for E. coli using the classic tris(2,2′-bipyridyl)ruthenium(II) and tripropylamine ECL reaction.555 These and other smartphone-based devices have utilized different strategies to apply potential without a bulky potentiostat: 1.5 V AA batteries,554 drawing energy from NFC,556 and drawing energy from the USB port of the phone.553,555
7.6 SPR assays
There have been very few smartphone-based SPR assays reported to date. Examples include an array-format magnetic NP-enhanced sandwich immunoassay on a gold chip for the detection of multiple protein biomarkers for acute kidney injury (LODs in the range of 0.2–0.7 ng mL−1),45 a label-free immunoassay for CA125 and CA15-3 cancer biomarkers using an array of microfluidic channels with a surface layer of AuNPs at high density and a smartphone spectrometer,557 and a label-free binding assay on a gold nanoplasmonic chip for measuring autoantibody levels for diagnosis of rheumatoid arthritis.558 The latter approach also developed a solar-powered mini-centrifuge to pair with the smartphone device to isolate plasma from whole blood.8. Microfluidic devices
8.1 Chip-based microfluidics
Microfluidic-like chips are a popular format for sample containment with smartphone-based measurements and devices. Discrete wells, simple channels, and inlets to chambers are common designs with PDMS-on-glass chips and hard polymer chips. Many assays that use these designs derive little or no benefit from fluid flow. Rather, these formats are adopted for reasons such as simplicity, compact size, relatively low cost, small sample volumes, confinement of sample within the focal plane for imaging, and prevention of spills. Several of the smartphone-based assays highlighted in the preceding sections utilized sample containment of this nature. Here, in this section, the focus is on examples of smartphone-based assays where a microfluidic chip provided benefits or functions beyond simple sample containment. | ||
| Fig. 42 Wearable fluorescent sensor for the detection of Zn2+, Cl−, and Na+ (aq) in sweat. (A) Smartphone device and optics module for visualizing the microfluidic patch. Visualization of the patch under (B) ambient illumination and (C) blue light illumination. (D) The blue light is filtered from the phone flashlight (left) and the green emission is linear with respect to analyte concentration (right). Adapted with permission from ref. 40 Copyright (2018) Royal Society of Chemistry. | ||
Another motivation for adopting a microfluidic chip is facilitating isolation of an analyte. For example, a spiral microfluidic channel with a herringbone structure was adopted to improve influenza virus capture efficiency by antibodies immobilized on the channel walls.560 Subsequent steps completed a silver-enhanced colorimetric sandwich assay with AuNPs. In some cases, such as the detection of Schistosoma haematobium eggs in urine samples,561 channel dimensions can be tapered to trap large-sized analytes with dimensions greater than about 20 μm. A benefit of this approach is concentration of analyte in the defined imaging zone, but it does not provide molecular specificity. The immunocapture of cells (plus a wash step) within a microfluidic chip provides this specificity. For example, a microfluidic channel with antibody-conjugated pillars was used to assess CD64 expression on neutrophils (Fig. 43).173 With the effect of flow, cells with higher expression levels were captured earlier in the pillar channel, enabling both counting of the cells (via a fluorescent nuclear stain) and an estimate of the distribution of relative CD64 levels.
 | ||
| Fig. 43 Isolation of neutrophils based on CD64 expression. (A) Schematic of the microfluidic chip and summary of the workflow for the analysis (top), and schematic and photograph of the chip (bottom). (B) Design and photograph of the smartphone-based imaging device. (C) Examples of smartphone fluorescence images of captured cells. High-expression results in cells being captured earlier in the chip (bottom image). The scale bars are 1 mm. (D) Correlation between flow cytometry measurements and smartphone measurements. Adapted from ref. 173 Copyright (2019) Royal Society of Chemistry. | ||
The above examples of single-channel microfluidic immunoassays are multi-step and therefore require multiple volumes (e.g. sample followed by washes and reagents or immunoconjugates) to be introduced to the chip by the user. Other assays have pseudo-automated multiple steps through multi-channel microfluidic chips and multiple syringe pumps. Examples include a colorimetric enzyme-linked assay for miRNA pre-extracted from serum,562 and an AuNP-based colorimetric assay for Salmonella.563 Alternatively, solutions for a colorimetric sandwich ELISA have been added in sequence, via micropipette, to microfluidic channels and discharged via centrifugal flow with a portable device.564 A low-tech approach reported for a multistep fluorogenic ELISA of Dengue fever NS1 antigen was self-priming siphon fluidics (Fig. 44).565 Capture antibodies were immobilized within capillaries, and separate reservoirs for the sequences of reagents and washes were needed for the assay. Each of these approaches still requires multiple user actions, which is sub-optimal, but gains precision versus the manual introduction of solutions.
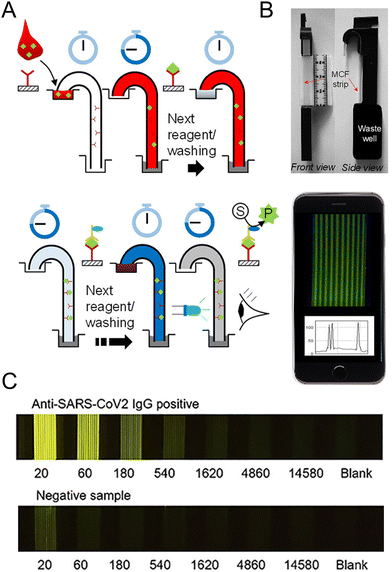 | ||
| Fig. 44 Gravity-driven self-priming siphon microfluidics for fluorogenic ELISA-based detection of dengue fever NS1 antigen and quantification of anti-SARS-CoV-2 IgG in serum spiked samples. (A) Workflow for the assay. (B) Photograph of the siphon device with a microcapillary film (MCF) strip (top) and example of a smartphone-captured fluorescence image (bottom). (C) Detection of IgG anti-viral antibodies in positive and negative samples at various dilution factors. Figure adapted from ref. 565 Copyright 2021 American Chemical Society. This publication is licensed under CC-BY 4.0. | ||
Other microfluidic approaches have housed reagents on-chip. Examples of varying complexity include a PDMS chip with two reagent reservoirs, a mixing channel, and micromechanical actuation for chromogenic detection of gaseous formaldehyde;566 a five-stage and valved microfluidic chip with pre-loaded reagents for an immunomagnetic bead-based colorimetric ELISA;567 and colorimetric ELISA using an AI-controlled chip with a micropump and valve, and process monitoring via the smartphone camera to initiate conditional actions, such as bubble removal and starting and stopping filling of the assay chamber (Fig. 45).90
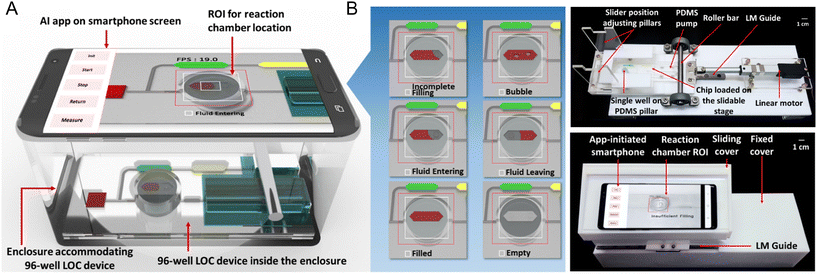 | ||
| Fig. 45 Automated colorimetric ELISA executed by an AI-controlled microfluidic chip with monitoring using the smartphone camera. (A) Rendering of the smartphone-based platform and app. (B) Diagrammatic representations of the fill states (left) and photographs of the device (right). Adapted with permission from ref. 90 Copyright 2022 American Chemical Society. | ||
Another technique that smartphone-based assays have borrowed from benchtop assays and general lab-on-a-chip development is magnetic capture. A few examples highlighted Sections 7.2, 7.4, and 7.6 have referred to immunomagnetic methods, which frequently use a fixed or actuated permanent magnet to hold analyte-linked magnetic particles in place for washing steps. One example not mentioned so far is an on-chip colorimetric ELISA for Zika virus antigen with a Arduino-controlled, motor-actuated magnet system.93 A more exotic particle trapping method that has been reported is contour-mode acoustic streaming tweezers, which enabled multiple sequential steps within a single channel for a microparticle-supported sandwich immunoassay with CL detection of PSA (LOD of 0.2 ng mL−1).513
Despite the greater number of proof-of-concept studies, there is good potential for simplification and utility of these assays beyond a lab. For example, an LFIA was developed for ochratoxin A in wine and coffee (LODs of 0.1–0.3 ng mL−1), where sample was injected into an analytical cartridge with the LFIA-ready membrane and a fluidic component that dispensed the required reagents via “button pushing” with in-cartridge reagent pouches and “switch flipping” with on-cartridge flow control valves (Fig. 46).528
 | ||
| Fig. 46 LFIA-ready membrane in a cartridge with finger-actuated reagent pouches for the CL-based detection of ochratoxin A (OTA) in wine and coffee samples. (A) Schematic overview of LFIA analysis. Sample is loaded, transferred, and mixed with various reagents on-chip before being injected onto the LFIA strip. (B) Photographs of the physical device and chip. (C) Example of a smartphone-acquired CL image of the LFIA and a calibration plot for OTA obtained in wine and instant coffee. Adapted with permission from ref. 528 Copyright 2021 Elsevier B.V. | ||
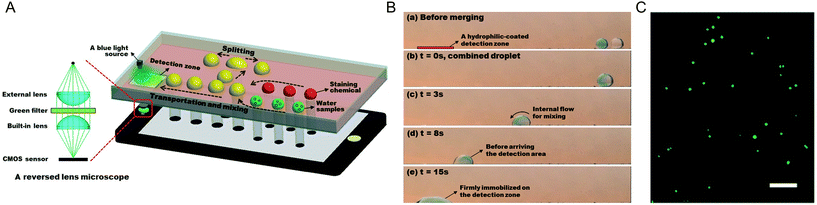 | ||
| Fig. 47 Smartphone display-actuated optoelectrowetting droplet microfluidics for on-chip sample preparation and water quality detection. (A) Schematic overview of the digital microfluidic operations and (B) video frames showing these operations. (C) Smartphone-acquired fluorescent image of marine water sample showing target algae cells. The scale bar is 100 μm. Adapted from ref. 568 Copyright 2018 Royal Society of Chemistry. | ||
A subset of microfluidic approaches have only addressed nucleic acid amplification and detection. Examples include a chamber-based microfluidic chip for digital PCR with a portable device for thermocycling;94 digital droplet LAMP, using both channel-based droplet generation462,569 and a pump-free open droplet array;458 and a non-digital silicon microfluidic chip with multiple channels in which LAMP primers and a dsDNA-sensitive dye were pre-loaded.570 The silicon chip was paired with a portable smartphone-based device for heating and fluorescence detection of up to five different equine respiratory viruses (one genetic sequence per channel) from pre-processed horse nasal swab samples.570
A subsequent study bridged the gap between sample and detection by devising a microfluidic cartridge for processing whole blood for Zika virus detection (LOD of 270 copies per μL; Fig. 48).460 Reagent solutions for chemical lysis of the virus particles to release RNA, and for reverse transcription and isothermal amplification, were transported in multiple stages to different regions of the chip to execute the processing steps. The user manually rotated a built-in threaded screw syringe and actuated two “keys” to move sliding valves into the proper positions. The processed sample was ultimately transferred to a multichannel silicon microfluidic chip for LAMP and detection with a smartphone-based device.460 Another example was a PMDS microfluidic chip with integrated chitosan-modified capillaries to pre-concentrate RNA for PCR amplification and detection.461 The device had a Peltier chip and valves operated by an air-pump and a vacuum generator. In combination with negative pressure manually applied by a syringe draw, the valves enabled on-chip lysis, nucleic acid isolation, and PCR amplification. The device was relatively large and plugged into a mains socket. A third example of a user-driven two-stage approach combined a basket with a DNA extraction disc at the bottom, a block module with a wash chamber for DNA purification, a reaction chamber with a rotary valve for target DNA amplification via RPA, and a microfluidic channel with immobilized oligonucleotide probes for capture of amplified targets (Fig. 49).571 AuNPs and silver enhancement were used for labeling and colorimetric signal enhancement for detection of genes isolated from bacteria in milk samples and urine, human genes isolated from blood samples, and more. This example was also notable because it used heat generated by the smartphone to reach the 37 °C needed for amplification.
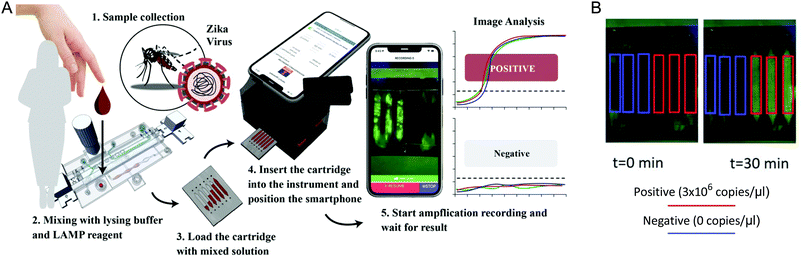 | ||
| Fig. 48 Point-of-care blood testing for Zika virus with smartphone-based detection. (A) Assay workflow with a multi-stage manually actuated microfluidic chip for sample processing and transfer to a detection cartridge for smartphone-based fluorescence imaging. (B) Smartphone images for in-cartridge detection of virus spiked into whole blood. Adapted from ref. 460 Copyright 2022 The Royal Society of Chemistry. | ||
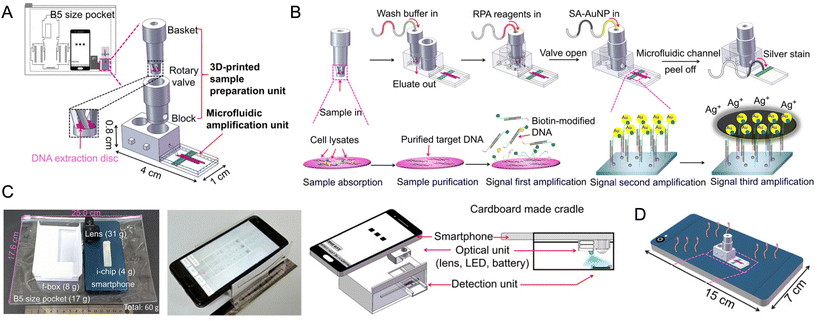 | ||
| Fig. 49 Smartphone-based colorimetric detection of target DNA sequences using a pocket-sized basket-and-microfluidic system for sample preparation and assay execution with a foldable box. (A) Design of the device. (B) Assay workflow. (C) Photographs of the smartphone-based foldable-box device. (D) Illustration of how the smartphone is used to provide heat for DNA amplification. Figure adapted from ref. 571 under a Creative Commons CC BY 4.0 license. | ||
A single chip design for both isolation and amplification is also possible. One such example was a finger-actuated microfluidic chip with four buttons, each of which was pushed repeatedly by the user to dispense and flow buffers and reagents through microfluidic channels.572 In this manner, and with assistance from an external magnet, the chip executed an immunomagnetic isolation of a pathogenic bacterium, a nucleic acid extraction and purification with a second type of magnetic nanoparticle, and on-chip amplification for detection with a smartphone.
8.2 Paper-based microfluidics
Many of the examples of smartphone-based assays highlighted in earlier sections have utilized paper as a substrate. The paper has been homogeneous in some cases, but more often has been patterned with one or more channels using wax or another method to function as a μPAD. The simplest examples of μPADs direct a single aliquot of sample to multiple test zones. The analysis of sweat is an example of such an application, where one study designed a paper microfluidic device to adhere to skin and detect pH, glucose, lactate, and uric acid through indicator dyes and chromogenic reactions with reagents and enzymes pre-deposited in the test zones (Fig. 50).573 This format has also been used to run replicate measurements for CL detection.532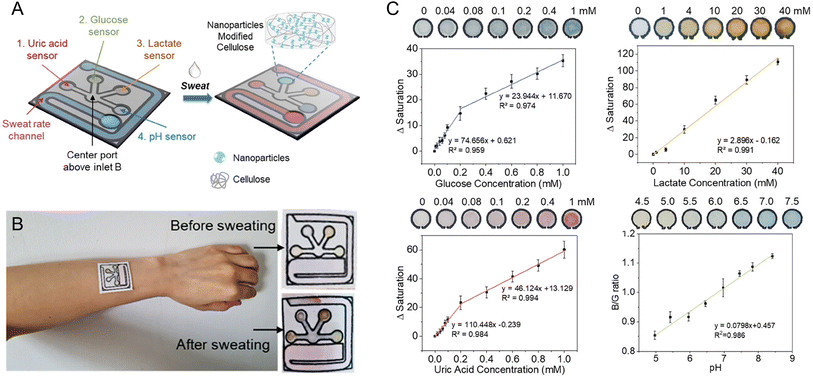 | ||
| Fig. 50 Wearable μPAD for the analysis of sweat biomarkers: uric acid, glucose, lactate and pH. (A) Design of the μPAD. The NP detection region is highlighted. (B) After sweat exposure, a color change is visualized. (C) Calibration curves (based on HSV color saturation (S) or a B/G channel ratio) for the analytes from smartphone imaging of the μPAD. Adapted with permission from ref. 573 Copyright 2023 Wiley-VCH GmbH. | ||
Other μPADs have sought to derive function beyond sample distribution. For example, a paper-based pump-free rapid mixer design was applied to the colorimetric detection of organo-phosphate pesticide.574 Multiple assays have also extended the paper channel beyond the test zone to enable on-PAD washing steps: the fluorimetric detection of aqueous metal ions (Cd2+, As3+, Pb2+, Hg2+) via DNA-based probes (e.g. aptamers, DNAzyme, thymine-rich oligonucleotide);575 the detection of a pesticide via a fluorescence recovery mechanism with dye-labeled aptamer;576 and magnetic-based ELISA for a malaria biomarker, Plasmodium falciparum lactate dehydrogenase.577 More sophisticated multi-step assay capability has been achieved through a μPAD that incorporates a mechanically actuated paper-based rotary valve system to execute a colorimetric ELISA for the β-amyloid peptide 1–42 biomarker of Alzheimer's disease (Fig. 51).96 Reagents were pre-loaded on the μPAD, such that the user added the sample to the test zone and buffer to the inlet zone, and the rotary motion separated binding, washing, and color development steps. Manual actuation has been utilized for a multi-step assay, such as in the case of determining serum phosphate levels from a single drop of whole blood using malachite green as a colorimetric indicator.578 Serum was isolated from blood cells via flow through a suitable membrane, and a mechanical mechanism dipped a serum-impregnated collection pad into reagent solution to develop a colorimetric signal.
 | ||
| Fig. 51 Automated platform for detection of Alzheimer's disease biomarkers. (A) Design of μPAD platform (left) and the smartphone-based device (right). (B) Workflow of the colorimetric ELISA assay. (C) Calibration curve in terms of grayscale intensity versus Aβ 1–42 concentration (top) and comparison of the result to a traditional analytical method for artificial plasma samples (bottom). Figure adapted with permission from ref. 96 Copyright 2024 Elsevier B.V. All rights reserved. | ||
Foldable μPADs have also been a means of executing multiple assay steps in a user-friendly format. In one example, sputum samples that were either positive or negative for infection by Pseudomonas aeruginosa were liquified with a bacterial catalase and hydrogen peroxide, and the liquid sample was transferred to the μPAD.579 The μPAD was folded to bring pre-loaded AuNP-antibody conjugates into contact with the sample, the sample spot was washed, and the residual color of bound AuNPs provided a signal for the detected bacteria. A method for the detection of viral RNA was based on the release of Ag+ (aq) ions from an oligonucleotide probe upon hybridization with target RNA, where the Ag+ inhibited urease and prevented a pH and color change from turnover of its substrate in the presence a phenol red indicator dye (Fig. 52).580 Sample was added to the μPAD, which was folded in sequence to separate the steps of binding with probe and the chromogenic reaction. Different regions on the device were used for multiplexed screening for MERS-CoV, SARS-CoV, and SARS-CoV-2. Similarly, another study pre-dried reagents for CL-based glucose detection (LOD 10 μM) onto a foldable μPAD. This format kept the reagents separated until needed for the assay, which reduced the required user steps to only pipetting to add sample and standard solution, a fold, and adding transport buffer.531
 | ||
| Fig. 52 Origami-based μPAD for assaying and multiplexed screening of SARS-CoV-2 variants with single-nucleotide resolution. (A and B) Design and workflow for the assay. (C) Design of the smartphone-based imaging device. (D) Examples of smartphone-acquired images and green-area-ratio values as the metric for analysis. Each dot on the device corresponds to a different targeted gene and mutation. Figure adapted with permission from ref. 580 Copyright 2022, the author(s), under exclusive license to Springer Nature Limited. | ||
An interesting capability that has emerged with smartphone-based detection using paper test strips is quantitation by distance measurements. One report took advantage of the change in surface tension from immunoagglutination to detect SARS-CoV-2 (LOD 1–10 fg mL−1) nucleocapsid via a change in the capillary flow velocity in a patterned paper channel, where flow was tracked using dye-doped polystyrene NPs.581 Another report measured IL2-positive and total natural killer cell concentrations in buffy coat blood samples via their migration distance along a paper channel. The binding of fluorescent polymer NP-antibody conjugates slowed the cell migration to a degree proportional to their CD56 level, such that the fluorescence intensity profile along the channel reflected the relative distribution of CD56 expression levels in the sample.487 Other distance-based assays have been based on analyte depletion rather than velocity. In one case, an indicator dye was deposited in a paper channel in the pattern of a repeating QR code. The number of QR codes that appeared in a fully readable form along the length of the channel determined whether the analyte (Cu2+) concentration was low, medium, or high within the dynamic range of the assay (0.4–3.2 mM).582 In another case, a rapid blood grouping analysis was done through depletion of the relevant red blood cell type through immunocapture within a spoke pattern of paper channels with colorimetric contrast augmented by bromocresol green (Fig. 53).161 The layout of the μPAD was such that result was readable as a QR code via the smartphone camera. Distance measurements are not unique to paper devices, as demonstrated by smartphone readout of the radial travel of DNA motor system immobilized on 5 μm beads across a DNA modified gold chip.583 This format was able to detect SARS-CoV-2 spiked in artificial saliva and exhaled breath condensate with a sensitivity of 1000 copies per mL within 15 minutes.
