Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
A combined digital microfluidic test for assessing infection and immunity status for viral disease in saliva†
N. Sathishkumar‡
 ab,
Jose Gilberto Camacho Valenzuela‡
ab,
Jose Gilberto Camacho Valenzuela‡ bc,
Nguyen H. Leab,
Anthony K. C. Yongab,
Martin A. Rossottid,
Joshua Dahmerab,
Alexandros A. Sklavounos
bc,
Nguyen H. Leab,
Anthony K. C. Yongab,
Martin A. Rossottid,
Joshua Dahmerab,
Alexandros A. Sklavounos ab,
Martin Plantee,
Daniel Brassard
ab,
Martin Plantee,
Daniel Brassard fg,
Lidija Malic
fg,
Lidija Malic fgh,
Anna N. Moraitisi,
Ruzica Bigaj,
Imane El Idrissij,
Jamshid Tanhadk,
Jean Labrecquei,
Teodor Veresfg and
Aaron R. Wheeler
fgh,
Anna N. Moraitisi,
Ruzica Bigaj,
Imane El Idrissij,
Jamshid Tanhadk,
Jean Labrecquei,
Teodor Veresfg and
Aaron R. Wheeler *abcg
*abcg
aDepartment of Chemistry, University of Toronto, 80 St. George Street, Toronto, Ontario M5S 3H6, Canada. E-mail: aaron.wheeler@utoronto.ca
bDonnelly Centre for Cellular and Biomolecular Research, University of Toronto, 160 College Street, Toronto, Ontario M5S 3E1, Canada
cInstitute of Biomedical Engineering, University of Toronto, 164 College Street, Toronto, Ontario M5S 3G9, Canada
dHuman Health Therapeutics Research Centre, Life Sciences Division, National Research Council Canada, 100 Sussex Drive, Ottawa, Ontario K1A 0R6, Canada
eHuman Health Therapeutics Research Centre, Life Sciences Division, National Research Council Canada, 6100 Royalmount Avenue, Montreal, Quebec H4P 2R2, Canada
fMedical Devices Research Center, Life Sciences Division, National Research Council Canada, 75 de Mortagne Blvd, Boucherville, Quebec J4B 6Y4, Canada
gCentre for Research and Applications in Fluidic Technologies (CRAFT), University of Toronto, 5 King's College Road, Toronto, ON M5S 1A8, Canada
hDepartment of Biomedical Engineering, McGill University, 775 Rue University, Suite 316, Montreal, Quebec H3A 2B4, Canada
iMedical Devices Research Center, Life Sciences Division, National Research Council Canada, 6100 Royalmount Avenue, Montreal, Quebec H4P 2R2, Canada
jFoundation for Innovative New Diagnostics (FIND), Chemin du Pommier 40, 1218 Grand-Saconnex, Geneva, Switzerland
kDepartment of Biochemistry, Microbiology and Immunology, Faculty of Medicine, University of Ottawa, 451 Smyth Road, Ottawa, Ontario K1H 8M5, Canada
First published on 27th May 2025
Abstract
Population assessments of infection and immunity status are critical for public health response to infectious disease. Most microfluidic tools are developed to assess one or the other – few assess both. This study introduces a multiplexed, fully automated digital microfluidic (DMF) platform designed to detect viral protein as a proxy for infection status and host IgG and IgA antibodies as a marker for immunity status. SARS-CoV-2 and patient saliva were used as a model system to evaluate the concept. Specifically, the infection assay relied on nanobody-based capture and detection agents specific to SARS-CoV-2 trimeric spike protein, with a limit of detection (LOD) of 3.8 ng mL−1 in saliva. And the immunity relied on monoclonal antibodies for host IgG and IgA specific to SARS-CoV-2 spike S1 domain, with LODs of 4.8 ng mL−1 and 13.3 ng mL−1 in saliva, respectively. Clinical validation in saliva samples from human subjects experiencing symptoms (n = 14) showed strong correlation with PCR and commercial ELISA, achieving 100% sensitivity and 100% specificity for infection detection and 100% sensitivity with 91.7% and 90.9% specificity for host IgG and IgA, respectively. These results highlight potential applications for the new system as a portable diagnostic tool for outbreak surveillance and public health management, as a step toward preparing for the next global pandemic.
Introduction
The COVID-19 pandemic highlighted the critical role of diagnostic tests in public health policy, which is vital for controlling disease spread and managing outbreaks. The urgency created by the pandemic spurred substantial advancements in portable microfluidic systems,1 which have the potential to revolutionize infectious disease diagnosis. The new systems that have been developed include nucleic acid amplification tests (NAATs) and viral antigen tests for detecting infection,2–10 and human immunoglobulin (Ig) G/M/A tests for assessing immunity.11–14 Detecting both active infection and immune response allows for epidemiological monitoring of infection and immunity status within populations. This enables targeted interventions and optimized resource allocation by identifying individuals at high risk and low risk of infection, which can guide public health policy and outbreak management. Thus, it is widely understood15–17 that preparedness for future infectious disease outbreaks will require integrated tools accessing infection and immunity simultaneously. There are few such tools described in the literature, which motivated the work described here.Integrating infection and immunity tests requires a single sample type. Among the various candidates, saliva presents advantages as it contains viral RNA, viral antigens, and host antibodies that can be useful for assessing active infection and immune response.18 With particular reference to the COVID-19 pandemic, saliva testing has been shown to provide comparable accuracy to nasopharyngeal swabs for detecting SARS-CoV-2 infection,19 and to plasma in the detection of host IgG and IgA responses to infection.20 These characteristics, along with the ease of collection and the potential for self-sampling, make saliva a particularly attractive choice for the work described here.
In scouring the literature, we have been able to find just three previous reports21–23 of combined microfluidic tests for infection and immunity for SARS-CoV-2 in saliva. These studies represent important milestones in the field, but each suffer from limitations that have precluded widespread adoption and use. For example, Torrente-Rodríguez et al.21 report a paper-based sensor that can detect viral antigen and host IgM/IgG in saliva, but the assay is manual, requiring a long series of pipetting steps to load sample and reagents onto each reaction spot for analysis. On the other hand, Najjar et al.22 present a more automated system for assessing infection/immune response, with integrated, microfluidic processing of saliva samples for detection of viral RNA and (in parallel) host IgG or IgM. But each assay is quite long (up to 120 min), the system requires a built-in heater (increasing the complexity of the instrument), and despite the impressive automation of subsequent steps, the user initiates the assay by loading separate aliquots of sample into the device for the immunity (15 μL diluted saliva) and infection (280 μL diluted saliva) tests. Likewise, Moakhar et al.23 describe a system for simultaneous detection of SARS-CoV-2 antigen and host IgG/IgM with similarly impressive levels of microfluidic automation, but the user must collect and load two types of samples (700 μL saliva for the immunity tests and 3 μL blood for the infection test), making it ill-suited for applications in the field. Finally, none of the aforementioned studies21–23 describe the use of built-in negative controls, which are widely recognized24–26 to be a critical component of portable diagnostics.
