Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Advances of dual-organ and multi-organ systems for gut, lung, skin and liver models in absorption and metabolism studies
Konstanze
Brandauer†
 ,
Sophie
Schweinitzer†
,
Sophie
Schweinitzer†
 ,
Alexandra
Lorenz†
,
Alexandra
Lorenz†
 ,
Judith
Krau߆
,
Judith
Krau߆
 ,
Silvia
Schobesberger†
,
Silvia
Schobesberger†
 ,
Martin
Frauenlob
and
Peter
Ertl
,
Martin
Frauenlob
and
Peter
Ertl
 *
*
Faculty of Technical Chemistry, TU Wien, Getreidemarkt 9, 1060 Vienna, Austria. E-mail: peter.ertl@tuwien.ac.at
First published on 13th February 2025
Abstract
Drug development is a costly and timely process with high risks of failure during clinical trials. Although in vitro tissue models have significantly advanced over the years, thus fostering a transition from animal-derived models towards human-derived models, failure rates still remain high. Current cell-based assays are still not able to provide an accurate prediction of the clinical success or failure of a drug candidate. To overcome the limitations of current methods, a variety of microfluidic systems have been developed as powerful tools that are capable of mimicking (micro)physiological conditions more closely by integrating physiological fluid flow conditions, mechanobiological cues and concentration gradients, to name only a few. One major advantage of these biochip-based tissue cultures, however, is their ability to seamlessly connect different organ models, thereby allowing the study of organ-crosstalk and metabolic byproduct effects. This is especially important when assessing absorption, distribution, metabolism, and excretion (ADME) processes of drug candidates, where an interplay between various organs is a prerequisite. In the current review, a number of in vitro models as well as microfluidic dual- and multi-organ systems are summarized with a focus on absorption (skin, lung, gut) and metabolism (liver). Additionally, the advantage of multi-organ chips in identifying a drug's on and off-target toxicity is discussed. Finally, the potential high-throughput implementation and modular chip design of multi-organ-on-a-chip systems within the pharmaceutical industry is highlighted, outlining the necessity of reducing handling complexity.
1 Introduction
In drug development, nine out of ten drugs still fail during clinical trials, while up to 15 years pass for the successful candidate to enter the market.1 For anticancer drugs the success rate was even less with 3.4% in the study time of 2000 to 2015.2 Overall, the most abundant reasons for failure were reported to be a lack of efficacy (52%) and safety (24%) for the reporting period 2013–2015.3 Especially failure rates in the more expensive clinical phases II and III dramatically increase research and development costs, thus posing a great financial burden for companies.4 To minimize the risk of late-stage failure, it is indispensable to increase the predictive validity already in preclinical studies since animal models or 2D cell culture models often fail to accurately predict drug metabolism and the metabolites' toxicity on the human body. Consequently, advanced human cell-based in vitro models, such as organ-on-a-chip systems, have been developed to recapitulate key physiological functions and thereby provide better prediction.5 By establishing multi-organ chips, either by connecting single organ modules or by combining several organ models on a single chip, inter-organ communication can be resembled, thereby facilitating a better replication of physiological processes in the human body.6 Additionally, the simulation of a circulating stream is key in studying the effects of drug metabolites on other tissues, thereby identifying possible secondary drug toxicity early on that reduces costs and attrition rates during clinical trials.7,8 The Food and Drug Administration (FDA) recently paved the way for the use of these microphysiological systems (MPS) in drug safety and effectiveness testing as an alternative to animal testing before entering clinical trials, underlining the potential for broad applicability.9Besides the need for better predictive models, the number of drug candidates has risen with the introduction of artificial intelligence in pharmaceutical research and development.10 As a result, the demand for pharmacokinetic studies with reliable and physiological-relevant in vitro models to investigate absorption, distribution, metabolism, and excretion (ADME) of drug candidates has significantly increased in recent years.11 Additionally, nanoparticles as advanced drug delivery systems have also been intensively studied for pharmaceutical applications, however, their behavior within the human body is hardly understood.12 Besides their application in drug delivery, their widespread usage in various industrial fields, including food additives and cosmetics, further highlights the need for reliable multi-organ models to study their ADME effects on the human body.13 As a potential technological solution that can also cope with the increasing number of drug candidates, miniaturized human-derived multi-organ platforms are capable of emulating physiologically-relevant metabolism.14 However, handling complexity needs to be reduced and/or operation automated to enable high-throughput screening with these platforms.
In attempt to summarize recent dual- and multi-organ-on-a-chip advances and highlight strategies to improve drug development, this review describes in detail the three main absorption organs (intestine, lung, and skin) involved in different administration routes (oral, pulmonary, and transdermal) and the metabolism organ (liver) as well as their already established in vitro models. Since the liver is the main metabolizing organ, available biochip systems connecting a liver model to various absorption organ models are compiled, and their ability to study a drug's toxicity on target and non-target organs is critically reviewed and discussed. Since the application of microfluidic multi-organ systems in the pharmaceutical industry is still in its infancy, high-throughput organ-on-a-chip systems that offer reduced handling complexity are highlighted and current unmet needs for a transition from the academic to the industrial sector are discussed.
2 Relevant organs in drug absorption and metabolism and their in vitro models
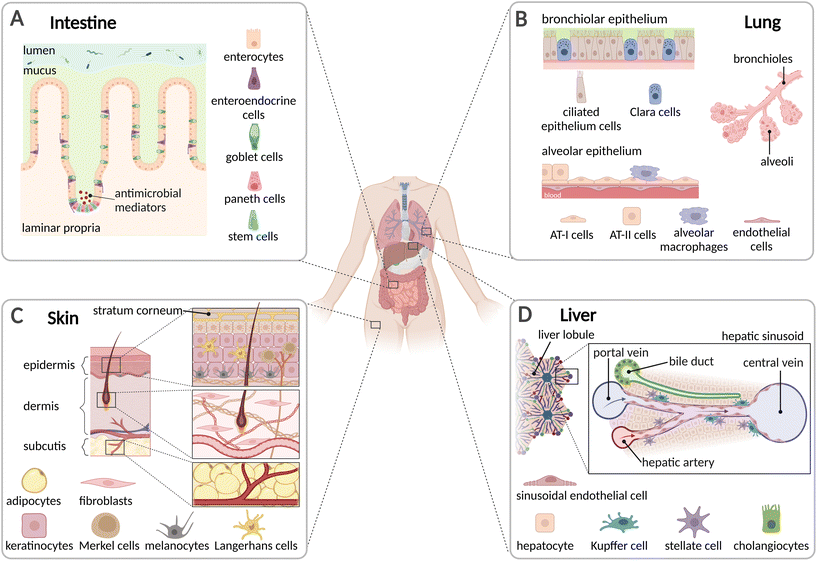
Before elaborating the advances of in vitro models, the following chapter discusses the organs' structural architecture and primary functions. It highlights organ-specific cell types and biophysical properties, such as characteristic flow rate or shear stress, that define the microenvironment. Understanding these key properties is essential for an accurate replications of organ functions in in vitro models.2.1 Organ architecture
2.2 From 2D to 3D: different cell types and culturing techniques in in vitro organ models
In vitro organ models which are of 2D and 3D spatial arrangement, are composed of various cell types, each with different functions leading to unique advantages and disadvantages. While commercial cell lines are easy to culture, reproducible, and long-lasting, they are often derived from cancerous tissues.44 In turn, primary cells represent more closely the in vivo phenotype, but have a limited lifespan, high donor variability, complex culturing conditions and access is limited through national and international ethical guidelines to obtain donor tissue. Although induced pluripotent stem cell (iPSC) technology can be used to differentiate into various human cellular phenotypes, it requires complex and labor-intensive differentiation protocols underlying strict quality control. A key advantage of iPSCs, however, is their potential to be produced from a living human as a source of autologous cells for personalized medical treatments showing higher success rates and fewer side effects in drug screening.45Next to the choice of representative cell types, the cell culturing methods need to be considered, where often, for historical reasons and functionality, the implementation of 2D culture protocols is used for high-throughput screening applications (Fig. 2).46 To investigate tissue interactions while keeping simplicity and the possibility for high-throughput sampling, Transwell® systems, a two-reservoir setup separated by a porous membrane, are employed. By cultivating cells on the membrane, also considered as 2.5D cultivation, barrier formation, function and permeability can be investigated.47,48 Advancing from 2.5D cell culture, scaffolds derived from biological as well as non-biological materials have been routinely employed over the years to provide structural support for the cultivation of cells in a 3D spatial arrangement. Hydrogels, consisting of ECM or of synthetic origin, have been commonly used to embed cells in 3D, thereby resembling the host tissue more closely. Advancing from of this manual labor-intense work, bioprinting applications were developed, where cell-laden hydrogels are dispensed in a controlled and automated manner attempting to generate 3D functional human tissues.49 However, these systems have limitations in relation to up-scaling, drug adsorption on scaffolds, complex handling, and batch-to-batch variations of the used embedding matrix.50 The 3D matrices gave rise to a promising alternative, derived from pluripotent stem cells (iPSCs or embryonic stem cells) or primary donor tissues, namely organoids. These cell constructs are self-organized aggregates of a diverse range of differentiated cells and residual stem cells, resembling organ regions in a functional and cytoarchitectural manner. In the case of liver and intestinal organoids, the resemblance is so extensive that they are histologically indistinguishable from native tissue.46 These organ models were capable to demonstrate the toxicity of drugs, that had been FDA-approved but then were retracted from the market due to complications in patients, while preclinical models including 2D in vitro studies and animal studies failed to do so.51 However, generation and maintenance of organoids are both cost and time-intensive. Their 3D architecture with a not easily accessible lumen hinders barrier permeability assays, and reduced nutrient access of the inner cells often leads to necrotic cores of the aggregates.47 Furthermore, organoids still mainly model just one type of tissue and lack vascular, immunological, and direct connections to other organs, which are the base of the synergistic human body.52
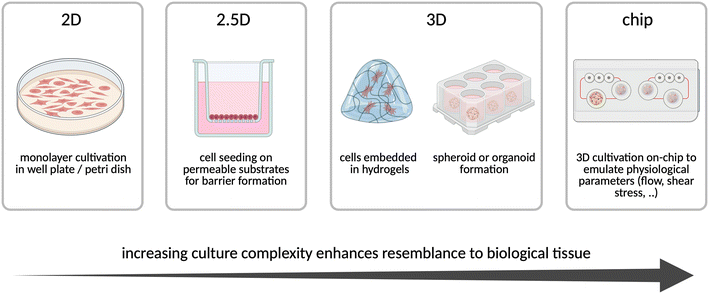 | ||
| Fig. 2 Overview of cell culture models sorted from left to right based on increasing cultivation complexity but enhanced resemblance of tissue from 2D to microfluidic organ models. | ||
Besides the abovementioned advances of these models, they still lack key factors of the cells' in vivo environment. In the human body, several cell types, such as endothelial cells or intestinal epithelial cells, are exposed to fluid flow and associated mechanical stress, while other cells require polarization facilitated via nutrient gradients to form in vivo like tissue structures for example in the cartilage. While these parameters cannot be addressed in conventional static cell culture models, microfluidic systems allow the control of the microenvironment in higher precision. These platforms incorporate one or several organ models connected through microchannels or porous membranes under consideration of mimicking the physiological environment.53 The integration of chemical and electrical sensors furthermore allows non-invasive monitoring of parameters such as pH, oxygen, glucose levels or transepithelial electrical resistance (TEER).46,53,54
Hereafter, the progress from simple to more advanced in vitro organ models is summarized based on selected articles, highlighting drug absorbing and metabolizing organs. The articles were selected because of their relevance for pharmacological and toxicological studies.