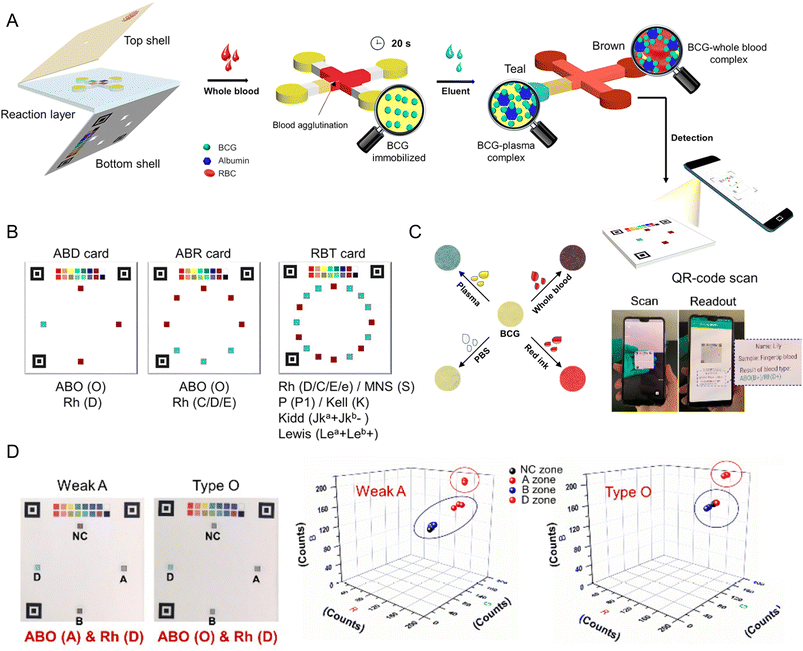 | ||
| Fig. 53 Antibody-based blood typing on a μPAD via QR code patterns. (A) Design and workflow of the assay. (B) Different assay formats for application to different blood-typing requirements. Reference color blocks and position detection boxes ensure consistency in interpretation. (C) Bromocresol green dyes provides optical contrast for smartphone imaging. (D) Smartphone images of cards (left) and 3D plots from RGB analysis (right) for two samples of two different blood types. Adapted with permission from ref. 161 Copyright 2021 American Chemical Society. | ||
9. Perspective: challenges and opportunities
As highlighted in this review, and in other recent reviews,584–586 smartphones have clearly emerged as capable and versatile platforms for molecular analyses based on optical measurements. The built-in camera is useful for quantitative readout of changes in optical extinction, color, and luminescence induced by molecular recognition and other selective chemistries. Numerous measurements have been successfully ported from laboratory instruments to smartphone-based devices, and qualitative methods for point-of-need testing have been made quantitative. The convergence of smartphones with microfluidics, NPs, and other complementary materials and technologies holds great potential for the lab-on-a-chip concept and better democratization of chemical and biological assays. What challenges and opportunities for innovation stand between this conceptualization and its practical realization?9.1 Pre-detection bottleneck: sample processing
About 1 in 6 smartphone-based methods and devices reported to date have been sample-to-answer approaches (Fig. 14). Most assays and devices have either been limited to proof of concept for smartphone detection or adopted one or more standard laboratory methods for processing authentic samples prior to smartphone detection. The challenge of the “world-to-chip” interface is well known in microfluidics587 and the literature reflects an analogous challenge with smartphone-based analyses. It will be essential for processing of crude samples to be rapid and automated, simple and safe, or unnecessary for the most impactful translations of smartphone technology to authentic analytical applications. This aspect of translation is minimized or unacknowledged in much of the literature. Although studies that focus on advances in detection are important, there is a need for more studies that address the bottleneck of sample processing.The above considerations provide many opportunities for innovation in stripping down laboratory assay methods to their bare minimum and enabling shortcuts through advanced optical materials, selective recognition agents, and clever optical designs. Examples include the development of chromogenic and fluorogenic reagents that have sufficient selectivity and efficiency to react with analyte in crude samples, and the development of FRET probes with fluorescent acceptors. The nanometer-scale distance dependence of FRET enables mix-and-measure protocols and ratiometric detection that will minimize the impact of inconsistent dilution and excitation intensity.588 FRET probes based on NP materials that have high brightness,589 exhibit upconversion PL,590 and support time-gated measurements591 will also mitigate challenges from sample autofluorescence. Although smartphone cameras do not support near-IR detection, the red RGB camera channel can be leveraged with similar effectiveness, and chips with integrated waveguides and short optical path lengths will facilitate optical interrogation of non-transparent samples. For crude samples that start opaque, sample chips that integrate membranes and other porous materials will ideally provide the cleanup needed for optical detection. There is also great potential in building reference markers into test strips and chips to enable compensation of variation in sample optical properties and transfer complexity from hardware to software.
Conventionally, the reuse of chips has been eschewed in point-of-need testing due to the less favorable business economics and obstacles such as sample carryover, replenishment of pre-loaded reagents, and deteriorating performance with each cycle of use. Two factors may encourage reconsideration of this aversion: the inability of disposable paper-based microfluidic devices to support the full scope of technical capabilities that will be needed to translate many assays from the bench to the field; and a future focus on sustainability and the different economics of equitable access, particularly in the case of technologies that offer high-value capability that is built on non-abundant, expensive, and non-green materials. In this scenario, the reuse of chips will require fast and reliable cleaning and decontamination using reagents that pose a minimal hazard to people and the environment. It is a prime opportunity for open chip configurations of (opto)electrowetting for digital microfluidics, both in terms of the disparate capabilities versus paper microfluidics and the potential to be wiped clean and sterile.
9.2 Pre-detection bottleneck: calibration
Nearly all of the smartphone-based assays reported to date require calibration or training that links optical signals with the quantity or identity of an analyte. Unfortunately, calibration is a laboratory procedure that is not practical for non-laboratory users of a portable device, but quantitative capability remains important. Alternative strategies are therefore needed.One possible strategy is to shift the onus of calibration to the production process. Such factory calibration will require reproducible manufacturing and persistence over long storage periods. It also requires excellent consistency between the readout devices used by the factory and user, and a means of compensating for variation in sample optical properties will be useful for some authentic applications. Detection based on ratio between two colors of luminescence, or on ratio of luminescence intensity between test and reference zones, is expected to provide the best tolerance of variation in sample optical properties and between readout devices. Detection based on changes from one brightfield color to another should also have more tolerance of device variability than detection that is solely based on an absolute intensity of luminescence or the tint of a color.
Digital assays,592–594 which are an emerging trend in commercial diagnostics and in the general analytical literature, are an exciting alternative strategy. These assays count analyte molecules and are thus quantitative without requiring user calibration. Most digital assays are based on fluorescence and utilize either enzymatic amplification with fluorogenic reactions or the sophisticated and expensive hardware necessary for single-molecule detection. Enzymatic amplification adds reagents to assays with the potential challenges of cold storage, limited shelf life, special digitization chips, extra assay steps, and the associated additional costs. The typical hardware for single-molecule detection is the opposite of what is needed for a portable low-cost device. Here, there are opportunities for innovation in developing ultrabright fluorescent materials and optical configurations that will enable enzyme-free single-molecule detection with a smartphone-based device. Studies have demonstrated that such capability is a viable possibility for single fluorescent NPs and QDs,86 multi-dye DNA origami,29 and single dyes.83 Remaining needs are simplification of hardware and methodology, and approaches to sampling and counting that produces results with suitable accuracy and precision.
Single-molecule detection is also not a strict necessity for quantitation by counting. Assay formats that have highly efficient mass transport of analyte to an interface with capture probes (e.g. microfluidic channels, microporous materials) are capable of depleting analyte from a flowing sample solution. A series of discrete clusters of capture probes along the direction of flow are then able to report on the quantity of analyte via the number of these clusters that have detectable luminescence or exhibit a color change. There are a myriad of unrealized opportunities for adapting binding assays to this quantitative format.
9.3 Multitasking
When the objective is to enable molecular assays outside of a laboratory, it follows that most users will not want a different smartphone-based device for every analyte or type of assay of interest. The ideal technology will be a single measurement platform that is able to utilize a variety of test strips or chips to execute multiple types of assays for a variety of analytes. This review has already highlighted how a broad scope of assays has been implemented across common test formats like clear plastic tubes, μPADs, LFIA membranes, and microfluidic chips. Assay formats have arguably been more consistent than the designs and optical configurations of the smartphone-based devices used for their readout. Here, there is great opportunity to create single devices that are reconfigurable through interchangeable modules that are matched to different test formats and different illumination configurations (Fig. 54). Smartphone-integrated devices are arguably more readymade for multitasking than linked and dongle devices because of the multitude of on-board features—including a trend of multiple camera modules with different optics—that can be accessed on-demand without bespoke reengineering.9.4 Cost
The previous subsections outlined objectives that are important and technically challenging to achieve. Researchers must also achieve these objectives on the cheap. Integrating a smartphone into a complex device that costs thousands of dollars will largely defeat the original purpose of adopting the smartphone. Practically, the smartphone must be the most expensive component of any smartphone-based device. In this vein, we propose that the total cost of all components in a device, except the smartphone, should be a multiple, f, of the cost of the smartphone, where 0.01 < f < 0.5. The value of f should align with the intended use case, such that non-smartphone costs ranges from $1 to $600 USD. Here, the opportunity is less innovation and more ingenuity: finding materials and components that are mass produced for consumers for non-scientific reasons and repurposing those things to meet a technical need in a smartphone-based device. One such example already highlighted is replacing research-grade optical filters with low-cost color films.196,197 Indeed, one of the tenants of “frugal science” is leveraging everyday items for scientific function.8The cost of the test substrate, chips, cartridges, and reagents must also be affordable. For some contexts, an affordable cost will be on the order of dollars per test; for other contexts, it will be on the order of cents per test. The simplification of test platforms is one opportunity for minimizing cost per test. A complete set of reagents freeze-dried or tableted595,596 into a simple disposable tube or a single-chamber chip will be more economical than multiple reagents loaded at different sites in a multichannel or multi-chamber chip. Affinity and capture agents that can self-conjugate with optical materials and interfaces, with plug-and-play substitution of different agents for different analytes, will minimize preparatory steps and enable multiple test kits to be produced using the same workflow, both reducing costs. Tetrameric antibody complexes (TACs) with fluorescent NPs such as QDs,597 Pdots,598 and QD assemblies485,486 are an example that approaches this ideal.
A reality of the need for low cost and simple operation is that some assays and applications will simply not be practically adaptable to a smartphone-integrated device for the foreseeable future. It is here that adaption of laboratory methods to portable detection is not sufficient, and that completely new methods of analysis must be developed for some analytes and samples.
9.5 Regulatory approval
For clinical applications, in vitro diagnostic tests and devices require regulatory approval. Laboratories that serve testing needs in clinical and many other contexts (e.g. food, environmental) also require or benefit from accreditations and certifications. Regulatory processes are generally slower than the pace of change in smartphone features and technical specifications, and are predicated, in part, on consistency. With an ever-changing distribution of smartphone models available for purchase and in the hands of consumers, there is natural doubt that regulatory approval of a smartphone-integrated device will be possible.Another frequent concern is data security and privacy. Of course, challenges with the security of health data already exist with computer systems and are managed. The main difference with the smartphone is that its small size makes it easier to steal or lose. Fortunately, smartphones already support security features (e.g. data encryption, device tracking, remote memory wiping). At present, security patches and OS upgrades for devices are at the discretion of the manufacturer: Apple guarantees a minimum of five years, and support for Android phones varies between 2–7 years depending on the manufacturer. Given the trend of increasing consumer concern about privacy, we speculate that the industry (and legislation) will continually take steps to enhance data security. Ultimately, regulatory approval is far more likely to be a barrier to the adoption of smartphone-based devices than security and privacy concerns.
For brevity, to address the question of regulatory approval, the discussion will be limited to the context of in vitro diagnostic tests for health applications. We also note that the regulatory hurdle is only an unknown for devices that integrate smartphones. Linked and dongle devices bypass this challenge because the smartphone does not generate the analytical data: there is a single design of a bespoke device to be approved.
So, for a smartphone-integrated device, what are some of the possible pathways to approval?
One pathway starts from the premise that the users for smartphone-integrated diagnostic devices will be professionals and not an arbitrary consumer. For example, a primary care physician or nurse in a rural community will adopt a smartphone-based device to enable local diagnostic capability that would otherwise only be available in a clinical lab in a city hundreds of kilometers away. The professional will also use a smartphone dedicated to diagnostic use and not their personal smartphone. In this scenario, the device can be distributed with one (or a few) specific model(s) of smartphone, providing consistency that will facilitate approval. The caveat is that the approval process may need to be repeated at multi-year intervals to enable upgrades of smartphone technology.
Other potential pathways to approval are predicated on recognition of the positive societal and global impacts of more accessible diagnostics and equitable quality of health care. For example, regulatory agencies might grant limited approval that would enable smartphone-based diagnostic testing in rural and remote communities without a clinical lab, but require that more rigorous clinical lab testing is done prior to medical interventions in the event of a disease indication. Even this limited approval would enhance health care in such communities. Likewise, with the trend of multiple camera modules per smartphone and growing interest in ethical and socially responsible smartphone technology (e.g. Fairphone), manufacturers might become inclined (or legislated) to include one diagnostic camera module per smartphone, which would have invariant specifications and data processing between manufacturers and models. This step would provide the consistency to facilitate regulatory approval, and the engineering challenge of accommodating different dimensions of phones is surmountable. Another scenario is that a new pandemic motivates an emergency adoption of smartphone-based methods as a rapidly deployable means of global surveillance, which then opens the door to developing a framework for approval of non-emergency use of the technology. Certainly, the two proactive scenarios are more appealing than this reactive scenario.
Another possible pathway to regulatory approval—and one over which researchers have the most control—is the development of smart tests and smart apps that identify and correct for sources of variability to yield consistent quantitative results (or declare an invalid test) regardless of smartphone model, device platform, and sample source.
9.6 Smart-tests and smart-apps for smartphones
If smartphone-integrated devices are to reach their full potential in enabling globally accessible and democratized molecular analyses, then the technology must embrace variability while also ensuring accuracy and precision in the analytical data. Reconciling this contradiction is well within the capabilities of data processing augmented by ML and AI because, to date, smartphone cameras have not advanced in ways that change their fundamental operating principles. Imagine test strips and chips that integrate reference markers for brightfield colors and tints, and for luminescence colors and intensities. Analysis of the data from these markers will enable corrections for variation in the color temperature of brightfield illumination, the intensity and uniformity of excitation light for luminescence, the intrinsic color or autofluorescence of a crude or minimally processed sample, and differences in the RGB channel responses of different smartphone cameras. QR codes printed on tests can store information about the prescribed data for the reference markers, the parameters from factory calibration of a test, and thresholds for when the smartphone should declare a test result invalid. NFC can be implemented to ensure that a reconfigurable smartphone-integrated device has the correct modules inserted for a given test. In other words, the collective ability of a smartphone camera, device, and sample to produce a reliable result from a test can be evaluated in real-time, and corrections implemented when the variable parameters fall within tolerances. As described in the review, some studies have already implemented these concepts on a more limited and individual basis. More dimensions of this concept, working in concert, will bring about smart-tests and smart-apps as another potential pathway to regulatory approval and the most globally impactful and democratized diagnostic and analytical technologies.10. Conclusions
Smartphones have been widely adopted as optical detectors and components of prototype devices for portable molecular analyses based on colorimetry and multiple types of luminescence. These devices utilize various combinations of the smartphone camera, flash, and display in tandem with other optics and optoelectronic components, and have achieved practical detection limits in assays of ions and small molecules, immunoassays, nucleic acid amplification tests, cytometry, and more. Although molecular indicator dyes, stains, and fluorophores have been adequate or well suited to many of these methods, the potential for optically active and catalytic NPs to enhance analytical performance has also been demonstrated. For either class of material, smartphone compatible assay formats have included simple sample cells, microwell arrays, lateral flow membranes, test strips, μPADs, and multiple types of microfluidic chips. In short, smartphone-based detection is now a proven concept.Among the next challenges and opportunities in developing smartphones as analytical tools are the elimination of sample processing for end users, achieving quantitative capability without the need for user-driven calibration, building multitasking devices, and engineering tolerance for operational variability between authentic samples, users, and devices without loss of accuracy and precision. If the field is able to address these challenges, then smartphone-based molecular analyses will be more than a passing fad in research and development and will instead evolve to be a bona fide tool in society and industry. Collectively, the design of digital assays and ultrabright materials, embedding smart features in test platforms and device modules, and quality-ensuring ML- and AI-powered software are a prospective route to regulatory approvals and the realization of smartphones as devices that democratize chemical and biological analysis with global benefits.