Here, we sought to develop a fully automated microfluidic diagnostic for SARS-CoV-2 infection and immunity that (i) only requires the user to load a single aliquot of a single sample and (ii) includes controls necessary for the results to be trusted in applications outside of the lab. The new system (Fig. 1) relies on digital microfluidics27 and is designed to test saliva samples for the presence of viral antigen and host IgG and IgA in minutes. In this work, we describe the selection and use of highly sensitive and specific nanobody pairs28 as capture and detection agents for a magnetic particle-mediated immunoassay to detect SARS-CoV-2 trimeric spike protein, while in parallel, spike S1 domain-coated magnetic beads are used to pull down and detect host IgG and IgA in human saliva samples. We describe how the assays were optimized and automated, and report their application to evaluate clinical saliva samples compared to reference test results. We propose that the new digital microfluidic methods described here offer a robust, user-friendly alternative for comprehensive surveillance, which could form the basis for tools that can contribute to effective public health management in the face of current and future infectious disease threats.
Experimental
Reagents and materials
Dynabeads™ M-270 Epoxy, Dynabeads™ M-280 Streptavidin, and Dynabeads MyOne Streptavidin T1 beads and Antibody Coupling Kit were purchased from Thermo Fisher Scientific (Mississauga, ON, Canada). Biotinylated recombinant SARS-CoV-2 spike S1 domain was obtained from Bioss (Woburn, MA, USA). Human (‘host’) anti-SARS-CoV-2 antibodies IgG (IgG3, Kappa) and IgA (IgA1, Kappa) were obtained from Absolute Antibody (Woburn, MA, USA). Horseradish peroxidase (HRP) conjugated goat anti-human IgG and IgA antibodies were obtained from Southern Biotech (Birmingham, AL, USA). Polyclonal unconjugated goat anti-human IgG was obtained from Southern Biotech. A recombinant trimeric SARS-CoV-2 spike glycoprotein29 (Reference Material SMT1-1, Wuhan; molecular weight 551 kDa) was obtained from the National Research Council Canada. Heavy-chain-only antibodies (VHHs) specific to SARS-CoV-2 were produced in-house as described previously.28 Inactivated cultured SARS-CoV-2 viral samples were provided by the National Microbiology Laboratory (NML) of the Public Health Agency of Canada. Pre-COVID pooled saliva was obtained from MyBioSource (San Diego, CA, USA). Streptavidin-polyHRP conjugate was obtained from Sigma Aldrich (Oakville, ON, Canada). All other reagents, including 1-Step™ TMB (3,3′,5,5′-tetramethylbenzidine) ELISA Substrate, SuperSignal™ ELISA Pico Chemiluminescent Substrate, and SuperBlock™ Blocking Buffer in phosphate-buffered saline (PBS), were obtained from Thermo Fisher Scientific (Mississauga, ON, Canada).Chromium and photoresist-coated glass slides (3 in × 3 in) were purchased from the Telic Company (Valencia, CA, USA). ITO-coated glass slides (25 mm × 75 mm × 0.7 mm) were sourced from Riley Supplies (Richmond Hill, ON, Canada). Parylene-C dimer was supplied by Specialty Coating System (Indianapolis, IN, USA). FluoroPel 1101 V and PFC110 solvent were purchased from Cytonix, LLC (Beltsville, MD, USA). Tetronic 90R4 (BASF Corp., Germany) was generously donated by BASF Corporation (Wyandotte, MI, USA). Saliva samples (n = 14) from Moldovan patients showing COVID-19 symptoms were collected by the Foundation for Innovative New Diagnostics (FIND, Geneva, Switzerland) and were stored at −80 °C until use.
Fabrication and operation of digital microfluidic devices
DMF devices, consisting of two plates separated by a spacer, were fabricated at the Centre for Research and Applications in Fluidic Technologies (CRAFT), following previously described methods30,31 involving UV photolithography and wet etching. Specifically, bottom plates were produced from 3 in ×3 in chromium-coated glass substrates. Patterned bottom plates were coated with a ∼5 μm layer of parylene-C via chemical vapour deposition at the Toronto Nanofabrication Center (TNFC), followed by spin coating a 1% (w/w) solution of FluoroPel PFC 1101 V dissolved in PFC110 at 2000 rpm for 30 seconds. Coated bottom plates were then baked in a dry oven at 110 °C for 15 minutes. Each bottom plate featured an array of seventy-eight offset-cross-shaped actuation electrodes (2.8 mm × 2.8 mm), six rectangular mixing electrodes (5.7 mm × 2.4 mm), six square waste electrodes (2.8 mm × 2.0 mm), ten rectangular dispensing electrodes (5.2 mm × 2.4 mm), and ten reservoir electrodes (10 mm × 6.7 mm), with inter-electrode gaps that ranged from 30–80 μm. Top plates were prepared by dip-coating ITO-coated glass slides with FluoroPel (the same solution indicated for spin-coating) and then heating them in a dry oven at 160 °C for 15 minutes. Devices were assembled by sandwiching the top and bottom plates with spacers formed from two layers of double-sided tape (3 M Co., St. Paul, MN, USA) with a thickness of ∼180 μm. Two layers of conductive copper adhesive tape (3 M Co.) of ∼180 μm thickness was placed onto the ground electrodes connecting the top and bottom plate. The volume of a single unit droplet, defined as a droplet that covers one actuation electrode, was approximately 1.3 μL. Each device was interfaced through pogo-pin connectors to the open-source digital microfluidic control system, DropBot.32 The DropBot instrument used was custom-built, including an integrated, automated magnetic lens and photomultiplier tube (PMT), described in detail elsewhere.33 Droplets were actuated by applying a force of ∼25 μN mm−1 using the open-source MicroDrop 3.0 software, under conditions determined to be below the saturation force34 for all the liquids used.Nanobody selection and characterization
In initial experiments, a panel of VHH antibodies specific to SARS-CoV-2 (ref. 28) was tested in pairs with inactivated virus in an agglutination assay to identify promising candidates, which included ‘VHH 11’ and ‘VHH 1d’. Monovalent (site-specifically biotinylated) and bivalent (fused to the human IgG1 Fc region) versions of these antibodies were then evaluated for their stability and binding properties in saliva as described in ESI† Note S1 and Fig. S1. Ultimately, the pair of reagents, referenced here as “capture nanobody” (VHH 11-Fc) and “detection nanobody” (VHH 1d-biotin) were selected for microfluidic assay development.Preparation of beads for SARS-CoV-2 infection assays
Dynabead™ M-270 Epoxy beads were coated with capture nanobodies using a modified version of the Dynabeads® Antibody Coupling Kit protocol. Briefly, 6 mg of magnetic beads was dispersed in 1 mL of C1 buffer and then pelleted, disposing of the supernatant. A 7.5 μL aliquot of an aqueous solution of capture nanobody (4 mg mL−1) was mixed with 292.5 μL of C1 buffer, and the mixture was used to suspend the pellet of magnetic beads referenced above to achieve a final concentration of 5 μg of nanobody per milligram of beads. 300 μL of C2 buffer was added to the mixture and incubated at 37 °C for 16 to 24 hours without allowing the beads to settle using a Roto-Mini™ Plus Rotator (Benchmark, NJ, USA). After incubation, the beads were washed by pelleting and resuspending in three wash buffers in series (HB, LB, and SB) as recommended by the manufacturer. Finally, the 6 mg of beads was resuspended in 600 μL of SB wash buffer and stored at 4 °C until use.Off-chip SARS-CoV-2 infection assays