Interestingly, comparative analysis of MucilAir™, a commercially available ALI culture using primary cells, demonstrated that those primary cultures were more resilient to toxins compared to lung cell lines.71 Likewise, EpiAirway™ was observed to be more resilient than A549 cells.72 These commercial models can additionally be utilized in preliminary drug discovery research, and toxicity screening.73 However, although ALI cultures are reproducible in vitro systems, they lack the 3D structure, the flow dynamics, and the mechanical stretch of the human lung. By combining ALI with a dynamic culture condition, including fluid flow and mechanical stretch, lung-on-chip systems overcome these limitations, creating models that better replicate features of the human lung.74 The first lung-on-chip model was developed by Huh et al., stimulating natural breathing by applying a vacuum to the side chambers, which are separated by a thin wall from the cell-containing microchannels and cause mechanical stretching of the PDMS membrane.75 This model was later applied to mimic the drug-induced pulmonary edema found in cancer patients. The study found that while increased barrier permeability emerged with IL-2 exposure alone, it was significantly enhanced when IL-2 exposure was combined with mechanical stretching to simulate physiological breathing movements. Performing a drug study, potential new therapies were identified by demonstrating that angiopoietin-1 and transient receptor potential vanilloid 4 restored barrier function.76 However, these investigations are limited by the use of the cancer cell line NCI-H441. Advancements to those “first-generation” lung-on-chip models have been made by replacing conventional membranes with stretchable biological membranes77 or hydrogels78 or by using more physiological cyclic stretch instead of unidirectional stretching.79 Detailed information about the application of lung models for drug and toxicity screenings can be found in the review of Moreira et al.80 Lastly, iPSCs advance lung-on-a-chip models, however, they have not yet been applied for drug screenings in microfluidic systems.81
With a wide range of commercially available models such as EpiSkin™, EpiDerm™, SkinEthic™ and KeraSkin™, RHE are the most used in vitro skin models for drug testing and still the only approved model for skin irritation testing by the Organization for Economic Co-operation and Development (OECD).92,93 Schäfer-Korting et al. conducted a validation study regarding the absorption of nine compounds including the drugs clotrimazole and digoxin with SkinEthic™, EpiSkin™ and EpiDerm™ in comparison to excised human and pig skin showing the feasibility of RHE models for skin penetration studies despite a higher barrier permeability.82 To demonstrate the physiological skin barrier and crosstalk between the skin layers more accurately, open-source or commercial full-thickness models like Phenion® (Henkel) have been applied for permeation studies with hydrophilic and lipophilic compounds such as caffeine and testosterone.88,94 Schmidt et al. developed an advanced three-layered in vitro skin model with isolated mature adipocytes preventing the dermal-mediated contraction of the epidermis by using a filter membrane as an artificial basal layer. The model was able to distinguish between heptanal and 2-propanol as irritating and non-irritating agents with a correlating cytokine expression.90,95
Regarding transdermal drug delivery and absorption, it is critical to incorporate vascularization and perfusion into in vitro skin models as drugs are distributed mainly over the capillaries in the dermis.96,97 For this reason, skin-on-a-chip models have emerged in recent years that allow for dynamic cultivation of in vitro models or ex vivo skin biopsies.98 Ataç et al. demonstrated the positive influence of shear stress by prolonged cultivation times of EpiDermFT™ and ex vivo skin on their microfluidic platform including two circuits with micropumps and wells for incorporating permeable inserts.99 Multiple PDMS-based pumpless microfluidic devices for drug testing have been established that incorporate patient-derived full-thickness skin models and were tested through doxorubicin exposure or antioxidant α-lipoic acid with regard to skin barrier integrity.100,101 The importance for vascularization in drug screening was shown by Salameh et al., developing a full-thickness perfusable skin model that was vascularized with HUVEC cells and additional microvascularization by self-assembly of HUVEC cells in the same layer. This model has shown improved barrier performance compared to commercial RHE models and has been tested for topical and systemic application of caffeine, minoxidil and benzo-a-pyrene.87 iPSC-derived skin models are considered as a promising tool, but due to their laborious culture conditions they have not been applied for drug screening yet.102
3 Microfluidic dual-and multi-organ studies for drug testing
Some drugs are specifically activated through metabolism in a non-target tissue (e.g. the liver) to be effective when reaching the target organ, or contrastingly, are inactivated by other organs. Consequently, single organ studies are not representative for studying ADME processes and might lead to false conclusions about drug candidates in pharmacological studies.112,113 Therefore, to investigate potential off-target toxicity of drugs and their metabolites, MPS incorporating several organ models have been developed.114 In this chapter, dual-organ systems including liver and an absorption organ will be described and summarized (Table 1), to give an overview of existing models representing absorption and drug metabolism in the liver. After that, the importance and advances of off-target toxicity models are highlighted, based on recent developments utilizing multi-organ chips. Finally, unmet needs are summarized, directing a potential path towards broad applicability of multi-organ MPS in drug development.| Organ models | Cell type(s) | Complexity | Material | Modularity | Drug | Ref. |
|---|---|---|---|---|---|---|
| Liver–gut | Caco-2-BBE, HT29-MTX, MoDC | 2.5D | PSU | + | Diclofenac, hydrocortisone | Tsamandouras et al.115 |
| PHH, KCs | 3D | |||||
| Caco2 | 2.5D | PDMS | — | Phenacetin | Bricks et al.116 | |
| HepG2 | 2D | |||||
| Caco-2, HepG2, PHH/PRH | 2.5D | PC, PDMS | + | Paracetamol | Prot et al.117 | |
| Caco-2 | 2.5D | Veroclear polymer | + | — | Esch et al.118 | |
| PHH, NPC | 3D | |||||
| Caco-2 | 2.5D | PFPE | — | Midazolam | Wang et al.119 | |
| PHH | 2D | |||||
| Caco-2, HT29 | 2.5D | — | — | Mycophenolate mofetil/mycophenolic acid | Milani et al.113 | |
| PHH | 3D | |||||
| Caco-2, HepaRG, HUVEC | 2.5D | PBT | — | Irinotecan | Lucchetti et al.112 | |
| THP-1 | 2D | |||||
| Liver–skin | Skin explant | 2.5D | PDMS, PC, glass (HUMIMIC Chip2) | + | Troglitazone | Wagner et al.120 |
| HepaRG, HHSteC | 3D | |||||
| Skin explant | 2.5D | Troglitazone | Maschmeyer et al.121 | |||
| HepaRG, HHSteC | 3D | |||||
| HDMEC | 2D | |||||
| Primary nHEK, nHDF | 2.5D | Terbinafine | Tavares et al.122 | |||
| HepaRG, HHSteC | 3D | |||||
| EpiDerm™ | 2.5D | Permethrin, hyperforin | Kühnl et al.123 | |||
| HepaRG, HHSteC | 3D | |||||
| PhenionFT™, SkinEthic™ | 2.5D | 4-Amino-2-hydroxytoluene | Brandmair et al.124 | |||
| HepaRG, HHSteC | 3D | |||||
| EpiDerm™ | 2.5D | Genistein | Tao et al.125 | |||
| HepaRG, HHSteC | 3D | |||||
| nHEK | 3D | PDMS, glass | — | Tretinoin, acetaminophen, camphor | Lee et al.126 | |
| hiPSC HEPs | 3D | |||||
| Liver–lung | MucilAir™ | 2.5D | PDMS, PC, glass (HUMIMICChip3plus) | + | Aflatoxin B1 | Schimek et al.127 |
| HepaRG | 3D | |||||
| NHBE | 2.5D | PEEK | + | Aflatoxin B1 | Bovard et al.128 | |
| HepaRG | 3D | |||||
| A549 | 2.5D | PMMA, PC | + | Curcumin | Miller et al.129 | |
| HepG2 C3A, MDA-MB-231 | 3D | |||||
| HepG2, A549 | 2D | PDMS | — | Irinotecan | Shinha et al.130 | |
| PHH, KCs | 3D | PEI, Kapton® polyimide | + | — | Coppeta et al.131 | |
| NHBE | 2.5D |
3.1 Combined absorption and metabolism studies
3.2 Advances through dual- and multi-organ models in drug testing
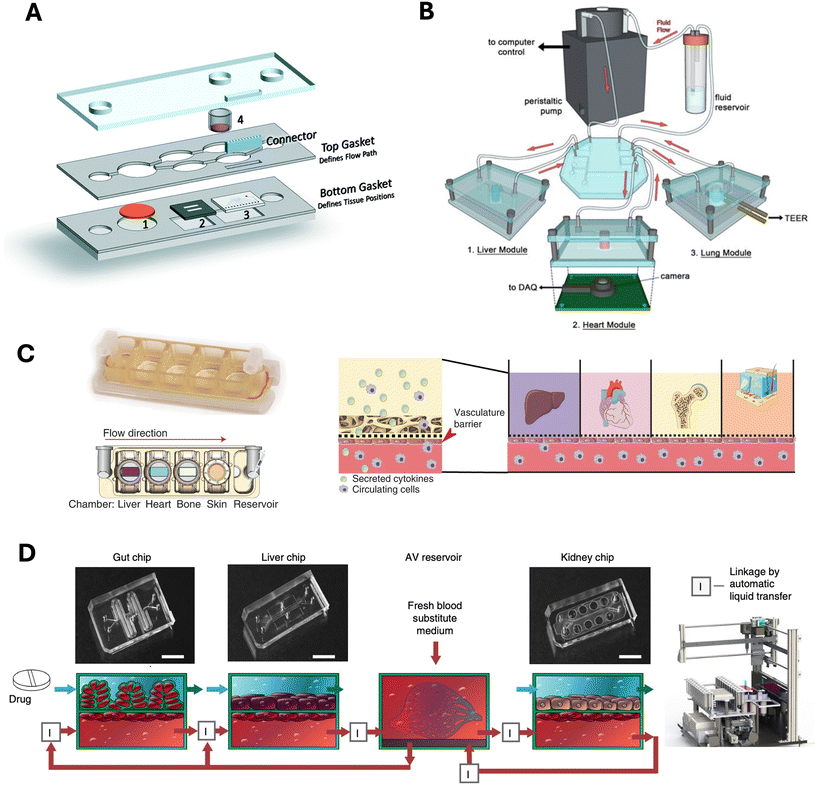 | ||
| Fig. 3 Selected examples of multi-organ chips. A) Multi-organ chip with liver, heart, and skin compartment for drug toxicity studies. Reproduced with permission from Pires de Mello et al.,144 Royal Society of Chemistry. B) Liver, lung, and heart module connected via tubing to study drug response. Reprinted with permission from Skardal et al.,146 Springer Nature Limited. C) Modular multi-organ chip with individual compartments for liver, heart, bone, and skin with a vascular barrier for crosstalk. Reprinted with permission from Ronaldson-Bouchard et al.,147 Springer Nature Limited. D) Organ modules connected via liquid transfer between the endothelium channels. Reprinted with permission from Herland et al.,148 Springer Nature Limited. | ||
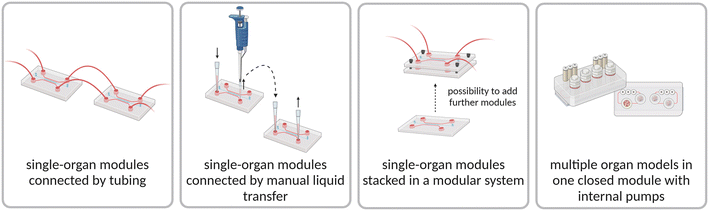
3.3 Advancing the operation of multi-organ chips