Abbreviations
| 3D | Three-dimensional |
| A-GPS | Assisted (or augmented) global positioning system |
| ABS | Acrylonitrile butadiene styrene |
| AFB1 | Aflatoxin B1 |
| AgNP | Silver nanoparticle |
| AI | Artificial intelligence |
| ALS | Ambient light sensor |
| API | Application programming interface |
| AuNP | Gold nanoparticle |
| AuNR | Gold nanorod |
| B | Blue |
| BCG | Bromocresol green |
| BL | Bioluminescence |
| BODIPY | 4,4-Difluoro-4-bora-3a,4a-diaza-s-indacene |
| BPA | Bisphenol A |
| BRET | Bioluminescence resonance energy transfer |
| Cas | CRISPR-associated protein |
| CD | Compact disk |
| CFA | Color filter array |
| CFU | Colony-forming unit |
| CIE | International Commission on Illumination |
| CK-MB | Creatine kinase-MB |
| CL | Chemiluminescence |
| CMOS | Complimentary metal-oxide-semiconductor |
| CRET | Chemiluminescence resonance energy transfer |
| CRISPR | Clustered regularly interspaced short palindromic repeats |
| Cy5 | Cyanine 5 |
| DAB | 3,3′-Diaminobenzidine |
| DAPI | 4′,6-Diamidino-2-phenylindole |
| DC | Direct current |
| DLP | Digital light processing |
| DNA | Deoxyribonucleic acid |
| DNG | Digital negative |
| dsDNA | Double-stranded deoxyribonucleic acid |
| DTNB | 5,5′-Dithiobis(2-nitrobenzoic acid) |
| DVD | Digital versatile disk |
| ECL | Electrochemiluminescence |
| ELISA | Enzyme-linked immunosorbent assay |
| EphA2 | Ephrin type-A receptor 2 |
| FDM | Fused deposition modelling |
| FFF | Fused filament fabrication |
| FLFIA | Fluorescence lateral flow immunoassay |
| FPGA | Field programmable gate array |
| FPS | Frames per second |
| FRET | Förster resonance energy transfer |
| FWHM | Full width at half maximum |
| G | Green |
| GDP | Gross domestic product |
| GPIO | General-purpose input/output |
| GPS | Global positioning system |
| GUD | Beta-D-glucuronidase |
| H | Hue |
| HBV | Hepatitis B virus |
| HCA | Hierarchical cluster analysis |
| HCV | Hepatitis C virus |
| HEIC | High efficiency image container |
| HiLo | Hybrid illumination |
| HIV | Human immunodeficiency virus |
| HPTS | 8-Hydroxypyrene-1,3,6-trisulfonic acid |
| HPV | Human papillomavirus |
| HRP | Horseradish peroxidase |
| HSB | Hue-saturation-brightness |
| HSV | Hue-saturation-value |
| IDE | Integrated development environment |
| IR | Infrared |
| ISO | International Organization for Standardization |
| JPEG | Joint Photographic Experts Group |
| LAMP | Loop-mediated isothermal amplification |
| LCD | Liquid crystal display |
| LDA | Linear discriminant analysis |
| LED | Light-emitting diode |
| LFA | Lateral flow assay |
| LFIA | Lateral flow immunosorbent assay |
| LiDAR | Light detection and ranging |
| LLC | Luminescent lanthanide complex |
| LM | Latex microsphere |
| LOC | Lab on a chip |
| LOD | Limit of detection |
| LTE | Long term evolution |
| MCU | Microcontroller unit |
| MERS-CoV | Middle East respiratory syndrome coronavirus |
| miRNA | Micro ribonucleic acid |
| ML | Machine learning |
| MOF | Metal–organic framework |
| MSLA | Masked stereolithography |
| NADH | Nicotinamide adenine dinucleotide |
| NC | Nitrocellulose |
| NFC | Near-field communication |
| NIR | Near-infrared |
| NN | Neural network |
| NP | Nanoparticle |
| NR | Nanorod |
| OLED | Organic light-emitting diode |
| OS | Operating system |
| PCA | Principal component analysis |
| PCR | Polymerase chain reaction |
| PDMS | Polydimethylsiloxane |
| Pdots | Semiconducting polymer dots |
| PES | Polyethersulfone |
| PL | Photoluminescence |
| PLA | Polylactic acid |
| PLNP | Persistent-luminescent nanoparticle |
| PMMA | Polymethyl methacrylate |
| PSA | Prostate-specific antigen |
| PTSA | 1,3,6,8-Pyrenetetrasulfonic acid |
| QD | Quantum dot |
| qPCR | Quantitative polymerase chain reaction |
| QR | Quick response |
| R | Red |
| RAM | Random-access memory |
| RCA | Rolling circle amplification |
| RFLFIA | Ratiometric fluorescence lateral flow immunoassay |
| RGB | Red, green, blue |
| RNA | Ribonucleic acid |
| ROI | Region of interest |
| ROX | Carboxyrhodamine |
| RPA | Recombinase polymerase amplification |
| rRNA | Ribosomal ribonucleic acid |
| RT | Reverse transcription |
| RTF-EXPAR | Reverse transcription-free exponential amplification reaction |
| S | Saturation |
| SARS-CoV-2 | Severe acute respiratory syndrome coronavirus 2 |
| SBC | Single-board computer |
| SDK | Software development kit |
| SLA | Stereolithography |
| SPR | Surface plasmon resonance |
| SVM | Support vector machine |
| TAC | Tetrameric antibody complex |
| TAMRA | Carboxytetramethylrhodamine |
| TMB | 3,3′,5,5′-Tetramethylbenzidine |
| TNFα | Tumor necrosis factor |
| ToF | Time-of-flight |
| TRRS | Tip-ring ring-sleeve |
| TTA | Triplet–triplet annihilation |
| UCNP | Upconversion nanoparticle |
| USB | Universal serial bus |
| USB-C | Universal serial bus type-C |
| USD | United states dollar |
| UV | Ultraviolet |
| UVB | Ultraviolet B |
| UVC | Ultraviolet C |
| V | Value |
| W | White |
| Y | Yellow |
| ZIF | Zeolitic imidazolate framework |
| μPAD | Micro paper analytical device |
Data availability
The data supporting the meta-analysis in this review article have been uploaded as ESI† in a Microsoft Excel spreadsheet file format (.xlsx).Author contributions
Daina V. Baker: writing – original draft preparation; formal analysis (data for Fig. 14); investigation (data for Fig. 2–5, 7); visualization (adaptation of figures from the literature, Fig. 1, 6, 13). Jasmine Bernal-Escalante: writing – original draft preparation; formal analysis (data for Fig. 14); investigation (data for Fig. 2–5, 7); visualization (adaptation of figures from the literature). Christine Traaseth: writing – original draft preparation; formal analysis (data for Fig. 14); investigation (data for Fig. 2–5, 7); visualization (adaptation of figures from the literature). Yihao Wang: writing – original draft preparation; visualization (adaptation of figures from the literature, creation of device models in Fig. 1, 9, 54A). Michael V. Tran: writing – original draft preparation. Seth Keenan: writing – original draft preparation. W. Russ Algar: conceptualization; writing – review & editing; visualization; supervision; funding acquisition.Conflicts of interest
There are no conflicts to declare.Acknowledgements
We thank the Natural Sciences and Engineering Research Council of Canada (NSERC), Canadian Institutes of Health Research (CIHR), Canada Foundation for Innovation (CFI), British Columbia Knowledge Development Fund (BCKDF), and the University of British Columbia (UBC) for support of our research program. JBE is grateful for a Miguel Romero Sánchez Fellowship in Chemistry from UBC. SK is grateful for an NSERC Canada Graduate Scholarship, an Ed Shuter Scholarship, and a Gladys Estella Laird Research Fellowship. We thank Zhujun Xiao and Rupsa Gupta for helpful discussion, and Ghinwa Darwish for providing the QD PL emission spectra data.References
- E. Samiei, M. Tabrizian and M. Hoorfar, Lab Chip, 2016, 16, 2376–2396 RSC.
- S. Surappa, P. Multani, U. Parlatan, P. D. Sinawang, J. Kaifi, D. Akin and U. Demirci, Lab Chip, 2023, 23, 2942–2958 RSC.
- N. Azizipour, R. Avazpour, D. H. Rosenzweig, M. Sawan and A. Ajji, Micromachines, 2020, 11, 599 CrossRef PubMed.
- V. Narayanamurthy, Z. E. Jeroish, K. S. Bhuvaneshwari, P. Bayat, R. Premkumar, F. Samsuri and M. M. Yusoff, RSC Adv., 2020, 10, 11652–11680 RSC.
- U. A. Gurkan, D. K. Wood, D. Carranza, L. H. Herbertson, S. L. Diamond, E. Du, S. Guha, J. D. Paola, P. C. Hines, I. Papautsky, S. S. Shevkoplyas, N. J. Sniadecki, V. K. Pamula, P. Sundd, A. Rizwan, P. Qasba and W. A. Lam, Lab Chip, 2024, 24, 1867–1874 RSC.
- 68% of the world population projected to live in urban areas by 2050, says UN, https://www.un.org/uk/desa/68-world-population-projected-live-urban-areas-2050-says-un, (accessed 4 November 2024) Search PubMed.
- Urban Development: Overview, https://www.worldbank.org/en/topic/urbandevelopment/overview, (accessed 4 November 2024) Search PubMed.
- G. Byagathvalli, E. J. Challita and M. S. Bhamla, Ind. Eng. Chem. Res., 2021, 60, 15874–15884 CrossRef CAS PubMed.
- M. Monteiro and A. C. Martí, Am. J. Phys., 2022, 90, 328–343 CrossRef.
- P. Taylor, Number of mobile (cellular) subscriptions worldwide from 1993 to 2023, https://www.statista.com/statistics/262950/global-mobile-subscriptions-since-1993/, (accessed 4 November 2024) Search PubMed.
- F. Laricchia, Penetration rate of smartphones in selected countries 2022, https://www.statista.com/statistics/539395/smartphone-penetration-worldwide-by-country/, (accessed 4 November 2024) Search PubMed.
- Smartphone owners are now the global majority, New GSMA report reveals, https://www.gsma.com/newsroom/press-release/smartphone-owners-are-now-the-global-majority-new-gsma-report-reveals/, (accessed 4 November 2024) Search PubMed.
- P. Taylor, Mobile coverage rate worldwide from 2011 to 2029, by technology, https://www.statista.com/statistics/1016292/mobile-coverage-by-technology-worldwide/, (accessed 4 November 2024).
- N. O. Theonest, K. Ngowi, E. R. Kussaga, A. Lyimo, D. Kuchaka, I. Kiwelu, D. Machuve, J.-M. Vianney, J. Reboud, B. T. Mmbaga, J. M. Cooper and J. Buza, PLOS Digit. Health, 2024, 3, e0000565 CrossRef PubMed.
- The Mobile Economy 2022, https://www.gsma.com/solutions-and-impact/connectivity-for-good/mobile-economy/wp-content/uploads/2022/02/280222-The-Mobile-Economy-2022.pdf, (accessed 4 November 2024) Search PubMed.
- F. Laricchia, Number of smartphones sold to end users worldwide from 2007 to 2023, https://www.statista.com/statistics/263437/global-smartphone-sales-to-end-users-since-2007/, (accessed 4 November 2024) Search PubMed.
- G. R. D. Prabhu and P. L. Urban, Chem. Rev., 2020, 120, 9482–9553 CrossRef CAS PubMed.
- G. Niezen, P. Eslambolchilar and H. Thimbleby, BMJ Innov., 2016, 2, 78–83 CrossRef PubMed.
- J. M. Pearce, Sci. Public Policy, 2016, 43, 192–195 CrossRef.
- M. D. M. Dryden, R. Fobel, C. Fobel and A. R. Wheeler, Anal. Chem., 2017, 89, 4330–4338 CrossRef CAS PubMed.
- S. Lehtola and A. J. Karttunen, WIREs Comput. Mol. Sci., 2022, 12, e1610 CrossRef CAS.
- A. M. Chagas, PLoS Biol., 2018, 16, e3000014 CrossRef PubMed.
- R. M. Sarıyer, A. D. Edwards and S. H. Needs, Biosensors, 2023, 13, 948 CrossRef PubMed.
- J. J. Davis, S. W. Foster and J. P. Grinias, J. Chromatogr. A, 2021, 1638, 461820 CrossRef CAS PubMed.
- I. Nuñez, T. Matute, R. Herrera, J. Keymer, T. Marzullo, T. Rudge and F. Federici, PLoS One, 2017, 12, e0187163 CrossRef PubMed.
- S. Majumder and M. J. Deen, ACS Sens., 2019, 19, 2164 CrossRef PubMed.
- Y. Xia, J. Hu, S. Zhao, L. Tao, Z. Li, T. Yue and J. Kong, Biosens. Bioelectron.: X, 2022, 11, 100195 CAS.
- S. Tominaga, S. Nishi and R. Ohtera, ACS Sens., 2021, 21, 4985 CrossRef PubMed.
- C. Vietz, M. L. Schütte, Q. Wei, L. Richter, B. Lalkens, A. Ozcan, P. Tinnefeld and G. P. Acuna, ACS Omega, 2019, 4, 637–642 CrossRef CAS PubMed.
- T. Krishnan, H.-N. Wang and T. Vo-Dinh, ACS Sens., 2021, 21, 8044 CrossRef CAS PubMed.
- X. Ruan, V. Hulubei, Y. Wang, Q. Shi, N. Cheng, L. Wang, Z. Lyu, W. C. Davis, J. N. Smith, Y. Lin and D. Du, Biosens. Bioelectron., 2022, 208, 114190 CrossRef CAS PubMed.
- E. V. Woodburn, K. D. Long and B. T. Cunningham, IEEE Sens. J., 2019, 19, 508–514 CAS.
- W. Cao, Y. Deng, T. Pan, H. Hao and M. Wang, IEEE Photonics J., 2019, 11, 6802708 Search PubMed.
- M. Schäfer, V. Bräuler and R. Ulber, Sens. Actuators, B, 2018, 255, 1902–1910 CrossRef.
- A. S. Paterson, B. Raja, V. Mandadi, B. Townsend, M. Lee, A. Buell, B. Vu, J. Brgoch and R. C. Willson, Lab Chip, 2017, 17, 1051–1059 RSC.
- M. Hermann, P. Agrawal, I. Koch and R. Oleschuk, Lab Chip, 2019, 19, 654–664 RSC.
- M. R. Muir and A. Innes, Anal. Methods, 2024, 16, 5571–5583 RSC.
- E. Petryayeva and W. R. Algar, Anal. Bioanal. Chem., 2016, 408, 2913–2925 CrossRef CAS PubMed.
- R. Gupta, W. J. Peveler, K. Lix and W. R. Algar, Anal. Chem., 2019, 91, 10955–10960 CrossRef CAS PubMed.
- Y. Sekine, S. B. Kim, Y. Zhang, A. J. Bandodkar, S. Xu, J. Choi, M. Irie, T. R. Ray, P. Kohli, N. Kozai, T. Sugita, Y. Wu, K. Lee, K.-T. Lee, R. Ghaffari and J. A. Rogers, Lab Chip, 2018, 18, 2178–2186 RSC.
- F. He, X. Lv, X. Li, M. Yao, K. Li and Y. Deng, Microchem. J., 2022, 179, 107551 CrossRef CAS.
- S. Jain, A. Paliwal, V. Gupta and M. Tomar, Biosens. Bioelectron., 2022, 201, 113919 CrossRef CAS PubMed.
- Y. Liu, Q. Liu, S. Chen, F. Cheng, H. Wang and W. Peng, Sci. Rep., 2015, 5, 12864 CrossRef CAS PubMed.
- L. Lu, Z. Jiang, Y. Hu, H. Zhou, G. Liu, Y. Chen, Y. Luo and Z. Chen, Opt. Express, 2019, 27, 25420–25427 CrossRef CAS PubMed.
- Q. Zhang, Y. Li, Q. Hu, R. Xie, W. Zhou, X. Liu and Y. Wang, Lab Chip, 2022, 22, 4941–4949 RSC.
- P. Preechaburana and S. Amloy, Eur. J. Phys., 2021, 42, 045302 CrossRef.
- S. Kheireddine, A. S. Perumal, Z. J. Smith, D. V. Nicolau and S. Wachsmann-Hogiu, Lab Chip, 2019, 19, 825–836 RSC.
- N. Phadungcharoen, N. Pengwanput, A. Nakapan, U. Sutitaphan, P. Thanomklom, N. Jongudomsombut, A. Chinsriwongkul and T. Rojanarata, Talanta, 2020, 219, 121271 CrossRef CAS PubMed.
- S. Kheireddine, Z. J. Smith, D. V. Nicolau and S. Wachsmann-Hogiu, Biomed. Opt. Express, 2019, 10, 4369–4380 CrossRef PubMed.
- D. Rabha, M. A. Rather, M. Mandal and P. Nath, Opt. Lasers Eng., 2022, 151, 106931 CrossRef.
- J. Yu, X. Qin, D. Wang, C. Lin, X. Xian and N. Tao, Anal. Chem., 2019, 91, 6632–6637 CrossRef CAS PubMed.
- J. Chan, A. Raghunath, K. E. Michaelsen and S. Gollakota, Proc. ACM Interact. Mob. Wearable Ubiquitous Technol., 2022, 6, 1–27 CrossRef.
- M. Bouazizi, C. Ye and T. Ohtsuki, 2021 IEEE Global Communications Conference, 2021, pp. 1–6 Search PubMed.
- Y. Liu, G. W. Wornell, W. T. Freeman and F. Durand, Sci. Adv., 2024, 10, eadj3608 CrossRef PubMed.
- D. Hatiboruah, D. Y. Devi, N. D. Namsa and P. Nath, J. Biophotonics, 2020, 13, e201960159 CrossRef PubMed.
- X. Chen, C. Ma, Q. Kang, Y. Chen and D. Shen, New J. Chem., 2021, 45, 2529–2535 RSC.
- Q. Fu, C. Zhang, J. Xie, Z. Li, L. Qu, X. Cai, H. Ouyang, Y. Song, D. Du, Y. Lin and Y. Tang, Anal. Chim. Acta, 2019, 1092, 126–131 CrossRef CAS PubMed.
- Y. Zhao, M. Yang, Q. Fu, H. Ouyang, W. Wen, Y. Song, C. Zhu, Y. Lin and D. Du, Anal. Chem., 2018, 90, 7391–7398 CrossRef CAS PubMed.
- L. Huang, W. Xiao, T. Xu, H. Chen, Z. Jin, Z. Zhang, Q. Song and Y. Tang, Sens. Actuators, B, 2021, 327, 128893 CrossRef CAS.
- H. Xu, A. Xia, J. Luo, M. Gao, R. Liao, F. Li, Q. Zhong, W. Zhang, Y. Wang, J. Cui, W. Fu, K. Chang, M. Gan, W. Jiang and M. Chen, Sens. Actuators, B, 2020, 308, 127750 CrossRef CAS.
- E. Zhang, Q. Zeng, Y. Xu, J. Lu, C. Li, K. Xiao, X. Li, J. Li, T. Li, C. Li and L. Zhang, Lab Chip, 2024, 24, 4639–4648 RSC.
- Y. Castillo-Escario, I. Ferrer-Lluis, J. M. Montserrat and R. Jané, in 2019 41st Annual International Conference of the IEEE Engineering in Medicine and Biology Society (EMBC), 2019, pp. 4982–4985 Search PubMed.
- H. Nakano, K. Hirayama, Y. Sadamitsu, A. Toshimitsu, H. Fujita, S. Shin and T. Tanigawa, J. Clin. Sleep Med., 2014, 10, 73–78 CrossRef PubMed.
- L. Montanini, N. Sabino, S. Spinsante and E. Gambi, 2018 IEEE International Conference on Consumer Electronics, 2018, pp. 1–4 Search PubMed.
- Phyphox sensor database, https://phyphox.org/sensordb/ (accessed 15 January 2025) Search PubMed.
- A. Sherif, Market share of mobile operating systems worldwide from 2009 to 2024, by quarter, https://www.statista.com/statistics/272698/global-market-share-held-by-mobile-operating-systems-since-2009/ (accessed 4 November 2024) Search PubMed.
- B. C. Gross, J. L. Erkal, S. Y. Lockwood, C. Chen and D. M. Spence, Anal. Chem., 2014, 86, 3240–3253 CrossRef CAS PubMed.
- C. Schubert, M. C. van Langeveld and L. A. Donoso, Br. J. Ophthalmol., 2014, 98, 159–161 CrossRef PubMed.
- T. Rayna and L. Striukova, Technol. Forecast. Soc. Change, 2016, 102, 214–224 CrossRef.
- R. Su, F. Wang and M. C. McAlpine, Lab Chip, 2023, 23, 1279–1299 RSC.
- N. Bhattacharjee, A. Urrios, S. Kanga and A. Folch, Lab Chip, 2016, 16, 1720–1742 RSC.
- S. Waheed, J. M. Cabot, N. P. Macdonald, T. Lewis, R. M. Guijt, B. Paull and M. C. Breadmore, Lab Chip, 2016, 16, 1993–2013 RSC.
- G. D. Berglund and T. S. Tkaczyk, Opt. Express, 2019, 27, 30405–30420 CrossRef CAS PubMed.
- N. Vaidya and O. Solgaard, Microsyst. Nanoeng., 2018, 4, 18 CrossRef PubMed.
- J. Christopher, L. M. Rooney, M. Donnachie, D. Uttamchandani, G. McConnell and R. Bauer, Biomed. Opt. Express, 2024, 15, 2224–2237 CrossRef PubMed.
- V. R. Pereira and B. S. Hosker, PLoS Biol., 2019, 17, e3000321 CrossRef PubMed.
- Y. Jung, Y. Heo, J. J. Lee, A. Deering and E. Bae, J. Microbiol. Methods, 2020, 168, 105800 CrossRef CAS PubMed.
- X. Hu, L. Huang, S. Wang, R. Ahmed, P. Li, U. Demirci and Z. Zhang, Sens. Actuators, B, 2023, 376, 132956 CrossRef CAS.
- J. Shin, S. Kim, T. Yoon, C. Joo and H.-I. Jung, Biosens. Bioelectron., 2019, 136, 106–111 CrossRef CAS PubMed.
- S. Li, X. Zhong, Y. Xu, Y. Zheng, X. Shi, F. Li, S. Guo and J. Yang, Food Chem., 2021, 360, 130019 CrossRef CAS PubMed.
- K. Ming, J. Kim, M. J. Biondi, A. Syed, K. Chen, A. Lam, M. Ostrowski, A. Rebbapragada, J. J. Feld and W. C. W. Chan, ACS Nano, 2015, 9, 3060–3074 CrossRef CAS PubMed.
- B. Ning, T. Yu, S. Zhang, Z. Huang, D. Tian, Z. Lin, A. Niu, N. Golden, K. Hensley, B. Threeton, C. J. Lyon, X.-M. Yin, C. J. Roy, N. S. Saba, J. Rappaport, Q. Wei and T. Y. Hu, Sci. Adv., 2021, 7, eabe3703 CrossRef CAS PubMed.
- K. Trofymchuk, V. Glembockyte, L. Grabenhorst, F. Steiner, C. Vietz, C. Close, M. Pfeiffer, L. Richter, M. L. Schütte, F. Selbach, R. Yaadav, J. Zähringer, Q. Wei, A. Ozcan, B. Lalkens, G. P. Acuna and P. Tinnefeld, Nat. Commun., 2021, 12, 950 CrossRef CAS PubMed.
- M. V. Tran, K. Susumu, I. L. Medintz and W. R. Algar, Anal. Chem., 2019, 91, 11963–11971 CrossRef CAS PubMed.
- Z. Xiao, G. H. Darwish, K. Susumu, I. L. Medintz and W. R. Algar, ACS Meas. Sci. Au, 2022, 2, 57–66 CrossRef CAS PubMed.
- Q. Wei, G. Acuna, S. Kim, C. Vietz, D. Tseng, J. Chae, D. Shir, W. Luo, P. Tinnefeld and A. Ozcan, Sci. Rep., 2017, 7, 2124 CrossRef PubMed.
- H. K. Kondaveeti, N. K. Kumaravelu, S. D. Vanambathina, S. E. Mathe and S. Vappangi, Comput. Sci. Rev., 2021, 40, 100364 CrossRef.
- J. M. Pearce, HardwareX, 2020, 8, e00139 CrossRef PubMed.