Off-chip SARS-CoV-2 infection assays were carried out using a 9-step procedure. (1) A 2 μL aliquot of capture nanobody-coated magnetic bead suspension was added to each tube in a strip of eight tubes. (2) Each sample was pelleted and resuspended in a 50 μL aliquot of PBS SuperBlock™. (3a) A 2 μL aliquot of the sample (SARS-CoV-2 trimeric spike protein spiked in PBS SuperBlock™ or pooled pre-COVID-19 saliva) was added to each tube. (3b) A 2 μL aliquot of secondary reagent (detection nanobody solution, 20 μg mL−1) was added to each tube. (3c) The contents were then mixed using a rotator for 10 minutes. (4) The beads in each tube were then washed three times by pelleting and resuspending in 50 μL aliquots of PBS SuperBlock™. (5) A 2 μL aliquot of aqueous streptavidin-polyHRP solution (typically 5 ng mL−1, see Note S2, Fig. S2†) was added to each tube and mixed for 30 minutes with a rotator. (6) The beads in each tube were then washed three times by pelleting and resuspending in 50 μL aliquots of PBS SuperBlock™. (7) A 5 μL aliquot of aqueous 1-Step TMB™ solution was added to each tube and mixed for 5 minutes. (8) The reactions were quenched by adding a 0.5 μL aliquot of 1 M H2SO4 to each tube. (9) The beads in each tube were then pelletized, and a 2 μL aliquot of the supernatant in each tube was collected for optical density measurement at 450 nm using a NanoDrop spectrophotometer (Thermo Fisher, Mississauga, ON). Calibration curves were generated by fitting with four-parameter logistics (4PL) curve functions. The limits of detection (LOD) and quantification (LOQ) were determined as the concentrations corresponding to the position on the curve of the mean signal generated from blank measurements plus 3 (LOD) or 10 (LOQ) times the standard deviation of the blank measurements (Note 2, Fig. S3†).Preparation of beads for SARS-CoV-2 immunity assays
Dynabeads® M-280 streptavidin-coated paramagnetic beads were selected for implementing immunity assays after optimization (see Note 3, Fig. S4†). Site-specifically biotinylated SARS-CoV-2 spike S1 domain protein (Bioss, MA, USA) was buffer-exchanged into DPBS at 1 μg μL−1 using Zeba™ spin desalting columns (7 kDa MWCO) and immobilized on the beads following a modified version of the manufacturer's protocol. Briefly, 250 μL of bead stock suspension was washed with PBS containing 0.02% (v/v) Tween-20, then incubated overnight at 4 °C with 20 μL of the biotinylated S1 solution. After washing, beads were blocked using 4% (w/v) Blocker™ BSA in PBS with 0.02% (v/v) Tween-20 and incubated for 2 hours. Final washes were performed with PBS containing 0.01% (v/v) Tween-20, and the functionalized beads were stored at 4 °C in PBS with 0.01% (v/v) Tween-20. Prior to each assay, the beads were equilibrated by washing with PBS SuperBlock™ containing 0.01% Tetronic 90R4.Off-chip SARS-CoV-2 immunity assays
Off-chip immunity assays for host antibodies were implemented using 9-step protocols that were similar to the one described above but with the following changes. In step (1), spike S1 domain protein-immobilized beads were used. In step (3a), a 2 μL aliquot of sample – host antibody (IgG or IgA) spiked in PBS SuperBlock™ or pooled pre-COVID-19 saliva – was added to each tube. In many assays, there was no step (3b), and in step (3c), incubation was typically 5 min for IgG and 10 min for IgA (see Note S3, Fig. S5†). In step (5), an aqueous solution of HRP conjugated to anti-human IgG or IgA (50 ng mL−1) was added to the beads. Finally, calibration curves (with 4PL fitting) and LODs/LOQs were generated as above (Note S3, Fig. S6†). Upon observation of reactivity in IgA assays in saliva (Note S3, Fig. S7†), step (3b) was added to the procedure for such samples – the addition of a 2 μL aliquot of goat anti-human IgG (1 mg mL−1).DMF SARS-CoV-2 infection assays
All reagents for this and all other DMF procedures were supplemented with Tetronic 90R4 surfactant to a final concentration of 0.01% (w/v). A 16-step protocol was developed. (1) A double-unit droplet of capture nanobody-coated magnetic bead suspension was dispensed from a reservoir, and the beads were separated using a magnetic lens. (2a) A double-unit droplet of sample (pooled pre-COVID-19 saliva spiked with SARS-CoV-2 trimeric spike protein) was dispensed onto the array. (2b) A double-unit droplet of secondary reagent (detection nanobody solution, 0.1 μg mL−1) was dispensed onto the array. The two solutions were merged. (2c) The droplet was used to resuspend the beads and then to actively mix them for 10 minutes. (3) The beads were immobilized, and the supernatant droplet was moved to waste. (4–7) The beads were washed four times, in each case resuspending them in a fresh double-unit droplet of PBS SuperBlock™, immobilizing the beads and driving the supernatant droplet to waste. (8) A double-unit droplet of tertiary reagent (streptavidin-polyHRP solution, 20 ng mL−1) was dispensed, used to resuspend the beads, and then to actively mix them for 5 minutes. (9–14) The beads were washed six times as above. (15) A double-unit droplet of an aqueous mixture of luminol and H2O2 (a 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 mixture of regents from the SuperSignal™ ELISA Pico kit, formed immediately prior to assay) was dispensed and mixed with the beads for one minute. (16) The droplet was moved to the detection region, and the chemiluminescence signal was recorded by the PMT (biased at 1000 V) for 300 seconds (and the mean PMT response during this period was recorded as “signal”), after which the droplet was driven to the waste reservoir. Typically, steps 1–14 of the procedure above were applied in parallel to five test-solutions (four samples and a negative control comprising pooled, pre-COVID-19 saliva with no spiked analyte) in zones A-E on the device. Steps 15–16 were then performed serially for each of the solutions, and each of the signals was corrected by subtracting a PMT measurement with no droplet present. A calibration curve was prepared and fit with a linear function to generate LOD and LOQ as described above.
1 mixture of regents from the SuperSignal™ ELISA Pico kit, formed immediately prior to assay) was dispensed and mixed with the beads for one minute. (16) The droplet was moved to the detection region, and the chemiluminescence signal was recorded by the PMT (biased at 1000 V) for 300 seconds (and the mean PMT response during this period was recorded as “signal”), after which the droplet was driven to the waste reservoir. Typically, steps 1–14 of the procedure above were applied in parallel to five test-solutions (four samples and a negative control comprising pooled, pre-COVID-19 saliva with no spiked analyte) in zones A-E on the device. Steps 15–16 were then performed serially for each of the solutions, and each of the signals was corrected by subtracting a PMT measurement with no droplet present. A calibration curve was prepared and fit with a linear function to generate LOD and LOQ as described above.