Despite the advances of multi-organ chips in drug toxicity studies, standardization and throughput still need to be improved to increase the use in routine drug screening studies.155 Thereby, automation and high-throughput capability of multi-organ chips are of high interest, since they reduce the cell culture handling complexity while simultaneously increasing parallelization. At the same time, the used technology needs to provide standardized operation, quality control steps to increase the predictiveness and comparability of experimental results. Among other advances, in this chapter we highlight two promising advances in the operation of multi-organ chips. Firstly, modular system configurations are introduced as powerful tool since they allow separate cultivation of different organ models before assembly, enabling individual quality control of the organ (disease) before starting multi-organ studies. Secondly, advances towards high-throughput and automation of cultivation processes are summarized.Another approach has been the design of platforms based on the dimensions of a microtiter plate, enabling the use of pipetting robots and allowing readouts to be performed with standard laboratory equipment, such as a plate reader. As an example, Azizgolshani et al. developed an organ-on-chip platform for high-throughput screening based on a standard plate facilitating 96 cultivations. This platform can be applied for co-culture studies and has integrated electrical sensors for barrier function measurements and an optical sensor for dissolved oxygen determination.164 In a follow up study, the developed platform was used for co-culturing endothelial cells and pericytes to study the permeability as well as the response to shear stress and TNF-α stimuli.165 However, this platform has not been used for drug screening applications yet. In another study, Hou et al. developed a high-throughput platform based on a 384-well microplate consisting of two chips for cultivating liver and tumor microtissue separately and assembling them for compound screening later. The feasibility of the developed platform was verified with anticancer drugs such as the CPT-11, epirubicin, adriamycin and plumbagin.162 Important to highlight are the high-throughput capabilities of the OrganoPlate®, housing 96 chips on a well plate format. Each chip consists of a connected organ and endothelium compartment with perfusion induced by placing the plate on a rocker. The design based on a well plate allows automation with a liquid handler.110 By automizing the cultivation of organ-chip models, reproducibility and throughput are increased and thereby the time to identify lead drug candidates is reduced, leading to a broader applicability in the industry.46
4 Conclusion
Recent advances in the development of miniaturized and automated multi-organ chips have opened new avenues in the understanding of drug absorption and metabolism by investigating complex organ-organ interactions. It is crucial to emphasize that multi-organ chips, especially those incorporating a liver model, are essential for drug screening studies since the hepatic tissue plays a key role in drug metabolism and its metabolites can significantly affect other tissues. Thereby, not only the drug's hepatotoxicity but also the effect of a drug and/or its metabolites on off-target organs, such as the heart, can be evaluated. Furthermore, the constant crosstalk between different organs enables to further understand signaling as a response to drug exposure. However, besides the many promising organs-on-a-chip developments, limitations such as complex cultivation procedures for establishing and maintaining different organ models remain. The main drawback is their moderate throughput, which will continue to impede the transition from academic settings to the industrial sector. To make these systems more feasible for industrial usage and high-throughput screening applications, design adaptations along with automated liquid handlers or modular systems are needed. The separate cultivation of modular systems allows not only quality control before assembly and drug testing, but also fosters parallelization and throughput. Additionally, further improvement of the biological model, for example through vascularization of organ models, enhances the translation to the human body. Consequently, continuous improvements on the technological and biological side of multi-organ systems will result in broader applicability in drug testing, better predictability of drug candidates and thereby reduce animal testing in drug development.Data availability
No primary research results, software or code have been included and no new data were generated or analysed as part of this review.Conflicts of interest
There are no conflicts to declare.Acknowledgements
This work was supported by the ActiTOX project (H2020-MSCA-RISE-2018, project no.: 823981). Illustrations were created in http://Biorender.com. The authors acknowledge TU Wien Bibliothek for financial support through its Open Access Funding Program.References
- D. Sun, W. Gao, H. Hu and S. Zhou, Why 90% of Clinical Drug Development Fails and How to Improve It?, Acta Pharm. Sin. B, 2022, 12(7), 3049–3062, DOI:10.1016/J.APSB.2022.02.002.
- C. H. Wong, K. W. Siah and A. W. Lo, Estimation of Clinical Trial Success Rates and Related Parameters, Biostatistics, 2019, 20(2), 273–286, DOI:10.1093/BIOSTATISTICS/KXX069.
- R. K. Harrison, Phase II and Phase III Failures: 2013–2015, Nat. Rev. Drug Discovery, 2016, 15(12), 817–818, DOI:10.1038/NRD.2016.184.
- N. Franzen, W. H. van Harten, V. P. Retèl, P. Loskill, J. van den Eijnden-van Raaij and M. IJzerman, Impact of Organ-on-a-Chip Technology on Pharmaceutical R&D Costs, Drug Discovery Today, 2019, 24(9), 1720–1724, DOI:10.1016/J.DRUDIS.2019.06.003.
- B. Zhang, A. Korolj, B. F. L. Lai and M. Radisic, Advances in Organ-on-a-Chip Engineering, Nat. Rev. Mater., 2018, 3(8), 257–278, DOI:10.1038/s41578-018-0034-7.
- N. Picollet-D'hahan, A. Zuchowska, I. Lemeunier and S. Le Gac, Multiorgan-on-a-Chip: A Systemic Approach To Model and Decipher Inter-Organ Communication, Trends Biotechnol., 2021, 39(8), 788–810, DOI:10.1016/J.TIBTECH.2020.11.014.
- M. Malik, Y. Yang, P. Fathi, G. J. Mahler and M. B. Esch, Critical Considerations for the Design of Multi-Organ Microphysiological Systems (MPS), Front. Cell Dev. Biol., 2021, 9, 2353, DOI:10.3389/FCELL.2021.721338/BIBTEX.
- Y. I. Wang, C. Carmona, J. J. Hickman and M. L. Shuler, Multiorgan Microphysiological Systems for Drug Development: Strategies, Advances, and Challenges, Adv. Healthcare Mater., 2018, 7(2), 1701000, DOI:10.1002/ADHM.201701000.
- A. Stewart, D. Denoyer, X. Gao and Y. C. Toh, The FDA Modernisation Act 2.0: Bringing Non-Animal Technologies to the Regulatory Table, Drug Discovery Today, 2023, 28(4), 103496, DOI:10.1016/J.DRUDIS.2023.103496.
- M. A. Sellwood, M. Ahmed, M. H. S. Segler and N. Brown, Artificial Intelligence in Drug Discovery, Future Med. Chem., 2018, 10(17), 2025–2028, DOI:10.4155/fmc-2018–0212.
- S. H. Lee and J. H. Sung, Microtechnology-Based Multi-Organ Models, Bioengineering, 2017, 4(2), 46, DOI:10.3390/BIOENGINEERING4020046.
- A. Zhang, K. Meng, Y. Liu, Y. Pan, W. Qu, D. Chen and S. Xie, Absorption, Distribution, Metabolism, and Excretion of Nanocarriers in Vivo and Their Influences, Adv. Colloid Interface Sci., 2020, 284, 102261, DOI:10.1016/j.cis.2020.102261.
- M. Li, K. T. Al-Jamal, K. Kostarelos and J. Reineke, Physiologically Based Pharmacokinetic Modeling of Dermal Absorption, ASC Nano, 2010, 4(11), 6303–6317, DOI:10.3109/9781841848570.037.
- L. H. Chi, A. D. Burrows and R. L. Anderson, Can Preclinical Drug Development Help to Predict Adverse Events in Clinical Trials?, Drug Discovery Today, 2022, 27(1), 257–268, DOI:10.1016/J.DRUDIS.2021.08.010.
- H. J. Freeman and A. B. R. Thomson, The Small Intestine, Clin. Gastroenterol., 2006, 14(4), 689–723, DOI:10.1097/00000441-196608000-00027.
- K. Thelen and J. B. Dressman, Cytochrome P450-Mediated Metabolism in the Human Gut Wall, J. Pharm. Pharmacol., 2010, 61(5), 541–558, DOI:10.1211/JPP.61.05.0002.
- M. Herath, S. Hosie, J. C. Bornstein, A. E. Franks and E. L. Hill-Yardin, The Role of the Gastrointestinal Mucus System in Intestinal Homeostasis: Implications for Neurological Disorders, Front. Cell. Infect. Microbiol., 2020, 10, 248, DOI:10.3389/fcimb.2020.00248.
- F. M. Gribble and F. Reimann, Enteroendocrine Cells: Chemosensors in the Intestinal Epithelium, Annu. Rev. Physiol., 2016, 78, 277–299, DOI:10.1146/annurev-physiol-021115–105439.
- N. Zeitouni, J. Fandrey, H. Y. Naim and M. von Köckritz-Blickwede, Measuring Oxygen Levels in Caco-2 Cultures, Hypoxia, 2015, 53, DOI:10.2147/hp.s85625.
- H. J. Kim, D. Huh, G. Hamilton and D. E. Ingber, Human Gut-on-a-Chip Inhabited by Microbial Flora That Experiences Intestinal Peristalsis-like Motions and Flow, Lab Chip, 2012, 12(12), 2165–2174, 10.1039/c2lc40074j.
- M. Ghadiri, P. M. Young and D. Traini, Strategies to Enhance Drug Absorption via Nasal and Pulmonary Routes, Pharmaceutics, 2019, 11(3), 113, DOI:10.3390/PHARMACEUTICS11030113.
- P. Nighot and T. Ma, Endocytosis of Intestinal Tight Junction Proteins: In Time and Space, Inflammatory Bowel Dis., 2021, 27(2), 283–290, DOI:10.1093/IBD/IZAA141.
- N. Noureddine, M. Chalubinski and P. Wawrzyniak, The Role of Defective Epithelial Barriers in Allergic Lung Disease and Asthma Development, J. Asthma Allergy, 2022, 15, 487–504, DOI:10.2147/JAA.S324080.
- E. Fröhlich, Non-Cellular Layers of the Respiratory Tract: Protection against Pathogens and Target for Drug Delivery, Pharmaceutics, 2022, 14(5), 992, DOI:10.3390/pharmaceutics14050992.
- E. Fröhlich and S. Salar-Behzadi, Toxicological Assessment of Inhaled Nanoparticles: Role of in Vivo, Ex Vivo, in Vitro, and in Silico Studies, Int. J. Mol. Sci., 2014, 15(3), 4795–4822, DOI:10.3390/ijms15034795.
- M. R. Lascia, Polonelli, L, Pulm. Drug Delivery, 2018, 1–67 Search PubMed.