- K. de Haan, H. Ceylan Koydemir, Y. Rivenson, D. Tseng, E. Van Dyne, L. Bakic, D. Karinca, K. Liang, M. Ilango, E. Gumustekin and A. Ozcan, NPJ Digit. Med., 2020, 3, 1–9 CrossRef PubMed.
- N. H. Bhuiyan, J. H. Hong, M. J. Uddin and J. S. Shim, Anal. Chem., 2022, 94, 3872–3880 CrossRef CAS PubMed.
- Z. Zhang, S. Zhao, L. Jiang, J. Wu, W. Zhao, X. Guo, N. Peng and F. Hu, Analyst, 2022, 147, 4876–4887 RSC.
- L. Zhu, S. Li, W. Liu, J. Chen, Q. Yu, Z. Zhang, Y. Li, J. Liu and X. Chen, Biosens. Bioelectron., 2021, 187, 113284 CrossRef CAS PubMed.
- M. A. Kabir, H. Zilouchian, M. Sher and W. Asghar, Diagnostics, 2020, 10, 42 CrossRef CAS PubMed.
- T. Gou, J. Hu, W. Wu, X. Ding, S. Zhou, W. Fang and Y. Mu, Biosens. Bioelectron., 2018, 120, 144–152 CrossRef CAS PubMed.
- V. D. Nguyen, H. Q. Nguyen, K. H. Bui, Y. S. Ko, B. J. Park and T. S. Seo, Biosens. Bioelectron., 2022, 195, 113632 CrossRef CAS PubMed.
- S. Duan, T. Cai, F. Liu, Y. Li, H. Yuan, W. Yuan, K. Huang, K. Hoettges, M. Chen, E. G. Lim, C. Zhao and P. Song, Anal. Chim. Acta, 2024, 1308, 342575 CrossRef CAS PubMed.
- J. J. Lee, J. Kang and C. Kim, J. Hazard. Mater., 2024, 465, 133168 CrossRef CAS PubMed.
- X. Xu, L. Cai, S. Liang, Q. Zhang, S. Lin, M. Li, Q. Yang, C. Li, Z. Han and C. Yang, Lab Chip, 2023, 23, 1169–1191 RSC.
- M. Yafia, A. Ahmadi, M. Hoorfar and H. Najjaran, Micromachines, 2015, 6, 1289–1305 CrossRef.
- S. K. Thio and S.-Y. Park, Lab Chip, 2022, 22, 3987–4006 RSC.
- Y. Li, S. Cai, H. Shen, Y. Chen, Z. Ge and W. Yang, Biomicrofluidics, 2022, 16, 031502 CrossRef CAS PubMed.
- P. Zhang, H. Bachman, A. Ozcelik and T. J. Huang, Annu. Rev. Anal. Chem., 2020, 13, 17–43 CrossRef CAS PubMed.
- E. M. Khalaf, H. Sanaan Jabbar, R. Mireya Romero-Parra, G. R. L. Al-Awsi, H. S. Budi, A. S. Altamimi, M. Abdulfadhil Gatea, K. T. Falih, K. Singh and K. A. Alkhuzai, Microchem. J., 2023, 190, 108692 CrossRef CAS.
- N. Convery and N. Gadegaard, Micro Nano Eng., 2019, 2, 76–91 CrossRef.
- M. N. H. Z. Alam, F. Hossain, A. Vale and A. Kouzani, Int. J. Precis. Eng. Manuf., 2017, 18, 1287–1296 CrossRef.
- J. A. Cataño, S. Farthing, Z. Mascarenhas, N. Lake, P. K. D. V. Yarlagadda, Z. Li and Y.-C. Toh, Micromachines, 2023, 14, 930 CrossRef PubMed.
- T. Ching, J. Vasudevan, H. Y. Tan, C. T. Lim, J. Fernandez, Y.-C. Toh and M. Hashimoto, HardwareX, 2021, 10, e00202 CrossRef PubMed.
- S. B. Park and J. H. Shin, HardwareX, 2024, 19, e00550 CrossRef PubMed.
- M. R. Behrens, H. C. Fuller, E. R. Swist, J. Wu, M. M. Islam, Z. Long, W. C. Ruder and R. Steward, Sci. Rep., 2020, 10, 1543 CrossRef CAS PubMed.
- A. Jönsson, A. Toppi and M. Dufva, HardwareX, 2020, 8, e00115 CrossRef PubMed.
- A. S. Samokhin, J. Anal. Chem., 2020, 75, 416–421 CrossRef CAS.
- S. B. Park and J. H. Shin, Sens. Actuators, B, 2024, 405, 135289 CrossRef CAS.
- S. Baas and V. Saggiomo, HardwareX, 2021, 10, e00219 CrossRef PubMed.
- E. Noviana, T. Ozer, C. S. Carrell, J. S. Link, C. McMahon, I. Jang and C. S. Henry, Chem. Rev., 2021, 121, 11835–11885 CrossRef CAS PubMed.
- A. Olanrewaju, M. Beaugrand, M. Yafia and D. Juncker, Lab Chip, 2018, 18, 2323–2347 RSC.
- S. Nishat, A. T. Jafry, A. W. Martinez and F. R. Awan, Sens. Actuators, B, 2021, 336, 129681 CrossRef CAS.
- S. Altundemir, A. K. Uguz and K. Ulgen, Biomicrofluidics, 2017, 11, 041501 CrossRef CAS PubMed.
- T. Narahari, J. Dahmer, A. Sklavounos, T. Kim, M. Satkauskas, I. Clotea, M. Ho, J. Lamanna, C. Dixon, D. G. Rackus, S. J. R. da Silva, L. Pena, K. Pardee and A. R. Wheeler, Lab Chip, 2022, 22, 1748–1763 RSC.
- X. Liu, D. Ma, H. Ye, Y. Hou, X. Bai, Y. Xing, X. Cheng, B. Lin and Y. Lu, TrAC, Trends Anal. Chem., 2023, 166, 117153 CrossRef CAS.
- E. Samiei, M. Tabrizian and M. Hoorfar, Lab Chip, 2016, 16, 2376–2396 RSC.
- S. L. S. Freire, Sens. Actuators, A, 2016, 250, 15–28 CrossRef CAS.
- K. Choi, A. H. C. Ng, R. Fobel and A. R. Wheeler, Annu. Rev. Anal. Chem., 2012, 5, 413–440 CrossRef CAS PubMed.
- R. Fobel, C. Fobel and A. R. Wheeler, Appl. Phys. Lett., 2013, 102, 193513 CrossRef.
- V. D. Nguyen, H. Q. Nguyen, K. H. Bui, Y. S. Ko, B. J. Park and T. S. Seo, Biosens. Bioelectron., 2022, 195, 113632 CrossRef CAS PubMed.
- H. Jiang, D. Jiang, X. Liu and J. Yang, Sens. Actuators, B, 2021, 349, 130785 CrossRef CAS.
- W. Xu, M. Althumayri, A. Mohammad and H. C. Koydemir, Biosens. Bioelectron., 2023, 242, 115755 CrossRef CAS PubMed.
- L. Yang, Z. Zhang and X. Wang, Micromachines, 2022, 13, 552 CrossRef PubMed.
- S. Ghosh, K. Aggarwal, V. T. U., T. Nguyen, J. Han and C. H. Ahn, Microsyst. Nanoeng., 2020, 6, 1–18 CrossRef PubMed.
- A. Agha, W. Waheed, N. Alamoodi, B. Mathew, F. Alnaimat, E. Abu-Nada, A. Abderrahmane and A. Alazzam, Macromol. Mater. Eng., 2022, 307, 2200053 CrossRef CAS.
- Y.-J. Juang and Y.-J. Chiu, Polymers, 2022, 14, 2028 CrossRef CAS PubMed.
- W. R. Algar, M. Massey, K. Rees, R. Higgins, K. D. Krause, G. H. Darwish, W. J. Peveler, Z. Xiao, H.-Y. Tsai, R. Gupta, K. Lix, M. V. Tran and H. Kim, Chem. Rev., 2021, 121, 9243–9358 CrossRef CAS PubMed.
- L. A. Marquez and H. B. Dunford, Biochemistry, 1997, 36, 9349–9355 CrossRef CAS PubMed.
- M. Taniguchi and J. S. Lindsey, Photochem. Photobiol., 2018, 94, 290–327 CrossRef CAS PubMed.
- S. K. Gupta, R. M. Kadam and P. K. Pujari, Coord. Chem. Rev., 2020, 420, 213405 CrossRef CAS.
- S. Li, Y. Zhang, Q. Wang, A. Lin and H. Wei, Anal. Chem., 2022, 94, 312–323 CrossRef CAS PubMed.
- D. Jiang, D. Ni, Z. T. Rosenkrans, P. Huang, X. Yan and W. Cai, Chem. Soc. Rev., 2019, 48, 3683–3704 RSC.
- E. Petryayeva, W. R. Algar and I. L. Medintz, Appl. Spectrosc., 2013, 67, 215–252 CrossRef CAS PubMed.
- H. Zhong, T. Mirkovic and G. D. Scholes, in Comprehensive Nanoscience and Technology, ed. D. L. Andrews, G. D. Scholes and G. P. Wiederrecht, Academic Press, Amsterdam, 2011, pp. 153–201 Search PubMed.
- E. Petryayeva and W. R. Algar, Anal. Chem., 2014, 86, 3195–3202 CrossRef CAS PubMed.
- E. Petryayeva and W. R. Algar, Anal. Chem., 2013, 85, 8817–8825 CrossRef CAS PubMed.
- E. Petryayeva and W. R. Algar, Analyst, 2015, 140, 4037–4045 RSC.
- K. M. Tsoi, Q. Dai, B. A. Alman and W. C. W. Chan, Acc. Chem. Res., 2013, 46, 662–671 CrossRef CAS PubMed.
- C. E. Bradburne, J. B. Delehanty, K. B. Gemmill, B. C. Mei, H. Mattoussi, K. Susumu, J. B. Blanco-Canosa, P. E. Dawson and I. L. Medintz, Bioconjugate Chem., 2013, 24, 1570–1583 CrossRef CAS PubMed.
- E. Oh, R. Liu, A. Nel, K. B. Gemill, M. Bilal, Y. Cohen and I. L. Medintz, Nat. Nanotechnol., 2016, 11, 479–486 CrossRef CAS PubMed.
- R. Gupta, W. J. Peveler, K. Lix and W. R. Algar, Anal. Chem., 2019, 91, 10955–10960 CrossRef CAS PubMed.
- K. Sun, Y. Yang, H. Zhou, S. Yin, W. Qin, J. Yu, D. T. Chiu, Z. Yuan, X. Zhang and C. Wu, ACS Nano, 2018, 12, 5176–5184 CrossRef CAS PubMed.
- H. Chen, J. Yu, X. Men, J. Zhang, Z. Ding, Y. Jiang, C. Wu and D. T. Chiu, Angew. Chem., Int. Ed., 2021, 60, 12007–12012 CrossRef CAS PubMed.
- F. Wang and X. Liu, Chem. Soc. Rev., 2009, 38, 976–989 RSC.
- S. Wen, J. Zhou, K. Zheng, A. Bednarkiewicz, X. Liu and D. Jin, Nat. Commun., 2018, 9, 2415 CrossRef PubMed.
- S. S. Kanani, H.-Y. Tsai and W. R. Algar, Anal. Chem., 2023, 95, 13258–13265 CrossRef CAS PubMed.
- S. Wu, Y. Li, W. Ding, L. Xu, Y. Ma and L. Zhang, Nano-Micro Lett., 2020, 12, 70 CrossRef CAS PubMed.
- Y. Yakimova, European Parliament, Long-awaited common charger for mobile devices will be a reality in 2024, https://www.europarl.europa.eu/news/en/press-room/20220930IPR41928/long-awaited-common-charger-for-mobile-devices-will-be-a-reality-in-2024, (accessed 2024/11/04) Search PubMed.
- J. Hou, M. Li and Y. Song, Nano Today, 2018, 22, 132–144 CrossRef CAS.
- L. Hu and M. J. Serpe, Polymers, 2012, 4, 134–149 CrossRef CAS.
- J. Seo, J. Kang, J. Kim, H. Han, M. Park, M. Shin and K. Lee, ACS Biomater. Sci. Eng., 2024, 10, 4035–4045 CrossRef CAS PubMed.
- G. Chen, C. Fang, H. H. Chai, Y. Zhou, W. Yun Li and L. Yu, Sens. Actuators, B, 2019, 281, 253–261 CrossRef CAS.
- M. Moslemzadeh, A. Larki and K. Ghanemi, Microchem. J., 2020, 159, 105583 CrossRef CAS.
- V. A. O. P. da Silva, R. C. de Freitas, P. R. de Oliveira, R. C. Moreira, L. H. Marcolino-Júnior, M. F. Bergamini, W. K. T. Coltro and B. C. Janegitz, Measurement, 2020, 164, 108085 CrossRef.
- S. Apichai, N. Thunyajaroen, T. Prajongsangsri, P. Tananchai, T. Pattananandecha, F. Ogata, N. Kawasaki, K. Grudpan and C. Saenjum, Sustainable Chem. Pharm., 2022, 29, 100808 CrossRef CAS.
- A. Shahvar, D. Shamsaei and M. Saraji, Measurement, 2020, 150, 107068 CrossRef.
- H. Zhang, R. Liu, Q. Li, X. Hu, L. Wu, Y. Zhou, G. Qing, R. Yuan, J. Huang, W. Gu, Y. Ye, C. Qi, M. Han, X. Chen, X. Zhu, Y. Deng, L. Zhang, H. Chen, H. Zhang, W. Gao, Y. Liu and Y. Luo, ACS Nano, 2021, 15, 7649–7658 CrossRef CAS PubMed.
- H. Choi, J. H. Seo and S. Weon, J. Hazard. Mater., 2023, 460, 132510 CrossRef CAS PubMed.
- K. Zhang, J. Zhang, F. Wang and D. Kong, ACS Sens., 2021, 6, 2261–2269 CrossRef CAS PubMed.
- Y. Sun, J. Wang, Q. Lu, J. Zhang, Y. Li, Y. Pang, C. Yang, Q. Wang and D. Kong, ACS Sens., 2024, 9, 1515–1524 CrossRef CAS PubMed.
- T. Taccioli, E. Ragusa, T. Pomili, P. Gastaldo and P. P. Pompa, IEEE Sens. J., 2023, 23, 29869–29876 CAS.
- B. Berg, B. Cortazar, D. Tseng, H. Ozkan, S. Feng, Q. Wei, R. Y.-L. Chan, J. Burbano, Q. Farooqui, M. Lewinski, D. Di Carlo, O. B. Garner and A. Ozcan, ACS Nano, 2015, 9, 7857–7866 CrossRef CAS PubMed.
- Z. Wu, C. Wang, B. Liu, C. Liang, J. Lu, J. Li, X. Tang, C. Li and T. Li, ACS Sens., 2022, 7, 1985–1995 CrossRef CAS PubMed.
- R. Deng, X. Chao, H. Li, X. Li, Z. Yang and H.-Z. Yu, Analyst, 2023, 148, 735–741 RSC.
- Y. Yano-Ozawa, N. Lobsiger, Y. Muto, T. Mori, K. Yoshimura, Y. Yano, W. J. Stark, M. Maeda, T. Asahi, A. Ogawa and T. Zako, RSC Adv., 2021, 11, 11984–11991 RSC.
- X. Huang, Y. Yan, L. Zhang, L. Yuan, Y. Tang, X. Jiang, W. Zhu, Y. Yuan, J. Nie and Y. Zhang, Anal. Bioanal. Chem., 2024, 416, 1821–1832 CrossRef CAS PubMed.
- Q. Gao, J. Wan, X. Chen, X. Mo, Y. Sun, J. Zou, J. Nie and Y. Zhang, RSC Adv., 2021, 11, 39306–39310 RSC.
- S. Chung, L. E. Breshears, A. Gonzales, C. M. Jennings, C. M. Morrison, W. Q. Betancourt, K. A. Reynolds and J.-Y. Yoon, Nat. Protoc., 2021, 16, 1452–1475 CrossRef CAS PubMed.
- T. Ghonge, H. C. Koydemir, E. Valera, J. Berger, C. Garcia, N. Nawar, J. Tiao, G. L. Damhorst, A. Ganguli, U. Hassan, A. Ozcan and R. Bashir, Analyst, 2019, 144, 3925–3935 RSC.
- M. A. Sami, M. Tayyab, P. Parikh, H. Govindaraju and U. Hassan, Analyst, 2021, 146, 2531–2541 RSC.
- L. Zhang, K. S. Chen and H.-Z. Yu, ACS Appl. Mater. Interfaces, 2020, 12, 7665–7672 CrossRef CAS PubMed.
- C. Jurischka, F. Dinter, A. Efimova, R. Weiss, J. Schiebel, C. Schulz, B. Fayziev, P. Schierack, T. Fischer and S. Rödiger, Clin. Hemorheol. Microcirc., 2020, 75, 57–84 CAS.
- A. Piruska, I. Nikcevic, S. H. Lee, C. Ahn, W. R. Heineman, P. A. Limbach and C. J. Seliskar, Lab Chip, 2005, 5, 1348–1354 RSC.
- W. E. van der Veer and D. Wolpert, Laser Focus World, Optics for microscopy: Fluorescence of colored-glass filters can be stronger than expected, https://www.laserfocusworld.com/optics/article/16554918/optics-for-microscopy-fluorescence-of-colored-glass-filters-can-be-stronger-than-expected, (accessed 2024/11/04) Search PubMed.
- Y. Zhang, R. Zhang, X. Yang, H. Qi and C. Zhang, J. Pharm. Anal., 2018, 9, 9–19 CrossRef PubMed.
- G. Giagu, A. Fracassa, A. Fiorani, E. Villani, F. Paolucci, G. Valenti and A. Zanut, Microchim. Acta, 2024, 191, 359 CrossRef CAS PubMed.
- Y. Liu, P. Li, N. Zhang, S. Chen, Z. Liu and J. Guang, IEEE Sens. J., 2019, 19, 11955–11960 CAS.
- E. S. da Silva, L. C. de Souza and L. C. Oliveira, IEEE Sens. J., 2021, 21, 16613–16620 CAS.
- C. Xiao, J. Eriksson, A. Suska, D. Filippini and W. C. Mak, Anal. Chim. Acta, 2022, 1201, 339606 CrossRef CAS PubMed.
- S. Bhaskar and S. S. Ramamurthy, ACS Appl. Nano Mater., 2019, 2, 4613–4625 CrossRef CAS.
- S. Bhaskar, N. C. S. S. Kowshik, S. P. Chandran and S. S. Ramamurthy, Langmuir, 2020, 36, 2865–2876 CrossRef CAS PubMed.
- A. Rai, S. Bhaskar, N. Reddy and S. S. Ramamurthy, ACS Sustainable Chem. Eng., 2021, 9, 14959–14974 CrossRef CAS.
- S. Bhaskar, A. Rai, K. M. Ganesh, R. Reddy, N. Reddy and S. S. Ramamurthy, Langmuir, 2022, 38, 12035–12049 CrossRef CAS PubMed.
- R. Matsuda and F. Okiharu, In Vitro Cell. Dev. Biol.: Anim., 2024, 60, 740–747 CrossRef PubMed.
- B. Jin, A. R. Jean, A. V. Maslov and V. N. Astratov, Laser Photonics Rev., 2023, 17, 2300146 CrossRef.
- A. V. Maslov, B. Jin and V. N. Astratov, Sci. Rep., 2023, 13, 6688 CrossRef CAS PubMed.
- X. Guo, J. Chang, W. Chen, Y. Hu, N. Ma and J. Zhang, Appl. Opt., 2023, 62, 5236–5243 CrossRef PubMed.
- Y. Zheng, H. Yin, C. Zhou, W. Zhou, Z. Huan and W. Ma, Biosensors, 2023, 13, 978 CrossRef PubMed.
- D. Sakharkar, A. Jain, A. Parakh and R. R. Khandelwal, IEEE Sens. Lett., 2023, 7, 1–4 Search PubMed.
- Z. Jiao, M. Pan, K. Yousaf, D. Doveiko, M. Maclean, D. Griffin, Y. Chen and D. D. U. Li, J. Microsc., 2024, 296, 10–23 CrossRef PubMed.
- K. C. Lee, K. Lee, J. Jung, S. H. Lee, D. Kim and S. A. Lee, ACS Photonics, 2021, 8, 1307–1315 CrossRef CAS.
- S. Kim, K. Sosnowski, D. S. Hwang and J.-Y. Yoon, ACS ES&T Eng., 2024, 4, 186–195 Search PubMed.
- M. A. Schaefer, H. N. Nelson, J. L. Butrum, J. R. Gronseth and J. H. Hines, Sci. Rep., 2023, 13, 2722 CrossRef CAS PubMed.
- B. Dai, Z. Jiao, L. Zheng, H. Bachman, Y. Fu, X. Wan, Y. Zhang, Y. Huang, X. Han, C. Zhao, T. J. Huang, S. Zhuang and D. Zhang, Light: Sci. Appl., 2019, 8, 75 CrossRef PubMed.
- C. Song, Y. Yang, X. Tu, Z. Chen, J. Gong and C. Lin, IEEE Sens. J., 2021, 21, 1229–1235 CAS.