DMF SARS-CoV-2 immunity assays
DMF immunity assays for host antibodies were implemented using 16-step protocols that were similar to the one described above but with the following changes. In step (1), spike S1 protein-coated beads were used. For host IgG assays, in step (2a), the sample was a double-unit droplet of IgG spiked in pooled pre-COVID-19 saliva, there was no step (2b), and in step (2c), the beads were actively mixed for 7 minutes. For host IgA assays, in step (2a) the sample was a single-unit droplet of IgA spiked in pooled pre-COVID-19 saliva, in step (2b) the secondary reagent was a single-unit droplet of pretreatment solution (polyclonal goat anti-human IgG, 1 mg mL−1 in PBS SuperBlock™) and upon merging, the combined droplet was actively mixed for 2 min, and in step (2c), the beads were actively mixed for 7 minutes. In step (8), the tertiary reagent was a double-unit droplet of a solution of HRP conjugated to anti-human IgG or IgA (typically 50 ng mL−1 in PBS SuperBlock™). As with the infection assays, steps 1–14 were applied in parallel to five test-solutions, steps 15–16 were performed serially, and calibration curves were generated (fit with a linear function) to generate LODs and LOQs.DMF combined SARS-CoV-2 infection and immunity assays
Combined DMF infection and immunity assays were implemented using a 16-step protocol that was similar to those described above, but with the following changes. In step (1), three double-unit droplets of spike S1 protein-coated paramagnetic bead suspension were dispensed from a reservoir and moved to zones A, C and D on the device. Likewise, two double-unit droplets of capture-nanobody-coated bead suspension were dispensed from a reservoir and moved to zones B and E. In step (2a), two double-unit droplets of negative control (pooled saliva collected pre-COVID-19 diluted 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 10 in PBS SuperBlock™) and three double-unit droplets of sample (diluted saliva from a symptomatic COVID-19 patient) were dispensed from their reservoirs and delivered to zones A–B and C–E, respectively. In step 2(b), one double-unit droplet of goat anti-human IgG (1 mg mL−1) and two double-unit droplets of detection nanobody solution (0.1 μg mL−1) were dispensed from their reservoirs and delivered to zones D and B&E, respectively, where they were merged with their respective sample/control droplets and then actively mixed for 2 minutes. In step 2(c), the droplets in each zone were used to resuspend the beads and then to actively mix them for 10 minutes. In step (8), two double-unit droplets of HRP conjugated to anti-human IgG (50 ng mL−1), one double-unit droplet of HRP conjugated to anti-human IgA (50 ng mL−1), and two double-unit droplets of streptavidin-polyHRP (20 ng mL−1) were dispensed from their reservoirs and delivered to zones A&C, D, and B&E respectively. As described above, steps 1–14 were performed in parallel, steps 15–16 were performed serially, and signals were recorded and processed. Signal-to-background (S/B) ratios were generated for each sample result by dividing its corrected PMT signal by the corrected PMT signal of its corresponding negative control (noting that the negative control for IgG was used as the ‘background’ for both IgG and IgA assays). The signals associated with the LOD and LOQ from each assay were also divided by background (to generate SLOD/B and SLOQ/B, respectively). Samples with S/B > SLOQ/B were deemed ‘positive’, those with SLOD/B < S/B < SLOQ/B were deemed ‘undefined’, and those with S/B < SLOD/B were deemed ‘negative’. Finally, all data were normalized by dividing by SLOQ/B for display.
10 in PBS SuperBlock™) and three double-unit droplets of sample (diluted saliva from a symptomatic COVID-19 patient) were dispensed from their reservoirs and delivered to zones A–B and C–E, respectively. In step 2(b), one double-unit droplet of goat anti-human IgG (1 mg mL−1) and two double-unit droplets of detection nanobody solution (0.1 μg mL−1) were dispensed from their reservoirs and delivered to zones D and B&E, respectively, where they were merged with their respective sample/control droplets and then actively mixed for 2 minutes. In step 2(c), the droplets in each zone were used to resuspend the beads and then to actively mix them for 10 minutes. In step (8), two double-unit droplets of HRP conjugated to anti-human IgG (50 ng mL−1), one double-unit droplet of HRP conjugated to anti-human IgA (50 ng mL−1), and two double-unit droplets of streptavidin-polyHRP (20 ng mL−1) were dispensed from their reservoirs and delivered to zones A&C, D, and B&E respectively. As described above, steps 1–14 were performed in parallel, steps 15–16 were performed serially, and signals were recorded and processed. Signal-to-background (S/B) ratios were generated for each sample result by dividing its corrected PMT signal by the corrected PMT signal of its corresponding negative control (noting that the negative control for IgG was used as the ‘background’ for both IgG and IgA assays). The signals associated with the LOD and LOQ from each assay were also divided by background (to generate SLOD/B and SLOQ/B, respectively). Samples with S/B > SLOQ/B were deemed ‘positive’, those with SLOD/B < S/B < SLOQ/B were deemed ‘undefined’, and those with S/B < SLOD/B were deemed ‘negative’. Finally, all data were normalized by dividing by SLOQ/B for display.
The procedure outlined above was used for some experiments. In other experiments, a revised ‘double-assay negative control’ was used for IgG and IgA assays, described in Note S4, Fig. S8.†
Clinical samples
Clincial samples were processed and handled according to Protocol #57265 approved by the University of Toronto Research Ethics Board. Saliva samples from COVID-19 patients were diluted 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 10 in PBS SuperBlock™ (off-chip) and then evaluated by the 16-step DMF SARS-CoV-2 combined assay procedure. DMF data were then compared with reference test results. For infection assays, reference test results were interpreted from polymerase chain reaction (PCR) data generated as described previously.35 For immunity assays, reference test results were generated using commercial well-plate-based ELISA kits. Briefly, saliva samples were diluted 1
10 in PBS SuperBlock™ (off-chip) and then evaluated by the 16-step DMF SARS-CoV-2 combined assay procedure. DMF data were then compared with reference test results. For infection assays, reference test results were interpreted from polymerase chain reaction (PCR) data generated as described previously.35 For immunity assays, reference test results were generated using commercial well-plate-based ELISA kits. Briefly, saliva samples were diluted 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 100 in sample diluent and then were evaluated by IEQ-CoVS1RBD-IgG and IE-CoVS1RBD-IgA (RayBiotech Life, GA, USA), according to the manufacturer's instructions. Signals were measured using a multiwell plate reader (PHERAstar, BMG Labtech, Germany), and samples were classified based on readings relative to the control solutions included in the kits.
100 in sample diluent and then were evaluated by IEQ-CoVS1RBD-IgG and IE-CoVS1RBD-IgA (RayBiotech Life, GA, USA), according to the manufacturer's instructions. Signals were measured using a multiwell plate reader (PHERAstar, BMG Labtech, Germany), and samples were classified based on readings relative to the control solutions included in the kits.
Results and discussion
Infection and immunity assay development
The overall goal of this project was to develop an integrated microfluidic platform capable of assessing infection and immunity status in patient saliva for SARS-CoV-2. As a first step towards this goal (prior to porting to microfluidics), off-chip assays were developed, optimized, and assessed.In developing our test for COVID-19 infection, we sought a viral protein that can serve as a proxy for the presence of viral particles in a “rapid antigen test”. There are many such tests on the market, often relying on SARS-CoV-2 nucleocapsid protein or spike protein. We chose the latter for this work, given how tests for spike protein and its constituents have been shown36 to have reduced rates of false-positive results (but in principle, a similar system could be developed for nucleocapsid in the future). With this in mind, we used the reference material SMT1-1,29 a well-characterized trimeric spike glycoprotein, as the antigen in this work. A pair of camelid nanobodies with high specificity for this antigen was selected from a panel of similar reagents,28 and the pair (i.e., the “capture” and the “detection” nanobodies) was tested to confirm stability and binding in saliva (Note S1, Fig. S1†).
Methods were then developed to reproducibly modify paramagnetic beads with the capture nanobody, and a 9-step assay procedure relying on absorbance detection was developed (Fig. 2A) and optimized (Note S2, Fig. S2†). An initial calibration curve in buffer was generated (Note S2, Fig. S3†), and the assay was then tested in a dilution series of analyte spiked in pooled pre-COVID-19 saliva. The performance in saliva (Fig. 2B) was similar to that found in buffer, with LOD and LOQ of 88.5 ng mL−1 and 399.6 ng mL−1, respectively. While these values are ∼3–5× higher than those reported for commercial rapid antigen tests for Spike protein37–39 and for literature reports of tests for Spike protein in saliva,40–42 they were nevertheless considered suitable as a starting-point for the development of more sensitive microfluidic test, described below.