- C. Carvalho, N. Daum and C. M. Lehr, Carrier Interactions with the Biological Barriers of the Lung: Advanced in Vitro Models and Challenges for Pulmonary Drug Delivery, Adv. Drug Delivery Rev., 2014, 75, 129–140, DOI:10.1016/j.addr.2014.05.014.
- B. Button, R. C. Boucher and University of North Carolina Virtual Lung Group, Role of Mechanical Stress in Regulating Airway Surface Hydration and Mucus Clearance Rates, Respir. Physiol. Neurobiol., 2008, 163(1–3), 189–201, DOI:10.1016/J.RESP.2008.04.020.
- V. Phatale, K. K. Vaiphei, S. Jha, D. Patil, M. Agrawal and A. Alexander, Overcoming Skin Barriers through Advanced Transdermal Drug Delivery Approaches, J. Controlled Release, 2022, 351, 361–380, DOI:10.1016/j.jconrel.2022.09.025.
- N. Tiwari, E. R. Osorio-Blanco, A. Sonzogni, D. Esporrín-Ubieto, H. Wang and M. Calderón, Nanocarriers for Skin Applications: Where Do We Stand?, Angew. Chem., Int. Ed., 2022, 61(3), e202107960, DOI:10.1002/anie.202107960.
- M. Vestita, P. Tedeschi and D. Bonamonte, Anatomy and Physiology of the Skin, in Textbook of Plastic and Reconstructive Surgery: Basic Principles and New Perspectives, 2022, pp. 3–13, DOI:10.1007/978-3-030-82335-1_1.
- J. E. Lai-Cheong and J. A. McGrath, Structure and Function of Skin, Hair and Nails, Medicine, 2017, 45(6), 347–351, DOI:10.1016/j.mpmed.2017.03.004.
- C. Gorzelanny, C. Mess, S. W. Schneider, V. Huck and J. M. Brandner, Skin Barriers in Dermal Drug Delivery: Which Barriers Have to Be Overcome and How Can We Measure Them?, Pharmaceutics, 2020, 12(7), 684, DOI:10.3390/pharmaceutics12070684.
- U. Blume-Peytavi and A. Vogt, Human Hair Follicle: Reservoir Function and Selective Targeting, Br. J. Dermatol., 2011, 165(s2), 13–17, DOI:10.1111/j.1365–2133.2011.10572.x.
- A. J. Foster, B. Chouhan, S. L. Regan, H. Rollison, S. Amberntsson, L. C. Andersson, A. Srivastava, M. Darnell, J. Cairns, S. E. Lazic, K. J. Jang, D. B. Petropolis, K. Kodella, J. E. Rubins, D. Williams, G. A. Hamilton, L. Ewart and P. Morgan, Integrated in Vitro Models for Hepatic Safety and Metabolism: Evaluation of a Human Liver-Chip and Liver Spheroid, Arch. Toxicol., 2019, 93(4), 1021–1037, DOI:10.1007/s00204-019-02427-4.
- A. Corsini and M. Bortolini, Drug-Induced Liver Injury: The Role of Drug Metabolism and Transport, J. Clin. Pharmacol., 2013, 53(5), 463–474, DOI:10.1002/jcph.23.
- B. Corrado, V. De Gregorio, G. Imparato, C. Attanasio, F. Urciuolo and P. A. Netti, A Three-Dimensional Microfluidized Liver System to Assess Hepatic Drug Metabolism and Hepatotoxicity, Biotechnol. Bioeng., 2019, 116(5), 1152–1163, DOI:10.1002/BIT.26902.
- E. Trefts, M. Gannon and D. H. Wasserman, The Liver, Curr. Biol., 2017, 27(21), R1147–R1151, DOI:10.1016/j.cub.2017.09.019.
- F. Tasnim, J. Xing, X. Huang, S. Mo, X. Wei, M. H. Tan and H. Yu, Generation of Mature Kupffer Cells from Human Induced Pluripotent Stem Cells, Biomaterials, 2019, 192, 377–391, DOI:10.1016/j.biomaterials.2018.11.016.
- Y. Mizuguchi, S. Specht, K. Isse, J. G. Lunz and A. J. Demetris, Biliary Epithelial Cells, Molecular pathology of liver diseases, 2011, vol. 5, pp. 27–51, DOI:10.1007/978-1-4419-7107-4_4.
- K. Tsaioun, B. J. Blaauboer and T. Hartung, Evidence-Based Absorption, Distribution, Metabolism, Excretion (ADME) and Its Interplay with Alternative Toxicity Methods, ALTEX, 2016, 33(4), 343–358, DOI:10.14573/altex.1610101.
- J. Poisson, S. Lemoinne, C. Boulanger, F. Durand, R. Moreau, D. Valla and P. E. Rautou, Liver Sinusoidal Endothelial Cells: Physiology and Role in Liver Diseases, J. Hepatol., 2017, 66(1), 212–227, DOI:10.1016/J.JHEP.2016.07.009.
- S. Prill, D. Bavli, G. Levy, E. Ezra, E. Schmälzlin, M. S. Jaeger, M. Schwarz, C. Duschl, M. Cohen and Y. Nahmias, Real-Time Monitoring of Oxygen Uptake in Hepatic Bioreactor Shows CYP450-Independent Mitochondrial Toxicity of Acetaminophen and Amiodarone, Arch. Toxicol., 2016, 90(5), 1181–1191, DOI:10.1007/s00204-015-1537-2.
- P. Mirabelli, L. Coppola and M. Salvatore, Cancer Cell Lines Are Useful Model Systems for Medical Research, Cancers, 2019, 11(8), 1098, DOI:10.3390/cancers11081098.
- V. E. J. M. Palasantzas, I. Tamargo-Rubio, K. Le, J. Slager, C. Wijmenga, I. H. Jonkers, V. Kumar, J. Fu and S. Withoff, IPSC-Derived Organ-on-a-Chip Models for Personalized Human Genetics and Pharmacogenomics Studies, Trends Genet., 2023, 39(4), 268–284, DOI:10.1016/J.TIG.2023.01.002.
- Y. Zhao, S. Landau, S. Okhovatian, C. Liu, R. Xing, Z. Lu, B. Fook, L. Lai, Q. Wu, J. Kieda, K. Cheung, S. Rajasekar, K. Jozani, B. Zhang and M. Radisic, Integrating Organoids and Organ-on-a-Chip Devices, Nat. Rev. Bioeng., 2024, 2(7), 588–608, DOI:10.1038/s44222-024-00207-z.
- S. M. Jung and S. Kim, In Vitro Models of the Small Intestine for Studying Intestinal Diseases, Front. Microbiol., 2022, 12, 767038, DOI:10.3389/FMICB.2021.767038/BIBTEX.
- T. Däullary, F. Imdahl, O. Dietrich, L. Hepp, T. Krammer, C. Fey, W. Neuhaus, M. Metzger, J. Vogel, A. J. Westermann, A. E. Saliba and D. Zdzieblo, A Primary Cell-Based in Vitro Model of the Human Small Intestine Reveals Host Olfactomedin 4 Induction in Response to Salmonella Typhimurium Infection, Gut Microbes, 2023, 15(1), 2186109, DOI:10.1080/19490976.2023.2186109.
- J. Zhang, E. Wehrle, M. Rubert and R. Müller, 3D Bioprinting of Human Tissues: Biofabrication, Bioinks, and Bioreactors, Int. J. Mol. Sci., 2021, 22(8), 3971, DOI:10.3390/IJMS22083971.
- T. Messelmani, L. Morisseau, Y. Sakai, C. Legallais, A. Le Goff, E. Leclerc and R. Jellali, Liver Organ-on-Chip Models for Toxicity Studies and Risk Assessment, Lab Chip, 2022, 22(13), 2423–2450, 10.1039/D2LC00307D.
- A. Skardal, J. Aleman, S. Forsythe, S. Rajan, S. Murphy, M. Devarasetty, N. Pourhabibi Zarandi, G. Nzou, R. Wicks, H. Sadri-Ardekani, C. Bishop, S. Soker, A. Hall, T. Shupe and A. Atala, Drug Compound Screening in Single and Integrated Multi-Organoid Body-on-a-Chip Systems, Biofabrication, 2020, 12(2), 025017, DOI:10.1088/1758–5090/AB6D36.
- J. Taelman, M. Diaz and J. Guiu, Human Intestinal Organoids: Promise and Challenge, Front. Cell Dev. Biol., 2022, 10, 854740, DOI:10.3389/FCELL.2022.854740/BIBTEX.
- C. M. Leung, P. de Haan, K. Ronaldson-Bouchard, G. A. Kim, J. Ko, H. S. Rho, Z. Chen, P. Habibovic, N. L. Jeon, S. Takayama, M. L. Shuler, G. Vunjak-Novakovic, O. Frey, E. Verpoorte and Y. C. Toh, A Guide to the Organ-on-a-Chip, Nat. Rev. Methods Primers, 2022, 2(1), 1–29, DOI:10.1038/s43586-022-00118-6.
- P. Schuller, M. Rothbauer, S. R. A. Kratz, G. Höll, P. Taus, M. Schinnerl, J. Genser, N. Bastus, O. H. Moriones, V. Puntes, B. Huppertz, M. Siwetz, H. Wanzenböck and P. Ertl, A Lab-on-a-Chip System with an Embedded Porous Membrane-Based Impedance Biosensor Array for Nanoparticle Risk Assessment on Placental Bewo Trophoblast Cells, Sens. Actuators, B, 2020, 312, 127946, DOI:10.1016/J.SNB.2020.127946.
- S. Jalili-Firoozinezhad, F. S. Gazzaniga, E. L. Calamari, D. M. Camacho, C. W. Fadel, A. Bein, B. Swenor, B. Nestor, M. J. Cronce, A. Tovaglieri, O. Levy, K. E. Gregory, D. T. Breault, J. M. S. Cabral, D. L. Kasper, R. Novak and D. E. Ingber, A Complex Human Gut Microbiome Cultured in an Anaerobic Intestine-on-a-Chip, Nat. Biomed. Eng., 2019, 3(7), 520–531, DOI:10.1038/s41551-019-0397-0.
- N. Panse and P. M. Gerk, The Caco-2 Model: Modifications and Enhancements to Improve Efficiency and Predictive Performance, Int. J. Pharm., 2022, 624, 122004, DOI:10.1016/J.IJPHARM.2022.122004.
- K. D. Boger, A. E. Sheridan, A. L. Ziegler and A. T. Blikslager, Mechanisms and Modeling of Wound Repair in the Intestinal Epithelium, Tissue Barriers, 2023, 11(2), 2087454, DOI:10.1080/21688370.2022.2087454.
- B. Yi, K. Y. Shim, S. K. Ha, J. Han, H. H. Hoang, I. Choi, S. Park and J. H. Sung, Three-Dimensional in Vitro Gut Model on a Villi-Shaped Collagen Scaffold, BioChip J., 2017, 11(3), 219–231, DOI:10.1007/S13206-017-1307-8.
- J. Creff, L. Malaquin and A. Besson, In Vitro Models of Intestinal Epithelium: Toward Bioengineered Systems, J. Tissue Eng., 2021, 12 DOI:10.1177/2041731420985202.