- H. Zhang, W. Zhang, Z. Zuo and J. Yang, Microsc. Res. Tech., 2024, 87, 1521–1533 CrossRef PubMed.
- R. Koohkan, M. Kaykhaii, M. Sasani and B. Paull, ACS Omega, 2020, 5, 31450–31455 CrossRef CAS PubMed.
- M. Sargazi and M. Kaykhaii, Spectrochim. Acta, Part A, 2020, 227, 117672 CrossRef CAS PubMed.
- F. Matinrad, M. Kompany-Zareh, N. Omidikia and M. Dadashi, Anal. Chim. Acta, 2020, 1129, 98–107 CrossRef CAS PubMed.
- L. Kong, Y. Gan, T. Liang, L. Zhong, Y. Pan, D. Kirsanov, A. Legin, H. Wan and P. Wang, Anal. Chim. Acta, 2020, 1093, 150–159 CrossRef CAS PubMed.
- R. O. Hassan, H. O. Othman and D. S. Ali, Spectrochim. Acta, Part A, 2023, 302, 123009 CrossRef CAS PubMed.
- E. Vidal, A. S. Lorenzetti, C. D. Garcia and C. E. Domini, Anal. Chim. Acta, 2021, 1151, 338249 CrossRef CAS PubMed.
- R. Bogucki, M. Greggila, P. Mallory, J. Feng, K. Siman, B. Khakipoor, H. King and A. W. Smith, J. Chem. Educ., 2019, 96, 1527–1531 CrossRef CAS.
- A. Bayram, N. Horzum, A. U. Metin, V. Kılıç and M. E. Solmaz, IEEE Sens. J., 2018, 18, 5948–5955 CAS.
- K. D. Long, H. Yu and B. T. Cunningham, Biomed. Opt. Express, 2014, 5, 3792–3806 CrossRef CAS PubMed.
- D. Jian, B. Wang, H. Huang, X. Meng, C. Liu, L. Xue, F. Liu and S. Wang, Biosens. Bioelectron., 2019, 143, 111632 CrossRef CAS PubMed.
- E. Pituła, M. Koba and M. Śmietana, Opt. Laser Technol., 2021, 140, 107067 CrossRef.
- L.-J. Wang, Y.-C. Chang, R. Sun and L. Li, Biosens. Bioelectron., 2017, 87, 686–692 CrossRef CAS PubMed.
- M. A. Hossain, J. Canning, K. Cook and A. Jamalipour, Opt. Lett., 2016, 41, 2237–2240 CrossRef PubMed.
- L.-J. Wang, N. Naudé, Y.-C. Chang, A. Crivaro, M. Kamoun, P. Wang and L. Li, J. Biophotonics, 2018, 11, e201700382 CrossRef PubMed.
- X. Hong, T. Lu, L. Fruzyna and B. Yu, Sci. Rep., 2019, 9, 15713 CrossRef PubMed.
- C. B. Sánchez, R. P. D. Redondo, A. F. Vilas and A. M. S. Bermúdez, Comput. Appl. Eng. Educ., 2019, 27, 371–379 CrossRef.
- S. Dhawan, Int. J. Electron. Commun. Technol., 2011, 2, 22–26 Search PubMed.
- A. K. Jain, Proc. IEEE, 1981, 69, 349–389 Search PubMed.
- K. Marlapalli, R. S. B. P. Bandlamudi, R. Busi, V. Pranav and B. Madhavrao, in Communication Software and Networks, ed. S. C. Satapathy, V. Bhateja, M. Ramakrishna Murty, N. Gia Nhu and J. Kotti, Springer, Singapore, 2021, pp. 271–279 Search PubMed.
- X. Bao, S. Jiang, Y. Wang, M. Yu and J. Han, Analyst, 2018, 143, 1387–1395 RSC.
- A. Y. Mutlu, V. Kılıç, G. K. Özdemir, A. Bayram, N. Horzum and M. E. Solmaz, Analyst, 2017, 142, 2434–2441 RSC.
- P. Siribunbandal, Y.-H. Kim, T. Osotchan, Z. Zhu and R. Jaisutti, ACS Omega, 2022, 7, 18714–18721 CrossRef CAS PubMed.
- G. H. Darwish, D. V. Baker and W. R. Algar, ACS Sens., 2023, 8, 4686–4695 CrossRef CAS PubMed.
- S. Duan, T. Cai, J. Zhu, X. Yang, E. G. Lim, K. Huang, K. Hoettges, Q. Zhang, H. Fu, Q. Guo, X. Liu, Z. Yang and P. Song, Anal. Chim. Acta, 2023, 1248, 340868 CrossRef CAS PubMed.
- S. Mondal, S. Park, T. H. Vo, J. Choi, V. H. M. Doan, D. T. Phan, C.-S. Kim, B. Lee and J. Oh, Mater. Chem. Phys., 2022, 287, 126289 CrossRef CAS.
- E. Ghohestani, J. Tashkhourian, H. Sharifi, N. M. Bojanowski, K. Seehafer, E. Smarsly, U. H. F. Bunz and B. Hemmateenejad, Analyst, 2022, 147, 4266–4274 RSC.
- M. Adampourezare, B. Nikzad, S. Sajedi-Amin and E. Rahimpour, BMC Chem., 2024, 18, 80 CrossRef CAS PubMed.
- F. Wang, Y. Lu, J. Yang, Y. Chen, W. Jing, L. He and Y. Liu, Analyst, 2017, 142, 3177–3182 RSC.
- L. Guo, T. Wang, Z. Wu, J. Wang, M. Wang, Z. Cui, S. Ji, J. Cai, C. Xu and X. Chen, Adv. Mater., 2020, 32, 2004805 CrossRef CAS PubMed.
- Z. Wang, Y. Dong, X. Sui, X. Shao, K. Li, H. Zhang, Z. Xu and D. Zhang, npj Flexible Electron., 2024, 8, 1–11 CrossRef.
- J. Cruz, R. Sáez-Hernández, S. Armenta, A. E. Morales-Rubio and M. L. Cervera, Talanta, 2024, 276, 126217 CrossRef CAS PubMed.
- J.-Q. Du, W.-C. Luo, J.-T. Zhang, Q.-Y. Li, L.-N. Bao, M. Jiang, X. Yu and L. Xu, Sens. Actuators, B, 2024, 417, 136173 CrossRef CAS.
- S. K. R, K. R. Ashok Kumar, K. Vijayakrishna, A. Sivaramakrishna, C. V. S. Brahmmananda Rao, N. Sivaraman and S. K. Sahoo, Inorg. Chem., 2018, 57, 15270–15279 CrossRef CAS PubMed.
- D. S. Lim, S. Y. Park, K. S. Hwang and S.-K. Chang, Tetrahedron Lett., 2018, 59, 1819–1822 CrossRef CAS.
- T. Anand and S. K. Sahoo, Phys. Chem. Chem. Phys., 2019, 21, 11839–11845 RSC.
- L. K. Shaji, J. Jose, R. Bhaskar, R. S. Kumar, V. Vetriarasu, S. G. Bhat and S. K. A. Kumar, Inorg. Chem. Commun., 2023, 147, 110252 CrossRef CAS.
- D. Aydin, S. N. K. Elmas and F. N. Arslan, Food Chem., 2023, 402, 134439 CrossRef CAS PubMed.
- R. Kaushik, R. Sakla, N. Kumar, A. Ghosh, V. D. Ghule and D. A. Jose, Sens. Actuators, B, 2021, 328, 129026 CrossRef CAS.
- X. Liu, Z. Chen, R. Gao, C. Kan and J. Xu, Sens. Actuators, B, 2021, 340, 129958 CrossRef CAS.
- I. Lewińska, M. Ścibisz and Ł. Tymecki, Anal. Chim. Acta, 2024, 1308, 342639 CrossRef PubMed.
- L. Bokelmann, O. Nickel, T. Maricic, S. Pääbo, M. Meyer, S. Borte and S. Riesenberg, Nat. Commun., 2021, 12, 1467 CrossRef CAS PubMed.
- Q. Tang, J. Hu, S. Li, S. Lin, Y. Tu and X. Gui, Int. J. Food Sci. Technol., 2022, 57, 6867–6880 CrossRef CAS.
- K. Lee, S. Baek, D. Kim and J. Seo, Food Packag. Shelf Life, 2019, 19, 40–46 CrossRef.
- L. R. Magnaghi, G. Alberti, B. M. Pazzi, C. Zanoni and R. Biesuz, New J. Chem., 2022, 46, 19460–19467 RSC.
- Y. Cao, Y. Liu, F. Li, S. Guo, Y. Shui, H. Xue and L. Wang, Microchem. J., 2019, 150, 104176 CrossRef CAS.
- K. Chayavanich, P. Thiraphibundet and A. Imyim, Spectrochim. Acta, Part A, 2020, 226, 117601 CrossRef CAS PubMed.
- L. Placer, I. Lavilla, F. Pena-Pereira and C. Bendicho, Sens. Actuators, B, 2023, 377, 133109 CrossRef CAS.
- N. Peamaroon, J. Jakmunee and N. Moonrungsee, J. Anal. Test., 2021, 5, 379–386 CrossRef.
- M. D. Yilmaz, Microchem. J., 2024, 196, 109554 CrossRef.
- H. Zhang, X. Jiang, Q. Yu, X. Cui, Y. Liu, P.-L. Tremblay and T. Zhang, Spectrochim. Acta, Part A, 2024, 323, 124930 CrossRef CAS PubMed.
- S. Erdemir and S. Malkondu, Talanta, 2020, 207, 120278 CrossRef CAS PubMed.
- A. Kathiravan, S. Sengottiyan, T. Puzyn, P. Gopinath, K. Ramasubramanian, P. Ayyappan Susila and M. Asha Jhonsi, Anal. Methods, 2022, 14, 518–525 RSC.
- K. Kalinowska, W. Wojnowski and M. Tobiszewski, Anal. Methods, 2023, 15, 1395–1401 RSC.
- J. Câmara Cardozo, I. D. Barbosa Segundo, E. R. V. P. Galvão, D. R. da Silva, E. V. dos Santos and C. A. Martínez-Huitle, Sci. Rep., 2023, 13, 11082 CrossRef PubMed.
- A. P. Vargas, F. Gámez, J. Roales, T. Lopes-Costa and J. M. Pedrosa, Chemosensors, 2021, 9, 40 CrossRef CAS.
- A. Maiti, T. Sultana, B. Rajbanshi, B. Bhaumik, N. Roy and M. Nath Roy, Microchem. J., 2024, 199, 109977 CrossRef CAS.
- A. Shen, X. Hao, L. Zhang, M. Du, M. Li, J. Yuan, X. Du, S. Ma, Y. Zhao, L. Hou, Z. Li and Y. Yang, Dyes Pigm., 2022, 207, 110674 CrossRef CAS.
- Y. Zhang, Y. Cai, F. Dong, L. Bian, H. Li, J. Wang, J. Du, X. Qi and Y. He, Anal. Bioanal. Chem., 2019, 411, 8063–8071 CrossRef CAS PubMed.
- R. Silva Lamarca, N. da Costa Luchiari, A. Francielli Bonjorno, J. Passaretti Filho, A. Alves Cardoso and P. Clairmont Feitosa de Lima Gomes, Anal. Methods, 2019, 11, 3697–3705 RSC.
- S. Jantra, L. Waiysuksri, P. Rattanamunee, P. Rashatasakhon, M. Sukwattanasinitt and S. Wacharasindhu, Dyes Pigm., 2023, 214, 111210 CrossRef CAS.
- C. Vakh, Z. Mallabaeva and M. Tobiszewski, Spectrochim. Acta, Part A, 2024, 315, 124238 CrossRef CAS PubMed.
- H.-A. H. Abd-ElSalam, G. N. N. Saleh, K. G. Waked, O. A. Refaeey, K. S. Poules, H. H. Georgey and E. S. Elzanfaly, Chem. Pap., 2024, 78, 6607–6615 CrossRef CAS.
- I. Lewińska, M. Speichert, M. Granica and Ł. Tymecki, Sens. Actuators, B, 2021, 340, 129915 CrossRef.
- X.-X. Zhang, Y.-Z. Song, F. Fang and Z.-Y. Wu, Anal. Bioanal. Chem., 2018, 410, 2665–2669 CrossRef CAS PubMed.
- D. Merli, A. Profumo, S. Tinivella and S. Protti, Forensic Chem., 2019, 14, 100167 CrossRef CAS.
- A. Procida and K. C. Honeychurch, J. Forensic Sci., 2022, 67, 1697–1703 CrossRef CAS PubMed.
- Ö. Berfin Mercan, V. Kılıç and M. Şen, Sens. Actuators, B, 2021, 329, 129037 CrossRef.
- T.-T. Wang, C. Kit Lio, H. Huang, R.-Y. Wang, H. Zhou, P. Luo and L.-S. Qing, Talanta, 2020, 206, 120211 CrossRef CAS PubMed.
- E. Yüzer, V. Doğan, V. Kılıç and M. Şen, Sens. Actuators, B, 2022, 371, 132489 CrossRef.
- M. Domínguez, D. Moraru, S. Lasso, I. Sanz-Vicente, S. de Marcos and J. Galbán, Anal. Bioanal. Chem., 2024, 416, 7317–7323 CrossRef PubMed.
- Y. Jin Chi, B. Ryu, S. Ahn and W.-G. Koh, Sens. Actuators, B, 2023, 396, 134601 CrossRef.
- H. Wu, J. Chen, Y. Yang, W. Yu, Y. Chen, P. Lin and K. Liang, Anal. Bioanal. Chem., 2022, 414, 1759–1772 CrossRef CAS PubMed.
- D. Wang, D. Liu, H. Duan, Y. Xu, Z. Zhou and P. Wang, J. Agric. Food Chem., 2020, 68, 9252–9259 CrossRef CAS PubMed.
- L. Li, Z. Liu, H. Zhang, W. Yue, C.-W. Li and C. Yi, Sens. Actuators, B, 2018, 254, 337–346 CrossRef CAS.
- N. Rafat, L. Brewer, N. Das, D. J. Trivedi, B. K. Kaszala and A. Sarkar, ACS Sens., 2023, 8, 534–542 CrossRef CAS PubMed.
- X. Wang, J. Li, D. Jian, Y. Zhang, Y. Shan, S. Wang and F. Liu, Sens. Actuators, B, 2021, 329, 129173 CrossRef CAS.
- C. Chen, P.-W. Wang, Y.-C. Yen, H.-L. Lin, Y.-C. Fan, S.-M. Wu and C.-F. Chen, Sens. Actuators, B, 2019, 282, 251–258 CrossRef CAS.
- L. Zhong, J. Sun, Y. Gan, S. Zhou, Z. Wan, Q. Zou, K. Su and P. Wang, Anal. Sci., 2019, 35, 133–140 CrossRef CAS PubMed.
- X. Li, Y. Cheng, R. Xu, Z. Zhang, X. Qi, L. Chen and M. Zhu, Talanta, 2022, 247, 123567 CrossRef CAS PubMed.
- Y.-P. Hsu, H.-W. Yang, N.-S. Li, Y.-T. Chen, H.-H. Pang and S.-T. Pang, ACS Sens., 2020, 5, 928–935 CrossRef CAS PubMed.
- G. Papadakis, A. K. Pantazis, N. Fikas, S. Chatziioannidou, V. Tsiakalou, K. Michaelidou, V. Pogka, M. Megariti, M. Vardaki, K. Giarentis, J. Heaney, E. Nastouli, T. Karamitros, A. Mentis, A. Zafiropoulos, G. Sourvinos, S. Agelaki and E. Gizeli, Sci. Rep., 2022, 12, 3775 CrossRef CAS PubMed.
- J. Song, B. Cha, J. Moon, H. Jang, S. Kim, J. Jang, D. Yong, H.-J. Kwon, I.-C. Lee, E.-K. Lim, J. Jung, H. G. Park and T. Kang, ACS Nano, 2022, 16, 11300–11314 CrossRef CAS PubMed.
- F. Li, Y. Hu, A. Zhao, Y. Xi, Z. Li and J. He, Microchim. Acta, 2020, 187, 425 CrossRef CAS PubMed.
- L. Xue, N. Jin, R. Guo, S. Wang, W. Qi, Y. Liu, Y. Li and J. Lin, ACS Sens., 2021, 6, 2883–2892 CrossRef CAS PubMed.
- B. Zhang, R. Zhou, H. Zhang, D. Cai, X. Lin, Y. Lang, Y. Qiu, X. Shentu, Z. Ye and X. Yu, Foods, 2022, 11, 2900 CrossRef CAS PubMed.
- J. Liu, Y. Xing, B. Xue and X. Zhou, Biosens. Bioelectron., 2022, 205, 114099 CrossRef CAS PubMed.
- X. Zhang, D. Wu, Y. Wu and G. Li, Biosens. Bioelectron., 2021, 172, 112776 CrossRef CAS PubMed.
- T. D. Tran, P. T. Nguyen, T. N. Le and M. I. Kim, Biosens. Bioelectron., 2021, 182, 113187 CrossRef CAS PubMed.
- Z. Zhu, X. Wang, N. Wang, C. Zeng, L. Zhang, J. Fan, X. Yang, P. Li, H. Yuan, Y. Feng, S. Huo and X. Lu, Anal. Bioanal. Chem., 2024, 416, 4417–4426 CrossRef CAS PubMed.
- E. Ko, W. Hur, S. E. Son, G. H. Seong and D. K. Han, Microchim. Acta, 2021, 188, 382 CrossRef CAS PubMed.
- Y.-T. Zhuang, S. Chen, R. Jiang, Y.-L. Yu and J.-H. Wang, Anal. Chem., 2019, 91, 5346–5353 CrossRef CAS PubMed.
- D.-H. Park, J.-M. Heo, W. Jeong, Y. H. Yoo, B. J. Park and J.-M. Kim, ACS Appl. Mater. Interfaces, 2018, 10, 5014–5021 CrossRef CAS PubMed.
- T.-T. T. Nguyen, B. T. Huy and Y.-I. Lee, ACS Omega, 2019, 4, 12665–12670 CrossRef CAS PubMed.
- L. Engel, I. Benito-Altamirano, K. R. Tarantik, C. Pannek, M. Dold, J. D. Prades and J. Wöllenstein, Sens. Actuators, B, 2021, 330, 129281 CrossRef CAS.
- P. Escobedo, C. E. Ramos-Lorente, A. Ejaz, M. M. Erenas, A. Martínez-Olmos, M. A. Carvajal, C. García-Núñez, I. de Orbe-Payá, L. F. Capitán-Vallvey and A. J. Palma, Sens. Actuators, B, 2023, 376, 133001 CrossRef CAS.
- J. R. P. Albuquerque, C. N. Makara, V. G. Ferreira, L. C. Brazaca and E. Carrilho, RSC Adv., 2024, 14, 23392–23403 RSC.
- F. Li, Y. Hu, Z. Li, J. Liu, L. Guo and J. He, Anal. Bioanal. Chem., 2019, 411, 6497–6508 CrossRef CAS PubMed.
- S. Chunta, P. Jarujamrus, A. Prakobkij, S. Khongwichit, N. Ditcharoen, S. Pencharee and M. Amatatongchai, Microchim. Acta, 2024, 191, 402 CrossRef CAS PubMed.
- A. K. Yetisen, R. Moreddu, S. Seifi, N. Jiang, K. Vega, X. Dong, J. Dong, H. Butt, M. Jakobi, M. Elsner and A. W. Koch, Angew. Chem., Int. Ed., 2019, 58, 10506–10513 CrossRef CAS PubMed.
- A. T. Güntner and F. M. Schenk, Nanoscale, 2023, 15, 3967–3977 RSC.
- T. Zhao, X. Liang, X. Guo, X. Yang, J. Guo, X. Zhou, X. Huang, W. Zhang, Y. Wang, Z. Liu, Z. Jiang, H. Zhou and H. Zhou, Food Chem., 2023, 404, 134768 CrossRef CAS PubMed.
- E. M. Materón, F. R. Gómez, M. B. Almeida, F. M. Shimizu, A. Wong, K. B. R. Teodoro, F. S. R. Silva, M. J. A. Lima, M. K. S. C. Angelim, M. E. Melendez, N. Porras, P. M. Vieira, D. S. Correa, E. Carrilho, O. N. Oliveira, R. B. Azevedo and D. Goncalves, ACS Appl. Mater. Interfaces, 2022, 14, 54527–54538 CrossRef PubMed.
- S. Ngernpimai, S. Srijampa, P. Thongmee, S. Teerasong, T. Puangmali, W. Maleewong, A. Chompoosor and P. Tippayawat, Bioconjugate Chem., 2022, 33, 2103–2112 CrossRef CAS PubMed.
- J. Zhang, Y. Wang, X. Zhao, M. Chen, Y. Peng, J. Bai, S. Li, D. Han, S. Ren, K. Qin, S. Li, T. Han and Z. Gao, Anal. Chem., 2021, 93, 16922–16931 CrossRef CAS PubMed.
- F. Liu, R. Chen, W. Song, L. Li, C. Lei and Z. Nie, Anal. Chem., 2021, 93, 3517–3525 CrossRef CAS PubMed.