As a next step, we sought to identify antibodies produced in the host (i.e., the infected patient) in response to infection as a proxy for immunity status. Literature reports20,43,44 indicate that immunoglobulin A and G are the primary host antibodies found in human saliva in SARS-CoV-2 infection, where IgA appears early (detectable 0–15 days post-symptoms) but then its concentration reduces over time, while IgG appears later (detectable 15–30 days post-symptoms) and persists for longer periods, presumably contributing to long-term immunity against the disease. These analytes were selected for monitoring in the work described here.
Methods were developed to reproducibly modify paramagnetic beads with SARS-CoV-2 spike S1 protein, and a 9-step assay procedure relying on absorbance detection was developed (Fig. 2C and E) and optimized (Note S3, Fig. S4 and S5†) for the detection of recombinant human IgG and IgA specific to SARS-CoV-2 spike protein. Initial calibration curves in the buffer were generated (Note S3, Fig. S6†), and the assay was then tested in a dilution series of the two analytes in pooled pre-COVID-19 saliva. A key step that was found to be required for reliable IgA detection in saliva was the addition of a pre-treatment step (exposure to polyclonal anti-human IgG) to compensate for the presence of cross-reactive species45 (such as host IgG) in the saliva (Note 3, Fig. S7†). With this measure in place, the performance of the tests in saliva (Fig. 2D and F) was similar to that observed in buffer, with LODs and LOQs of 6.6 and 20.2 ng mL−1 for IgG and 5.2 and 9.8 ng mL−1 for IgA, respectively. Notably, these LOD values were lower than the thresholds required for clinical detection of human IgG and IgA for patients with immunity to SARS-CoV-2.20
On-chip SARS-CoV-2 infection and immunity assays
After developing off-chip assays, the optimized procedures were transformed for use “on-chip” with digital microfluidics. To improve the analytical sensitivity for these tests, the enzyme-substrate for each of the assays was changed from one relying on absorbance to one relying on chemiluminescence. In addition, a surfactant additive was included in the procedure that allows for extended droplet manipulation with minimal protein adsorption and fouling.46 Devices with layout similar to what has been described previously30,31 were adapted for use with a new 16-step assay protocol (Fig. 3A, Movie S1†). The protocol was developed to run five assays in parallel (typically, four spiked samples plus one negative control). In practice, after loading sample and reagents, the process was completely automated – all dispensing/metering, reagent delivery, bead pelleting and resuspension, chemiluminescence detection, etc. – is ‘hands-free,’ in a process that requires ∼70 minutes to complete for five assays. | ||
| Fig. 3 On-chip SARS-CoV-2 infection and immunity assays in saliva. (A) Frame from Movie S1† showing five droplets (four spiked-saliva samples and one negative control, with blue dye added to guide the eye) being processed for an infection assay on a DMF device. The inset comprises frames from the same movie illustrating how (i) the sample droplet is driven to the magnetic beads (red arrow), (ii) the beads are resuspended and mixed (yellow/red arrow), and (iii) the supernatant is removed from the beads (red arrow). Variations of this process were repeated 16 times to complete the assay. (B) Plot of chemiluminescence in arbitrary units (a.u., blue markers) as a function of the concentration of trimeric spike protein standard spiked in pre-COVID-19 saliva. (C) Plot of chemiluminescence (blue markers) as a function of the concentration of recombinant human IgG specific to SARS-CoV-2 spike protein spiked in pre-COVID-19 saliva. (D) Plot of chemiluminescence (blue markers) as a function of the concentration of recombinant human IgA specific to SARS-CoV-2 spike protein spiked in pooled pre-COVID-19 saliva. The red lines in (B–D) represent linear fits to the data, and the error bars represent ± one standard deviation for n = 3 replicates per condition. | ||
A calibration curve for SARS-CoV-2 trimeric spike protein in pooled pre-COVID-19 saliva was generated using the automated microfluidic technique, as shown in Fig. 3B. The data were linear (with R2 = 0.9668), and the LOD and LOQ were determined to be 3.8 and 5.6 ng mL−1, respectively. This performance compares favourably to LODs reported for commercial ELISA kits for spike protein intended for use with nasal swab samples [23 ng mL−1 (ref. 37), 31 ng mL−1 (ref. 38), and 6.3 ng mL−1 (ref. 39)], and for literature reports of tests for spike protein in saliva [19 ng mL−1 (ref. 40), 28.9 ng mL−1 (ref. 41) and 39.1 ng mL−1 (ref. 42)]. This performance is likely appropriate for standard assessments of the infection state for COVID-19, but if improved analytical sensitivity is needed in the future, a recently reported, ultrasensitive ‘digital detection’ assay implemented by DMF47 for SARS-CoV-2 spike protein might be used in its place.
Calibration curves for host IgG and host IgA in pooled pre-COVID-19 saliva generated using the automated technique are shown in Fig. 3C and D. The data for the analysis of IgG and IgA were linear (with R2 = 0.9500 and R2 = 0.9875), the LODs were determined to be 4.8 ng mL−1 and 13.3 ng mL−1, and the LOQs were determined to be 18.0 ng mL−1 and 53 ng mL−1, respectively. As was the case for the off-chip assays, these detection limits are comfortably below the thresholds required for clinical detection of human IgG and IgA for patients with immunity to SARS-CoV-2.20
In all of the assays and in each of the individual replicates that were run, the negative control samples (pooled, pre-COVID-19 saliva) generated signals that were at least 3-fold lower than those for the reported LOQs. Thus, assuming the LOQ signal as a cutoff, these control signals might be used in the future as they are in a commercial product, where a control signal greater than or equal to the LOQ indicates an error in the assay, such that the results should not be trusted.
Combined DMF SARS-CoV-2 infection and immunity assays
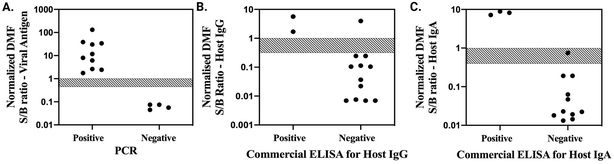
After verifying adequate performance for on-chip infection and immunity assays, separately, the protocols were combined into a unified procedure in which a single patient's saliva sample was loaded onto the device, where it was automatically aliquoted into three sub-samples that were separately driven through infection (spike protein detection) and immunity (host IgG and IgA detection) assay protocols. In parallel, pooled pre-COVID-19 saliva was automatically aliquoted into two sub-samples, which were processed as negative controls. To fit the footprint of the device used here, a combined IgG/IgA control was developed for the immunity assay negative control (Note S4, Fig. S8†) (in the future, a slightly larger device might be developed and used if separate IgG and IgA negative controls are needed). As with the individual assays, the entire suite of tests – assays for viral antigen, host IgG, host IgA, and two negative controls – was completely automated, requiring ∼70 min to complete.The new method was applied to evaluating a panel of 14 saliva samples from patients exhibiting COVID-19 symptoms. DMF assay results were then compared to reference test data – PCR results provided by the sample collectors for infection assays, and commercial well-plate ELISA data for immunity assays. Infection assay results are shown in Fig. 4A. As indicated, the DMF assay perfectly discriminated between the 10 positive and 4 negative samples determined by PCR, representing 100% sensitivity and specificity for this dataset. Host IgG data are shown in Fig. 4B. The DMF assay correctly identified 2 positives and 11 negatives determined by commercial ELISA; however, 1 well-plate negative was incorrectly assigned to be positive by the DMF system. Finally, host IgA data are shown in Fig. 4C. The DMF assay correctly identified 3 positives and 10 negatives determined by commercial ELISA, but mis-assigned one sample as undefined. Thus, the sensitivity/specificity for the host IgG and IgA assays were 100%/91.7% and 100%/90.9%, respectively. Interestingly, the mis-assigned IgA and IgG result were from the same patient sample.