- S. A. Jelinsky, M. Derksen, E. Bauman, C. S. Verissimo, W. T. M. van Dooremalen, J. L. Roos, C. H. Barón, C. Caballero-Franco, B. G. Johnson, M. G. Rooks, J. Pott, B. Oldenburg, R. G. J. Vries, S. F. Boj, M. T. Kasaian, F. Pourfarzad and C. V. Rosadini, Molecular and Functional Characterization of Human Intestinal Organoids and Monolayers for Modeling Epithelial Barrier, Inflammatory Bowel Dis., 2023, 29(2), 195–206, DOI:10.1093/IBD/IZAC212.
- A. Fedi, C. Vitale, G. Ponschin, S. Ayehunie, M. Fato and S. Scaglione, In Vitro Models Replicating the Human Intestinal Epithelium for Absorption and Metabolism Studies: A Systematic Review, J. Controlled Release, 2021, 335, 247–268, DOI:10.1016/J.JCONREL.2021.05.028.
- M. Kasendra, A. Tovaglieri, A. Sontheimer-Phelps, S. Jalili-Firoozinezhad, A. Bein, A. Chalkiadaki, W. Scholl, C. Zhang, H. Rickner, C. A. Richmond, H. Li, D. T. Breault and D. E. Ingber, Development of a Primary Human Small Intestine-on-a-Chip Using Biopsy-Derived Organoids, Sci. Rep., 2018, 8(1), 1–14, DOI:10.1038/s41598-018-21201-7.
- B. Rothen-Rutishauser, M. Gibb, R. He, A. Petri-Fink and C. M. Sayes, Human Lung Cell Models to Study Aerosol Delivery – Considerations for Model Design and Development, Eur. J. Pharm. Sci., 2023, 180, 106337, DOI:10.1016/J.EJPS.2022.106337.
- D. F. Lee, M. I. Lethem and A. B. Lansley, A Comparison of Three Mucus-Secreting Airway Cell Lines(Calu-3, SPOC1 and UNCN3T) for Use as Biopharmaceutical Models of the Nose and Lung, Eur. J. Pharm. Biopharm., 2021, 167, 159–174, DOI:10.1016/j.ejpb.2021.07.016.
- N. Sibinovska, S. Žakelj, R. Roškar and K. Kristan, Suitability and Functional Characterization of Two Calu-3 Cell Models for Prediction of Drug Permeability across the Airway Epithelial Barrier, Int. J. Pharm., 2020, 585, 119484, DOI:10.1016/j.ijpharm.2020.119484.
- N. Petpiroon, W. Netkueakul, K. Sukrak, C. Wang, Y. Liang, M. Wang, Y. Liu, Q. Li, R. Kamran, K. Naruse, S. Aueviriyavit and K. Takahashi, Development of Lung Tissue Models and Their Applications, Life Sci., 2023, 334, 122208, DOI:10.1016/J.LFS.2023.122208.
- P. R. Tata, H. Mou, A. Pardo-Saganta, R. Zhao, M. Prabhu, B. M. Law, V. Vinarsky, J. L. Cho, S. Breton, A. Sahay, B. D. Medoff and J. Rajagopal, Dedifferentiation of Committed Epithelial Cells into Stem Cells in Vivo, Nature, 2013, 503(7475), 218–223, DOI:10.1038/nature12777.
- R. E. Rayner, P. Makena, G. L. Prasad and E. Cormet-Boyaka, Optimization of Normal Human Bronchial Epithelial(NHBE) Cell 3D Cultures for in Vitro Lung Model Studies, Sci. Rep., 2019, 9(1), 500, DOI:10.1038/S41598-018-36735-Z.
- A. A. Pezzulo, T. D. Starner, T. E. Scheetz, G. L. Traver, A. E. Tilley, B. G. Harvey, R. G. Crystal, P. B. McCray and J. Zabner, The Air-Liquid Interface and Use of Primary Cell Cultures Are Important to Recapitulate the Transcriptional Profile of in Vivo Airway Epithelia, Am. J. Physiol., 2011, 300(1), 25–31, DOI:10.1152/AJPLUNG.00256.2010.
- P. S. Hiemstra, G. Grootaers, A. M. van der Does, C. A. M. Krul and I. M. Kooter, Human Lung Epithelial Cell Cultures for Analysis of Inhaled Toxicants: Lessons Learned and Future Directions, Toxicol. In Vitro, 2018, 47, 137–146, DOI:10.1016/J.TIV.2017.11.005.
- I. M. Kooter, M. Gröllers-Mulderij, M. Steenhof, E. Duistermaat, F. A. A. van Acker, Y. C. M. Staal, P. C. Tromp, E. Schoen, C. F. Kuper and E. van Someren, Cellular Effects in an In Vitro Human 3D Cellular Airway Model and A549/BEAS-2B In Vitro Cell Cultures Following Air Exposure to Cerium Oxide Particles at an Air–Liquid Interface, Applied In Vitro Toxicology, 2016, 2(1), 56–66, DOI:10.1089/AIVT.2015.0030.
- J. Zavala, B. Obrien, K. Lichtveld, K. G. Sexton, I. Rusyn, I. Jaspers and W. Vizuete, Assessment of Biological Responses of EpiAirway 3-D Cell Constructs versus A549 Cells for Determining Toxicity of Ambient Air Pollution, Inhalation Toxicol., 2016, 28(6), 251–259, DOI:10.3109/08958378.2016.1157227.
- K. Zscheppang, J. Berg, S. Hedtrich, L. Verheyen, D. E. Wagner, N. Suttorp, S. Hippenstiel and A. C. Hocke, Human Pulmonary 3D Models For Translational Research, Biotechnol. J., 2018, 13(1), 1700341, DOI:10.1002/BIOT.201700341.
- A. Doryab, J. Groll, A. Doryab and J. Groll, Biomimetic In Vitro Lung Models: Current Challenges and Future Perspective, Adv. Mater., 2023, 35(13), 2210519, DOI:10.1002/ADMA.202210519.
- D. Huh, B. D. Matthews, A. Mammoto, M. Montoya-Zavala, H. Yuan Hsin and D. E. Ingber, Reconstituting Organ-Level Lung Functions on a Chip, Science, 2010, 328(5986), 1662–1668, DOI:10.1126/science.1188302.
- D. Huh, D. C. Leslie, B. D. Matthews, J. P. Fraser, S. Jurek, G. A. Hamilton, K. S. Thorneloe, M. A. McAlexander and D. E. Ingber, A Human Disease Model of Drug Toxicity–Induced Pulmonary Edema in a Lung-on-a-Chip Microdevice, Sci. Transl. Med., 2012, 4(159), 159ra147, DOI:10.1126/SCITRANSLMED.3004249.
- P. Zamprogno, S. Wüthrich, S. Achenbach, G. Thoma, J. D. Stucki, N. Hobi, N. Schneider-Daum, C. M. Lehr, H. Huwer, T. Geiser, R. A. Schmid and O. T. Guenat, Second-Generation Lung-on-a-Chip with an Array of Stretchable Alveoli Made with a Biological Membrane, Commun. Biol., 2021, 4(1), 168, DOI:10.1038/S42003-021-01695-0.
- M. Humayun, C. W. Chow and E. W. K. Young, Microfluidic Lung Airway-on-a-Chip with Arrayable Suspended Gels for Studying Epithelial and Smooth Muscle Cell Interactions, Lab Chip, 2018, 18(9), 1298–1309, 10.1039/C7LC01357D.
- J. D. Stucki, N. Hobi, A. Galimov, A. O. Stucki, N. Schneider-Daum, C. M. Lehr, H. Huwer, M. Frick, M. Funke-Chambour, T. Geiser and O. T. Guenat, Medium Throughput Breathing Human Primary Cell Alveolus-on-Chip Model, Sci. Rep., 2018, 8(1), 1–13, DOI:10.1038/s41598-018-32523-x.
- A. Moreira, M. Müller, P. F. Costa and Y. Kohl, Advanced In Vitro Lung Models for Drug and Toxicity Screening: The Promising Role of Induced Pluripotent Stem Cells, Adv. Biol., 2022, 6(2), 2101139, DOI:10.1002/ADBI.202101139.
- S. Yadav, K. Fujimoto, T. Takenaga, S. Takahashi, Y. Muramoto, R. Mikawa, T. Noda and S. Gotoh, Yokokawa Isogenic IPSC-Derived Proximal and Distal Lung-on-Chip Models: Tissue- and Virus-Specific Immune Responses in Human Lungs, bioRxiv, 2023, preprint, DOI:10.1101/2023.11.24.568532.
- M. Schäfer-Korting, U. Bock, W. Diembeck, H. J. Düsing, A. Gamer, E. Haltner-Ukomadu, C. Hoffmann, M. Kaca, H. Kamp, S. Kersen, M. Kietzmann, H. C. Korting, H. U. Krächter, C. M. Lehr, M. Liebsch, A. Mehling, C. Müller-Goymann, F. Netzlaff, F. Niedorf, M. K. Rübbelke, U. Schäfer, E. Schmidt, S. Schreiber, H. Spielmann, A. Vuia and M. Weimer, The Use of Reconstructed Human Epidermis for Skin Absorption Testing: Results of the Validation Study, ATLA, Altern. Lab. Anim., 2008, 36(2), 161–187, DOI:10.1177/026119290803600207.
- S. H. Mathes, H. Ruffner and U. Graf-Hausner, The Use of Skin Models in Drug Development, Adv. Drug Delivery Rev., 2014, 69–70, 81–102, DOI:10.1016/j.addr.2013.12.006.
- E. Hofmann, A. Schwarz, J. Fink, L.-P. Kamolz and P. Kotzbeck, Modelling the Complexity of Human Skin In Vitro, Biomedicines, 2023, 11(3), 794, DOI:10.3390/biomedicines11030794.
- E. Footner, K. Firipis, E. Liu, C. Baker, P. Foley, R. M. I. Kapsa, E. Pirogova, C. O'Connell and A. Quigley, Layer-by-Layer Analysis of In Vitro Skin Models, ACS Biomater. Sci. Eng., 2023, 9(11), 5933–5952, DOI:10.1021/acsbiomaterials.3c00283.
- E. Footner, K. Firipis, E. Liu, C. Baker, P. Foley, R. M. I. Kapsa, E. Pirogova, C. O'Connell and A. Quigley, Layer-by-Layer Analysis of In Vitro Skin Models, ACS Biomater. Sci. Eng., 2023, 9(11), 5933–5952, DOI:10.1021/acsbiomaterials.3c00283.
- S. Salameh, N. Tissot, K. Cache, J. Lima, I. Suzuki, P. A. Marinho, M. Rielland, J. Soeur, S. Takeuchi, S. Germain and L. Breton, A Perfusable Vascularized Full-Thickness Skin Model for Potential Topical and Systemic Applications, Biofabrication, 2021, 13(3), 035042, DOI:10.1088/1758–5090/abfca8.
- P. Zoio, S. Lopes-Ventura, J. Marto and A. Oliva, Open-Source Human Skin Model with an in Vivo-like Barrier for Drug Testing, ALTEX, 2022, 39(3), 405–418, DOI:10.14573/altex.2111182.