- L. Ma, L. Yin, X. Li, S. Chen, L. Peng, G. Liu, S. Ye, W. Zhang and S. Man, Biosens. Bioelectron., 2022, 195, 113646 CrossRef CAS PubMed.
- S. Tang, T. Qi, D. Xia, M. Xu, M. Xu, A. Zhu, W. Shen and H. K. Lee, Anal. Chem., 2019, 91, 5888–5895 CrossRef CAS PubMed.
- T. Qi, M. Xu, Y. Yao, W. Chen, M. Xu, S. Tang, W. Shen, D. Kong, X. Cai, H. Shi and H. K. Lee, Talanta, 2020, 220, 121388 CrossRef CAS PubMed.
- N. Villarino, F. Pena-Pereira, I. Lavilla and C. Bendicho, ACS Sens., 2022, 7, 839–848 CrossRef CAS PubMed.
- Z. Li, R. Paul, T. B. Tis, A. C. Saville, J. C. Hansel, T. Yu, J. B. Ristaino and Q. Wei, Nat. Plants, 2019, 5, 856–866 CrossRef CAS PubMed.
- A. W. Kahandal, L. Sharma, V. Sirdeshmukh, A. Kulkarni and C. K. Tagad, Int. J. Environ. Sci. Technol., 2023, 20, 9077–9088 CrossRef CAS.
- B.-W. Liu, P.-C. Huang and F.-Y. Wu, Anal. Bioanal. Chem., 2020, 412, 8051–8059 CrossRef PubMed.
- C.-Y. Wen, Y. Chen, R.-S. Liu, J. Huang, D. Wang, Z. Cao, B. Meteku and J. Zeng, J. Mater. Chem. C, 2021, 9, 4661–4669 RSC.
- S. Bhaskar, P. Jha, C. Subramaniam and S. S. Ramamurthy, Phys. E, 2021, 132, 114764 CrossRef CAS.
- H.-J. Fu, L. Luo, Y. Wang, C.-L. Wang, H. Wang, Y.-D. Shen, H.-T. Lei, N. Hildebrandt and Z.-L. Xu, ACS Appl. Nano Mater., 2022, 5, 12915–12925 CrossRef CAS.
- T. Zhang, H.-B. Wang, Z.-T. Zhong, C.-Q. Li, W. Chen, B. Liu and Y.-D. Zhao, Microchem. J., 2020, 157, 105038 CrossRef CAS.
- Y. Wu, Y. Hu, N. Jiang, R. Anantharanjit, A. K. Yetisen and M. F. Cordeiro, Lab Chip, 2022, 22, 3521–3532 RSC.
- A. Roberts, D. Prakashan, H. Dhanze, R. K. Gandham, S. Gandhi and G. T. Sharma, Nanoscale Adv., 2022, 4, 3966–3977 RSC.
- A. Roda, S. Cavalera, F. D. Nardo, D. Calabria, S. Rosati, P. Simoni, B. Colitti, C. Baggiani, M. Roda and L. Anfossi, Biosens. Bioelectron., 2021, 172, 112765 CrossRef CAS PubMed.
- D. P. Hainsworth, A. Gangula, S. Ghoshdastidar, R. Kannan and A. Upendran, Am. J. Ophthalmol., 2020, 213, 306–319 CrossRef CAS PubMed.
- N. Yang, L.-L. Xie, C. Pan, M.-F. Yuan, Z.-H. Tao and H.-P. Mao, J. Food Process Eng., 2019, 42, e12976 CrossRef CAS.
- C. Rodriguez-Quijada, C. Lyons, M. Sanchez-Purra, C. Santamaria, B. M. Leonardo, S. Quinn, M. F. Tlusty, M. Shiaris and K. Hamad-Schifferli, ACS Omega, 2023, 8, 19494–19502 CrossRef CAS PubMed.
- J. P. Devadhasan, J. Gu, P. Chen, S. Smith, B. Thomas, M. Gates-Hollingsworth, D. Hau, S. Pandit, D. AuCoin and F. Zenhausern, Anal. Chem., 2021, 93, 9337–9344 CrossRef CAS PubMed.
- Y. Uwamino, S. Tanaka, A. Shibata, T. Kurafuji, H. Ishihara, Y. Sato and H. Matsushita, Diagn. Microbiol. Infect. Dis., 2024, 108, 116166 CrossRef CAS PubMed.
- G. Yang, J. Li, S. Zhang, H. Ouyang, C. Jiang and H. Pan, J. Immunol. Methods, 2023, 523, 113574 CrossRef CAS PubMed.
- S. B. Park and J. H. Shin, BioChip J., 2022, 16, 480–489 CrossRef CAS PubMed.
- T. Kosawatphat, A. Yakoh, S. Rengpipat, N. Khongchareonporn, O. Chailapakul, S. Chaiyo and N. Praphairaksit, Anal. Lett., 2022, 55, 2517–2530 CrossRef CAS.
- C. Kim, Y. K. Yoo, S. I. Han, J. Lee, D. Lee, K. Lee, K. S. Hwang, K. H. Lee, S. Chung and J. H. Lee, Lab Chip, 2017, 17, 2451–2458 RSC.
- R. Tian, J. Ji, Y. Zhou, Y. Du, X. Bian, F. Zhu, G. Liu, S. Deng, Y. Wan and J. Yan, Biosens. Bioelectron., 2020, 160, 112218 CrossRef CAS PubMed.
- H. Tong, C. Cao, M. You, S. Han, Z. Liu, Y. Xiao, W. He, C. Liu, P. Peng, Z. Xue, Y. Gong, C. Yao and F. Xu, Biosens. Bioelectron., 2022, 213, 114449 CrossRef CAS PubMed.
- R. Thakur, F. Akram, V. Rastogi, A. Mitra, R. Nawani, V. Av, S. K. Dubey and C. Shakher, IEEE Sens. J., 2020, 20, 14491–14500 CAS.
- G.-R. Han, H. J. Koo, H. Ki and M.-G. Kim, ACS Appl. Mater. Interfaces, 2020, 12, 34564–34575 CrossRef CAS PubMed.
- P. Phangwipas, B. Thangavel and J. H. Shin, Chemosensors, 2023, 11, 36 CrossRef CAS.
- G. Zhao, S. Liu, G. Li, W. Fang, Y. Liao, R. Li, L. Fu and J. Wang, Talanta, 2023, 253, 123925 CrossRef CAS PubMed.
- S. Lee, Y. K. Yoo, S. I. Han, D. Lee, S.-Y. Cho, C. Park, D. Lee, D. S. Yoon and J. H. Lee, Analyst, 2023, 148, 6001–6010 RSC.
- M. Colombo, L. Bezinge, A. R. Tapia, C.-J. Shih, A. J. de Mello and D. A. Richards, Sens. Diagn., 2023, 2, 100–110 RSC.
- K. H. Foysal, S. E. Seo, M. J. Kim, O. S. Kwon and J. W. Chong, Sensors, 2019, 19, 4812 CrossRef CAS PubMed.
- S. Zhang, X. Jiang, S. Lu, G. Yang, S. Wu, L. Chen and H. Pan, Sensors, 2023, 23, 6401 CrossRef CAS PubMed.
- S. Lee, S. Kim, D. S. Yoon, J. S. Park, H. Woo, D. Lee, S.-Y. Cho, C. Park, Y. K. Yoo, K.-B. Lee and J. H. Lee, Nat. Commun., 2023, 14, 2361 CrossRef CAS PubMed.
- S. Kumar, T. Ko, Y. Chae, Y. Jang, I. Lee, A. Lee, S. Shin, M.-H. Nam, B. S. Kim, H. S. Jun and S. Seo, Biosensors, 2023, 13, 623 CrossRef PubMed.
- S. Arumugam, J. Ma, U. Macar, G. Han, K. McAulay, D. Ingram, A. Ying, H. H. Chellani, T. Chern, K. Reilly, D. A. M. Colburn, R. Stanciu, C. Duffy, A. Williams, T. Grys, S.-F. Chang and S. K. Sia, Commun. Med., 2023, 3, 1–14 CrossRef PubMed.
- N. Cheng, Y. Song, M. M. A. Zeinhom, Y.-C. Chang, L. Sheng, H. Li, D. Du, L. Li, M.-J. Zhu, Y. Luo, W. Xu and Y. Lin, ACS Appl. Mater. Interfaces, 2017, 9, 40671–40680 CrossRef CAS PubMed.
- F. D. Nardo, E. Alladio, C. Baggiani, S. Cavalera, C. Giovannoli, G. Spano and L. Anfossi, Talanta, 2019, 192, 288–294 CrossRef PubMed.
- Z. Lu, Y. Huang, X. Xiao, Y. Luo, J. Liu, J. Yin, C. Liu, S. Shan, D. Liu and W. Lai, Sens. Actuators, B, 2023, 383, 133580 CrossRef CAS.
- J. Chen, P. Luo, Z. Liu, Z. He, Y. Pang, H. Lei, Z. Xu, H. Wang and X. Li, Anal. Chim. Acta, 2022, 1221, 340138 CrossRef CAS PubMed.
- D. Quesada-González, A. Sena-Torralba, W. P. Wicaksono, A. de la Escosura-Muñiz, T. A. Ivandini and A. Merkoçi, Biosens. Bioelectron., 2019, 132, 132–135 CrossRef PubMed.
- F. Ghorbanizamani, H. Moulahoum and S. Timur, IEEE Sens. J., 2022, 22, 1146–1153 CAS.
- S. Teepoo, J. Jantra, K. Panapong and D. T. Ajayi, Anal. Chim. Acta, 2024, 1285, 342031 CrossRef CAS PubMed.
- G. M. S. Ross, G. I. Salentijn and M. W. F. Nielen, Biosensors, 2019, 9, 143 CrossRef CAS PubMed.
- R. Shu, Y. Liang, S. Liu, L. Dou, T. Bu, S. Wang, X. Lan, D. Zhang, J. Sun, M. Zhu and J. Wang, Biosens. Bioelectron., 2023, 219, 114807 CrossRef CAS PubMed.
- M. Mao, X. Chen, Y. Cai, H. Yang, C. Zhang, Y. Zhang, Z. Wang and C. Peng, Sens. Actuators, B, 2023, 378, 133148 CrossRef CAS.
- X. Zhu, J. Tang, X. Ouyang, Y. Liao, H. Feng, J. Yu, L. Chen, Y. Lu, Y. Yi and L. Tang, J. Hazard. Mater., 2024, 465, 133178 CrossRef CAS PubMed.
- Q. He, H. Yang, J. Pan, X. Cui, D. Shen, S. A. Eremin, Y. Fang and S. Zhao, ACS Appl. Bio Mater., 2020, 3, 8849–8856 CrossRef CAS PubMed.
- D. Y. Kong, N. S. Heo, J. W. Kang, J. B. Lee, H. J. Kim and M. I. Kim, Anal. Bioanal. Chem., 2022, 414, 3257–3265 CrossRef CAS PubMed.
- F. Feng, Q. Fu, F. Cao, Y. Yuan, R. Kong, D. Ji and H. Liu, ChemBioChem, 2024, 25, e202300575 CrossRef CAS PubMed.
- S. Agarwal, C. Warmt, J. Henkel, L. Schrick, A. Nitsche and F. F. Bier, Anal. Bioanal. Chem., 2022, 414, 3177–3186 CrossRef CAS PubMed.
- T. Sukonta, S. Senapin, S. Taengphu, P. Hannanta-anan, M. Kitthamarat, P. Aiamsa-at and T. Chaijarasphong, Aquaculture, 2022, 560, 738538 CrossRef CAS.
- P.-R. Li, Z.-X. Wang, Z.-K. Xu, J. Wang, B. Li, X. Shen and Z.-L. Xu, J. Agric. Food Chem., 2024, 72, 14000–14010 CrossRef CAS PubMed.
- M. Liu, X. Li, J. Xu, S. Zhou, L. Deng, D. Men, Y. Duan, D. Huo and C. Hou, Microchem. J., 2024, 205, 111313 CrossRef CAS.
- L. Yin, N. Duan, S. Chen, Y. Yao, J. Liu and L. Ma, Sens. Actuators, B, 2021, 347, 130586 CrossRef CAS.
- S. H. Needs, H. M. I. Osborn and A. D. Edwards, J. Microbiol. Methods, 2021, 187, 106199 CrossRef CAS PubMed.
- D. H. Jung, Y. Kim, H. H. Cho, B. Lee, S.-J. Suh, J. H. Heo and J. H. Lee, Chem. Eng. J., 2022, 450, 138281 CrossRef CAS.
- S. Fu, P. Zuo and B.-C. Ye, Biotechnol. J., 2021, 16, 2000126 CrossRef CAS PubMed.
- C. Chang, H. Yang, W. Bi, C. Huang and Z. Xu, ACS Sens., 2023, 8, 543–554 CrossRef CAS PubMed.
- C. Imashiro, Y. Tokuoka, K. Kikuhara, T. G. Yamada, K. Takemura and A. Funahashi, IEEE Access, 2020, 8, 170033–170043 Search PubMed.
- D. Rabha, A. Sarmah and P. Nath, J. Microsc., 2019, 276, 13–20 CrossRef CAS PubMed.
- R. Shrestha, R. Duwal, S. Wagle, S. Pokhrel, B. Giri and B. B. Neupane, PLoS Neglected Trop. Dis., 2020, 14, e0008560 CrossRef PubMed.
- M. K. Kanakasabapathy, H. J. Pandya, M. S. Draz, M. K. Chug, M. Sadasivam, S. Kumar, B. Etemad, V. Yogesh, M. Safavieh, W. Asghar, J. Z. Li, A. M. Tsibris, D. R. Kuritzkes and H. Shafiee, Lab Chip, 2017, 17, 2910–2919 RSC.
- S. Ilyas, M. Sher, E. Du and W. Asghar, Biosens. Bioelectron., 2020, 165, 112417 CrossRef CAS PubMed.
- M. K. Aslan, Y. Ding, S. Stavrakis and A. J. deMello, Anal. Chem., 2023, 95, 14526–14532 CrossRef CAS PubMed.
- M. Cesaretti, J. Gal, C. Bouveyron, A. Diaspro, E. Fontas, A. Antonini, R. Anty, A. Iannelli and S. Patouraux, Microsc. Res. Tech., 2020, 83, 1025–1031 CrossRef CAS PubMed.
- W. Sirithanaphol, N. Incharoen, U. Rompsaithong, P. Kiatsopit, S. Lumbiganon and J. Chindaprasirt, Heliyon, 2021, 7, e07189 CrossRef PubMed.
- A. F. Sampaio, L. Rosado and M. J. M. Vasconcelos, IEEE Access, 2021, 9, 152188–152205 Search PubMed.
- V. Mosiichuk, A. Sampaio, P. Viana, T. Oliveira and L. Rosado, Appl. Sci., 2023, 13, 9850 CrossRef CAS.
- F. Veronese, F. Branciforti, E. Zavattaro, V. Tarantino, V. Romano, K. M. Meiburger, M. Salvi, S. Seoni and P. Savoia, Diagnostics, 2021, 11, 451 CrossRef PubMed.
- H. Li, Y. Liang, Y. Liu, X. Xian, Y. Xue, H. Huang, Q. Yao and W. Liu, Comput. Electron. Agric., 2023, 214, 108276 CrossRef.
- P. Treepong and N. Theera-Ampornpunt, Curr. Res. Food Sci., 2023, 7, 100574 CrossRef PubMed.
- M. Rolfe, S. Hayes, M. Smith, M. Owen, M. Spruth, C. McCarthy, A. Forkan, A. Banerjee and R. K. Hocking, J. Hazard. Mater., 2024, 463, 132853 CrossRef CAS PubMed.
- Y. Zhang, X. Song, J. Xie, J. Hu, J. Chen, X. Li, H. Zhang, Q. Zhou, L. Yuan, C. Kong, Y. Shen, J. Wu, L. Fang and Q. Dai, Nat. Commun., 2023, 14, 4118 CrossRef CAS PubMed.
- S. Assaad, D. Dov, R. Davis, S. Kovalsky, W. T. Lee, R. Kahmke, D. Rocke, J. Cohen, R. Henao, L. Carin and D. E. Range, Mod. Pathol., 2023, 36, 100129 CrossRef PubMed.
- M. Y. Lu, D. F. K. Williamson, T. Y. Chen, R. J. Chen, M. Barbieri and F. Mahmood, Nat. Biomed. Eng., 2021, 5, 555–570 CrossRef PubMed.
- M. K. Kanakasabapathy, P. Thirumalaraju, C. L. Bormann, H. Kandula, I. Dimitriadis, I. Souter, V. Yogesh, S. K. S. Pavan, D. Yarravarapu, R. Gupta, R. Pooniwala and H. Shafiee, Lab Chip, 2019, 19, 4139–4145 RSC.
- J. S. Prusty and A. Kumar, Rend. Fis. Acc. Lincei., 2021, 32, 163–180 CrossRef.
- A. Jamir, S. Longkumer, V. K. Roy, R. K. Kharwar and P. P. Pankaj, Multimed. Tools Appl., 2024, 83, 16197–16203 CrossRef.
- M. Ren, Y. Dong, J. Wang, J. Lin, L. Qu, Y. Zhou and Y. Chen, Anal. Chim. Acta, 2023, 1278, 341687 CrossRef CAS PubMed.
- G. M. e Silva, J. A. Garcia, J. de A. Garitta, D. G. F. Cunha, N. R. Finkler, E. M. Mendiondo and F. Ghiglieno, J. Environ. Manage., 2022, 323, 116214 CrossRef PubMed.
- W. Xiao, Z. Deng, J. Huang, Z. Huang, M. Zhuang, Y. Yuan, J. Nie and Y. Zhang, Anal. Chem., 2019, 91, 15114–15122 CrossRef CAS PubMed.
- K. Yuan, Y. Sun, F. Liang, F. Pan, M. Hu, F. Hua, Y. Yuan, J. Nie and Y. Zhang, RSC Adv., 2022, 12, 23379–23386 RSC.
- F. Pan, F. Hua, Y. Yan, X. Huang, L. Yuan, Y. Tang, Y. Yuan, J. Nie and Y. Zhang, Microchem. J., 2023, 190, 108707 CrossRef CAS.
- X. Mo, J. Huang, Y. Sun, X. Chen, Y. Deng, C. Liu, W. Jin, J. Nie and Y. Zhang, Microchem. J., 2022, 174, 107075 CrossRef CAS.
- J. Huang, X. Mo, H. Fu, Y. Sun, Q. Gao, X. Chen, J. Zou, Y. Yuan, J. Nie and Y. Zhang, Sens. Actuators, B, 2021, 344, 130218 CrossRef CAS.
- Y. Sun, K. Yuan, X. Mo, X. Chen, Y. Deng, C. Liu, Y. Yuan, J. Nie and Y. Zhang, Talanta, 2022, 238, 122999 CrossRef CAS PubMed.
- M. Hu, W. Xiao, Y. Chen, Q. He, K. Yuan, X. Huang, W. Jin, J. Nie and Y. Zhang, Photochem. Photobiol. Sci., 2023, 22, 631–640 CrossRef CAS PubMed.
- F. Hua, F. Pan, J. Yang, Y. Yan, X. Huang, Y. Yuan, J. Nie, H. Wang and Y. Zhang, Anal. Bioanal. Chem., 2023, 415, 2705–2713 CrossRef CAS PubMed.
- X. Chen, Y. Sun, X. Mo, Q. Gao, Y. Deng, M. Hu, J. Zou, J. Nie and Y. Zhang, RSC Adv., 2021, 11, 36859–36865 RSC.
- F.-J. Cao, H.-H. Cheng, S.-X. Ma, F. Jiao and D.-M. Dong, Sens. Biosensing Res., 2022, 38, 100533 CrossRef.
- S. İ. Dönmez, S. H. Needs, H. M. I. Osborn and A. D. Edwards, Sens. Actuators, B, 2020, 323, 128645 CrossRef.
- A. S. Day, T.-H. Ulep, B. Safavinia, T. Hertenstein, E. Budiman, L. Dieckhaus and J.-Y. Yoon, Biosens. Bioelectron., 2021, 179, 113099 CrossRef CAS PubMed.
- I.-J. Doh, B. Dowden, V. Patsekin, B. Rajwa, J. P. Robinson and E. Bae, Sensors, 2022, 22, 2646 CrossRef PubMed.
- M. Fang, Y. Wang, T. Yang, J. Zhang, H. Yu, Z. Luo, B. Su and X. Lin, ACS Sens., 2024, 9, 2010–2019 CrossRef CAS PubMed.
- H. Nguyen, I. Misbah and W.-C. Shih, IEEE Sens. J., 2020, 20, 6685–6691 CAS.
- W. Lu, C. Lin, J. Yang, X. Wang, B. Yao and M. Wang, Anal. Bioanal. Chem., 2019, 411, 5383–5391 CrossRef CAS PubMed.
- M. Li, Y. Gao, Y. Zhang, S. Gong, X. Tian, Y. Yang, X. Xu, Z. Wang and S. Wang, Spectrochim. Acta. Part A, 2021, 262, 120135 CrossRef CAS PubMed.
- Y. Li, X. Jiang, Y. Li, X. Yan, L. Tang, X. Sun, K. Zhong, X. Li and J. Li, Food Chem., 2024, 458, 140239 CrossRef CAS PubMed.