Saliva is known to be a ‘tricky’ matrix for diagnostics,48–50 with substantial variations between samples arising from differing concentrations of mucins and other constituents, as well as variations in food residue, differences in oral hygiene, and a strong dependence on how the samples are collected. Thus, it was not surprising that in running experiments with patient samples, we observed the viscosities and surface tensions to be quite heterogeneous, causing them (in some cases) to behave differently than what was expected from the pooled, pre-COVID-19 saliva used in controls and in method development. With some tweaks to the protocol (e.g., including a sample dilution step prior to analysis), these differences were found to be surmountable in the work described here. In future work, it may be interesting to closely examine how saliva heterogeneity might be best handled in digital microfluidic systems.
As indicated in the introduction, the new method joins three previous reports21–23 describing microfluidic infection and immunity assays for SARS-CoV-2 in saliva. The new method has several differences relative to those described in previous reports, including the demonstration of built-in negative controls, which are widely understood24–26 to be important in portable diagnostics. Furthermore, the new method is substantially more automated than the method described by Torrente-Rodríguez et al.21 (which requires a long series of user pipetting steps), and is moderately more automated than the techniques described by Najjar et al.22 and Moakhar et al.,23 which require the user to manually load separate samples into the system (one each for the infection and immunity assays), unlike the method introduced here that automatically sub-divides the saliva after a single load step. Most importantly, the four methods (described in the three previous reports and the current one) together illustrate the creative work that the microfluidics community is doing to prepare for future large-scale outbreaks and pandemics.
More tests with additional patient samples are needed, but the data in Fig. 4 are promising, suggesting that the new technique, which relies on a technology that has been rigorously validated for operation in remote settings,33,51 could be a powerful tool for surveillance of infection and immunity for this disease. But like any report of a new method, it is a work-in-progress, and there is a long list of potential improvements that might be made in the future. For example, devices might be expanded to also enable built-in positive controls, which are also known to be useful in portable applications.24–26,52,53 Likewise, future work might be implemented in inexpensive, inkjet-printed, roll-coated DMF devices,54 and/or the devices might be manufactured bearing dried reagents55 to further reduce the level of user intervention that is required. Finally, NAAT methods are routinely implemented in DMF format8,9 and might be incorporated into future methods, and/or active-matrix DMF devices56 might be developed to evaluate many patient samples simultaneously.
Conclusion
The DMF platform described here integrates the detection of SARS-CoV-2 trimeric spike protein and host antibodies (IgG/IgA) into a single, fully automated workflow. Key performance metrics demonstrate high sensitivity and accuracy, with limits of detection (LODs) of 3.8 ng mL−1 for spike protein, 4.8 ng mL−1 for host IgG, and 13.3 ng mL−1 for host IgA—values that compare favorably to existing ELISA-based and saliva-based assays. Clinical validation in human saliva samples (n = 14) showed strong concordance with PCR and well-plate-based ELISA, achieving 100% sensitivity and 100% specificity for the infection assay, and 100% sensitivity with 91.7% and 90.9% specificity for host IgG and IgA, respectively. The advantages of this system, including non-invasive sampling, minimal reagent consumption, and suitability for portable applications in the field—make it a compelling potential solution for rapid, decentralized diagnostics, as we prepare for the next global pandemic.Data availability
The raw data associated with this manuscript is available from the corresponding author upon request.Author contributions
N. S. and J. G. C. V. contributed equally to this study. M. A. R. and J. T. developed, modified, and characterized nanobodies targeting the SARS-CoV-2 trimeric spike protein. A. A. S., A. Y., J. D., J. L., and T. V. performed and oversaw the selection of nanobody pairs for the infection assay. D. B., L. M., M. P., and A. M. advised on assay development, performance targets, and the importance of control samples. J. G. C. V. and A. A. S. conducted optimization studies for selecting and functionalizing paramagnetic beads for the immunity assay. N. S. designed, developed, and optimized both off-chip and on-chip SARS-CoV-2 infection assays. N. S., J. G. C. V., and A. A. S. designed, developed, and optimized the off-chip and on-chip SARS-CoV-2 immunity assay. N. S., J. G. C. V., N. L., and A. Y. carried out all on-chip experiments and tested the clinical samples. N. S. conceptualized the multiplexed immunity and infection assay, designed the clinical study, analyzed the data and contributed to visualization. J. D. and A. A. S. designed, built, and tested the DMF hardware, and modified the control software. R. B. and I. E. I. acquired and provided clinical samples along with PCR results. N. S. conducted ELISA assays on clinical samples using a commercial ELISA kit. A. R. W. secured funding for the study and provided supervision. N. S., J. G. C. V., and A. R. W. wrote the manuscript, with contributions from all authors.Conflicts of interest
M. A. R. and J. T. declare the following competing interests. National Research Council Canada has filed a patent (PCT/IB2022/053756, “Antibodies that bind SARS-CoV-2 spike protein”) that includes the two VHHs described here with M. A. R. and J. T. named as inventors. All other authors declare no competing interests.Acknowledgements
We thank Professor Jennifer Gommerman and Baweleta Isho (University of Toronto) for illuminating discussions about salivary immunity markers and diagnostics. We thank the NRC for the Pandemic Response Challenge Program (PR-015.1), the Centre for Research and Applications in Fluidic Technologies (Project Award), the Natural Sciences and Engineering Research Council of Canada (ALLRP 554378-20), and the University of Toronto (Temerty KT Grant and Connaught COVID-19 Fund) for funding. Finally, J. G. C. V. acknowledges support from the CONACyT Mexican Scholarship under Award Number 757699, and A. R. W. thanks the Canada Research Chair (CRC) foundation for a CRC.References
- T. Lehnert and M. A. M. Gijs, Lab Chip, 2024, 24, 1441–1493 RSC.
- E. A. Tarim, B. Karakuzu, C. Oksuz, O. Sarigil, M. Kizilkaya, M. K. A. A. Al-Ruweidi, H. C. Yalcin, E. Ozcivici and H. C. Tekin, Emergent Mater., 2021, 4, 143–168 CrossRef CAS PubMed.
- A. Basiri, A. Heidari, M. F. Nadi, M. T. P. Fallahy, S. S. Nezamabadi, M. Sedighi, A. Saghazadeh and N. Rezaei, Rev. Med. Virol., 2021, 31, e2154 CrossRef PubMed.
- S. Kim, E. Yee, E. A. Miller, Y. Hao, D. M. Y. Tay, K.-J. Sung, H. Jia, J. M. Johnson, M. Saeed, C. R. Mace, D. Yüksel Yurt and H. D. Sikes, ACS Appl. Mater. Interfaces, 2021, 13, 38990–39002 CrossRef CAS PubMed.
- L. Malic, D. Brassard, D. Da Fonte, C. Nassif, M. Mounier, A. Ponton, M. Geissler, M. Shiu, K. J. Morton and T. Veres, Lab Chip, 2022, 22, 3157–3171 RSC.