- F. F. Schmidt, S. Nowakowski and P. J. Kluger, Improvement of a Three-Layered in Vitro Skin Model for Topical Application of Irritating Substances, Front. Bioeng. Biotechnol., 2020, 8, 1–11, DOI:10.3389/fbioe.2020.00388.
- B. Huber, A. Link, K. Linke, S. A. Gehrke, M. Winnefeld and P. J. Kluger, Integration of Mature Adipocytes to Build-Up a Functional Three-Layered Full-Skin Equivalent, Tissue Eng., Part C, 2016, 22(8), 756–764, DOI:10.1089/ten.tec.2016.0141.
- J. Kober, A. Gugerell, M. Schmid, L.-P. Kamolz and M. Keck, Generation of a Fibrin Based Three-Layered Skin Substitute, BioMed Res. Int., 2015, 2015(1), 170427, DOI:10.1155/2015/170427.
- Test No. 439: In Vitro Skin Irritation: Reconstructed Human Epidermis Test Method, OECD Guidelines for the Testing of Chemicals, 2021, Section 4, DOI:10.1787/9789264242845-en.
- H. Zhao, Z. Chen, X. Kang, B. Yang, P. Luo, H. Li and Q. He, The Frontline of Alternatives to Animal Testing: Novel in Vitro Skin Model Application in Drug Development and Evaluation, Toxicol. Sci., 2023, 196(2), 152–169, DOI:10.1093/toxsci/kfad093.
- K. Ackermann, S. Lombardi Borgia, H. C. Korting, K. R. Mewes and M. Schäfer-Korting, The Phenion® Full-Thickness Skin Model for Percutaneous Absorption Testing, Skin Pharmacol. Physiol., 2009, 23(2), 105–112, DOI:10.1159/000265681.
- F. F. Schmidt, S. Nowakowski and P. J. Kluger, Improvement of a Three-Layered in Vitro Skin Model for Topical Application of Irritating Substances, Front. Bioeng. Biotechnol., 2020, 8, 1–11, DOI:10.3389/fbioe.2020.00388.
- N. Mori, Y. Morimoto and S. Takeuchi, Skin Integrated with Perfusable Vascular Channels on a Chip, Biomaterials, 2017, 116, 48–56, DOI:10.1016/j.biomaterials.2016.11.031.
- S. Salameh, N. Tissot, K. Cache, J. Lima, I. Suzuki, P. A. Marinho, M. Rielland, J. Soeur, S. Takeuchi, S. Germain and L. Breton, A Perfusable Vascularized Full-Thickness Skin Model for Potential Topical and Systemic Applications, Biofabrication, 2021, 13(3), 035042, DOI:10.1088/1758-5090/abfca8.
- N. Ismayilzada, C. Tarar, S. R. Dabbagh, B. K. Tokyay, S. A. Dilmani, E. Sokullu, H. E. Abaci and S. Tasoglu, Skin-on-a-Chip Technologies towards Clinical Translation and Commercialization, Biofabrication, 2024, 16(4), 042001, DOI:10.1088/1758-5090/ad5f55.
- B. Ataç, I. Wagner, R. Horland, R. Lauster, U. Marx, A. G. Tonevitsky, R. P. Azar and G. Lindner, Skin and Hair On-a-Chip: In Vitro Skin Models versus Ex Vivo Tissue Maintenance with Dynamic Perfusion, Lab Chip, 2013, 13(18), 3555–3561, 10.1039/C3LC50227A.
- K. Kim, J. Kim, H. Kim and G. Y. Sung, Effect of α-Lipoic Acid on the Development of Human Skin Equivalents Using a Pumpless Skin-on-a-Chip Model, Int. J. Mol. Sci., 2021, 22(4), 2160, DOI:10.3390/ijms22042160.
- H. E. Abaci, K. Gledhill, Z. Guo, A. M. Christiano and M. L. Shuler, Pumpless Microfluidic Platform for Drug Testing on Human Skin Equivalents, Lab Chip, 2015, 15(3), 882–888, 10.1039/C4LC00999A.
- Z.-X. Hong, S.-T. Zhu, H. Li, J.-Z. Luo, Y. Yang, Y. An, X. Wang and K. Wang, Bioengineered Skin Organoids: From Development to Applications, Mil. Med. Res., 2023, 10(1), 40, DOI:10.1186/s40779-023-00475-7.
- K. S. Vellonen, M. Malinen, E. Mannermaa, A. Subrizi, E. Toropainen, Y. R. Lou, H. Kidron, M. Yliperttula and A. Urtti, A Critical Assessment of in Vitro Tissue Models for ADME and Drug Delivery, J. Controlled Release, 2014, 190, 94–114, DOI:10.1016/j.jconrel.2014.06.044.
- C. C. Bell, D. F. G. Hendriks, S. M. L. Moro, E. Ellis, J. Walsh, A. Renblom, L. Fredriksson Puigvert, A. C. A. Dankers, F. Jacobs, J. Snoeys, R. L. Sison-Young, R. E. Jenkins, Å. Nordling, S. Mkrtchian, B. K. Park, N. R. Kitteringham, C. E. P. Goldring, V. M. Lauschke and M. Ingelman-Sundberg, Characterization of Primary Human Hepatocyte Spheroids as a Model System for Drug-Induced Liver Injury, Liver Function and Disease, Sci. Rep., 2016, 6, 1–13, DOI:10.1038/srep25187.
- S. D. Collins, G. Yuen, T. Tu, M. A. Budzinska, K. Spring, K. Bryant and N. A. Shackel, In Vitro Models of the Liver: Disease Modeling, Drug Discovery and Clinical Applications, Hepatocell. Carcinoma, 2019, 47–67, DOI:10.15586/hepatocellularcarcinoma.2019.
- C. Eilenberger, M. Rothbauer, E. K. Ehmoser, P. Ertl and S. Küpcü, Effect of Spheroidal Age on Sorafenib Diffusivity and Toxicity in a 3D HepG2 Spheroid Model, Sci. Rep., 2019, 9(1), 1–11, DOI:10.1038/s41598-019-41273-3.
- J. W. Yang, D. Khorsandi, L. Trabucco, M. Ahmed, A. Khademhosseini, M. R. Dokmeci, J. Y. Ye and V. Jucaud, Liver-on-a-Chip Integrated with Label-Free Optical Biosensors for Rapid and Continuous Monitoring of Drug-Induced Toxicity, Small, 2024, 2403560, DOI:10.1002/SMLL.202403560.
- Y. Wang, H. Wang, P. Deng, W. Chen, Y. Guo, T. Tao and J. Qin, In Situ Differentiation and Generation of Functional Liver Organoids from Human IPSCs in a 3D Perfusable Chip System, Lab Chip, 2018, 18(23), 3606–3616, 10.1039/C8LC00869H.
- L. Ewart, A. Apostolou, S. A. Briggs, C. V. Carman, J. T. Chaff, A. R. Heng, S. Jadalannagari, J. Janardhanan, K. J. Jang, S. R. Joshipura, M. M. Kadam, M. Kanellias, V. J. Kujala, G. Kulkarni, C. Y. Le, C. Lucchesi, D. V. Manatakis, K. K. Maniar, M. E. Quinn, J. S. Ravan, A. C. Rizos, J. F. K. Sauld, J. D. Sliz, W. Tien-Street, D. R. Trinidad, J. Velez, M. Wendell, O. Irrechukwu, P. K. Mahalingaiah, D. E. Ingber and J. W. Scannell, Levner, D. Performance Assessment and Economic Analysis of a Human Liver-Chip for Predictive Toxicology, Commun. Med., 2022, 2(1), 1–16, DOI:10.1038/s43856-022-00209-1.
- K. M. Bircsak, R. DeBiasio, M. Miedel, A. Alsebahi, R. Reddinger, A. Saleh, T. Shun, L. A. Vernetti and A. Gough, A 3D Microfluidic Liver Model for High Throughput Compound Toxicity Screening in the OrganoPlate®, Toxicology, 2021, 450, 152667, DOI:10.1016/J.TOX.2020.152667.
- E. Moradi, S. Jalili-Firoozinezhad and M. Solati-Hashjin, Microfluidic Organ-on-a-Chip Models of Human Liver Tissue, Acta Biomater., 2020, 116, 67–83, DOI:10.1016/J.ACTBIO.2020.08.041.
- M. Lucchetti, K. Oluwasegun Aina, L. Grandmougin, C. Jäger, P. Pérez Escriva, E. Letellier, A. S. Mosig, P. Wilmes, M. Lucchetti, L. Grandmougin, C. Jäger, P. Wilmes, K. O. Aina, A. S. Mosig, P. Pérez Escriva and E. Letellier, An Organ-on-Chip Platform for Simulating Drug Metabolism Along the Gut–Liver Axis, Adv. Healthcare Mater., 2024, 2303943, DOI:10.1002/ADHM.202303943.
- N. Milani, N. Parrott, D. Ortiz Franyuti, P. Godoy, A. Galetin, M. Gertz and S. Fowler, Application of a Gut–Liver-on-a-Chip Device and Mechanistic Modelling to the Quantitative in Vitro Pharmacokinetic Study of Mycophenolate Mofetil, Lab Chip, 2022, 22(15), 2853–2868, 10.1039/D2LC00276K.
- C. W. McAleer, C. J. Long, D. Elbrecht, T. Sasserath, L. R. Bridges, J. W. Rumsey, C. Martin, M. Schnepper, Y. Wang, F. Schuler, A. B. Roth, C. Funk, M. L. Shuler and J. J. Hickman, Multi-Organ System for the Evaluation of Efficacy and off-Target Toxicity of Anticancer Therapeutics, Sci. Transl. Med., 2019, 11(497), eaav1386, DOI:10.1126/SCITRANSLMED.AAV1386.
- N. Tsamandouras, W. L. K. Chen, C. D. Edington, C. L. Stokes, L. G. Griffith and M. Cirit, Integrated Gut and Liver Microphysiological Systems for Quantitative In Vitro Pharmacokinetic Studies, AAPS J., 2017, 19(5), 1499–1512, DOI:10.1208/S12248-017-0122-4/FIGURES/8.
- T. Bricks, P. Paullier, A. Legendre, M. J. Fleury, P. Zeller, F. Merlier, P. M. Anton and E. Leclerc, Development of a New Microfluidic Platform Integrating Co-Cultures of Intestinal and Liver Cell Lines, Toxicol. In Vitro, 2014, 28(5), 885–895, DOI:10.1016/J.TIV.2014.02.005.
- J. M. Prot, L. Maciel, T. Bricks, F. Merlier, J. Cotton, P. Paullier, F. Y. Bois and E. Leclerc, First Pass Intestinal and Liver Metabolism of Paracetamol in a Microfluidic Platform Coupled with a Mathematical Modeling as a Means of Evaluating ADME Processes in Humans, Biotechnol. Bioeng., 2014, 111(10), 2027–2040, DOI:10.1002/BIT.25232.
- M. B. Esch, H. Ueno, D. R. Applegate and M. L. Shuler, Modular, Pumpless Body-on-a-Chip Platform for the Co-Culture of GI Tract Epithelium and 3D Primary Liver Tissue, Lab Chip, 2016, 16(14), 2719–2729, 10.1039/C6LC00461J.