- E. Garrido, G. Hernández-Sigüenza, E. Climent, M. D. Marcos, K. Rurack, P. Gaviña, M. Parra, F. Sancenón, V. Martí-Centelles and R. Martínez-Máñez, Sens. Actuators, B, 2023, 377, 133043 CrossRef CAS.
- Z. Jiao, Z. Guo, X. Huang, J. Yang, J. Huang, Y. Liu, G. Liu, P. Zhang, C. Song and B. Z. Tang, ACS Sens., 2021, 6, 2845–2850 CrossRef CAS PubMed.
- M. Xu, W. Huang, D. Lu, C. Huang, J. Deng and T. Zhou, Anal. Methods, 2019, 11, 4267–4273 RSC.
- E. Teknikel, Spectrochim. Acta. Part A, 2024, 322, 124807 CrossRef CAS PubMed.
- W.-Y. Zhang, T. Tian, L.-J. Peng, H.-Y. Zhou, H. Zhang, H. Chen and F.-Q. Yang, Biosensors, 2022, 12, 893 CrossRef CAS PubMed.
- Y. Zhu, J. Zhang, J. Song, J. Yang, Z. Du, W. Zhao, H. Guo, C. Wen, Q. Li, X. Sui and L. Zhang, Adv. Funct. Mater., 2020, 30, 1905493 CrossRef CAS.
- Nandini and D. A. Jose, Sens. Actuators, B, 2024, 417, 136070 CrossRef CAS.
- D. Hatiboruah, T. Das, N. Chamuah, D. Rabha, B. Talukdar, U. Bora, K. U. Ahamad and P. Nath, Measurement, 2020, 154, 107507 CrossRef.
- C. Zhang, J. P. Kim, M. Creer, J. Yang and Z. Liu, Biosens. Bioelectron., 2017, 97, 164–168 CrossRef CAS PubMed.
- Y. Sun, M. Wei, R. Liu, H. Wang, H. Li, Q. Kang and D. Shen, Talanta, 2019, 194, 452–460 CrossRef CAS PubMed.
- Z. Lu, M. Chen, T. Liu, C. Wu, M. Sun, G. Su, X. Wang, Y. Wang, H. Yin, X. Zhou, J. Ye, Y. Shen and H. Rao, ACS Appl. Mater. Interfaces, 2023, 15, 9800–9812 CrossRef CAS PubMed.
- L. Luo, J. Li, X. Bi, P. Jiang, L. Li, G. Qiao and T. You, J. Hazard. Mater., 2024, 476, 134967 CrossRef CAS PubMed.
- R. Gu, X. Li, Y. Meng, Z. Li, H. Nie, X. Wang and D. Xiao, Analyst, 2022, 147, 4228–4236 RSC.
- P. Jia, Q. Wu, B. Sun and L. Wang, Small, 2023, 19, 2304096 CrossRef CAS PubMed.
- F. Liu, T. Lei, Y. Zhang, Y. Wang and Y. He, Anal. Chim. Acta, 2021, 1184, 339026 CrossRef CAS PubMed.
- W. Yang, L. Ye, Y. Wu, X. Wang, S. Ye, Y. Deng, K. Huang, H. Luo, J. Zhang and C. Zheng, J. Hazard. Mater., 2024, 470, 134038 CrossRef CAS PubMed.
- K. G. Shah, V. Singh, P. C. Kauffman, K. Abe and P. Yager, Anal. Chem., 2018, 90, 6967–6974 CrossRef CAS PubMed.
- P. Preechakasedkit, K. Osada, Y. Katayama, N. Ruecha, K. Suzuki, O. Chailapakul and D. Citterio, Analyst, 2018, 143, 564–570 RSC.
- J. Wang, C. Jiang, J. Yuan, L. Tong, Y. Wang, D. Zhuo, L. Huang, W. Ni, J. Zhang, M. Huang, D. Li, B. Su and J. Hu, Anal. Chem., 2022, 94, 10865–10873 CrossRef CAS PubMed.
- B. Peng, J. Liang, Y. Wang, G. He, X. Zhang, C. Lu, Q. Song, Y. Zhang, G. Li, Y. Hao and Y. Tang, Sens. Actuators, B, 2022, 369, 132376 CrossRef CAS.
- Y. Wu, Y. Zhang, Y. Hu, N. Jiang, R. P. Patel, A. K. Yetisen and M. F. Cordeiro, Adv. Mater. Technol., 2024, 9, 2400238 CrossRef CAS.
- C. Lee, J. Noh, S. E. O'Neal, A. E. Gonzalez, H. H. Garcia and S. Handali, PLoS Neglected Trop. Dis., 2019, 13, e0007746 CrossRef PubMed.
- S. Bock, H.-M. Kim, J. Kim, J. An, Y.-S. Choi, X.-H. Pham, A. Jo, K. Ham, H. Song, J.-W. Kim, E. Hahm, W.-Y. Rho, S. H. Lee, S. Park, S. Lee, D. H. Jeong, H.-Y. Lee and B.-H. Jun, Nanomaterials, 2021, 12, 33 CrossRef PubMed.
- T. Zhong, J. Li, S. Zhang and H. Pan, Appl. Biochem. Microbiol., 2023, 59, 946–958 CrossRef CAS.
- J. Liu, B. Wang, H. Huang, D. Jian, Y. Lu, Y. Shan, S. Wang and F. Liu, Food Chem., 2021, 335, 127596 CrossRef CAS PubMed.
- Z. Rong, Q. Wang, N. Sun, X. Jia, K. Wang, R. Xiao and S. Wang, Anal. Chim. Acta, 2019, 1055, 140–147 CrossRef CAS PubMed.
- H. Chen, Y. Ding, J. Li, L. Huang, G. González-Sapienza, B. D. Hammock, M. Wang and X. Hua, Anal. Chem., 2022, 94, 7358–7367 CrossRef CAS PubMed.
- J. Wang, C. Jiang, J. Jin, L. Huang, W. Yu, B. Su and J. Hu, Angew. Chem., Int. Ed., 2021, 133, 13152–13159 CrossRef.
- Y. Zhu, L. Ao, S. Chu, Y. Liao, J. Wang, J. Hu and L. Huang, Adv. Funct. Mater., 2024, 34, 2316147 CrossRef CAS.
- L. Huang, Z. Shi, D. Li, C. Jiang, X. Wang, M. Huang, J. Wang and B. Su, ACS Appl. Nano Mater., 2022, 6, 86–95 CrossRef.
- V. K. Rajendran, P. Bakthavathsalam, P. L. Bergquist and A. Sunna, Sens. Actuators, B, 2019, 298, 126849 CrossRef CAS.
- Z. Rong, Z. Bai, J. Li, H. Tang, T. Shen, Q. Wang, C. Wang, R. Xiao and S. Wang, Biosens. Bioelectron., 2019, 145, 111719 CrossRef CAS PubMed.
- C. Lu, W. Xiao, Y. Su, X. Zhang, Y. Chen, K. Lu, P. Teng, J. Liang, H. Yang, Q. Song, Y. Tang and D. Cao, ACS Sens., 2023, 8, 1950–1959 CrossRef CAS PubMed.
- Y. Tian, X. Hu, J. Jiang, X. Tang, Z. Tian, Z. Zhang and P. Li, Foods, 2023, 12, 431 CrossRef CAS PubMed.
- H. Jiang, D. Wu, L. Song, Q. Yuan, S. Ge, X. Min, N. Xia, S. Qian and X. Qiu, SLAS Technol., 2017, 22, 122–129 CrossRef PubMed.
- Z. Liu, Q. Hua, J. Wang, Z. Liang, J. Li, J. Wu, X. Shen, H. Lei and X. Li, Biosens. Bioelectron., 2020, 158, 112178 CrossRef CAS PubMed.
- Y. Gong, Y. Zheng, B. Jin, M. You, J. Wang, X. Li, M. Lin, F. Xu and F. Li, Talanta, 2019, 201, 126–133 CrossRef CAS PubMed.
- J. Guo, S. Chen, S. Tian, K. Liu, X. Ma and J. Guo, Talanta, 2021, 230, 122335 CrossRef CAS PubMed.
- H. He, B. Liu, S. Wen, J. Liao, G. Lin, J. Zhou and D. Jin, Anal. Chem., 2018, 90, 12356–12360 CrossRef CAS PubMed.
- M. You, M. Lin, Y. Gong, S. Wang, A. Li, L. Ji, H. Zhao, K. Ling, T. Wen, Y. Huang, D. Gao, Q. Ma, T. Wang, A. Ma, X. Li and F. Xu, ACS Nano, 2017, 11, 6261–6270 CrossRef CAS PubMed.
- Y. Zhang, L. Wang, W.-L. Wang, C. Yang, Y. Feng and X. Shi, Talanta, 2021, 232, 122427 CrossRef CAS PubMed.
- Y. Cao, T. Bu, H. Wu, J. Xi, Y. Wang, C. Xuan, P. Jia, B. Zheng, J. Zhao, Y. Zhuang and L. Wang, Anal. Chem., 2023, 95, 16585–16592 CrossRef CAS PubMed.
- A. N. Danthanarayana, E. Finley, B. Vu, K. Kourentzi, R. C. Willson and J. Brgoch, Anal. Methods, 2020, 12, 272–280 RSC.
- S. Zhang, J. Xiong, S. Wang, Z. Li, L. Qin, B. Sun, Z. Wang, X. Liu, Y. Zheng and H. Jiang, J. Hazard. Mater., 2024, 469, 134068 CrossRef CAS PubMed.
- W. Wu, Y. Li, P. Song, Q. Xu, D. Lei, J. Wang, B. Fu and W. Kong, J. Hazard. Mater., 2024, 465, 133103 CrossRef CAS PubMed.
- R. Paul, E. Ostermann, Y. Chen, A. C. Saville, Y. Yang, Z. Gu, A. E. Whitfield, J. B. Ristaino and Q. Wei, Biosens. Bioelectron., 2021, 187, 113312 CrossRef CAS PubMed.
- P. Teengam, N. Nisab, N. Chuaypen, P. Tangkijvanich, T. Vilaivan and O. Chailapakul, Biosens. Bioelectron., 2021, 189, 113381 CrossRef CAS PubMed.
- V. K. Rajendran, P. Bakthavathsalam, P. L. Bergquist and A. Sunna, Biosens. Bioelectron., 2019, 134, 68–75 CrossRef CAS PubMed.
- X. Qiu, J. I. Shu, O. Baysal, J. Wu, S. Qian, S. Ge, K. Li, X. Ye, N. Xia and D. Yu, Microfluid. Nanofluid., 2019, 23, 1–8 CrossRef CAS.
- B. Shi, Y. Li, D. Wu and W. Wu, Analyst, 2020, 145, 2767–2773 RSC.
- W. Chen, H. Yu, F. Sun, A. Ornob, R. Brisbin, A. Ganguli, V. Vemuri, P. Strzebonski, G. Cui, K. J. Allen, S. A. Desai, W. Lin, D. M. Nash, D. L. Hirschberg, I. Brooks, R. Bashir and B. T. Cunningham, Anal. Chem., 2017, 89, 11219–11226 CrossRef CAS PubMed.
- R. R. G. Soares, A. S. Akhtar, I. F. Pinto, N. Lapins, D. Barrett, G. Sandh, X. Yin, V. Pelechano and A. Russom, Lab Chip, 2021, 21, 2932–2944 RSC.
- J. Wang, G. Jing, W. Huang, L. Xin, J. Du, X. Cai, Y. Xu, X. Lu and W. Chen, Anal. Chem., 2022, 94, 18083–18091 CrossRef CAS PubMed.
- P. Mao, L. Cao, Z. Li, M. You, B. Gao, X. Xie, Z. Xue, P. Peng, C. Yao and F. Xu, Analyst, 2021, 146, 6960–6969 RSC.
- B. P. Sullivan, Y.-S. Chou, A. T. Bender, C. D. Martin, Z. G. Kaputa, H. March, M. Song and J. D. Posner, Lab Chip, 2022, 22, 2352–2363 RSC.
- A. M. Jankelow, H. Lee, W. Wang, T.-H. Hoang, A. Bacon, F. Sun, S. Chae, V. Kindratenko, K. Koprowski, R. A. Stavins, D. D. Ceriani, Z. W. Engelder, W. P. King, M. N. Do, R. Bashir, E. Valera and B. T. Cunningham, Analyst, 2022, 147, 3838–3853 RSC.
- X. Zhu, J. Zhao, A. Hu, J. Pan, G. Deng, C. Hua, C. Zhu, Y. Liu, K. Yang and L. Zhu, Micromachines, 2020, 11, 186 CrossRef PubMed.
- S.-A. Hsieh, D. Shamsaei, D. R. Eitzmann and J. L. Anderson, Anal. Chem., 2022, 94, 11949–11956 CrossRef CAS PubMed.
- M. Moazeni, P. Berger and C. Padeste, Micro Nano Eng., 2023, 19, 100184 CrossRef CAS.
- Y. Zhu, X. Gu, Q. Tang, W. Jiang, R. Xia, J. Zhang, H. Ji, Y. Qin and L. Wu, Anal. Chem., 2024, 96, 14116–14124 CrossRef CAS PubMed.
- B. Jin, C. Ma, C. Zhang, H. Yin, G. Zhao, J. Hu and Z. Li, Microchim. Acta, 2024, 191, 177 CrossRef CAS PubMed.
- T. Tian, Z. Qiu, Y. Jiang, D. Zhu and X. Zhou, Biosens. Bioelectron., 2022, 196, 113701 CrossRef CAS PubMed.
- M. Lin, H. Yue, T. Tian, E. Xiong, D. Zhu, Y. Jiang and X. Zhou, Anal. Chem., 2022, 94, 8277–8284 CrossRef CAS PubMed.
- Z. Lei, L. Lian, L. Zhang, C. Liu, S. Zhai, X. Yuan, J. Wei, H. Liu, Y. Liu, Z. Du, I. Gul, H. Zhang, Z. Qin, S. Zeng, P. Jia, K. Du, L. Deng, D. Yu, Q. He and P. Qin, ACS Omega, 2023, 8, 32555–32564 CrossRef CAS PubMed.
- P. Fozouni, S. Son, M. D. de L. Derby, G. J. Knott, C. N. Gray, M. V. D'Ambrosio, C. Zhao, N. A. Switz, G. R. Kumar, S. I. Stephens, D. Boehm, C.-L. Tsou, J. Shu, A. Bhuiya, M. Armstrong, A. R. Harris, P.-Y. Chen, J. M. Osterloh, A. Meyer-Franke, B. Joehnk, K. Walcott, A. Sil, C. Langelier, K. S. Pollard, E. D. Crawford, A. S. Puschnik, M. Phelps, A. Kistler, J. L. DeRisi, J. A. Doudna, D. A. Fletcher and M. Ott, Cell, 2021, 184, 323–333 CrossRef CAS PubMed.
- H. De Puig, R. A. Lee, D. Najjar, X. Tan, L. R. Soenksen, N. M. Angenent-Mari, N. M. Donghia, N. E. Weckman, A. Ory, C. F. Ng, P. Q. Nguyen, A. S. Mao, T. C. Ferrante, G. Lansberry, H. Sallum, J. Niemi and J. J. Collins, Sci. Adv., 2021, 7, eabh2944 CrossRef PubMed.
- J. Arizti-Sanz, C. A. Freije, A. C. Stanton, B. A. Petros, C. K. Boehm, S. Siddiqui, B. M. Shaw, G. Adams, T.-S. F. Kosoko-Thoroddsen, M. E. Kemball, J. N. Uwanibe, F. V. Ajogbasile, P. E. Eromon, R. Gross, L. Wronka, K. Caviness, L. E. Hensley, N. H. Bergman, B. L. MacInnis, C. T. Happi, J. E. Lemieux, P. C. Sabeti and C. Myhrvold, Nat. Commun., 2020, 11, 5921 CrossRef CAS PubMed.
- J. Liu, N. Li, L. Zhang, Y. Lu, M. Shen, Y. Zhang, L. Feng, J. Jing, J. Cheng and Y. Xu, Small Methods, 2024, 8, 2400030 CrossRef CAS PubMed.
- T. Yu, S. Zhang, R. Matei, W. Marx, C. L. Beisel and Q. Wei, AIChE J., 2021, 67, e17365 CrossRef CAS.
- Y. Zhu, J. Wang, H. Xie, H. Liu, S. Liu, D. He, P. Mi, S. He, J. Wang and Y. Sun, ACS Appl. Mater. Interfaces, 2022, 14, 10212–10226 CrossRef CAS PubMed.
- X. Peng, X. Mei, X. Liu, G. Zhang and Y. Li, Anal. Chem., 2024, 96, 13252–13259 CrossRef CAS PubMed.
- C. M. Green, J. Spangler, K. Susumu, D. A. Stenger, I. L. Medintz and S. A. Díaz, ACS Nano, 2022, 16, 20693–20704 CrossRef CAS PubMed.
- T. Treebupachatsakul, C. Lochotinunt, T. Teechot, N. Pensupa and S. Pechprasarn, IEEE Sens. J., 2022, 22, 12473–12484 CAS.
- S. Hong, D.-W. Zheng, Q.-L. Zhang, W.-W. Deng, W.-F. Song, S.-X. Cheng, Z.-J. Sun and X.-Z. Zhang, Chem. Sci., 2020, 11, 4403–4409 RSC.
- M. N. Tahir, Y. Xie, M. A. Sami, R. Punde and U. Hassan, IEEE Sens. Lett., 2024, 8, 1–4 Search PubMed.
- M. V. Bills, B. T. Nguyen and J.-Y. Yoon, IEEE Sens. J., 2019, 19, 7822–7828 CAS.
- J. W. Snow, H. C. Koydemir, D. K. Karinca, K. Liang, D. Tseng and A. Ozcan, Lab Chip, 2019, 19, 789–797 RSC.
- C. Goenka, W. Lewis, L. R. Chevres-Fernández, A. Ortega-Martínez, E. Ibarra-Silva, M. Williams and W. Franco, Lasers Surg. Med., 2019, 51, 201–207 CrossRef PubMed.
- Y. Liu, A. M. Rollins, R. M. Levenson, F. Fereidouni and M. W. Jenkins, Commun. Biol., 2021, 4, 1–14 CrossRef PubMed.
- O. Ormachea, A. Villazón, P. Rodriguez and M. Zimic, Biosensors, 2022, 12, 960 CrossRef CAS PubMed.
- G. H. Darwish, J. Asselin, M. V. Tran, R. Gupta, H. Kim, D. Boudreau and W. R. Algar, ACS Appl. Mater. Interfaces, 2020, 12, 33530–33540 CrossRef CAS PubMed.
- K. Rees, G. H. Darwish and W. R. Algar, ACS Appl. Mater. Interfaces, 2023, 15, 18672–18684 CrossRef CAS PubMed.
- R. Zenhausern, A. S. Day, B. Safavinia, S. Han, P. E. Rudy, Y.-W. Won and J.-Y. Yoon, Biosens. Bioelectron., 2022, 200, 113916 CrossRef CAS PubMed.
- S. Kim, A. Romero-Lozano, D. S. Hwang and J.-Y. Yoon, J. Hazard. Mater., 2021, 413, 125338 CrossRef CAS PubMed.
- S. Chung, L. E. Breshears, S. Perea, C. M. Morrison, W. Q. Betancourt, K. A. Reynolds and J.-Y. Yoon, ACS Omega, 2019, 4, 11180–11188 CrossRef CAS PubMed.
- T.-H. Ulep, R. Zenhausern, A. Gonzales, D. S. Knoff, P. A. Lengerke Diaz, J. E. Castro and J.-Y. Yoon, Biosens. Bioelectron., 2020, 153, 112042 CrossRef CAS PubMed.
- S. Kim, P. Akarapipad, B. T. Nguyen, L. E. Breshears, K. Sosnowski, J. Baker, J. L. Uhrlaub, J. Nikolich-Žugich and J.-Y. Yoon, Biosens. Bioelectron., 2022, 200, 113912 CrossRef CAS PubMed.
- Y. Liang, A. Zhou, C. S. Bever, L. W. Cheng and J.-Y. Yoon, Microchim. Acta, 2022, 189, 322 CrossRef CAS PubMed.
- Y. Liang, A. Zhou and J.-Y. Yoon, ACS Omega, 2022, 7, 30064–30073 CrossRef CAS PubMed.
- S. Kim, K. Samanta, B. T. Nguyen, S. Mata-Robles, L. Richer, J.-Y. Yoon and M. Gomes-Solecki, Sci. Rep., 2023, 13, 7546 CrossRef CAS PubMed.
- Q. Deng, Z. Lan, L. Xu, Z. Zhu and X. Shu, Sens. Actuators, B, 2021, 343, 130086 CrossRef CAS.
- J. Luo and Z. Zhu, Anal. Chem., 2024, 96, 11115–11120 CrossRef CAS PubMed.
- C.-H. Huang, Y. I. Park, H.-Y. Lin, D. Pathania, K. S. Park, M. Avila-Wallace, C. M. Castro, R. Weissleder and H. Lee, ACS Nano, 2019, 13, 11698–11706 CrossRef CAS PubMed.