- H. Ehzari, M. Safari, R. Hallaj and M. Amiri, Microchem. J., 2024, 197, 109779 CrossRef CAS.
- S. F. Khan, P. Rathod, V. K. Gupta, P. B. Khedekar and R. V. Chikhale, Anal. Chem., 2024, 96, 8124–8146 CrossRef CAS PubMed.
- M. Ho, N. Sathishkumar, A. A. Sklavounos, J. Sun, I. Yang, K. P. Nichols and A. R. Wheeler, Lab Chip, 2024, 24, 63–73 RSC.
- C. Dong, F. Li, Y. Sun, D. Long, C. Chen, M. Li, T. Wei, R. P. Martins, T. Chen and P.-I. Mak, Lab Chip, 2024, 24, 3850–3862 RSC.
- A. C. Mora, A. J. Tierney, A. K. Sogn, P. T. Lawrence and E. Tzavaras, ChemRxiv, 2025, preprint, DOI:10.26434/chemrxiv-2025-b40mr.
- H. Hartanto, M. Wu, M. L. Lam and T.-H. Chen, Biomicrofluidics, 2020, 14, 061507 CrossRef CAS PubMed.
- M. Tayyab, M. A. Sami, H. Raji, S. Mushnoori and M. Javanmard, IEEE Sens. J., 2021, 21, 4007–4017 CAS.
- O. J. Kweon, S. Yoon, K. W. Choe, H. Kim, Y. K. Lim and M.-K. Lee, Sci. Rep., 2024, 14, 18200 CrossRef CAS PubMed.
- H. Bae, D. Lee, H. Kim, Y. S. Jee, J. Kim, B. Min, Y.-N. Park, J.-S. Seo, J. Lee, S. Lee, J. Kim, H.-S. Jo, K. Na, S. Chung and I. B. Suh, Sens. Actuators, B, 2024, 403, 135093 CrossRef CAS.
- P. Mlcochova, D. Collier, A. Ritchie, S. M. Assennato, M. Hosmillo, N. Goel, B. Meng, K. Chatterjee, V. Mendoza, N. Temperton, L. Kiss, L. C. James, K. A. Ciazynska, X. Xiong, J. A. G. Briggs, J. A. Nathan, F. Mescia, L. Bergamaschi, H. Zhang, P. Barmpounakis, N. Demeris, R. Skells, P. A. Lyons, J. Bradley, S. Baker, J. P. Allain, K. G. C. Smith, R. Bousfield, M. Wilson, D. Sparkes, G. Amoroso, E. Gkrania-Klotsas, S. Hardwick, A. Boyle, I. Goodfellow and R. K. Gupta, Cell Rep. Med., 2020, 1, 100099 CrossRef CAS PubMed.
- M. Stone, J. Bainbridge, A. M. Sanchez, S. M. Keating, A. Pappas, W. Rountree, C. Todd, S. Bakkour, M. Manak, S. A. Peel, R. W. Coombs, E. M. Ramos, M. K. Shriver, P. Contestable, S. V. Nair, D. H. Wilson, M. Stengelin, G. Murphy, I. Hewlett, T. N. Denny and M. P. Busch, J. Clin. Microbiol., 2018, 56, e02045-17 CrossRef PubMed.
- C. Aira, G. González-García, J. Martínez-Cano, N. de la Roja, M. Giammarioli, F. Feliziani, Ž. Šteingolde, J. Buitkuviene, P. Václavek, D. Glišić, C. Gallardo, P. Sastre, M. García-Durán, P. Rueda and A. Fresco-Taboada, Vaccines, 2024, 12, 307 CrossRef CAS PubMed.
- A. L. Wyllie, J. Fournier, A. Casanovas-Massana, M. Campbell, M. Tokuyama, P. Vijayakumar, J. L. Warren, B. Geng, M. C. Muenker, A. J. Moore, C. B. F. Vogels, M. E. Petrone, I. M. Ott, P. Lu, A. Venkataraman, A. Lu-Culligan, J. Klein, R. Earnest, M. Simonov, R. Datta, R. Handoko, N. Naushad, L. R. Sewanan, J. Valdez, E. B. White, S. Lapidus, C. C. Kalinich, X. Jiang, D. J. Kim, E. Kudo, M. Linehan, T. Mao, M. Moriyama, J. E. Oh, A. Park, J. Silva, E. Song, T. Takahashi, M. Taura, O.-E. Weizman, P. Wong, Y. Yang, S. Bermejo, C. D. Odio, S. B. Omer, C. S. Dela Cruz, S. Farhadian, R. A. Martinello, A. Iwasaki, N. D. Grubaugh and A. I. Ko, N. Engl. J. Med., 2020, 383, 1283–1286 CrossRef PubMed.
- L. M. Czumbel, S. Kiss, N. Farkas, I. Mandel, A. Hegyi, Á. Nagy, Z. Lohinai, Z. Szakács, P. Hegyi, M. C. Steward and G. Varga, Front. Med., 2020, 7, 465 CrossRef PubMed.
- B. Isho, K. T. Abe, M. Zuo, A. J. Jamal, B. Rathod, J. H. Wang, Y. Durocher, A. J. McGeer, J. L. Gommerman and A. C. Gingras, et al., Sci. Immunol., 2020, 5, eabe5511 CrossRef PubMed.
- R. M. Torrente-Rodríguez, H. Lukas, J. Tu, J. Min, Y. Yang, C. Xu, H. B. Rossiter and W. Gao, Matter, 2020, 3, 1981–1998 CrossRef PubMed.
- D. Najjar, J. Rainbow, S. Sharma Timilsina, P. Jolly, H. de Puig, M. Yafia, N. Durr, H. Sallum, G. Alter, J. Z. Li, X. G. Yu, D. R. Walt, J. A. Paradiso, P. Estrela, J. J. Collins and D. E. Ingber, Nat. Biomed. Eng., 2022, 6, 968–978 CrossRef CAS PubMed.
- R. Siavash Moakhar, C. del Real Mata, M. Jalali, H. Shafique, A. Sanati, J. de Vries, J. Strauss, T. AbdElFatah, F. Ghasemi, M. McLean, I. I. Hosseini, Y. Lu, S. G. Yedire, S. S. Mahshid, M. A. Tabatabaiefar, C. Liang and S. Mahshid, Adv. Sci., 2022, 9, 2204246 CrossRef CAS PubMed.
- M. Lipsitch, E. T. Tchetgen and T. Cohen, Epidemiology, 2010, 21, 383–388 CrossRef PubMed.
- V. Gubala, L. F. Harris, A. J. Ricco, M. X. Tan and D. E. Williams, Anal. Chem., 2012, 84, 487–515 CrossRef CAS PubMed.
- J. Hedges, Insight into the importance of controls in molecular diagnostics assays, https://clinlabint.com/insight-into-the-importance-of-controls-in-molecular-diagnostics-assays/, (accessed 19 December 2024).
- X. Xu, L. Cai, S. Liang, Q. Zhang, S. Lin, M. Li, Q. Yang, C. Li, Z. Han and C. Yang, Lab Chip, 2023, 23, 1169–1191 RSC.
- M. A. Rossotti, H. van Faassen, A. T. Tran, J. Sheff, J. K. Sandhu, D. Duque, M. Hewitt, X. Wen, J. Bavananthasivam, S. Beitari, K. Matte, G. Laroche, P. M. Giguère, C. Gervais, M. Stuible, J. Guimond, S. Perret, G. Hussack, M.-A. Langlois, Y. Durocher and J. Tanha, Commun. Biol., 2022, 5, 933 CrossRef CAS PubMed.