- M. Wang, Y. Sasaki, R. Sakagami, T. Minamikawa, M. Tsuda, R. Ueno, S. Deguchi, R. Negoro, K. So, Y. Higuchi, R. Yokokawa, K. Takayama and F. Yamashita, Perfluoropolyether-Based Gut-Liver-on-a-Chip for the Evaluation of First-Pass Metabolism and Oral Bioavailability of Drugs, ACS Biomater. Sci. Eng., 2024, 10, 4635–4644, DOI:10.1021/ACSBIOMATERIALS.4C00605.
- I. Wagner, E.-M. Materne, S. Brincker, U. Süßbier, C. Frädrich, M. Busek, F. Sonntag, D. A. Sakharov, E. V. Trushkin, A. G. Tonevitsky, R. Lauster and U. Marx, A Dynamic Multi-Organ-Chip for Long-Term Cultivation and Substance Testing Proven by 3D Human Liver and Skin Tissue Co-Culture, Lab Chip, 2013, 13(18), 3538–3547, 10.1039/C3LC50234A.
- I. Maschmeyer, T. Hasenberg, A. Jaenicke, M. Lindner, A. K. Lorenz, J. Zech, L. A. Garbe, F. Sonntag, P. Hayden, S. Ayehunie, R. Lauster, U. Marx and E. M. Materne, Chip-Based Human Liver-Intestine and Liver-Skin Co-Cultures--A First Step toward Systemic Repeated Dose Substance Testing in Vitro, Eur. J. Pharm. Biopharm., 2015, 95(Pt A), 77–87, DOI:10.1016/J.EJPB.2015.03.002.
- R. S. N. Tavares, T. P. Tao, I. Maschmeyer, S. S. Maria-Engler, M. Schäfer-Korting, A. Winter, C. Zoschke, R. Lauster, U. Marx and L. R. Gaspar, Toxicity of Topically Applied Drugs beyond Skin Irritation: Static Skin Model vs. Two Organs-on-a-Chip, Int. J. Pharm., 2020, 589, 119788, DOI:10.1016/j.ijpharm.2020.119788.
- J. Kühnl, T. P. Tao, K. Brandmair, S. Gerlach, T. Rings, U. Müller-Vieira, J. Przibilla, C. Genies, C. Jaques-Jamin, A. Schepky, U. Marx, N. J. Hewitt and I. Maschmeyer, Characterization of Application Scenario-Dependent Pharmacokinetics and Pharmacodynamic Properties of Permethrin and Hyperforin in a Dynamic Skin and Liver Multi-Organ-Chip Model, Toxicology, 2021, 448, 152637, DOI:10.1016/j.tox.2020.152637.
- K. Brandmair, T.-P. Tao, S. Gerlach, J. Przibilla, A. Schepky, U. Marx, N. J. Hewitt, J. Kühnl and I. Maschmeyer, Suitability of Different Reconstructed Human Skin Models in the Skin and Liver Chip2 Microphysiological Model to Investigate the Kinetics and First-Pass Skin Metabolism of the Hair Dye, 4-Amino-2-Hydroxytoluene, J. Appl. Toxicol., 2024, 44(3), 333–343, DOI:10.1002/jat.4542.
- T.-P. Tao, K. Brandmair, S. Gerlach, J. Przibilla, A. Schepky, U. Marx, N. J. Hewitt, I. Maschmeyer and J. Kühnl, Application of a Skin and Liver Chip2 Microphysiological Model to Investigate the Route-Dependent Toxicokinetics and Toxicodynamics of Consumer-Relevant Doses of Genistein, J. Appl. Toxicol., 2024, 44(2), 287–300, DOI:10.1002/jat.4540.
- J. S. Lee, J. Kim, B. Cui, S. K. Kim, S.-A. Cho, S. An and S.-W. Cho, Hybrid Skin Chips for Toxicological Evaluation of Chemical Drugs and Cosmetic Compounds, Lab Chip, 2022, 22(2), 343–353, 10.1039/D1LC00550B.
- K. Schimek, S. Frentzel, K. Luettich, D. Bovard, I. Rütschle, L. Boden, F. Rambo, H. Erfurth, E. M. Dehne, A. Winter, U. Marx and J. Hoeng, Human Multi-Organ Chip Co-Culture of Bronchial Lung Culture and Liver Spheroids for Substance Exposure Studies, Sci. Rep., 2020, 10(1), 1–13, DOI:10.1038/s41598-020-64219-6.
- D. Bovard, A. Sandoz, K. Luettich, S. Frentzel, A. Iskandar, D. Marescotti, K. Trivedi, E. Guedj, Q. Dutertre, M. C. Peitsch and J. Hoeng, A Lung/Liver-on-a-Chip Platform for Acute and Chronic Toxicity Studies, Lab Chip, 2018, 18(24), 3814–3829, 10.1039/C8LC01029C.
- P. G. Miller, C. Y. Chen, Y. I. Wang, E. Gao and M. L. Shuler, Multiorgan Microfluidic Platform with Breathable Lung Chamber for Inhalation or Intravenous Drug Screening and Development, Biotechnol. Bioeng., 2020, 117(2), 486–497, DOI:10.1002/BIT.27188.
- K. Shinha, W. Nihei, T. Ono, R. Nakazato and H. Kimura, A Pharmacokinetic-Pharmacodynamic Model Based on Multi-Organ-on-a-Chip for Drug-Drug Interaction Studies, Biomicrofluidics, 2020, 14(4) DOI:10.1063/5.0011545.
- J. R. Coppeta, M. J. Mescher, B. C. Isenberg, A. J. Spencer, E. S. Kim, A. R. Lever, T. J. Mulhern, R. Prantil-Baun, J. C. Comolli and J. T. Borenstein, A Portable and Reconfigurable Multi-Organ Platform for Drug Development with Onboard Microfluidic Flow Control, Lab Chip, 2016, 17(1), 134–144, 10.1039/C6LC01236A.
- L. Boeri, L. Izzo, L. Sardelli, M. Tunesi, D. Albani and C. Giordano, Advanced Organ-on-a-Chip Devices to Investigate Liver Multi-Organ Communication: Focus on Gut, Microbiota and Brain, Bioengineering, 2019, 6(4), 91, DOI:10.3390/BIOENGINEERING6040091.
- T. Bricks, P. Paullier, A. Legendre, M. J. Fleury, P. Zeller, F. Merlier, P. M. Anton and E. Leclerc, Development of a New Microfluidic Platform Integrating Co-Cultures of Intestinal and Liver Cell Lines, Toxicol. In Vitro, 2014, 28(5), 885–895, DOI:10.1016/J.TIV.2014.02.005.
- J. M. Prot, L. Maciel, T. Bricks, F. Merlier, J. Cotton, P. Paullier, F. Y. Bois and E. Leclerc, First Pass Intestinal and Liver Metabolism of Paracetamol in a Microfluidic Platform Coupled with a Mathematical Modeling as a Means of Evaluating ADME Processes in Humans, Biotechnol. Bioeng., 2014, 111(10), 2027–2040, DOI:10.1002/BIT.25232.
- M. B. Esch, H. Ueno, D. R. Applegate and M. L. Shuler, Modular, Pumpless Body-on-a-Chip Platform for the Co-Culture of GI Tract Epithelium and 3D Primary Liver Tissue, Lab Chip, 2016, 16(14), 2719–2729, 10.1039/C6LC00461J.
- I. Maschmeyer, T. Hasenberg, A. Jaenicke, M. Lindner, A. K. Lorenz, J. Zech, L. A. Garbe, F. Sonntag, P. Hayden, S. Ayehunie, R. Lauster, U. Marx and E. M. Materne, Chip-Based Human Liver-Intestine and Liver-Skin Co-Cultures--A First Step toward Systemic Repeated Dose Substance Testing in Vitro, Eur. J. Pharm. Biopharm., 2015, 95(Pt A), 77–87, DOI:10.1016/J.EJPB.2015.03.002.
- I. Wagner, E.-M. Materne, S. Brincker, U. Süßbier, C. Frädrich, M. Busek, F. Sonntag, D. A. Sakharov, E. V. Trushkin, A. G. Tonevitsky, R. Lauster and U. Marx, A Dynamic Multi-Organ-Chip for Long-Term Cultivation and Substance Testing Proven by 3D Human Liver and Skin Tissue Co-Culture, Lab Chip, 2013, 13(18), 3538–3547, 10.1039/C3LC50234A.
- J. Hübner, M. Raschke, I. Rütschle, S. Gräßle, T. Hasenberg, K. Schirrmann, A. Lorenz, S. Schnurre, R. Lauster, I. Maschmeyer, T. Steger-Hartmann and U. Marx, Simultaneous Evaluation of Anti-EGFR-Induced Tumour and Adverse Skin Effects in a Microfluidic Human 3D Co-Culture Model, Sci. Rep., 2018, 8(1), 15010, DOI:10.1038/s41598-018-33462-3.
- J. Theobald, M. A. Abu el Maaty, N. Kusterer, B. Wetterauer, M. Wink, X. Cheng and S. Wölfl, In Vitro Metabolic Activation of Vitamin D3 by Using a Multi-Compartment Microfluidic Liver-Kidney Organ on Chip Platform, Sci. Rep., 2019, 9(1), 1–11, DOI:10.1038/s41598-019-40851-9.
- Q. Huang, T. Yang, Y. Song, W. Sun, J. Xu, Y. Cheng, R. Yin, L. Zhu, M. Zhang, L. Ma, H. Li and H. Zhang, A Three-Dimensional(3D) Liver–Kidney on a Chip with a Biomimicking Circulating System for Drug Safety Evaluation, Lab Chip, 2024, 24(6), 1715–1726, 10.1039/D3LC00980G.
- P. Mamoshina, B. Rodriguez and A. Bueno-Orovio, Toward a Broader View of Mechanisms of Drug Cardiotoxicity, Cell Rep. Med., 2021, 2(3), 100216, DOI:10.1016/J.XCRM.2021.100216.
- Y. Baert, I. Ruetschle, W. Cools, A. Oehme, A. Lorenz, U. Marx, E. Goossens and I. Maschmeyer, A Multi-Organ-Chip Co-Culture of Liver and Testis Equivalents: A First Step toward a Systemic Male Reprotoxicity Model, Hum. Reprod., 2020, 35(5), 1029–1044, DOI:10.1093/HUMREP/DEAA057.
- C. Oleaga, A. Riu, S. Rothemund, A. Lavado, C. W. McAleer, C. J. Long, K. Persaud, N. S. Narasimhan, M. Tran, J. Roles, C. A. Carmona-Moran, T. Sasserath, D. H. Elbrecht, L. Kumanchik, L. R. Bridges, C. Martin, M. T. Schnepper, G. Ekman, M. Jackson, Y. I. Wang, R. Note, J. Langer, S. Teissier and J. J. Hickman, Investigation of the Effect of Hepatic Metabolism on Off-Target Cardiotoxicity in a Multi-Organ Human-on-a-Chip System, Biomaterials, 2018, 182, 176–190, DOI:10.1016/J.BIOMATERIALS.2018.07.062.