- Y. Wang, S. Sadeghi, A. Velayati, R. Paul, Z. Hetzler, E. Danilov, F. S. Ligler and Q. Wei, PNAS Nexus, 2023, 2, pgad313 CrossRef PubMed.
- B. Bruininks and L. B. F. Juurlink, J. Chem. Educ., 2022, 99, 2168–2174 CrossRef CAS.
- M. Xiao, N. Xu, A. He, Z. Yu, B. Chen, B. Jin, L. Jiang and C. Yi, iScience, 2023, 26, 106553 CrossRef CAS PubMed.
- H. Yu, Y. Tan and B. T. Cunningham, Anal. Chem., 2014, 86, 8805–8813 CrossRef CAS PubMed.
- M. A. Hossain, J. Canning, S. Ast, K. Cook, P. J. Rutledge and A. Jamalipour, Opt. Lett., 2015, 40, 1737–1740 CrossRef PubMed.
- D. Hatiboruah, S. Biswas, D. Sarma and P. Nath, Sens. Actuators, A, 2022, 341, 113586 CrossRef CAS.
- S. Shukla, A. N. Sah, D. Hatiboruah, S. Ahirwar, P. Nath and A. Pradhan, Sci. Rep., 2022, 12, 11192 CrossRef CAS PubMed.
- Q. Deng, Y. Liu, Z. Zhu and X. Shu, Opt. Lett., 2022, 47, 3427–3430 CrossRef CAS PubMed.
- S. Shukla, B. S. Deo, C. Vishwakarma, S. Mishra, S. Ahirwar, A. N. Sah, K. Pandey, S. Singh, S. N. Prasad, A. K. Padhi, M. Pal, P. K. Panigrahi and A. Pradhan, J. Biophotonics, 2024, 17, e202300468 CrossRef CAS PubMed.
- Y. Sun, L. Shi, Q. Wang, L. Mi and T. Li, Anal. Chem., 2019, 91, 3652–3658 CrossRef CAS PubMed.
- L. Shi, Y. Sun, L. Mi and T. Li, ACS Sens., 2019, 4, 3219–3226 CrossRef CAS PubMed.
- L. Mi, Y. Sun, L. Shi and T. Li, ACS Appl. Mater. Interfaces, 2020, 12, 7879–7887 CrossRef CAS PubMed.
- Y. Zhou, S. Xie, B. Liu, C. Wang, Y. Huang, X. Zhang and S. Zhang, Anal. Chem., 2023, 95, 3332–3339 CrossRef CAS PubMed.
- P. M. Kalligosfyri, A. Sevastou, I. K. Kyriakou, S. S. Tragoulias, D. P. Kalogianni and T. K. Christopoulos, Anal. Chim. Acta, 2019, 1088, 123–130 CrossRef CAS PubMed.
- H. Chen, Y. Feng, F. Liu, C. Tan, N. Xu, Y. Jiang and Y. Tan, Biosens. Bioelectron., 2024, 247, 115929 CrossRef CAS PubMed.
- X. Chen, Y. Ning, S. Pan, B. Liu, Y. Chang, W. Pang and X. Duan, ACS Sens., 2021, 6, 2386–2394 CrossRef CAS PubMed.
- D. Liu, C. Ju, C. Han, R. Shi, X. Chen, D. Duan, J. Yan and X. Yan, Biosens. Bioelectron., 2021, 173, 112817 CrossRef CAS PubMed.
- F. Li, L. Guo, Z. Li, J. He and H. Cui, Anal. Chem., 2020, 92, 6827–6831 CrossRef CAS PubMed.
- S. Wang, F. Qu, R. Zhang, T. Jin, T. Zheng, J. Shu and H. Cui, Anal. Chem., 2023, 95, 12497–12504 CrossRef CAS PubMed.
- Y. Lai, J. Huang, D. Tang, X. Chen, L. Hou, S. Zhao and T. Lin, Biosens. Bioelectron., 2024, 263, 116602 CrossRef CAS PubMed.
- H. Chen, Z. Zhuang, N. Xu, Y. Feng, K. Fang, C. Tan and Y. Tan, Biosensors, 2024, 14, 135 CrossRef CAS PubMed.
- X. Yu, Y. Ma, S. Liu, C. Qi, W. Zhang, W. Xiang, Z. Li, K. Yang, S. Duan, X. Du, J. Yu, Y. Xie, Z. Wang, W. Jiang, L. Zhang and X. Lin, Anal. Chim. Acta, 2023, 1281, 341899 CrossRef CAS PubMed.
- H. Wu, Y. Fang, L. Tian, X. Liu, X. Zhou, X. Chen, H. Gao, H. Qin and Y. Liu, ACS Sens., 2023, 8, 3205–3214 CrossRef CAS PubMed.
- X. Tao, L. Yue, T. Tian, Y. Zhang, X. Zhou and E. Song, Anal. Chem., 2024, 96, 9270–9277 CrossRef CAS PubMed.
- A. Sevastou, S. S. Tragoulias, D. P. Kalogianni and T. K. Christopoulos, Anal. Bioanal. Chem., 2020, 412, 5663–5669 CrossRef CAS PubMed.
- A. Shahvar, M. Saraji and D. Shamsaei, Sens. Actuators, B, 2018, 255, 891–894 CrossRef CAS.
- M. J. Juachon, J. G. Regala, J. M. Marquez and M. X. Bailon, Open Chem., 2019, 17, 270–278 CAS.
- C. Wang, Z. Wu, M. Hao, B. Chen, J. Zhu, X. Cui and T. Wang, Sens. Actuators, B, 2023, 388, 133840 CrossRef CAS.
- R. S. Rafee, H. R. Pouretedal and S. Damiri, Luminescence, 2024, 39, e4775 CrossRef CAS PubMed.
- H. A. J. Al Lawati, J. Hassanzadeh, N. Bagheri and I. Al Lawati, Talanta, 2021, 234, 122648 CrossRef CAS PubMed.
- M. Zangheri, F. Di Nardo, D. Calabria, E. Marchegiani, L. Anfossi, M. Guardigli, M. Mirasoli, C. Baggiani and A. Roda, Anal. Chim. Acta, 2021, 1163, 338515 CrossRef CAS PubMed.
- J. Ma, Y. Guan, F. Xing, Y. Wang, X. Li, Q. Yu and X. Yu, Food Chem., 2023, 413, 135654 CrossRef CAS PubMed.
- Z. Li, Y. Xi, A. Zhao, J. Jiang, B. Li, X. Yang, J. He and F. Li, Anal. Bioanal. Chem., 2021, 413, 3541–3550 CrossRef CAS PubMed.
- D. Calabria, M. Zangheri, I. Trozzi, E. Lazzarini, A. Pace, M. Mirasoli and M. Guardigli, Biosensors, 2021, 11, 381 CrossRef CAS PubMed.
- S. Rink, A. Duerkop and A. J. Baeumner, Anal. Sens., 2023, 3, e202200111 CAS.
- J. Hassanzadeh, H. A. J. Al Lawati and N. Bagheri, Talanta, 2024, 276, 126219 CrossRef CAS PubMed.
- K. Kourentzi, K. Brosamer, B. Vu and R. C. Willson, Acc. Chem. Res., 2024, 57, 1372–1383 CrossRef CAS PubMed.
- M. Chabi, B. Vu, K. Brosamer, M. Smith, D. Chavan, J. C. Conrad, R. C. Willson and K. Kourentzi, Analyst, 2023, 148, 839–848 RSC.
- Y. Li, L. Zhou, W. Ni, Q. Luo, C. Zhu and Y. Wu, Anal. Chem., 2019, 91, 14838–14841 CrossRef CAS PubMed.
- D. Chang, K. T. Kim, E. Lindberg and N. Winssinger, ACS Sens., 2020, 5, 807–813 CrossRef CAS PubMed.
- L. Zhou, L. Zhang, L. Yang, W. Ni, Y. Li and Y. Wu, Biosens. Bioelectron., 2021, 173, 112824 CrossRef CAS PubMed.
- M. Hattori, N. Sugiura, T. Wazawa, T. Matsuda and T. Nagai, Anal. Chem., 2021, 93, 13520–13526 CrossRef CAS PubMed.
- Y. Li, P. Yang, N. Lei, Y. Ma, Y. Ji, C. Zhu and Y. Wu, Anal. Chem., 2018, 90, 11495–11502 CrossRef CAS PubMed.
- K. Tomimuro, K. Tenda, Y. Ni, Y. Hiruta, M. Merkx and D. Citterio, ACS Sens., 2020, 5, 1786–1794 CrossRef CAS PubMed.
- M. M. Calabretta, D. Gregucci, M. Guardigli and E. Michelini, Biosens. Bioelectron., 2024, 261, 116454 CrossRef CAS PubMed.
- Y. Ding, X. Hua, H. Chen, F. Liu, G. González-Sapien and M. Wang, Anal. Chem., 2018, 90, 2230–2237 CrossRef CAS PubMed.
- J. Li, N. Wang, M. Xiong, M. Dai, C. Xie, Q. Wang, K. Quan, Y. Zhou and Z. Qing, Anal. Chem., 2023, 95, 7142–7149 CrossRef CAS PubMed.
- M.-Y. Lu, W.-C. Kao, S. Belkin and J.-Y. Cheng, Sensors, 2019, 19, 3882 CrossRef CAS.
- R. Zhang, Y. Wang, H. Deng, S. Zhou, Y. Wu and Y. Li, Anal. Chim. Acta, 2023, 1273, 341538 CrossRef CAS PubMed.
- Y. Itoh, M. Hattori, T. Wazawa, Y. Arai and T. Nagai, ACS Sens., 2021, 6, 889–895 CrossRef CAS PubMed.
- M. M. Calabretta, R. Álvarez-Diduk, E. Michelini, A. Roda and A. Merkoçi, Biosens. Bioelectron., 2020, 150, 111902 CrossRef CAS PubMed.
- A. Lopreside, L. Montali, B. Wang, A. Tassoni, M. Ferri, M. M. Calabretta and E. Michelini, Biosens. Bioelectron., 2021, 194, 113569 CrossRef CAS PubMed.
- M. N. Hossain, R. Ishida, M. Hattori, T. Matsuda and T. Nagai, Sensors, 2020, 20, 3164 CrossRef CAS PubMed.
- E. Michelini, M. M. Calabretta, L. Cevenini, A. Lopreside, T. Southworth, D. M. Fontaine, P. Simoni, B. R. Branchini and A. Roda, Biosens. Bioelectron., 2019, 123, 269–277 CrossRef CAS PubMed.
- A. Lopreside, M. M. Calabretta, L. Montali, M. Ferri, A. Tassoni, B. R. Branchini, T. Southworth, M. D'Elia, A. Roda and E. Michelini, Anal. Bioanal. Chem., 2019, 411, 4937–4949 CrossRef CAS PubMed.
- M. Bhaiyya, P. K. Pattnaik and S. Goel, Microchim. Acta, 2022, 189, 79 CrossRef CAS PubMed.
- D. Calabria, E. Lazzarini, A. Pace, I. Trozzi, M. Zangheri, S. Cinti, M. Difonzo, G. Valenti, M. Guardigli, F. Paolucci and M. Mirasoli, Biosens. Bioelectron., 2023, 227, 115146 CrossRef CAS PubMed.
- S. Li, J. Liu, Z. Chen, Y. Lu, S. S. Low, L. Zhu, C. Cheng, Y. He, Q. Chen, B. Su and Q. Liu, Sens. Actuators, B, 2019, 297, 126811 CrossRef CAS.
- J. Totoricaguena-Gorriño, M. Dei, A. F. Alba, N. Peřinka, L.-R. Rubio, J. L. Vilas-Vilela and F. J. del Campo, ACS Sens., 2022, 7, 1544–1554 CrossRef PubMed.
- Z. Fan, Z. Geng, W. Fang, X. Lv, Y. Su, S. Wang and H. Chen, Sensors, 2020, 20, 446 CrossRef PubMed.
- P. Dutta, T.-Y. Su, A.-Y. Fu, M.-C. Chang, Y.-J. Guo, I.-J. Tsai, P.-K. Wei, Y.-S. Chang, C.-Y. Lin and Y.-J. Fan, Chem. Eng. J., 2022, 434, 133864 CrossRef CAS.
- Y. Zhou, A. Cui, D. Xiang, Y. Luan, Q. Wang, J. Huang, J. Liu, X. Yang and K. Wang, Sens. Actuators, B, 2024, 398, 134734 CrossRef CAS.
- Y. Xia, Y. Chen, Y. Tang, G. Cheng, X. Yu, H. He, G. Cao, H. Lu, Z. Liu and S.-Y. Zheng, ACS Sens., 2019, 4, 3298–3307 CrossRef CAS.
- M. Armstrong, A. R. Harris, M. V. D'Ambrosio, J. T. Coulibaly, S. Essien-Baidoo, R. K. D. Ephraim, J. R. Andrews, I. I. Bogoch and D. A. Fletcher, Am. J. Trop. Med. Hyg., 2022, 106, 1442–1449 CAS.
- X. Zeng, L. Wang, C. Liu, J. Zhang, H.-W. Shi, W. Shen, D. Kong, C. Huang, H. K. Lee and S. Tang, Talanta, 2024, 272, 125838 CrossRef CAS PubMed.
- Y. Man, M. Ban, A. Li, X. Jin, Y. Du and L. Pan, Food Chem., 2021, 354, 129578 CrossRef CAS PubMed.
- F. Li, Y. Zheng, J. Wu, L. Zhao, L. Shui, Q. Pu and S. Liu, Talanta, 2019, 203, 83–89 CrossRef CAS PubMed.
- N. M. Reis, S. H. Needs, S. M. Jegouic, K. K. Gill, S. Sirivisoot, S. Howard, J. Kempe, S. Bola, K. Al-Hakeem, I. M. Jones, T. Prommool, P. Luangaram, P. Avirutnan, C. Puttikhunt and A. D. Edwards, ACS Sens., 2021, 6, 4338–4348 CrossRef CAS PubMed.
- X.-L. Guo, Y. Chen, H.-L. Jiang, X.-B. Qiu and D.-L. Yu, Sensors, 2018, 18, 3141 CrossRef PubMed.
- T. Li, N. Yang, X. Pan, X. Zhang and L. Xu, Biosens. Bioelectron., 2024, 244, 115794 CrossRef CAS PubMed.
- D. Jiang, S. Lee, S. W. Bae and S.-Y. Park, Lab Chip, 2018, 18, 532–539 RSC.
- E. Ditchendorf, I. Ahmed, J. Sepate and A. Priye, Sensors, 2023, 23, 8310 CrossRef CAS PubMed.
- F. Sun, A. Ganguli, J. Nguyen, R. Brisbin, K. Shanmugam, D. L. Hirschberg, M. B. Wheeler, R. Bashir, D. M. Nash and B. T. Cunningham, Lab Chip, 2020, 20, 1621–1627 RSC.
- H. Xu, A. Xia, D. Wang, Y. Zhang, S. Deng, W. Lu, J. Luo, Q. Zhong, F. Zhang, L. Zhou, W. Zhang, Y. Wang, C. Yang, K. Chang, W. Fu, J. Cui, M. Gan, D. Luo and M. Chen, Sci. Adv., 2020, 6, eaaz7445 CrossRef CAS PubMed.
- Y. Shang, G. Xing, X. Liu, H. Lin and J.-M. Lin, Anal. Chem., 2022, 94, 16787–16795 CrossRef CAS PubMed.
- X. T. Zheng, W. P. Goh, Y. Yu, L. Sutarlie, D. Y. Chen, S. C. L. Tan, C. Jiang, M. Zhao, T. Ba, H. Li, X. Su and L. Yang, Adv. Healthcare Mater., 2024, 13, 2302173 CrossRef CAS PubMed.
- I. Jang, D. B. Carrão, R. F. Menger, A. R. Moraes de Oliveira and C. S. Henry, ACS Sens., 2020, 5, 2230–2238 CrossRef CAS PubMed.
- M. Yuan, C. Li, Y. Zheng, H. Cao, T. Ye, X. Wu, L. Hao, F. Yin, J. Yu and F. Xu, Talanta, 2024, 266, 125112 CrossRef CAS PubMed.
- S. Liu, J. Zhao, J. Wu, L. Wang, C. Yao, J. Hu and H. Zhang, Food Chem., 2025, 463, 141205 CrossRef CAS PubMed.
- K. Arias-Alpízar, A. Sánchez-Cano, J. Prat-Trunas, E. Sulleiro, P. Bosch-Nicolau, F. Salvador, I. Oliveira, I. Molina, A. Sánchez-Montalvá and E. Baldrich, Biosensors, 2022, 12, 680 CrossRef PubMed.
- A. Ray, S. Esparza, D. Wu, M. R. Hanudel, H.-A. Joung, B. Gales, D. Tseng, I. B. Salusky and A. Ozcan, Analyst, 2020, 145, 1841–1848 RSC.
- A. Clemente, A. Alba-Patiño, E. Rojo-Molinero, S. M. Russell, M. Borges, A. Oliver and R. de la Rica, ACS Sens., 2020, 5, 3956–3963 CrossRef CAS PubMed.
- T. Zhang, R. Deng, Y. Wang, C. Wu, K. Zhang, C. Wang, N. Gong, R. Ledesma-Amaro, X. Teng, C. Yang, T. Xue, Y. Zhang, Y. Hu, Q. He, W. Li and J. Li, Nat. Biomed. Eng., 2022, 6, 957–967 CrossRef CAS PubMed.
- P. Akarapipad, K. Kaarj, L. E. Breshears, K. Sosnowski, J. Baker, B. T. Nguyen, C. Eades, J. L. Uhrlaub, G. Quirk, J. Nikolich-Žugich, M. Worobey and J.-Y. Yoon, Biosens. Bioelectron., 2022, 207, 114192 CrossRef CAS PubMed.
- A. Katoh, K. Maejima, Y. Hiruta and D. Citterio, Analyst, 2020, 145, 6071–6078 RSC.
- S. Piranej, L. Zhang, A. Bazrafshan, M. Marin, G. B. Melikian and K. Salaita, ACS Cent. Sci., 2024, 10, 1332–1347 CrossRef CAS PubMed.
- S. Banik, S. K. Melanthota, Arbaaz, J. M. Vaz, V. M. Kadambalithaya, I. Hussain, S. Dutta and N. Mazumder, Anal. Bioanal. Chem., 2021, 413, 2389–2406 CrossRef CAS PubMed.
- I. Hussain and A. K. Bowden, Biomed. Opt. Express, 2021, 12, 1974–1998 CrossRef PubMed.
- C. S. Wood, M. R. Thomas, J. Budd, T. P. Mashamba-Thompson, K. Herbst, D. Pillay, R. W. Peeling, A. M. Johnson, R. A. McKendry and M. M. Stevens, Nature, 2019, 566, 467–474 CrossRef CAS PubMed.
- M. L. Cunha, S. S. da Silva, M. C. Stracke, D. L. Zanette, M. N. Aoki and L. Blanes, Anal. Chem., 2022, 94, 41–58 CrossRef CAS PubMed.
- W. R. Algar, N. Hildebrandt, S. S. Vogel and I. L. Medintz, Nat. Methods, 2019, 16, 815–829 CrossRef CAS PubMed.
- N. Hildebrandt, C. M. Spillmann, W. R. Algar, T. Pons, M. H. Stewart, E. Oh, K. Susumu, S. A. Díaz, J. B. Delehanty and I. L. Medintz, Chem. Rev., 2017, 117, 536–711 CrossRef CAS PubMed.
- Z. Li, H. Yuan, W. Yuan, Q. Su and F. Li, Coord. Chem. Rev., 2018, 354, 155–168 CrossRef CAS.
- X. Qiu, J. Xu, M. Cardoso Dos Santos and N. Hildebrandt, Acc. Chem. Res., 2022, 55, 551–564 CrossRef CAS PubMed.
- C.-W. Kuo and A. M. Smith, Acc. Chem. Res., 2023, 56, 1031–1042 CrossRef CAS PubMed.
- D. C. Duffy, Lab Chip, 2023, 23, 818–847 RSC.
- W. Yin, J. Zhuang, J. Li, L. Xia, K. Hu, J. Yin and Y. Mu, Small, 2023, 19, 2303398 CrossRef CAS PubMed.
- V. Y. C. Li, B. Udugama, P. Kadhiresan and W. C. W. Chan, Anal. Chem., 2022, 94, 17102–17111 CrossRef CAS PubMed.
- B. Udugama, P. Kadhiresan, A. Samarakoon and W. C. W. Chan, J. Am. Chem. Soc., 2017, 139, 17341–17349 CrossRef CAS PubMed.
- K. Rees, M. V. Tran, M. Massey, H. Kim, K. D. Krause and W. R. Algar, Bioconjugate Chem., 2020, 31, 861–874 CrossRef CAS PubMed.
- K. Lix, M. V. Tran, M. Massey, K. Rees, E. R. Sauve, Z. M. Hudson and W. R. Algar, ACS Appl. Bio Mater., 2020, 3, 432–440 CrossRef CAS.
Footnotes |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d4lc00966e |
| ‡ These authors contributed equally. |
| This journal is © The Royal Society of Chemistry 2025 |