- B. B. Stocks, M.-P. Thibeault, J. D. Schrag and J. E. Melanson, Anal. Bioanal. Chem., 2022, 414, 3561–3569 CrossRef CAS PubMed.
- C. Dixon, J. Lamanna and A. R. Wheeler, Lab Chip, 2020, 20, 1845–1855 RSC.
- A. H. C. Ng, M. Lee, K. Choi, A. T. Fischer, J. M. Robinson and A. R. Wheeler, Clin. Chem., 2015, 61, 420–429 CrossRef CAS PubMed.
- R. Fobel, C. Fobel and A. R. Wheeler, Appl. Phys. Lett., 2013, 102, 193513 CrossRef.
- A. K. Knipes, A. Summers, A. A. Sklavounos, P. A. Rota, A. R. Wheeler and H. M. Scobie, et al., PLoS One, 2022, 17, e0278749 CrossRef CAS PubMed.
- I. Swyer, R. Fobel and A. R. Wheeler, Langmuir, 2019, 35, 5342–5352 CrossRef CAS PubMed.
- A. Berger, M. T. N. Nsoga, F. J. Perez-Rodriguez, Y. A. Aad, P. Sattonnet-Roche, A. Gayet-Ageron, C. Jaksic, G. Torriani, E. Boehm, I. Kronig, J. A. Sacks, M. de Vos, F. J. Bausch, F. Chappuis, A. Renzoni, L. Kaiser, M. Schibler and I. Eckerle, PLoS One, 2021, 16, e0248921 CrossRef CAS PubMed.
- M. Barlev-Gross, S. Weiss, A. Ben-Shmuel, A. Sittner, K. Eden, N. Mazuz, I. Glinert, E. Bar-David, R. Puni, S. Amit, O. Kriger, O. Schuster, R. Alcalay, E. Makdasi, E. Epstein, T. Noy-Porat, R. Rosenfeld, H. Achdout, O. Mazor, T. Israely, H. Levy and A. Mechaly, Anal. Bioanal. Chem., 2021, 413, 3501–3510 CrossRef CAS PubMed.
- Acrobiosystems, SARS-CoV-2 Spike Trimer (B.1.1.529) Specific ELISA Kit.
- Acrobiosystems, SARS-CoV-2 Spike Trimer (B.1.617.2) Specific ELISA Kit.
- Acrobiosystems, SARS-CoV-2 Spike Trimer (XBB.1.5) ELISA Kit.
- L. Fabiani, M. Saroglia, G. Galatà, R. De Santis, S. Fillo, V. Luca, G. Faggioni, N. D'Amore, E. Regalbuto, P. Salvatori, G. Terova, D. Moscone, F. Lista and F. Arduini, Biosens. Bioelectron., 2021, 171, 112686 CrossRef CAS PubMed.
- X. Yu, B. Pan, C. Zhao, D. Shorty, L. N. Solano, G. Sun, R. Liu and K. S. Lam, ACS Sens., 2023, 8, 2159–2168 CrossRef CAS PubMed.
- N. Farsaeivahid, C. Grenier, S. Nazarian and M. L. Wang, Sensors, 2023, 23, 43 Search PubMed.
- A. Varadhachary, D. Chatterjee, J. Garza, R. P. Garr, C. Foley, A. Letkeman, J. Dean, D. Haug, J. Breeze, R. Traylor, A. Malek, R. Nath and L. Linbeck, medRxiv, 2020, preprint, DOI:10.1101/2020.08.07.20170258.
- V. P. Costantini, K. Nguyen, Z. Lyski, S. Novosad, A. C. Bardossy, A. K. Lyons, P. Gable, P. K. Kutty, J. D. Lutgring, A. Brunton, N. J. Thornburg, A. C. Brown, L. C. McDonald, W. Messer and J. Vinjé, J. Immunol., 2022, 208, 1500–1508 CrossRef CAS PubMed.
- A. H. C. Ng, M. Lee, K. Choi, A. T. Fischer, J. M. Robinson and A. R. Wheeler, Clin. Chem., 2015, 61, 420–429 CrossRef CAS PubMed.
- M. Ho, A. Au, R. Flick, T. V. Vuong, A. A. Sklavounos, I. Swyer, C. M. Yip and A. R. Wheeler, ACS Appl. Mater. Interfaces, 2023, 15, 6326–6337 CrossRef CAS PubMed.
- N. H. Le, N. Sathishkumar, A. Salari, R. Manning, R. E. Meyer, C. W. Kan, A. D. Wiener, M. A. Rossotti, S. Decombe, R. P. S. de Campos, M. D. Chamberlain, J. Tanha, N. R. Pollock, D. C. Duffy and A. R. Wheeler, Lab Chip, 2025 10.1039/D5LC00103J.
- N. Jehmlich, K. H. D. Dinh, M. Gesell-Salazar, E. Hammer, L. Steil, V. M. Dhople, C. Schurmann, B. Holtfreter, T. Kocher and U. Völker, J. Periodontal Res., 2013, 48, 392–403 CrossRef CAS PubMed.
- Y. Li, Y. Ou, K. Fan and G. Liu, Theranostics, 2024, 14, 6969–6990 CrossRef CAS PubMed.
- S. Kumari, M. Samara, R. Ampadi Ramachandran, S. Gosh, H. George, R. Wang, R. P. Pesavento and M. T. Mathew, Biomed. Mater. & Devices, 2024, 2, 121–138 Search PubMed.
- A. H. C. Ng, R. Fobel, C. Fobel, J. Lamanna, D. G. Rackus, A. Summers, C. Dixon, M. D. M. Dryden, C. Lam, M. Ho, N. S. Mufti, V. Lee, M. A. M. Asri, E. A. Sykes, M. D. Chamberlain, R. Joseph, M. Ope, H. M. Scobie, A. Knipes, P. A. Rota, N. Marano, P. M. Chege, M. Njuguna, R. Nzunza, N. Kisangau, J. Kiogora, M. Karuingi, J. W. Burton, P. Borus, E. Lam and A. R. Wheeler, Sci. Transl. Med., 2018, 10, eaar6076 CrossRef PubMed.
- C. J. Mattocks, M. A. Morris, G. Matthijs, E. Swinnen, A. Corveleyn, E. Dequeker, C. R. Müller, V. Pratt, A. Wallace and for the E. V. Group, Eur. J. Hum. Genet., 2010, 18, 1276–1288 Search PubMed.
- Y.-D. Ma, K.-H. Li, Y.-H. Chen, Y.-M. Lee, S.-T. Chou, Y.-Y. Lai, P.-C. Huang, H.-P. Ma and G.-B. Lee, Lab Chip, 2019, 19, 3804–3814 RSC.
- C. Dixon, A. H. C. Ng, R. Fobel, M. B. Miltenburg and A. R. Wheeler, Lab Chip, 2016, 16, 4560–4568 RSC.
- A. A. Sklavounos, J. Lamanna, D. Modi, S. Gupta, A. Mariakakis, J. Callum and A. R. Wheeler, Clin. Chem., 2021, 67, 1699–1708 CrossRef PubMed.
- B. Zhang, J. Fu, M. Du, K. Jin, Q. Huang, J. Li, D. Wang, S. Hu, J. Li and H. Ma, Lab Chip, 2024, 24, 2193–2201 RSC.
Footnotes |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d5lc00308c |
| ‡ Equal contribution. |
| This journal is © The Royal Society of Chemistry 2025 |