- C. P. P. de Mello, C. Carmona-Moran, C. W. McAleer, J. Perez, E. A. Coln, C. J. Long, C. Oleaga, A. Riu, R. Note, S. Teissier, J. Langer and J. J. Hickman, Microphysiological Heart–Liver Body-on-a-Chip System with a Skin Mimic for Evaluating Topical Drug Delivery, Lab Chip, 2020, 20(4), 749–759, 10.1039/C9LC00861F.
- F. Yin, X. Zhang, L. Wang, Y. Wang, Y. Zhu, Z. Li, T. Tao, W. Chen, H. Yu and J. Qin, HiPSC-Derived Multi-Organoids-on-Chip System for Safety Assessment of Antidepressant Drugs, Lab Chip, 2021, 21(3), 571–581, 10.1039/D0LC00921K.
- A. Skardal, S. V. Murphy, M. Devarasetty, I. Mead, H. W. Kang, Y. J. Seol, Y. S. Zhang, S. R. Shin, L. Zhao, J. Aleman, A. R. Hall, T. D. Shupe, A. Kleensang, M. R. Dokmeci, S. Jin Lee, J. D. Jackson, J. J. Yoo, T. Hartung, A. Khademhosseini, S. Soker, C. E. Bishop and A. Atala, Multi-Tissue Interactions in an Integrated Three-Tissue Organ-on-a-Chip Platform, Sci. Rep., 2017, 7(1), 1–16, DOI:10.1038/s41598-017-08879-x.
- K. Ronaldson-Bouchard, D. Teles, K. Yeager, D. N. Tavakol, Y. Zhao, A. Chramiec, S. Tagore, M. Summers, S. Stylianos, M. Tamargo, B. M. Lee, S. P. Halligan, E. H. Abaci, Z. Guo, J. Jacków, A. Pappalardo, J. Shih, R. K. Soni, S. Sonar, C. German, A. M. Christiano, A. Califano, K. K. Hirschi, C. S. Chen, A. Przekwas and G. Vunjak-Novakovic, A Multi-Organ Chip with Matured Tissue Niches Linked by Vascular Flow, Nat. Biomed. Eng., 2022, 6(4), 351–371, DOI:10.1038/s41551-022-00882-6.
- A. Herland, B. M. Maoz, D. Das, M. R. Somayaji, R. Prantil-Baun, R. Novak, M. Cronce, T. Huffstater, S. S. F. Jeanty, M. Ingram, A. Chalkiadaki, D. Benson Chou, S. Marquez, A. Delahanty, S. Jalili-Firoozinezhad, Y. Milton, A. Sontheimer-Phelps, B. Swenor, O. Levy, K. K. Parker, A. Przekwas and D. E. Ingber, Quantitative Prediction of Human Pharmacokinetic Responses to Drugs via Fluidically Coupled Vascularized Organ Chips, Nat. Biomed. Eng., 2020, 4(4), 421–436, DOI:10.1038/s41551-019-0498-9.
- C. W. McAleer, C. J. Long, D. Elbrecht, T. Sasserath, L. R. Bridges, J. W. Rumsey, C. Martin, M. Schnepper, Y. Wang, F. Schuler, A. B. Roth, C. Funk, M. L. Shuler and J. J. Hickman, Multi-Organ System for the Evaluation of Efficacy and off-Target Toxicity of Anticancer Therapeutics, Sci. Transl. Med., 2019, 11(497), eaav1386, DOI:10.1126/SCITRANSLMED.AAV1386.
- C. Oleaga, C. Bernabini, A. S. T. Smith, B. Srinivasan, M. Jackson, W. Mclamb, V. Platt, R. Bridges, Y. Cai, N. Santhanam, B. Berry, S. Najjar, N. Akanda, X. Guo, C. Martin, G. Ekman, M. B. Esch, J. Langer, G. Ouedraogo, J. Cotovio, L. Breton, M. L. Shuler and J. J. Hickman, Multi-Organ Toxicity Demonstration in a Functional Human in Vitro System Composed of Four Organs, Sci. Rep., 2015, 6(1), 20030, DOI:10.1038/srep20030.
- T. Satoh, S. Sugiura, K. Shin, R. Onuki-Nagasaki, S. Ishida, K. Kikuchi, M. Kakiki and T. Kanamori, A Multi-Throughput Multi-Organ-on-a-Chip System on a Plate Formatted Pneumatic Pressure-Driven Medium Circulation Platform, Lab Chip, 2017, 18(1), 115–125, 10.1039/C7LC00952F.
- C. D. Edington, W. L. K. Chen, E. Geishecker, T. Kassis, L. R. Soenksen, B. M. Bhushan, D. Freake, J. Kirschner, C. Maass, N. Tsamandouras, J. Valdez, C. D. Cook, T. Parent, S. Snyder, J. Yu, E. Suter, M. Shockley, J. Velazquez, J. J. Velazquez, L. Stockdale, J. P. Papps, I. Lee, N. Vann, M. Gamboa, M. E. Labarge, Z. Zhong, X. Wang, L. A. Boyer, D. A. Lauffenburger, R. L. Carrier, C. Communal, S. R. Tannenbaum, C. L. Stokes, D. J. Hughes, G. Rohatgi, D. L. Trumper and M. Cirit, Griffith, L. G. Interconnected Microphysiological Systems for Quantitative Biology and Pharmacology Studies, Sci. Rep., 2018, 8(1), 1–18, DOI:10.1038/s41598-018-22749-0.
- Q. Ramadan, R. S. Fardous, R. Hazaymeh, S. Alshmmari and M. Zourob, Pharmacokinetics-On-a-Chip: In Vitro Microphysiological Models for Emulating of Drugs ADME, Adv. Biol., 2021, 5(9), 2100775, DOI:10.1002/ADBI.202100775.
- S. Fowler, W. L. K. Chen, D. B. Duignan, A. Gupta, N. Hariparsad, J. R. Kenny, W. G. Lai, J. Liras, J. A. Phillips and J. Gan, Microphysiological Systems for ADME-Related Applications: Current Status and Recommendations for System Development and Characterization, Lab Chip, 2020, 20(3), 446–467, 10.1039/C9LC00857H.
- C. Ma, Y. Peng, H. Li and W. Chen, Organ-on-a-Chip: A New Paradigm for Drug Development, Trends Pharmacol. Sci., 2021, 42(2), 119–133, DOI:10.1016/J.TIPS.2020.11.009.
- J. Wu, H. Fang, J. Zhang and S. Yan, Modular Microfluidics for Life Sciences, J. Nanobiotechnol., 2023, 21(1), 85, DOI:10.1186/S12951-023-01846-X.
- X. Lai, M. Yang, H. Wu and D. Li, Modular Microfluidics: Current Status and Future Prospects, Micromachines, 2022, 13(8), 1363, DOI:10.3390/MI13081363.
- L. J. Y. Ong, T. Ching, L. H. Chong, S. Arora, H. Li, M. Hashimoto, R. Dasgupta, P. K. Yuen and Y. C. Toh, Self-Aligning Tetris-Like (TILE) Modular Microfluidic Platform for Mimicking Multi-Organ Interactions, Lab Chip, 2019, 19(13), 2178–2191, 10.1039/C9LC00160C.
- D. J. Carvalho, A. M. Kip, A. Tegel, M. Stich, C. Krause, M. Romitti, C. Branca, B. Verhoeven, S. Costagliola, L. Moroni and S. Giselbrecht, A Modular Microfluidic Organoid Platform Using LEGO-Like Bricks, Adv. Healthcare Mater., 2024, 13(13), 2303444, DOI:10.1002/ADHM.202303444.
- I. Koh and M. Hagiwara, Modular Tissue-in-a-CUBE Platform to Model Blood-Brain Barrier (BBB) and Brain Interaction, Commun. Biol., 2024, 7(1), 1–13, DOI:10.1038/s42003-024-05857-8.
- Q. Sun, J. Pei, Q. Li, K. Niu and X. Wang, Reusable Standardized Universal Interface Module (RSUIM) for Generic Organ-on-a-Chip Applications, Micromachines, 2019, 10(12), 849, DOI:10.3390/MI10120849.
- Y. Hou, X. Ai, L. Zhao, Z. Gao, Y. Wang, Y. Lu, P. Tu and Y. Jiang, An Integrated Biomimetic Array Chip for High-Throughput Co-Culture of Liver and Tumor Microtissues for Advanced Anticancer Bioactivity Screening, Lab Chip, 2020, 20(14), 2482–2494, 10.1039/D0LC00288G.
- R. Novak, M. Ingram, S. Marquez, D. Das, A. Delahanty, A. Herland, B. M. Maoz, S. S. F. Jeanty, M. R. Somayaji, M. Burt, E. Calamari, A. Chalkiadaki, A. Cho, Y. Choe, D. B. Chou, M. Cronce, S. Dauth, T. Divic, J. Fernandez-Alcon, T. Ferrante, J. Ferrier, E. A. FitzGerald, R. Fleming, S. Jalili-Firoozinezhad, T. Grevesse, J. A. Goss, T. Hamkins-Indik, O. Henry, C. Hinojosa, T. Huffstater, K. J. Jang, V. Kujala, L. Leng, R. Mannix, Y. Milton, J. Nawroth, B. A. Nestor, C. F. Ng, B. O'Connor, T. E. Park, H. Sanchez, J. Sliz, A. Sontheimer-Phelps, B. Swenor, G. Thompson, G. J. Touloumes, Z. Tranchemontagne, N. Wen, M. Yadid, A. Bahinski, G. A. Hamilton, D. Levner, O. Levy, A. Przekwas, R. Prantil-Baun, K. K. Parker and D. E. Ingber, Robotic Fluidic Coupling and Interrogation of Multiple Vascularized Organ Chips, Nat. Biomed. Eng., 2020, 4(4), 407–420, DOI:10.1038/s41551-019-0497-x.
- H. Azizgolshani, J. R. Coppeta, E. M. Vedula, E. E. Marr, B. P. Cain, R. J. Luu, M. P. Lech, S. H. Kann, T. J. Mulhern, V. Tandon, K. Tan, N. J. Haroutunian, P. Keegan, M. Rogers, A. L. Gard, K. B. Baldwin, J. C. de Souza, B. C. Hoefler, S. S. Bale, L. B. Kratchman, A. Zorn, A. Patterson, E. S. Kim, T. A. Petrie, E. L. Wiellette, C. Williams, B. C. Isenberg and J. L. Charest, High-Throughput Organ-on-Chip Platform with Integrated Programmable Fluid Flow and Real-Time Sensing for Complex Tissue Models in Drug Development Workflows, Lab Chip, 2021, 21(8), 1454–1474, 10.1039/D1LC00067E.
- M. T. Rogers, A. L. Gard, R. Gaibler, T. J. Mulhern, R. Strelnikov, H. Azizgolshani, B. P. Cain, B. C. Isenberg, N. J. Haroutunian, N. E. Raustad, P. M. Keegan, M. P. Lech, L. Tomlinson, J. T. Borenstein, J. L. Charest and C. Williams, A High-Throughput Microfluidic Bilayer Co-Culture Platform to Study Endothelial-Pericyte Interactions, Sci. Rep., 2021, 11(1), 1–14, DOI:10.1038/s41598-021-90833-z.
Footnote |
| † Equally contributing authors. |
| This journal is © The Royal Society of Chemistry 2025 |